Abstract
The germinal center (GC) is a microanatomical compartment wherein high affinity antibody-producing B cells are selectively expanded. B cells proliferate and mutate their antibody genes in the dark zone (DZ) of the GC, and are then selected by T cells in the light zone (LZ) on the basis of affinity. Here we show that T cell help regulates the speed of cell cycle phase transitions and DNA replication of GC B cells. Genome sequencing and single-molecule analyses revealed that T cell help shortens S phase by regulating replication fork progression while preserving the relative order of replication origin activation. Thus, high affinity GC B cells are selected by a mechanism that involves prolonged dwell time in the DZ where selected cells undergo accelerated cell cycles.
Antibodies elicited during T cell-dependent immune responses undergo significant increases in affinity over time (1). This phenomenon, known as affinity maturation, takes place in the germinal center (GC), where antigen-specific B cells diversify their antibodies by somatic hypermutation (2) and undergo selective clonal expansion (3–7). Together, these events are essential to the development of effective antibody responses.
GC B cells bearing antibody variants with higher affinity are selectively expanded during iterative rounds of migration between the DZ, where they proliferate and hypermutate, and the LZ, where they capture antigen displayed on the surface of follicular dendritic cells (8–11). By binding and internalizing more antigen in the LZ, high affinity clones present more peptide-major histocompatibility complex II (MHCII) and thereby elicit greater help from CD4+ T follicular helper cells (11, 12). The magnitude of T cell help determines how long B cells reside in the DZ, providing selected cells more time to proliferate and expand in between rounds of competition in the LZ (13). Whether this mechanism alone explains how high affinity B cells are selected remains unknown.
To explore additional mechanisms that could contribute to selection, we employed an adoptive transfer model in which antigen presentation by a subset of GC B cells can be acutely and selectively increased (11, 14, 15). B cells carrying a knock-in antigen receptor specific for the hapten 4-hydroxy-3-nitrophenylacetyl (NP) (B1–8hi) were transferred into ovalbumin (OVA)-primed wild-type mice that were boosted with NP-OVA. Whereas the majority of transferred B1–8hi B cells were DEC205−/− (~85%), a subset (~15%) of the B1–8hi B cells were DEC205+/+ (10, 16). DEC205 is an endocytic