Highlight
A JAZ7 T-DNA activation mutant confers increased JA-sensitivity, up-regulated defense and JA-mediated gene expression, and increased susceptibility to two pathogens that disrupt host JA-responses, Fusarium oxysporum and Pst DC3000.
Key words: Defense, ERF-associated amphiphilic repressor, fungal pathogen, methyl jasmonate, Pseudomonas syringae, TOPLESS, transcriptional repression.
Abstract
In Arabidopsis, jasmonate (JA)-signaling plays a key role in mediating Fusarium oxysporum disease outcome. However, the roles of JASMONATE ZIM-domain (JAZ) proteins that repress JA-signaling have not been characterized in host resistance or susceptibility to this pathogen. Here, we found most JAZ genes are induced following F. oxysporum challenge, and screening T-DNA insertion lines in Arabidopsis JAZ family members identified a highly disease-susceptible JAZ7 mutant (jaz7-1D). This mutant exhibited constitutive JAZ7 expression and conferred increased JA-sensitivity, suggesting activation of JA-signaling. Unlike jaz7 loss-of-function alleles, jaz7-1D also had enhanced JA-responsive gene expression, altered development and increased susceptibility to the bacterial pathogen Pst DC3000 that also disrupts host JA-responses. We also demonstrate that JAZ7 interacts with transcription factors functioning as activators (MYC3, MYC4) or repressors (JAM1) of JA-signaling and contains a functional EAR repressor motif mediating transcriptional repression via the co-repressor TOPLESS (TPL). We propose through direct TPL recruitment, in wild-type plants JAZ7 functions as a repressor within the JA-response network and that in jaz7-1D plants, misregulated ectopic JAZ7 expression hyper-activates JA-signaling in part by disturbing finely-tuned COI1-JAZ-TPL-TF complexes.
Received 5 November 2015; Revised 13 January 2016; Accepted 20 January 2016
Introduction
The root-infecting fungal pathogen Fusarium oxysporum is responsible for vascular wilt disease in over 100 different plant species, including bananas (Musa spp.), cotton (Gossypium spp.), grain legumes and horticultural crops such as tomato (Lycopersicum esculentum) (Di Pietro et al., 2003; Agrios, 2005; Berrocal-Lobo and Molina, 2008). This pathogen also infects Arabidopsis (Arabidopsis thaliana) where the pathogen-host interaction can be readily studied in a model system.
Contrasting roles for jasmonate (JA) signaling and JA-mediated defense in Arabidopsis resistance to F. oxysporum have been proposed (Kidd et al., 2009; Thatcher et al., 2009). Firstly, activation of JA-mediated defense responses promotes resistance to this pathogen, most likely due to direct antimicrobial activities. Increased resistance to F. oxysporum can be achieved in transgenic plants through the over-expression of JA-responsive defense gene expression (e.g. thionins; Thi2.1) (Epple et al., 1997; Chan et al., 2005), or manipulation of transcription factors that activate JA-mediated defenses (e.g. defensins and chitinases; PDF1.2, CHIB). For example, mutation of MYC2, a key regulator of downstream JA-defense signaling, mutation of LBD20, a MYC2-regulated transcription factor, or overexpression of the Ethylene Response Factors ERF1 and AtERF2, activators of JA-defenses, results in up-regulated expression of a specific subset of JA-dependent defense genes and increased resistance to F. oxysporum (Berrocal-Lobo et al., 2002; Anderson et al., 2004; McGrath et al., 2005; Thatcher et al., 2012a). Secondly, non-defensive aspects of JA-signaling such as JA-mediated senescence appear to promote susceptibility to this pathogen (Berrocal-Lobo and Molina 2004; McGrath et al., 2005; Kidd et al., 2009; Thatcher et al., 2009, 2012a). It is proposed that in wild-type plants both defensive and non-defensive aspects of JA-signaling are activated following F. oxysporum infection but that non-defensive aspects have greater contribution to disease outcome (Thatcher et al., 2009).
