Highlight
We demonstrate that 12-oxo-phytodienoic acid acts together with ABA in a complex regulatory network to promote seed dormancy and repress germination in Arabidopsis.
Key words: Abscisic acid, dormancy, germination, MFT, OPDA, RGL2.
Abstract
We previously demonstrated that the oxylipin 12-oxo-phytodienoic acid (OPDA) acts along with abscisic acid to regulate seed germination in Arabidopsis thaliana, but the mechanistic details of this synergistic interaction remain to be elucidated. Here, we show that OPDA acts through the germination inhibition effects of abscisic acid, the abscisic acid-sensing ABI5 protein, and the gibberellin-sensing RGL2 DELLA protein. We further demonstrate that OPDA also acts through another dormancy-promoting factor, MOTHER-OF-FT-AND-TFL1 (MFT). Both abscisic acid and MFT positively feed back into the OPDA pathway by promoting its accumulation. These results confirm the central role of OPDA in regulating seed dormancy and germination in A. thaliana and underline the complexity of interactions between OPDA and other dormancy-promoting factors such as abscisic acid, RGL2, and MFT.
Introduction
Seed dormancy prevents germination until conditions are favourable for plant growth. In Arabidopsis thaliana (Arabidopsis), dormancy can be released by exposing imbibed seeds to low temperatures (i.e. stratification) or by an extended period of dry seed storage (i.e. after-ripening). Plant hormones are chemically diverse compounds, which occur in plants at low concentrations and regulate plant growth, development, and responses to external stimuli (Davies, 2010). Among these, the two hormones that are of the utmost importance in seed dormancy and germination regulation are abscisic acid (ABA) and gibberelic acid (GA). ABA plays a key role in induction of primary dormancy during Arabidopsis seed development and acts as a germination repressor, while GA promotes germination and the alleviation of dormancy (Finkelstein et al., 2008; Graeber et al., 2012; Holdsworth et al., 2008). Both biosynthesis and catabolism determine the final amount of ABA and GA in the seed, and the interactions between these two antagonistic hormones, at the metabolism and signalling levels, are crucial in the regulation of germination.
The transcription factors ABI3, ABI4, and ABI5 are important components of the ABA signal transduction pathway involved in germination regulation (Clerkx et al., 2003; Finkelstein, 1994; Finkelstein and Lynch, 2000; Finkelstein et al., 1998; Lopez-Molina and Chua, 2000; Lopez-Molina et al., 2002; Xu et al., 2014). ABI5 is a basic leucine-zipper (bZIP) transcription factor (Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000); in Arabidopsis seeds it is expressed in the embryo and micropylar endosperm (Penfield et al., 2006a). ABA causes an increase in ABI5 transcript levels and protein abundance, and also activates ABI5 via phosphorylation (Lopez-Molina et al., 2001; Piskurewicz et al., 2008). Integral to GA signal transduction are the growth-repressing DELLA proteins. GA binds to the receptor GA INSENSITIVE DWARF1 (GID1) and enhances GID1-DELLA interaction, leading to DELLA degradation by the ubiquitin-proteasome pathway (Murase et al., 2008; Sun, 2010). DELLA proteins repress germination, and of the five DELLA genes in Arabidopsis, RGA-LIKE2 (RGL2) is the major DELLA involved in seed germination (Cao et al., 2005; Lee et al., 2002; Penfield et al., 2006b; Piskurewicz and Lopez-Molina, 2009; Piskurewicz et al., 2008; Tyler et al., 2004). RGL2 accumulation can also lead to an increase in ABA levels via regulation of the RING-zinc finger protein-encoding gene XERICO, which has been shown to promote accumulation of ABA in an as yet unknown manner (Ko et al., 2006; Piskurewicz et al., 2008; Zentella et al., 2007).
FLOWERING-LOCUS-T (FT), TERMINAL-FLOWER1 (TFL1), and MOTHER-OF-FT-AND-TFL1 (MFT) belong to the phosphatidyl ethanolamine-binding protein family of proteins in Arabidopsis. While FT and TFL1 are involved in regulation of flowering time control, MFT has been shown to be a regulator of germination and dormancy in different species (Li et al., 2014; Nakamura et al., 2011; Vaistij et al., 2013; Xi and Yu, 2010; Xi et al., 2010). Data reported by Vaistij et al. (2013) and Nakamura et al. (2011) indicate that MFT promotes dormancy, and this is consistent with the fact that MFT expression is promoted by RGL2 and ABI5 in Arabidopsis (Xi et al., 2010). However, MFT has also been found to negatively regulate ABA signalling (Xi et al., 2010).
Jasmonic acid (JA) and its precursor 12-oxo-phytodienoic acid (OPDA) are oxylipins derived from α-linolenic acid (Wasternack and Hause, 2013). JA regulates biotic and abiotic stress responses, as well as plant growth and developmental processes (Balbi and Devoto, 2008; Santino et al., 2013; Wasternack, 2014; Wasternack et al., 2013). OPDA has also been shown to possess distinct, JA-independent, signalling properties in different species, and its involvement has been reported in biotic and abiotic stress responses, embryo development, and seed germination (Bosch et al., 2014; Dave and Graham, 2012; Dave et al., 2011; Goetz et al., 2012; Guo et al., 2014; Park et al., 2013; Ribot et al., 2008; Satoh et al., 2014; Savchenko and Dehesh, 2014; Savchenko et al., 2014; Taki et al., 2005). A role for OPDA in repressing germination in Arabidopsis was proposed based on analysis of comatose (cts) seeds disrupted in the activity of the ATP binding cassette (ABC) transporter CTS, also known as PXA1 or PED3 (Footitt et al., 2002; Hayashi et al., 2002; Russell et al., 2000; Zolman et al., 2001). CTS is involved in the transport of OPDA into the peroxisome, where JA biosynthesis occurs. cts-2/pxa1-1 seeds accumulate high levels of OPDA, leading to the low germination rates observed for this mutant (Dave et al., 2011). Moreover, exogenous OPDA leads to an increase in ABI5 protein abundance and inhibition of the germination of wild-type (WT) seeds (Dave et al., 2011).