receptor expressed by GC B cells that delivers antigen to MHCII processing compartments (14). Targeting DEC205 with an antibody that is fused at its C terminus to OVA (αDEC–OVA), but not the irrelevant control antigen Plasmodium falciparum circumsporozoite protein (αDEC-CS) (17), increases the amount of cognate peptide-MHCII displayed on the surface of B1–8hi DEC205+/+ GC B cells, leading to their selective expansion (11–13).
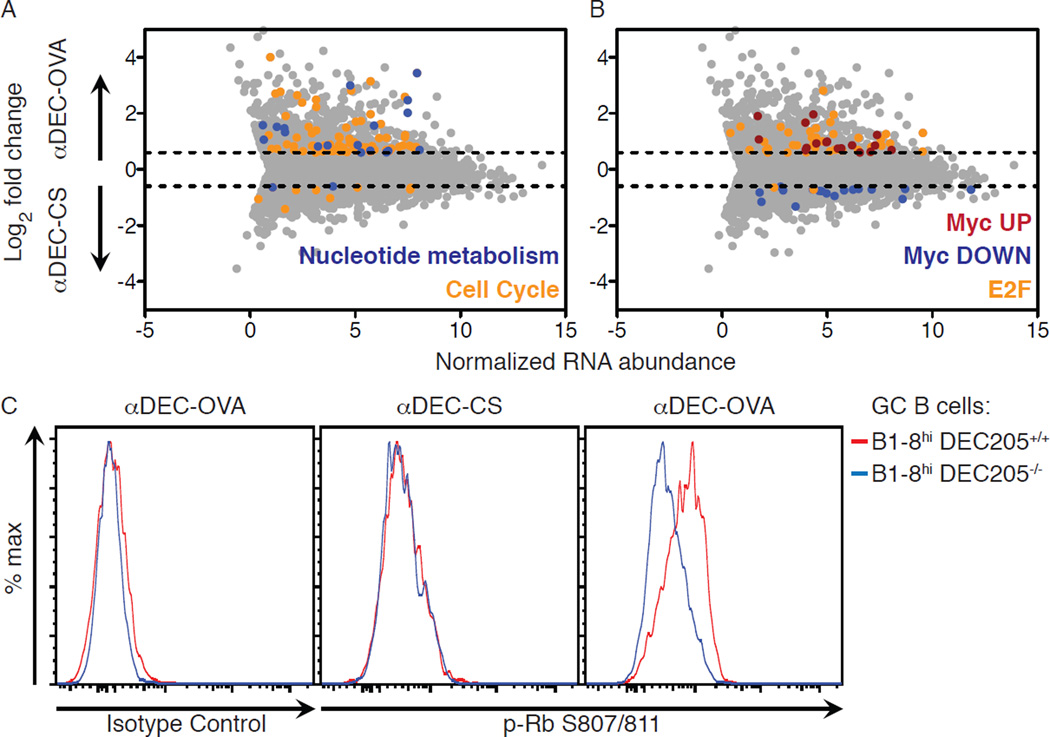
To determine whether B cells receiving high levels of T cell help show a specific change in gene expression, we compared DZ cells in the G1 phase of the cell cycle from αDEC-OVA and control αDEC-CS treated GCs using a fluorescent ubiquitination-based cell cycle indicator (Fuccitg) (fig. S1) (18, 19). RNA sequencing revealed that T cell-mediated selection produced a statistically significant increase in gene expression programs associated with the cell cycle, metabolism, including the metabolism of nucleotides, and genes downstream of c-Myc and the E2F transcription factors (Fig. 1A and B and fig. S2). Finding an increase in expression of c-Myc target genes is in agreement with the observation that c-Myc is induced by T cell help in the GC (20, 21). E2F transcription factors are principal drivers of the cell cycle and are activated by cyclin-dependent kinase (CDK) phosphorylation of the retinoblastoma (Rb) protein (22, 23). Consistent with this, Rb was highly phosphorylated in GC B cells receiving enhanced T cell help (Fig. 1C). E2F and c-Myc are crucial drivers of cell cycle phase transitions; moreover, their activation regulates nucleotide metabolism and controls DNA replication dynamics (23–26), suggesting that T cell help might control the cell cycle dynamics of selected GC B cells in vivo.
Figure 1. T cell help regulates cell cycle and metabolic gene expression programs in selected GC B cells.
(A, B) RNA sequencing analysis showing genes up- or down-regulated by a fold-change of at least 0.6 (log2) upon treatment with αDEC-OVA or αDEC-CS. For clarity, enriched gene sets according to curated reactome gene sets (A) and transcription factor target genes (B) are shown separately. (C) Histograms showing intracellular levels of Rb phosphorylation in B1–8hi DEC205+/+ and B1–8hi DEC205−/− GC B cells from mice treated 2 days earlier with αDEC-OVA or αDEC-CS. Results represent two (A, B) or three (C) independent experiments with 4–5 mice per condition for each experiment.
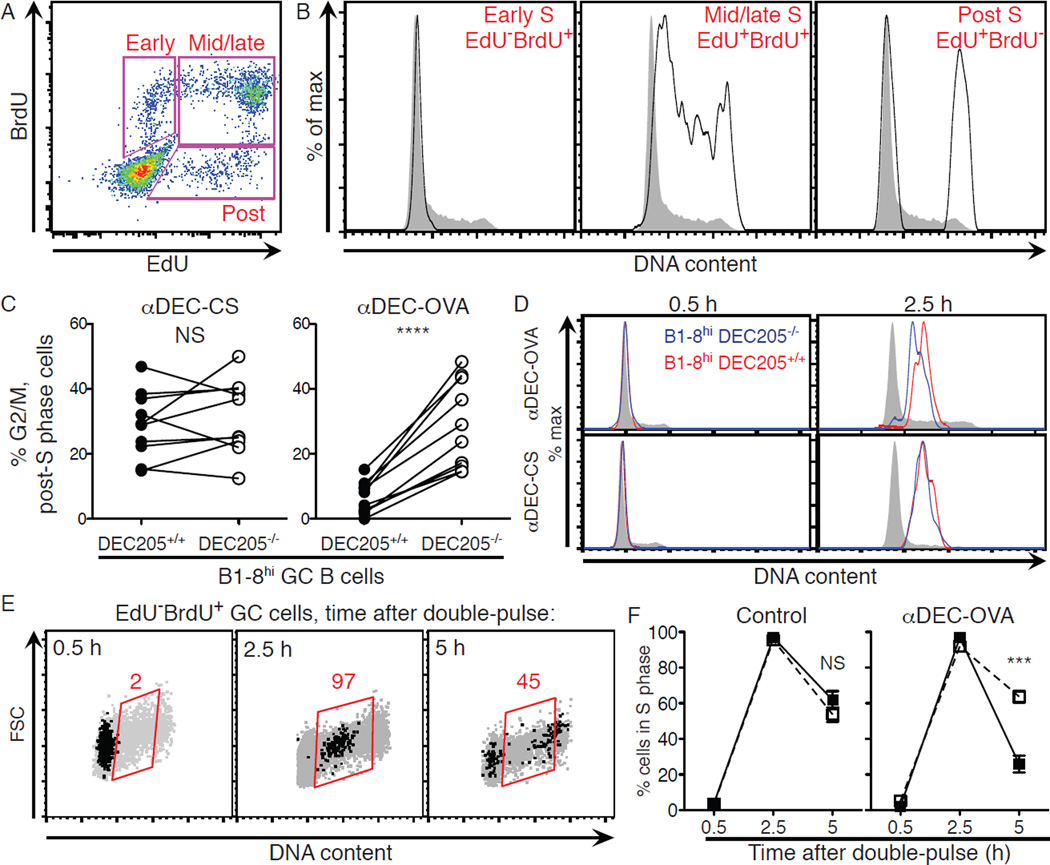
