Abstract
The human connectome refers to a comprehensive description of the brain’s structural and functional connections in terms of brain networks. As the field of brain connectomics has developed, data acquisition, subsequent processing and modeling, and ultimately the representation of the connectome have become better defined and integrated with network science approaches. In this way, the human connectome has provided a way to elucidate key features of not only the healthy brain but also diseased brains. The field has quickly evolved, offering insights into network disruptions that are characteristic for specific neurodegenerative disorders. In this paper, we provide a brief review of the field of brain connectomics, as well as a more in-depth survey of recent studies that have provided new insights into brain network pathologies, including those found in Alzheimer’s disease (AD), patients with mild cognitive impairment (MCI), and finally in people classified as being “at risk”. Until the emergence of brain connectomics, most previous studies had assessed neurodegenerative diseases mainly by focusing on specific and dispersed locales in the brain. Connectomics-based approaches allow us to model the brain as a network, which allows for inferences about how dynamic changes in brain function would be affected in relation to structural changes. In fact, looking at diseases using network theory gives rise to new hypotheses on mechanisms of pathophysiology and clinical symptoms. Finally, we discuss the future of this field and how understanding both the functional and structural connectome can aid in gaining sharper insight into changes in biological brain networks associated with cognitive impairment and dementia.
Keywords: Brain connectomics, Neurodegenerative disease, Network science, Cognitive impairment
Introduction
The average life expectancy has steadily increased with respect to each previous generation. As the mean age rises, experimental evidence indicates a greater decline of sensory, motor, and cognitive functions in the elderly population [1]. Given that abnormal aging can result from a variety of factors (such as genes or environment or lifestyle), cognitive impairment associated with aging is considered a systemic problem. Therefore, effective prediction of successful aging versus cognitive decline becomes difficult. Aiming to face this complexity, interest in early markers of cognitive impairment has grown, since it is widely regarded as an early marker of dementia. Recently, hope has been placed in identifying biomarkers that predict risk for developing cognitive impairment. Given advances in non-invasive neuroimaging techniques and in network analysis, the emerging field of brain connectomics is increasingly well situated to link cognitive impairment with structural and/or functional network changes. These changes may be reflected in a number of topological features that cover global and local aspects of the brain organization and communicability.
For the reader’s convenience, a glossary is presented for terms related to network science. Throughout this review words in italics will refer to items present in Table 1 which includes a glossary of network indices.
Table 1.
Glossary for network terms in alphabetical order: network indices
| Index | SC or FC measure | Definition |
|---|---|---|
| Cluster | SC and FC | Refers to a group of nodes that have denser connections with each other than with the rest of the network |
| Edge | SC and FC | Refers to a connection between nodes. Can either be weighted or binary |
| Fractional anisotropy (FA) | SC | A scalar value between 0 (indicating unrestricted diffusivity) and 1 that describes the degree of anisotropy of a diffusion process |
| Functional connectome | FC | A comprehensive map of correlated brain regions as measured by BOLD signals acquired using RS-fMRI studied as a network by means of a network science and graph theory |
| Global efficiency | SC | Related to the idea of the average shortest path is that of global efficiency, which is the inverse of the average shortest path. It is the overall information transfer efficiency across the whole network |
| Hub | SC and FC | A highly connected node. These nodes are relevant for efficient network communication, and damage to these nodes might be especially disruptive for network integrity |
| Independent component analysis (ICA) | FC | A statistical method that aims at decomposing multivariate signal into subcomponents. It has become useful in identifying resting-state networks by separating a signal into unique spatial and time components |
| K-core | SC and FC | A measure that assesses the overall connectedness of a network by identifying the most highly interconnected sub-networks within a global network |
| Modularity | FC | Modularity is revealed by subdividing the network into densely connected groups of nodes |
| Network | SC and FC | Graph comprising a set of nodes joined by a set of connections referred to as edges |
| Network core | SC and FC | Much like a cluster, a network core is a set of nodes that are highly and mutually interconnected |
| Node | SC and FC | The point at which two (or more) edges meet |
| Nodal degree | SC and FC | The amount of points at which two (or more) edges meet |
| Path/fiber length | SC | Describes the (shortest) path length between two nodes |
| Radial diffusivity | SC and FC | Derived from eigenvalues from the diffusion tensor and attempts to better capture white matter pathology by being sensitive to change in myelin in white matter |
| Rich club | SC | A collection of highly connected hubs, thought to be important for integration of information |
| Streamline | SC | A representation of a direct connection obtained through tractography (usually referring to white matter tracts) |
| Structural connectome | SC | A comprehensive map of anatomical white matter connections in the brain studied as a network by means of network science and graph theory |
The Human Connectome
The concept of the human brain as a large-scale complex network termed “connectome” was originally introduced in 2005 [2]. The key proposal was to model the brain as a network, with the different gray matter regions acting as nodes, and their structural connectivity (SC) through white matter fiber bundles as edges. As the first drafts of the human connectome became available [3] an increasing interest in applying graph theory to human neuroimaging data (specifically to diffusion weighted imaging (DWI)) emerged. This helped to uncover topological features of the structural connectome such as small world-ness, highly connected hubs, and modularity. It also allowed modeling and visualization of the most prominent white fiber tracts constructed from bundles of streamlines connecting different brain regions, an important first step in identifying the basic layout of the brain’s SC and its relation to functional connectivity (FC) [4]. Interestingly, the basic idea of the brain as a structural network of connected neurons with functional implications was proposed years, if not decades, before the idea of the connectome. The foundational writings of Santiago Ramon y Cajal were a first antecedent of the neuronal networks we now know [5]. His work and intricate circuit diagrams unveiled individual neurons and their synapses. In comparison, the field of brain connectomics works at a much coarser grain scale and aims to model and assess the complex interactions between neural populations (as divided by regions) to understand the behavior of the system overall. In turn, brain connectomics uses network science to handle the complex methodologies needed to understand the dynamic interactions of different brain regions both functionally and structurally and how these interactions might influence cognition.
