Abstract
Heparan sulfate (HS) represents a major class of glycans that perform central physiological functions. Emerging HS and glycosaminoglycan microarray techniques are used to interrogate the structure and function relationship to develop novel therapeutic agents. Availability of HS with specific sulfation patterns has been a limiting factor, and impedes the accuracy of HS glycomics studies. Although organic synthesis provides oligosaccharides, these may not fully represent the biological functions of polysaccharides. Here, we present a study for developing an enzyme-based approach to synthesize a polysaccharide library with different sulfation patterns. Using different combinations of biosynthetic enzymes, we synthesized eight unique polysaccharides. We discovered that polysaccharides without iduronic acid residue displayed strong binding affinity to antithrombin and high anti-Xa and anti-IIa activities. The enzyme-based synthetic approach could become a general method for discovering new HS structures with unique biological functions.
Heparin, a commonly used anticoagulant drug, is a special form of heparan sulfate (HS). HS or heparin contains glucuronic/iduronic acid and glucosamine carrying sulfo groups. A large body of evidence has demonstrated that this highly sulfated polysaccharide plays a role in regulating embryonic development, inflammatory response, assisting viral/bacterial infections and blood coagulation [1]. The wide range of biological functions of HS attracts considerable interests to exploit heparin or heparin-like molecules for the development of anticancer and antiviral drugs in addition to the use for anticoagulant purposes [2, 3]. The uniquely distributed sulfation pattern of the HS polysaccharide is believed to regulate its functional specificity [4–6]. An approach for synthesizing a polysaccharide with specific sulfation patterns will aid to decipher the structure and function of HS for the development of HS-based drugs [7].
Chemical synthesis has been the major route to obtain structurally defined heparin and HS oligosaccharides [8]. The most important example is the synthesis of the antithrombin-binding pentasaccharide. Currently, a synthetic anticoagulant pentasaccharide has been marketed worldwide under the trade name Arixtra. However, chemical synthesis of non-analog heparin and HS oligosaccharides, larger than hexasaccharides, is extremely difficult if not impossible based on currently available methods for carbohydrate synthesis. While a number of groups continue to pursue the synthesis of heparin [9–11], it has become clear that chemical synthesis with current technology alone will be incapable of generating most larger oligosaccharide structures. The availability of an in vitro system to synthesize HS polysaccharides using the biosynthetic enzymes offers a promising alternative approach [12, 13].
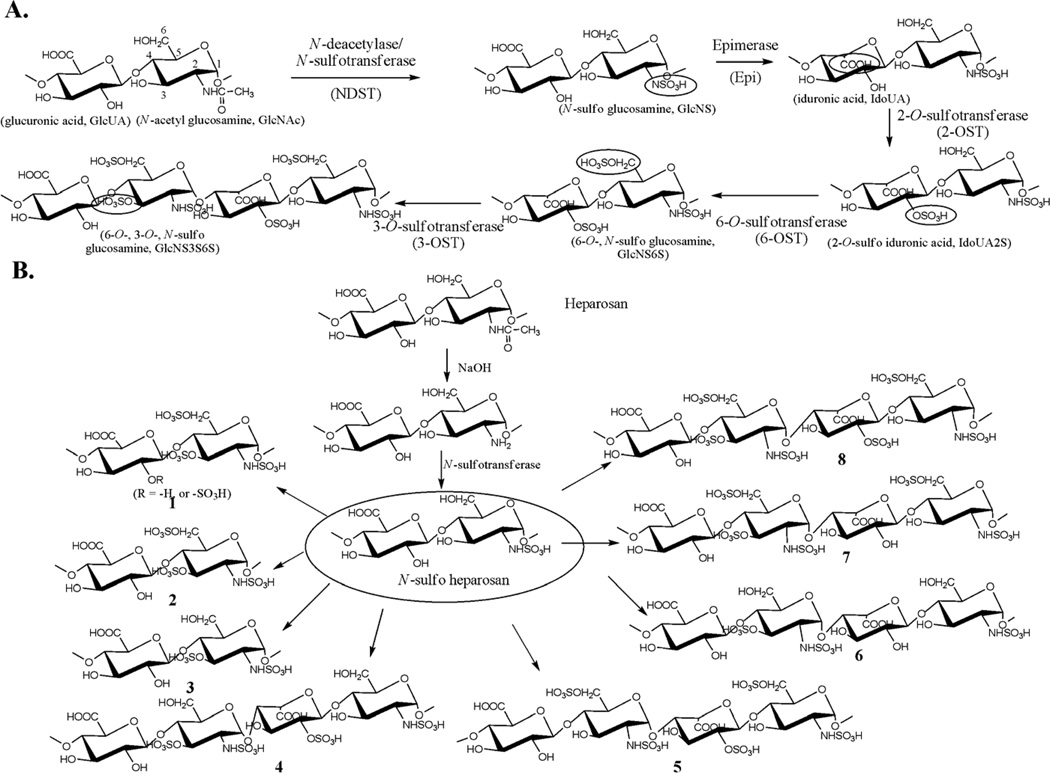
The biosynthesis of HS involves a series of specialized sulfotransferases and other enzymes. Control of the sulfation pattern mainly depends on the substrate specificities of the enzymes involved in the biosynthetic pathway. HS is initially synthesized as a linear copolymer of glucuronic acid (GlcUA) and N-acetylated glucosamine (GlcNAc), which then undergoes various modifications. These modifications are carried out by a series of biosynthetic enzymes, including glucosaminyl N-deacetylase/N-sulfotransferase, which converts the GlcNAc unit to an N-sulfo glucosamine (GlcNS) unit (Figure 1A). After the N-sulfation, C5-epimerase converts GlcUA unit to an iduronic acid (IdoUA) unit. The resultant polysaccharide is further modified by 2-O-sulfotransferase (2-OST), 6-O-sulfotransferase (6-OST) and 3-O-sulfotransferase (3-OST) to introduce the 2-O-sulfo group to IdoUA/GlcUA, and the 6-O-sulfo/3-O-sulfo groups to the glucosamine unit, respectively (Figure 1A) [14]. 6-OST is present in three isoforms, and 3-OST is present in seven isoforms.
Figure 1. Biosynthetic pathway of HS and the synthetic scheme of polysaccharide variants.
Panel A shows the reactions catalyzed by HS biosynthetic enzymes. The reaction sites at each modification step are circled. The names and abbreviations for the saccharide units are displayed underneath the sugar unit. Panel B shows the steps involved in the synthesis of a library of HS-like polysaccharides. The enzymes used for each compound are shown in Table 1.