Upstream of the MYC2 and ERF transcription factors in the JA-signaling pathway is the F-box protein CORONATINE INSENSITIVE 1 (COI1), which together with JASMONATE ZIM DOMAIN (JAZ) proteins, perceives the JA-signal and forms part of the Skp1/Cullin/F-box (SCF) E3 ubiquitin ligase complex SCFCOI1-JAZ (Yan et al., 2009; Sheard et al. 2010). JAZ proteins provide the connection between perception of the JA signal in the SCFCOI1-JAZ receptor complex, and downstream transcriptional regulators such as MYC2. In the absence of JA or under low JA levels, JAZ proteins repress transcriptional activators such as MYC2, MYC3 and MYC4, and/or MYC-like transcriptional repressors such as bHLH003/JA-ASSOCIATED MYC2-LIKE 3 (JAM3), bHLH013/JAM2 and bHLH017/JAM1, thereby interfering with the expression of JA-responsive genes. Upon increased JA levels, the ubiquitin-mediated degradation of JAZ proteins leads to the release of these transcription factors from repression (Chini et al., 2007; Thines et al., 2007; Katsir et al., 2008; Melotto et al., 2008; Fernandez-Calvo et al., 2011; Nakata and Ohme-Takagi, 2013; Nakata et al., 2013; Sasaki-Sekimoto et al., 2013, 2014; Song et al., 2013; Fonseca et al., 2014). Although JAZ proteins characterized to date function as repressors of JA-responses, apart from JAZ5, JAZ6, JAZ7, JAZ8 and the non-conventional JAZ13, most do not contain known repression motifs. They form repressor complexes by recruiting the co-repressor TOPLESS (TPL) and TPL-related proteins. This recruitment is mediated through binding of the JAZ ZIM domain to the adaptor protein NINJA (novel interactor of JAZ), which contains an ERF-associated amphiphilic repressor (EAR) motif to recruit TPL (Kagale et al., 2010; Pauwels et al., 2010; Arabidopsis Interactome Mapping Consortium, 2011; Causier et al., 2012; Shyu et al. 2012). For recent reviews and updates on JAZ proteins and JA-signaling, see Kazan and Manners (2012), Wager and Browse (2012), Wasternack and Hause (2013) and Sasaki-Sekimoto et al. (2014).
Mutation of COI1 and subsequent lack of JA-induced defenses results in enhanced susceptibility to most fungal necrotrophs (e.g. Botrytis cinerea, Alternaria brassicicola, Thomma et al., 1998). Interestingly however, COI1 confers susceptibility to F. oxysporum with the coi1 mutant displaying a near-immune like resistance to this pathogen (Thatcher et al., 2009). coi1-mediated resistance to F. oxysporum is therefore independent of JA-dependent defense gene expression but correlates with compromised non-defensive aspects of JA-dependent responses such as reduced expression of some senescence and oxidative-stress associated genes. Other mutants with compromised JA-defenses but strong resistance to F. oxysporum include pft1 carrying a mutation in the MED25 gene encoding a subunit of the RNA polymerase II-interacting MEDIATOR complex (Kidd et al., 2009; Cevik et al., 2012). These results imply F. oxysporum hijacks the host JA-signaling pathway to promote disease symptom development.
The key role of JAZ repressors in linking COI1 and downstream transcriptional responses suggests these proteins may also play key roles in mediating disease outcome to F. oxysporum. Indeed, JAZ expression is induced by other pathogens (Pseudomonas syringae pv tomato, Pst), herbivory and wounding (Chini et al., 2007; Thines et al., 2007; Chung et al. 2008; Demianski et al., 2012). Potential redundancy amongst the 13 JAZ family members has, however, hampered the determination of functional roles for individual members. C-terminal truncated versions of some JAZ proteins generated from alternate splicing, or in domain-deletion mutants, results in a reduced capacity to bind COI1 leading to stabilization of the JAZ protein. These mutations confer phenotypes such as reduced JA-sensitivity, compromised resistance to herbivory, and/or increased resistance to Pst (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Chung et al., 2008; Chung and Howe, 2009). Further, Chung et al. (2011) found all JAZs except JAZ1, JAZ7 and JAZ8 contain a conserved intron that if retained, modifies the COI1-binding motif, inhibiting COI1-mediated degradation and producing dominant JAZ repressors. Altered JA-responses from overexpression or removal of JAZ proteins has only been observed for overexpression of JAZ8 and the recently identified JAZ13 (both resulting in reduced JA-sensitivity) or T-DNA or RNAi knockdown lines of jaz1 or jaz10 (resulting in increased JA-sensitivity and/or increased resistance to the fungal pathogen Botrytis cinerea) (Yan et al., 2007; Grunewald et al., 2009; Cerrudo et al., 2012; Demianski et al., 2012; Shyu et al., 2012; Leone et al., 2014; Thireault et al., 2015).