The Structural and Functional Connectomes
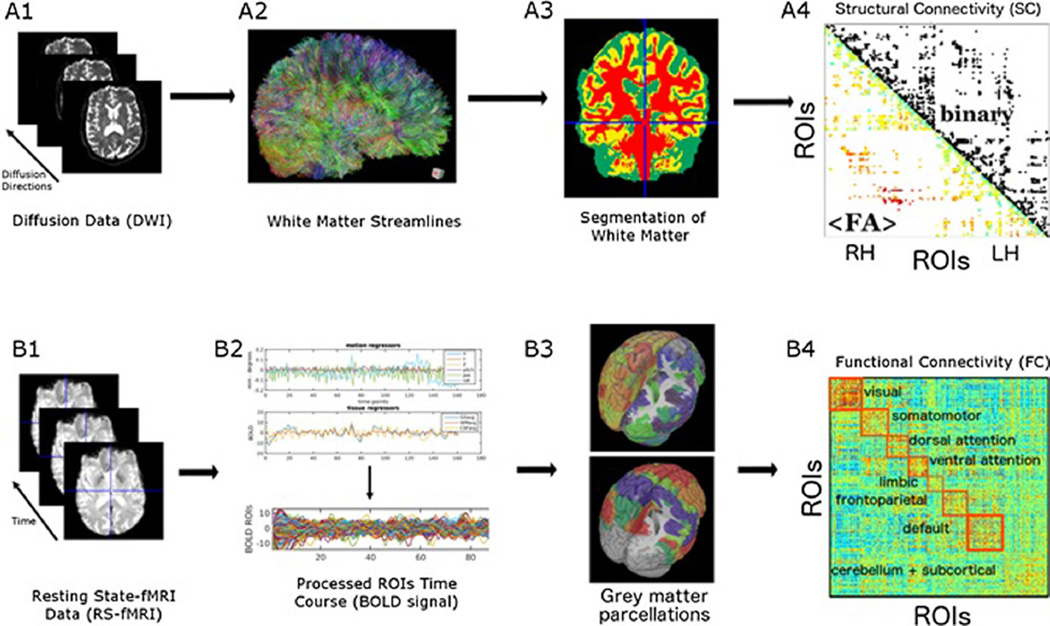
Using DWI to assess SC by modeling white matter fiber tracts, SC information can be represented as a binary network where nodes represent brain regions and edges represent the presence or absence of fibers connecting those regions. To further elaborate this network representation, it is possible to gather fiber descriptors such as number of streamlines, fiber integrity as measured by fractional anisotropy (FA), the spatial trajectory of fibers, the estimated length of these fibers (in millimeters, fiber length), and composites such as fiber density [3, 6] between pairs of brain regions. This collection of fiber descriptors allows weighted representations of SC (see Fig. 1a, b), which more fully defines the physiological effects and capacity for plasticity changes within the structural connectome. In contrast to structural connections, functional connections refer to statistical dependencies among time series of neuronal activity or blood oxygen level dependent (BOLD) signals, often expressed simply as linear Pearson correlations. Functional connections are time-dependent and can fluctuate on time scales as fast as seconds (fMRI) or even hundreds of milliseconds (EEG, MEG). Recent work has shown that functional connections exhibit dynamic changes during rest as well as reconfiguration in the context of different stimuli and tasks [7].
Fig. 1.
(A) Top panel denotes basic steps for processing of diffusion data. (A1) Different directions are taken in the scanner and combined; (A2) “streamlines” are obtained from this data and visualized using tractography (the more directions that can be obtained, the more accurate the data); (A3) using segmentation masks “streamlines” that fall within white matter are isolated; (A4) structural connectivity matrices can be obtained from the data in both binary and weighted form. The binary form is represented in the upper triangular where a dark dot denotes the presence of fibers connecting pairs of regions. The weighted form is represented in the lower triangular and denotes average fractional anisotropy (FA) values. (B) Lower panel denotes basic processing steps for RS-fMRI data. (B1) shows extracted time course from fMRI images; (B2) top shows BOLD signal isolated by regressing out nuisance variables, shown on the bottom; (B3) top shows an example of a gray matter parcellation, whereas bottom shows seven most prominent resting-state networks as reported by Yeo et al.; (B4) example of a resting-state functional connectivity matrix with rows and columns ordered according to those resting-state networks. Red squares highlight each of the RSNs
Systematic assessment of resting-state FC has led to the concept of the “functional connectome.” In a landmark study employing resting-state functional MRI (RS-fMRI), Biswal and colleagues found that looking at BOLD signal fluctuations that occur at a lower frequency (<0.1 Hz) revealed patterns of coherent spontaneous fluctuations among spatially remote and distinct brain regions [8]. This seminal work spurred further studies to confirm the existence of synchronous spontaneous fluctuations between different areas in the brain, revealing coherent components that were highly consistent among individuals [8–12]. Low frequency, spontaneous (BOLD) fluctuations that occur at rest are referred to as intrinsic brain activity and its correlated components represent so-called resting-state networks (RSNs) [11, 13]. Note that RSNs refer to components or sub-networks within the cortex; hence, the broad term “network” denotes here a set of regions or even voxels showing high functional coherence within the sub-network and low functional coherence with the rest of the system. There has recently been significant work demonstrating the validity and biological underpinnings of using temporal correlations found in RSNs to infer FC [14–18]. By studying the relationships between spontaneous BOLD signals in different/distinct brain regions, multiple RSNs have been identified (see Fig. 1d–f). RSN analysis allows for a functional based assessment of network differences between subjects or clinical groups, while greatly reducing the dimensionality of the approach, from thousands of voxels to a few prominent sub-networks. For instance, differential connectivity between subjects with impaired cognition and normal cognition in the default mode network (DMN, one of the earlier [19] and most well-studied networks) suggests that FC is sensitive to pathology and its assessment may improve diagnostic methods.
The Human Connectome and Normal Aging
Research on structural and functional brain networks has jointly given rise to the field of brain connectomics. Network science and graph theory have furthered this advancement by providing a suitable framework to quantify and give objective meaning to SC/FC or brain connectomics changes. For instance, using these tools, Betzel and colleagues examined how the connectome changes with aging and found that indeed both SC and FC evolve or “re-organize” during aging [20]. According to the study, several RSNs FC decreased with healthy aging, while other RSNs experienced an increase in FC. Changes were also observed in SC, including an overall decrease in the density of anatomical connections between brain regions and an overall loss of white matter fiber tracts as age progressed. This paper added evidence that FC and SC are closely related and encourage further exploration of how age-related changes in SC and FC correlate with changes in behavior and cognition.
Further research on the aging connectome found that the topology of important “hubs” or highly connected regions in the structural connectome of older adults tended to remain similar to the young adult population, with only subtle gender differences in terms of fiber length [21]. The preserved hub regions included subcortical structures such as the thalamus, striatum, and the amygdala, as well as cortical regions such as the anterior cingulate, insula, and precentral gyrus, which were consistent with other adult structural network studies [22, 23]. This indicates that there is some topological network stability across normal aging, which has strong implications for healthy large-scale network communication and might support the preservation of memory and executive functions in normal aging.
Using Connectomics to Study Alzheimer’s Disease
As dementia continues to become more prevalent, the field of brain connectomics is quickly evolving to not only study human connectome disruptions that occur as a function of normal aging but disruptions in neurodegenerative disorders as well [24, 25]. Many brain connectome studies have already begun generating exciting results regarding the dysfunction occurring in neurodegenerative disorders. Relative to studies focalized in a particular region of interest or in a relatively small targeted neural circuit, brain connectomics provides amore systemic view by modeling whole brain networks. In this way, brain connectomics can assess whole brain organizational changes or disruptions. AD is often thought of as a disconnection disorder in that a lesion or plaque in one region of the brain can disrupt healthy communication between other brain regions resulting in an eventual overall reduction of connections required for healthy cognitive functioning [26]. As a result, changes in brain networks have begun to be looked at extensively in AD [27–33]. Analyses of SC and FC in AD have given rise to new hypotheses of the underlying pathophysiology of AD. Further, they can also be used to understand new correlations with clinical and cognitive symptoms that cannot be explained from a single region or connection [33]. Table 2 summarizes a list of recent studies focused on using SC and/or FC in those at risk for and with AD.
Table 2.