In this article, we reported an enzyme-based combinatorial approach to synthesize the polysaccharides with different sulfation patterns (Figure 1B). From the products, we discovered that polysaccharides without the IdoUA residue displayed strong binding affinity to antithrombin and high anti-Xa and anti-IIa activities. Further, these polysaccharides have no activity in promoting cell proliferation, thus providing evidence for the synthesis of a functionally specific anticoagulant polysaccharide. Chemical synthesis of the IdoUA residue requires 10 steps [15]. Although the synthesis of IdoUA can be achieved by epimerase, the product contains a mixture of IdoUA and GlcUA because the reaction catalyzed by epimerase is reversible [16]. Consequently, the product has greater structural heterogeneity, increasing the untoward effects of an anticoagulant drug. Anticoagulant polysaccharide without the IdoUA residues could decrease the structural heterogeneity and reduce the complexity in the synthesis of HS-based anticoagulant drug. Because HS is also involved in tumor growth and viral infections, the enzyme-based synthetic approach could be used to prepare the HS structures displaying anticancer and antiviral activities.
Results and Discussion
In an effort to establish the synthesis of HS polysaccharide library, we have expressed essentially all HS biosynthetic enzymes in E. coli in large quantities [4, 7]. We started the synthesis from a capsular polysaccharide of E. coli K5 strain, known as heparosan, which is like a nonsulfated and unepimerized HS (Fig. 1). In addition, heparosan can be converted into anticoagulant HS using HS biosynthetic enzymes [12, 17]. Because N-sulfation is critical for the subsequent O-sulfations and epimerization [18], we decided to prepare polysaccharides with different types of O-sulfation patterns using N-sulfo heparosan as a starting material (Figure 1B). Eight different polysaccharides were prepared, which differ by the type of sulfations such as 2-O-, 3-O-, and 6-O-sulfations as well as the level of IdoUA residues as illustrated in Figure 1B. The overall sulfation level among the products was between 0.9 and 1.4 nmoles of O-sulfo groups per µg of polysaccharide, namely 0.39 to 0.61 O-sulfo groups per disaccharide unit as measured based on the specific [35S]radioactivity (Table 1). Compound 6 was an exception because it contained only 3-O-sulfation, which is an inherently low abundant sulfation type in the HS isolated from a natural source [19]. Subjecting the synthetic polysaccharides to an antithrombin (AT)-affinity column, we found that a significant portion of compounds 1, 2, 7, and 8 bound to AT, while 3, 4, 5, and 6 showed much less or no binding to AT (Table 1). We noted that 1, 2, 7 and 8 all carry 3-O- and 6-O-sulfo groups, while 3, 4, 5, and 6 either lacked 3-O-sulfo or 6-O-sulfo groups. Thus, we concluded that both 3-O-sulfation and 6-O-sulfation of the glucosamine unit are critical for binding to AT. In addition, 2-O-sulfation of IdoUA is not essential, provided that 7 binds to AT. These conclusions are largely consistent with previously published results of chemically synthesized pentasaccharides [20] as well as enzymatically synthesized polysaccharides [12, 21]. The binding constants (Kds) of the synthetic polysaccharides and AT were determined to be in the range of 27 to 57 nM (Table 1), which are very similar to that of previously characterized anticoagulant HS [17].
Table 1.
Summary of the synthetic polysaccharides and the results of their bindings to antithrombin (AT)
| Compounds | Modification enzymes | Structural features1 |
O-[35S]sulfation level (nmoles /µg polysaccharide) |
Binding to antithrombin affinity column (%)2 |
Binding constant to antithrombin (Kd) (nM)3 |
|---|---|---|---|---|---|
| 1 | 2-OST, 6-OST-1, 6-OST-3 and 3-OST-1 | No iduronic acid residue | 1.4 | 51% | 27 nM |
| 2 | 6-OST-1, 6-OST-3 and 3-OST-1 | No iduronic acid, no 2-O-sulfation | 1.1 | 52% | 57 nM |
| 3 | 3-OST-1 and 2-OST | No iduronic acid, no 6-O-sulfation | 0.9 | 1.0% | Not determined |
| 4 | Epi, 2-OST and 3-OST-1 | No 6-O-sulfation | 1.3 | 0.7% | Not determined |
| 5 | Epi, 2-OST, 6-OST-1 and 6-OST-3 | No 3-O-sulfation | 1.5 | 0.6% | Not determined |
| 6 | Epi and 3-OST-1 | No 2-O- and 6-O-sulfations | 0.4 | 5.1% | Not determined |
| 7 | Epi, 6-OST-1, 6-OST-3 and 3-OST-1 | No 2-O-sulfation | 1.3 | 29% | 25 nM |
| 8 | Epi, 6-OST-1, 6-OST-3, 3-OST-1 and 2-OST | Very similar to the HS isolated from tissues | 1.5 | 35% | 29 nM |
These structural features are compared to heparin and HS isolated from natural sources.
The percentage of the 35S-labeled polysaccharide bound to AT-affinity column was determined incubating AT and the polysaccharides. The complex of AT and polysaccharide was captured by using ConA-agarose column.
The binding affinity of the polysaccharides and AT was determined using affinity coelectrophoresis.
The synthetic polysaccharides displayed excellent anticoagulant activity as measured by anti-Xa and anti-IIa activities (Table 2). The IC50 values of compounds 1, 2, 7, and 8 for Xa are 2 to 2.5 fold higher than that of unfractionated heparin, while about 2-fold lower than that of Lovenox, a low molecular weight heparin drug manufactured by Aventis. A similar trend was observed for the anti-IIa activity of these compounds. The N-sulfo heparosan 6-O-sulfate was used as a negative control showing no inhibitory effect on the activity of Xa and IIa.
Table 2.
Anti-Xa and Anti-IIa activities of the synthetic polysaccharides
| Compounds | Anti-Xa activity, IC50 (ng/ml) |
Anti-IIa activity, IC50 (ng/ml) |
|---|---|---|
| Heparin | 25 | 8 |
| 1 | 50 | 18 |
| 2 | 50 | 20 |
| 7 | 45 | 16 |
| 8 | 40 | 15 |
| N-sulfo heparosan 6-O-sulfatea | >5,000 | >3,000 |
| Lovenoxb | 90 | 50 |
The synthesis of N-sulfo heparosan 6-O-sulfate is displayed in Supplementary Figure 2.
Lovenox was purchased from a local pharmacy.