Here, we show that ABA, ABI5, RGL2, and MFT are key components of the OPDA pathway that represses germination. We also show that there is a complex network of feedback interactions by which these components downstream of OPDA affect OPDA levels in seeds.
Materials and methods
Plant and growth conditions
Arabidopsis thaliana WT and mutant plants used were in the Columbia-0 (Col), Landsberg erecta (Ler) and Wassilewskija (Ws) ecotypes as stated. The mutants used in this study have been described previously: pxa1-1 (Zolman et al., 2001), cts-2 (Footitt et al., 2002), aos (Park et al., 2002), opr3-1 (Stintzi and Browse, 2000), rgl2-1 (Lee et al., 2002), aba1-1 (Koornneef et al., 1982), mft-2 (Xi et al., 2010), and ga1-3 (Sun and Kamiya, 1994). Plants were grown in controlled environment growth cabinets at 22 °C with 16h white light at 80–100 µmol m−2 s−1 light intensity. Seeds were harvested from brown siliques that had begun to dehisce, and were size-sieved using a 250 µm mesh sieve. Experiments performed with freshly harvested seeds were conducted within 24h of harvest. For the experiments with after-ripened seeds, freshly harvested seeds were dry stored for 6–8 weeks. For stratification, seeds were imbibed on water agar plates and stratified in the dark at 4 °C for 3 days before being placed in growth cabinets.
Seed germination assays
Germination assays were performed on 0.9% (w/v) water agar plates, with 50–100 sterilized seeds, and plates were placed in a controlled environment growth cabinet with continuous light (150 µmol m−2 s−1) at 20 °C. In experiments where germination assays were conducted with OPDA (Larodan), ABA (Sigma), Norflurazon (Sigma), and Paclobutrazol (Sigma), the appropriate amounts of these compounds were included in the water agar medium. Germination was scored as radicle emergence after 7 days of incubation.
Hormone and oxylipin analysis
ABA, GA, OPDA, and JA-isoleucine (JA-Ile) conjugate were extracted and quantified from dry seed (80–100mg dry weight) and imbibed seed (300–400mg fresh weight) according to the protocol described for GA and ABA quantification (Dave et al., 2011), with d2-GA4, d6-ABA and Prostaglandin A1 included as internal standards for GA4, ABA, and OPDA/JA-Ile, respectively.
Gene expression analysis
RNA extractions from developing seeds and imbibed seeds, cDNA synthesis, and quantitative PCR analysis were performed according to methods described previously (Penfield et al., 2005; Vaistij et al., 2013). cDNA amplifications were normalized to ACTIN2 and relative gene expression was calculated as a ratio between the value of each sample and their respective controls. The primers used are described in Supplementary Table 1 at JXB online.
Statistical analysis
In all Figures the data represent the mean of three biological replicates±standard deviation. Differences between controls and samples stated in the text are statistically significant as determined by Student’s t-test (P≤0.05).
Results and discussion
Seed dormancy is affected in OPDA biosynthetic and catabolic mutants
We demonstrated previously that Arabidopsis pxa1-1/cts-2 mutant seeds are highly dormant due to an over-accumulation of OPDA (Dave et al., 2011). We also showed that seeds of the aos mutant (disrupted in an early step in the oxylipin pathway prior to OPDA; Park et al., 2002) and of the opr3-1 mutant (unable to convert OPDA into JA; Stintzi and Browse, 2000) have lower and higher levels of OPDA, respectively, than WT seeds (Dave et al., 2011). Based on these observations, we hypothesized that seeds of these mutants should also have altered dormancy levels correlating with their OPDA levels in seeds. To test this hypothesis, germination levels of freshly harvested seeds were determined. We found that aos seeds, which are deficient in OPDA accumulation, indeed germinated faster and at a higher frequency than WT (Col) seeds (Fig. 1). In contrast, opr3-1 seeds, which contain more OPDA, germinated at a lower frequency than their respective WT (Ws) seeds (Fig. 1). These observations further confirm that levels of dormancy positively correlate with OPDA accumulation in freshly harvested seeds.
Fig. 1.
Seeds affected in OPDA metabolism have altered dormancy. Germination profiles of WT Col and Ws, aos (in Col background), and opr3-1 (in Ws background) freshly harvested, unstratified seeds from 1 to 7 days after imbibition (1D–7D) on water agar plates under constant light. In this and all other Figures, error bars represent the standard deviation of three independent measurements.