Summary of connectivity studies included in review
| Studies | Diagnosis | Modality | Network | Main findings | Significance |
|---|---|---|---|---|---|
| Sporns et al., 2005 | Healthy controls | DWI | Structural | Research proposal that introduces the idea of displaying connections between brain regions as matrix | Introduces the term “human connectome” and the research strategy to achieve it |
| Biswal et al., 1995 | Healthy controls | RS-fMRI | Functional | Confirm the existence of synchronous spontaneous fluctuations between primary and higher order somatosensory areas in the brain | First to identify resting-state networks |
| Betzel et al., 2014 | Healthy aging adults | DWI/RS-fMRI | Structural/functional | SC and FC evolve/re-organize with healthy aging | Indicates the close relationship between SC and FC and adds evidence to how age-related connectivity changes |
|
Rose et al. 2000 Bozzali et al., 2002 Kiuchi et al., 2009 Ukmar et al., 2008 |
MCI and AD | DWI | Structural | Disruption of the white matter in important hub regions: cingulum bundle tract, corpus callosum, and the superior longitudinal fasciculus | Indicates a re-organization of important “hub” regions as a result of disease state |
| Douaud et al., 2011 | MCI and mild AD | DTI | Structural | MCI show higher FA values in hub region, centrum semiovale region than controls | Diffusion MRI has the ability to detect sensitive changes in SC between disease states |
| Wang et al., 2015 | MCI and AD | DTI/RS-fMRI | Structural/functional | Found disease stage specific changes in FC and SC. Further analysis showed FC changes (>coupling in anterior, <posterior brain regions) were positively correlated with SC changes (< FA, >MD) demonstrating an association between the two | Demonstrate specific patterns of interhemispheric changes between MCI and AD and the relationship between FC and SC |
| Liu et al., 2014 | AD | RS-fMRI | Functional | Patients with severe AD were found to have reduced amplitude fMRI oscillations and reduced strength of FC, specifically in long-distance FC compared to mild AD patients (disease not as progressed) | Progression of disease can be modeled through looking at connectomics |
| Daianu et al., 2013 | MCI and AD | DTI | Structural | Network nodal degree, normalized characteristic path length, and efficiency decreased with AD, while normalized small-worldness increased | Connectivity metrics can help understand whole brain network breakdown as cognitive impairment increases |
| Daianu et al., 2015 | MCI and AD | DTI | Structural | AD patients were found to have an undisturbed structural core network | Demonstrates possible preservation and plasticity in diseased brains |
| Dzezga et al., 2015 | MCI | MRI/PET | Functional | Significant disruptions of whole brain connectivity were found in amyloid-positive patients with and without MCI in typical cortical hubs | Indicate that connectomics can capture early functional consequences of emerging molecular AD pathology before clinical symptoms |
| Brown et al., 2011 | APOE ε4 carriers (at risk) | DTI | Structural | APOE ε4 carriers demonstrated an accelerated age-related loss of mean local interconnectivity compared to non-carriers as well as age-related loss in mean cortical thickness | Highlights a genotype-specific brain network pattern that can potentially affect the rate and/or spatial distribution of AD-related pathology |
| Wang et al., 2015 | AD APOE ε4 carriers and AD APOE ε4 non-carriers | RS-fMRI | Functional | Whole brain graph analyses revealed that APOE ε4 significantly disrupted whole brain topological organization | Highlights how connectomics can help uncover genotype-specific abnormal functional brain network pattern in a preclinical population |
| Chen et al., 2015 | HC (at risk) | DTI/RS-fMRI | Structural/functional | Reduced global efficiency was found in WM and marginally in FC (FC dysfunctions mainly in medial temporal areas) in APOE ε4 carriers | Connectomics helped demonstrate ε4-specific abnormal structural and functional patterns despite no clinical diagnosis, which may potentially serve as biomarkers for early detection before the onset of the disease |
The Connectome in AD and MCI
Analyzing the structural connectome of AD patients has shown disruptions in connectivity in the cingulum bundle tract, the corpus callosum, and the superior longitudinal fasciculus ([34–37]; for additional information, see [38]). Patients with mild cognitive impairment (MCI), which is considered a prodromal stage of AD, have also been shown to exhibit many of these same disruptions, although not as severely as seen in AD. This further supports MCI as an intermediate state between healthy aging and AD ([39–41]; additionally, see review [42]). Another study also found significant abnormalities in a crossing fiber region known as the centrum semiovale (close to an important hub region, [6]) only in MCI patients compared to cognitively healthy age-matched controls [39]. When looking at whole brain diffusion differences, MCI patients were found to have a higher mode of FA values in this region than cognitively healthy age-matched controls. Mode of FA, in this case, refers to the shape of diffusion ranging from −1 (indicating less directionality) to +1 (indicating more directionality). The study also revealed that their patient population (AD) had higher FA values in these regions. Together, this evidence suggests a selective degeneration in crossing fiber pathways. This result is important for two reasons: (a) it demonstrates that diffusion MRI is able to detect subtle changes in the structural connectome between disease states and (2) it highlights the need for further interpretation of both increases and decreases in SC with regards to the underlying neuropathology. Overall, changes in the fiber tracts provide evidence of a disruption of whole brain organization in prodromal and clinical AD through damage to hub regions.
Important findings have also been reported regarding FC in AD. Many studies have already found significant FC changes in AD patients [43, 44]. For instance, patients with severe AD showed reduced amplitude of fMRI oscillations, as well as reduced FC strength, specifically in long-distance FC connections, when compared to mild and moderate AD patients [45]. A reduction in clustering and modularity was also observed in patients with AD compared to cognitively healthy age-matched controls, indicating a less segregated or modular nature of connectivity, and hence a more disorganized way in which brain regions communicate [46–48]. Finally, studies using graph theory to assess SC have also allowed us to better understand the relationship of SC and FC as AD progresses [31].
Other metrics have also been used to measure the way different pathways break down in AD. One such metric is the k-core, which is a measure that assesses the overall connectedness of a network by identifying the most highly interconnected sub-networks within a global network [6]. As opposed to looking at a whole connectivity matrix that may include unreliable connections making it difficult to pull out meaningful disease effects, using the k-core can enhance detection of changes that may be caused by the disease. This is accomplished by looking at deeper highly connected sub-networks or cores. In AD patients, k-core analysis revealed that there was a complete loss of all core networks in the left hemisphere indicating a dramatic unilateral change in brain network topology as disease progresses [49, 50]. This finding suggests that there is a loss of healthy organizational structure in the brains of AD patients. Similarly, Daianu and colleagues assessed structural cores of the brain in AD patients and found that structural brain connectivity was affected according to a variety of network metrics that describe the topological organization of the brain [51]. Disrupted functional whole brain connectivity in cortical hubs was also reported in patients with MCI [48]. Evaluation of other SC measures revealed a similar story; nodal degree decreased as the disease progressed, as did path length and efficiency [50, 51]. It has recently been shown that not all structural core networks are disrupted in AD. Specifically, AD more strongly affected low-degree brain regions rather than the high-degree nodes that form a rich club, leaving this low-degree network better preserved [52]. However, more research is needed to better understand the SC and FC changes in AD and their impact on clinical and cognitive decline.
The Connectome in Populations at Risk for AD
AD is believed to have a prolonged prodromal and preclinical phase initially characterized by the development of silent pathologic changes, including the development of amyloid-beta (Aβ) plaques and neurofibrillary tangles caused by hyperphosphorylated tau, with no clinical symptoms [53–56]. Thus, cognitively normal individuals with significant amyloid and tau pathology (assessed either by measuring cerebrospinal fluid (CSF) levels or by positron emission tomography (PET) techniques) are at high risk for progression to AD [57–60]. Other changes in brain network structure and function may also be present in the preclinical phase of AD. For instance, one group set out to measure FC in the DMN using independent component analysis (ICA) in a distinct preclinical group (marked by individuals who have cognitive complaints yet have normal psychometric performances) along with an MCI group and healthy controls. It was found that the preclinical group, identified as informant-verified cognitive complainers (CC), demonstrated lower connectivity in the DMN, a RSN known to be disrupted in AD, compared to cognitively healthy age-matched controls without complaints yet have higher connectivity compared to the MCI group [61]. In another study by the same group, diffusion tensor imaging revealed white matter changes between diagnostic groups as measured by FA values in regions associated with AD. Their main findings revealed lower FA values in parahippocampal WM among the MCI group compared to cognitively healthy age-matched controls, as well as intermediate FA values in the CC group falling between MCI and cognitively healthy age-matched controls without complaints. These studies added evidence to the existence of a preclinical intermediate state using connectivity measures [62]. Another group, Drzezga and colleagues, used CSF measures to show that asymptomatic individuals with increased amyloid burden demonstrated subtle whole brain FC disruptions and hypometabolism (as measured through [18F]fluorodeoxyglucose PET) [48]. While the pathology of this preclinical category is still unclear, these studies indicate that connectomics can capture early functional and structural changes. This may be a consequence of emerging molecular AD pathology.