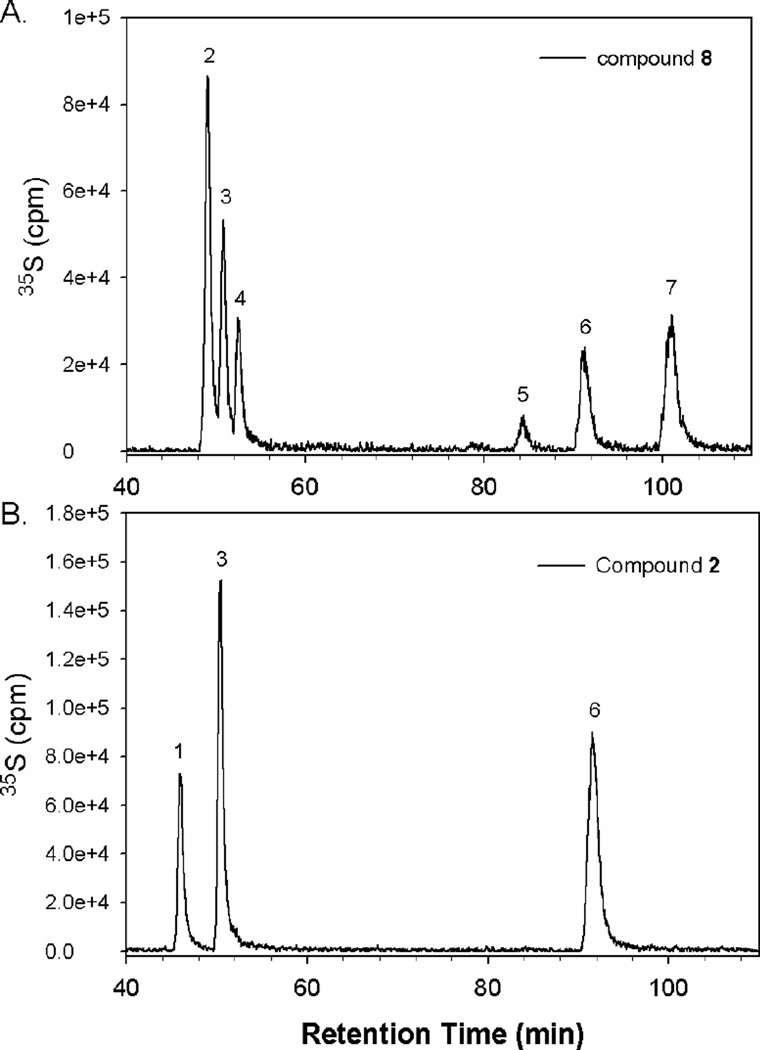
Among the synthesized AT-binding polysaccharides, compound 2 has the simplest structure because it should contain neither IdoUA nor 2-O-sulfo groups. To further elucidate the structure of the synthesized polysaccharides, we conducted the disaccharide analysis of compound 2 and compared the disaccharide composition of compound 8. The resultant disaccharides were resolved on reversed-phase ion pairing (RPIP) HPLC (Figure 2). Indeed, six disaccharides were observed in compound 8, while only three disaccharides were observed in compound 2. In addition, we noted that the ratio of the three disaccharides in 2, including GlcUA-AnMan3S, GlcUA-AnMan6S, and GlcUA-AnMan3S6S, is about 1:2:1 (Table 3), suggesting that this polysaccharide consists of a repeating tetrasaccharide unit with a structure of –GlcUA-GlcNS6S-GlcUA-GlcNS3S± 6S-. The results from disaccharide analysis also concluded that the IdoUA residue is not important for the polysaccharide to bind to AT.
Figure 2. RPIP-HPLC chromatograms of the disaccharide analysis of compound 8 (Panel A) and 2 (Panel B).
The 35S-labeled polysaccharides were degraded by nitrous acid at pH 1.5 followed by sodium borohydride reduction. The resultant 35S-labeled disaccharides were desalted on a BioGel P-2 column, and then resolved by reversed-phase ion pairing HPLC chromatography. The elution positions were identified by coeluting with 35S-labeled or 3H-labeled disaccharide standards, where 1 represents GlcUA-AnMan3S, 2 represents IdoUA2S-AnMan, 3 represents GlcUA-AnMan6S, 4 represents IdoUA-AnMan6S, 5 represents IdoUA-AnMan3S6S, 6 represents GlcUA-AnMan3S6S, and 7 represents IdoUA2S-AnMan6S.
Table 3.
Compositional analysis of compound 2 and 8
| Disaccharides | Compound 2 (mol/mol %) |
Compound 8 (mol/mol %) |
|---|---|---|
| GlcUA-AnMan3S | 24.7% | N.D.* |
| IdoUA2S-AnMan | N.D. | 35.9% |
| GlcUA-AnMan6S | 48.8% | 23.3% |
| IdoUA-AnMan6S | N.D. | 15.9% |
| IdoUA-AnMan3S6S | N.D. | 2.2% |
| GlcUA-AnMan3S6S | 26.2% | 8.9% |
| IdoUA2S-AnMan6S | N.D. | 13.4% |
Not detectable
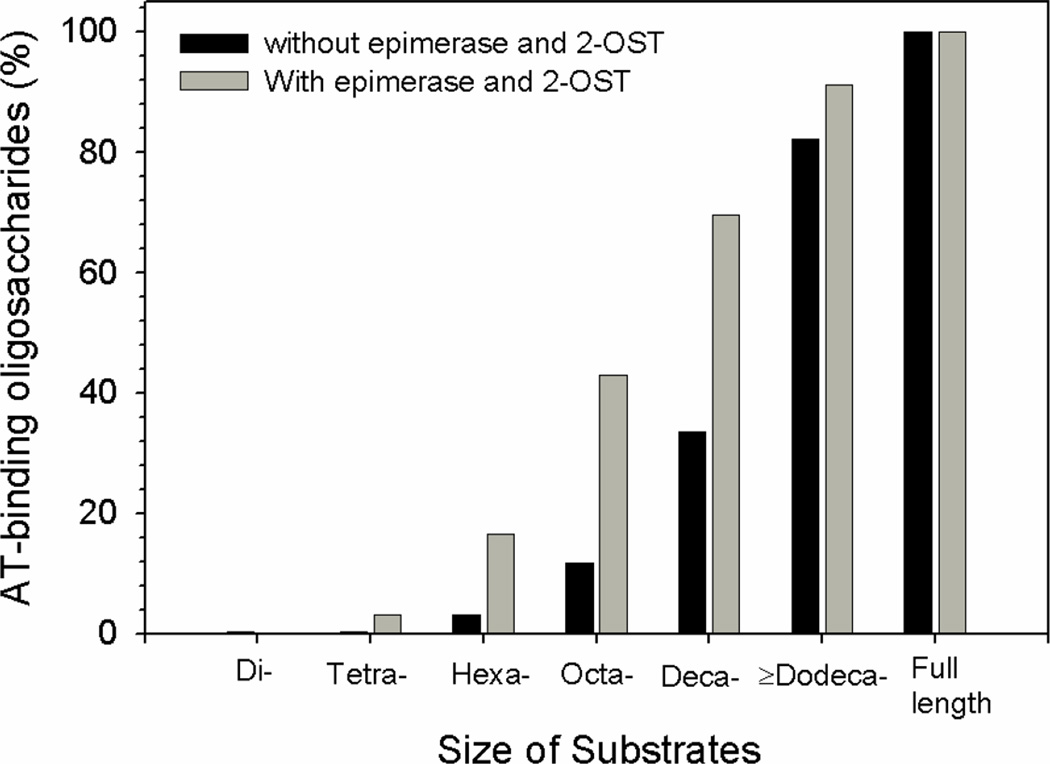
The fact that compound 2 exhibits high binding affinity to AT and strong anticoagulant activity appeared to be contradictory to previously published results. The IdoUA residue has been implicated to be essential for the binding to AT because it provides the conformational flexibility [22]. In the pentasaccharides, the IdoUA residue exists in either a chair (1C4) or a skew boat (2S0) conformation [22]. Furthermore, the unique chair conformation (2S0) of IdoUA2S in a synthetic pentasaccharide was proved to be essential for the binding to AT [23]. We believed that necessity in the conformational flexibility for AT-binding affinity is size dependent. To test this hypothesis, we compared the AT-binding oligosaccharides’ synthetic efficiency using the mixture of enzymes with or without the capability of synthesizing IdoUA2S. In one preparation, the oligosaccharides (for the synthesis of oligosaccharides without the IdoUA2S residue) were incubated with the mixture of enzymes containing 6-OST-1, 6-OST-3 and 3-OST-1. In another preparation, the oligosaccharides (for the synthesis of oligosaccharides with the IdoUA2S residue) were incubated with a mixture