To examine cell cycle progression, mice were pulsed sequentially with the nucleoside analog 5-ethynyl-2’-deoxyuridine (EdU) followed 1 hour later by 5-bromo-2’-deoxyuridine (BrdU) and GC B cells were then stained for DNA content (Fig. 2A and fig. S3) (13). At 0.5 hours after the BrdU pulse, early S phase cells were EdU−BrdU+ labeled and had replicated only a small amount of their genome, making their DNA content similar to that of G1 cells (Fig. 2A and B). By contrast, mid/late-S phase cells were EdU+BrdU+ labeled, and post-S phase cells (EdU+BrdU− labeled) were either in G2/M phase or in the G1 phase of the next cell cycle (Fig. 2A and B). Under control conditions (αDEC-CS), B1–8hi DEC205+/+ and B1–8hi DEC205−/− post-S phase GC B cells were similarly distributed between G2/M and G1, indicating equivalent rates of progression through the G2/M phases of the cell cycle (Fig. 2C). By contrast, inducing selection by αDEC–OVA resulted in rapid progression through G2/M and return to G1 (Fig. 2C). Thus, T cell-mediated selection accelerates progression through G2/M.
Figure 2. T cell help regulates progression through S and G2/M phases during selection.
(A) Mice were injected with EdU intravenously followed 1 hour later by BrdU and then analyzed by flow cytometry 0.5 hours later. (B) Representative histograms displaying DNA content among all GC B cells (grey) and gated populations (black) shown in (A). (C) Mean fraction of cells in G2/M as determined by DNA content among EdU+BrdU− (post-S phase) B1–8hi DEC205+/+ and B1–8hi DEC205−/− GC B cells treated with αDEC-OVA or αDEC-CS (control) 2 days earlier. Lines connect indicated cell populations from the same animal. (D) Histograms showing progressive accumulation of DNA content among EdU−BrdU+ GC B cells from 0.5 to 2.5 hours after EdU/BrdU double-pulse. (E) Flow cytometry plots showing time course of cell cycle progression of EdU−BrdU+ cells (black dots) at 0.5, 2.5, and 5 hours following double-pulse labeling. Grey dots represent all GC B cells in the same mice. Red values represent fraction in S phase gate. (F) Mean fraction of GC B cells in S phase among EdU−BrdU+ B1–8hi DEC205+/+ (black squares) and B1–8hi DEC205−/− (white squares) cells as determined by DNA content at 0.5, 2.5 and 5 hours after double-pulse labeling in mice treated 2 days earlier with αDEC-OVA or either PBS or αDEC-CS (control). Error bars = SEM. Two-tailed paired t-test was used in (C) and two-tailed Mann-Whitney test in (F). **** p < 0.0001. *** p = 0.0002. Experiments represent 2–3 independent experiments with 7–10 mice total for each time point and condition.