Genetic markers have also helped identify asymptomatic people with high risk for AD. The major genetic risk factor for late-onset AD is the ε4 allele of the apolipoprotein E (APOE) gene on chromosome 19 [63]. An early study demonstrated that asymptomatic APOE ε4 carriers exhibited an increase in the magnitude and extent of brain activation during a verbal memory task in brain regions known to degenerate in AD. Furthermore, these same subjects were re-tested 2 years later and brain activation in the previously identified areas correlated with the degree of memory decline [64]. More recently, cognitively intact carriers of the APOE ε4 allele have been linked to accelerated age-related decline in local interconnectivity of structural brain networks [65], as well as disruption in the functional connectome [66], relative to APOE ε4 non-carriers. Cognitively normal APOE ε4 carriers also show differences in large-scale brain functional networks (i.e., decrease in nodal efficiency in medial temporal areas) and structural connections (aberrant regional topological patterns in temporal lobe and other regions) [67]. Interestingly, Wang and colleagues found reduced efficiency in whole brain topological organization, as well as decreased intramodular connectivity, within the posterior DMN and executive control network in APOE ε4 carriers with AD relative to non-carriers [66]. Impaired functional hubs and their rich club connections with other RSNs were also found in APOE ε4 AD patients indicating differential brain network organization as a function of APOE ε4 variants in AD. These studies explore the connectome by combining different imaging modalities and a network science approach suggest that these tools can help elucidate changes that would otherwise be undetectable in a high-risk, preclinical population.
Recent Developments
While the continued development and refinement of RS-fMRI [68–70] and DWI methods [71] hold great promise for understanding biological changes associated with normal aging and dementia so can integrating genomic data. The combination of improved neuroimaging methods to measure biological processes and genetic information from across the genome is likely to provide a more complete picture of health and disease. Genetic analysis may also be incorporated with DWI data. Indeed, the diffusion properties of white matter, placement of fiber tracts, and overall SC networks are thought to have strong heritability [72]. White matter integrity (as measured by FA) was found to show a strong genetic influence, specifically in the bilateral frontal and parietal lobes and the left occipital lobe. Results from a larger family-based study from Kochunov and colleagues supported high heritability of white matter diffusion properties, showing that average measures for FA and radial diffusivity were highly linked to genetics [73]. Overall, these studies suggest that the microstructure of cerebral white matter is at least partially genetically determined. Imaging genetics with DWI data has also been applied to dementia research. Jahanshad et al. evaluated the influence of gene variants or “polymorphisms” on SC and found that variations in a gene known as SPON1 affected SC in an older adult population with varying degrees of cognitive impairment [74]. Furthermore, older adults who carried the variant associated with increased SC had less severe dementia scores and a lower risk of AD. This discovery of SPON1 variants associated with brain connectivity suggests a new neurogenetic pathway with links to dementia severity that can be further explored.
Genomic data may also help elucidate the biological basis for the synchronous low frequency fluctuations that are suggested to give rise to brain networks using RS-fMRI. One study found that expression of 136 genes (later identified to be involved in ion channel formation) were significantly associated with RS-fMRI measures in humans and significantly associated with axonal connectivity in a rodent model [75]. Additionally, Richiardi and colleagues demonstrated that functional brain networks as defined by RS-fMRI are significantly correlated with gene expression data. Their data revealed that functional networks are associated with a set of genes that code for ion channels and other important synaptic functions (i.e., dopamine neurotransmitter release), further supporting a strong biological foundation of RSNs. The results from these studies provide additional evidence that integrating gene expression and brain connectivity measures can provide insight into the molecular mechanisms underlying not only functional networks but also structural networks. Other studies have found that an altered expression of genes associated with AD can translate to altered FC at the molecular level [76]. These studies integrating RS-fMRI and genomics suggest that the connectome may offer a rich and promising target for genetic analysis.
Another future direction of brain connectomics is through the integration of different technologies, specifically studies that integrate multiple imaging modalities [77]. Multimodal imaging refers to the integration of data from more than one imaging modality, such as DWI, RS-fMRI, EEG, and PET. This integration typically occurs at the level of statistical analysis where each modality has an influence over the end result. Additionally, the data can be integrated in a more exploratory fashion, where the comparison is more qualitative than quantitative. For example, data analysis has been treated in this way with disorders such as autism spectrum disorder (ASD). ASD has been conceptualized as a breakdown of neural communication and connectivity. Integration of multimodal imaging techniques has shown significant promise in elucidating brain connectomics in ASD. Over the last decade, theoretical accounts of ASD have shifted towards emphasizing the breakdown of neural communication and connectivity much like AD. In light of this shift, the use of multimodal imaging has begun to show promise. Studies in ASD show convergence of altered SC and FC within the same regions, including regions of the medial prefrontal cortex and posterior cingulate/precuneus that are core members of specific RSNs [78–80]. Additionally, a convergence in altered function, volume, and connectivity of frontal regions involved in executive function, language, motor, and sensorimotor functions was also found in ASD [81–83]. Finally, a study revealed age-related differences in the coupling of function and structure in healthy controls that did not occur in ASD patients.
Multimodal neuroimaging are also useful for achieving the best possible spatial and temporal resolution (i.e., EEG and fMRI [84]) as well as for using data from one modality to constrain the interpretation of another modality. In the latter case, the purpose of multimodal imaging would not be to merge data but to assist with informed judgment of the data within a broader context. For instance, EEG data measuring participant alertness has been used to inform interpretation of fMRI data [84]. Ultimately, multimodal imaging has the potential to help obtain a more comprehensive physiological view of brain processes both in healthy adults and patients.
As a complement to the challenges and opportunities of this technological integration, the area of brain connectomics is also evolving from a theoretical point of view. Multimodal imaging naturally allows us to elaborate on multiplex or multi-layered networks [85, 86], where one system is not represented by a unique adjacency or connectivity matrix, but by more than one, attending to different features or to the different nature of their connections. Another emergent area of research consists of network morphospaces [87–90] where networks occupy a space in n-dimensional systems that obey to different trade-offs of network characteristics. This incorporation of information from theoretical concepts [91] into brain connectomics is expanding the way we understand communication within networks [92, 93] and in the human brain [94, 95].
