Highlight
The banana transcriptional repressor MaDof23 and transcriptional activator MaERF9 may act antagonistically in regulating 10 ripening-related genes associated with cell wall degradation and aroma formation.
Key words: Banana, Dof, ERF, fruit ripening, Musa acuminate, protein interaction, transcriptional regulation.
Abstract
The DNA binding with one finger (Dof) proteins, a family of plant-specific transcription factors, are involved in a variety of plant biological processes. However, little information is available on their involvement in fruit ripening. We have characterized 25 MaDof genes from banana fruit (Musa acuminata), designated as MaDof1–MaDof25. Gene expression analysis in fruit subjected to different ripening conditions revealed that MaDofs were differentially expressed during different stages of ripening. MaDof10, 23, 24, and 25 were ethylene-inducible and nuclear-localized, and their transcript levels increased during fruit ripening. Moreover, yeast two-hybrid and bimolecular fluorescence complementation analyses demonstrated a physical interaction between MaDof23 and MaERF9, a potential regulator of fruit ripening reported in a previous study. We determined that MaDof23 is a transcriptional repressor, whereas MaERF9 is a transcriptional activator. We suggest that they might act antagonistically in regulating 10 ripening-related genes, including MaEXP1/2/3/5, MaXET7, MaPG1, MaPME3, MaPL2, MaCAT, and MaPDC, which are associated with cell wall degradation and aroma formation. Taken together, our findings provide new insight into the transcriptional regulation network controlling banana fruit ripening.
Introduction
Transcriptional regulation of gene expression in higher plants plays a pivotal role in influencing or controlling many important biological processes, such as growth and development, signal transduction, and environmental stress responses (Riechmann and Ratcliffe, 2000). Transcription factors (TFs) are important regulators of gene expression that are able to bind to specific cis-regulatory elements in the promoter region, and activate or repress the transcription of their target genes (Wray et al., 2003). In plants, about 7% of all genes encode putative TFs (Udvardi et al., 2007). Arabidopsis genomes include at least 1533 TFs, and tomato (Solanum lycopersicum) at least 998 (Riechmann and Ratcliffe, 2000; Zhang et al., 2010; Sato et al., 2012). In banana (Musa acuminata), 3155 putative TFs have been identified among the 36 542 predicted protein-coding genes (D’Hont et al., 2012). TFs are grouped into different families based on conserved structural domains. A wide range of TF families have been identified in plants, including WRKY, NAM/ATAF1/CUC2 (NAC), basic leucine zipper (bZIP), APETALA 2/ethylene-responsive element binding factor (AP2/ERF), basic helix-loop-helix, and Cys2(C2)His2-type zinc fingers (ZFs) (Lindemose et al., 2013; Yamasaki et al., 2013).
The DNA binding with one finger (Dof) proteins, which were first found in maize (Zea mays), are a family of plant-specific TFs of the ZF super-family, containing a highly conserved DNA binding Dof domain in the N-terminal region (Yanagisawa and Izui, 1993; Gupta et al., 2015). The Dof domain is composed of 50–56 amino acid residues, structured as a C2/C2 ZF, mediating both DNA–protein and protein–protein interactions (Yanagisawa, 2002). The DNA binding sequence of all Dof proteins, except a pumpkin Dof protein (AOBP) that recognizes an AGTA DNA motif, includes the AAAG motif or its reverse-oriented sequence CTTT in the plant gene promoters (Diaz et al., 2002; Yanagisawa, 2002; Gupta et al., 2015). Since the first Dof TF was isolated from maize, Dof TFs have been identified in other plants, including Arabidopsis, rice (Oryza sativa), poplar (Populus trichocarpa), barley (Hordeum vulgare), bread wheat (Triticum aestivum), maize, sorghum (Sorghum bicolor), tomato, soybean (Glycine max), Chinese cabbage (Brassica rapa L. ssp. pekinensis), and potato (Solanum tuberosum) (Yanagisawa, 2002, 2004; Cai et al., 2013; Guo and Qiu, 2013; Noguero et al., 2013; Gupta et al., 2015; Ma et al., 2015; Venkatesh and Park, 2015). Moreover, genetic and molecular studies have suggested that Dof TFs are involved in many plant-specific physiological processes, such as light-responsiveness, tissue differentiation, seed development or germination, metabolic regulation, and phytochrome and phytohormone signalling (Noguero et al., 2013; Gupta et al., 2015). Recently, Dof TFs were reported to be involved in biotic and abiotic stress responses, possibly via the induction of various stress-responsive genes, such as COR15, RD29A, and RD10 (Corrales et al., 2014; Gupta et al., 2015; Sasaki et al., 2015). Given the diverse function of Dof TFs in plant biological processes, it is important to gain insights into their involvement in fruit ripening.
Banana is one of the most important fruit crops in the world, serving as part of a well-balanced human diet and a staple food for more than 400 million people in many tropical and subtropical countries (D’Hont et al., 2012; Hölscher et al., 2014). Unfortunately, banana is a typical climacteric fruit; once ripening is initiated, the shelf-life is limited owing to its rapid softening and susceptibility to diseases. This restricts the handling and transportation of the fruit, which can cause significant losses to both farmers and consumers (Bapat et al., 2010; Shan et al., 2012; Xiao et al., 2013). Therefore, understanding the ripening mechanism of banana fruit is critical for improving its postharvest life. Several ripening-related TFs, including MADS-box (Elitzur et al., 2010; Choudhury et al., 2012; Liu et al., 2013), EIN3/EIL (Mbéguié-A-Mbéguié et al., 2008), NAC (Shan et al., 2012), ERF (Xiao et al., 2013), and LBD (Ba et al., 2014a), as well as their involvement in the transcriptional control of ripening, have been reported. Fruit ripening is a complex and highly coordinated developmental process that involves dramatic changes in colour, flavour, aroma, texture, and nutritional content of the flesh, and is controlled by transcriptional regulatory networks involving several TFs (Giovannoni, 2004; Klee and Giovannoni, 2011; Martel et al., 2011; Gapper et al., 2013; Seymour et al., 2013a, b; Cherian et al., 2014). Whether extensive cooperation exists between the different types of fruit ripening-related TFs remains obscure.
There are 74 Dof genes in the banana genome (Plant Transcription Factor Database, http://planttfdb.cbi.pku.edu.cn), and 25 of them are differentially expressed during the ripening stages as revealed by an RNA sequencing analysis (D’Hont et al., 2012). However, the regulatory mechanism of Dofs involved in banana fruit ripening is largely unknown. We have therefore isolated and characterized these 25 Dof genes, designated MaDof1–MaDof25, from banana fruit and analysed their expression patterns under three different ripening conditions. Moreover, we investigated the interactions of MaDofs with a previously identified ripening-related AP2/ERF TF, MaERF9, as well as their involvement in the transcriptional regulation of 11 ripening-related genes. These included genes responsible for softening, such as expansins (MaEXP1/2/3/5), xyloglucan endotransglycosylases (MaXET7), polygalacturonase (MaPG1), pectin methylesterase (MaPME3), pectate lyase (MaPL2), and galactosidase (MaGAL), and genes related to aroma formation, such as branched-chain amino acid transaminase (MaCAT) and pyruvate decarboxylase (MaPDC). Our findings expand our understanding of the transcriptional regulatory network of banana fruit ripening.
Materials and methods
Plant materials and treatments
Banana (Musa acuminata, AAA group, cv. Cavendish) fruit at the 70–80% plump stage were harvested from a commercial plantation near Guangzhou, south-eastern China. Three conditions, comprising natural ripening, ethylene-induced ripening (100 µL L–1 ethylene, 18h), and 1-methylcyclopropene (1-MCP)-delayed ripening (0.5 µl L–1 1-MCP, 18h), were considered as described previously (Shan et al., 2012). After each treatment, fruit were held at 22°C and 90% relative humidity until the production of climacteric ethylene and complete ripening. Sample taking, as well as the recording of ethylene production and fruit firmness at each sampling time, were described previously in Shan et al. (2012). All of the samples were frozen in liquid nitrogen and stored at −80°C for further use.
In silico analysis
The whole genome sequence of M. acuminata was used to identify Dof TFs (http://banana-genome.cirad.fr/) (D’Hont et al., 2012). On the basis of gene annotation and bioinformatics and RNA sequencing analyses, 25 Dof genes named MaDof1 to MaDof25 were identified. Total RNA was extracted using the method of Wan and Wilkins (1994) and the cDNA was obtained using PrimeScript® RT Reagent Kit with gDNA Eraser (TaKaRa). The sequences of MaDof1 to MaDof25 were further verified by recloning and resequencing. Gene sequences were subjected to a homology search in the National Center for Biotechnology Information database. Multiple alignments were analysed by CLUSTALW (version 1.83) and GeneDoc software, and a phylogenetic tree of Dof proteins was constructed using the UPGMA method in the MEGA5.
