Abstract
The Middle East respiratory syndrome coronavirus (MERS-CoV) is a novel enzootic betacoronavirus that was first described in September 2012. The clinical spectrum of MERS-CoV infection in humans ranges from an asymptomatic or mild respiratory illness to severe pneumonia and multi-organ failure; overall mortality is around 35.7%. Bats harbour several betacoronaviruses that are closely related to MERS-CoV but more research is needed to establish the relationship between bats and MERS-CoV. The seroprevalence of MERS-CoV antibodies is very high in dromedary camels in Eastern Africa and the Arabian Peninsula. MERS-CoV RNA and viable virus have been isolated from dromedary camels, including some with respiratory symptoms. Furthermore, near-identical strains of MERS-CoV have been isolated from epidemiologically linked humans and camels, confirming inter-transmission, most probably from camels to humans. Though inter-human spread within health care settings is responsible for the majority of reported MERS-CoV cases, the virus is incapable at present of causing sustained human-to-human transmission. Clusters can be readily controlled with implementation of appropriate infection control procedures. Phylogenetic and sequencing data strongly suggest that MERS-CoV originated from bat ancestors after undergoing a recombination event in the spike protein, possibly in dromedary camels in Africa, before its exportation to the Arabian Peninsula along the camel trading routes. MERS-CoV serosurveys are needed to investigate possible unrecognized human infections in Africa. Amongst the important measures to control MERS-CoV spread are strict regulation of camel movement, regular herd screening and isolation of infected camels, use of personal protective equipment by camel handlers and enforcing rules banning all consumption of unpasteurized camel milk and urine.
Keywords: MERS-CoV, Coronavirus, Middle East, Animal, Dromedary, Camel, Bat, Zoonosis
Introduction
The Middle East respiratory syndrome coronavirus (MERS-CoV) was first isolated from a 60-year man who died in a hospital in Jeddah, Saudi Arabia, in June 2012 with severe pneumonia and multi-organ failure.1 Thus far, the majority of MERS-CoV cases have originated in countries in the Middle East, including Saudi Arabia, the United Arab Emirates (UAE), Qatar, Oman, Kuwait and Iran.2 Clinical illness associated with MERS-CoV ranges from mild upper respiratory symptoms to fulminant pneumonia and multi-system failure.3, 4−5 Human-to-human transmission of MERS-CoV is well documented in family clusters, community settings and more often in health care settings.3, 6–8 Larger hospital outbreaks have been driven by a combination of late recognition, over-crowding and inadequate infection control precautions.5, 9, 10 However, MERS-CoV inter-human transmissibility is thought to be relatively limited.11−13
Up to 12 August 2015, a total of 1401 laboratory-confirmed MERS-CoV infections, including 500 associated deaths, have been reported to the World Health Organization.14 As with many emerging viral infections, a zoonotic source was suspected soon after the identification of MERS-CoV.15 We herein review the available evidence that associates MERS-CoV with animal sources and the probable directions of transmission.
Host Susceptibility
MERS-CoV entry into their host cells is mediated by binding of a receptor-binding domain on their spike (S) proteins to specific cellular receptors known as dipeptidyl peptidase 4 (DPP4).16 DPP4 is expressed on the epithelial and endothelial cells of most human organs; an observation that might explain the multi-system clinical spectrum of MERS-CoV infection.18 DPP4 of small animals such as mice, ferrets and hamsters do not support MERS-CoV cell entry, and hence, such animals are not susceptible to MERS-CoV infection.19 However, mice are susceptible to MERS-CoV once transduced with recombinant adenovirus expressing human DPP4 receptors.20 On the other hand, experimental inoculation of rhesus macaques and marmosets resulted in viral replication, cytopathic cellular changes and mild to severe respiratory illness.21–24−24 MERS-CoV can also be detected in lungs of inoculated rabbits, but without any associated symptoms or histopathological changes.25 Macaques and marmosets have already proved useful animal models for the investigation of potential MERS-CoV therapeutic agents.26, 27 Notably, MERS-CoV can utilize DPP4 expressed on cell lines derived from goats, sheep and cows making these animals’ potential reservoirs or intermediate hosts for MERS-CoV.19, 28 However, MERS-CoV antibodies have never been identified in any such animals.29–33−34
Bats as Putative Origin of MERS-CoV
Bats are known natural reservoirs for several emerging viral infections in humans including rabies, Nipah virus, Hendra virus and Ebola virus.35 Several features enable bats to be efficient sources of emerging human viral infections. As an extremely diverse species with a long evolutionary history, bats have co-evolved with a variety of viruses.36 Their lack of B-cell-mediated immune responses allows them to carry viruses without showing overt signs of illness.37 Low metabolic rate and suppressed immune response during bats’ hibernation result in delayed viral clearance.38 Bats live closely together in extremely large numbers facilitating stable circulation of viruses amongst them.39, 40 Furthermore, bats are capable of flying and hence carrying potentially infectious pathogens over considerable distances.41 A pertinent feature of bats is that they chew fruits to absorb their sugars and spit out the remains.42 The discarded fruits can be contaminated with viruses from the oral cavity, urine and faeces providing a ready source for transmission to other potential hosts such animals and humans.37, 42
Severe acute respiratory syndrome coronavirus (SARS-CoV), which emerged in China in 2003 and caused over 8000 human infections, originated in horseshoe bats (Rhinolophus sinicus) and was transmitted to humans via palm civets as intermediate hosts.43−46 Moreover, MERS-CoV belongs to Betacoronavirus clade c, along with bat coronaviruses HKU4 and HKU5.47, 48 It is therefore not surprising that initial efforts to identify the origins of MERS-CoV focused on bats.41, 49
Throat swabs, urine, faeces and serum samples were collected from wild bats in Saudi Arabia including the area where the first MERS-CoV patient had lived and worked. Several coronaviruses were identified in 227 of 1003 samples. A 190-nucleotide fragment of the RNA-dependent RNA polymerase (RdRp) region of MERS-CoV genome was detected in one faecal pellet from an Egyptian tomb bat (Taphozous perforates).50 The sequenced amplification product was identical to that of the MERS-CoV sequence obtained from the first index human case.1 Unfortunately, the quality of the samples deteriorated when the cold chain was interrupted for more than 48 hours during their transport from Saudi Arabia to Columbia University in the United States, and it was hence not possible to produce the full genomic sequence of the isolate.50