Finally, another innovation in modeling FC is the emergence of dynamic RS-fMRI. Until recently, most RS-fMRI studies looked at data in a stationary framework to interpret results. While studies employing this framework have uncovered large-scale properties of brain function, the resulting characterization ultimately represents an average across complex spatiotemporal phenomena. Quantifying changes in FC metrics over a longer scan (10 min or longer) may provide greater insight into fundamental properties of brain networks and how they change over time [96]. Recent studies have demonstrated that correlations among brain regions, both within and between networks, indeed evolve over time [7, 97–99]. These results suggest that resting brain activity can be divided into subsets or “communities” of brain regions that strongly interact for a time but that these interactions are not static. For instance, low frequency BOLD signal fluctuations revealed synchronized communities reoccurred intermittently in time and across scanning sessions during RS-fMRI scans [100, 101]. In addition, the synchronized communities constitute components of previously defined RSNs known to be engaged in sensory-motor or cognitive function. As discussed earlier, altered static FC in RSNs has been found in AD patients. Thus, it would be of great interest to see if using a non-static or “sliding-window” approach would provide an even more accurate description of FC changes in AD. One of the first reports demonstrating RS-fMRI changes in AD patients beyond the traditional average FC metrics [102] reported impairments in the dynamics of spontaneous activity by examining time varying changes of a modularity metric [103]. Using dynamic FC representations, the authors reported differences in the “dwell time” within different sub-network configurations of the DMN between AD and cognitively healthy age-matched controls. In other words, less time was spent in brain states with strong posterior DMN region contributions and more time in states characterized by anterior DMN region contributions in AD patients. This observation has the potential to lead to a better understanding of the large-scale characterizations of AD and potentially a more accurate prognostic picture. This early work offers great promise in revealing aspects of dynamic FC at a macroscopic scale.
Conclusion
The increased sensitivity and whole brain perspective of SC and FC measures assessed using diffusion weighted MRI and RS-fMRI, respectively, have already provided numerous insights into the specific disturbances of network organization that occur in the diseased brain. For instance, normal cognitive aging results in changes in brain network features but are mainly localized in hub regions in the frontal, parietal, and occipital lobes. However, it is important to note that, overall, brain networks have been shown to have high topological global efficiency despite aging. While significant topological modification is observed with brain maturation, the general structure of the connectome remains stable over time. This review has considered changes in both the structural and functional connectome in the context of AD-associated neurodegeneration as the primary example at three distinct stages, i.e., manifest disease, MCI prodrome, and the preclinical at-risk stage. At the disease stage, brain connectomic techniques have afforded a way to model the structural and functional failing integrity of specific neural subsystems and offered new insights into how degenerative processes may spread through interconnected networks via central regions. Specifically, AD patients have shown disruptions in SC in important hub regions, interhemispheric regions, and fiber crossing regions. These changes are characterized by a reduced network nodal degree, change in white matter path lengths, and a decreased global efficiency of networks. During the MCI stages, evidence of the impact of intermediate neurodegeneration has been shown with regard to several network features including abnormal loss of white matter and significant changes in functional network organization. At the earliest stage, there is significant evidence of emerging changes reflected by differences between at-risk populations and cognitively healthy age-matched controls. These changes have been found primarily amid the interconnectivity between major RSNs, as well as differences in whole brain organization and network structure suggesting that connectomics level analysis is sensitive to subtle changes in brain networks before the onset of clinical symptoms. However, research on brain connectomics in disorders of cognitive aging remains at an early stage of development and new findings on changes in network features in preclinical and prodromal neurodegenerative diseases including non-AD and mixed types of disease will undoubtedly augment our knowledge in important ways relevant for clinical translation.
The present review also examines more recent developments such as the relationship to genetic markers, other multimodal imaging methods, and the emergence of dynamic FC which has aided the field in characterizing patient–specific abnormalities. The convergence of this information is likely to be beneficial for design of targeted interventions especially in heterogeneous diseases like AD and dementias of mixed etiology. Selective regional pathology plays an important role in the manifestations of different neurodegenerative diseases; and characterizing the progression to and from these brain regions during the advancement of neurodegeneration is important. Connectomics analysis is likely to be useful for enhancement of clinical trial designs by incorporating information about regional brain networks at baseline and after interventions. Connectomics analyses will also likely contribute to clinical trials by providing a dynamic metric that can (a) help identify subtle changes early in the disease course and (b) provide more comprehensive metrics to assess the impact of targeted therapeutic strategies.
In this review, we have illustrated the emerging utility of brain connectomics in providing new quantitative network features that can be used to detect specific structural and functional disruptions within brain circuitry including identification of white matter tracts and their integrity, the strength of functional connections between regions, and network disruption in unique RSNs. The future of brain connectomics holds significant promise in several areas. First, connectome studies may permit identification of early pathological changes that can predict disease progression. Second, due to the non-invasive nature of RS-fMRI and diffusion MRI, it is possible to collect data more easily and at more time-points to address long-term cognitive and behavioral changes and relate them to changes in the structural and functional connectome. Third, such studies offer insight into the unique characterization of different diseases identifying neurodegenerative signatures based on network features. To elaborate on the idea of a unique signature or “fingerprint,” brain connectomics may be well-suited to reveal individual differences in brain networks and to help adapt therapeutic approaches centered on a person’s baseline topological features and assess how treatment changes network topology, whether positively or negatively in terms of adverse side effects. In this way, connectome analysis may constitute a new approach that can guide the brain repair and recovery of brain networks especially as disease modifying interventions are developed.
Connectome-wide analyses hold promise of an unbiased means of characterizing brain network disturbances during manifest disease and in early preclinical and prodromal stages before onset of significant observable cognitive differences. However, this promise is likely to be realized with further improvements in imaging technology, statistical methodology, and integration of large-scale multimodal and dynamic data sets. Improving image quality will permit more precision in tracking changes in brain networks with disease progression, as well as to help resolve the directionality of fiber tracts with regards to SC. For SC, this will involve developing more accurate fiber reconstruction and tissue segmentation methodologies and also defining measures of connectivity that have a clear biological interpretation.
Further development of appropriate statistical methods is needed to allow better handling of complex data models, and the integration of multiple multimodal data sets as existing methodology is limited. Lastly, learning to account for the distinctive nature of dynamic data will be essential for the mapping of BOLD fluctuations in psychological states to their corresponding neurobiological states.
The young field of brain connectomics has been successful in generating evidence for aberrant network properties in early stage neurodegenerative diseases, demonstrating the potential for improved prediction of cognitive impairment, prior to onset of significant clinical symptoms. However, more clinical research is needed to refine methodology and validate the initial observations from patients with various neurodegenerative diseases at all stages. Eventually, follow-up studies linking in-vivo connectomics changes to post-mortem analyses of specific proteinopathies underlying many dementing disorders will be important. MRI sequences and post-processing methods will continue to evolve bringing significant improvement in accuracy and reliability, capacity for mapping more subtle changes in connectivity. All of these developments will facilitate the translation of intriguing research findings on connectome changes in cognitive aging and early stage disease to a level of development where they can contribute along with clinical assessment and other –omics and biomarkers to diagnosis and treatment monitoring as part of an evolving precision medicine.
Acknowledgments
The authors’ research was supported, in part, by grants from the National Institutes of Health (NIH R01 AG19771, P30 AG10133, R01 LM011360, K01 AG049050, R01 AT009036 and the Indiana CTSI: U54 RR025761, RR027710, and RR020128), as well as by NSF IIS-1117335, an Alzheimer’s Association New Investigator Research Grant (SLR), the Indiana University Network Science Institute (IUNI), the Health-Indiana University School of Medicine Strategic Research Initiative, and the Indiana Clinical and Translational Science Institute (JAC, SLR). JAC also acknowledges partial support from Grant Numbers TL1 TR001107 and UL1 TR001108 (A. Shekhar, PI) from the National Institutes of Health, National Center for Advancing Translational Sciences, Clinical and Translational Sciences Award.
Andrew Saykin reports support from multiple NIH grants, and an investigator initiated grant from Eli Lilly.
Footnotes
This article is part of the Topical Collection on Geropsychiatry and Cognitive Disorders of Late Life
Compliance with Ethics Standards
Human and Animal Rights and Informed Consent Deidentified human MRI data was included in Figure 1. Source scans were collected under an IRB-approved protocol at the Indiana University School of Medicine. No other data from human subjects was collected for this review article.