In this report, we examined the roles of JAZ family members during the Arabidopsis-F. oxysporum interaction through the characterization of JAZ gene expression, and the analysis of Arabidopsis JAZ T-DNA insertion lines. We identified a unique JAZ7 allele that confers increased susceptibility to F. oxysporum and Pst. Interestingly, additional work revealed the T-DNA inserted into the JAZ7 promoter in this mutant caused constitutive JAZ7 expression (jaz7-1D), conferring activation of JA-signaling and increased JA-sensitivity. However, we demonstrate JAZ7 contains a functional EAR repressor motif, recruiting the co-repressor TPL and repressing transcriptional activation. Further, JAZ7 interacted with both transcriptional activators and repressors of JA-signaling. Based on these results, we propose the misregulated JAZ7 expression in jaz7-1D plants resulting from the JAZ7 T-DNA promoter insertion activates JA-signaling conferring increased JA-sensitivity through recruitment of TPL to specific transcriptional regulators, and disturbing the function of proteins acting within the multi-protein COI1-JAZ-TPL-TF complex.
Materials and methods
Plant material and growth conditions
Unless otherwise specified, all experiments were conducted with the A. thaliana Columbia-0 (Col-0) accession grown under a short daylight regime (8h light:16h dark) at 21ºC as described previously (Thatcher et al., 2009). The T-DNA insertion mutants (Alonso et al., 2003; Woody et al., 2007) coi1 (SALK_035548), jaz7-1D (SALK_040835), jaz7-1 (WiscDsLox7H11) and other jaz insertion lines (Supplementary Table S1 available at JXB online) were obtained from the Arabidopsis Biological Resource Centre (ABRC) or the Nottingham Arabidopsis Stock Centre (NASC). T-DNA mutants were confirmed for correct loci insert and homozygous state. Backcrossed, double or triple jaz insertion lines were all confirmed by PCR. For generation of JAZ7 overexpression (35S:JAZ7) lines, the JAZ7 CDS was amplified using JAZ7-HindIII-F and JAZ7-EcoRI-R, cloned into pKEN (McGrath et al., 2005), mobilized into Agrobacterium AGL1 and transformed into Col-0 by floral dipping. Transgenic plants were selected on 50mg l−1 Pestanal (glufosinate-ammonium) (Riedel-de Haen, Seelze, Germany) and resulting T3 lines used in subsequent experiments. For generation of 35S:CUC1-JAZ7 EAR plants the JAZ7 EAR domain with an added stop codon was first cloned into pKEN (McGrath et al., 2005) by annealing and fill-in of the two primers JAZ7-EAR-HIII-Sma-F and JAZ7_EAR_STOP-EcoRI-R to produce 35S:JAZ7 EAR pKEN. The length of the JAZ7 EAR domain was based on the SRDXEAR sequence (Hiratsu et al., 2003). The CUC1 CDS was amplified using CUC1-HIII-F and CUC1-STOPdel-Sma-R, which has the CUC1 stop codon removed, cloned into 35S:JAZ7 EAR pKEN to make 35S:CUC1-JAZ7 EAR pKEN, mobilized into Agrobacterium AGL1, and transformed into Col-0. Primers for the generation of transgenic plants are listed in Supplementary Table S2.
Pathogen assays
Root-dip inoculations on 3–4-week-old plants with a 1×106 cell ml−1 spore suspension of F. oxysporum strain Fo5176 (Thatcher et al., 2012b ) were performed as described previously (Thatcher et al., 2009). Pseudomonas syringae assays were performed with the strain P. syringae pv. tomato DC3000 (Pst DC3000) and syringe infiltrated into leaves at 5×106 cells ml−1. Infiltrated plants were incubated at 28°C (16h light/8h dark) under a clear plastic dome and bacterial growth quantified as previously described (Gleason et al., 2011). Alternaria brassicicola assays were performed as described in Gleason et al. (2011) using 5×106 spores ml−1 of the isolate UQ4273.
F. oxysporum culture filtrate assay
F. oxysporum culture filtrate assays were performed as per Thatcher et al. (2012a) on 15 leaves per line.
Flowering time
Flowering time experiments were performed according to Kidd et al. (2009) under short-day conditions (8h light/16h dark).
MeJA root elongation inhibition assays
Seeds of wild-type, jaz7-1D, jaz7-1 or JAZ7-OX lines were surface sterilized and plated onto MS media in either the presence or absence of 50 µM MeJA. Root length was measured on 7-day-old seedlings using ImageJ (Schneider et al., 2012).
Quantitative RT-PCR
Quantitative-RT-PCR (qRT-PCR) experiments were performed on tissue collected after control, F. oxysporum (see ‘Pathogen assays’) or MeJA treatment (see ‘Microarray analysis’). Three biological replicates were taken for all experiments comprising tissue pooled from 5–30 plants. RNA extraction, cDNA synthesis and Q-RT-PCR were conducted as described by McGrath et al. (2005) using an Applied Biosystems 7900HT Fast Real-Time PCR System (Foster City, CA) or by Thatcher et al. (2015) using a CFX384 (Bio-Rad) system. Absolute gene expression levels relative to the previously validated reference genes β-actin 2, β-actin 7 and β-actin 8 (At1g49240, At3g18780 and At5g09810, respectively) were used for each cDNA sample using the equation: relative ratio gene of interest/actin=(Egene-Ct gene)/(Eactin-Ct actin) where Ct is the cycle threshold value. The gene specific primer sequences are listed in Supplementary Table S3.