Away from the Arabian Peninsula, novel MERS-CoV-related coronaviruses were detected in slit-faced bats (Nycteris gambiensis) from Ghana and pipistrelle bats (Pipistrellus pipistrellus, P. kuhlii, P. nathusii, P. pipistrellus and P. pygmaeus) from Germany, the Netherlands, Romania and Ukraine.51 The 816-nucleotide RdRp amino acid sequence of the novel Pipistrellus and Nycteris bat viruses differed from that of MERS-CoV by only 1.8% and 7.5%, respectively. Novel betacoronaviruses closely related to MERS-CoV have also been identified from Asian particoloured bats (Vespertilio superans) in China, serotine bats (Eptesicus serotinus) in Italy and broad-eared bat (Nyctinomops laticaudatus) in Mexico, in addition to bat guano fertilizer from Thailand.52−55
More recently, a novel betacoronavirus named NeoCoV was identified in a vesper bat (Neoromicia capensis) from South Africa. The sequenced 816-nucleotide RdRp fragment from NeoCoV differed from that of MERS-CoV by only one amino acid.56 The close relatedness of MERS-CoV and various bat viruses allows speculation that its ancestors might exist in Old World bats.57
Though S protein of the bat coronavirus HKU4 can recognize human DPP4, it is not activated by human proteases and therefore cannot mediate viral entry into human cells.58, 59 However, the introduction of two mutations which are already present in MERS-CoV S protein, S746R and N762A, into HKU4 S protein enabled its activation by human proteases and entry into human cells.60 It had been previously shown that two mutations in S protein allowed SARS-CoV to be transmitted from civets to humans.61 A similar event in bat coronaviruses could explain the emergence of MERS-CoV and its ability to cross the species barrier between bats and humans, directly or through an intermediate host.62, 63
Dromedary Camels as Reservoirs for MERS-CoV
Camels are large mammals with distinctive humps. Two species of camels are in current existence; the two-humped bactrians (Camelus bactrianus) and the one-humped dromedaries (Camelus dromedarius).64 Bactrian camels inhabit Central Asia and constitute around five per cent of the world’s camel population.64 On the other hand, the majority of dromedary camels are found in Eastern Africa, from where they are exported to countries in the Arabian Peninsula.64, 65 Saudi Arabia, UAE, Qatar and Yemen have the largest populations of dromedary camels in the Middle East.66 In this region, camels have prominent economic, cultural and recreational significance. In addition to being a source of milk and meat, dromedary camels are involved in racing, parades and annual festivals.67 Consumption of unpasteurized camel milk is not uncommon, and camel urine is widely believed to have medicinal benefits.67 Socio-economic development and rapid urbanization in the region has resulted in camel farms becoming gradually concentrated in close proximity to major cities.65, 68 There are therefore ample opportunities in the Middle East for direct human contact with camels and their products.
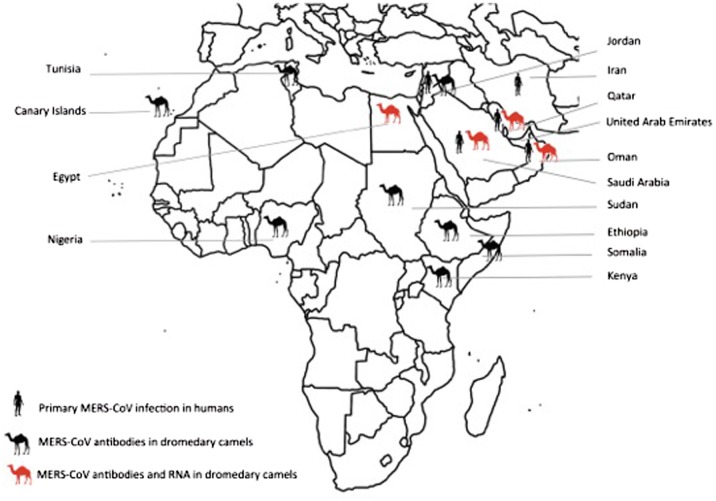
Multiple lines of evidence implicate dromedary camels in the emergence and transmission of MERS-CoV. Firstly, MERS-CoV antibodies are highly prevalent in dromedary camels from across the Arabian Peninsula, North Africa and Eastern Africa (Figure 1 and Table 1).29–31, 34, 70 MERS-CoV antibodies were detected in stored camel sera dating as far back as early 1990s, suggesting that MERS-CoV may have been circulating in dromedaries for over 20 years before it was first recognized as a cause of human infection.32,71,72 The prevalence of MERS-CoV antibodies is significantly higher in older camels compared with those aged two years or less.31, 32 Interestingly, despite the high overall prevalence of MERS-CoV seropositivity in dromedary camels from Kenya, no MERS-CoV antibodies were detected in those from the North Eastern region where dromedaries have been raised largely in isolation from those in the rest of the country.71 Similarly, all dromedaries raised at the Dubai Central Veterinary Research Laboratory, which had no contact with other camels, were seronegative for MERS-CoV.75 Also noteworthy is that 15 of 105 dromedary camels from the Canary Islands were seropositive for MERS-CoV. Although the majority of camels in the herd were born and bred in the Canary Islands, the group included three camels that were imported from Morocco.29 In contrast, no MERS-CoV antibodies were detected in dromedary camels from Australia, Canada, the United States of America, Germany, the Netherlands or Japan.29, 34 The high prevalence of MERS-CoV seropositivity in Africa and the Middle East suggests that animal movement has facilitated the transmission and circulation of MERS-CoV amongst dromedary camels in these regions. MERS-CoV antibodies have neither been found in Mongolian or Dutch bactrian camels, nor in South American camelids such as lamas, alpacas and guanacos.29
Figure 1.
Countries with primary human MERS-CoV infections and MERS-CoV antibody and RNA detection in dromedary camels.
Table 1.
Summary of studies reporting prevalence of MERS-CoV antibodies in dromedary camels
| Location | Sampling year(s) | Camel age | Number | % Positive* |
|---|---|---|---|---|
| Saudi Arabia31 | 2012–2013 | <1 to >5 years | 310 | 90.3 |
| Saudi Arabia32 | 1992–1996 | NA | 132 | 93.2 |
| 2004–2010 | NA | 132 | 81.1 | |
| 2013 | NA | 203 | 73.9 | |
| Saudi Arabia85 | 2013 | NA | 9 | 100 |
| Qatar89 | 2013 | NA | 14 | 100 |
| Qatar79 | 2014 | 76 aged ≤1 year | 105 | 97 |
| 29 aged >1 year | ||||
| UAE34 | 2005 | NA | 11 | 81.8 |
| UAE75 | 2003 | Adult | 151 | 100 |
| 2013 | 2–8 years | 500 | 97.2 | |
| UAE74 | 2014 | <1 year | 108 | 85.2 |
| 2–4 years | 340 | 96.5 | ||
| >4 years | 310 | 96.1 | ||
| Unknown | 85 | 80.0 | ||
| All ages | 843 | 93.2 | ||
| UAE83 | 2015 | 4–10 years | 8 | 100 |
| Oman29 | 2013 | 8–12 years | 50 | 100 |
| Jordan30 | 2013 | 3–14 months | 11 | 100 |
| Egypt82 | 2013 | >6 years | 52 | 92.3 |
| Egypt33 | 2013 | 5–7 years | 110 | 98.2 |
| Egypt103 | 1997 | >6 years | 43 | 81.4 |
| Tunisia69 | 2009 | ≤2 years | 46 | 30 |
| >2 years | 158 | 54 | ||
| Ethiopia69 | 2010–2011 | ≤2 years | 31 | 93 |
| >2 years | 157 | 97 | ||
| Nigeria69 | 2010–2011 | >2 years | 358 | 94 |
| Somalia103 | 1983–1984 | NA | 86 | 83.7 |
| Sudan103 | 1984 | NA | 60 | 86.7 |
| Kenya71 | 1992–2013 | NA | 774 | 29.5 |
| Canary Islands29 | 2013 | 17 aged ≤4 years | 105 | 14.3 |
| 88 aged >4 years |
NA: not available; UAE: United Arab Emirates.