of enzymes containing epimerase and 2-OST as well as 6-OST-1, 6-OST-3 and 3-OST-1. The oligosaccharide substrates with structures of ΔUA-(GlcNS-GlcUA)n-GlcNS-, where n = 0, 1, 2, 3, 4 and ≥5, were prepared by subjecting N-sulfo heparosan to partial depolymerization with heparin lyase III followed by fractionation on gel permeation chromatography (Supplementary Figure 1). After the modifications, the AT-binding 35S-labeled oligosaccharides were captured by the AT-affinity column, and the data is shown in Figure 3. In the absence of epimerase and 2-OST, 35S-labeled octa- and decasaccharides showed substantial binding to AT, while 35S-labeled hexasaccharide showed very low yield. In contrast, using the same oligosaccharide substrates in the presence of epimerase and 2-OST, 35S-labeled hexasaccharides displayed significant binding to AT. We noted that the amount of AT-binding hexasaccharides and octasaccharides is about 4 to 5 fold higher than when the substrates were modified with a mixture of enzymes containing epimerase and 2-OST. These results suggest that the contribution of the IdoUA2S on the AT-binding affinity may be less essential when the size of the oligosaccharides is larger than an octasaccharide.
Figure 3. Enzymatic synthesis of AT-binding oligosaccharides.
The oligosaccharide substrates (1 µg) or the full length N-sulfo heparosan (1µg) were incubated with a mixture of enzymes, including 6-OST-1 (20 µg), 6-OST-3 (20 µg) and 3-OST-1 (10 µg) and [35S]PAPS (30 µM, 285,000 cpm/nmole) at 37°C for 2 hrs. Alternatively, the oligosaccharides (1 µg) or full length N-sulfo heparosan (1 µg) were preincubated with epimerase (8 µg) at 37°C for 30 min followed by incubating with mixture of 6-OST-1 (20 µg), 6-OST-3 (20 µg), 2-OST-1 (40 µg) and 3-OST-1 (10 µg) in the presence of [35S]PAPS. The resultant 35S-labeled oligosaccharides were incubated with AT, and the complex of AT and 35S-labeled oligosaccharides were captured by ConA-agarose column. The amount of 35S-labeled products using full length N-sulfo heparosan was defined to be 100%.
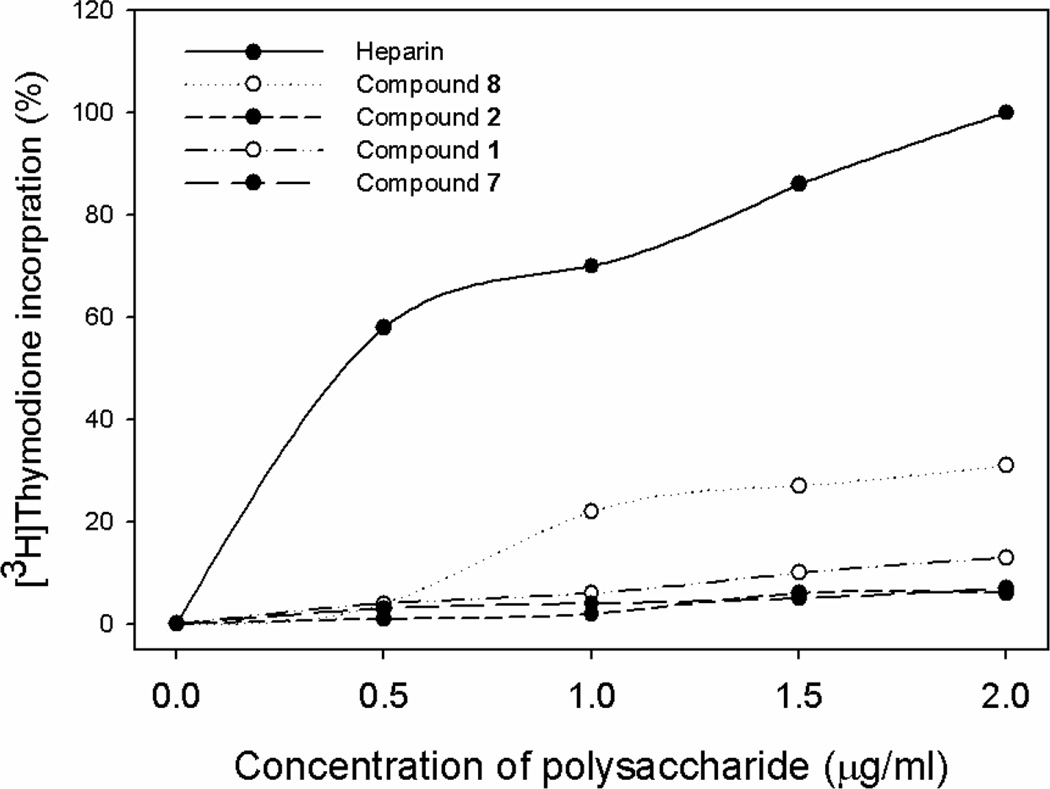
Heparin and HS form a ternary complex with fibroblast growth factors (FGFs) and fibroblast growth factor receptors (FGFR) to stimulate cell proliferation, however, this interaction clearly is not related to the anticoagulant activity of HS. Here, we tested whether the synthetic polysaccharides permit the separation of the anticoagulant activity and cell proliferation activity using BaF3 cells model system. The BaF3 cells overexpressing FGFR1 normally depend on IL-3 for growth. In the absence of IL-3, the cell proliferation depends on the addition of both FGF and heparin or HS [24]. The cells receiving heparin and compound 8 showed an increase in [3H]thymidine incorporation (Figure 3). Compound 8 displayed a lower extent in stimulating BaF3 cell growth than that of heparin since its structure is closer to HS than heparin [7]. We also found that compound 1 showed modest activity in stimulating cell proliferation. Compound 2 and 7 had no detectable activity in stimulating cell proliferation, suggesting that we have successfully separated the anticoagulant activity and cell proliferation activity by removing IdoUA2S and IdoUA residues from the polysaccharide.