Gene expression analysis
Quantitative real-time PCR (qRT-PCR) were used to analyse gene expression. The sequences of all primers used for qRT-PCR are listed in Supplementary Table S1. All qRT-PCR analyses were normalized using the cycle threshold value of MaRPS2 (ribosomal protein 2) as the reference gene (Chen et al., 2011). qRT-PCR was carried out in a Bio-Rad CFX96 Real-Time PCR System using the SYBR®Green PCR Supermix Kit (Bio-Rad Laboratories) following the manufacturer’s instructions. The relative expression levels of target genes were calculated with the formula 2-ΔΔCT. Three independent biological replicates were used.
Subcellular localization assay
The coding regions of MaDofs without the stop codon were amplified (primers are listed in Supplementary Table S1), and subcloned into the pEAQ vector containing the gene for green fluorescent protein (GFP; kindly supplied by Dr George P. Lomonossoff). The fusion constructs and the control GFP vector were electroporated into Agrobacterium tumefaciens strain GV3101 using the Gene Pulser XcellTM Electroporation System (Bio-Rad, CA). A tobacco (Nicotiana benthamiana) leaf infiltration assay for subcellular localization was performed (Sainsbury et al., 2009). After infiltration, plants were incubated at 22°C with a 16h photoperiod for at least 48h before analysis. GFP fluorescence signals were observed with a fluorescence microscope (Zeiss Axioskop 2 Plus). All transient expression assays were repeated at least three times.
Transcriptional activation analysis in yeast cells
The coding regions of MaDofs were cloned into the pGBKT7 [GAL4 DNA-binding domain (GAL4BD)] vector (Clontech, USA) to create the pGBKT7-MaDof constructs (primers are listed in Supplementary Table S1). pGBKT7-MaDofs, the positive control pGBKT7-53 + pGADT7-T, and the negative control pGBKT7 plasmids were transformed into the AH109 yeast strain using the lithium acetate method. The transformed strains were streaked onto plates with minimal medium without tryptophan (SD/−Trp) or without tryptophan, histidine, and adenine (SD/−Trp−His−Ade) plates, and the transactivation activity of each protein was evaluated according to their growth status and the activity of α-galactosidase (α-Gal).
Yeast two-hybrid assay
A yeast two-hybrid (Y2H) assay was performed using the Matchmaker™ GoldYeast Two-Hybrid System (Clontech). The coding sequences of MaDof23 and MaERF9 were subcloned into the pGBKT7 or pGADT7 vector to fuse with the BD and activation domain (AD), respectively, to create the bait and prey (primers are listed in Supplementary Table S1). The bait and prey constructs were then co-transformed into yeast strain Gold Y2H using the lithium acetate method, and yeast cells were grown on DDO medium [minimal media double dropouts (SD/−Leu−Trp)] for 3 d. Transformed colonies were plated onto QDO medium [minimal media quadruple dropouts (SD/−Leu−Trp−Ade−His but containing 125 μm aureobasidin A)] containing 4mg mL−1 X-α-Gal to test the possible interaction between MaDof23 and MaERF9 according to their growth status and blue colour development.
Bimolecular fluorescence complementation assay
To create constructs for a bimolecular fluorescence complementation (BiFC) assay, the full-length coding sequences, without their stop codons, of MaDof23 in fusion with YNE and MaERF9 in fusion with YCE were cloned into the pEAQ-HT vector (Sainsbury et al., 2009). The resulting constructs were then electroporated into A. tumefaciens strain GV3101 and co-infiltrated into tobacco leaves. Infected tissues were analysed at 48h after infiltration. YFP fluorescence was captured using the Confocal Spectral Microscope Imaging System (Leica TCS SP5), with an argon blue laser at 488nm, a beam splitter for excitation at 500nm, and a spectral detector set between 515nm and 540nm. Primers used for generating the constructs are listed in Supplementary Table S1.
Promoter isolation and analysis
Genomic DNA was extracted from banana leaves using the DNeasy Plant Mini Kit (Qiagen). The promoters of the 11 ripening-related genes, including MaEXP1/2/3/5, MaXET7, MaPG1, MaPME3, MaPL2, and MaGAL associated with cell wall degradation (Trivedi and Nath 2004; Sane et al., 2007; Mbéguié-A-Mbéguié et al., 2009; Asif et al., 2014), and MaCAT and MaPDC associated with aroma formation (Yang et al., 2011), were isolated using a Genome Walker Kit (Clontech) with nest PCR according to the manufacturer’s instructions (see specific primers in Supplementary Table S1). Conserved cis-element motifs of promoter were predicted using PLACE (http://www.dna.affrc.go.jp/PLACE/signalscan.html) and Plant-CARE (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) databases.
Dual-luciferase reporter assay
To analyse the in vivo transcriptional activities of MaDof23 and MaERF9, the coding sequence of MaDof23 or MaERF9 was inserted into the constructed pBD vector driven by the 35S promoter with the translation enhancer Ω sequence as the effector. The double reporter vector included a native GAL4-LUC (Firefly luciferase), and an internal control REN (Renilla luciferase) driven by a 35S promoter, which was modified based on the pGreenII 0800-LUC reporter vector (Hellens et al., 2005). GAL4-LUC contains five copies of the GAL4 binding element and 35S promoter or a minimal TATA region of the 35S promoter of cauliflower mosaic virus (CaMV), and these sequences are located upstream of the LUC.
To assay the binding activity of MaDof23 or MaERF9 to the promoters of the 11 ripening-related genes mentioned above, the promoters were cloned into a pGreenII 0800-LUC double-reporter vector, while MaDof23 or MaERF9 was cloned into the pEAQ vector as the effectors. All primers used for generating constructs for the transient expression assay are listed in Supplementary Table S1.
The constructed effector and reporter plasmids were co-transformed into tobacco leaves by A. tumefaciens strain GV3101. After 2 d, LUC and REN luciferase activities were measured using a Dual-Luciferase Assay kit (Promega) on the Luminoskan Ascent Microplate Luminometer (Thermo). The results were calculated using the ratio of LUC to REN. At least six biological repeats were assayed for each combination.
Statistical analysis
The study was carried out using a completely randomized design. In figures, data have been plotted as means ± standard errors (SE). Statistical comparisons of the mean values was performed using a one-way ANOVA, followed by Duncan’s multiple range test at the 0.05 or 0.01 confidence levels using the DPS7.05 software (Zhejiang University, Hangzhou, China). A complete linkage hierarchical clustering of the 25 MaDof genes in banana fruit pulp during ethylene-induced ripening was generated using the MeV 4.9 clustering algorithm according to their gene expression levels.
Results
Sequence analysis of 25 Dof genes and their deduced protein products in banana fruit
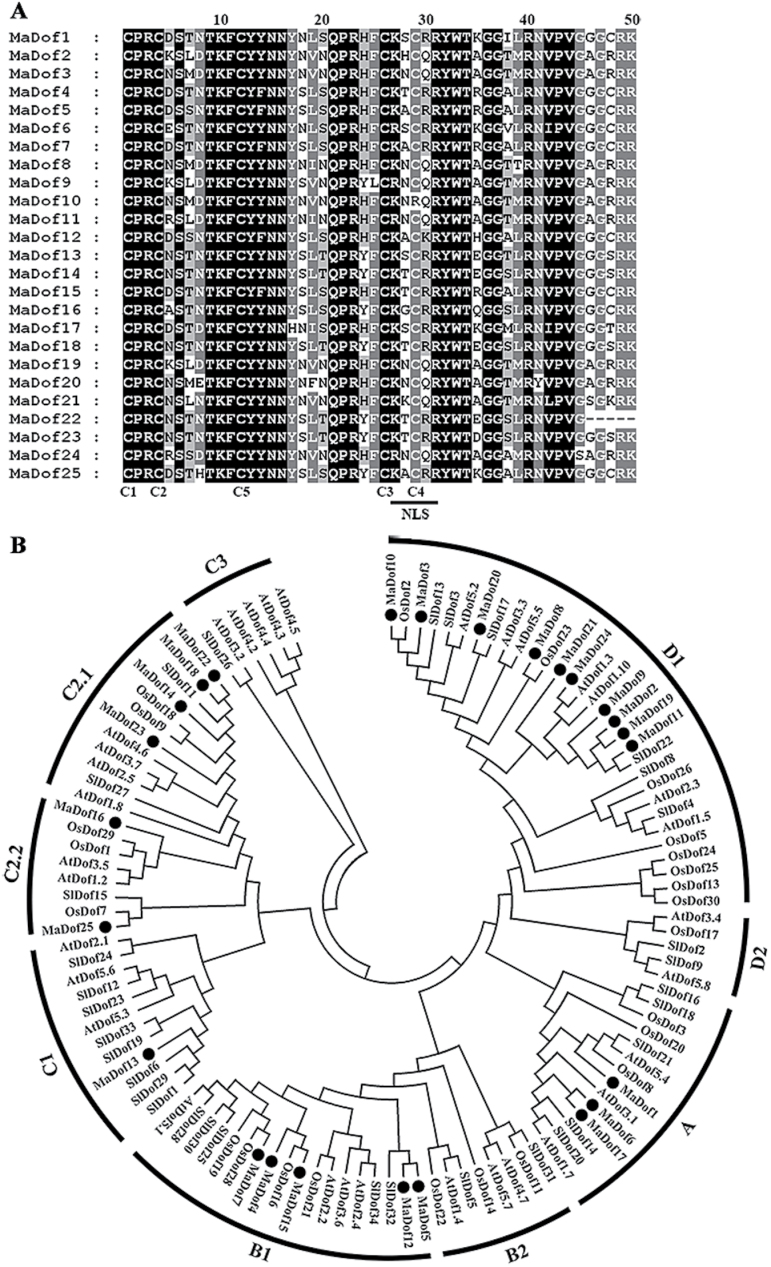
Twenty-five Dof genes named MaDof1–MaDof25 were isolated from banana fruit. The deduced amino acid residues of MaDof1–MaDof25 varied from 182 to 511 amino acids with the isoelectric point (pI) from 5.29 to 9.93, and the molecular mass from 19.43kDa to 54.68kDa (Supplementary Table S2). The sequence similarities of the MaDofs varied from 6.4% (MaDof4 and MaDof7) to 72.6% (MaDof1 and MaDof6) (Supplementary Table S3). Alignments of MaDofs showed a highly conserved DBD, designated the Dof domain and located in the N-terminal region, that consisted of 50–56 amino acid residues structured as a C2/C2 ZF (Fig. 1A), which is a defining character of the Dof TF family (Cai et al., 2013). A nuclear localization signal (NLS), overlapping partly with the highly conserved Dof domain (Krebs et al., 2010; Noguero et al., 2013), has also been identified in all MaDof proteins (Fig. 1A), suggesting the nuclear localization of MaDof proteins.