Conflict of Interest Olaf Sporns, Shannon L Risacher, Joaquin Goñi, and Joey Contreras have no relevant conflicts to report.
References
- 1.Hofer SM, Berg S, Era P. Evaluating the interdependence of aging-related changes in visual and auditory acuity, balance, and cognitive functioning. Psychol Aging. 2003;18(2):285–305. doi: 10.1037/0882-7974.18.2.285. [DOI] [PubMed] [Google Scholar]
- 2.Sporns O, Tononi G, Kotter R. The human connectome: a structural description of the human brain. PLoS Comput Biol. 2005;1(4):e42. doi: 10.1371/journal.pcbi.0010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hagmann P, et al. Mapping human whole-brain structural networks with diffusion MRI. PLoS One. 2007;2(7):e597. doi: 10.1371/journal.pone.0000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cole MW, et al. Intrinsic and task-evoked network architectures of the human brain. Neuron. 2014;83(1):238–251. doi: 10.1016/j.neuron.2014.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.S RyC. Histology of the nervous system. Oxford University Press; 1909. [Google Scholar]
- 6.Hagmann P, et al. Mapping the structural core of human cerebral cortex. PLoS Biol. 2008;6(7):e159. doi: 10.1371/journal.pbio.0060159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Allen EA, et al. Tracking whole-brain connectivity dynamics in the resting state. Cereb Cortex. 2014;24(3):663–676. doi: 10.1093/cercor/bhs352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biswal B, et al. Functional connectivity in the motor cortex of resting human brain using echo-planar MRI. Magn Reson Med. 1995;34(4):537–541. doi: 10.1002/mrm.1910340409. [DOI] [PubMed] [Google Scholar]
- 9.Beckmann CF, et al. Investigations into resting-state connectivity using independent component analysis. Philos Trans R Soc Lond B Biol Sci. 2005;360(1457):1001–1013. doi: 10.1098/rstb.2005.1634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Power JD, et al. Functional network organization of the human brain. Neuron. 2011;72(4):665–678. doi: 10.1016/j.neuron.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith SM, et al. Correspondence of the brain's functional architecture during activation and rest. Proc Natl Acad Sci U S A. 2009;106(31):13040–13045. doi: 10.1073/pnas.0905267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yeo BT, et al. The organization of the human cerebral cortex estimated by intrinsic functional connectivity. J Neurophysiol. 2011;106(3):1125–1165. doi: 10.1152/jn.00338.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox MD, Raichle ME. Spontaneous fluctuations in brain activity observed with functional magnetic resonance imaging. Nat Rev Neurosci. 2007;8(9):700–711. doi: 10.1038/nrn2201. [DOI] [PubMed] [Google Scholar]
- 14.de Pasquale F, et al. Temporal dynamics of spontaneous MEG activity in brain networks. Proc Natl Acad Sci U S A. 2010;107(13):6040–6045. doi: 10.1073/pnas.0913863107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Foster BL, et al. Intrinsic and task-dependent coupling of neuronal population activity in human parietal cortex. Neuron. 2015;86(2):578–590. doi: 10.1016/j.neuron.2015.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li JM, et al. Functional connectivity arises from a slow rhythmic mechanism. Proc Natl Acad Sci U S A. 2015;112(19):E2527–E2535. doi: 10.1073/pnas.1419837112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sadaghiani S, et al. Intrinsic connectivity networks, alpha oscillations, and tonic alertness: a simultaneous electroencephalography/functional magnetic resonance imaging study. J Neurosci. 2010;30(30):10243–10250. doi: 10.1523/JNEUROSCI.1004-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brookes MJ, et al. Investigating the electrophysiological basis of resting state networks using magnetoencephalography. Proc Natl Acad Sci U S A. 2011;108(40):16783–16788. doi: 10.1073/pnas.1112685108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fox MD, et al. The human brain is intrinsically organized into dynamic, anticorrelated functional networks. Proc Natl Acad Sci U S A. 2005;102(27):9673–9678. doi: 10.1073/pnas.0504136102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Betzel RF, et al. Changes in structural and functional connectivity among resting-state networks across the human lifespan. Neuroimage. 2014;102P2:345–357. doi: 10.1016/j.neuroimage.2014.07.067. [DOI] [PubMed] [Google Scholar]
- 21.Perry A, et al. The organisation of the elderly connectome. Neuroimage. 2015;114:414–426. doi: 10.1016/j.neuroimage.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 22.van den Heuvel MP, Sporns O. Rich-club organization of the human connectome. J Neurosci. 2011;31(44):15775–15786. doi: 10.1523/JNEUROSCI.3539-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van den Heuvel MP, Sporns O. Network hubs in the human brain. Trends Cogn Sci. 2013;17(12):683–696. doi: 10.1016/j.tics.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 24.Weiler M, et al. Default mode, executive function, and language functional connectivity networks are compromised in mild Alzheimer's disease. Curr Alzheimer Res. 2014;11(3):274–282. doi: 10.2174/1567205011666140131114716. [DOI] [PubMed] [Google Scholar]
- 25.Kelly C, et al. Characterizing variation in the functional connectome: promise and pitfalls. Trends Cogn Sci. 2012;16(3):181–188. doi: 10.1016/j.tics.2012.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Delbeuck X, Van der Linden M, Collette F. Alzheimer's disease as a disconnection syndrome? Neuropsychol Rev. 2003;13(2):79–92. doi: 10.1023/a:1023832305702. [DOI] [PubMed] [Google Scholar]
- 27.Crossley NA, et al. The hubs of the human connectome are generally implicated in the anatomy of brain disorders. Brain. 2014;137(Pt 8):2382–2395. doi: 10.1093/brain/awu132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dai Z, et al. Identifying and mapping connectivity patterns of brain network hubs in Alzheimer's disease. Cereb Cortex. 2014 doi: 10.1093/cercor/bhu246. [DOI] [PubMed] [Google Scholar]
- 29.Prescott JW, et al. The Alzheimer structural connectome: changes in cortical network topology with increased amyloid plaque burden. Radiology. 2014;273(1):175–184. doi: 10.1148/radiol.14132593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Thomas JB, et al. Functional connectivity in autosomal dominant and late-onset Alzheimer disease. JAMA Neurol. 2014;71(9):1111–1122. doi: 10.1001/jamaneurol.2014.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, et al. Interhemispheric functional and structural disconnection in Alzheimer's disease: a combined resting-state fMRI and DTI study. PLoS One. 2015;10(5):e0126310. doi: 10.1371/journal.pone.0126310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiang J, et al. An abnormal resting-state functional brain network indicates progression towards Alzheimer's disease. Neural Regen Res. 2013;8(30):2789–2799. doi: 10.3969/j.issn.1673-5374.2013.30.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sepulcre J, Johnson KA, Sperling R. Tau and AB deposits relate to distinctive functional connectivity disruptions in the elderly brain. Alzheimer's & Dementia. The Journal of the Alzheimer's Association. 2014;10(4):159–160. [Google Scholar]
- 34.Bozzali M, et al. White matter damage in Alzheimer's disease assessed in vivo using diffusion tensor magnetic resonance imaging. J Neurol Neurosurg Psychiatry. 2002;72(6):742–746. doi: 10.1136/jnnp.72.6.742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kiuchi K, et al. Abnormalities of the uncinate fasciculus and posterior cingulate fasciculus in mild cognitive impairment and early Alzheimer's disease: a diffusion tensor tractography study. Brain Res. 2009;1287:184–191. doi: 10.1016/j.brainres.2009.06.052. [DOI] [PubMed] [Google Scholar]