The excellent activity and less structural heterogeneity of compound 2 led us to conduct the synthesis on a larger scale. We repeated the synthesis of 2 from deacetylated heparosan using sequential modifications with N-sulfotransferase (NST), 6-OST-1 and 6-OST-3 and 3-OST-1 as described in the Supplementary Figure 2. In this synthesis, we coupled a PAPS regeneration system to use p-nitrophenol sulfate (PNPS) as a sulfo donor in replace of PAPS, which potentially reduces the cost of the synthesis by more than 1000-fold [7, 25]. Disaccharide analysis on the intermediate compounds, including N-sulfo heparosan and N-sulfo heparosan 6-O-sulfate, revealed that N-sulfation reached to 75.6% and 6-O-sulfation reached 85% (Supplementary Table 2). The 3-O-sulfation carried out by 3-OST-1 went to about 90% completion, and was estimated to be 0.5 3-O-sulfo groups/disaccharide based on the modification using [35S]PAPS. The final product, renamed as Recomparin (for recombinant heparin), showed anti-Xa activity at the IC50 value of 25 ng/ml, which is very close to the value of heparin at 19 ng/ml (Supplementary Figure 3). Taken together, our results show that the enzyme-based approach is fully capable of synthesizing milligram scales of this anticoagulant polysaccharide.
In summary, we report a novel approach to prepare HS polysaccharides with different sulfation patterns. The compounds are used to investigate the contribution of the distribution of the sulfo groups to the biological functions. Utilizing the in vitro HS biosynthetic system, we can prepare a variety of HS structures by including or excluding certain enzymes. Unlike chemical sulfonation approach, our method permits the synthesis of polysaccharides that are restricted with certain types of sulfations due to the high sulfotransferases’ substrate specificities. More importantly, this approach permits the synthesis of biologically active polysaccharides that may mimic the action of HS under physiological conditions. Using this approach, we have found that anticoagulant polysaccharides do not require the IdoUA residue. The characterization of the precise structure of the anticoagulant structure without IdoUA is currently underway. We hope that the new structure will aid to simplify the synthesis of anticoagulant drug with reduced untoward effects. Due to the versatility and flexibility of the enzymatic synthesis, this approach will become an increasingly important tool for identifying the lead structures for the development of HS-based therapeutic agents.
Significance
HS is a major component on the mammalian cell surface and in the extracellular matrix with a wide range of biological functions. There are considerable interests for developing HS-based drugs to inhibit tumor growth, bacterial viral infections and bacterial infections and modulate inflammatory responses. It is clear that an enzymatic approach is a viable method for generating the polysaccharides with different sulfation patterns and biological functions. The synthesis can be conducted in parallel to produce a large number of compounds by varying the levels of individual enzymes. The next challenge for HS/heparin research is the development of a technique to prepare “recombinant polysaccharides” with more precise structures so as to achieve high functional selectivity. In the coming years of the glycomics era, continuing efforts on improving the enzymatic synthesis of HS will lead to the development of novel therapeutic reagents.
Methods
Expression of HS biosynthetic enzymes
Expressions of HS biosynthetic enzymes, including NST, epimerase, 2-OST, 6-OST-1and 3-OST-1, were carried out in E. coli as described previously [7, 26–28]. The expression of 6-OST-3 was also carried out in E. coli. Briefly, the catalytic domain of mouse 6-OST-3 (Pro121–Pro450) was cloned into a pMalc2x vector (New England BioLab) from mouse brain Quick clone cDNA library (BD Biosciences). The expression was carried out in Origami-B cells (Novagen) carrying pGro7 (Takara, Japan) plasmid expressing chaperonin proteins GroEL and GroES of E. coli. Transformed cells were grown in 6 liters of LB medium supplemented with 2 mg/ml glucose, 12.5 µg/ml tetracycline, 15 µg/ml kanamycin, 35 µg/ml chloramphenicol, and 50 µg/ml carbenicillin at 37 °C. When the A600 reached 0.4–0.7, isopropyl-thiogalactopyranoside (0.15 mM) and L-arabinose (1 mg/ml) were added to induce the expression of 6-OST-3 and chaperonin proteins, respectively. The cells were allowed to shake overnight at 22 °C. Purification was carried out using an amylose-agarose (New England BioLab) column following the protocols provided by manufacturer.
Preparation of enzymatically modified polysaccharides
N-sulfo heparosan was prepared by incubating chemically deacetylated heparosan with purified NST and the components for PAPS regeneration system as described below. N-sulfo heparosan (150 µg) was incubated with [35S]PAPS (120 µM, 4.5 ×103 cpm/nmole) and the combination of epimerase (2.7 mg if included), 2-OST (8 mg if included), 6-OST-1 (1.2 mg if included), 6-OST-3 (1.4 mg if included) and 3-OST-1 (0.9 mg if included) in 50 mM MES and 1% Triton X-100, pH 7.0. The reactions were incubated at 37°C for 2 hrs with shaking. The resultant 35S-labeled polysaccharides were purified by DEAE chromatography [7].