To examine S phase dynamics, we followed GC B cells 2.5 and 5 hours after the EdU/BrdU double-pulse described above. At 2.5 hours, EdU−BrdU+ cells, which were in early S phase at 0.5 hours, had progressed into mid/late-S phase as determined by DNA content (Fig. 2D and E). With only these 2 additional hours to replicate their genomes, selected GC B cells accumulated more DNA content and had therefore replicated their genomes at a faster rate than control cells obtained from αDEC-CS treated mice (Fig. 2D and fig. S4). After 5 hours, nearly half of control cells that were labeled in early S phase (EdU−BrdU+) had completed S phase and were in G2/M or G1 (Fig. 2E). Selection significantly accelerated progression through S phase, since a far greater fraction of B1–8hi DEC205+/+ GC cells targeted with αDEC– OVA completed DNA replication in 5 hours than did co-transferred B1–8hi DEC205−/− GC cells (Fig. 2F). We conclude that T cells induce accelerated progression of selected GC B cells through the S and G2/M phases of the cell cycle.
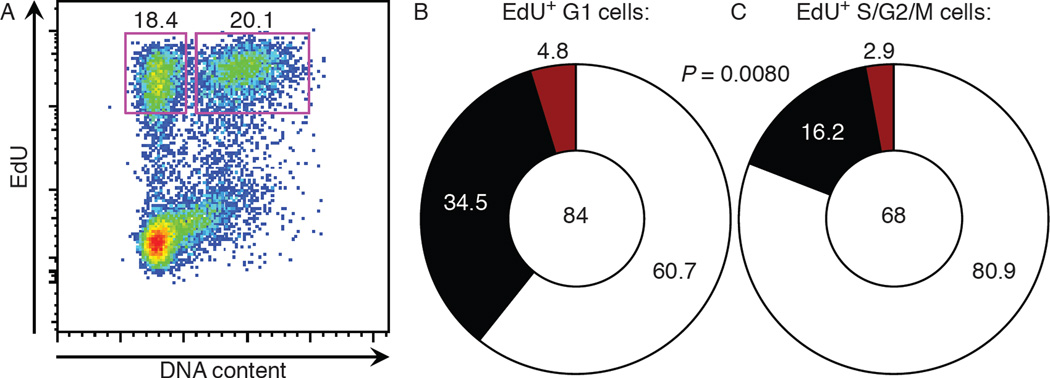
To determine whether accelerated progression through the cell cycle is also a feature of polyclonal GC responses, we examined the response to NP in C57BL/6 mice. High affinity clones in anti-NP GCs carry a tryptophan to leucine mutation at position 33 (W33L) or a lysine to arginine mutation at position 59 (K59R) in the VH186.2 antibody gene (27, 28). Two weeks after immunization with NP-OVA, mice were pulsed with EdU followed 30 minutes later by a large dose of BrdU to inhibit further EdU uptake (fig. S5). After 3.25 hours, EdU+ GC cells were separated into 2 distinct populations based on DNA content (Fig. 3A): (1) EdU+ cells in S/G2/M that represent a mixed population that initiated S phase at the time of EdU injection and/or progressed slowly through S/G2/M; (2) EdU+ cells in G1 that were close to completing S phase during the EdU injection and/or progressed rapidly through S/G2/M. If high affinity GC B cells progress through the cell cycle at an accelerated rate, then they should be enriched in the EdU+ G1 population. Sequencing the VH186.2 genes of these cells revealed that EdU+ G1 cells were significantly enriched in high affinity clones compared to EdU+ S/G2/M cells (Fig. 3B and C). Thus, affinity-enhancing mutations in the polyclonal GC are associated with rapid progression through S/G2/M.
Figure 3. Affinity-enhancing mutations in polyclonal GCs are associated with accelerated S/G2/M progression.
(A) WT mice were immunized with NP-OVA and 14 days later were administered EdU for 0.5 hours before BrdU. At 3.25 hours after BrdU administration, EdU+ G1 cells and EdU+ S/G2/M GC B cells were sorted and analyzed for affinity-enhancing mutation in VH186.2 genes. (B, C) Pie charts show frequency of W33L+ (black) and K59R+ (red) clones among VH186.2 sequences within EdU+ G1 (B) and EdU+ S/G2/M (C) GC B cells. One sequence was doubly positive for the W33L and K59R mutations and was counted within the black slice in (C). Total number of clones analyzed is shown in center. P value was determined using Fisher’s exact test. Results are pooled from 2 independent experiments with 10 mice each.