Microarray analysis
Four independent biological replicates each consisting of shoot material from 20 wild-type and jaz7-1D plants were harvested 6h after mock or MeJA treatments. Treatment involved enclosing trays of 4-week-old soil-grown plants under clear plastic covers with a treated cotton ball attached to the inside of the cover, either 1ml of mock solution (100% ethanol) or 1ml of 5% MeJA dissolved in 100% ethanol, and sealing each tray in two layers of opaque plastic bags. Total RNA was extracted (RNeasy Plant Mini Kit, Qiagen), then labeled, hybridized, washed and scanned by the Australian Genome Research Facility (AGRF) (Melbourne, Australia) onto 16 ATH1 GeneChip arrays and the resulting data analyzed using GenespringGX 7.3.1 (Agilent) as previously described (Dombrecht et al., 2007). Briefly, the raw CEL files were normalized using the RMA algorithm, and then the resulting expression values were normalized per chip to the median across all chips. The microarray data was also analyzed using a two-way analysis of variance (ANOVA; P<0.05) on the entire dataset with the inclusion of the Benjamini and Hochberg false discovery rate (FDR) (microarray data is deposited under accession number GSE61884 at the NCBI Gene Expression Omnibus). Gene Ontology (GO) term enrichment analysis was performed using agriGO v1.2 (Du et al., 2010) using the default FDR (P≤0.05) determined P-value significance. Functional annotations of genes and AGI symbols were sourced from TAIR9 datasets.
Y2H assays
For Y2H experiments, JAZ7, JAZ5, JAZ8, MYC2, MYC3, MYC4, TPL and JAM1 were PCR-amplified from Arabidopsis cDNA (accession Col-0) using the primers in Supplementary Table S2 followed by second amplification with pAttB1 and pAttB2, and cloned into the pDONRZeo plasmid (Invitrogen). The JAZ7mEAR motif was generated by mutating the conserved leucine residues of the EAR motif to alanine using the QuickChange II Site Directed Mutagenesis Kit (Agilent Technologies) following the manufacturer’s recommendations. The entry clones were then recombined with the Gateway-compatible yeast two-hybrid (Y2H) vectors derived from pGADT7 and pGBKT7 (Clontech). Y2H assays were performed using Clontech’s Matchmaker system with strain AH109.
Co-IP assays
JAZ7, JAZ7mEAR, JAZ5 and JAZ8 were constructed as described for Y2-H assays except JAZ7-R2, JAZ8-R2 or JAZ5-R2 (Supplementary Table S2) were used as reverse primers in order to remove stop codons and cloned into pDONRZeo plasmid (Invitrogen). The entry clones were recombined with the Gateway-compatible pEarleyGate 101 (with 35S promoter and C-terminal YFP fusion tag), and GFP cloned into pEarleyGate100 used as control (Earley et al., 2006). TPL was amplified from Col-0 gDNA and cloned into pICH47742 (with 35S-promoter and C-terminal 4×Myc fusion tag) using the Golden Gate assembly method (Engler et al., 2008). Five-week-old N. benthamiana plants were used for Agrobacterium tumefaciens-mediated transient expression of indicated constructs as described previously (Cevik and Kazan, 2013). Co-immunoprecipitation experiments were carried out as described previously (Sohn et al., 2012). Leaf samples were harvested 2 d post-inoculation, and total protein extracts were incubated with 20 μl GFP-affinity matrix (Chromotek) for immunoprecipitation. HRP-conjugated anti-GFP antibody (Santa Cruz) and anti-c-Myc antibody (Santa Cruz) were used for immunoblot analyses.
Transcriptional activation assays
Full length MYC3 and MYC4 were used to construct effector plasmids by fusing with the yeast GAL4 DNA-binding domain (GAL4DB)-coding region under the control of the CaMV35S promoter into the pGAL4DBGW vector (Cevik et al., 2012). Full length JAZ7, JAZ7mEAR and JAZ8 were cloned into the Gateway-compatible pJIT60 vector. pGAL4UAS:GUS was used as the reporter plasmid. The plasmid p2X35S:fLUC was used as a control to normalize reporter gene activity. Transcription activation assays by particle bombardment into rosette leaves of 5–6-week-old Col-0 plants were carried out as described by Cevik et al. (2012). Luminescence and fluorescence were determined with a Varioskan Flash multiplate reader (Thermo Scientific).