Fig. 1.
(A) Multiple sequence alignment of the conserved Dof domain of 25 MaDof proteins. Identical and similar amino acids are indicated by black and grey shading, respectively. Gaps were introduced to optimize alignment. The NLS motif, and conserved cysteine residues (C1–C5) within the Dof domain are indicated in the alignment. (B) Phylogenetic tree of Dofs. Banana MaDofs were aligned with the Arabidopsis, rice, and tomato Dof family. The multiple alignment was made using ClustalW, and the phylogenetic tree was constructed with MEGA 5.0 using a bootstrap test of phylogeny with minimum evolution test and default parameters.
Phylogenetic analysis of MaDofs and Dofs from other plants, including Arabidopsis, rice, and tomato, revealed that MaDof sequences were clustered into six of the nine subfamilies of Dof proteins (Cai et al., 2013) (Fig. 1B). These six subfamilies, including A, B1, C1, C2.1, C2.2, and D1, contained 3, 5, 1, 4, 2, and 10 MaDofs, respectively (Fig. 1B).
To gain more insight into the diversity of the 25 MaDof proteins selected in this study, we analysed putative motifs using the program MEME. Fifteen distinct motifs were identified, and annotated by InterProScan (Supplementary Fig. S1). Motif 1, uniformly observed in all of the MaDof proteins, is the conserved Dof domain. As expected, most of the closely related members in the phylogenetic tree had common motif compositions, suggesting functional similarities among the Dof proteins within the same subfamily (Supplementary Fig. S2).
Differential expression of MaDof genes in fruit pulp during ripening
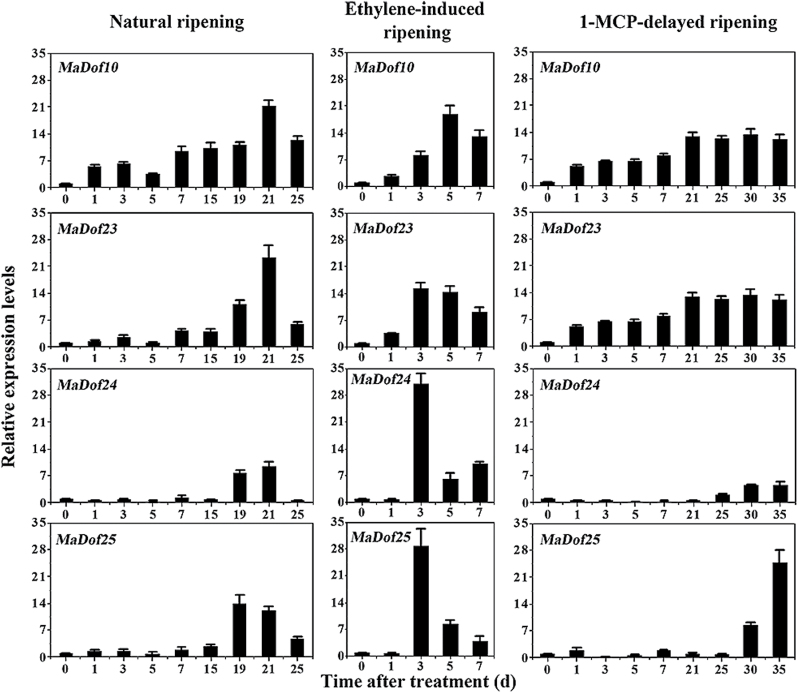
To investigate the possible role of MaDof genes in relation to banana fruit ripening, we first detected their mRNA accumulations in fruit pulp during ethylene-induced ripening using qRT-PCR. We observed differential expression patterns of MaDofs during banana fruit ripening. Among the 25 MaDof genes, MaDof2, 3, 10, 20, 23, 24, and 25 were obviously induced after ethylene treatment, reaching their maximum level at 3 or 5 d of storage, and then decreasing thereafter (Supplementary Fig. S3; Fig. 2). In contrast, transcripts of the other 18 MaDofs exhibited only slight changes (Supplementary Fig. S3). Based on the phylogenetic tree and expression profiles of MaDofs, we selected four MaDof genes from three different subfamilies, that is, MaDof10 and 24 (subfamily D1), MaDof23 (subfamily C2.1), and MaDof25 (subfamily C2.2), and measured their expressions in fruit pulp under two different ripening conditions: natural and 1-MCP-delayed ripening. Their transcript levels in fruit pulp under natural or 1-MCP-delayed ripening also clearly increased following ripening (Fig. 2).
Fig. 2.
Expressions of MaDof10, 23, 24, and 25 in pulp during three ripening behaviours: natural (control), ethylene-induced, and 1-MCP-delayed ripening. The expression levels of each gene are represented as a ratio relative to the harvest time (0 d of control), which was set at 1. Each value represents the mean ± SE of three biological replicates. The physiological data related to fruit ripening and softening, including changes in fruit firmness and ethylene production in banana fruit subjected to three different ripening conditions, have been described in Shan et al. (2012).
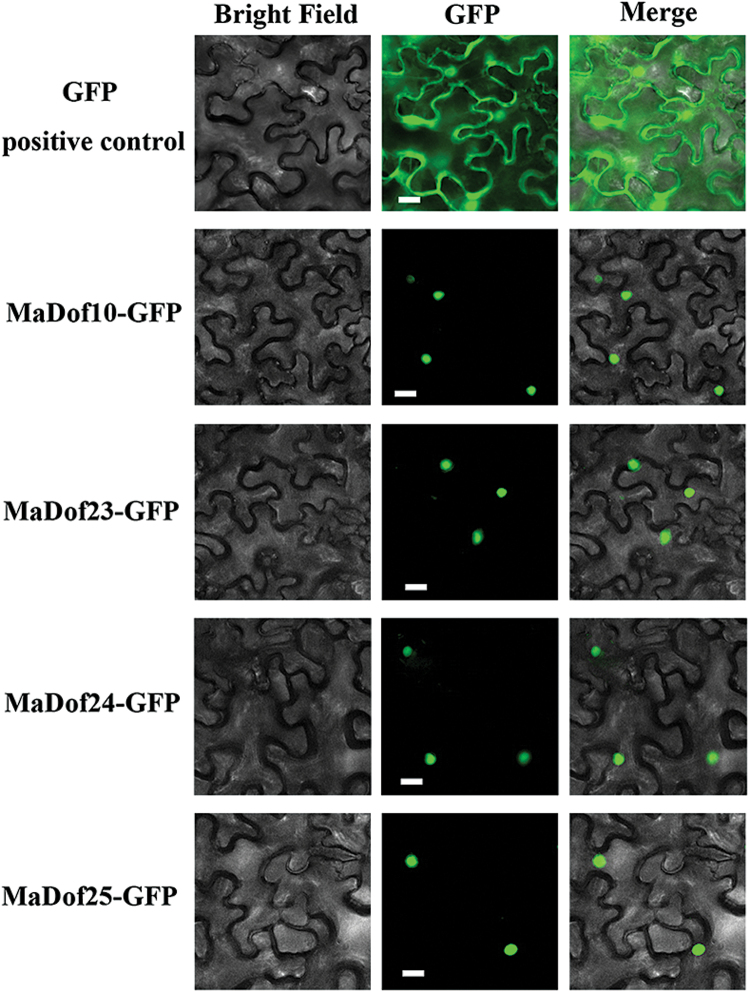
Nuclear localization of MaDof10, 23, 24, and 25
Potential NLS sequences were predicted for MaDof10, 23, 24, and 25 based on the sequence analysis (Fig. 1A). To validate the subcellular localizations of MaDof10, 23, 24 and 25, the full-length coding sequences of these proteins were fused in frame with GFP. The fluorescence from the transient expression of these constructs in tobacco leaf epidermal cells was localized exclusively in the nucleus (Fig. 3). By contrast, fluorescence from the GFP-only control was distributed throughout the entire cell. We also observed similar localization when GFP-MaDof10, 23, 24, or 25 were transiently expressed in tobacco BY2 protoplasts (Supplementary Fig. S4).