OPDA acts through the ABA pathway
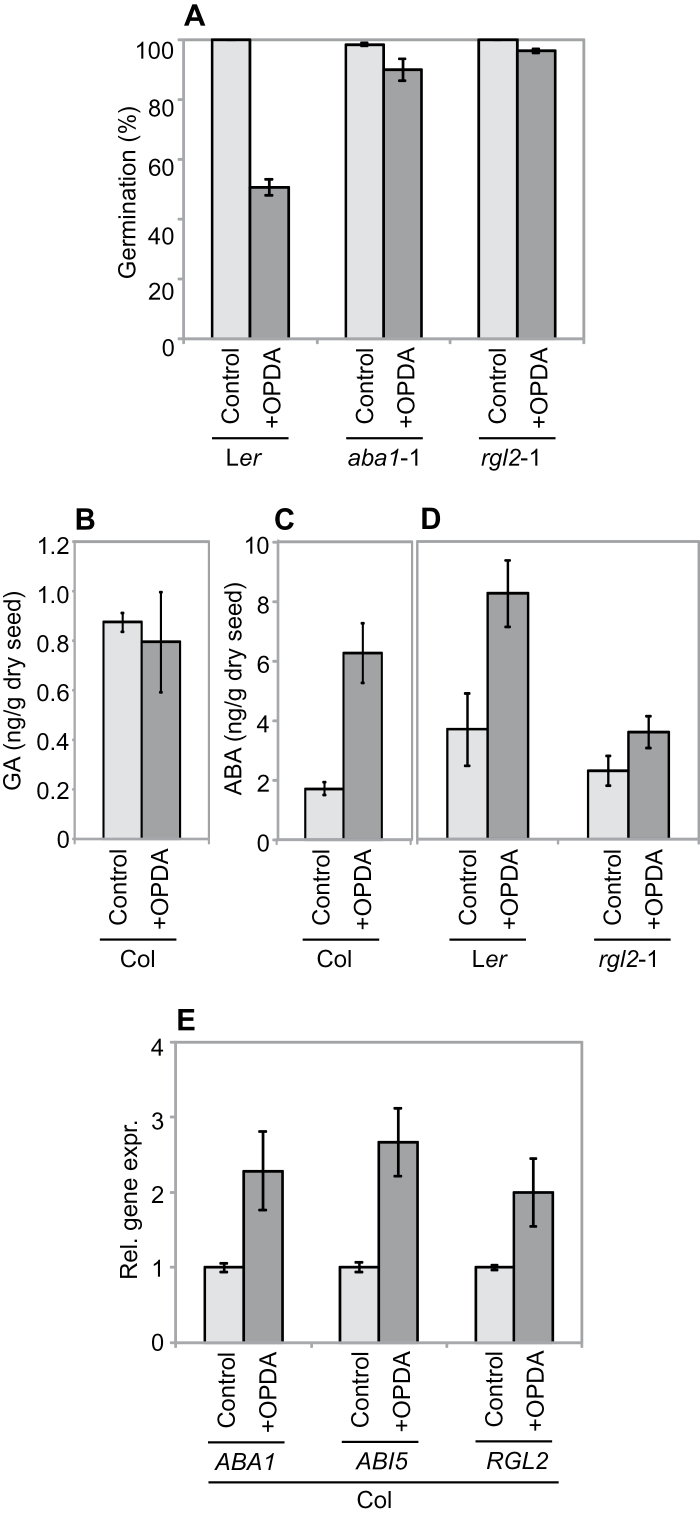
We showed in our previous work that exogenously applied OPDA and ABA synergistically inhibit germination of after-ripened seeds (Dave et al., 2011). Here, we extended our analyses by performing germination assays using seeds of the ABA-biosynthesis mutant aba1-1 and the GA-signalling mutant rgl2-1. As shown for Col and Ws (Dave et al., 2011), germination of after-ripened Ler seeds was inhibited by exogenously applied OPDA (Fig. 2A). However, OPDA was almost ineffective when germination assays were performed with aba1-1 and rgl2-1 seeds (Fig. 2A). The ABA-dependent effect of OPDA was also observed when WT (Col) seeds were treated jointly with OPDA and the indirect ABA biosynthesis inhibitor Norflurazon, to mimic the aba1-1 mutation (Supplementary Fig. S1). We also measured GA and ABA accumulation in OPDA-treated seeds and found that while GA levels were unchanged (Fig. 2B), ABA accumulation was increased in both Col and Ler WT seeds (Fig. 2C, D). However, the ABA increase was strongly reduced in rgl2-1 mutant seeds (Fig. 2D). This is in accordance with previous observations showing that RGL2 indirectly promotes ABA biosynthesis (Lee et al., 2010). Taken together, these observations demonstrate that exogenously applied OPDA triggers ABA accumulation.
Fig. 2.
The ABA biosynthetic pathway is required for OPDA-driven inhibition of germination. (A) Germination profiles [7 days after imbibition (DAI)] of after-ripened WT (Ler), aba1-1 and rgl2-1 seeds on water agar (Control) plates and water agar supplemented with 10 µM OPDA (+OPDA). (B–D) GA (GA4) and ABA levels in after-ripened WT (Col and Ler) and rgl2-1 seeds on Control or +OPDA plates measured 2 DAI. (E) Relative gene expression of ABA1, ABI5, and RGL2 in after-ripened seeds.
We then investigated whether elevated levels of endogenous OPDA in seeds also correlated with increased amounts of ABA. To do this, we quantified ABA in OPDA over-accumulating cts-2 seeds (Dave et al., 2011) and found that, as with OPDA-treated seeds, their ABA levels were increased (Supplementary Fig. S2).
In water-stressed Arabidopsis roots, induction of the JA-Ile conjugate, but not OPDA, is required for ABA accumulation in an NCED3-dependent manner (De Ollas et al., 2015). NCED3 is a key gene regulating ABA biosynthesis under stress conditions (Nambara and Marion-Poll, 2005). One possible explanation for the germination-inhibiting effect of OPDA is that OPR3 converts OPDA into JA/JA-Ile, which in turn promotes ABA biosynthesis to inhibit germination. However, opr3-1 seeds, which over-accumulate OPDA but not JA/JA-Ile (Dave et al., 2011), are more dormant than WT seeds (Fig. 1). Additionally, exogenously applied OPDA represses germination of opr3-1 seeds as well as WT control seeds (Supplementary Fig. S3). Hence, in the germination inhibition response, it is OPDA rather than JA/JA-Ile that leads to the increase in ABA to inhibit germination. Thus, from our own work and that of others, it seems that both JA/JA-Ile and OPDA are able to affect ABA biosynthesis, but which compound does so depends on the tissue, stress stimuli, and developmental stage. Drought also induces OPDA and ABA accumulation, and these two compounds cooperatively regulate stomatal aperture in a JA-independent manner; however, in this case the response initiated by OPDA is unlikely to be via its effect on ABA biosynthesis (Savchenko et al., 2014). Thus, a mode of action for OPDA whereby it acts along with ABA, but not through a direct effect on ABA biosynthesis, is also possible.
