Highlight
Suppression of the phytoglobin GLB2 enhances jasmonic acid which promotes the accumulation of auxin and the formation of somatic embryos in Arabidopsis
Key words: Auxin, PGB2, jasmonic acid, nitric oxide, phytoglobin, somatic embryogenesis.
Abstract
Previous studies have shown that the beneficial effect of suppression of the Arabidopsis phytoglobin 2 gene, PGB2, on somatic embryogenesis occurs through the accumulation of nitric oxide (NO) within the embryogenic cells originating from the cultured explant. NO activates the expression of Allene oxide synthase (AOS) and Lipoxygenase 2 (LOX2), genes encoding two key enzymes of the jasmonic acid (JA) biosynthetic pathway, elevating JA content within the embryogenic tissue. The number of embryos in the single aos1-1 mutant and pgb2-aos1-1 double mutant declined, and was not rescued by increasing levels of NO stimulating embryogenesis in wild-type tissue. NO also influenced JA responses by up-regulating PLANT DEFENSIN 1 (PDF1) and JASMONATE-ZIM-PROTEIN (JAZ1), as well as down-regulating MYC2. The NO and JA modulation of MYC2 and JAZ1 controlled embryogenesis. Ectopic expression of JAZ1 or suppression of MYC2 promoted the formation of somatic embryos, while repression of JAZ1 and up-regulation of MYC2 reduced the embryogenic performance. Sustained expression of JAZ1 induced the transcription of several indole acetic acid (IAA) biosynthetic genes, resulting in higher IAA levels in the embryogenic cells. Collectively these data fit a model integrating JA in the PGB2 regulation of Arabidopsis embryogenesis. Suppression of PGB2 increases JA through NO. Elevated levels of JA repress MYC2 and induce JAZ1, favoring the accumulation of IAA in the explants and the subsequent production of somatic embryos.
Introduction
Found in all nucleated organisms, hemoglobins are important Fe heme-containing proteins fulfilling a variety of tasks. Initially characterized in vertebrates in relation to their ability to bind and transport oxygen and other ligands such as CO2, and NO, hemoglobin-like compounds were also found expressed in the nodules of legumes containing nitrogen-fixing bacteria where they served to prevent inactivation of nitrogenase by binding oxygen (reviewed in Smagghe et al., 2009). Additional plant hemoglobin-like coumpounds were subsequently found to be more widely distributed in plants (Hill, 2012) and were ascribed the name, nonsymbiotic hemoglobins to distinguish them from leghemoglobin. The types (Garrocho-Villegas et al., 2007) and function (Hill, 2014) of nonsymbiotic hemoglobins have expanded considerably to the point that a more specific name, phytoglobin (Pgb), has been introduced.
From a phylogenetic perspective, oxygen binding activity, and the expression profile, three types of plant Pgbs have been described (Hunt et al., 2001). Class 1 and 2 Pgbs possess a 3-on-3 α-helical loop surrounding the heme moiety, while class 3 Pgbs are similar to truncated bacterial globins (Watts et al., 2001). The majority of the studies are centered on class 1 and 2 which are characterized by a very high oxygen binding affinity: an approximate K m of 2nM for members of class 1 and 150nM for members of class 2 (Hoy and Hargrove, 2008; Dordas, 2009). The high affinity for oxygen exhibited by class 1 Pgbs discounts their roles in oxygen sensing and transport (Hill, 1998). The most plausible function of plant Pgbs is to scavenge NO, as demonstrated in several developmental and stress responses (reviewed in Hill, 2012).
Three Pgb genes categorized in their respective classes have been identified in Arabidopsis: PGB1, PGB2, and PGB3. While no information is available on the function of PGB3, several studies have provided evidence for the role of PGB1 and PGB2 during hypoxia. Due to its ability to scavenge NO efficiently under hypoxic conditions, PGB1 exercises a protective role during abiotic stress (Perazzolli et al., 2004). Arabidopsis roots grown under low oxygen conditions rapidly expressed PGB1, and plants overexpressing PGB1 exhibited a higher survival rate resulting from a depletion in cellular NO (Hunt et al., 2002). The role of protection from hypoxia observed for PGB1 was also documented with other class 1 Pgbs. Hypoxic roots of alfalfa overexpressing a class 1 Pgb developed fewer aerenchyma and exhibited enhanced growth due to sustained NO-scavenging mechanisms (Dordas et al., 2003). Opposite results were observed in plants suppressing the barley Pgb (Dordas et al., 2003).
Like PGB1, overexpression of PGB2 increased NO scavenging (Hebelstrup and Jensen, 2008; Hebelstrup et al., 2012) and enhanced plant survival under hypoxic conditions (Hebelstrup et al., 2006). Expression of PGB2 is favored by cytokinin and low temperatures (Trevaskis et al., 1997; Hunt et al., 2001), conditions not affecting PGB1 and denoting differential control mechanisms and possibly functions of the two Pgbs. The preferential expression of PGB2 in immature and developing organs, such as somatic embryos, leaflets, and immature seeds and fruits (Hendriks et al., 1998; Hunt et al., 2002; Wang et al., 2003), is suggestive of a function for the gene to accommodate the high energy demand of the organs. In developing Arabidopsis seeds, overexpression of PGB2 has been associated with improved oil accumulation through the maintenance of a high energy status (Vigeolas et al., 2011).
A recurring theme emerging from these studies is the mediation of NO in many Pgb-regulated events. The prominent role of NO as a signal molecule in many physiological responses, in conjunction with the expression of Pgbs under normoxic conditions (Hill, 2012), suggests a possible involvement of Pgbs in fundamental developmental processes. Major phenotypic defects were observed in Arabidopsis plants with altered PGB1 and PGB2 expression (Hebelstrup et al., 2006). Independent evidence suggests a control of meristem function by Pgbs. While overexpression of either PGB1 or PGB2 encourages the vegetative–reproductive transition of the shoot meristem, the repression of PGB1 affects the time of flowering (Hebelstrup and Jensen, 2008). Meristem formation in vitro was also affected by Pgbs, with the overexpression of both PGB1 and PGB2 favoring the formation of shoots through the activation of auxin and cytokinin perception (Wang et al., 2011). By modulating NO emission, PGB1 and PGB2 also regulate hyponastic responses during flooding, an observation integrating Pgbs in long-range plant signaling mechanisms (Hebelstrup et al., 2013).
While the participation of Pgbs in developmental processes has mainly been investigated post-embryonically, Pgbs might play a central role during plant embryogenesis. Phytoglobin genes are expressed during embryo development (Smagghe et al., 2007), and when hemoglobin is applied exogenously it influences somatic embryogenesis, the ability of somatic cells to produce embryos (Jayabalan et al., 2004). A more direct involvement of Pgbs during embryo formation was demonstrated by Elhiti et al. (2013) using Arabidopsis somatic embryogenesis. Suppression of PGB2 increased embryogenesis by elevating NO levels at the sites of the explants forming somatic embryos. Accumulation of NO suppresses MYC2 (Elhiti et al., 2013), a basic helix–loop–helix (bHLH) domain-containing transcription factor which represses the biosynthesis of auxin (Dombrecht et al., 2007), the inductive signal which initiates the embryogenic process (Raghavan, 2004). As a result of this mechanism, PGB2-suppressed cells accumulate more auxin and produce a large number of somatic embryos (Elhiti et al., 2013). While representing a valid framework integrating Pgb signaling in plant embryogenesis, this model is most probably incomplete in terms of the number of intermediates transducing the PGB2 response. During post-embryonic growth, both NO and MYC2 operate at the interphase of a variety of transduction pathways often involving hormones, predominantly jasmonic acid (JA) (Chen et al., 2011; Mur et al., 2013). While the link between MYC2 and JA signaling has been well established during plant–pathogen interactions and insect predation (Lorenzo and Solano, 2005), the relationship between NO and JA is far from clear. JA synthesis is repressed by NO in some systems but induced in others (Orozco-Cardenas and Ryan, 2002). A rapid induction in JA level following inoculation with Botryts cinerea was observed in Arabidopsis plants accumulating NO through suppression of Pgb (Hebelstrup et al., 2012). Based on these observations, it cannot be excluded that JA plays a key role during embryogenesis, possibly as an integrated component of the PGB2 regulatory mechanisms.
In an effort to establish a relationship between PGB2, NO, and JA in the regulation of embryo formation, we used the well-characterized Arabidopsis somatic embryogenesis system (Elhiti et al., 2013). Formation of somatic embryos in Arabidopsis is a two-step process (Fig. 1A). The first involves culturing early cotyledonary zygotic embryos on an auxin-containing induction medium which stimulates the formation of embryogenic tissue. Production of somatic embryos from the embryogenic tissue is then initiated by the removal of auxin (Bassuner et al., 2007). Our results suggest that JA is a key component of PGB2 regulation of embryogenesis in a model including NO and several JA-responsive intermediates.
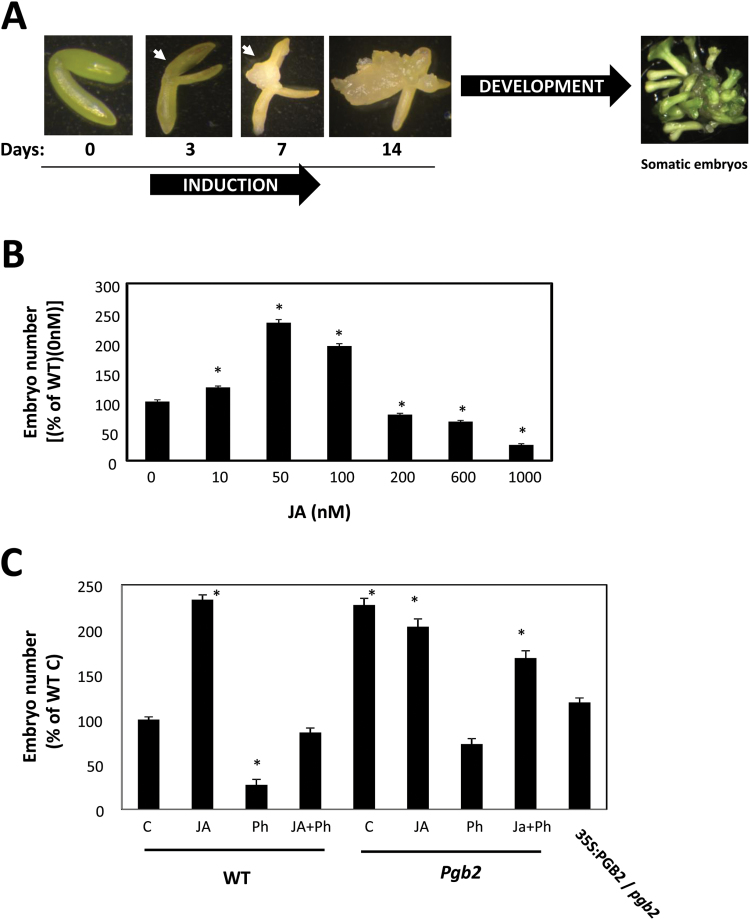
Fig. 1.