Fig. 3.
Subcellular localization of MaDof10, 23, 24, and 25 in tobacco leaves. MaDof10, 23, 24, and 25 fused with the GFP or GFP positive control were infiltrated into tobacco leaves via A. tumefaciens strain GV3101. After 48h of infiltration, GFP fluorescence signals were visualized using a fluorescence microscope. Merge indicates a digital merge of bright field and fluorescent images. Images were taken in a dark field for green fluorescence, while the outline of the cell and the Merged images were photographed in a bright field. Scale bars, 25 μm.
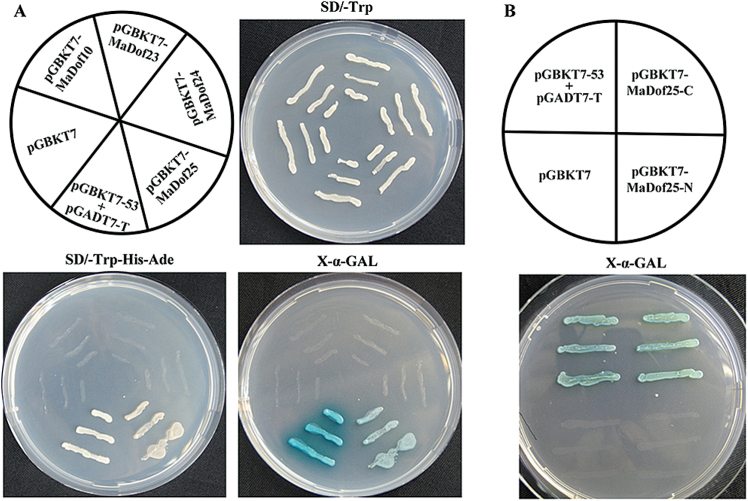
Transcriptional activation ability of MaDof10, 23, 24, and 25 proteins in yeast
To investigate the abilities of MaDof10, 23, 24, and 25 to activate transcription, we performed a transient expression assay using a GAL4-responsive reporter system in yeast cells. The full-length coding regions of MaDof10, 23, 24, and 25 were fused to the GAL4BD to generate pGBKT7-MaDof10, 23, 24, and 25 fusion plasmids (Fig. 4). The plasmids were then transformed into yeast strain AH109 and the transformants were assayed for their ability to activate transcription from the GAL4 upstream activation sequence and to promote yeast growth in medium lacking Trp, His, and Ade. The transformants containing pGBKT7-53 + pGADT7-T and pGBKT7 vectors (empty) were used as positive and negative controls, respectively. As shown in Fig. 4, the transformed yeast cells harbouring pGBKT7-MaDof25 and pGBKT7-53 + pGADT7-T (positive control) grew well in SD/−Trp−His−Ade and showed α-Gal activity, whereas cells containing pGBKT7-MaDof10, -MaDof23, -MaDof24, or pGBKT7 alone (negative control) showed no α-Gal activity. To further identify the transcriptional activation domain of MaDof25, the N- and C-terminal regions of MaDof25 were fused to the GAL4BD. The yeast cells harbouring pGBKT7-MaDof25-C showed α-Gal activity (Fig. 4). These results indicate that the MaDof25 C-terminal region is able to activate transcription.
Fig. 4.
Transcriptional activation of MaDof10, 23, 24, and 25 in yeast. The coding regions of MaDof10, 23, 24, and 25, as well as C- and N-terminal derivatives of MaDof25, were cloned into the pGBKT7 (GAL4BD) vector to create the pGBKT7-MaDof10, 23, 24, 25, 25-N, and 25-C constructs. All of the constructs mentioned above, together with the positive control (pGBKT7-53 + pGADT7-T) and negative control (pGBKT7) were transformed into yeast strain AH109. Yeast clones transformed with different constructs were grown on SD plates without tryptophan (SD/−Trp) or without tryptophan, histidine, and adenine but containing 125 μM aureobasidin A (SD/−Trp−His−Ade) for 3 d at 30°C. Transcription activation was monitored by the detection of yeast growth and an α-Gal assay.
Interaction between MaDof23 and MaERF9
In a previous study, we reported that MaERF9 might act as transcriptional activator in the regulation of fruit ripening by activating the expression of ethylene biosynthetic genes (Xiao et al., 2013). Because the canonical recognition sequence for Dof TFs is very short, it is highly likely that they function in association with other TFs and in combination generate the necessary promoter specificity (Noguero et al., 2013). Thus, we investigated whether MaDof10, 23, 24, and 25 can interact with MaERF9. First we used the GAL4 transcription activation-based Y2H system. Full-length sequences of MaDof10, 23, and 24 and the N-terminus of MaDof25 were fused with the GAL4BD (BD-MaDof10, 23, 24, and 25-N), and MaERF9 was ligated with the activation domain (AD-MaERF9), to create the bait and prey, respectively, given that MaERF9 has transcription-activating activity in yeast (data not shown). As shown in Fig. 5A, co-expression of BD-MaDof23 with AD-MaERF9 resulted in strong activation of α-Gal activity, indicating that only MaDof23 interacted with MaERF9 in yeast cells, whereas MaDof10, 24, and 25 did not.
Fig. 5.
In vitro and in vivo interaction between MaDof23 and MaERF9. (A) A Y2H assay for the interaction between MaDof23 and MaERF9. The coding regions of MaDof10, 23, 24, and 25-N were fused with pGBKT7 (BD) and the coding region of MaERF9 with pGADT7 (AD) vectors as indicated, and co-transformed into the yeast strain Gold Y2H. The ability of yeast cells to grow on QDO medium (SD/−Leu−Trp−Ade−His but containing 125 μm aureobasidin A), and to turn blue in QDO medium containing 4mg mL−1 X-α-Gal, was scored as a positive interaction. (B) BiFC in tobacco leaf epidermal cells showing the interaction between MaDof23 and MaERF9 in living cells. MaDof23 and MaERF9 were fused with the N-terminus of YFP (YNE) or the C-terminus of YFP (YCE), as indicated, and co-transfected into N. benthamiana leaves by A. tumefaciens infiltration. Expressions of MaDof23 or MaERF9 alone were used as negative controls. YFP indicates fluorescence of YFP; Merge indicates a digital merge of bright field and fluorescent images. Scale bar, 30 μm.
To further substantiate that MaDof23 interacts with MaERF9 in plant cells, we examined their interaction by BiFC assay. Both MaDof23 and MaERF9 were fused to the N-terminal 174-amino acid portion of yellow fluorescent protein (YFP) as well as the C-terminal 66-amino acid portion of YFP, in the pEAQ vector. The corresponding constructs were co-transfected into N. benthamiana leaves by A. tumefaciens infiltration. As shown in Fig. 5B, co-expression of MaDof23-YNE and MaERF9-YCE, or MaDof23-YCE and MaERF9-YNE reconstituted a functional YFP in the nucleus, whereas co-expression with the negative control combinations failed to generate YFP fluorescence. These data together confirm that MaDof23 physically interacts with MaERF9.