To gain more insight into the mechanism of OPDA-driven ABA accumulation in seeds, we analysed the expression of ABA1 and RGL2. We found that exogenously applied OPDA promotes the expression of both genes (Fig. 2E). Thus, OPDA has the ability to regulate the expression of genes involved in ABA production, but it remains unknown how OPDA operates to achieve this. Work from other laboratories has shown that expression of the ABA-signalling gene ABI5 is up-regulated by endogenous and exogenous ABA (Lopez-Molina et al., 2001; Okamoto et al., 2010; Piskurewicz et al., 2008). This prompted us to assess whether exogenously applied OPDA has a similar effect on ABI5 expression, and we found that it does (Fig. 2E). Hence, OPDA not only triggers ABA accumulation but can also increase ABA sensitivity. Furthermore, the importance of ABI5 in the OPDA-imposed dormancy pathway has been demonstrated in planta by studying ped3 mutant seeds, which are deficient in the same ABC transporter as cts-2/pxa1-1. The ped3 seeds are highly dormant, have increased ABI5 expression, and are rescued by the abi5 mutation (Kanai et al., 2010). However, the regulation of ABI5 by OPDA does not occur exclusively at the level of gene expression, since OPDA also increases ABI5 protein stability (Dave et al., 2011). Taken together, these observations show that OPDA promotes ABA biosynthesis by increasing ABA1 and RGL2 expression, and ABA sensitivity by increasing ABI5 gene expression and ABI5 protein stability.
ABA regulates OPDA accumulation
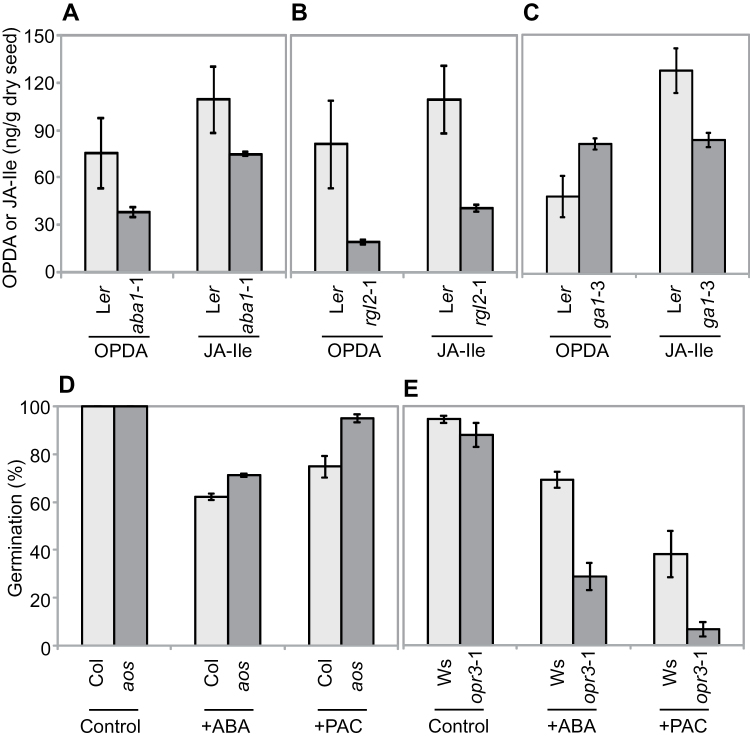
It has been reported that Arabidopsis seedlings infected with the fungus Phytium irregulare accumulate JA and OPDA in an ABA-biosynthesis-dependent manner (Adie et al., 2007). This led us to investigate a possible feedback of ABA and GA into oxylipin biosynthesis in seeds. Thus, OPDA and JA-Ile levels were measured in freshly harvested dry seeds of ABA/GA metabolism and signalling mutants. We found that aba1-1 and rgl2-1 seeds accumulate less OPDA and JA-Ile than WT (Ler) controls (Fig. 3A, B). However, GA-deficient ga1-3 mutant seeds, which also accumulated lower levels of JA-Ile, contained a small but significant increase in OPDA levels (Fig. 3C). We also assessed whether the germination-inhibiting effects of ABA and Paclobutrazol (a GA-biosynthesis inhibitor) were dependent on OPDA by analysing aos and opr3-1 mutants. We found a small but significant reduction in the effect of ABA and Paclobutrazol on after-ripened aos mutant seeds, which lack OPDA (Fig. 3D). Consistent with this result, opr3-1 seeds, which accumulate OPDA, were found to be hypersensitive to ABA and Paclobutrazol (Fig. 3E).
Fig. 3.
ABA affects the oxylipin pathway. (A–C) OPDA and JA-Ile levels in WT (Ler), aba1-1, rgl2-1, and ga1-3 freshly harvested dry seeds. (D–E) Germination profiles (7 DAI) of WT (Col), aos, and opr3-1 freshly harvested, stratified (3 days in the dark at 4 °C), seeds on water agar (Control) plates and water agar supplemented with 2 µM ABA (+ABA) or 1 µM Paclobutrazol (+PAC).