Results
JAZ genes are differentially expressed both locally and systemically in response to F. oxysporum
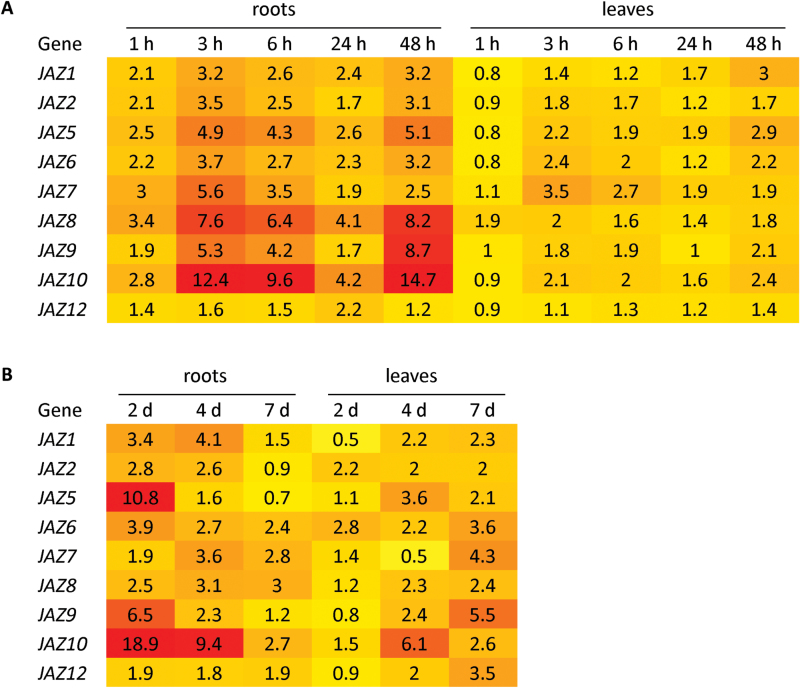
To examine the role of JAZ genes in the Arabidopsis-F. oxysporum interaction, we first determined if their expression was responsive to pathogen infection. As F. oxysporum infects through the roots and spreads upwards into leaf tissues where the disease symptoms manifest, we examined root and leaf tissues separately, and initially sampled 1–48h post-inoculation (Fig. 1A) as JAZ expression can be rapidly induced by JA signals. Most JAZ genes exhibited higher inductions over control treatments in roots compared to leaves, where expression peaked at 3h post-inoculation, then rose again at 48h post-inoculation. The largest inductions of 5- to 15-fold were observed for JAZ5, JAZ7, JAZ8, JAZ9 and JAZ10. The expression of JAZ3, JAZ4 and JAZ11 did not differ from those observed in control treated samples (data not shown). These results suggest some JAZ members may have roles in mediating responses to early stages of F. oxysporum infection, particularly in root tissues.
Fig. 1.
Differential JAZ gene expression is induced after F. oxysporum inoculation. Heat map of JAZ gene expression in roots or leaves of F. oxysporum inoculated wild-type plants over (A) a 1–48h or (B) 2–7 d time-course. Expression is relative to control treatment. JAZ3, JAZ4 and JAZ11 expression did not differ between inoculation or control treatments and are not shown. Values were determined by quantitative RT-PCR from three biological replicates consisting of pools of 10–20 plants.
As F. oxysporum disease symptoms become evident in the leaves of wild-type Col-0 plants at 7–10 d post-inoculation, we were also interested to determine the persistence of JAZ expression at later time-points. In this later time-course (2, 4 and 7 d post-inoculation), fold-inductions were again typically higher in root tissues with JAZ6, JAZ7, JAZ9, JAZ10 and JAZ12 also highly induced in leaf tissues at 7 d post-inoculation (Fig. 1B). As with the earlier time-course, expression of JAZ3, JAZ4 or JAZ11 was non-responsive (data not shown). In summary, the differential expression patterns of multiple JAZ genes in response to F. oxysporum suggests that at least some members may have roles in modulating host responses to this pathogen.