In vivo transcriptional activities of MaDof23 and MaERF9
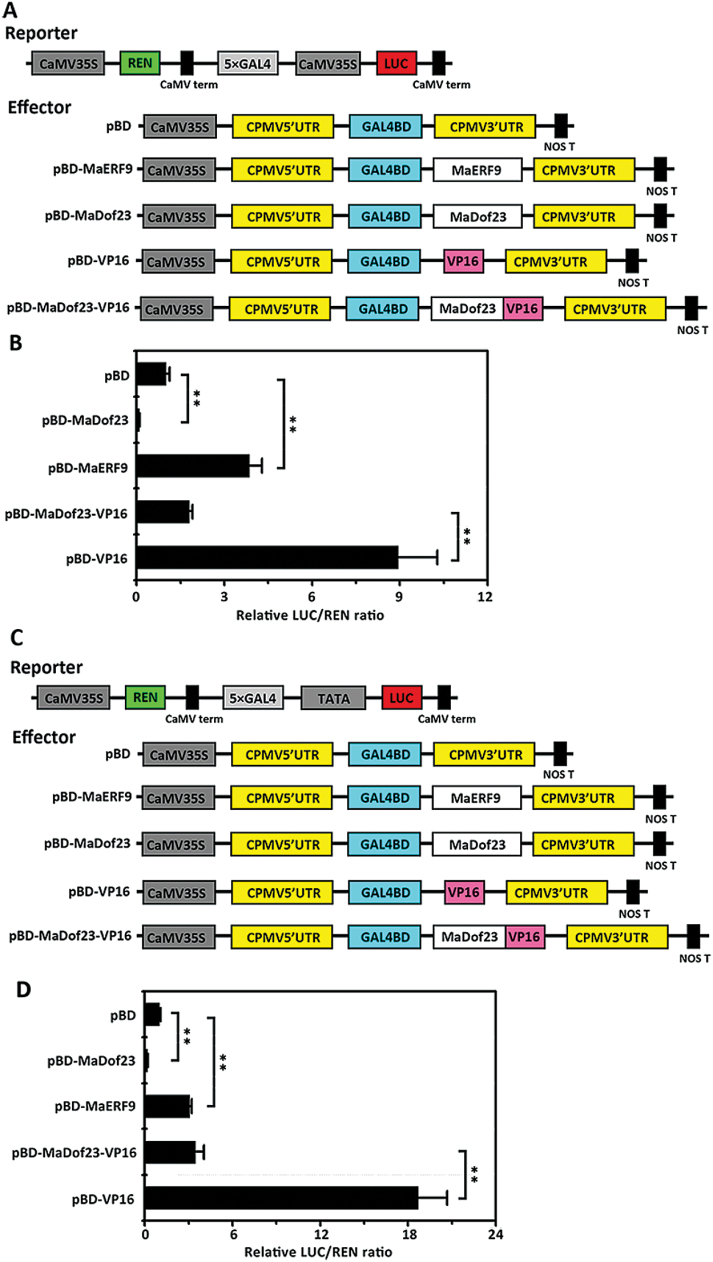
To investigate the transcriptional activities of MaDof23 and MaERF9 in vivo, we performed a dual-luciferase assay. The double-reporter vector contained 5 × GAL4 and CaMV35S fused to LUC and REN driven by CaMV35S, and the effector plasmids contained the MaDof23 or MaERF9 gene fused to the GAL4BD driven by the CaMV35S (Fig. 6A). These constructs were co-expressed in tobacco leaves by Agrobacterium transfection. As shown in Fig. 6B, compared with the GAL4BD (empty, pBD) control, pBD-MaDof23 significantly repressed the expression of the LUC reporter, and the LUC to REN ratio of MaDof23 was 11.9% that of the pBD control. pBD-MaERF9, together with the transcriptional activator control pBD-VP16, strongly activated the LUC reporter gene, and the LUC to REN ratio of MaERF9 was 1.37-fold higher than that of the pBD control (Fig. 6B). However, the activation of pBD-VP16 was significantly inhibited by MaDof23 (pBD-MaDof23-VP16) (Fig. 6B), in a manner similar to that of the repression domain of ERF3 (Ohta et al., 2001). Similarly, when the LUC reporter gene was fused with 5 × GAL4 and the minimal TATA region, we also observed that pBD-MaDof23 remarkably repressed, and pBD-MaERF9 activated, the expression of the LUC reporter compared with pBD alone (Fig. 6C, D). The activation of pBD-VP16 was also drastically impaired by MaDof23 (Fig. 6D). These in vivo data demonstrate that MaDof23 may act as a transcriptional repressor, and MaERF9 as a transcriptional activator.
Fig. 6.
Transcriptional repression or activation ability of MaDof23 or MaERF9 in tobacco leaves. (A and C) Diagram of the various constructs used in this assay. The double-reporter plasmids contained LUC luciferase fused with 5 × GAL4 and CaMV35S or 5 × GAL4 and the minimal TATA region of CaMV35S, and REN luciferase driven by CaMV35S. The effector plasmids contained the MaDof23 or MaERF9 genes fused to GAL4BD driven by the CaMV35S. pBD was used as a negative control, while the GAL4BD fused with VP16 activation domain was used as positive control. (B and D) The dual REN/LUC reporter and effectors were co-transformed into tobacco leaves using A. tumefaciens strain GV3101. After 48h of infiltration, LUC and REN luciferase activities were assayed, and the transcription repression or activation ability of MaDof23 or MaERF9 is indicated by the ratio of LUC to REN. The ratio of LUC to REN of the pBD vector was used as a calibrator (set as 1). Each value represents the means of six biological replicates, and vertical bars represent the SE. Asterisks indicate a statistically significant difference compared with pBD by one-way ANOVA; *P < 0.05, **P < 0.01. This figure is available in colour at JXB online.
Antagonistic action of MaDof23 and MaERF9 in transcriptionally regulating 10 ripening-related genes
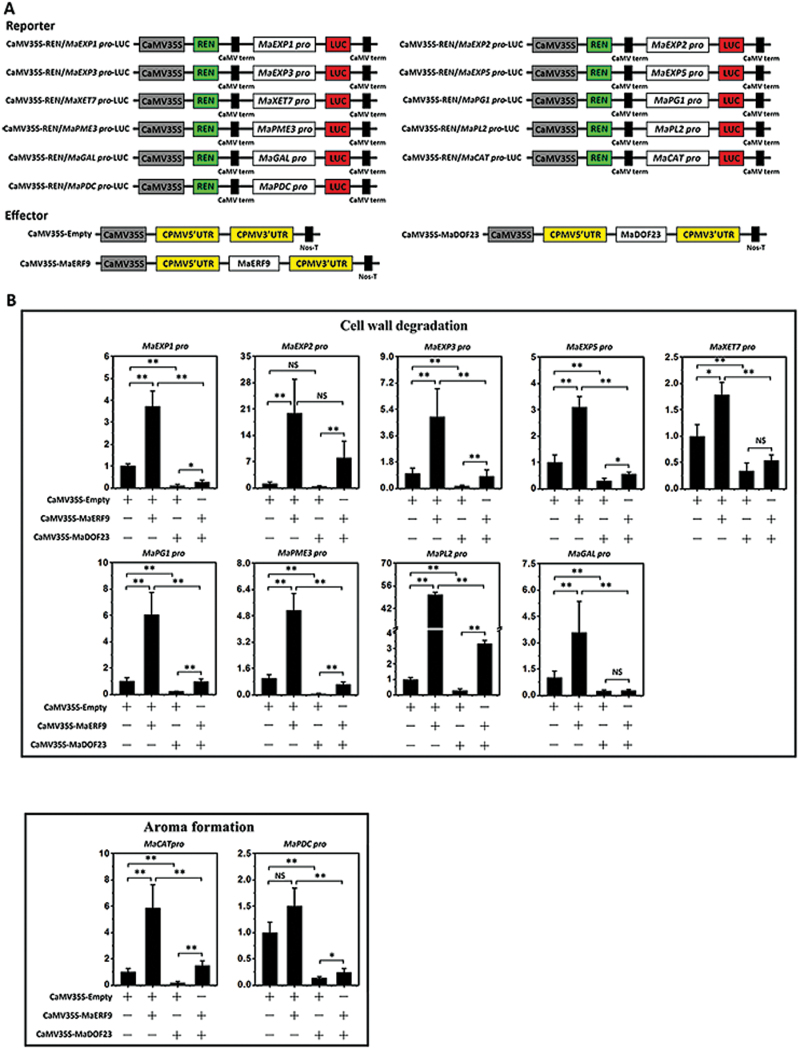
It is well documented that Dof TFs preferentially bind to the core sequence 5′-(T/A)AAAG-3′ (or its reversibly orientated sequence, CTTT) and the and ERF TFs to the GCC-box in their target promoters (Hao et al., 1998; Diaz et al., 2002; Yanagisawa 2002). Sequence analysis identified the (T/A)AAAG elements and GCC-box in the promoters of all 11 ripening-related genes, including MaEXP1/2/3/5, MaXET7, MaPG1, MaPME3, MaPL2, MaGAL, MaCAT, and MaPDC (Supplementary Fig. S5), suggesting that these 11 ripening-related genes might be direct targets of MaDof23 and MaERF9. Moreover, the physical interaction between MaDof23 and MaERF9 led us to investigate whether they might mutually interfere in regulating target genes. To this end, we performed additional transient expression assays using the dual-luciferase reporter system. In this experiment, a dual-luciferase reporter plasmid harbouring the promoters of 11 ripening-related genes fused to LUC, together with an effector plasmid expressing MaDof23 or MaERF9 under the control of the CaMV35S promoter (Fig. 7A), were transiently co-expressed in the leaves of N. benthamiana. Compared with the control, co-expression of MaDof23 with each of the 11 promoters significantly repressed the LUC to REN ratio. In contrast, co-expression of MaERF9 enhanced the LUC to REN ratio (Fig. 7B), suggesting that MaDof23 trans-represses and MaERF9 trans-activates these ripening-related genes. Moreover, except for MaGAL, the trans-activation of MaERF9 or the trans-repression of MaDof23 of the other 10 gene promoters was impaired when MaDof23 and MaERF9 were co-expressed (Fig. 7B). Collectively, these data reveal that MaDof23 and MaERF9 might act antagonistically in the transcriptional regulation of ripening-related genes.
Fig. 7.