Cross-talk between oxylipin (specifically JA) signalling and GA signalling has been reported previously in non-seed systems (Hou et al., 2010; Navarro et al., 2008; Qi et al., 2014; Wild et al., 2012; Yang et al., 2012), but interaction between OPDA and GA at the level of biosynthesis has not been shown before. It is possible that the GA and OPDA interactions are indirect via ABA, since GA triggers degradation of RGL2, which in turn indirectly promotes accumulation of ABA and oxylipins (Figs 2E and 3B). Taken together, these observations demonstrate that the germination-inhibitory effects of ABA and Paclobutrazol are (at least partially) augmented by OPDA, and that there is a feedback from ABA and GA to regulate OPDA accumulation.
OPDA and MFT interact reciprocally to inhibit germination
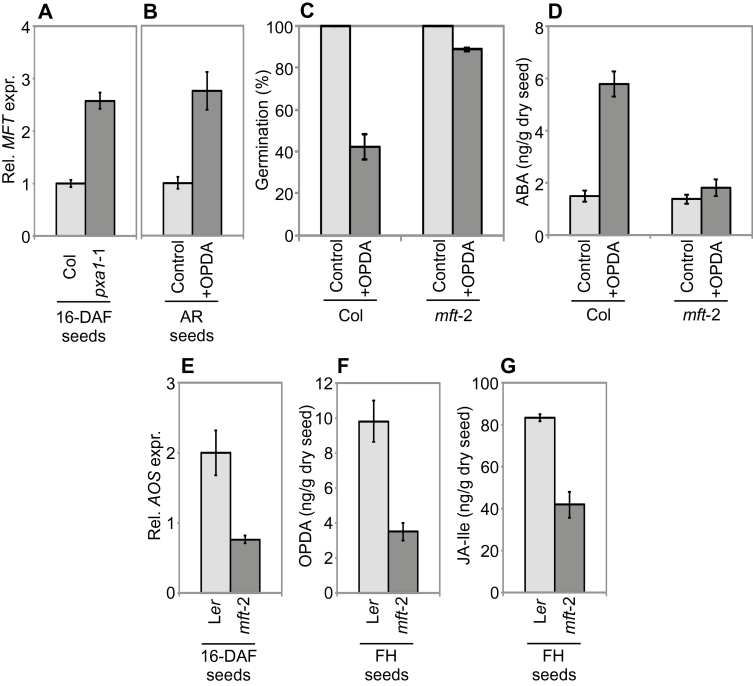
We reported previously that MFT plays a major role in promoting dormancy in Arabidopsis seeds (Vaistij et al., 2013). This prompted us to assess whether the OPDA and MFT pathways are linked. Analysis of a transcriptomic dataset derived from pxa1-1 developing seeds (Dave et al., 2011) revealed that MFT expression is higher in the mutant compared with WT. This observation was validated in independent samples of developing pxa1-1 seeds (Fig. 4A). Furthermore, MFT expression was also found to be elevated in OPDA-treated after-ripened WT seeds (Fig.4B). Thus, high endogenous or exogenous OPDA levels induce expression of the dormancy-promoting MFT gene. Interestingly, we also found that mft-2 after-ripened seeds are almost insensitive to OPDA (Fig. 4C), which shows that MFT is required for OPDA-induced inhibition of germination. In order to gain more insight into the interactions between OPDA and MFT, we assessed ABA levels in OPDA-treated mft-2 after-ripened seeds, and found that they were lower than the levels observed in WT (Col) controls (Fig. 4D). This suggests that in order to trigger ABA biosynthesis, in addition to RGL2 (Fig. 2D), OPDA also requires MFT. We also investigated possible MFT-to-OPDA feedback interactions. To do this, we assessed AOS expression and found that it is down-regulated in mft-2 developing seeds (Fig. 4E). Whether MFT regulates AOS expression directly or indirectly remains to be determined but, consistent with AOS expression levels, both OPDA and JA-Ile levels were found to be lower in mft-2 freshly harvested dry seeds than levels in WT seeds (Fig. 4F, G).
Fig. 4.
Interactions between OPDA and MFT. (A, B) Relative MFT expression in WT (Col) and pxa1-1 developing seeds 16 days after flowering (16-DAF) (A), and in after-ripened (AR) WT (Col) seeds on water agar (Control) plates and water agar supplemented with 10 µM of OPDA (+OPDA) (panel B). (C) Germination profiles (7 DAI) of WT (Col) and mft-2 after-ripened seeds on Control and +OPDA plates. (D) ABA levels in WT (Col) and mft-2 after-ripened seeds imbibed on Control and +OPDA plates measured 2 DAI. (E) Relative AOS expression in 16-DAF WT (Col) and mft-2 seeds. (F, G) OPDA and JA-Ile levels in WT (Ler) and mft-2 freshly harvested (FH) dry seeds.