To prepare the enzymatically modified oligosaccharides, the N-[35S]sulfo oligosaccharide substrates (1 µg, 1,500 cpm) were incubated with a mixture of 6-OST-1 (20 µg), 6-OST-3 (20 µg) and 3-OST-1 (10 µg) and [35S]PAPS (50 µM, 2.85 × 105 cpm/nmole) in 100 µl of 50 mM MES and 1% Triton X-100. Alternatively, the oligosaccharide substrates (1 µg) were incubated with epimerase (7 µg) at 37°C for 30 min before it was incubated with a mixture of 2-OST (40 µg), 6-OST-1 (20 µg), 6-OST-3 (20 µg) and 3-OST-1 (10 µg). The reactions were terminated by heating at 100°C for 2 min. The resultant oligosaccharides were mixed with AT to determine the bindings using ConA-agarose column as described below.
Disaccharide analysis of polysaccharides
The 35S-labeled HS was degraded with nitrous acid at pH 1.5, followed by reduction with sodium borohydride [29]. The resultant 35S-labeled disaccharides were resolved by a C18 reversed phase column (0.46 × 25 cm) (Vydac) under reversed phase ion pairing HPLC conditions. The identities of the disaccharides were determined by coeluting with appropriate 35S-labeled disaccharide standards [30].
Determination of the amount of polysaccharides
The amount of the synthetic polysaccharides was estimated by a colorimetric method using alcian as described [31], where the standard curve was generated using HS isolated from bovine kidney. To measure the concentration of heparosan, N-sulfo heparosan, and N-sulfo heparosan 6-O-sulfate, the polysaccharides were depolymerized to disaccharides by a mixture of heparin lyase I, II and III. The amount of the resultant disaccharides was determined by coeluting with disaccharide standards (from Seikagaku) on RPIP-HPLC as described above using UV 232 nm detection.
Antithrombin-binding
Approximately 1×105 cpm of [35S]-labeled compound was incubated with 5 µg of human AT (Cutter Biological) in 50 µl binding buffer containing 10 mM Tris-HCl, pH 7.5, 150 mM NaCl, 1 mM Mn2+, 1 mM Mg2+, 1 mM Ca2+, 10 µM dextran sulfate, 0.0004% Triton X-100, and 0.02% sodium azide for 30 min at room temperature. Concanavalin A (ConA)-Sepharose (Sigma, 50 µl of 1:1 slurry) was then added and the reaction was shaken at room temperature for 1 h. The beads were then washed by 3 × 1 ml binding buffer, and the bound polysaccharide was eluted with 1 M NaCl.
Anti-Xa and anti-IIa assays
Assays were based on two previous methods [32, 33]. Briefly, factor Xa (Enzyme Research Laboratories, South Bend, IN) and thrombin (Sigma) were diluted to 1 unit/ml and 8 units/ml with PBS containing 1 mg/ml BSA, respectively. AT was diluted with PBS containing 1 mg/ml BSA to give a stock solution at the concentration of 27 µM. The chromogenic substrates, S-2765 (for factor Xa assay) and S-2238 (for thrombin assay), were from Diapharma and prepared at 1 mM with PBS. The synthesized polysaccharides or heparin was dissolved in a buffer containing 50 mM Tris-HCl, pH 8.4, 7.5 mM Na2EDTA, 175 mM NaCl at various concentrations (1 ng/ml to 10,000 ng/ml). The reaction mixture, which consisted of 25 µl of AT stock solution and 25 µl of the solution containing polysaccharide, was incubated at 37°C for 2 min. Factor Xa (25 µl) or thrombin (25 µl) was added. After incubating 37°C for 4 min, 25 µl of S-2765 or S-2238 was added. The absorbance of the reaction mixture was measured at 405 nm continuously for 10 min. The absorbance values were plotted against the reaction time. The initial reaction rates as a function of concentration were used to calculate the IC50 values.
Affinity coelectrophoresis
The dissociation constant (Kd) of each sample and AT was determined using affinity co-electrophoresis [34]. Approximately 4000–5000 cpm of antithrombin-binding 35S-labeled polysaccharide was loaded per lane with zones of AT at concentrations 0, 8, 16, 60 and 120 nM. The gel was run at 400mA for 2 hours, dried and then analyzed on a PhosphoImager (Amersham Biosciences, Storm 860). The retardation coefficient was calculated at R = (M0−M)/M, where M0 is the mobility of the polysaccharide through the zone without AT, and M is the mobility of the polysaccharide through each separation zone. The retardation coefficient was then plotted against the retardation coefficient divided by its respective concentration of AT. The slope of the line represents −1/Kd.
FGF2/FGFR1C-mediated proliferation assay
The BaF3 cells ectopically expressing FGFR1C has been previously described [24]. The BaF3-FGFR1c cells were maintained in RPMI 1640 media (Sigma, St. Louis, MO) supplemented with 10% fetal bovine serum, 0.5 ng/ml IL-3 (PeproTech Inc., Rocky Hill, NJ), 2 mM L-glutamine, penicillin (50 IU/ml) and streptomycin (50 µg/ml), and 50 µM β-mercaptoethanol. For mitogenic assays, BaF3 FGFR1c cells were washed three times with RPMI 1640 media to remove IL-3 and resuspended in the growth media lacking IL-3. About 30,000 cells were plated per well in a 96-well plate in media containing various concentrations of heparin, compounds 1, 2, 7, and 8 and 2 nM of FGF-2 (PeproTech) in a total volume of 200 µl. The cells were then incubated at 37°C for 40 h. To each well, an additional 50 µl of growth media containing 1 µCi of [3H]thymidine was added. Cells were harvested after 4–5 h by centrifugation. The incorporation of [3H]thymidine into the DNA was determined by scintillation counting.
PAPS regeneration system
The reactions involved in the PAPS regeneration system are shown in Supplementary Figure 2. N-terminal (His)6 tagged AST-IV was expressed in E. coli and purified as described by Burkat and colleagues [25] at a yield ≈ 50 mg/liter of bacterial culture.
Preparation of N-sulfo heparosan
Heparosan was isolated from E. coli K5 strain using a DEAE column [35]. The deacetylated heprosan was achieved under alkaline conditions [36]. The deacetylated heparosan was incubated with purified NST and coupled with a PAPS regeneration system. Briefly, 2.5 mg purified AST-IV was incubated with 40 µM PAP and 1 mM p-nitrophenol sulfate (PNPS) in 40 ml of 50 mM MES, pH 7.0, 1% Triton X-100, 1 mM MgCl2, and 1 mM MnCl2 at 25°C for 15 min. The reaction mixture was mixed with N-sulfotransferase (10 mg), 5 mg of deacetylated heparosan added and rotated at 25°C for 24 h. The N-sulfo heparosan was recovered by a DEAE column. The resultant product was subjected to a mixture of heparin lyases digestion followed by a disaccharide analysis to assess the level of N-sulfation. After two rounds of the modifications, the N-sulfation level reached to 80%.