Once a GC B cell has entered the cell cycle, S phase occupies most of the time it takes for the cell to divide (Fig. 2) (13). We therefore sought to understand how T cell help accelerates progression through S phase. Control of S phase length is a well-documented phenomenon during embryonic development (29). In this context, S phase control operates at the level of initiation of DNA replication; rapid S phases are due to an increased number of synchronously fired replication origins (30–33).
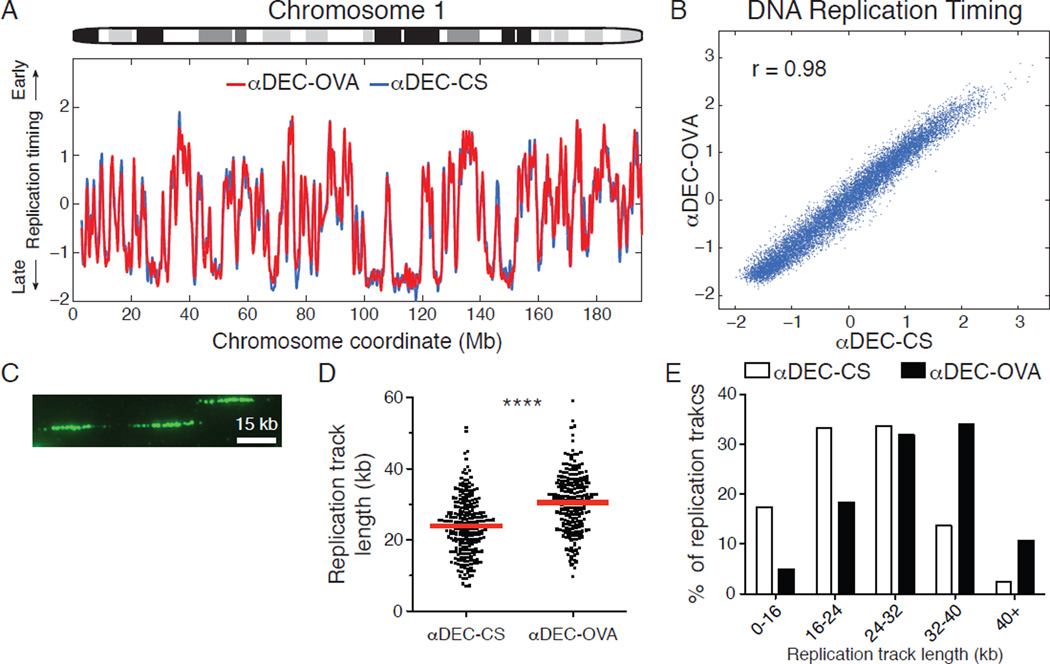
To evaluate the dynamics of DNA replication initiation, we sorted and sequenced the genomes of B1–8hi DEC205+/+ GC B cells in G1 and S phase under control conditions or during their selective expansion (fig. S6A and B). In a population of S phase cells, genomic sites that replicate early have higher copy number than regions that are replicated later during S phase. Thus, by analyzing the relative copy number of DNA sequences along chromosomes in S phase cells compared to G1 phase cells, we obtained genome-wide profiles of DNA replication timing (34, 35). The relative timing of DNA replication among selected and non-selected GC B cells was essentially identical, with the same locations and activation times of origins throughout the genome (r = 0.98, Fig. 4A and B and fig. S6C). Thus, T cell help accelerates S phase by proportionally condensing the replication timing program, while maintaining the overall dynamics of DNA replication albeit in a shorter timescale.
Figure 4. T cell help accelerates S phase by regulating the speed of DNA replication elongation.
(A) Replication timing profile of chromosome 1 was determined by analyzing the ratio of DNA copy number between sorted S and G1 phase B1–8hi DEC205+/+ GC B cells treated with αDEC-CS or αDEC-OVA 2 days earlier. (B) Genome-wide correlation between replication timing in the two conditions. (C) Representative IdU-labeled DNA fibers are shown. (D) Replication track lengths from αDEC-CS or αDEC-OVA treated B1–8hi DEC205+/+ GC B cells. Each data point represents an IdU-labeled track, and red lines represent mean values. Results are pooled from 2 independent experiments and involve over 220 measured fibers in total for each condition. (E) Distribution of replication track lengths. **** p < 0.0001, Two-tailed Mann-Whitney test.