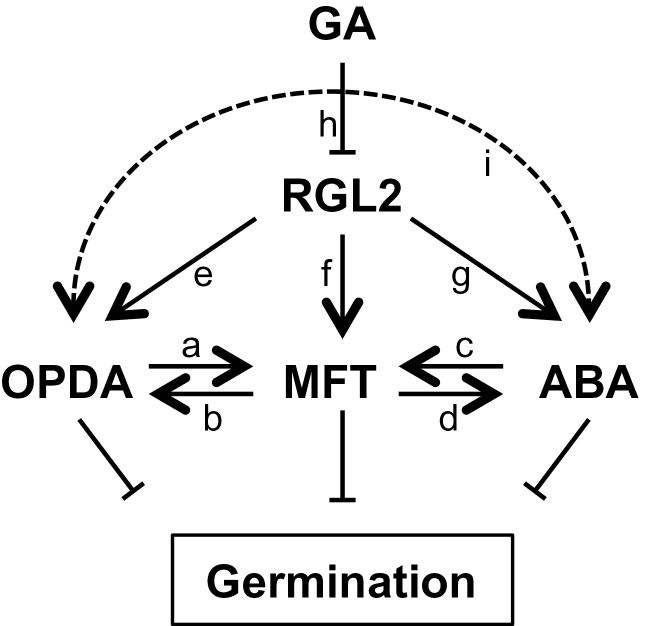
Our results indicate that at least one role of the phosphatidyl ethanolamine-binding protein MFT and the DELLA protein RGL2 in the OPDA response is to elevate ABA levels. DELLA proteins are known to promote ABA accumulation by inducing XERICO expression, and XERICO protein promotes ABA accumulation by an unknown mechanism (Ko et al., 2006; Piskurewicz et al., 2008; Zentella et al., 2007). MFT is also capable of influencing ABA biosynthesis by promoting ABA1 expression (Li et al., 2014). Previous work has shown that MFT also influences ABA sensing, since it negatively regulates ABI5 and, in turn, MFT gene expression is promoted by ABI5 and RGL2 (Xi et al., 2010). Our data demonstrate that in terms of germination, OPDA acts through MFT to promote both biosynthesis of and sensitivity towards ABA. Furthermore, RGL2 and MFT positively feed back on this regulation by promoting OPDA accumulation. However, we cannot exclude the possibility that OPDA and ABA also interact independently of MFT, or that OPDA directly inhibits germination independently of both ABA and MFT (Fig. 5).
Fig. 5.
Model of interactions between the germination repressors OPDA, MFT, and ABA. OPDA promotes MFT gene expression (a; Fig. 4A, 4B) and MFT protein is required for OPDA accumulation (b; Fig. 4F). ABA also promotes MFT expression (c; Xi et al., 2010) and MFT is required for OPDA-driven ABA accumulation (d; Fig. 4D). RGL2 is necessary for OPDA accumulation (e; Fig. 3B), MFT expression (f; Xi et al., 2010), and OPDA-driven ABA accumulation (g; Fig. 2D). GA triggers RGL2 protein degradation (h; Murase et al., 2008). The reciprocal positive feedback between OPDA and ABA (Figs 2C and 3A) may happen through MFT (a and d; c and b) or another indirect pathway (i). OPDA and MFT may per se have ABA-independent germination inhibitory effects. For simplicity, the involvement of ABI5 has not been depicted in this model.
In conclusion, cross-talk between different hormone pathways is known to occur in order to regulate physiological processes at various developmental stages and in response to external stimuli. In this work we have confirmed the role of OPDA as a potent dormancy-promoting/germination-inhibiting factor in Arabidopsis. We established that it acts in conjunction (but possibly not exclusively) with ABA, ABI5, RGL2, and MFT via a complex network of positive and negative feedback interactions. Further research is required to understand how OPDA is regulated by external stimuli and what other physiological processes can be influenced by OPDA in Arabidopsis.
Supplementary data
Figure S1. OPDA requires ABA to inhibit germination.
Figure S2. OPDA and ABA levels are increased in cts-2 seeds.
Figure S3 . opr3-1 seeds are sensitive to OPDA.
Table S1. Sequence of primers used for gene expression analyses.
Acknowledgements
This work was supported by the Biotechnology and Biological Sciences Research Council (grant number BB/J00216X/1). We thank Dr Tony Larson for his assistance with measurement of phytohormones and oxylipins, the departmental horticultural service for plant care, and Thiago Barros-Galvão and Judith Mitchell for comments on and proofreading of the manuscript, respectively.
References
- Adie BA, Pérez-Pérez J, Pérez-Pérez MM, Godoy M, Sánchez-Serrano JJ, Schmelz EA, Solano R. 2007. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defences in Arabidopsis. The Plant Cell 19, 1665–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbi V, Devoto A. 2008. Jasmonate signalling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios. New Phytologist 177, 301–318. [DOI] [PubMed] [Google Scholar]
- Bosch M, Wright LP, Gershenzon J, Wasternack C, Hause B, Schaller A, Stintzi A. 2014. Jasmonic acid and its precursor 12-oxophytodienoic acid control different aspects of constitutive and induced herbivore defenses in tomato. Plant Physiology 166, 396–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D, Hussain A, Cheng H, Peng J. 2005. Loss of function of four DELLA genes leads to light-and-gibberellin-independent seed germination in Arabidopsis . Planta 223, 105–113. [DOI] [PubMed] [Google Scholar]
- Clerkx EJ, Vries HB, Ruys GJ, Groot SP, Koornneef M. 2003. Characterization of green seed, an enhancer of abi3-1 in Arabidopsis that affects seed longevity. Plant Physiology 132, 1077–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave A, Graham IA. 2012. Oxylipin signalling: a distinct role for the jasmonic acid precursor 12-oxo-phytodienoic acid (OPDA). Frontiers in Plant Science 3, 42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dave A, Hernández ML, He Z, Andriotis VME, Vaistij FE, Larson TR, Graham IA. 2011. 12-Oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis . The Plant Cell 23, 583–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies PJ. 2010. The plant hormones: their nature, occurrence, and function. In Davies PJ, ed., Plant hormones: biosynthesis, signal transduction, action! Dordrecht: Springer, 1–15. [Google Scholar]
- De Ollas C, Arbona V, Gómez-Cadenas A. 2015. Jasmonoyl isoleucine accumulation is needed for abscisic acid build-up in roots of Arabidopsis under water stress conditions. Plant Cell & Environment 38, 2157–2170. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR. 1994. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. The Plant Journal 5, 765–771. [Google Scholar]
- Finkelstein RR, Ariizumi T, Steber C. 2008. Molecular aspects of seed dormancy. Annual Reviews of Plant Biology 59, 387–415. [DOI] [PubMed] [Google Scholar]