Effects of altered levels of jasmonic acid (JA) on Arabidopsis somatic embryogenesis. (A) Arabidopsis somatic embryos are generated through a two-step process. Dissected zygotic embryos are initially plated on 2,4-D-containing induction medium required for the formation of embryogenic tissue. After 14 d, the explants are transferred onto a hormone-free development medium which stimulates the production of somatic embryos. Fully developed somatic embryos can be obtained after 9 d on development medium. Arrows indicate the formation of the embryogenic tissue. (B) Changes in embryo number in the wild type (WT) line cultured with increasing levels of JA applied to the induction medium. Values are means ±SE of at least three biological replicates. An asterisk indicates statistically significant differences (P≤0.005) from the control (JA=0nM) value set at 100%. (C) Effects of applications of JA (50nM) and/or Phenidone (Ph) on the number of somatic embryos produced by the WT, pgb2, and 35S:PGB2/pgb2 lines. Values are means ±SE of at least three biological replicates. An asterisk indicates statistically significant differences (P≤0.005) from value of the WT (C) set at 100%. (This figure is available in colour at JXB online.)
Materials and methods
Plant materials
The Arabidopsis (Columbia) mutant lines ami1-1 (SALK_069970) (Elhiti et al. 2013), jaz1-1 (SALK 011957) (Demianski et al., 2012), and aos1-1 (SALK 017756) (Park et al., 2002), and the pASA1:GUS reporter line (CS16701), were obtained from the Arabidopsis Biological Resource Center (ABRC). The following lines were received as gifts: the pgb2 knock-out line (referred to as glb2 in Hebelstrup et al., 2008); the myc2-1 mutant and the 35S:MYC2 line (Dombrecht et al., 2007); the 35S:JAZ1 line (Thines et al., 2007); the pJAZ1:GUS-GFP line (Gutierrez et al., 2012); the 35S:MYC2-GUS line (Zhai et al., 2013); the pYUC4:GUS line (Eklund et al., 2011); and the pPDF1.2:GUS line (Koorneef et al., 2008). pgb2-aos1-1 double mutant lines were generated by crossing (Supplementary Fig. S1 at JXB online).
Growth conditions and induction of somatic embryogenesis
Arabidopsis seeds were sterilized (70% ethanol+0.5% Triton X-100 for 15min followed by 95% ethanol for 15min) and plated on germination medium (half-strength MS; Murashige and Skoog, 1962). The plates were kept at 4 °C in the dark for 2–3 d and then transferred to a growth cabinet (20–22 °C, 16h light/8h dark photoperiod). Plants were grown until siliques were formed, ~21–28 d.
Somatic embryogenesis was promoted using a modified method based on that described by Bassuner et al. (2007). Immature zygotic embryos were plated on induction medium containing 2,4-dichlorophenoxyacetic acid (2,4-D) for 14 d, followed by transfer onto hormone-free development medium. Fully developed somatic embryos were counted after 9 d.
Chemical treatments
The NO scavenger 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO) and the NO donor sodium nitroprusside (SNP) were applied as specified in Elhiti et al. (2013). Applications were performed by dispensing 10 μl of a 10 µM solution directly on the explants every other day throughout culture in the induction medium.
JA (Sigma) was dissolved in water and added to the culture medium at different concentrations as reported in the text. The JA inhibitor 1-phenyl-3-pyrazolidinone (Phenidone, Ph) was applied at a concentration of 10nM.
Total RNA isolation and quantitative real-time PCR analysis
Total RNA was extracted with TRIzol reagent (Invitrogen), treated with DNase I (RNase-free, Promega), and utilized for cDNA synthesis with the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems).
Quantitative real-time PCR was performed as described in Elhiti et al. (2010) using the primers listed in Supplementary Table S1. The relative level of gene expression was analyzed with the 2−∆∆Ct method described by Livak and Schmittgen (2001) using UBQ10 (AT4G05320) as a reference (Czechowski et al., 2005; Hong et al., 2010).
β-Glucuronidase assays
β-Glucuronidase (GUS) histochemical staining assay was performed as described by Sieburth and Meyerowitz (1997). The somatic embryos were examined and photographed using a dissecting microscope equipped with a Leica DC500 digital camera. A minimum of 20 samples per treatment were imaged.
IAA and JA immunolocalization
Immunolocalization of endogenous indole acetic acid (IAA) was carried out following the procedure used by Elhiti et al. (2013). Immunolocalization of endogenous JA was performed as described by Mielke et al. (2011), with some minor modifications. The plant material was fixed in 4% (w/v) 1-ethyl-3-(3–dimethylaminopropyl) carbodiimide (EDC) in phosphate-buffered saline (PBS) for 3h at room temperature. After dehydration in a graded ethanol series, the specimens were infiltrated in PEG-8 distearate containing low melting point wax (Electron Microscopy Sciences) at 45 °C. Sections (10 μm thickness) were incubated with anti-JA antibodies raised in rabbit (kindly donated by Professor House, IPK, Germany) diluted 1:1000 in PBS containing 5% (w/v) BSA and 1% (v/v) acetylated BSA (BSAac; Promega). The secondary goat anti-rabbit IgG antibody conjugated with AlexaFluor594 (Invitrogen) was used according to the manufacturer’s instructions at a 1:2000 dilution. Sections were analyzed by epifluorescence microscopy.
Statistical analysis
All experiments were performed using at least three biological replicates, and Tukey’s post-hoc test for multiple variance was used to compare differences among samples (Zar, 1999) (P=0.05) by the SPSS 14 statistical program.
Results
Repression of PGB2 increases embryogenesis through the NO-mediated elevation in JA level
Arabidopsis somatic embryogenesis is a two-step process (Fig. 1A). Dissected zygotic embryos were cultured on a 2,4-D solid induction medium for 14 d. After 3 d, the cotyledons of the explants started swelling and embryogenic tissue became apparent at day 7. During the following days on induction medium (days 7–14), the embryogenic tissue increased in size. Embryo production was stimulated by transferring the tissue onto a hormone-free development medium. As also reported previously (Elhiti et al., 2013), the term somatic embryos used in this study refers to fully developed embryos collected after 9 d on the development medium (Fig. 1A).
In wild-type (WT) tissue, inclusion of JA in the induction medium affected the number of somatic embryos in a dose-dependent fashion, with the most pronounced increase observed with 50nM JA (Fig. 1B). This concentration was used to compare the effects of JA manipulations on somatic embryos produced by the WT line and the Pgb2 knock-out (pgb2) line characterized by a superior embryogenic performance (Elhiti et al., 2013). In the WT, the JA stimulation of embryogenesis was reversed by Ph, a JA biosynthetic inhibitor (Farmer et al., 1994; Bruinsma et al., 2009), which strongly repressed embryo production when applied alone (Fig. 1C). The beneficial effect of JA on somatic embryogenesis was not observed in the pgb2 line. To prove that the increased number of embryos observed in the pgb2 line relative to the WT line was solely due to suppression of the gene, we overexpressed PGB2 in the mutant line. The resulting 35S:PGB2/pgb2 (Supplementary Fig. S2) had an embryonic yield comparable with the WT line (Fig. 1C)
The different embryonic behavior of the two lines following JA treatments was further examined in light of the following premises: PGB2 is an effective scavenger of NO (Hebelstrup et al., 2008) expressed at the sites of the Arabidopsis explants producing embryogenic tissue (Elhiti et al., 2013); NO accumulates preferentially in cells suppressing PGB2, and this accumulation is required for the enhanced embryogenic performance of the pgb2 line (Elhiti et al., 2013).
To establish an experimental baseline for our experiments, we confirmed previous results related to the effects of NO manipulations on somatic embryogenesis (Elhiti et al., 2013). Applications of the NO donor SNP increased embryo production in the WT line, whereas inclusion of the NO scavenger cPTIO compromised the embryogenic process in both WT and pgb2 lines (Fig. 2A). Compared with the WT, the number of somatic embryos more than doubled in the pgb2 line, an observation consistent with the higher level of NO accumulating in the latter (Elhiti et al., 2013). However, the beneficial effects of high NO levels (either by SNP or by suppression of PGB2) on somatic embryogenesis were JA dependent. In the WT line, depletion of JA by Ph in an NO-enriched environment (SNP+Ph) suppressed embryo production, whereas elevated JA levels promoted embryogenesis even in an environment depleted of NO (cPTIO+JA) (Fig. 2A). Similar results were also observed in the NO-accumulator pgb2 line where applications of JA reversed the inhibitory effect of cPTIO (Fig. 2A). Therefore, the beneficial effects of high NO levels (by SNP or suppression of PGB2) on embryogenesis are mediated by JA.
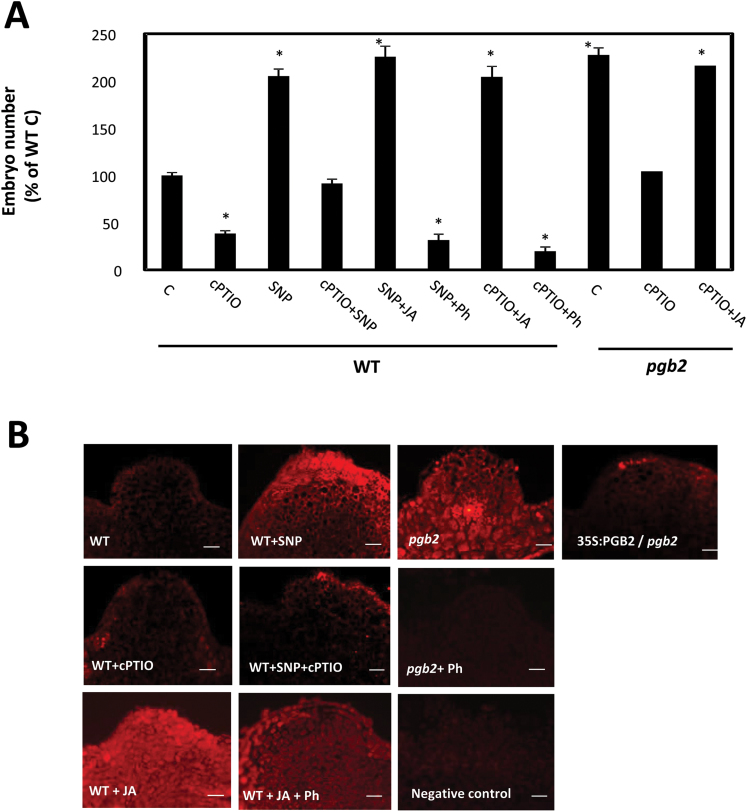
Fig. 2.