- 36.Rose SE, et al. Loss of connectivity in Alzheimer's disease: an evaluation of white matter tract integrity with colour coded MR diffusion tensor imaging. J Neurol Neurosurg Psychiatry. 2000;69(4):528–530. doi: 10.1136/jnnp.69.4.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ukmar M, et al. Evaluation of white matter damage in patients with Alzheimer's disease and in patients with mild cognitive impairment by using diffusion tensor imaging. Radiol Med. 2008;113(6):915–922. doi: 10.1007/s11547-008-0286-1. [DOI] [PubMed] [Google Scholar]
- 38.Xie T, He Y. Mapping the Alzheimer's brain with connectomics. Front Psychiatry. 2011;2:77. doi: 10.3389/fpsyt.2011.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Douaud G, et al. DTI measures in crossing-fibre areas: increased diffusion anisotropy reveals early white matter alteration in MCI and mild Alzheimer's disease. Neuroimage. 2011;55(3):880–890. doi: 10.1016/j.neuroimage.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhu D, et al. Connectome-scale assessments of structural and functional connectivity in MCI. Hum Brain Mapp. 2014;35(7):2911–2923. doi: 10.1002/hbm.22373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Risacher SL, Saykin AJ. Neuroimaging and other biomarkers for Alzheimer's disease: the changing landscape of early detection. Annu Rev Clin Psychol. 2013;9:621–648. doi: 10.1146/annurev-clinpsy-050212-185535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chua TC, et al. Diffusion tensor imaging in mild cognitive impairment and Alzheimer's disease: a review. Curr Opin Neurol. 2008;21(1):83–92. doi: 10.1097/WCO.0b013e3282f4594b. [DOI] [PubMed] [Google Scholar]
- 43.Allen G, et al. Reduced hippocampal functional connectivity in Alzheimer disease. Arch Neurol. 2007;64(10):1482–1487. doi: 10.1001/archneur.64.10.1482. [DOI] [PubMed] [Google Scholar]
- 44.Wang K, et al. Altered functional connectivity in early Alzheimer's disease: a resting-state fMRI study. Hum Brain Mapp. 2007;28(10):967–978. doi: 10.1002/hbm.20324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y, et al. Impaired long distance functional connectivity and weighted network architecture in Alzheimer's disease. Cereb Cortex. 2014;24(6):1422–1435. doi: 10.1093/cercor/bhs410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brier MR, et al. Functional connectivity and graph theory in preclinical Alzheimer's disease. Neurobiol Aging. 2014;35(4):757–768. doi: 10.1016/j.neurobiolaging.2013.10.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Buckner RL, et al. Cortical hubs revealed by intrinsic functional connectivity: mapping, assessment of stability, and relation to Alzheimer's disease. J Neurosci. 2009;29(6):1860–1873. doi: 10.1523/JNEUROSCI.5062-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Drzezga A, et al. Neuronal dysfunction and disconnection of cortical hubs in non-demented subjects with elevated amyloid burden. Brain. 2011;134(Pt 6):1635–1646. doi: 10.1093/brain/awr066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daianu M, et al. Left versus right hemisphere differences in brain connectivity: 4-tesla hardi tractography in 569 twins. Proc IEEE Int Symp Biomed Imaging. 2012;2012:526–529. doi: 10.1109/ISBI.2012.6235601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Daianu M, et al. Breakdown of brain connectivity between normal aging and Alzheimer's disease: a structural k-core network analysis. Brain Connect. 2013;3(4):407–422. doi: 10.1089/brain.2012.0137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daianu M, et al. Alzheimer's disease disrupts rich club organization in brain connectivity networks. Imaging: Proc IEEE Int Symp Biomed. 2013:266–269. doi: 10.1109/ISBI.2013.6556463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daianu M, et al. Rich club analysis in the Alzheimer's disease connectome reveals a relatively undisturbed structural core network. Hum Brain Mapp. 2015 doi: 10.1002/hbm.22830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Petrella JR. Neuroimaging and the search for a cure for Alzheimer disease. Radiology. 2013;269(3):671–691. doi: 10.1148/radiol.13122503. [DOI] [PubMed] [Google Scholar]
- 54.Braak H, et al. Stages of the pathologic process in Alzheimer disease: age categories from 1 to 100 years. J Neuropathol Exp Neurol. 2011;70(11):960–969. doi: 10.1097/NEN.0b013e318232a379. [DOI] [PubMed] [Google Scholar]
- 55.Price JL, et al. Neuropathology of nondemented aging: presumptive evidence for preclinical Alzheimer disease. Neurobiol Aging. 2009;30(7):1026–1036. doi: 10.1016/j.neurobiolaging.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Price JL, Morris JC. Tangles and plaques in nondemented aging and "preclinical" Alzheimer's disease. Ann Neurol. 1999;45(3):358–368. doi: 10.1002/1531-8249(199903)45:3<358::aid-ana12>3.0.co;2-x. [DOI] [PubMed] [Google Scholar]
- 57.Caselli RJ, Reiman EM. Characterizing the preclinical stages of Alzheimer's disease and the prospect of presymptomatic intervention. J Alzheimers Dis. 2013;33(Suppl 1):S405–S416. doi: 10.3233/JAD-2012-129026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mattsson N, et al. Predicting reduction of cerebrospinal fluid beta-amyloid 42 in cognitively healthy controls. JAMA Neurol. 2015;72(5):554–560. doi: 10.1001/jamaneurol.2014.4530. [DOI] [PubMed] [Google Scholar]
- 59.Moghekar A, et al. CSF biomarker changes precede symptom onset of mild cognitive impairment. Neurology. 2013;81(20):1753–1758. doi: 10.1212/01.wnl.0000435558.98447.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Vos SJ, et al. Preclinical Alzheimer's disease and its outcome: a longitudinal cohort study. Lancet Neurol. 2013;12(10):957–965. doi: 10.1016/S1474-4422(13)70194-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang Y, et al. Altered default mode network connectivity in older adults with cognitive complaints and amnestic mild cognitive impairment. J Alzheimers Dis. 2013;35(4):751–760. doi: 10.3233/JAD-130080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang Y, et al. Selective changes in white matter integrity in MCI and older adults with cognitive complaints. Biochim Biophys Acta. 2012;1822(3):423–430. doi: 10.1016/j.bbadis.2011.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Saunders AM, et al. Association of apolipoprotein E allele epsilon 4 with late-onset familial and sporadic Alzheimer's disease. Neurology. 1993;43(8):1467–1472. doi: 10.1212/wnl.43.8.1467. [DOI] [PubMed] [Google Scholar]