- Finkelstein RR, Lynch TJ. 2000. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. The Plant Cell 12, 387–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkelstein RR, Wang ML, Lynch TJ, Rao S, Goodman HM. 1998. The Arabidopsis abscisic acid response locus ABI4 encodes an APETALA2 domain protein. The Plant Cell 10, 1043–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Footitt S, Slocombe SP, Larner V, Kurup S, Wu Y, Larson T, Graham IA, Baker A, Holdsworth M. 2002. Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. The EMBO Journal 21, 2912–2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz S, Hellwege A, Stenzel I, et al. 2012. Role of cis-12-oxo-phytodienoic acid in tomato embryo development. Plant Physiology 158, 1715–1727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber K, Nakabayashi K, Miatton E, Leubner-Metzger G, Soppe WJ. 2012. Molecular mechanisms of seed dormancy. Plant Cell & Environment 35, 1769–1786. [DOI] [PubMed] [Google Scholar]
- Guo HM, Li HC, Zhou SR, Xue HW, Miao XX. 2014. Cis-12-oxo-phytodienoic acid stimulates rice defense response to a piercing-sucking insect. Molecular Plant 7, 1683–1692. [DOI] [PubMed] [Google Scholar]
- Hayashi M, Nito K, Takei-Hoshi R, Yagi M, Kondo M, Suenaga A, Yamaya T, Nishimura M. 2002. Ped3p is a peroxisomal ATP-binding cassette transporter that might supply substrates for fatty acid beta-oxidation. Plant and Cell Physiology 43, 1–11. [DOI] [PubMed] [Google Scholar]
- Holdsworth MJ, Bentsink L, Soppe WJ. 2008. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytologist 179, 33–54. [DOI] [PubMed] [Google Scholar]
- Hou X, Lee LY, Xia K, Yan Y, Yu H. 2010. DELLAs modulate jasmonate signaling via competitive binding to JAZs. Developmental Cell 19, 884–894. [DOI] [PubMed] [Google Scholar]
- Kanai M, Nishimura M, Hayashi M. 2010. A peroxisomal ABC transporter promotes seed germination by inducing pectin degradation under the control of ABI5 . The Plant Journal 62, 936–947. [DOI] [PubMed] [Google Scholar]
- Ko JH, Yang SH, Han KH. 2006. Upregulation of an Arabidopsis RING-H2 gene, XERICO, confers drought tolerance through increased abscisic acid biosynthesis. The Plant Journal 47, 343–355. [DOI] [PubMed] [Google Scholar]
- Koornneef M, Jorna ML, Brinkhorst-van der Swan DLC, Karssen CM. 1982. The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.). Theoretical and Applied Genetics 61, 385–393. [DOI] [PubMed] [Google Scholar]
- Lee KP, Piskurewicz U, Turecková V, Strnad M, Lopez-Molina L. 2010. A seed coat bedding assay shows that RGL2-dependent release of abscisic acid by the endosperm controls embryo growth in Arabidopsis dormant seeds. Proceedings of the National Academy of Sciences of the United States of America 107, 19108–19113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S, Cheng H, King KE, Wang W, He Y, Hussain A, Lo J, Harberd NP, Peng J. 2002. Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes & Development 16, 646–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Fan C, Zhang X, Wang X, Wu F, Hu R, Fu Y. 2014. Identification of a soybean MOTHER OF FT AND TFL1 homolog involved in regulation of seed germination. PLoS ONE 9, e99642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Chua NH. 2000. A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana . Plant Cell & Environment 41, 541–547. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, Chua NH. 2001. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis . Proceedings of the National Academy of Sciences of the United States of America 98, 4782–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L, Mongrand S, McLachlin DT, Chait BT, Chua NH. 2002. ABI5 acts downstream of ABI3 to execute an ABA dependent growth arrest during germination. The Plant Journal 32, 317–328. [DOI] [PubMed] [Google Scholar]
- Murase K, Hirano Y, Sun TP, Hakoshima T. 2008. Gibberellin-induced DELLA recognition by the gibberellin receptor GID1. Nature 456, 459–463. [DOI] [PubMed] [Google Scholar]
- Nakamura S, Abe F, Kawahigashi H, et al. 2011. A wheat homolog of MOTHER OF FT AND TFL1 acts in the regulation of germination. The Plant Cell 23, 3215–3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E, Marion-Poll A. 2005. Abscisic acid biosynthesis and catabolism. Annual Review of Plant Biology 56, 165–185. [DOI] [PubMed] [Google Scholar]
- Navarro L, Bari R, Achard P, Lisón P, Nemri A, Harberd NP, Jones JD. 2008. DELLAs control plant immune responses by modulating the balance of jasmonic acid and salicylic acid signalling. Current Biology 18, 650–655. [DOI] [PubMed] [Google Scholar]
- Okamoto M, Tatematsu K, Matsui A, et al. 2010. Genome-wide analysis of endogenous abscisic acid-mediated transcription in dry and imbibed seeds of Arabidopsis using tiling arrays. The Plant Journal 62, 39–51. [DOI] [PubMed] [Google Scholar]
- Park JH, Halitschke R, Kim HB, Baldwin IT, Feldmann KA, Feyereisen R. 2002. A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. The Plant Journal 31, 1–12. [DOI] [PubMed] [Google Scholar]
- Park SW, Li W, Viehhauser A, et al. 2013. Cyclophilin 20–3 relays a 12-oxo-phytodienoic acid signal during stress responsive regulation of cellular redox homeostasis. Proceedings of the National Academy of Sciences of the United States of America 110, 9559–9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S, Gilday AD, Halliday KJ, Graham IA. 2006b. DELLA-mediated cotyledon expansion breaks coat-imposed seed dormancy. Current Biology 16, 2366–2370. [DOI] [PubMed] [Google Scholar]
- Penfield S, Josse EM, Kannangara R, Gilday AD, Halliday KJ, Graham IA. 2005. Cold and light control seed germination through the bHLH transcription factor SPATULA. Current Biology 15, 1998–2006. [DOI] [PubMed] [Google Scholar]