Screening of JAZ T-DNA insertion lines identifies a JAZ7 line with increased susceptibility to F. oxysporum
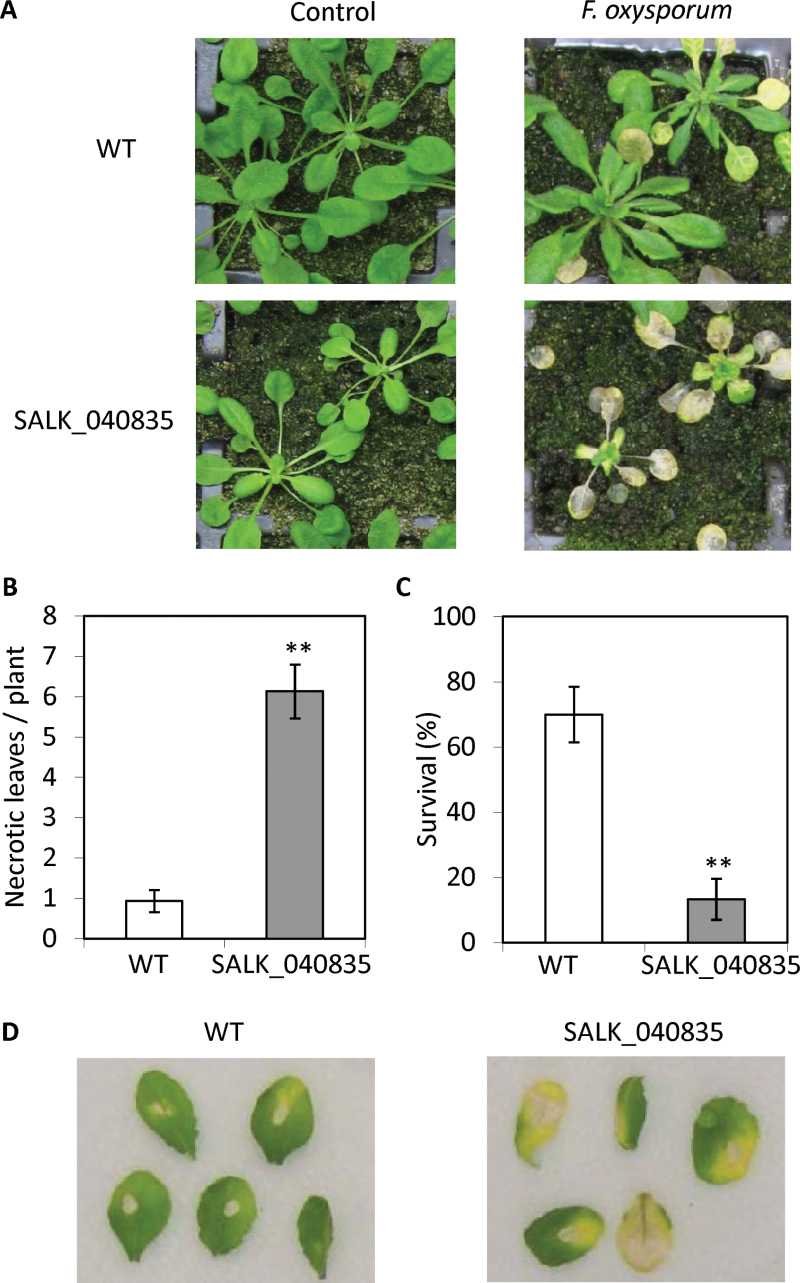
To investigate possible roles of JAZ proteins in F. oxysporum resistance, we screened publically available JAZ T-DNA insertion lines with F. oxysporum. With the exception of JAZ1, JAZ3, JAZ6, JAZ7, JAZ8 and JAZ11, the remaining T-DNA insertions are located within the transcribed regions of the JAZ genes (Supplementary Fig. S1; de Torres Zabala et al., 2015). Of the insertion lines screened, one line (SALK_040835) with a T-DNA insertion in the promoter of the JAZ7 gene (351bp upstream of transcription start site) displayed significantly (P<0.01, Student’s t-test) higher susceptibility to F. oxysporum than wild-type in three independent inoculation experiments (Fig. 2). As shown in Fig. 2A–C, both disease symptom development and plant death in inoculated SALK_040835 plants were significantly higher than those of wild-type plants. The SALK_040835 plants also had a smaller rosette size (Fig. 2A). No significantly altered resistance to F. oxysporum was evident in the remaining T-DNA insertion lines tested (Supplementary Fig. S2).
Fig. 2.
SALK_040835 is highly susceptible to F. oxysporum. Wild-type (WT) and SALK_040835 were inoculated with F. oxysporum and disease symptoms monitored over 21 d. (A) Representative images of WT and SALK_040835 plants 10 dpi or control treatment. (B) Necrotic leaves per plant at 10 d and (C) survival rates at 21 d post-inoculation. Values are averages ±SE (n=30). Asterisks indicate values that are significantly different (**, P<0.01; Student’s t-test) from WT. Similar results were obtained in independent experiments. (D) F. oxysporum culture filtrate was applied to detached WT and SALK_040835 leaves. Representative leaves are shown from three replicates 6 d post-treatment. Control treatments of potato dextrose broth (PDB) and H2O showed no phenotype (not shown). Similar results were obtained in an independent experiment.