- 64.Bookheimer SY, et al. Patterns of brain activation in people at risk for Alzheimer's disease. N Engl J Med. 2000;343(7):450–456. doi: 10.1056/NEJM200008173430701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Brown JA, et al. Brain network local interconnectivity loss in aging APOE-4 allele carriers. Proc Natl Acad Sci U S A. 2011;108(51):20760–20765. doi: 10.1073/pnas.1109038108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang J, et al. Apolipoprotein E epsilon4 modulates functional brain connectome in Alzheimer's disease. Hum Brain Mapp. 2015;36(5):1828–1846. doi: 10.1002/hbm.22740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen Y, et al. Disrupted functional and structural networks in cognitively normal elderly subjects with the APOE varepsilon4 allele. Neuropsychopharmacology. 2015;40(5):1181–1191. doi: 10.1038/npp.2014.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Power JD, et al. Spurious but systematic correlations in functional connectivity MRI networks arise from subject motion. Neuroimage. 2012;59(3):2142–2154. doi: 10.1016/j.neuroimage.2011.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Power JD, et al. Methods to detect, characterize, and remove motion artifact in resting state fMRI. Neuroimage. 2014;84:320–341. doi: 10.1016/j.neuroimage.2013.08.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Siegel JS, et al. Statistical improvements in functional magnetic resonance imaging analyses produced by censoring high-motion data points. Hum Brain Mapp. 2014;35(5):1981–1996. doi: 10.1002/hbm.22307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Coupe P, et al. An optimized blockwise nonlocal means denoising filter for 3-D magnetic resonance images. IEEE Trans Med Imaging. 2008;27(4):425–441. doi: 10.1109/TMI.2007.906087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chiang MC, et al. Genetics of brain fiber architecture and intellectual performance. J Neurosci. 2009;29(7):2212–2224. doi: 10.1523/JNEUROSCI.4184-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kochunov P, et al. Genetics of microstructure of cerebral white matter using diffusion tensor imaging. Neuroimage. 2010;53(3):1109–1116. doi: 10.1016/j.neuroimage.2010.01.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jahanshad N, et al. Genome-wide scan of healthy human connectome discovers SPON1 gene variant influencing dementia severity. Proc Natl Acad Sci U S A. 2013;110(12):4768–4773. doi: 10.1073/pnas.1216206110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Richiardi J, et al. Correlated gene expression supports synchronous activity in brain networks. Science. 2015;348(6240):1241–1244. doi: 10.1126/science.1255905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zeidan-Chulia F, et al. Altered expression of Alzheimer's disease-related genes in the cerebellum of autistic patients: a model for disrupted brain connectome and therapy. Cell Death Dis. 2014;5:e1250. doi: 10.1038/cddis.2014.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Brown JA, et al. The UCLA multimodal connectivity database: a web-based platform for brain connectivity matrix sharing and analysis. Front Neuroinform. 2012;6:28. doi: 10.3389/fninf.2012.00028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Schaer M, et al. Decreased frontal gyrification correlates with altered connectivity in children with autism. Front Hum Neurosci. 2013;7:750. doi: 10.3389/fnhum.2013.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Beacher FD, et al. Autism attenuates sex differences in brain structure: a combined voxel-based morphometry and diffusion tensor imaging study. AJNR Am J Neuroradiol. 2012;33(1):83–89. doi: 10.3174/ajnr.A2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Poustka L, et al. Fronto-temporal disconnectivity and symptom severity in children with autism spectrum disorder. World J Biol Psychiatry. 2012;13(4):269–280. doi: 10.3109/15622975.2011.591824. [DOI] [PubMed] [Google Scholar]
- 81.Thakkar KN, et al. Response monitoring, repetitive behaviour and anterior cingulate abnormalities in autism spectrum disorders (ASD) Brain. 2008;131(Pt 9):2464–2478. doi: 10.1093/brain/awn099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sahyoun CP, et al. Neuroimaging of the functional and structural networks underlying visuospatial vs. linguistic reasoning in high-functioning autism. Neuropsychologia. 2010;48(1):86–95. doi: 10.1016/j.neuropsychologia.2009.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Schmitz N, et al. Neural correlates of executive function in autistic spectrum disorders. Biol Psychiatry. 2006;59(1):7–16. doi: 10.1016/j.biopsych.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 84.Rosenkranz K, Lemieux L. Present and future of simultaneous EEG-fMRI. MAGMA. 2010;23(5–6):309–316. doi: 10.1007/s10334-009-0196-9. [DOI] [PubMed] [Google Scholar]
- 85.Mucha PJ, et al. Community structure in time-dependent, multiscale, and multiplex networks. Science. 2010;328(5980):876–878. doi: 10.1126/science.1184819. [DOI] [PubMed] [Google Scholar]
- 86.Bianconi G. Statistical mechanics of multiplex networks: entropy and overlap. Phys Rev E Stat Nonlin Soft Matter Phys. 2013;87(6):062806. doi: 10.1103/PhysRevE.87.062806. [DOI] [PubMed] [Google Scholar]
- 87.Avena-Koenigsberger A, et al. Using Pareto optimality to explore the topology and dynamics of the human connectome. Philos Trans R Soc Lond B Biol Sci. 2014;369:1653. doi: 10.1098/rstb.2013.0530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Avena-Koenigsberger A, et al. Network morphospace. J R Soc Interface. 2015;12(103) doi: 10.1098/rsif.2014.0881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Corominas-Murtra B, et al. On the origins of hierarchy in complex networks. Proc Natl Acad Sci U S A. 2013;110(33):13316–13321. doi: 10.1073/pnas.1300832110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Goni J, et al. Exploring the morphospace of communication efficiency in complex networks. PLoS One. 2013;8(3):e58070. doi: 10.1371/journal.pone.0058070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Shannon CE. The mathematical theory of communication. 1963 MD Comput. 1997;14(4):306–317. [PubMed] [Google Scholar]
- 92.Rosvall M, et al. Searchability of networks. Phys Rev E Stat Nonlin Soft Matter Phys. 2005;72(4 Pt 2):046117. doi: 10.1103/PhysRevE.72.046117. [DOI] [PubMed] [Google Scholar]
- 93.Rosvall M, Minnhagen P, Sneppen K. Navigating networks with limited information. Phys Rev E Stat Nonlin Soft Matter Phys. 2005;71(6 Pt 2):066111. doi: 10.1103/PhysRevE.71.066111. [DOI] [PubMed] [Google Scholar]
- 94.Goni J, et al. Resting-brain functional connectivity predicted by analytic measures of network communication. Proc Natl Acad Sci U S A. 2014;111(2):833–838. doi: 10.1073/pnas.1315529111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Kolchinsky A, et al. Multi-scale integration and predictability in resting state brain activity. Front Neuroinform. 2014;8:66. doi: 10.3389/fninf.2014.00066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hutchison RM, et al. Dynamic functional connectivity: promise, issues, and interpretations. Neuroimage. 2013;80:360–378. doi: 10.1016/j.neuroimage.2013.05.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.de Pasquale F, et al. A cortical core for dynamic integration of functional networks in the resting human brain. Neuron. 2012;74(4):753–764. doi: 10.1016/j.neuron.2012.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chang C, Glover GH. Time-frequency dynamics of resting-state brain connectivity measured with fMRI. Neuroimage. 2010;50(1):81–98. doi: 10.1016/j.neuroimage.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hutchison RM, et al. Resting-state networks show dynamic functional connectivity in awake humans and anesthetized macaques. Hum Brain Mapp. 2013;34(9):2154–2177. doi: 10.1002/hbm.22058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Leonardi N, Van De Ville D. On spurious and real fluctuations of dynamic functional connectivity during rest. Neuroimage. 2015;104:430–436. doi: 10.1016/j.neuroimage.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 101.Ponce-Alvarez A, et al. Resting-state temporal synchronization networks emerge from connectivity topology and heterogeneity. PLoS Comput Biol. 2015;11(2):e1004100. doi: 10.1371/journal.pcbi.1004100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jones DT, et al. Non-stationarity in the "resting brain's" modular architecture. PLoS One. 2012;7(6):e39731. doi: 10.1371/journal.pone.0039731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Rubinov M, Sporns O. Weight-conserving characterization of complex functional brain networks. Neuroimage. 2011;56(4):2068–2079. doi: 10.1016/j.neuroimage.2011.03.069. [DOI] [PubMed] [Google Scholar]