The highest proportion positive by any serological assay used in the study
The second line of evidence is the reported detection of MERS-CoV by RT-PCR in oro-nasal and faecal samples from dromedary camels in multiple locations in the Arabian Peninsula (Figure 1 and Table 2).32, 70, 78 The four of 110 dromedary camels in which MERS-CoV RNA was detected in Egypt were all imported from Sudan or Ethiopia for slaughter.82 Of particular concern is the particularly high rate of MERS-CoV RNA detection in camels presented for slaughter in Eastern Saudi Arabia and Qatar; the latter was in the vicinity of a market in Doha to which two prior human cases were linked.78 Overall, MERS-CoV is more commonly detected and in higher viral loads in nasal swabs than in faecal samples.32 Another notable observation is that sequences from MERS-CoV strains isolated several months apart within the same location are often identical to one another, indicating that MERS-CoV circulation in dromedary herds is very stable.73,74
Table 2.
Summary of MERS-CoV RT-PCR studies in dromedary camels
| Location | Sampling year(s) | Sample type | Number | Camel age | RT-PCR (% positive) |
|---|---|---|---|---|---|
| Saudi Arabia78 | 2013–2014 | NS | 36 | <4 years | 41.7 |
| NS | 60 | ≥4 years | 21.7 | ||
| LT | 28 | <4 years | 82.1 | ||
| LT | 63 | ≥4 years | 52.4 | ||
| Saudi Arabia32 | 2013 | B, NS and RS | 104 | ≤2 years | 34.6 |
| 2013 | 98 | >2 years | 15.3 | ||
| Saudi Arabia73 | 2013–2014 | B, NS, OS and RS | 27 | ≤1 year | 25.9 |
| 14 | >1 years | 14.3 | |||
| Saudi Arabia85 | 2013 | NS | 9 | NA | 22.2 |
| Saudi Arabia86 | 2013 | NS | 3 | ≤1 year | 33.3 |
| 6 | >1 year | 0 | |||
| Qatar89 | 2013 | B, NS and RS | 14 | NA | 35.7 |
| Qatar80 | 2014 | NS | 53 | NA | 1.8 |
| Qatar79 | 2014 | NS, OS, RS and LNT | 105 | 76 aged ≤1 year | 59.0 |
| 29 aged >1 year | |||||
| UAE81 | 2015 | NS | 7,803 | NA | 1.6 |
| UAE34 | 2005 | B | 11 | NA | 0 |
| UAE75 | 2013 | F | 182 | NA | 0 |
| UAE74 | 2014 | B, NS | 250 | >4 years | 0 |
| 344 | 2–4 years | 2.9 | |||
| 68 | <1 years | 35.3 | |||
| 209 | Unknown | 5.3 | |||
| 871 | All ages | 5.1 | |||
| UAE83 | 2015 | NS | 8 | 4–10 years | 100 |
| Oman70 | 2013 | CS, NS | 76 | NA | 6.6 |
| Egypt82 | 2013 | NS | 110 | >6 years | 3.6 |
B: blood; CS: conjunctival swab; F: faeces; LT: lung tissue; LNT: lymph node tissue; NA: not available; NS: nasal swab; OS: oral swab; RS: rectal swab; S: serum; UAE: United Arab Emirates
Interestingly, the prevalence of MERS-CoV RNA shedding is significantly higher in juvenile than in adult camels.32, 78 In one prospective study, MERS-CoV detection by RT-PCR in dromedary camels was highest in the period between the months of November and January, coinciding with the dromedaries’ calving season.78 This, along with the observed increased incidence of human MERS-CoV infection during the period between March and May, suggests that juvenile camels are an important source of new infections in camels and potentially humans.78
MERS-CoV RNA prevalence data also indicate that animal movement may facilitate the introduction of MERS-CoV into herds of dromedaries. For example, phylogenetic analysis of MERS-CoV strains that were detected in a herd of dromedary camels in Dubai showed that they were closely related to those circulating in Eastern Saudi Arabia, where some of the camels had recently grazed.74 In another example, a group of camels imported from Oman was screened by RT-PCR on arrival in UAE and was found positive for MERS-CoV.83 Moreover, MERS-CoV strains isolated from dromedary camels in Eastern Saudi Arabia were phylogenetically related to those isolated from camels several hundred kilometres away in Buraidah, central Saudi Arabia.73 Most significantly, phylogenetic analyses of partial and whole MERS-CoV genomes from dromedary camels show that they are clustered within human isolates, supporting possible camel–human inter-transmission.32, 70, 78, 84 RT-PCR was positive in camels that had prior evidence of MERS-CoV seropositivity, indicating that animal re-infection is possible.73,74,79,83
The third level of evidence is the demonstration of active MERS-CoV infection in dromedary camels through documented rises in anti-MERS-CoV antibody titres.85 MERS-CoV was also detected by RT-PCR in symptomatic camels.78, 85 Dromedaries with active MERS-CoV infection exhibited symptoms such muco-purulent nasal and lachrymal discharge, cough, sneezing, fever and loss of appetite.78, 85
It was initially argued that detection of MERS-CoV in camels by RT-PCR is not necessarily evidence of shedding of infectious virus and thus transmissibility of MERS-CoV between dromedaries and humans.87, 88 However, several groups have since been able to isolate viable MERS-CoV in cell cultures of nasal and faecal samples from dromedary camels.84 Moreover, the potential infectiousness of MERS-CoV recovered from dromedary camels was evident by its capability to cause ex-vivo infection in human respiratory cells and human hepatoma cells (Huh-7).80,90 Successful MERS-CoV cultures usually coincide with corresponding high viral loads in the same specimens.84
The fifth level of evidence is the successful experimental MERS-CoV infection of dromedary camels with resultant mild clinical infection manifesting as fever and rhinorrhea.91 Three MERS-CoV seronegative adult dromedary camels were inoculated via intranasal, intra-tracheal and conjunctival routes with a total dose of 107 50% tissue culture infective dose (TCID50) of MERS-CoV. Clinical symptoms appeared within 2 days and persisted for up to 2 weeks. Submucosal inflammation and necrosis were evident in the upper and lower respiratory tracts, but not the alveoli. MERS-CoV antibodies were detected within 14 days of inoculation. Shedding of infectious MERS-CoV in oral and nasal secretions, as determined by plaque assay titres, continued for up 5 and 7 days, respectively. MERS-CoV was detectable by quantitative PCR for up to 35 days.91 Under certain condition, MERS-CoV can survive on plastic and steel surfaces for up to 30 hours.92 Moreover, MERS-CoV RNA can be detected in milk expressed from MERS-CoV infected camels.93 Whether MERS-CoV is secreted in camel milk or it is contaminated from the animal’s other body secretions is probably immaterial as viable MERS-CoV can be recovered from dromedary camel milk stored at 22 degrees Celsius for up to 48 hours.94 It is therefore possible to envisage that in the absence of appropriate precautions, the environment surrounding a MERS-CoV infected camel can become extensively contaminated with viable, potentially infectious virus.