- Penfield S, Li Y, Gilday AD, Graham S, Graham IA. 2006a. Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. The Plant Cell 18, 1887–1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Jikumaru Y, Kinoshita N, Nambara E, Kamiya Y, Lopez-Molina L. 2008. The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. The Plant Cell 20, 2729–2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U, Lopez-Molina L. 2009. The GA-signaling repressor RGL3 represses testa rupture in response to changes in GA and ABA levels. Plant Signaling & Behavior 4, 63–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi T, Huang H, Wu D, Yan J, Qi Y, Song S, Xie D. 2014. Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. The Plant Cell 26, 1118–1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribot C, Zimmerli C, Farmer EE, Reymond P, Poirier Y. 2008. Induction of the Arabidopsis PHO1;H10 gene by 12-oxo-phytodienoic acid but not jasmonic acid via a CORONATINE INSENSITIVE1-dependent pathway. Plant Physiology 147, 696–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell L, Larner V, Kurup S, Bougourd S, Holdsworth M. 2000. The Arabidopsis COMATOSE locus regulates germination potential. Development 127, 3759–3767. [DOI] [PubMed] [Google Scholar]
- Santino A, Taurino M, De Domenico S, Bonsegna S, Poltronieri P, Pastor V, Flors V. 2013. Jasmonate signaling in plant development and defense response to multiple (a)biotic stresses. Plant Cell Reports 32, 1085–1098. [DOI] [PubMed] [Google Scholar]
- Satoh M, Tokaji Y, Nagano AJ, Hara-Nishimura I, Hayashi M, Nishimura M, Ohta H, Masuda S. 2014. Arabidopsis mutants affecting oxylipin signaling in photo-oxidative stress responses. Plant Physiology and Biochemistry 81, 90–95. [DOI] [PubMed] [Google Scholar]
- Savchenko T, Dehesh K. 2014. Drought stress modulates oxylipin signature by eliciting 12-OPDA as a potent regulator of stomatal aperture. Plant Signaling & Behavior 9, e28304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savchenko T, Kolla VA, Wang CQ, Nasafi Z, Hicks DR, Phadungchob B, Chehab WE, Brandizzi F, Froehlich J, Dehesh K. 2014. Functional convergence of oxylipin and abscisic acid pathways controls stomatal closure in response to drought. Plant Physiology 164, 1151–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi A, Browse J. 2000. The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proceedings of the National Academy of Sciences of the United States of America 97, 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TB, Kamiya Y. 1994. The Arabidopsis GA1 locus encodes the cyclase ent-kaurene synthetase A of gibberellin biosynthesis. The Plant Cell 6, 1509–1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun TP. 2010. Gibberellin-GID1-DELLA: a pivotal regulatory module for plant growth and development. Plant Physiology 154, 567–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taki N, Sasaki-Sekimoto Y, Obayashi T, et al. 2005. 12-oxo-phytodienoic acid triggers expression of a distinct set of genes and plays a role in wound-induced gene expression in Arabidopsis. Plant Physiology 139, 1268–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyler L, Thomas SG, Hu J, Dill A, Alonso JM, Ecker JR, Sun TP. 2004. DELLA proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiology 135, 1008–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij FE, Gan Y, Penfield S, Gilday AD, Dave A, He Z, Josse EM, Choi G, Halliday KJ, Graham IA. 2013. Differential control of seed primary dormancy in Arabidopsis ecotypes by the transcription factor SPATULA. Proceedings of the National Academy of Sciences of the United States of America 110, 10866–10871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C. 2014. Action of jasmonates in plant stress responses and development-applied aspects. Biotechnology Advances 32, 31–39. [DOI] [PubMed] [Google Scholar]
- Wasternack C, Forner S, Strnad M, Hause B. 2013. Jasmonates in flower and seed development. Biochimie 95, 79–85. [DOI] [PubMed] [Google Scholar]
- Wasternack C, Hause B. 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. an update to the 2007 review in Annals of Botany. Annals of Botany 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild M, Davière JM, Cheminant S, Regnault T, Baumberger N, Heintz D, Baltz R, Genschik P, Achard P. 2012. The Arabidopsis DELLA RGA-LIKE3 is a direct target of MYC2 and modulates jasmonate signaling responses. The Plant Cell 24, 3307–3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi W, Liu C, Hou X, Yu H. 2010. MOTHER OF FT AND TFL1 regulates seed germination through a negative feedback loop modulating ABA signaling in Arabidopsis . The Plant Cell 22, 1733–1748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi W, Yu H. 2010. MOTHER OF FT AND TFL1 regulates seed germination and fertility relevant to the brassinosteroid signaling pathway. Plant Signaling & Behavior 5, 1315–1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Li J, Gangappa SN, Hettiarachchi C, Lin F, Andersson MX, Jiang Y, Deng XW, Holm M. 2014. Convergence of Light and ABA signaling on the ABI5 promoter. PLoS Genetics 10, e1004197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DL, Yao J, Mei CS, et al. 2012. Plant hormone jasmonate prioritizes defense over growth by interfering with gibberellin signaling cascade. Proceedings of the National Academy of Sciences of the United States of America 109, E1192–E1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zentella R, Zhang ZL, Park M, et al. 2007. Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis . The Plant Cell 19, 3037–3057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolman BK, Silva ID, Bartel B. 2001. The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiology 127, 1266–1278. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.