Elevated c-myc, CDK activity and nucleotide metabolism can increase the speed of DNA replication fork progression in tissue culture cells, yet a physiological role for this mechanism has not been documented (26, 36, 37). Since T cell help produces similar molecular changes in selected GC B cells, we examined DNA replication forks by single-molecule analyses (38). For this purpose, αDEC-OVA or control αDEC-CS treated mice were pulsed with the nucleoside analog 5-iodo-2’deoxyuridine (IdU) (fig. S6D). GC B cells were then isolated by cell sorting and processed to measure the lengths of DNA replication tracks in individual molecules (Fig. 4C). Selection resulted in an increase in the average length of replication tracks and altered the distribution of replication track lengths, indicating that T cell help regulates DNA replication by increasing the speed of individual DNA replication forks (Fig. 4D and E, p < 0.0001).
The GC imparts protective immunity by selectively expanding high affinity B cells, yet the mechanisms whereby such clones come to dominate the GC population have been obscure (3, 4). T cells discern among GC B cells on the basis of cell surface peptide-MHCII levels and transduce signal(s) that regulate the amount of time selected cells spend proliferating in the DZ (8, 11–13). Thus, T cell help influences the choice that a recently divided GC B cell in G1 must make in the DZ: to either reenter another S phase or instead return to the LZ (13). Our results indicate that T cell-mediated selection extends beyond this mode of regulation, accelerating progression through the entire cell cycle in selected GC B cells. Thus, the regulatory effects transduced by T cells work in concert to induce selected GC B cells to spend a longer period of time in the DZ where they proliferate at a faster rate.
There are few examples of physiological regulation of cell cycle speed in eukaryotes, and, to our knowledge, none known that involve directed signaling by one cell type to another (29). High levels of replication factors and CDK activity in the early embryo synchronize initiation from a greater number of origins to increase the rate of cell division (31, 39). By contrast, during B cell selection, T cell help induces higher levels of critical regulators of the cell cycle (c-myc and CDK) and nucleotide metabolism to accelerate DNA replication elongation while preserving the temporal order and the number of initiation events throughout the genome. We conclude that selection in the GC involves dynamic regulation of genome replication in S phase, the longest phase of the committed cell cycle.
Supplementary Material
Acknowledgments
We thank A. Miyawaki and T. Kurosaki for mice; T. Eisenreich for help with mouse colony management; K. Yao for technical help; J. Hurwitz and A. Farina for advice; B. Zhang and C. Zhao and the Rockefeller University Genomics Resource Center for assistance with high throughput sequencing; K. Velinzon for assistance with cell sorting; all members of the Nussenzweig laboratory for discussion. The data presented in this manuscript are tabulated in the main paper and in the supplementary materials. Supported by NIH MSTP grant T32GM07739 and NIAID grant 1F30AI109903-01 (A.D.G.); NIH grants AI037526-19 and AI072529-06 (M.C.N.); and the NIH Center for HIV/AIDS Vaccine Immunology and Immunogen Discovery (CHAVI-ID) 1UM1 AI100663-01 (M.C.N). Z.S. is a Human Frontiers of Science Program Fellow (reference LT000340/2011-L). C.T.M is an EMBO fellow (ALTF 456-2014) and is supported by the European Commission FP7 (Marie Curie Actions, EMBOCOFUND2012, GA-2012-600394). M.C.N. is an HHMI investigator.