Camel–Human MERS-CoV Inter-transmission
The strongest direct evidence yet of transmissibility of MERS-CoV between dromedary camels and humans is the simultaneous isolation of near-identical MERS-CoV strains from epidemiologically linked humans and dromedary camels. In October 2013, a 61-year-old owner of a herd of dromedary camels in Qatar and his 23-year-old co-worker were diagnosed with laboratory-confirmed MERS-CoV infection. MERS-CoV was detected by RT-PCR in five of their 14 camels.89 Alignment of six MERS-CoV genomic fragments, covering 4.2 kb of the MERS-CoV genome, from the camels and the human cases showed that they differed by one nucleotide each in ORF1a and ORF4b regions.89
In November 2013, a 43-year-old man died in a hospital in Jeddah, Saudi Arabia, with severe MERS-CoV infection. He had owned a herd of nine dromedary camels, some of which were reported to have had recent respiratory symptoms. MERS-CoV was detected by PCR and culture of nasal specimens from one camel.85 Partial MERS-CoV sequence of 4.6 kb from the camel and the linked human isolates differed in only two positions.85 Whole MERS-CoV genome sequences obtained from viral cultures of the human and camel isolates were 100% identical.86 Importantly, 4-fold rise in MERS-CoV antibody titres was documented in the camels, indicating that active MERS-CoV infection was probably circulating in the dromedary herd.85 Later, rising MERS-CoV antibodies were documented in the patient, suggesting that MERS-CoV infection was transmitted from the camels to the human and not vice versa.86
In May 2015, MERS-CoV was detected in eight asymptomatic dromedary camels at entry into UAE from Oman. Two asymptomatic men, aged 29 and 33 years, who were in contact with the camels were found to be positive for MERS-CoV RNA in their respiratory samples.83 Partial sequences of MERS-CoV spike, ORF3-4a and nucleocapsid regions from the human and linked camels were identical.83 Within 4–8 days from diagnosis, both patients had undetectable MERS-CoV RNA. All dromedaries were seropositive for MERS-CoV antibodies. However, two juvenile camels had high MERS-CoV viral loads in their nasal specimens and were last to become RT-PCR negative.83 The sequence of events strongly suggests that MERS-CoV infection occurred in the dromedary camels before the human cases.
MERS-CoV seroprevalence studies in humans with close contact with camels have yielded inconsistent results. A national serosurvey in Saudi Arabia found prevalence of MERS-CoV antibodies that was 15 times higher in camel shepherds (P = 0.0004) and 23 times higher in slaughterhouse workers (P < 0.0001), compared with the general population.95 Likewise, individuals who had occupational exposure to dromedary camels in Qatar were seropositive for MERS-CoV but not those without such exposure.96 However, none of 191 persons who had close occupational contacts with MERS-CoV infected camels were seropositive for MERS-CoV.97 Similarly, MERS-CoV antibodies were not detected in any of 114 animal workers in contact with MERS-CoV RNA positive camels in Egypt.82 Slaughterhouse and other animal workers in Western and Southern Saudi Arabia were also seronegative for MERS-CoV antibodies.98,99 Collectively, the available data indicate that MERS-CoV is highly prevalent in dromedary camels in the Arabian Peninsula and that transmission of infection from camels to humans, although inefficient, does occur.
Connecting the Dots
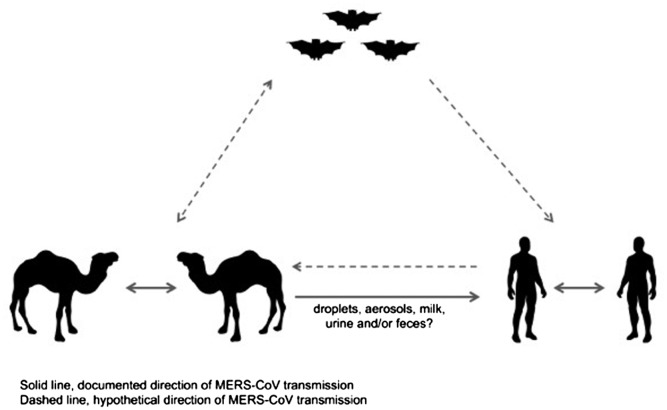
The sequenced full genomes of NeoCoV, the novel betacoronavirus that was isolated from vesper bats in South Africa and MERS-CoV strains from dromedary camels and humans confirm that they all belong to the same species.57 NeoCoV is at the root of their phylogenetic tree, with evidence of genetic evolution of MERS-CoV in camels before humans.57 Furthermore, the high similarity of NeoCoV and MERS-CoV with genetic divergence in NeoCoV spike gene suggests that a recombination event within this region may have resulted in the emergence of MERS-CoV.57 The high seroprevalence of MERS-CoV in camels in Eastern Africa indicates that such recombination event might have taken place in dromedaries, or another yet unidentified intermediate host, in Eastern Africa and that MERS-CoV followed the camel trading routes to emerge in humans in the Arabian Peninsula (Figure 2).41, 42 The fact that MERS-CoV antibodies have been detected in camel sera from Eastern Africa and the Arabian Peninsula dating back to the early 1990s supports such a hypothesis. Once established in dromedary camels, occasional transmission to humans is evident.
Figure 2.
Documented and theorized transmission directions of MERS-CoV between bats, camels and humans.