Antagonistic action of MaDof23 and MaERF9 in transcriptional regulation of 10 ripening-related genes including MaEXP1/2/3/5, MaXET7, MaPG1, MaPME3, and MaPL2 associated with cell wall degradation; and MaCAT and MaPDC associated with aroma formation. (A) Diagram of the various constructs used in this assay. The double-reporter plasmids contained the promoter of the 11 ripening-related genes fused to LUC luciferase and REN luciferase driven by CaMV35S. The effector plasmids contained MaDof23 or MaERF9 driven by CaMV35S. (B) The reporter and effector vectors were co-introduced into tobacco leaves using A. tumefaciens strain GV3101. After 48h of infiltration, LUC and REN luciferase activities were assayed, and the repression or activation of MaDof23 or MaERF9 to the promoter was showed by the ratio of LUC to REN. The ratio of LUC to REN of the empty vector plus promoter vector was used as a calibrator (set as 1). Each value represents the mean ± SE of six biological replicates. Asterisks indicate a statistically significant difference by one-way ANOVA; *P < 0.05, **P < 0.01. + or – indicates that the effector was present or absent in the combinations. This figure is available in colour at JXB online.
Discussion
Dofs constitute a large family of TFs that are associated with various biological processes unique to plants. The identification of Dof genes and their evolution have been reported in Arabidopsis and several crop species, such as rice (Lijavetzky et al., 2003), bread wheat (Shaw et al., 2009), sorghum (Kushwaha et al., 2011), tomato (Cai et al., 2013), and Chinese cabbage (Ma et al., 2015). However, little information is available on Dof TFs in fruits of substantial economic interest. We have identified 25 MaDof genes in banana based on the Musa genome (D’Hont et al., 2012). Similar to the previous reports (Cai et al., 2013; Ma et al., 2015), alignment of the 25 MaDof proteins showed that they shared a highly conserved Dof domain in the N-terminus (Fig. 1A), although they were highly diverse in length, ranging from 182 to 511 amino acids. The lowest similarity observed was 6.4% while the highest similarity observed was 72.6% (Supplementary Table S2, S3). The 25 MaDofs fell into six out of the nine different subgroups of the previously characterized Dof proteins (Cai et al., 2013; Ma et al., 2015) (Fig. 1B). In addition, except for the Dof domain, another 14 distinct motifs were identified, and were differentially distributed in different subgroups (Supplementary Figs S1 and S2), suggesting that different subgroups of Dof proteins may exhibit diverse functions (Cai et al., 2013; Ma et al., 2015).
The involvement of Dof TFs in various biological processes has been reported, including seed storage protein accumulation, carbon and nitrogen metabolism, light responses, plant morphology, guard cell development, seed germination, and stress responses (Noguero et al., 2013; Gupta et al., 2015). In this study, MaDofs exhibited differential expression patterns during banana fruit ripening. Among the 25 MaDof genes, MaDof10, 23, 24, and 25 were induced during ripening. Further, their expression was also upregulated following ethylene treatment (Fig. 2; Supplementary Fig. S3), indicating that their products might be associated with banana fruit ripening. This is the first evidence of a possible role for Dof TFs in fruit ripening. Dof proteins are suggested to act as TFs based on their nuclear localization and DNA-binding activity (Noguero et al., 2013; Gupta et al., 2015). Consistent with their role as TFs, all the 25 deduced MaDofs have basic amino acid regions that potentially serve as NLSs to target proteins to the nucleus (Fig. 1A). Nuclear localization was effectively confirmed by transient expression of MaDof10, 23, 24, and 25 in tobacco BY-2 protoplasts (Fig. 3). Interestingly, MaDof25 possessed transcriptional activation properties in yeast, while MaDof23 exhibited transcriptional repression activity in vivo (Figs 4 and 6), revealing that MaDofs may act as transcriptional activators or repressors involved in fruit ripening.
Over the last few years, more and more ripening-related TFs, such as MADS-box/RIN, NAC/NOR, CNR, and ERF, have been reported in various fruits (Elitzur et al., 2010; Yin et al., 2010; Klee and Giovannoni 2011; Seymour et al., 2011; Karlova et al., 2014). Extensive efforts have made to identify their downstream targets, especially for tomato MADS-box/RIN, which controls the expression of a wide range of ripening-related genes, including those involved in ethylene biosynthesis and perception, cell wall metabolism, carotenoid formation, aroma formation, and ATP generation (Seymour et al., 2013a, b). ERF TFs are also proposed to modulate fruit ripening by regulating cell wall-modifying genes (Yin et al., 2010). Many genes expressed in relation to banana ripening, particularly with regard to ethylene biosynthesis, pulp softening, and aroma development, have been isolated and characterized, including the MaACS and MaACO gene family (Huang et al., 2006; Inaba et al., 2007), associated with ethylene biosynthesis; MaEXPs, MaXET/XTHs, MaPME, MaPGs, and MaGALs (Trivedi and Nath 2004; Sane et al., 2007; Mbéguié-A-Mbéguié et al., 2009; Asif et al., 2014), associated with cell wall degradation; and MaCAT and MaPDC, associated with aroma formation (Yang et al., 2011). In our recent reports, we demonstrated that MaERFs, MaLBDs, and MaBSD1 TFs are involved in fruit ripening, at least in part via transcriptional regulation of MaACS, MaACO, MaEXP1, and MaEXP2 (Xiao et al., 2013; Ba et al., 2014a, b). However, the transcriptional regulatory networks of these ripening-related genes are still largely unknown. In the present work, we isolated the promoters of 11 ripening-related genes, including MaEXP1/2/3/5, MaXET7, MaPG1, MaPME3, MaPL2, MaGAL, MaCAT, and MaPDC (Supplementary Fig. S5). We showed that MaERF9 is an activator and MaDof23 a repressor of these ripening-related genes (Figs 6 and 7), implying that MaERF9 and MaDof23 may be playing opposite roles or acting antagonistically in the regulation of banana fruit ripening. Whether MaERF9 and MaDof23 regulate these 11 ripening-related genes via direct binding to the (T/A)AAAG elements or GCC-box in their promoters needs to be further investigated.
Often, many different TFs regulate the expression of a particular gene. Therefore, fine-tuning of these different types of TFs is necessary. This is thought to occur through the formation of enhanceosome or repressosome complexes that affect protein–protein and protein–DNA interactions (Martinez and Rao, 2012; Xu et al., 2013). Although multiple ripening-related TFs have been explored, only a few studies have shown them to cooperate in fruit ripening. Recently, RIN was found to interact with FUL1 and FUL2 (Shima et al., 2013), and in vitro protein binding assays showed that FUL homologs, RIN, and tomato AGAMOUS-LIKE1 form DNA binding complexes (Fujisawa et al., 2014). However, whether the tetramer complex of these MADS-box proteins increases or decreases the transcriptional activation of ripening-related genes is largely unknown. Previous studies suggested that Dof TFs form heterodimers with other TFs, such as bZIP, MYB, ZFP, and TCP, and are associated with hormone signalling, stress responses, endosperm development, floral organ abscission, and germination (Zhang et al., 1995; Washio 2003; Diaz et al., 2005; Wei et al., 2010; Rueda-Romero et al., 2012). In the present investigation, we demonstrated that the transcriptional repressor MaDof23 physically interacts with the transcriptional activator MaERF9 (Fig. 5). We found that the trans-activation of MaERF9 or the trans-repression of MaDof23 to its target gene promoters was impaired by their interaction (Fig. 7). Interestingly, ethylene induced MaDof23, but MaDof23 in turn repressed ripening-related genes (Figs 2 and 7). Similarly, SlZFP2 is induced by abscisic acid (ABA), but represses ABA biosynthesis during fruit development by directly suppressing the ABA biosynthesis genes, suggesting that S1ZEP2 acts as a repressor to fine-tune ABA biosynthesis during fruit development (Weng et al., 2015). It could be speculated that MaDof23 plays a role in fine-tuning the regulation of banana fruit ripening by negatively regulating the expression of ripening-related genes, possibly balancing the inductive effects of transcriptional activators such as MaERF9. In addition, it is possible that MaDof23 and MaEFRF9 competitively bind to the promoters of ripening-related genes via an unknown cis-element, leading to the antagonistic action between MaDof23 and MaERF9; however, this speculation need further investigation. Nevertheless, the interaction between MaDof23 and MaERF9 raises an interesting question of why plants use such an antagonistic interaction in fruit ripening. Given that Dofs are one of the largest TF families in plants, efforts should be made to elucidate whether other banana fruit MaDof TFs interact with MaERF9 to form enhanceosomes that synergistically regulate ripening-related genes.
In summary, we identified 25 banana fruit MaDof genes and found they were differentially expressed in pulp tissues during fruit ripening. Transient expression assays indicated that MaDof23 is a transcriptional repressor and physically interacts with MaERF9, a regulator of fruit ripening in banana. These proteins may act antagonistically to regulate the 10 ripening-related genes associated with cell wall degradation and aroma formation. Thus, our results have revealed that MaDofs regulate fruit ripening at least in part by the transcriptional regulation of specific ripening-related genes. Our findings provide new insights into the transcriptional regulatory network of fruit ripening in banana.
Supplementary data
Supplementary materials are available at JXB online.
Table S1. Summary of primers used in this study.
Table S2. Related information for each MaDof gene.
Table S3. Sequence similarities among the different MaDof genes.
Fig. S1. Multilevel consensus sequence identified by MEME among banana fruit MaDof proteins.
Fig. S2. Schematic distribution of conserved motifs in banana fruit MaDofs identified by MEME.
Fig. S3. Hierarchical clustering analysis of 25 MaDof expression profiles in ethylene-treated banana fruit pulp.