References and Notes
- 1.Eisen HN, Siskind GW. Variations in Affinities of Antibodies during the Immune Response. Biochemistry. 1964 Jul;3:996. doi: 10.1021/bi00895a027. [DOI] [PubMed] [Google Scholar]
- 2.Muramatsu M, et al. Class switch recombination and hypermutation require activation-induced cytidine deaminase (AID), a potential RNA editing enzyme. Cell. 2000 Sep 1;102:553. doi: 10.1016/s0092-8674(00)00078-7. [DOI] [PubMed] [Google Scholar]
- 3.Berek C, Berger A, Apel M. Maturation of the immune response in germinal centers. Cell. 1991 Dec 20;67:1121. doi: 10.1016/0092-8674(91)90289-b. [DOI] [PubMed] [Google Scholar]
- 4.Jacob J, Kelsoe G, Rajewsky K, Weiss U. Intraclonal generation of antibody mutants in germinal centres. Nature. 1991 Dec 5;354:389. doi: 10.1038/354389a0. [DOI] [PubMed] [Google Scholar]
- 5.Kocks C, Rajewsky K. Stepwise intraclonal maturation of antibody affinity through somatic hypermutation. Proceedings of the National Academy of Sciences of the United States of America. 1988 Nov;85:8206. doi: 10.1073/pnas.85.21.8206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rajewsky K. Clonal selection and learning in the antibody system. Nature. 1996 Jun 27;381:751. doi: 10.1038/381751a0. [DOI] [PubMed] [Google Scholar]
- 7.Victora GD, Nussenzweig MC. Germinal centers. Annual review of immunology. 2012;30:429. doi: 10.1146/annurev-immunol-020711-075032. [DOI] [PubMed] [Google Scholar]
- 8.Allen CD, Okada T, Tang HL, Cyster JG. Imaging of germinal center selection events during affinity maturation. Science. 2007 Jan 26;315:528. doi: 10.1126/science.1136736. [DOI] [PubMed] [Google Scholar]
- 9.Hauser AE, et al. Definition of germinal-center B cell migration in vivo reveals predominant intrazonal circulation patterns. Immunity. 2007 May;26:655. doi: 10.1016/j.immuni.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 10.Schwickert TA, et al. In vivo imaging of germinal centres reveals a dynamic open structure. Nature. 2007 Mar 1;446:83. doi: 10.1038/nature05573. [DOI] [PubMed] [Google Scholar]
- 11.Victora GD, et al. Germinal center dynamics revealed by multiphoton microscopy with a photoactivatable fluorescent reporter. Cell. 2010 Nov 12;143:592. doi: 10.1016/j.cell.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shulman Z, et al. Dynamic signaling by T follicular helper cells during germinal center B cell selection. Science. 2014 Aug 29;345:1058. doi: 10.1126/science.1257861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gitlin AD, Shulman Z, Nussenzweig MC. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature. 2014 May 29;509:637. doi: 10.1038/nature13300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jiang W, et al. The receptor DEC-205 expressed by dendritic cells and thymic epithelial cells is involved in antigen processing. Nature. 1995 May 11;375:151. doi: 10.1038/375151a0. [DOI] [PubMed] [Google Scholar]
- 15.Kamphorst AO, Guermonprez P, Dudziak D, Nussenzweig MC. Route of antigen uptake differentially impacts presentation by dendritic cells and activated monocytes. J Immunol. 2010 Sep 15;185:3426. doi: 10.4049/jimmunol.1001205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shih TA, Roederer M, Nussenzweig MC. Role of antigen receptor affinity in T cell-independent antibody responses in vivo. Nature immunology. 2002 Apr;3:399. doi: 10.1038/ni776. [DOI] [PubMed] [Google Scholar]
- 17.Boscardin SB, et al. Antigen targeting to dendritic cells elicits long-lived T cell help for antibody responses. The Journal of experimental medicine. 2006 Mar 20;203:599. doi: 10.1084/jem.20051639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sakaue-Sawano A, et al. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 2008 Feb 8;132:487. doi: 10.1016/j.cell.2007.12.033. [DOI] [PubMed] [Google Scholar]
- 19.Aiba Y, et al. Preferential localization of IgG memory B cells adjacent to contracted germinal centers. Proceedings of the National Academy of Sciences of the United States of America. 2010 Jul 6;107:12192. doi: 10.1073/pnas.1005443107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dominguez-Sola D, et al. The proto-oncogene MYC is required for selection in the germinal center and cyclic reentry. Nature immunology. 2012 Nov;13:1083. doi: 10.1038/ni.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calado DP, et al. The cell-cycle regulator c-Myc is essential for the formation and maintenance of germinal centers. Nature immunology. 2012 Nov;13:1092. doi: 10.1038/ni.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chellappan SP, Hiebert S, Mudryj M, Horowitz JM, Nevins JR. The E2F transcription factor is a cellular target for the RB protein. Cell. 1991 Jun 14;65:1053. doi: 10.1016/0092-8674(91)90557-f. [DOI] [PubMed] [Google Scholar]