In a large seroprevalence survey in Saudi Arabia conducted between December 2012 and December 2013, anti-MERS-CoV antibodies were detected in 0.15% of 10,009 samples. The authors extrapolated that around 45,000 individuals in Saudi Arabia could be seropositive for MERS-CoV.95 It therefore appears that human MERS-CoV infection had taken place in the region for some considerable time before it was identified. The recent identification of MERS-CoV infection in asymptomatic human contacts of MERS-CoV infected camels in UAE provides significant insight into the possible chain of events following such exposure.83 It is reasonable to theorize that infection from such asymptomatic individuals may be transmitted to others. A person admitted to a health care facility with unrecognized MERS-CoV infection can trigger clusters of various sizes. Indeed, the appearance of several community and hospital MERS-CoV clusters in the first half of the year 2013 without identifiable human or animal sources led to speculations that individuals with no or only mild respiratory symptoms might have a role in MERS-CoV transmissions.3, 4, 7 Memish et al. showed that MERS-CoV was detectable for up to 12 days in 30% of asymptomatic contacts.100 In another report, an asymptomatic health care worker had detectable MERS-CoV for over five weeks.101 Although MERS-CoV transmission from an asymptomatic individual is a strong probability, this has never been documented.12,13
MERS-CoV Control at the Animal–Human Interface
In countries where MERS-CoV is already established in dromedary camels, preventive strategies are unlikely to succeed without addressing such sources. Key elements for MERS-CoV control in animals should include the following:
-
(1)
Strict regulation of camel movement with imposition of requirement for MERS-CoV clearance prior to importation and transport of camels, including those that are presented for slaughter. Camels with detectable MERS-CoV RNA should be quarantined and tested at regular intervals.
-
(2)
Enforcing the use of personal protective equipment while handling dromedary camels.
-
(3)
Efforts to increase awareness amongst camel owners and the general public of the risks of consuming unpasteurized camel milk and urine. This may prove challenging, given the depth of customs and beliefs in some areas.
-
(4)
Accelerated development of safe and effective MERS-CoV vaccines for animal or human use.102
Conclusion
MERS-CoV is a zoonotic disease with bats and dromedary camels playing important parts in its emergence and epidemiology. Camel to human MERS-CoV transmission is well documented but is generally not very efficient. The exact mechanism of transmission is not clear, including whether other intermediate hosts are involved. Serosurveys in humans across Africa are urgently needed to investigate the possibility of unrecognized MERS-CoV infections in the continent. Furthermore, bats in Eastern Africa should be screened for betacoronaviruses that may provide better understanding of the genetic history of MERS-CoV. Finally, case-control studies of humans with sporadic MERS-CoV infection are urgently needed to identify risk factors and exposures that might explain the chains of transmission from camels and other possible zoonotic or environmental sources of human infections.
References
- 1.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus ADME, Fouchier RAM. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Eng J Med. 2012;367:1814–1820. [DOI] [PubMed] [Google Scholar]
- 2.World Health Organization Middle East respiratory syndrome coronavirus (MERS-CoV); Summary of Current Situation, Literature Update and Risk Assessment 7 July 2015. Geneva: WHO; 2015. [Google Scholar]
- 3.Memish ZA, Zumla AI, Al-Hakeem RF, Al-Rabeeah AA, Stephens GM. Family cluster of Middle East respiratory syndrome coronavirus infections. N Eng J Med. 2013;368:2487–2494. [DOI] [PubMed] [Google Scholar]
- 4.Omrani AS, Matin MA, Haddad Q, Al-Nakhli D, Memish ZA, Albarrak AM. A family cluster of Middle East Respiratory Syndrome Coronavirus infections related to a likely unrecognized asymptomatic or mild case. Int J Infect Dis. 2013;17:e668–e672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Oboho IK, Tomczyk SM, Al-Asmari AM, Banjar AA, Al-Mugti H, Aloraini MS, et al. 2014 MERS-CoV outbreak in Jeddah – a link to health care facilities. N Eng J Med. 2015;372:846–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Memish ZA, Cotten M, Watson SJ, Kellam P, Zumla A, Alhakeem RF, et al. Community case clusters of Middle East respiratory syndrome coronavirus in Hafr Al-Batin, Kingdom of Saudi Arabia: a descriptive genomic study. Int J Infect Dis. 2014;23:63–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Assiri A, McGeer A, Perl TM, Price CS, Al Rabeeah AA, Cummings DA, et al. Hospital outbreak of Middle East respiratory syndrome coronavirus. N Eng J Med. 2013;369:407–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guery B, Poissy J, el Mansouf L, Sejourne C, Ettahar N, Lemaire X, et al. Clinical features and viral diagnosis of two cases of infection with Middle East Respiratory Syndrome coronavirus: a report of nosocomial transmission. Lancet. 2013;381:2265–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Saad M, Omrani AS, Baig K, Bahloul A, Elzein F, Matin FA, et al. Clinical aspects and outcomes of 70 patients with Middle East respiratory syndrome coronavirus infection: a single-center experience in Saudi Arabia. Int J Infect Dis. 2014;29:301–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee SS, Wong NS. Probable transmission chains of MERS-CoV and the multiple generations of secondary infections in South Korea. Int J Infect Dis. 2015;38:65–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Breban R, Riou J, Fontanet A. Interhuman transmissibility of Middle East respiratory syndrome coronavirus: estimation of pandemic risk. Lancet. 2013;382:694–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drosten C, Meyer B, Muller MA, Corman VM, Al-Masri M, Hossain R, et al. Transmission of MERS-coronavirus in household contacts. N Eng J Med. 2014;371:828–835. [DOI] [PubMed] [Google Scholar]
- 13.Memish ZA, Al-Tawfiq JA, Makhdoom HQ, Al-Rabeeah AA, Assiri A, Alhakeem RF, et al. Screening for Middle East respiratory syndrome coronavirus infection in hospital patients and their healthcare worker and family contacts: a prospective descriptive study. Clin Microbiol Infect Dis. 2014;20:469–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.World Health Organization Middle East Respiratory Syndrome coronavirus (MERS-CoV) – Saudi Arabia 12 August 2015. Geneva: WHO; 2015. [Google Scholar]
- 15.Albarrak AM, Stephens GM, Hewson R, Memish ZA. Recovery from severe novel coronavirus infection. Saudi Med J. 2012;33:1265–1269. [PubMed] [Google Scholar]