The effects of nitric oxide (NO) on somatic embryogenesis are mediated by jasmonic acid (JA). (A) Effects of applications of the NO scavenger 2-(4-carboxyphenyl)-4,4,5,5-tetramethylimidazoline-1-oxyl-3-oxide (cPTIO), the NO-releasing agent sodium nitroprusside (SNP), JA, and Phenidone (Ph) on the number of somatic embryos produced by the WT and pgb2 lines. Values are means ±SE of at least three biological replicates. An asterisk indicates statistically significant differences (P≤0.005) from value of the WT (C) set at 100%. (B) Immunolocalization of JA on the embryogenic tissue arising from the cotyledons of the explants after 7 d on induction medium. Tissue was treated with SNP, cPTIO, JA, and/or Ph. Primary antibodies were omitted from the negative control. Scale bars=20 μm.
We further assessed if JA accumulated preferentially in cells with increasing levels of NO. While JA quantitation cannot be performed in our system due to the reduced size of the embryos, immunolocalization of JA was conducted across the cotyledons of the explants. These are the regions generating the embryogenic tissue (Fig. 1A) and accumulating NO following suppression of PGB2 (fig. 3A in Elhiti et al., 2013). An intense JA signal was observed in NO-enriched environments (WT tissue treated with SNP and pgb2 tissue) (Fig. 2B), while an experimental reduction in NO level by cPTIO reduced the fluorescence. Low JA signal was also observed in the 35S:PGB2/pgb2 line. Specificity of the antibody was verified using JA and/or Ph (Fig. 2B).
Collectively these results suggest that NO induces the accumulation of JA, and heightened levels of JA favor embryogenesis in a dose–response fashion, with exogenous applications increasing embryo number only in systems with reduced levels of endogenous JA (WT line), but not in others already enriched in JA (pgb2 line).
NO activates the expression of genes participating in JA biosynthesis
De novo biosynthesis of JA requires the co-ordinated expression of several genes including LYPOXYGENASE2 (LOX2) involved in the oxygenation of fatty acids to their hydroperoxy derivatives and ALLENE OXIDE SYNTHASE (AOS), forming unstable allene epoxides from the dehydratation of 1,3-hydroperoxy-octatrienoic acid (Wasterack and Hause, 2013). On day 3 and day 7 on induction medium (coinciding with the formation of embryogenic tissue Fig. 1A), the expression of both genes increased in NO-enriched environments (i.e. SNP-treated WT tissue and pgb2 tissue). Consistent with these observations, a reduction of NO level by cPTIO in both WT and pgb2 lines repressed the expression of both genes (Fig. 3A). Reintroduction of PGB2 in the pgb2 line reduced the expression levels of both LOX2 and AOS to WT values (Supplementary Fig. S3)
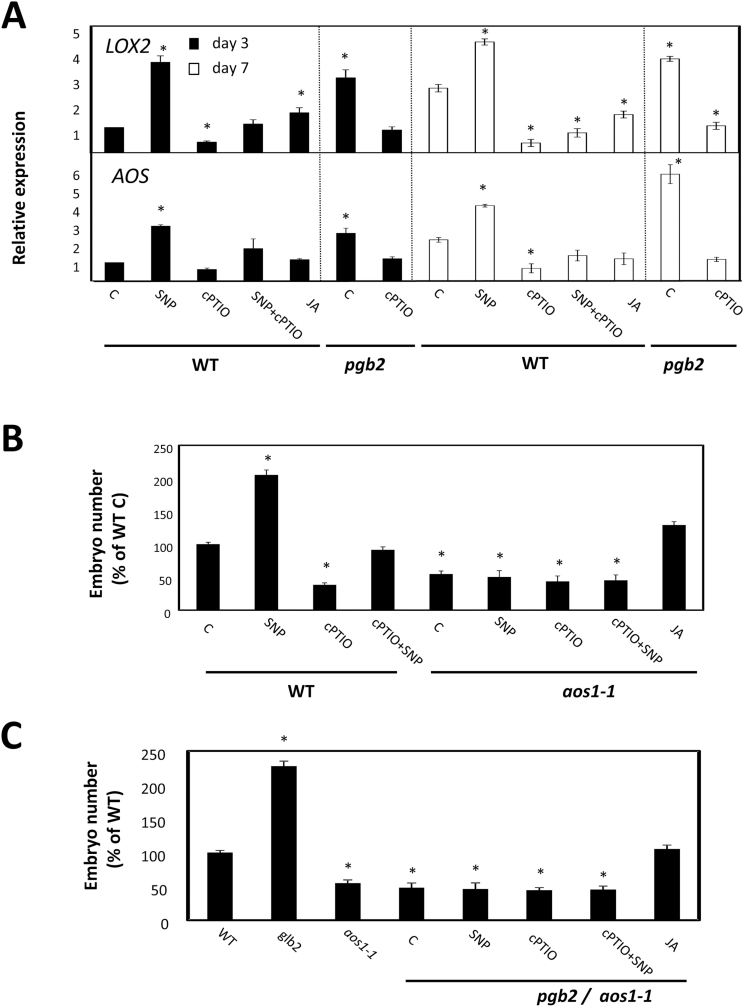
Fig. 3.
The effects of nitric oxide (NO) on jasmonic acid (JA) synthesis. (A) Expression level by quantitative (q)RT–PCR of the two JA biosynthetic genes [LIPOXYGENASE2 (LOX2) and ALLENE OXIDE SYNTHASE (AOS)] at days 3 and 7 in the induction medium of somatic embryogenesis. WT and pgb2 tissue was cultured in medium with altered levels of NO and JA using the pharmacological treatments described in Fig. 2. Values are means ±SE of at least three biological replicates and normalized to the value of the WT (C) of day 3 set at 1. Asterisks indicate statistically significant differences (P≤0.005) from the WT (C) value of the respective day in culture. (B) Number of somatic embryos generated from WT and aos plants. Explants were cultured on medium with altered levels of NO and JA. Values are means ±SE of at least three biological replicates. An asterisk indicates statistically significant differences (P≤0.005) from the WT (C) value set at 100%. (C) Number of somatic embryos generated from WT, pgb2 aos, and pgb2/aos double mutant plants. Explants were cultured on medium with altered levels of NO and JA. Values are means ±SE of at least three biological replicates. An asterisk indicates statistically significant differences (P≤0.005) from the WT value set at 100%.
The requirement for JA, and more specifically the expression of its biosynthetic gene AOS, as downstream components of the PGB2 and NO regulation of embryogenesis was further examined by analyzing the behavior in culture of the JA-deficient aos1-1 mutant (Park et al., 2002), and the pgb2-aos double mutant (Fig. 3B, C). Somatic embryo production was significantly reduced by the suppression of AOS independently from NO levels and PGB2 expression. This reduction was partially reversed by exogenous JA (Fig. 3B, C).
Evidence presented here suggests that NO is a transcriptional activator of JA biosynthesis, and this activation is required for the PGB2 regulation of somatic embryogenesis.
Regulation of embryogenesis by JA is mediated by JAZ1 and MYC2
JAZ1 is a JA-inducible nuclear-localized protein belonging to the larger family of the TIFY proteins (Vanholme et al., 2007) integrated in several JA biotic stress responses (Niu et al., 2010; Sasaki-Sekimoto et al., 2013). During somatic embryogenesis, JAZ1 expression is induced after 12h of JA application (Supplementary Fig. S4). On both day 3 and day 7 on induction medium, its expression increased in the pgb2 line and in response to pharmacological treatments elevating NO or JA levels, while it decreased following depletion of NO (by cPTIO) (Fig. 4A). Reintroduction of PGB2 in the pgb2 line reduced the expression levels of JAZ1 to WT values (Supplementary Fig. S3)
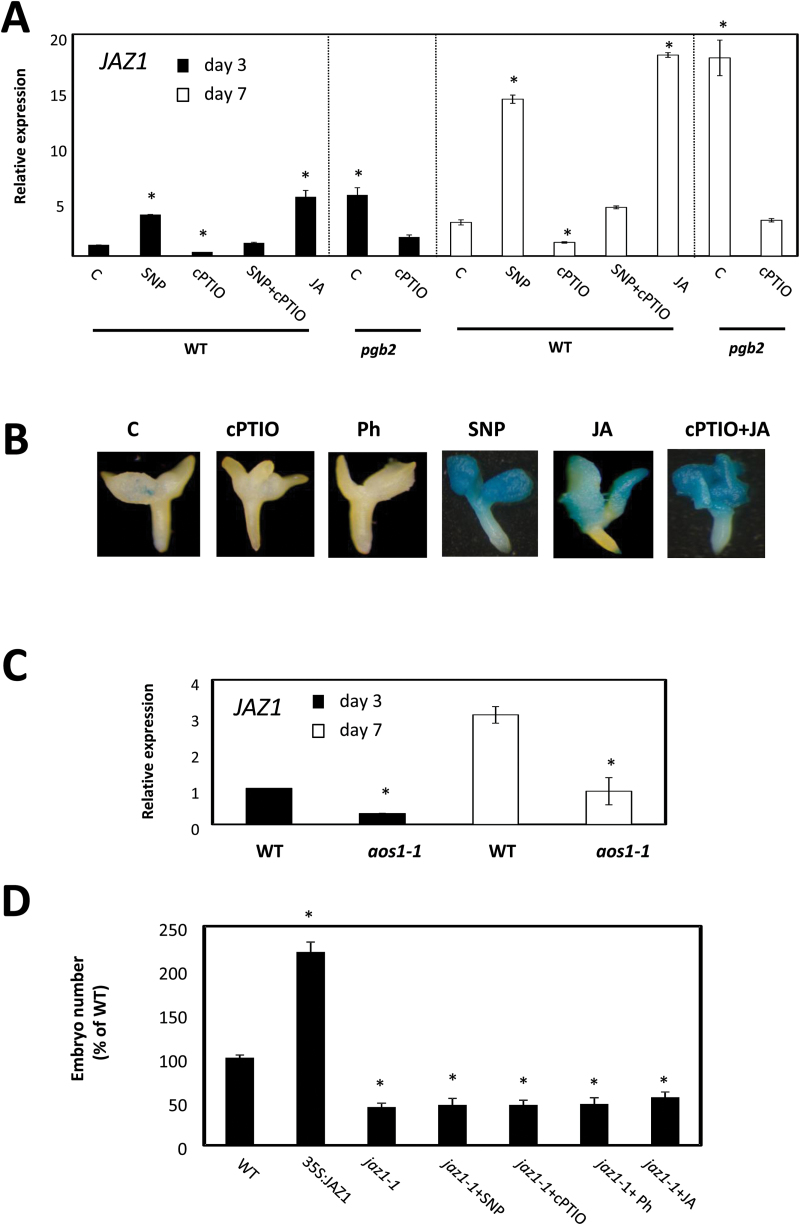
Fig. 4.