Fig. S4. Subcellular localization of MaDof10, 23, 24, and 25 in tobacco BY-2 protoplasts.
Fig. S5. Nucleotide sequences of the promoters of 11 ripening-related genes.
Acknowledgements
We thank Professor Seiichiro Hasezawa (Department of Integrated Biosciences, the University of Tokyo) for the generous gift of tobacco BY-2 suspension cells, Professor Shouyi Chen (Institute of Genetics and Developmental Biology, Chinese Academy of Sciences) and Professor Junping Gao (Department of Ornamental Horticulture, China Agricultural University) for the transient expression vectors, and Dr George P. Lomonossoff (Department of Biological Chemistry, John Innes Centre, Norwich Research Park) for the pEAQ vectors. We also thank Dr Prakash Lakshmanan (Sugar Research Australia) and Dr Yu-hai Cui (Southern Crop Protection and Food Research Centre, Agriculture and Agri-Food Canada) for constructive comments and critical language editing of the manuscript. This work was supported in part by the National Basic Research Program of China (grant No. 2013CB127104), National Natural Science Foundation of China (grant No. 31460535), China Postdoctoral Science Foundation (grant no. 2014M552209), and China Agriculture Research System (grant No. CARS-32-09).
References
- Asif MH, Lakhwani D, Pathak S, Gupta P, Bag SK, Nath P, Ttivedi PK. 2014. Transcriptome analysis of ripe and unripe fruit tissue of banana identifies major metabolic networks involved in fruit ripening process. BMC Plant Biology 14, 316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ba LJ, Shan W, Kuang JF, Feng BH, Xiao YY, Lu WJ, Chen JY. 2014a. The banana MaLBD (LATERAL ORGAN BOUNDARIES DOMAIN) transcription factors regulate EXPANSIN expression and are involved in fruit ripening. Plant Molecular Biology Reporter 32, 1103–1113. [Google Scholar]
- Ba LJ, Shan W, Xiao YY, Chen JY, Lu WJ, Kuang JF. 2014b. A ripening-induced transcription factor MaBSD1 interacts with promoters of MaEXP1/2 from banana fruit. Plant Cell Reports 33, 1913–1920. [DOI] [PubMed] [Google Scholar]
- Bapat VA, Trivedi PK, Ghosh A, Sane VA, Ganapathi TR, Nath P. 2010. Ripening of fleshy fruit: molecular insight and the role of ethylene. Biotechnology Advances 28, 94–107. [DOI] [PubMed] [Google Scholar]
- Cai XF, Zhang YY, Zhang CJ, Zhang TY, Hu TX, Ye J, Zhang JH, Wang TT, Li HX, Ye ZB. 2013. Genome-wide analysis of plant-specific Dof transcription factor family in tomato. Journal of Integrative Plant Biology 55, 552–566. [DOI] [PubMed] [Google Scholar]
- Chen L, Zhong HY, Kuang JF, Li JG, Lu WJ, Chen JY. 2011. Validation of reference genes for RT-qPCR studies of gene expression in banana fruit under different experimental conditions. Planta 234, 377–390. [DOI] [PubMed] [Google Scholar]
- Cherian S, Figueroa CR, Nair H. 2014. ‘Movers and shakers’ in the regulation of fruit ripening: a cross-dissection of climacteric versus non-climacteric fruit. Journal of Experimental Botany 65, 4705–4722. [DOI] [PubMed] [Google Scholar]
- Choudhury SR, Roy S, Nag A, Singh SK, Sengupta DN. 2012. Characterization of an AGAMOUS-like MADS Box protein, a probable constituent of flowering and fruit ripening regulatory system in banana. PLoS One 7, e44361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrales AR, Nebauer SG, Carrillo L, Fernández-Nohales P, Marqués J, Renau-Morata B, Medina J. 2014. Characterization of tomato Cycling Dof factors reveals conserved and new functions in the control of flowering time and abiotic stress responses. Journal of Experimental Botany 65, 995–1012. [DOI] [PubMed] [Google Scholar]
- D’Hont A, Denoeud F, Aury JM, et al. 2012. The banana (Musa acuminata) genome and the evolution of monocotyledonous plants. Nature 488, 213–217. [DOI] [PubMed] [Google Scholar]
- Diaz I, Vicente-Carbajosa J, Abraham Z, Martínez M, Isabel-La Moneda I, Carbonero P. 2002. The GAMYB protein from barley interacts with the DOF transcription factor BPBF and activates endosperm-specific genes during seed development. The Plant Journal 29, 453–464. [DOI] [PubMed] [Google Scholar]
- Diaz I, Martinez M, Isabel-LaMoneda I, Rubio-Somoza I, Carbonero P. 2005. The DOF protein, SAD, interacts with GAMYB in plant nuclei and activates transcription of endosperm-specific genes during barley seed development. Plant Journal 42, 652–662. [DOI] [PubMed] [Google Scholar]
- Elitzur T, Vrebalov J, Giovannoni JJ, Goldschmidt EE, Friedman H. 2010. The regulation of MADS-box gene expression during ripening of banana and their regulatory interaction with ethylene. Journal of Experimental Botany 61, 1523–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa M, Shima Y, Nakagawa H, Kitagawa M, Kimbara J, Nakano T, Kasumi T, Ito Y. 2014. Transcriptional regulation of fruit ripening by tomato FRUITFULL homologs and associated MADS box proteins. Plant Cell 26, 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gapper NE, McQuinn RP, Giovannoni JJ. 2013. Molecular and genetic regulation of fruit ripening. Plant Molecular Biology 82, 575–591. [DOI] [PubMed] [Google Scholar]
- Giovannoni JJ. 2004. Genetic regulation of fruit development and ripening. Plant Cell 16 (Suppl. ), S170–S180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Qiu LJ. 2013. Genome-wide analysis of the Dof transcription factor gene family reveals soybean-specific duplicable and functional characteristics. PLoS One 8, e76809. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gupta S, Malviya N, Kushwaha H, Nasim J, Bisht NC, Singh VK, Yadav D. 2015. Insights into structural and functional diversity of Dof (DNA binding with one finger) transcription factor. Planta 241, 549–562. [DOI] [PubMed] [Google Scholar]
- Hao D, Ohme-Takagi M, Sarai A. 1998. Unique mode of GCC box recognition by the DNA-binding domain of ethylene-responsive element-binding factor (ERF domain) in plant. Journal of Biological Chemistry 273, 26857–26861. [DOI] [PubMed] [Google Scholar]
- Hellens R, Allan A, Friel E, Bolitho K, Grafton K, Templeton M, Karunairetnam S, Gleave A, Laing W. 2005. Transient expression vectors for functional genomics, quantification of promoter activity and RNA silencing in plants. Plant Methods 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hölscher D, Dhakshinamoorthy S, Alexandrov T, et al. 2014. Phenalenone-type phytoalexins mediate resistance of banana plants (Musa spp.) to the burrowing nematode Radopholus similis . Proceedings of the National Academy of Sciences of the United States of America 111, 105–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang FC, Do YY, Huang PL. 2006. Genomic organization of a diverse ACC synthase gene family in banana and expression characteristics of the gene member involved in ripening of banana fruit. Journal of Agriculture and Food Chemistry 54, 3859–3868. [DOI] [PubMed] [Google Scholar]
- Inaba A, Liu X, Yokotani N, Yamane M, Lu WJ, Nakano R, Kubo Y. 2007. Differential feedback regulation of ethylene biosynthesis in pulp and peel tissues of banana fruit. Journal of Experimental Botany 258, 1047–1057. [DOI] [PubMed] [Google Scholar]
- Karlova R, Chapman N, David K, Angenent GC, Seymour GB, de Maagd RA. 2014. Transcriptional control of fleshy fruit development and ripening. Journal of Experimental Botany 65, 4527–4541. [DOI] [PubMed] [Google Scholar]
- Klee HJ, Giovannoni JJ. 2011. Genetics and control of tomato fruit ripening and quality attributes. Annual Review of Genetics 45, 41–59. [DOI] [PubMed] [Google Scholar]
- Krebs J, Mueller-Roeber B, Ruzicic S. 2010. A novel bipartite nuclear localization signal with an atypically long linker in DOF transcription factors. Journal of Plant Physiology 167, 583–586. [DOI] [PubMed] [Google Scholar]
- Kushwaha H, Gupta S, Singh VK, Rastogi S, Yadav D. 2011. Genome wide identification of Dof transcription factor gene family in sorghum and its comparative phylogenetic analysis with rice and Arabidopsis . Molecular Biology Report 38, 5037–5053. [DOI] [PubMed] [Google Scholar]
- Lijavetzky D, Carbonero P, Vicente-Carbajosa J. 2003. Genomewide comparative phylogenetic analysis of the rice and Arabidopsis Dof gene families. BMC Evolutionary Biology 3, 17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindemose S, O’Shea C, Jensen MK, Skriver K. 2013. Structure, function and networks of transcription factors involved in abiotic stress responses. International Journal of Molecular Sciences 14, 5842–5878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu JH, Zhang J, Jia CH, Zhang JB, Wang JS, Yang ZX, Xu BY, Jin ZQ. 2013. The interaction of banana MADS-box protein MuMADS1 and ubiquitin-activating enzyme E-MuUBA in post-harvest banana fruit. Plant Cell Reports 32, 129–137. [DOI] [PubMed] [Google Scholar]