The culture filtrate of F. oxysporum is known to trigger a leaf-chlorosis phenotype that is closely correlated with the F. oxysporum resistance/susceptibility phenotypes (Thatcher et al., 2009, 2012a ). We found culture filtrate-treated leaves from SALK_040835 developed an earlier and stronger senescence and chlorosis phenotype than leaves from wild-type plants (Fig. 2D). These results suggest JAZ7 may be involved in regulating plant defense responses to F. oxysporum.
The SALK_040835 line shows elevated JAZ7 expression
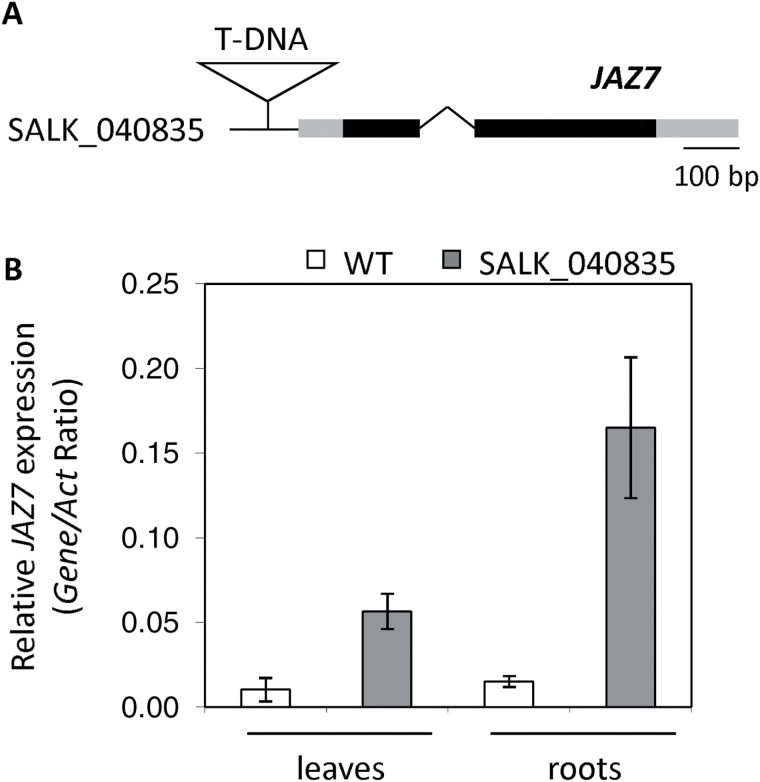
To determine how the T-DNA inserted into the promoter of JAZ7 (Fig. 3A) in SALK_040835 affects JAZ7 expression, we examined JAZ7 transcript levels in SALK_040835 and wild-type plants. Basal JAZ7 expression in the roots and leaves of SALK_040835 was 10.8- and 5.4-fold higher, respectively, than those of wild-type plants (Fig. 3B). This suggests SALK_040835 contains an activation-tagged JAZ7 allele. We therefore designated SALK_040835 as jaz7-1D. From the screening of over 30 plants, we were unable to isolate homozygous SALK_040835 lines suggesting jaz7-1D acts dominantly and that homozygous lines of this insertion mutant may be lethal, the latter of which we confirmed via detection of seed aborts in jaz7-1D siliques (Supplementary Fig. S3A). Independently, Yan et al. (2014) also recently reported SALK_040835C as a JAZ7 activation mutant and with small stature. Progeny from two other separately isolated SALK_040835 lines also showed small rosette size and increased susceptibility to F. oxysporum.
Fig. 3.
SALK_040835 shows elevated JAZ7 expression. (A) Schematic representation of the SALK_040835 T-DNA insertion line. The insertion (open triangle) lies upstream of the JAZ7 transcription start site. 5′ and 3′ UTR are shaded in gray, exons in black and the only intron as a removed segment. (B) JAZ7 expression was examined in the leaves and roots of wild-type (WT) and SALK_040835 plants. Values are averages ±SE of three biological replicates comprising 5–10 plants. Gene expression levels are relative to the internal control β-actin genes.