Nitric oxide (NO) and jasmonic acid (JA) affect JAZ1. (A) Expression level by quantitative (q)RT–PCR of JAZ1 at days 3 and 7 in the induction medium of somatic embryogenesis. WT and pgb2 tissue was cultured in medium with altered levels of NO and JA using the pharmacological treatments described in Fig. 2. Values are means ±SE of at least three biological replicates and are normalized to the value of the WT (C) of day 3 set at 1. An asterisk indicates statistically significant differences (P≤0.005) from the WT (C) value of the respective day in culture. (B) Localization patterns of JAZ1 by GUS staining at day 7 on induction medium. Tissue was subjected to the indicated pharmacological treatments. (C) Expression level by qRT–CR of JAZ1 in the WT and aos lines at days 3 and 7 on induction medium. Values are means ±SE of at least three biological replicates and are normalized to the value of the WT at day 3 set at 1. An asterisk indicates statistically significant differences (P≤0.005) from the WT value of the respective day in culture. (D) Number of somatic embryos generated from WT, 35S:JAZ1, and jaz1-1 plants. Explants were cultured on media with altered levels of NO and JA. Values are means ±SE of at least three biological replicates. An asterisk indicates statistically significant differences (P≤0.005) from the WT value set at 100%.
The regulation of JAZ1 during the induction of embryogenesis was also confirmed using the pJAZ1:GUS reporter line showing a staining pattern in the apical regions of the explants, including the embryogenic tissue, mediated by NO and JA levels (Fig. 4B).
To verify further the requirement for JA for JAZ1 expression during somatic embryogenesis, we used the JA-deficient aos1-1 line. Compared with the WT, the expression of JAZ1 was significantly repressed in the aos1-1 explants on both day 3 and day 7 on induction medium (Fig. 4C).
Modulations in JAZ1 expression have profound effects on somatic embryogenesis. While the constitutive expression of this gene in the 35S:JAZ1 line increased embryo production, its suppression in the jaz1-1 mutant repressed embryogenesis (Fig. 4D). The fact that this repression was not rescued by manipulating the levels of NO (by SNP or cPTIO) or JA (by Ph or JA) suggests that JAZ1 is a downstream component in the regulation of embryogenesis.
Another key executor of the JA response during biotic and abiotic stress responses is MYC2, a bHLH domain-containing transcription factor (Dombrecht et al., 2007). Expression of MYC2 shows a biphasic pattern in response to JA. While initial applications of JA increase MYC2 expression, prolonged exposure to JA repressed MYC2 expression (Supplementary Fig. S4).
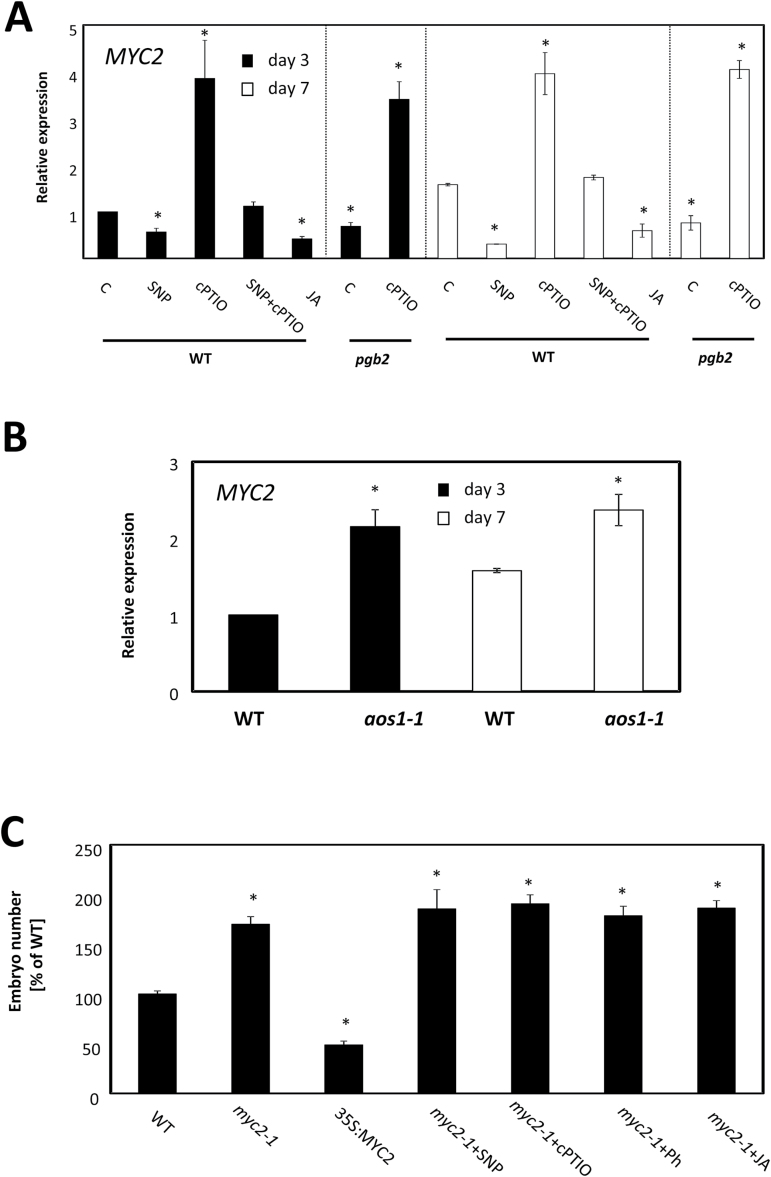
MYC2 is repressed in PGB2-suppressed cells, and this repression promotes auxin production and favors somatic embryogenesis (Elhiti et al., 2013). On both day 3 and day 7 on induction medium, MYC2 was down-regulated in high JA and NO environments (JA- or SNP-treated tissue and pgb2 tissue), while it was induced by cPTIO (Fig. 5A), or by the the overexpression of PGB2 in the pgb2 line (Supplementary Fig. S3). A significant induction of this gene also occurred in the JA-deficient aos1-1 explants on the same days in culture (Fig. 5B). Suppression in MYC2 in the myc2-1 mutant increased embryogenesis in a pattern consistent with previous studies (Elhiti et al., 2013). This increase, however, was not affected by pharmacological treatments altering the levels of NO or JA (Fig. 5C).
Fig. 5.
Nitric oxide (NO) and jasmonic acid (JA) affect MYC2. (A) Expression level by quantitative (q)RT–PCR of MYC2 at days 3 and 7 in the medium with altered levels of NO and JA using the pharmacological treatments described in Fig. 2. Values are means ±SE of at least three biological replicates and are normalized to the value of the WT (C) of day 3 set at 1. An asterisk indicates statistically significant differences (P≤0.005) from the WT (C) value of the respective day in culture. (B) Expression level by qRT–PCR of MYC2 in WT and aos lines at days 3 and 7 on induction medium. Values are means ±SE of at least three biological replicates and are normalized to the value of the WT at day 3 set at 1. An asterisk indicates statistically significant differences (P≤0.005) from the WT value of the respective day in culture. (C) Number of somatic embryos generated from WT, 35S:MYC2, and myc2-1 plants. Explants were cultured on media with altered levels of NO and JA. Values are means ±SE of at least three biological replicates. An asterisk indicates statistically significant differences (P≤0.005) from the WT value set at 100%.
These results suggest that transcriptional regulation of MYC2, like JAZ1, plays an important role in the NO and JA control of Arabidopsis somatic embryogenesis, and that increased embryo number is favored by the up-regulation of JAZ1 and the repression of MYC2.
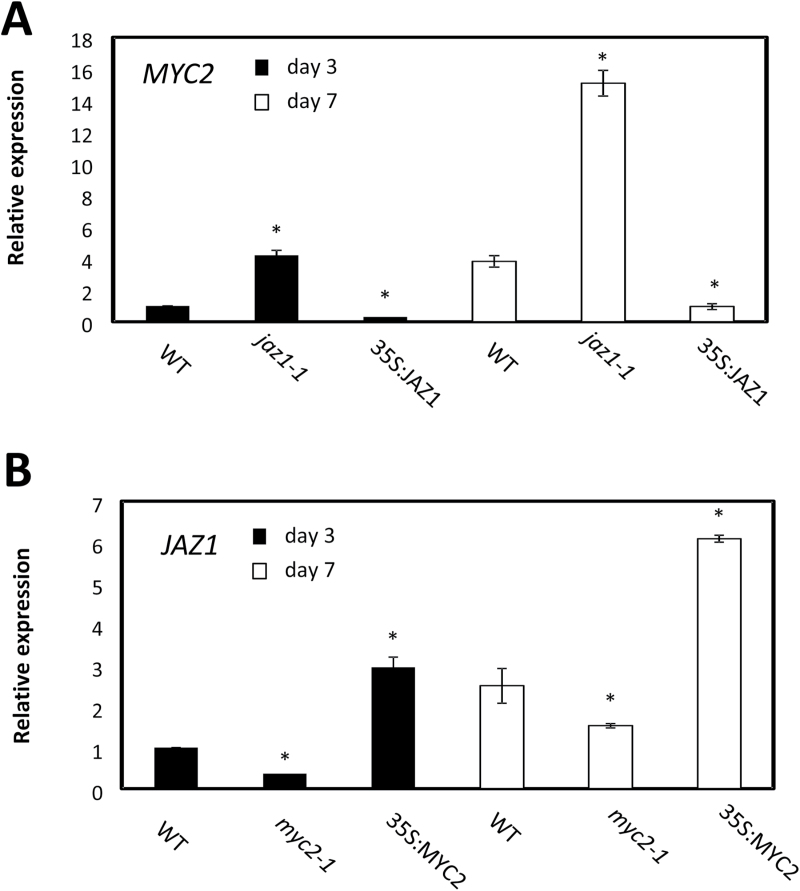
The regulation of JAZ1 and MYC2 during the JA response in plant–pathogen interaction is complicated by feedback loops influencing the expression of both (Chini et al., 2007; Thines et al., 2007; Chung et al., 2008; Kazan and Manners, 2013). To verify the presence of transcriptional regulatory mechanisms operating during somatic embryogenesis, the expression of the two genes was measured in lines suppressing or ectopically expressing MYC2 and JAZ1. On both day 3 and day 7 on induction medium, the expression of MYC2 was induced in the jaz1-1 line while it was repressed in the 35S:JAZ1 line. This pattern was in contrast to that observed for JAZ1 which was down-regulated in the myc2-1 line and up-regulated in the 35S:MYC2 line (Fig. 6).