Preparation of N-sulfo oligosaccharides
N-[35S]sulfo heparosan (1mg, 1.5 × 106 cpm) was mixed with 20 ng of purified heparin lyase III in 1ml of 50 mM sodium phosphate, pH 7.0. The digestion lasted for 18 hrs and was terminated by heating at 100°C for 2 min. The digested polysaccharide were resolved on a BioGel P-10 which was eluted with a buffer containing 25 mM Tris and 1000 mM NaCl, pH 7.4 at a flow rate of 2 ml/h.
Supplementary Material
Figure 4. The effect of the synthetic polysaccharides on FGF-2-dependent BaF3 FGFR1c cell proliferation.
BaF3 FGFR1c cells were seeded in 96-well plates with 2 nM FGF2 for control and 2 nM FGF2 plus various concentrations of synthetic polysaccharides and heparin. Cells were cultured for 40 hrs, followed by incubation in the media containing [3H]thymidine for 4 hrs. The cellular proliferation was determined by [3H]thymidine incorporation into the DNA.
Acknowledgments
The authors thank Heather Bethea (University of North Carolina) for reviewing the manuscript. The authors also thank Drs. David Ornitz and Lijuan Zhang (Washington University, St. Louis) for providing BaF3 cells expressing FGFR1c. This work is supported in part by National Institutes of Health grant AI50050.
Abbreviations
- HS
heparan sulfate
- AT
antithrombin
- PAPS
3’-phosphoadenosine 5’-phosphosulfate
- Epi
C5-epimerase
- 2-OST
2-O-sulfotransferase
- 6-OST-1 and -3 represents
6-O-sulfotransferase isoform 1 and isoform 3, respectively
- 3-OST-1
3-O-sulfotransferase isoform 1
- IdoUA
iduronic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare that they have no conflicting financial interests.
Reference
- 1.Esko JD, Selleck SB. Order out of chaos: Assembly of ligand binding sites in heparan sulfate. Ann. Rev. Biochem. 2002;71:435–471. doi: 10.1146/annurev.biochem.71.110601.135458. [DOI] [PubMed] [Google Scholar]
- 2.Fuster MM, Esko JD. The sweet and sour of cancer: Glycans as novel therapeutic targets. Nat. Rev Cancer. 2005;5:526–542. doi: 10.1038/nrc1649. [DOI] [PubMed] [Google Scholar]
- 3.Shriver Z, Raguram S, Sasisekharan R. Glycomics: A pathway to a class of new and improved therapeutics. Nat Rev Drug Discov. 2004:863–873. doi: 10.1038/nrd1521. [DOI] [PubMed] [Google Scholar]
- 4.Liu J, Pedersen LC. Anticoagulant heparan sulfate: Structural specificity and biosynthesis. Appl. Microbiol. Biotechnol. 2007;74:263–272. doi: 10.1007/s00253-006-0722-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kreuger J, Spillmann D, Li J-p, Lindahl U. Interactions between heparan sulfate and proteins: the concept of specificity. J. Cell Biol. 2006;174:323–327. doi: 10.1083/jcb.200604035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gama C, Tully SE, Sotogaku N, Clark PM, Rawat M, Vaidehi N, Goddard WA, Nishi A, Hsieh-Wilson LC. Sulfation patterns of glycosaminoglycans encode molecular recognition and activity. Nat Chem Biol. 2006;2:467–473. doi: 10.1038/nchembio810. [DOI] [PubMed] [Google Scholar]
- 7.Chen J, Avci FY, Muñoz EM, McDowell LM, Chen M, Pedersen LC, Zhang L, Linhardt RJ, Liu J. Enzymatically redesigning of biologically active heparan sulfate. J. Biol. Chem. 2005;280:42817–42825. doi: 10.1074/jbc.M504338200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Noti C, Seeberger PH. Chemical approaches to define the structure-activity relationship of heparin-like glycosaminoglycans. Chemistry & Biology. 2005;12:731–756. doi: 10.1016/j.chembiol.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Avci FY, Karst NA, Linhardt RJ. Curr. Pharmaceut. Des. 2003;9:2323–2335. doi: 10.2174/1381612033453929. [DOI] [PubMed] [Google Scholar]
- 10.de Paz JL, Noti C, Seeberger PH. Microarrays of synthetic heparin oligosaccharides. J. Am. Chem. Soc. 2006;128:2766–2767. doi: 10.1021/ja057584v. [DOI] [PubMed] [Google Scholar]
- 11.Lee JC, Lu XA, Kulkarni SS, Wen YS, Hung SC. Synthesis of heparin oligosaccharides. J. Am. Chem. Soc. 2004;126:476–477. doi: 10.1021/ja038244h. [DOI] [PubMed] [Google Scholar]
- 12.Balagurunathan K, Beeler DL, Lech M, Wu ZL, Rosenberg RD. Chemoenzymatic Synthesis of Classical and Non-classical Anticoagulant Heparan Sulfate Polysaccharides. J. Biol. Chem. 2003;278:52613–52621. doi: 10.1074/jbc.M305029200. [DOI] [PubMed] [Google Scholar]
- 13.Balagurunathan K, Lech MZ, Beeler DL, Wu ZL, Rosenberg RD. Enzymatic synthesis of antithrombin III-binding heparan sulfate pentasaccharide. Nat. Biotechnol. 2003;21:1343–1346. doi: 10.1038/nbt885. [DOI] [PubMed] [Google Scholar]
- 14.Feyerabend TB, Li J-P, Lindahl U, Rodewald H-R. Heparan sulfate C5-epimerase is essential for heparin biosynthesis in mast cells. Nat Chem Biol. 2006;2:195–196. doi: 10.1038/nchembio777. [DOI] [PubMed] [Google Scholar]
- 15.Orgueira HA, Bartollozzi A, Schell P, Litjens REJN, Palmacci ER, Seeberger PH. Modular synthesis of heparin oligosaccharides. Chemistry. 2003;9:140–169. doi: 10.1002/chem.200390009. [DOI] [PubMed] [Google Scholar]
- 16.Li J-p, Hagner-McWhirter A, Kjellen L, Palgi J, Jalkanen M, Lindahl U. Biosynthesis of Heparin/Heparan Sulfate. cDNA cloning and expression of D-glucuronyl C5-epimerase from bovine lung. J. Biol. Chem. 1997;272:28158–28163. doi: 10.1074/jbc.272.44.28158. [DOI] [PubMed] [Google Scholar]