- 23.Chen HZ, Tsai SY, Leone G. Emerging roles of E2Fs in cancer: an exit from cell cycle control. Nature reviews. Cancer. 2009 Nov;9:785. doi: 10.1038/nrc2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dang CV. MYC on the path to cancer. Cell. 2012 Mar 30;149:22. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dominguez-Sola D, Gautier J. MYC and the control of DNA replication. Cold Spring Harbor perspectives in medicine. 2014 Jun;4 doi: 10.1101/cshperspect.a014423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bester AC, et al. Nucleotide deficiency promotes genomic instability in early stages of cancer development. Cell. 2011 Apr 29;145:435. doi: 10.1016/j.cell.2011.03.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Allen D, Simon T, Sablitzky F, Rajewsky K, Cumano A. Antibody engineering for the analysis of affinity maturation of an anti-hapten response. The EMBO journal. 1988 Jul;7:1995. doi: 10.1002/j.1460-2075.1988.tb03038.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Furukawa K, Akasako-Furukawa A, Shirai H, Nakamura H, Azuma T. Junctional amino acids determine the maturation pathway of an antibody. Immunity. 1999 Sep;11:329. doi: 10.1016/s1074-7613(00)80108-9. [DOI] [PubMed] [Google Scholar]
- 29.Nordman J, Orr-Weaver TL. Regulation of DNA replication during development. Development. 2012 Feb;139:455. doi: 10.1242/dev.061838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blumenthal AB, Kriegstein HJ, Hogness DS. The units of DNA replication in Drosophila melanogaster chromosomes. Cold Spring Harbor symposia on quantitative biology. 1974;38:205. doi: 10.1101/sqb.1974.038.01.024. [DOI] [PubMed] [Google Scholar]
- 31.Collart C, Allen GE, Bradshaw CR, Smith JC, Zegerman P. Titration of four replication factors is essential for the Xenopus laevis midblastula transition. Science. 2013 Aug 23;341:893. doi: 10.1126/science.1241530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hyrien O, Maric C, Mechali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995 Nov 10;270:994. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- 33.McKnight SL, Miller OL., Jr Electron microscopic analysis of chromatin replication in the cellular blastoderm Drosophila melanogaster embryo. Cell. 1977 Nov;12:795. doi: 10.1016/0092-8674(77)90278-1. [DOI] [PubMed] [Google Scholar]
- 34.Koren A, et al. Genetic variation in human DNA replication timing. Cell. 2014 Nov 20;159:1015. doi: 10.1016/j.cell.2014.10.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koren A, et al. Differential relationship of DNA replication timing to different forms of human mutation and variation. American journal of human genetics. 2012 Dec 7;91:1033. doi: 10.1016/j.ajhg.2012.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Anglana M, Apiou F, Bensimon A, Debatisse M. Dynamics of DNA replication in mammalian somatic cells: nucleotide pool modulates origin choice and interorigin spacing. Cell. 2003 Aug 8;114:385. doi: 10.1016/s0092-8674(03)00569-5. [DOI] [PubMed] [Google Scholar]
- 37.Malinsky J, et al. The supply of exogenous deoxyribonucleotides accelerates the speed of the replication fork in early S-phase. Journal of cell science. 2001 Feb;114:747. doi: 10.1242/jcs.114.4.747. [DOI] [PubMed] [Google Scholar]
- 38.Jackson DA, Pombo A. Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. The Journal of cell biology. 1998 Mar 23;140:1285. doi: 10.1083/jcb.140.6.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Farrell JA, Shermoen AW, Yuan K, O'Farrell PH. Embryonic onset of late replication requires Cdc25 down-regulation. Genes & development. 2012 Apr 1;26:714. doi: 10.1101/gad.186429.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guo M, et al. A monoclonal antibody to the DEC-205 endocytosis receptor on human dendritic cells. Human immunology. 2000 Aug;61:729. doi: 10.1016/s0198-8859(00)00144-0. [DOI] [PubMed] [Google Scholar]
- 41.Yaffe E, et al. Comparative analysis of DNA replication timing reveals conserved large-scale chromosomal architecture. PLoS genetics. 2010 Jul;6:e1001011. doi: 10.1371/journal.pgen.1001011. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.