Fig. 6.
Transcriptional regulation of MYC2 and JAZ1. (A) Expression level by quantitative (q)RT–PCR of MYC2 at days 3 and 7 in the induction medium of somatic embryogenesis in the WT, jaz1-1, and 35S:JAZ1 lines. Values are means ±SE of at least three biological replicates and are normalized to the value of the WT of day 3 set at 1. An asterisk indicates statistically significant differences (P≤0.005) from the WT value of the respective day in culture. (B) Expression level by qRT–PCR of JAZ1 at days 3 and 7 in the induction medium of somatic embryogenesis in the WT, myc2-1, and 35S:MYC2 lines. Values are means ±SE of at least three biological replicates and are normalized to the value of the WT of day 3 set at 1. An asterisk indicates statistically significant differences (P≤0.005) from the WT value of the respective day in culture.
JA response modulates embryogenesis by altering IAA biosynthetic genes
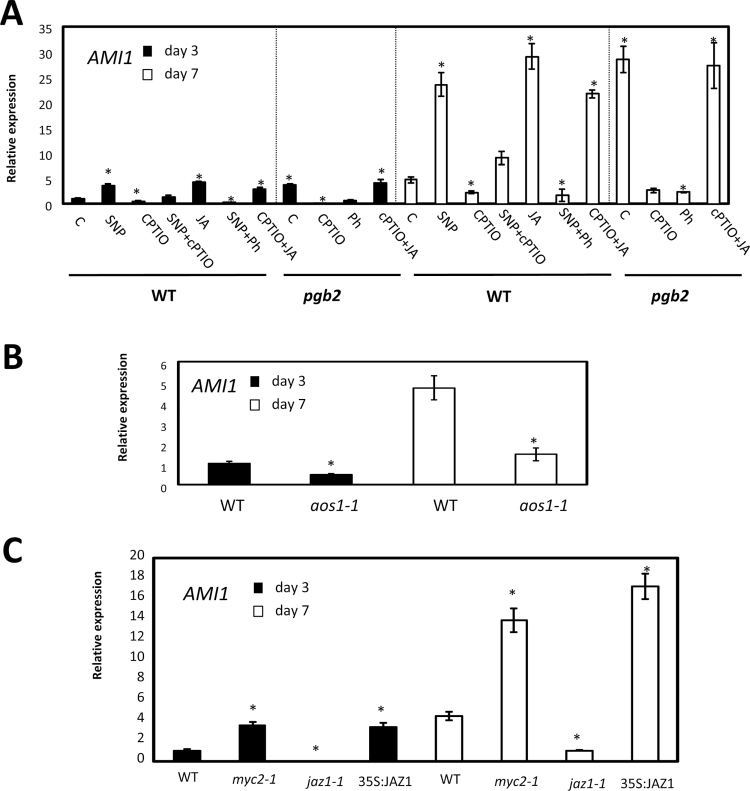
During the initial phases of somatic embryogenesis, auxin is the inductive signal promoting the re-differentiation of the somatic cells and formation of embryogenic tissue (Raghavan, 2004). Previous studies showed that suppression of PGB2 increases auxin levels in a process dependent on the concurrent suppression of MYC2, an inhibitor of IAA synthesis (Elhiti et al., 2013). To expand this model further, we assessed whether IAA biosynthesis was regulated by the JA response during embryogenesis. Production of IAA from indole-3-acetamide is modulated by AMIDASE1 (AMI1) (Pollman et al., 2003). On both days 3 and 7 on induction medium, AMI1 expression was up-regulated by all those treatments increasing NO (SNP or suppression of PGB2) or JA, while it was suppressed by low levels of NO (cPTIO) or JA (Ph) (Fig. 7A). In addition, a depletion of JA in an enriched NO environment (SNP+Ph) decreased the expression of this gene while an enrichment in JA level under low NO conditions (cPTIO+JA) had opposite effects (Fig. 7A). Reintroduction of PGB2 into the pgb2 line reduced AMI1 expression to WT values (Supplementary Fig. S3). The JA requirement for the sustained expression of AMI1 was also confirmed using the JA-deficient aos1-1 line in which AMI1 transcript levels were reduced on both day 3 and day 7 on induction medium (Fig. 7B). To assess further if AMI1 regulation was downstream of the JA response, transcription studies were conducted in lines with altered expression of JAZ1 and MYC2. The transcript levels of AMI1 were induced by suppression of MYC2 (myc2-1 mutant) or ectopic expression of JAZ1 (35S:JAZ1) (Fig. 7C). Besides AMI1, two other IAA biosynthetic genes: ANTHRANILATE SYNTHASE-α SUBUNIT (ASA1), encoding a component of the early enzyme converting chorismate to ribosyl anthralilate, and YUCCA (YUC4) converting indole-3-yl pyruvate to (indol-3-yl)acetate (Cheng et al., 2006; Zhao, 2011), show a similar JA-mediated transcriptional regulation (Supplementary Figs S3, S5, S6). The expression of both genes occurs mainly in the cotyledonary regions of the explant originating the embryogenic tissue (Supplementary Figs S5, S6).
Fig. 7.
Effects of jasmonic acid (JA) and nitric oxide (NO) on AMI1 expression. (A) Expression level by quantitative (q)RT–PCR of AMI1 at days 3 and 7 in the induction medium of somatic embryogenesis. WT and pgb2 tissue was cultured in medium with altered levels of NO and JA using the pharmacological treatments described in Fig. 2. Values are means ±SE of at least three biological replicates and are normalized to the value of the WT (C) of day 3 set at 1. An asterisk indicates statistically significant differences (P<0.005) from the WT (C) value of the respective day in culture. (B) Expression level by qRT–PCR of AMI1 in WT and aos lines at days 3 and 7 on induction medium. Values are means ±SE of at least three biological replicates and are normalized to the value of the WT of day 3 set at 1. An asterisk indicates statistically significant differences (P<0.005) from the WT value of the respective day in culture. (C) Expression level by qRT–PCR of AMI1 at days 3 and 7 in the induction medium of somatic embryogenesis in the WT, myc2-1, jaz1-1, and 35S:JAZ1 lines. Values are means ±SE of at least three biological replicates and are normalized to the value of the WT of day 3 set at 1. An asterisk indicates statistically significant differences (P<0.005) from the WT value of the respective day in culture.
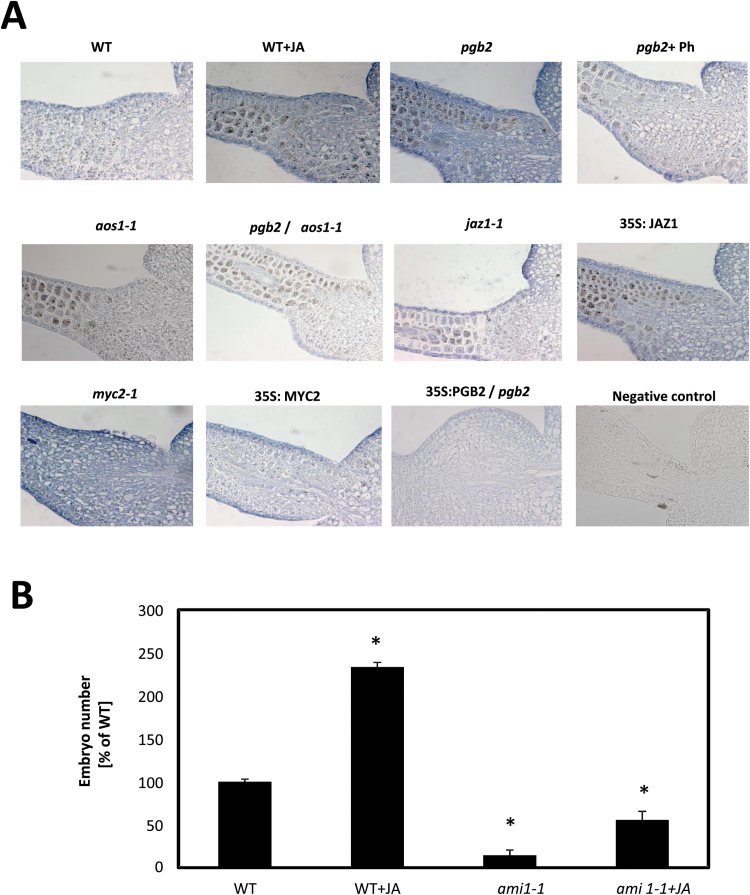
Immunolocalization of the auxin IAA in the embryogenic tissue arising from the cotyledons of the zygotic explants confirmed the localization pattern of the biosynthetic genes (Fig. 8A). Tissues with elevated JA levels (WT+JA or pgb2) or subjected to conditions enhancing the JA response (myc2-1 mutant or 35S:JAZ1) had strong IAA signals. A reduction in IAA level occurred either in tissue depleted in JA level (pgb2 +Ph, aos1-1, pgb2/aos1-1), following conditions repressing the JA response (jaz1-1 and 35S:MYC2), or by the reintroduction of PGB2 in the pgb2 line (Fig. 8A). The requirement for IAA production in the JA regulation of somatic embryogenesis was further verified using the ami1-1 mutant, which significantly limits the beneficial effect of JA on embryo number (Fig. 8B).
Fig. 8.
Auxin and somatic embryogenesis. (A) Immunolocalization of IAA along the cotyledons of explants collected at day 7 on induction medium. Treatments described in Figs 6 and 7 were utilized. Primary antibody was omitted from the negative control section. (B) Number of somatic embryos generated from the WT and the ami1-1 mutant treated with JA. Values are means ±SE of at least three biological replicates. An asterisk indicates statistically significant differences (P≤0.005) from the WT value set at 100%.