- 16.Belouzard S, Millet JK, Licitra BN, Whittaker GR. Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses. 2012;4:1011–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R, et al. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature. 2013;495:251–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zumla A, Hui DS, Perlman S. Middle East respiratory syndrome. Lancet. 2015;386:995–1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Doremalen N, Miazgowicz KL, Milne-Price S, Bushmaker T, Robertson S, Scott D, et al. Host species restriction of Middle East respiratory syndrome coronavirus through its receptor, dipeptidyl peptidase 4. J Virol. 2014;88:9220–9232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao J, Li K, Wohlford-Lenane C, Agnihothram SS, Fett C, Zhao J, et al. Rapid generation of a mouse model for Middle East respiratory syndrome. Proc Natl Acad Sci USA. 2014;111:4970–4975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.de Wit E, Rasmussen AL, Falzarano D, Bushmaker T, Feldmann F, Brining DL, et al. Middle East respiratory syndrome coronavirus (MERS-CoV) causes transient lower respiratory tract infection in rhesus macaques. Proc Natl Acad Sci USA. 2013;110:16598–16603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yao Y, Bao L, Deng W, Xu L, Li F, Lv Q, et al. An animal model of MERS produced by infection of rhesus macaques with MERS coronavirus. J Infect Dis. 2014;209:236–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munster VJ, de Wit E, Feldmann H. Pneumonia from human coronavirus in a macaque model. N Eng J Med. 2013;368:1560–1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Falzarano D, de Wit E, Feldmann F, Rasmussen AL, Okumura A, Peng X, et al. Infection with MERS-CoV causes lethal pneumonia in the common marmoset. PLoS Pathog. 2014;10:e1004250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Haagmans BL, van den Brand JM, Provacia LB, Raj VS, Stittelaar KJ, Getu S, et al. Asymptomatic Middle East respiratory syndrome coronavirus infection in rabbits. J Virol. 2015;89:6131–6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Falzarano D, de Wit E, Rasmussen AL, Feldmann F, Okumura A, Scott DP, et al. Treatment with interferon-alpha2b and ribavirin improves outcome in MERS-CoV-infected rhesus macaques. Nature Med. 2013;19:1313–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chan JF, Yao Y, Yeung ML, Deng W, Bao L, Jia L, et al. Treatment with lopinavir/ritonavir or interferon-beta1b improves outcome of MERS-CoV infection in a non-human primate model of common marmoset. J Infect Dis. 2015;212:1904–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Eckerle I, Corman VM, Muller MA, Lenk M, Ulrich RG, Drosten C. Replicative capacity of MERS coronavirus in livestock cell lines. Emerg Infect Dis. 2014;20:276–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reusken CB, Haagmans BL, Muller MA, Gutierrez C, Godeke GJ, Meyer B, et al. Middle East respiratory syndrome coronavirus neutralising serum antibodies in dromedary camels: a comparative serological study. Lancet Infect Dis. 2013;13:859–866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reusken CB, Ababneh M, Raj VS, Meyer B, Eljarah A, Abutarbush S, et al. Middle East Respiratory Syndrome coronavirus (MERS-CoV) serology in major livestock species in an affected region in Jordan, June to September 2013. Euro Surveill. 2013;18:20662. [DOI] [PubMed] [Google Scholar]
- 31.Hemida MG, Perera RA, Wang P, Alhammadi MA, Siu LY, Li M, et al. Middle East Respiratory Syndrome (MERS) coronavirus seroprevalence in domestic livestock in Saudi Arabia, 2010 to 2013. Euro Surveill. 2013;18:20659. [DOI] [PubMed] [Google Scholar]
- 32.Alagaili AN, Briese T, Mishra N, Kapoor V, Sameroff SC, Burbelo PD, et al. Middle East respiratory syndrome coronavirus infection in dromedary camels in Saudi Arabia. mBio. 2014;5:e00884-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Perera RA, Wang P, Gomaa MR, El-Shesheny R, Kandeil A, Bagato O, et al. Seroepidemiology for MERS coronavirus using microneutralisation and pseudoparticle virus neutralisation assays reveal a high prevalence of antibody in dromedary camels in Egypt, June 2013. Euro Surveill. 2013;18:pii 574. [DOI] [PubMed] [Google Scholar]
- 34.Alexandersen S, Kobinger GP, Soule G, Wernery U. Middle East respiratory syndrome coronavirus antibody reactors among camels in Dubai, United Arab Emirates, in 2005. Transbound Emerg Dis. 2014;61:105–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Han HJ, Wen HL, Zhou CM, Chen FF, Luo LM, Liu JW, et al. Bats as reservoirs of severe emerging infectious diseases. Virus Res. 2015;205:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karesh WB, Dobson A, Lloyd-Smith JO, Lubroth J, Dixon MA, Bennett M, et al. Ecology of zoonoses: natural and unnatural histories. Lancet. 2012;380:1936–1945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobson AP. What links bats to emerging infectious diseases? Science. 2005;310:628–629. [DOI] [PubMed] [Google Scholar]
- 38.George DB, Webb CT, Farnsworth ML, O'Shea TJ, Bowen RA, Smith DL, et al. Host and viral ecology determine bat rabies seasonality and maintenance. Proc Natl Acad Sci USA. 2011;108:10208–10213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Calisher CH, Childs JE, Field HE, Holmes KV, Schountz T. Bats: important reservoir hosts of emerging viruses. Clin Microbiol Rev. 2006;19:531–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luis AD, Hayman DTS, O’Shea TJ, Cryan PM, Gilbert AT, Pulliam JRC, et al. A comparison of bats and rodents as reservoirs of zoonotic viruses: are bats special? Proc R Soc Lond B Biol Sci. 2013;280:20122753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drexler JF, Corman VM, Drosten C. Ecology, evolution and classification of bat coronaviruses in the aftermath of SARS. Antiviral Res. 2014;101:45–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brook CE, Dobson AP. Bats as ‘special’ reservoirs for emerging zoonotic pathogens. Trends Microbiol. 2015;23:172–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Drosten C, Günther S, Preiser W, van der Werf S, Brodt H-R, Becker S, et al. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N Eng J Med. 2003;348:1967–1976. [DOI] [PubMed] [Google Scholar]
- 44.Ge X-Y, Li J-L, Yang X-L, Chmura AA, Zhu G, Epstein JH, et al. Isolation and characterization of a bat SARS-like coronavirus that uses the ACE2 receptor. Nature. 2013;503:535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guan Y, Zheng BJ, He YQ, Liu XL, Zhuang ZX, Cheung CL, et al. Isolation and characterization of viruses related to the SARS coronavirus from animals in Southern China. Science. 2003;302:276–278. [DOI] [PubMed] [Google Scholar]