- Ma J, Li MY, Wang F, Tang J, Xiong AS. 2015. Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genomics 16, 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martel C, Vrebalov J, Tafelmeyer P, Giovannoni JJ. 2011. The tomato MADS-box transcription factor RIPENING INHIBITOR interacts with promoters involved in numerous ripening processes in a COLORLESS NONRIPENING-dependent manner. Plant Physiology 157, 1568–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez GJ, Rao A. 2012. Immunology. Cooperative transcription factor complexes in control. Science 338, 891–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mbéguié-A-Mbéguié D, Hubert O, Fils-Lycaon B, Chillet M, Baurens FC. 2008. EIN3-like gene expression during fruit ripening of Cavendish banana (Musa acuminata cv. Grande naine). Physiologia Plantarum 133, 435–448. [DOI] [PubMed] [Google Scholar]
- Mbéguié-A-Mbéguié D, Hubert O, Baurens FC, Matsumoto T, Chillet M, Fils-Lycaon B, Sidibé-Bocs S. 2009. Expression patterns of cell wall-modifying genes from banana during fruit ripening and in relationship with finger drop. Journal of Experimental Botany 60, 2021–2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noguero M, Atif RM, Ochatt S, Thompson RD. 2013. The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Science 209, 32–45. [DOI] [PubMed] [Google Scholar]
- Ohta M, Matsui K, Hiratsu K, Shinshi H, Ohme-Takagi M. 2001. Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959–1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riechmann JL, Ratcliffe OJ. 2000. A genomic perspective on plant transcription factors. Current Opinion in Plant Biology 3, 423–434. [DOI] [PubMed] [Google Scholar]
- Rueda-Romero P, Barrero-Sicilia C, Gómez-Cadenas A, Carbonero P, Oñate-Sánchez L. 2012. Arabidopsis thaliana DOF6 negatively affects germination in non-after-ripened seeds and interacts with TCP14. Journal of Experimental Botany 63, 1937–1949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sainsbury F, Thuenemann EC, Lomonossoff GP. 2009. pEAQ: versatile expression vectors for easy and quick transient expression of heterologous proteins in plants. Plant Biotechnology Journal 7, 682–693. [DOI] [PubMed] [Google Scholar]
- Sane AVA, Sane AP, Nath P. 2007. Multiple forms of a-expansin genes are expressed during banana fruit ripening and development. Postharvest Biology and Technology 45, 184–192. [Google Scholar]
- Sasaki N, Matsumaru M, Odaira S, Nakata A, Nakata K, Nakayama I, Yamaguchi K, Nyunoya H. 2015. Transient expression of tobacco BBF1-related Dof proteins, BBF2 and BBF3, upregulates genes involved in virus resistance and pathogen defense. Physiological and Molecular Plant Pathology 89, 70–77. [Google Scholar]
- Sato S, Tabata S, Hirakawa H, et al. 2012. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour GB, Østergaard L, Chapman NH, Knapp S, Martin C. 2013a. Fruit development and ripening. Annual Review of Plant Biology 64, 219–241. [DOI] [PubMed] [Google Scholar]
- Seymour GB, Chapman NH, Chew BL, Rose JKC. 2013b. Regulation of ripening and opportunities for control in tomato and other fruits. Plant Biotechnology Journal 11, 269–278. [DOI] [PubMed] [Google Scholar]
- Seymour GB, Ryder CD, Cevik V, Hammond JP, Popovich A, King GJ, Vrebalov J, Giovannoni JJ, Manning K. 2011. A SEPALLATA gene is involved in the development and ripening of strawberry (Fragaria × ananassa Duch.) fruit, a non-climacteric tissue. Journal of Experimental Botany 62, 1179–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan W, Kuang JF, Chen L, Xie H, Peng HH, Xiao YY, Li XP, Chen WX, He QG, Chen JY, Lu WJ. 2012. Molecular characterization of banana NAC transcription factors and their interactions with ethylene signalling component EIL during fruit ripening. Journal of Experimental Botany 63, 5171–5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw LM, McIntyre CL, Gresshoff PM, Xue GP. 2009. Members of the Dof transcription factor family in Triticumae stivum are associated with light-mediated gene regulation. Functional & Integrative Genomics 9, 485–498. [DOI] [PubMed] [Google Scholar]
- Shima Y, Kitagawa M, Fujisawa M, Nakano T, Kato H, Kimbara J, Kasumi T, Ito Y. 2013. Tomato FRUITFULL homologues act in fruit ripening via forming MADS-box transcription factor complexes with RIN. Plant Molecular Biology 82, 427–438. [DOI] [PubMed] [Google Scholar]
- Trivedi PK, Nath P. 2004. MaExp1, an ethylene-induced expansin from ripening banana fruit. Plant Science 167, 1351–1358. [Google Scholar]
- Udvardi MK, Kakar K, Wandrey M, Montanari O, Murray J, Andriankaja A, Zhang JY, Benedito V, Hofer JM, Chueng F, Town CD. 2007. Legume transcription factors: global regulators of plant development and response to the environment. Plant Physiology 144, 538–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh J, Park SW. 2015. Genome-wide analysis and expression profiling of DNA-binding with one zinc finger (Dof) transcription factor family in potato. Plant Physiology and Biochemistry 94, 73–85. [DOI] [PubMed] [Google Scholar]
- Wan CY, Wilkins TA. 1994. A modified hot borate method significantly enhances the yield of high quality RNA from cotton (Gossypium hirsutum L.). Analytical Biochemistry 223, 7–12. [DOI] [PubMed] [Google Scholar]
- Washio K. 2003. Functional dissections between GAMYB and Dof transcription factors suggest a role for protein-protein associations in the gibberellin-mediated expression of the RAmy1A gene in the rice aleurone. Plant Physiology 133, 850–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei PC, Tan F, Gao XQ, Zhang XQ, Wang GQ, Xu H, Li LJ, Chen J, Wang XC. 2010. Overexpression of AtDOF4.7, an Arabidopsis DOF family transcription factor, induces floral organ abscission deficiency in Arabidopsis . Plant Physiology 153, 1031–1045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng L, Zhao F, Li R, Xu C, Chen K, Xiao H. 2015. The zinc finger transcription factor SlZFP2 negatively regulates abscisic acid biosynthesis and fruit ripening in tomato. Plant Physiology 167, 931–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA, Hahn MW, Abouheif E, Balhoff JP, Pizer M, Rockman MV, Romano LA. 2003. The evolution of transcriptional regulation in eukaryotes. Molecular Biology and Evolution 20, 1377–1419. [DOI] [PubMed] [Google Scholar]
- Xiao YY, Chen JY, Kuang JF, Shan W, Xie H, Jiang YM, Lu WJ. 2013. Banana ethylene response factors are involved in fruit ripening through their interactions with ethylene biosynthesis genes. Journal of Experimental Botany 64, 2499–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu ZY, Kim SY, Hyeon do Y, Kim DH, Dong T, Park Y, Jin JB, Joo SH, Kim SK, Hong JC, Hwang D, Hwang I. 2013. The Arabidopsis NAC transcription factor ANAC096 cooperates with bZIP-type transcription factors in dehydration and osmotic stress responses. Plant Cell 25, 4708–4724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamasaki K, Kigawa T, Seki M, Shinozaki K, Yokoyama S. 2013. DNA-binding domains of plant-specific transcription factors: structure, function, and evolution. Trends in Plant Science 18, 267–276. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S, Izui K. 1993. Molecular cloning of two DNA-binding proteins of maize that are structurally different but interact with the same sequence motif. Journal of Biological Chemistry 268, 16028–16036. [PubMed] [Google Scholar]
- Yanagisawa S. 2002. The Dof family of plant transcription factors. Trends in Plant Science 7, 555–560. [DOI] [PubMed] [Google Scholar]
- Yanagisawa S. 2004. Dof domain proteins: Plant-specific transcription factors associated with diverse phenomena unique to plants. Plant Cell Physiology 45, 386–391. [DOI] [PubMed] [Google Scholar]
- Yang XT, Song J, Fillmore S, Pang XQ, Zhang ZQ. 2011. Effect of high temperature on color, chlorophyll fluorescence and volatile biosynthesis in green-ripe banana fruit. Postharvest Biology and Technology 62, 246–257. [Google Scholar]
- Yin XR, Allan AC, Chen KS, Ferguson IB. 2010. Kiwifruit EIL and ERF genes involved in regulating fruit ripening. Plant Physiology 153, 1280–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang B, Chen W, Foley R, Buttner M, Singh K. 1995. Interactions between distinct types of DNA binding proteins enhance binding to ocs element promoter sequences. Plant Cell 7, 2241–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Jin J, Tang L, Zhao Y, Gu X, Gao G, Luo J. 2010. PlantTFDB 2.0: Update and improvement of the comprehensive plant transcription factor database. Nucleic Acids Research 39, 1114–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.