Collectively, these data suggest that increased NO levels, resulting either from pharmacological treatments or by suppression of PGB2, increase JA production through the transcription of two key biosynthetic enzymes, LOX2 and AOS, which activate the JA response (Fig. 9). Key events in this response are the induction of JAZ1 and the repression of MYC2 which promote the accumulation of IAA within the embryogenic tissue and the formation of somatic embryos.
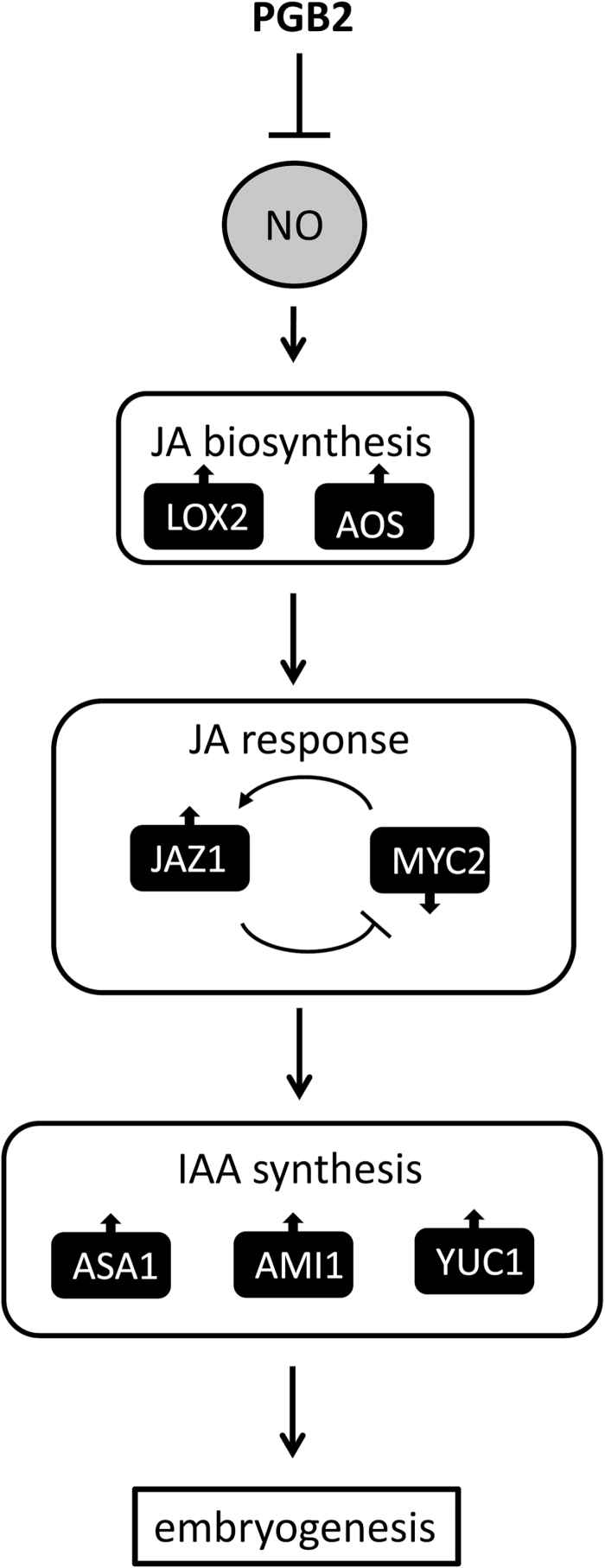
Fig. 9.
Suggested model describing the interaction among PGB2, NO, JA, and IAA during the induction phase of somatic embryogenesis in Arabidopsis.
Discussion
Generation of embryos in vitro is rendered possible by the inherent ability of plant cells to embark on novel developmental programs when subjected to specific changes in environmental conditions. This concept is best exemplified in Arabidopsis, where isolated zygotic embryos cultured on induction medium produce embryogenic tissue from the cotyledons; an event which becomes apparent after 3–7 d. The process is stimulated by the auxin 2,4-D, the inductive signal responsible for the de-differentiation of the cotyledon cells and proliferation of embryogenic cells (Raghavan, 2004). The embryogenic tissue is composed of immature embryos which are allowed to grow further on a development medium devoid of auxin (Fig. 1A). Production of somatic embryos in Arabidopsis is facilitated by the suppression of the phytoglobin PGB2, an effective NO scavenger (Hebelstrup et al., 2008). Explants suppressing PGB2 accumulate NO within the embryogenic tissue and this promotes the formation of somatic embryos (Elhiti et al., 2013). This enhanced embryogenic ability was solely due to the suppression of PGB2, as the reintroduction of PGB2 in the pgb2 line decreased the number of embryos to WT values (Fig. 1C)
In plants, NO participates in diverse physiological functions and operates at the interphase of many hormone-mediated responses (Neill et al., 2003; Wilson et al., 2008; Hancock et al., 2011; Freschi, 2013; Simontacchi et al., 2013). Involved in many developmental responses ranging from leaf expansion, root growth, and senescence (Wendehenne et al., 2001; Yadav et al., 2013), NO is tightly linked to JA in wounding and plant pathogen interaction (Grun et al., 2006; Wang et al., 2013). Having emerged as a downstream modulator in the JA response, NO has also been shown to contribute to JA synthesis (Mur et al., 2012). A rapid induction in JA level following inoculation with B. cinerea was observed in Arabidopsis plants accumulating NO (Hebelstrup et al., 2012). Interactions between NO and JA signaling have only been investigated during post-embryonic growth, with no information related to embryogenesis. Here we show that NO influences the number of somatic embryo produced by modulating the JA level and response, and that these effects can be integrated in the PGB2 regulation of somatic embryogenesis (Elhiti et al., 2013).
Reports on exogenous JA applications during in vitro embryogenesis are scarce and contradictory. While promoting the formation of protocorm bodies, the equivalent of somatic embryos in orchids (Teixeira da Silva, 2012), applications of JA repress callus growth and somatic embryogenesis in Medicago sativa (Kepezynska and Zielinska, 2006). These discrepancies might be ascribed, at least in part, to differences in endogenous JA content, as also observed in our system. Applications of JA (50nM) enhanced embryo production in the WT line, characterized by a low JA signal in the embryogenic tissue, but not in the pgb2 line accumulating high levels of JA and producing a higher number of embryos (Figs 1C, 2). In this latter line, a depletion in JA level by Ph, an effective lipoxygenase inhibitor of JA synthesis (Farmer et al., 1994; Bruinsma et al., 2009), reduces embryo number, and this reduction is not due to the toxic effect of the inhibitor since embryo production is re-established by JA+Ph (Fig. 1C). Therefore, the enhanced embryogenic performance of the pgb2 line is most probably the result of a higher JA content within the embryogenic tissue.
Jasmonic acid mediates the NO and PGB2 regulation of somatic embryogenesis. First, the promotive effect of high NO environments (by SNP or suppression of Pgb2) on embryo formation is abolished by depleting JA with Ph. Consistent with this result, the reduction in embryo number under conditions of low NO (by cPTIO) is fully reversed by applications of JA (Fig. 2A). Secondly, manipulations of the NO content influence JA levels in the embryogenic tissue, with SNP increasing the intensity of the signal and cPTIO reducing it (Fig. 2B). Thirdly, NO induces the expression of two important JA biosynthetic genes, LOX2 and AOS, at both day 3 and day 7 on the induction medium (Fig. 3A), corresponding to the appearance of the embryogenic tissue (arrows in Fig. 1A). Among the 13 LOX genes found in Arabidopsis, LOX2 is the major contributor (~75%) to the total JA produced after wounding (Bell et al., 1995; Schommer et al., 2008). A similar result was also obtained for AOS, which catalyzes the dehydration of the hydroperoxide to an unstable allene oxide in the JA biosynthetic pathway. Suppression of this gene almost completely abolishes the accumulation of JA following wounding (Park et al., 2002; Von Malek et al., 2002).
Unequivocal evidence for the involvement of JA as a downstream component of the NO mediation of somatic embryogenesis is apparent from the behavior of the JA-deficient aos1-1 line. Suppression of AOS reduces embryo production and this effect cannot be reversed by treatments altering NO levels, including SNP which under normal circumstances increases embryo production (Fig. 3B). The identical response observed in the pgb2/aos1-1 double mutant line, showing a reduction in embryogenesis independent of NO levels (Fig. 3C), confirms that JA is an integral mediator of NO generated by the suppression of PGB2.
The majority of JA responses observed during pollen development, wounding, and biotic stresses rely on a complicated interaction of downstream elements including JAZ1 and MYC2 (Kazan and Manners, 2013). In an uninduced state, the JA response is subdued by several repressors which are rapidly polyubiquinated and degraded via the 26S proteasome degradation pathway following JA treatments (Chini et al., 2007). A rise in JA induces MYC2, a bHLH domain-containing transcription factor (Dombrecht et al., 2007), which initiates a transcriptional cascade characterizing the ‘early JA response’. Within this cascade, genes participating in insect defense/wounding responses are up-regulated (Guerineau et al., 2003), while others regulating auxin biosynthesis and responses to pathogens are repressed (Turner et al., 2002; Wasternack and Hause, 2013) (diagram in Supplementary Fig. S7). The induction of MYC2 is transient, lasting only a few hours, as MYC2 also up-regulates its own repressor, JAZ1 (Chini et al., 2007). Suppression of MYC2 by JAZ1 triggers the ‘late JA response’ characterized by the repression of the genes VEGETATIVE STORAGE PROTEINS 1 and 2 (VSP1 and VSP2), induction of the PLANT DEFENSIN 1 (PDF1) gene through the repression of the AP2/ERF transcription factor ORA59 and ETHYLENE RESPONSIVE FACTOR1 (ERF1), and the restoration of auxin synthesis through the de-repression of PLETHORA 1 and 2 (PLT1 and PLT2; Supplementary Fig. S8) (Kazan and Manners, 2013). Therefore, the temporal separation of ‘early’ and ‘late’ JA responses is controlled by the MYC2–JAZ1 feedback regulatory mechanisms (diagram in Supplementary Fig. S7).