- 46.Lau SKP, Woo PCY, Li KSM, Huang Y, Tsoi H-W, Wong BHL, et al. Severe acute respiratory syndrome coronavirus-like virus in Chinese horseshoe bats. Proc Natl Acad Sci USA. 2005;102:14040–14045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woo PCY, Wang M, Lau SKP, Xu H, Poon RWS, Guo R, Comparative analysis of twelve genomes of three novel group 2c and group 2d coronaviruses reveals unique group and subgroup features. J Virol. 2007;81:1574–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Corman VM, Kallies R, Philipps H, Gopner G, Muller MA, Eckerle I, et al. Characterization of a novel betacoronavirus related to Middle East respiratory syndrome coronavirus in European hedgehogs. J Virol. 2014;88:717–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lelli D, Papetti A, Sabelli C, Rosti E, Moreno A, Boniotti MB. Detection of coronaviruses in bats of various species in Italy. Viruses. 2013;5:2679–2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Memish ZA, Mishra N, Olival KJ, Fagbo SF, Kapoor V, Epstein JH, et al. Middle East respiratory syndrome coronavirus in bats. Saudi Arabia. Emerg Infect Dis. 2013;19:1819–1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Annan A, Baldwin HJ, Corman VM, Klose SM, Owusu M, Nkrumah EE, et al. Human betacoronavirus 2c EMC/2012-related viruses in bats. Ghana and Europe. Emerg Infect Dis. 2013;19:456–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yang L, Wu Z, Ren X, Yang F, Zhang J, He G, et al. MERS-related betacoronavirus in vespertilio superans bats. China. Emerg Infect Dis. 2014;20:1260–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Benedictis P, Marciano S, Scaravelli D, Priori P, Zecchin B, Capua I, et al. Alpha and lineage C betaCoV infections in Italian bats. Virus Genes. 2014;48:366–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Anthony SJ, Ojeda-Flores R, Rico-Chavez O, Navarrete-Macias I, Zambrana-Torrelio CM, Rostal MK, et al. Coronaviruses in bats from Mexico. J Gen Virol. 2013;94:1028–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wacharapluesadee S, Sintunawa C, Kaewpom T, Khongnomnan K, Olival KJ, Epstein JH, et al. Group C betacoronavirus in bat guano fertilizer. Thailand. Emerg Infect Dis. 2013;19:1349–1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ithete NL, Stoffberg S, Corman VM, Cottontail VM, Richards LR, Schoeman MC, et al. Close relative of human Middle East respiratory syndrome coronavirus in bat. South Africa. Emerg Infect Dis. 2013;19:1697–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Corman VM, Ithete NL, Richards LR, Schoeman MC, Preiser W, Drosten C, et al. Rooting the phylogenetic tree of Middle East respiratory syndrome coronavirus by characterization of a conspecific virus from an African bat. J Virol. 2014;88:11297–11303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yang Y, Du L, Liu C, Wang L, Ma C, Tang J, et al. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc Natl Acad Sci USA. 2014;111:12516–12521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Q, Qi J, Yuan Y, Xuan Y, Han P, Wan Y, et al. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe. 2014;16:328–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yang Y, Liu C, Du L, Jiang S, Shi Z, Baric RS, et al. Two mutations were critical for bat-to-human transmission of Middle East Respiratory Syndrome Coronavirus. J Virol. 2015;89:9119–9123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li W, Zhang C, Sui J, Kuhn JH, Moore MJ, Luo S, et al. Receptor and viral determinants of SARS-coronavirus adaptation to human ACE2. EMBO J. 2005;24:1634–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lu G, Wang Q, Gao GF. Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol. 2015;23:468–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Graham RL, Baric RS. Recombination, reservoirs, and the modular spike: mechanisms of coronavirus cross-species transmission. J Virol. 2010;84:3134–3146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wikemedia Foundation Camel. San Diego, CA: Wikimedia; 2015. [Google Scholar]
- 65.French Agricultural Research and International Cooperation Organization Camels and dromedaries: a rapidly changing sector supported by dynamic research. Paris: CIRAD; 2013. [Google Scholar]
- 66.Gossner C, Danielson N, Gervelmeyer A, Berthe F, Faye B, Kaasik Aaslav K, et al. Human-dromedary camel interactions and the risk of acquiring zoonotic Middle East Respiratory Syndrome Coronavirus infection. Zoo Public Health. Epub 2014 Dec 27. doi: 10.1111/zph.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mackay IM, Arden KE. Middle East respiratory syndrome: an emerging coronavirus infection tracked by the crowd. Virus Res. 2015;202:60–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Abdallah HR, Faye F. Typology of camel farming system in Saudi Arabia. Emir J Food Agric. 2013;25:250–260. [Google Scholar]
- 69.Reusken CB, Messadi L, Feyisa A, Ularamu H, Godeke GJ, Danmarwa A, et al. Geographic distribution of MERS coronavirus among dromedary camels. Africa. Emerg Infect Dis. 2014;20:1370–1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nowotny N, Kolodziejek J. Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels, Oman, 2013. Euro Surveill. 2014;19:20781. [DOI] [PubMed] [Google Scholar]
- 71.Corman VM, Jores J, Meyer B, Younan M, Liljander A, Said MY, et al. Antibodies against MERS coronavirus in dromedary camels, Kenya, 1992–2013. Emerg Infect Dis. 2014;20:1319–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hemida MG, Perera RA, Al Jassim RA, Kayali G, Siu LY, Wang P, et al. Seroepidemiology of Middle East respiratory syndrome (MERS) coronavirus in Saudi Arabia (1993) and Australia (2014) and characterisation of assay specificity. Euro Surveill. 2014;19:20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hemida MG, Chu DK, Poon LL, Perera RA, Alhammadi MA, Ng HY, et al. MERS coronavirus in dromedary camel herd. Saudi Arabia. Emerg Infect Dis. 2014;20:1231–1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wernery U, Corman VM, Wong EY, Tsang AK, Muth D, Lau SK, et al. Acute Middle East respiratory syndrome coronavirus infection in livestock Dromedaries, Dubai, 2014. Emerg Infect Dis. 2015;21:1019–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Meyer B, Muller MA, Corman VM, Reusken CB, Ritz D, Godeke GJ, et al. Antibodies against MERS coronavirus in dromedary camels, United Arab Emirates, 2003 and 2013. Emerg Infect Dis. 2014;20:552–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shirato K, Azumano A, Nakao T, Hagihara D, Ishida M, Tamai K, et al. Middle East respiratory syndrome coronavirus infection not found in camels in Japan. Jpn J Infect Dis. 2015;68:256–258. [DOI] [PubMed] [Google Scholar]