- 17.Muñoz E, Xu D, Avci F, Kemp M, Liu J, Linhardt RJ. Enzymatic synthesis of heparin related polysaccharides on sensor chips: Rapis screening of heparin-protein interactions. Biochem. Biophys. Res. Commun. 2006;339:597–602. doi: 10.1016/j.bbrc.2005.11.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Conrad HE. Heparin-binding proteins. San Diego, CA: Academic Press; 1998. [Google Scholar]
- 19.Liu J, Thorp SC. Heparan sulfate and the roles in assisting viral infections. Med. Res. Rev. 2002;22:1–25. doi: 10.1002/med.1026. [DOI] [PubMed] [Google Scholar]
- 20.Atha DH, Lormeau J-C, Petitou M, Rosenberg RD, Choay J. Contribution of monosaccharide residues in heparin binding to antithrombin III. Biochemistry. 1985;24:6723–6729. doi: 10.1021/bi00344a063. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Lawrence R, Schwartz JJ, Bai X, Wei G, Esko JD, Rosenberg RD. The effect of precursor structures on the action of glucosaminyl 3-O-sulfotransferase-1 and the biosynthesis of anticoagulant heparan sulfate. J. Biol. Chem. 2001;276:28806–28813. doi: 10.1074/jbc.M100204200. [DOI] [PubMed] [Google Scholar]
- 22.Petitou M, van Boeckel CAA. A synthetic antithrombin III binding pentasaccharide is now a drug! What comes next? Angew. Chem. Int. Ed. 2004;43:3118–3133. doi: 10.1002/anie.200300640. [DOI] [PubMed] [Google Scholar]
- 23.Das S, Mallet J, J E, Driguez P, Duchaussoy P, Sizun P, Herault J, Herbert J, M P, P S. Synthesis of conformationally locked L-iduronic acid derivatives: direct evidence for a critical role of the skew-boat 2S0 conformer in the activation of antithrombin by heparin. Chemistry. 2001;7:4821–4834. doi: 10.1002/1521-3765(20011119)7:22<4821::aid-chem4821>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
- 24.Ornitz DM, Xu J, Colvin JS, McEwen DG, MacArthur CA, Coulier F, Gao G, Goldfarb M. Receptor specificity of the fibroblast growth factor family. J. Biol. Chem. 1996;271:15292–15297. doi: 10.1074/jbc.271.25.15292. [DOI] [PubMed] [Google Scholar]
- 25.Burkart MD, Izumi M, Chapman E, Lin C, Wong C. Regeneration of PAPS for the enzymatic synthesis of sulfated oligosaccharides. J. Org. Chem. 2000;65:5565–5574. doi: 10.1021/jo000266o. [DOI] [PubMed] [Google Scholar]
- 26.Kakuta Y, Li L, Pedersen LC, Pedersen LG, Negishi M. Heparan sulphate N-sulphotransferase activity: reaction mechanism and substrate recognition. Biochem. Soc. Trans. 2003;31(pt2):331–334. doi: 10.1042/bst0310331. [DOI] [PubMed] [Google Scholar]
- 27.Edavettal SC, Lee KA, Negishi M, Linhardt RJ, Liu J, Pedersen LC. Crystal structure and mutational analysis of heparan sulfate 3-O-sulfotransferase isoform 1. J. Biol. Chem. 2004;279:25789–25797. doi: 10.1074/jbc.M401089200. [DOI] [PubMed] [Google Scholar]
- 28.Muñoz E, Xu D, Kemp M, Zhang F, Liu J, Linhardt RJ. Affinity, kinetic and structural study of the interaction of 3-O-sulfotransferase isoform 1 with heparan sulfate. Biochemistry. 2006;45:5122–5128. doi: 10.1021/bi052403n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shively JE, Conrad HE. Formation of anhydrosugars in the chemical depolymerization of heparin. Biochemistry. 1976;15:3932–3942. doi: 10.1021/bi00663a005. [DOI] [PubMed] [Google Scholar]
- 30.Chen J, Duncan MB, Carrick K, Pope M, Liu J. Biosynthesis of 3-O-sulfated heparan sulfate: unique substrate specificity of heparan sulfate 3-O-sulfotransferase isoform 5. Glycobiology. 2003;13:785–794. doi: 10.1093/glycob/cwg101. [DOI] [PubMed] [Google Scholar]
- 31.Bjornsson S. Simultaneous preparation and quantitation of proteoglycans by precipitation with alcian blue. Anal. Biochem. 1993;210:282–291. doi: 10.1006/abio.1993.1197. [DOI] [PubMed] [Google Scholar]
- 32.Zhang L, Beeler DL, Lawrence R, Lech M, Liu J, Davis JC, Shriver Z, Sasisekharan R, Rosenberg RD. 6-O-sulfotransferase-1 represents a critical enzyme in the anticoagulant heparan sulfate biosynthetic pathway. J. Biol. Chem. 2001;276:42311–42321. doi: 10.1074/jbc.M101441200. [DOI] [PubMed] [Google Scholar]
- 33.Duncan MB, Chen J, Krise JP, Liu J. The biosynthesis of anticoagulant heparan sulphate by the heparan sulphate 3-O-sulphotransferase isoform 5. Biochim Biophys Acta. 2004;1671:34–43. doi: 10.1016/j.bbagen.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 34.Lee MK, Lander AD. Analysis affinity and strutural selectivity in the binding of proteins to glycosamineglycans: Development of a sensitive electrophoretc approach. Proc. Natl. Acad. Sci. USA. 1991;88:2768–2772. doi: 10.1073/pnas.88.7.2768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vann WF, Schimidt MA, Jann B, Jann K. The structure of the capsular polysaccharide (K5 antigen) of urinary-tract-infective Escherichia coli 010:K5:H4. Eur. J. Biochem. 1981;116:359–364. doi: 10.1111/j.1432-1033.1981.tb05343.x. [DOI] [PubMed] [Google Scholar]
- 36.Lindahl U, Li J, Kusche-Gullberg M, Salmivirta M, Alaranta S, Veromaa T, Emies J, Roberts I, Taylor C, Oreste P, Zoppetti G, Naggi A, Torri G, Casu B. Generation of "neoheparin" from E, Coli K5 capsular polysaccharide. J. Med. Chem. 2005;48:349–352. doi: 10.1021/jm049812m. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.