Several pieces of evidence presented here suggests that an ‘early’ and a ‘late’ JA response occurs during somatic embryogenesis (as denoted by the biphasic profile of MYC2 expression, and the late increase in JAZ1 expression following JA treatments; Supplementary Fig. S4). Furthermore, the increased number of embryos produced by the extended applications of JA in the induction medium is possibly the consequence of the ‘late’ response characterized by the repression of MYC2 (Fig. 5; Supplementary Fig. S4), the increase in JAZ1 expression (Fig. 4; Supplementary Fig. S4), and the characteristic expression profile of genes involved in insect defense, pathogen response, and auxin synthesis (Supplementary Fig. S7). Within the ‘late’ response, suppression of MYC2 and up-regulation of JAZ1, possibly through a feedback mechanism (with MYC2 inducing JAZ1 and JAZ1 repressing MYC2) (Fig. 6), are essential for enhanced embryogenic output. Suppression of MYC2 directly via JA or indirectly via NO (by SNP treatments or suppression of PGB2) (Fig. 5A) increases the number of Arabidopsis somatic embryos (Fig. 5C). Consistent with this behavior, treatments reducing JA levels (by cPTIO or by suppression of AOS) up-regulate MYC2 (Fig. 5B) and compromise embryogenesis. The inefficacy of NO and JA manipulations to reduce embryo yield in a myc2-1 background confirms MYC2 as a downstream component of the NO and JA response (Fig. 5C).
Unlike MYC2, a direct or indirect rise in JA (by SNP or suppression of PGB2) induces JAZ1 especially within the cotyledons of the explants producing embryogenic cells (Fig. 4B) and favors the production of somatic embryos (Fig. 4D). The expression of JAZ1 is repressed in environments depleted in JA (by cPTIO or by suppression of AOS), a condition inhibiting embryogenesis (Fig. 4A, C). As also observed for MYC2, NO and JA treatments have no effects on the embryogenic performance of the jaz1-1 line (Fig. 4D), thus placing JAZ1 as downstream of NO and JA.
The beneficial effects of MYC2 suppression and JAZ1 induction on somatic embryogenesis are linked to a rise in the level of auxin, the inductive signal triggering the de-differentiation of the cotyledon cells and the production of embryogenic tissue (Raghavan, 2004). Two key transcription factors linked to auxin production, PLETHORA 1 and 2 (Pinon et al., 2013), as well as several auxin biosynthetic genes including ASAI, converting chorismate to ribosyl anthranilate during the early steps of IAA synthesis, YUC4, converting indole-3-oyl pyruvate to (indol-3-oyl)acetate, and AMI1, producing IAA from indole-3-acetamide (Pollman et al. 2003), are induced in NO- and JA-enriched environments and repressed by conditions depleting NO and JA (Fig. 7; Supplementary Figs S5, S6, S8). The observation that their expression is increased by JA (regardless of the levels of NO, suppression of MYC2, or induction of JAZ1) places these genes downstream of the NO response mediated by JA. While the involvement of MYC2 in the regulation of these genes during embryogenesis confirms previous studies (Elhiti et al., 2013), that of JA and JAZ1 is novel and opens up new avenues for improving propagation methods in recalcitrant species. The embryogenic tissue-specific localization pattern of both ASA1 and YUC4 (Supplementary Figs S5, S6) suggests that regulation of these genes might be specific to the acquisition of embryogenic competence of the cotyledon cells, and might be related to the high levels of IAA observed in these areas (Fig. 8).
Collectively, these data suggest that mechanisms modulating late JA responses post-embryonically are integrated in the PGB2 and NO regulation of in vitro embryogenesis. In the proposed model (Fig. 9), a rise in NO due to suppression of PGB2 (or pharmacological treatments) induces the expression of the JA biosynthetic genes LOX2 and AOS and elevates JA along the cotyledons of the explants generating embryogenic tissue. JA evokes a response suppressing MYC2 and inducing JAZ1. This response, possibly reinforced by transcriptional feedback mechanisms between the two genes, up-regulates several auxin biosynthetic genes, leading to an increase in auxin levels. Auxin is required for the formation and proliferation of the embryogenic tissue, and ultimately for the successful production of the somatic embryos.
Supplementary data
Supplementary data are available at JXB online.
Figure S1. Characterization of the pgb2/aos1-1 double mutant lines.
Figure S2. Characterization of the 35S:PGB2/pgb2 line
Figure S3. Expression of jamsonic acid- and auxin-related genes in the 35S:PGB2/pgb2 line
Figure S4. Effects of JA applications on myc2-1 and jaz1-1.
Figure S5. Jasmonic acid (JA) and nitric oxide (NO) affect ASA1 expression and localization.
Figure S6. Jasmonic acid (JA) and nitric oxide (NO) affect YUC4 expression and localization.
Figure S7. Diagram showing the early and late JA responses.
Figure S8. Expression levels of PLETHORA 1 and 2 in day 7 explants.
Table S1. Primer list.
Acknowledgements
This work was supported by NSERC Discovery Grants to RDH and CS, and by a Fellowship to MM granted by Egypt’s Ministry of Higher Education and Research. The authors thank Mr Durnin for technical support and editing of the manuscript. The following people are also acknowledged for providing seed material: Dr Kim Hebelstrup, University of Aarhus, Denmark (pgb2 line); Professor Kazan, Commonwealth Scientific and Industrial Research Organization Plant Industry, Queensland, Australia (myc2 and 35S:MYC2 lines); Professor John Browse, Institute of Biological Chemistry, Washington State University, USA (35S:JAZ1 line); Professor Catherine Bellini, Umea University (pJAZ1:GUS-GFP line); Professor Chuanyou Li, Institute of Genetics and Developmental Biology, Chinese Academy of Science, China (35S:MYC2-GUS); Professor Eva Sundberg, Department of Plant Biology and Forest Genetics, Swedish University of Agriculture Science (pYUC4:GUS); and Professor Corné M.J. Pieterse, Department of Plant Microbe Interactions, Utrecht University (pPDF1.2:GUS). Professor House, Leibniz-Institut fur Pflanzenbiochemie, Germany, is also acknowledged for the anti-JA antibodies.
References
- Bassuner BM, Lam R, Lukowitz W, Yeung EC. 2007. Auxin and root initiation in somatic embryos of Arabidopsis. Plant Cell Reports 26, 1–11. [DOI] [PubMed] [Google Scholar]
- Bell E, Creelman RA, Mullet JE. 1995. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proceedings of the National Academy of Sciences, USA 92, 8675–8679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruinsma M, van Broekhoven S, Poelman EH, Posthumus MA, Mueller MJ, van Loon JJ, Dicke M. 2009. Inhibition of lipoxygenase affects induction of both direct and indirect defences against herbivorous insects. Oecologia 162, 393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Sun J, Zhai Q. 2011. The basic helix–loop–helix transcription factor MYC2 directly represses PLETHORA expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. The Plant Cell 23, 3335–3352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y. 2006. Auxin biosynthesis by the YUCCA flavin monooxygenases controls the formation of floral organs and vascular tissues in Arabidopsis. Genes and Development 20, 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernandez G, et al. 2007. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. [DOI] [PubMed] [Google Scholar]
- Chung HS, Koo AJ, Gao X, Jayanty S, Thines B, Jones AD, Howe GA. 2008. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiology 146, 952–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible WR. 2005. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiology 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demianski A, Chung K, Kunkel B. 2012. Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Molecular Plant Pathology 13, 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, et al. 2007. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. The Plant Cell 19, 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dordas C, Hasinoff BB, Igamberdiev AU, Manach N, Rivoal J, Hill RD. 2003. Expression of a stress-induced hemoglobin affects NO levels produced by alfalfa root cultures under hypoxic stress. The Plant Journal 35, 763–770. [DOI] [PubMed] [Google Scholar]
- Dordas C. 2009. Nonsymbiotic hemoglobins and stress tolerance in plants. Plant Science 176, 433–440. [DOI] [PubMed] [Google Scholar]
- Eklund M, Cierlik I, Staldal V, Claes A, Vestman D, Chandler J, Sundberg E. 2011. Expression of Arabidopsis SHORT INTERNODES/ STYLISH family genes in auxin biosynthesis zones of aerial organs is dependent on a GCC box-like regulatory element. Plant Physiology 157, 2069–2080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhiti M, Tahir M, Gulden RH, Khamiss K, Stasolla C. 2010. Modulation of embryo-forming capacity in culture through the expression of Brassica genes involved in the regulation of the shoot apical meristem. Journal of Experimental Botany 61, 4069–4085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elhiti M, Hebelstrup KH, Wang A, Li C, Cui Y, Hill RD, Stasolla C. 2013. Function of type-2 Arabidopsis hemoglobin in the auxin-mediated formation of embryogenic cells during morphogenesis. The Plant Journal 74, 946–958. [DOI] [PubMed] [Google Scholar]
- Farmer EE, Caldelari D, Pearce G, Walker-Simmons MK, Ryan CA. 1994. Diethyldithiocarbamic acid inhibits the octadecanoid signaling pathway for the wound induction of proteinase inhibitors in tomato leaves. Plant Physiology 106, 337–342. [Google Scholar]
- Freschi L. 2013. Nitric oxide and phytohormone interactions: current status and perspectives. Frontiers in Plant Science 4, 398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrocho-Villegas V, Gopalasubramaniam SK, Arredondo-Peter R. 2007. Plant hemoglobins: what we know six decades after their discovery. Gene 398, 78–85. [DOI] [PubMed] [Google Scholar]
- Grun S, Lindermayr C, Sell S, Durner J. 2006. Nitric oxide and gene regulation in plants. Journal of Expermental Botany 57, 507–516. [DOI] [PubMed] [Google Scholar]
- Guerineau F, Benjdia M, Zhou DX. 2003. A jasmonate-responsive element within the A. thaliana vsp1 promoter. Journal of Experimental Botany 54, 1153–1162. [DOI] [PubMed] [Google Scholar]
- Gutierrez L, Mongelard G, Floková K, et al. 2012. Auxin controls Arabidopsis adventitious root initiation by regulating jasmonic acid homeostasis. The Plant Cell 24, 2515–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock JT, Neill SJ, Wilson ID. 2011. Nitric oxide and ABA in the control of plant function. Plant Science 181, 555–559. [DOI] [PubMed] [Google Scholar]
- Hebelstrup KH, Hunt P, Dennis E, Jensen SB, Jensen EO. 2006. Hemoglobin is essential for normal growth of Arabidopsis organs. Physiologia Plantarum 127, 157–166. [Google Scholar]