- 77.Chan SM, Damdinjav B, Perera RA, Chu DK, Khishgee B, Enkhbold B, et al. Absence of MERS-coronavirus in bactrian camels, Southern Mongolia, November 2014. Emerg Infect Dis. 2015;21:1269–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Khalafalla AI, Lu X, Al-Mubarak AI, Dalab AH, Al-Busadah KA, Erdman DD. MERS-CoV in upper respiratory tract and lungs of dromedary camels, Saudi Arabia, 2013-2014. Emerg Infect Dis. 2015;21:1153–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Farag EA, Reusken CB, Haagmans BL, Mohran KA, Raj VS, Pas SD, et al. High proportion of MERS-CoV shedding dromedaries at slaughterhouse with a potential epidemiological link to human cases, Qatar 2014. Infect Ecol Epidemiol. 2015;5:28305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Raj VS, Farag EA, Reusken CB, Lamers MM, Pas SD, Voermans J, et al. Isolation of MERS coronavirus from a dromedary camel, Qatar, 2014. Emerg Infect Dis. 2014;20:1339–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yusof MF, Eltahir YM, Serhan WS, Hashem FM, Elsayed EA, Marzoug BA, et al. Prevalence of Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels in Abu Dhabi Emirate. United Arab Emirates. Virus Genes. 2015;50:509–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chu DK, Poon LL, Gomaa MM, Shehata MM, Perera RA, Abu Zeid D, et al. MERS coronaviruses in dromedary camels. Egypt. Emerg Infect Dis. 2014;20:1049–1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Al Hammadi AM, Chu DKW, Eltahir YM, Al Hosani F, Al Mulla M, Tarnini W, et al. Asymptomatic MERS-CoV infection in humans possibly linked to infected camels imported from Oman to United Arab Emirates, May 2015. Emerg Infect Dis. 2015;21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Briese T, Mishra N, Jain K, Zalmout IS, Jabado OJ, Karesh WB, et al. Middle East respiratory syndrome coronavirus quasispecies that include homologues of human isolates revealed through whole-genome analysis and virus cultured from dromedary camels in Saudi Arabia. mBio. 2014;5:e01146-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Memish ZA, Cotten M, Meyer B, Watson SJ, Alsahafi AJ, Al-Rabeeah AA, et al. Human infection with MERS coronavirus after exposure to infected camels, Saudi Arabia, 2013. Emerg Infect Dis. 2014;20:1012–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Azhar EI, El-Kafrawy SA, Farraj SA, Hassan AM, Al-Saeed MS, Hashem AM, et al. Evidence for camel-to-human transmission of MERS coronavirus. N Eng J Med. 2014;370:2499–2505. [DOI] [PubMed] [Google Scholar]
- 87.Nishiura H, Ejima K, Mizumoto K. Missing information in animal surveillance of MERS-CoV. Lancet Infect Dis. 2014;14:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Samara EM, Abdoun KA. Concerns about misinterpretation of recent scientific data implicating dromedary camels in epidemiology of Middle East respiratory syndrome (MERS). mBio. 2014;5:e01430-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Haagmans BL, Al Dhahiry SH, Reusken CB, Raj VS, Galiano M, Myers R, et al. Middle East respiratory syndrome coronavirus in dromedary camels: an outbreak investigation. Lancet Infect Dis. 2014;14:140–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Chan RW, Hemida MG, Kayali G, Chu DK, Poon LL, Alnaeem A, et al. Tropism and replication of Middle East respiratory syndrome coronavirus from dromedary camels in the human respiratory tract: an in-vitro and ex-vivo study. Lancet Respir Med. 2014;2:813–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Adney DR, van Doremalen N, Brown VR, Bushmaker T, Scott D, de Wit E, et al. Replication and shedding of MERS-CoV in upper respiratory tract of inoculated dromedary camels. Emerg Infect Dis. 2014;20:1999–2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van Doremalen N, Bushmaker T, Munster VJ. Stability of Middle East respiratory syndrome coronavirus (MERS-CoV) under different environmental conditions. Euro Surveill. 2013;18:20590. [DOI] [PubMed] [Google Scholar]
- 93.Reusken CB, Farag EA, Jonges M, Godeke GJ, El-Sayed AM, Pas SD, et al. Middle East respiratory syndrome coronavirus (MERS-CoV) RNA and neutralising antibodies in milk collected according to local customs from dromedary camels, Qatar, April 2014. Euro Surveill. 2014;19:20829. [DOI] [PubMed] [Google Scholar]
- 94.van Doremalen N, Bushmaker T, Karesh WB, Munster VJ. Stability of Middle East respiratory syndrome coronavirus in milk. Emerg Infect Dis. 2014;20:1263–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Müller MA, Meyer B, Corman VM, Al-Masri M, Turkestani A, Ritz D, et al. Presence of Middle East respiratory syndrome coronavirus antibodies in Saudi Arabia: a nationwide, cross-sectional, serological study. Lancet Infect Dis. 2015;15:559–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Reusken CB, Farag EA, Haagmans BL, Mohran KA, Godeke GJ, Raj S, et al. Occupational exposure to dromedaries and risk for MERS-CoV infection, Qatar, 2013–2014. Emerg Infect Dis. 2015;21:1422–1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hemida MG, Al-Naeem A, Perera RA, Chin AW, Poon LL, Peiris M. Lack of Middle East respiratory syndrome coronavirus transmission from infected camels. Emerg Infect Dis. 2015;21:699–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aburizaiza AS, Mattes FM, Azhar EI, Hassan AM, Memish ZA, Muth D, et al. Investigation of anti-Middle East respiratory syndrome antibodies in blood donors and slaughterhouse workers in Jeddah and Makkah, Saudi Arabia, fall 2012. J Infect Dis. 2014;209:243–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Memish ZA, Alsahly A, Masri MA, Heil GL, Anderson BD, Peiris M, et al. Sparse evidence of MERS-CoV infection among animal workers living in Southern Saudi Arabia during 2012. Influenza Other Respir Viruses. 2015;9:64–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Memish ZA, Assiri AM, Al-Tawfiq JA. Middle East respiratory syndrome coronavirus (MERS-CoV) viral shedding in the respiratory tract: an observational analysis with infection control implications. Int J Infect Dis. 2014;29:307–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Al-Gethamy M, Corman VM, Hussain R, Al-Tawfiq JA, Drosten C, Memish ZA. A case of long-term excretion and subclinical infection with Middle East respiratory syndrome coronavirus in a healthcare worker. Clin Infect Dis. 2015;60:973–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hotez PJ, Bottazzi ME, Tseng C-TK, Zhan B, Lustigman S, Du L, et al. Calling for rapid development of a safe and effective MERS vaccine. Microbes Infect. 2014;16:529–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Müller MA, Corman VM, Jores J, Meyer B, Younan M, Liljander A, et al. MERS coronavirus neutralizing antibodies in camels, Eastern Africa, 1983–1997. Emerg Infect Dis. 2014;20:2093–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]