- Hebelstrup KH, Jensen EO. 2008. Expression of NO scavenging hemoglobin is involved in the timing of bolting in Arabidopsis thaliana . Planta 227, 917–927. [DOI] [PubMed] [Google Scholar]
- Hebelstrup KH, Ostergaard-Ensen E, Hill RD. 2008. Bioimaging techniques for subcellular localization of plant hemoglobins and measurement of hemoglobin-dependent nitric oxide scavenging in planta. Methods in Enzymology 437, 595–604. [DOI] [PubMed] [Google Scholar]
- Hebelstrup KH, van Zanten M, Mandon J, Voesenek LA, Harren FJ, Cristescu SM, Moller IM, Mur LA. 2012. Hemoglobin modulates NO emission and hyponasty under hypoxia-related stress in Arabidopsis thaliana . Journal of Experimental Botany 63, 5581–5591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebelstrup KH, Jay KS, Simpson C, Schjoerring JK, Mandon J, Cristescu SM, Harren FJ, Christiansen MW, Mur LA, Igamberdiev AU. 2013. An assessment of the biotechnological use of hemoglobin modulation in cereals. Physiologia Plantarum 150, 593–603. [DOI] [PubMed] [Google Scholar]
- Hendriks T, Scheer I, Quillet MC, Randoux B, Delbreil B, Vasseur J, Hilbert JL. 1998. A nonsymbiotic hemoglobin gene is expressed during somatic embryogenesis in Cichorium. Biochimica et Biophysica Acta 1443, 193–197. [DOI] [PubMed] [Google Scholar]
- Hill RD. 1998. What are hemoglobins doing in plants? Canadian Journal of Botany 76, 707–712. [Google Scholar]
- Hill RD. 2012. Non-symbiotic haemoglobins – what’s happening beyond nitric oxide scavenging? AoB Plants 2012, pls004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SM, Bahn SC, Lyu A, Jung HS, Ahn JH. 2010. Identification and testing of superior reference genes for a starting pool of transcript normalization in Arabidopsis. Plant and Cell Physiology 51, 1694–1606. [DOI] [PubMed] [Google Scholar]
- Hoy JA, Hargrove MS. 2008. The structure and function of plant hemoglobins. Plant Physiology and Biochemistry 46, 371–379. [DOI] [PubMed] [Google Scholar]
- Hunt PW, Klok EJ, Trevaskis B, Watts RA, Ellis MH, Peacock WJ, Dennis ES. 2002. Increased level of hemoglobin 1 enhances survival of hypoxic stress and promotes early growth in Arabidopsis thaliana. Proceedings of the National Academy of Sciences, USA 99, 17197–17202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt PW, Watts RA, Trevaskis B, Llewelyn DJ, Burnell J, Dennis ES, Peacock WJ. 2001. Expression and evolution of functionally distinct haemoglobin genes in plants. Plant Molecular Biology 47, 677–692. [DOI] [PubMed] [Google Scholar]
- Jayabalan N, Anthony P, Davey MR, Power JB, Lowe KC. 2004. Hemoglobin promotes somatic embryogenesis in peanut cultivars. Artificial Cells, Blood Substitutes, and Immobilization Biotechnology 32, 147–157. [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM. 2013. MYC2: the master in action. Molecular Plant 6, 686–703. [DOI] [PubMed] [Google Scholar]
- Kepezynska E, Zielinska J. 2006. Regulation of Medicago sativa L. somatic embryos regeneration by gibberellin A3 and abscisic acid in relation to starch content and α-amylase activity. Plant Growth Regulation 49, 209–217. [Google Scholar]
- Koorneef A, Verhage A, Leon-Reyes A, Snetselaar R, Van Loon L, Pieterse C. 2008. Towards a reporter system to identify regulators of cross-talk between salicylate and jasmonate signaling pathways in Arabidopsis. Plant Signaling and Behavior 3, 543–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta DeltaC(T)) Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Solano R. 2005. Molecular players regulating the jasmonate signalling network. Current Opinion in Plant Biology 8, 532–540. [DOI] [PubMed] [Google Scholar]
- Mielke K, Forner S, Kramell R, Conrad U, Hause B. 2011. Cell-specific visualization of jasmonates in wounded tomato and Arabidopsis leaves using jasmonate-specific antibodies. New Phytologist 190, 1069–1080. [DOI] [PubMed] [Google Scholar]
- Mur LA, Sivakumaran A, Mandon J, Cristescu SM, Harren FJ, Hebelstrup KH. 2013. Hemoglobin modulates salicylate and jasmonate/ethylene-mediated resistance mechanisms against pathogens. Journal of Expermental Botany 63, 4375–4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F. 1962. A revised medium for rapid growth and bio-assays with tobacco tissue cultures. Physiologia Plantarum 15, 473–497. [Google Scholar]
- Neill SJ, Desikan R, Hancock JT. 2003. Nitric oxide signalling in plants. New Phytologist 159, 11–35. [DOI] [PubMed] [Google Scholar]
- Niu Y, Figueroa P, Browse J. 2010. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. Journal of Experimental Botany 62, 2143–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozco-Cardenas ML, Ryan CA. 2002. Nitric oxide negatively modulates wound signaling in tomato plants. Plant Physiology 130, 487–493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R. 2002. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. The Plant Journal 31, 1–12. [DOI] [PubMed] [Google Scholar]
- Perazzolli M, Dominici P, Romero-Puertas MC, Zago E, Zeier J, Sonoda M, Lamb C, Delledonne M. 2004. Arabidopsis nonsymbiotic hemoglobin AHb1 modulates nitric oxide bioactivity. The Plant Cell 16, 2785–2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinon V, Prasad K, Grigg SP, Sanchez-Perez GF, Scheres B. 2013. Local auxin biosynthesis regulation by PLETHORA transcription factors controls phyllotaxis in Arabidopsis. Proceedings of the National Academy of Sciences, USA 110, 1107–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollmann S, Neu D, Weiler EW. 2003. Molecular cloning and characterization of an amidase from Arabidopsis thaliana capable of converting indole-3-acetamide into the plant growth hormone, indole-3-acetic acid. Phytochemistry 62, 293–300. [DOI] [PubMed] [Google Scholar]
- Raghavan V. 2004. Role of 2,4-dichlorophenoxyacetic acid (2,4-D) in somatic embryogenesis on cultured zygotic embryos of Arabidopsis: cell expansion, cell cycling, and morphogenesis during continuous exposure of embryos to 2,4-D. American Journal of Botany 91, 1743–1756. [DOI] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y, Jikumaru Y, Obayashi T, Saito H, Masuda S, Kamiya Y, Ohta H, Shirasu K. 2013. Basic helix–loop–helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiology 163, 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schommer C, Palatnik JF, Aggarwal P, Chetelat A, Cubas P, Farmer EE, Nath U, Weigel D. 2008. Control of jasmonate biosynthesis and senescence by miR319 targets. PLoS Biology 6, 1991–2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sieburth LE, Meyerowitz EM. 1997. Molecular dissection of the AGAMOUS control region shows that cis elements for spatial regulation are located intragenically. The Plant Cell 9, 355–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simontacchi M, Garcia-Mata C, Bartoli CG, Santa-Maria GE, Lamattina L. 2013. Nitric oxide as a key component in hormone-regulated processes. Plant Cell Reports 32, 853–866. [DOI] [PubMed] [Google Scholar]
- Smagghe BJ, Blervacq AS, Blassiau C, Decottignies JP, Jacquot JP, Hargrove MS, Hilbert JL. 2007. Immunolocalization of non-symbiotic hemoglobins during somatic embryogenesis in chicory. Plant Signaling and Behavior 2, 43–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagghe BJ, Hoy JA, Percifield R. 2009. Review: correlations between oxygen affinity and sequence classifications of plant hemoglobins. Biopolymers 91, 1083–1096. [DOI] [PubMed] [Google Scholar]
- Teixeira da Silva JA. 2012. Jasmonic acid, but not salicylic acid, improves PLB formation of hybrid cymbidium. Plant Tissue Culture and Biotechnology 22, 187–192. [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. 2007. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. [DOI] [PubMed] [Google Scholar]
- Trevaskis B, Watts RA, Andersson C, Llewellyn D, Hargrove MS, Olson JS, Dennis ES, Peacock WJ. 1997. Two hemoglobin genes in Arabidopsis thaliana: the evolutionary origins of leghemoglobins. Proceedings of the National Academy of Sciences, USA 94, 12230–12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JG, Ellis C, Devoto A. 2002. The jasmonate signal pathway. The Plant Cell 14, S153–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanholme B, Grunewald W, Bateman A, Kohchi T, Gheysen G. 2007. The tify family previously known as ZIM. Trends in Plant Science 12, 239–244. [DOI] [PubMed] [Google Scholar]
- Vigeolas H, Huhn D, Geigenberger P. 2011. Non-symbiotic hemoglobin-2 leads to an elevated energy state and to a combined increase in polyunsaturated fatty acids and total oil content when over-expressed in developing seeds of transgenic Arabidopsis plants. Plant Physiology 155, 1435–1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Malek B, Graaff E, Schneitz K, Keller B. 2002. The Arabidopsis male-sterile mutant dde2-2 is defective in the ALLENE OXIDE SYNTHASE gene encoding one of the key enzymes of the jasmonic acid biosynthesis pathway. Planta 216, 187–192. [DOI] [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM. 2003. Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiology 132, 556–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Elhiti M, Hebelstrup KH, Hill RD, Stasolla C. 2011. Manipulation of hemoglobin expression affects Arabidopsis shoot organogenesis. Plant Physiology and Biochemistry 49, 1108–1116. [DOI] [PubMed] [Google Scholar]
- Wang Y, Loake GJ, Chu C. 2013. Cross-talk of nitric oxide and reactive oxygen species in plant programmed cell death. Plant Science 4, 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B. 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts RA, Hunt PW, Hvitved AN, Hargrove MS, Peacock WJ, Dennis ES. 2001. A hemoglobin from plants homologous to truncated hemoglobins of microorganisms. Proceedings of the National Academy of Sciences, USA 98, 10119–10124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendehenne D, Pugin A, Klessig DF, Durner J. 2001. Nitric oxide: comparative synthesis and signaling in animal and plant cells. Trends in Plant Science 6, 177–183. [DOI] [PubMed] [Google Scholar]
- Wilson ID, Neill SJ, Hancock JT. 2008. Nitric oxide synthesis and signalling in plants. Plant, Cell and Environment 31, 622–631. [DOI] [PubMed] [Google Scholar]
- Yadav S, David A, Baluska F, Bhatla SC. 2013. Rapid auxin-induced nitric oxide accumulation and subsequent tyrosine nitration of proteins during adventitious root formation in sunflower hypocotyls. Plant Signaling and Behavior 8, e23196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y. 2011. Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Molecular Plant 5, 334–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zar JH. 1999. Biostatistical analysis , 4th edn. Englewood Cliffs, NJ: Prentice-Hall, 208–228. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.