Recent re-sequencing of SALK T-DNA insertion lines (O’Malley et al., 2014, unpublished) suggests SALK_040835 may contain other insertions, and this raises the possibility that these additional insertions, if confirmed, may contribute to the jaz7-1D phenotypes. One insertion is proposed to be located within the promoter of At2g47780 (rubber elongation factor protein), one in the coding sequence of At2g47790 (GIGANTUS), and the others in intergenic regions. We therefore screened SALK_040835/jaz7-1D plants by PCR for insertions in At2g47780 and At2g47790 but were unable to identify any insertion in At2g47790, while all plants were heterozygous for the At2g47780 insertion. We also examined the Col-0 and SALK_040835C RNA sequencing data of Yan et al. (2014) to compare transcript levels of At2g47780 and At2g47790, and genes flanking the possible intergenic T-DNA insertions, but found no differential levels or truncated transcripts. Together, these results support the conclusion that the phenotypes observed in jaz7-1D are related to the JAZ7 promoter insertion.
A null mutation in JAZ7 does not affect resistance to F. oxysporum
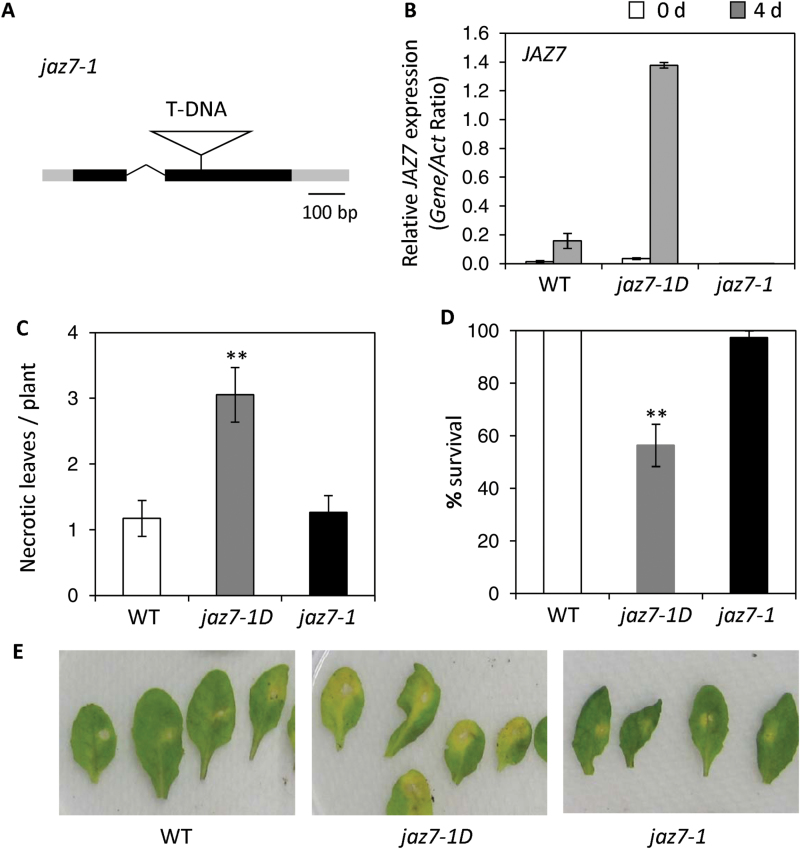
The finding that jaz7-1D contains an activation-tagged JAZ7 allele indicates the possibility that the increased expression of JAZ7 might be responsible for increased susceptibility to F. oxysporum in this line. To determine whether plants with null mutations in the JAZ7 gene could show an opposite F. oxysporum resistance phenotype, we isolated a homozygous jaz7 mutant (WiscDsLox7H11) designated as jaz7-1, where the T-DNA is inserted into the second exon of the JAZ7 gene (Fig. 4A). No detectable transcripts from the truncated jaz7-1 locus could be identified before or after inoculations with F. oxysporum in the jaz7-1 mutant (Fig. 4B, Supplementary Fig. S3). In contrast, JAZ7 transcript levels were hypersensitive to induction by F. oxysporum in the activation tagged jaz7-1D mutant (Fig. 4B). Compared to wild-type plants, jaz7-1 did not exhibit altered resistance to F. oxysporum in either disease or culture filtrate assays (Fig. 4C–E). The absence of any pathogen-associated phenotype in jaz7-1 is consistent with the view that null mutations in most JAZ-encoding genes do not produce JA-related phenotypes (e.g. Thines et al., 2007) possibly due to the functional redundancy within this gene family. We also screened jaz7-1 in double and triple jaz mutant lines, as well as other combinations of jaz mutants in F. oxysporum disease assays (Supplementary Table S1; de Torres Zabala et al., 2015). Most of the JAZ insertion lines we used have been previously characterized for loss-of-function or reduced transcript expression, and we further confirmed this for jaz2 (SALK_025279), jaz5 (SALK_053775) and jaz10 (SAIL_92_D08). Although further experiments need to be conducted to determine if JAZ transcript levels are affected in the remaining jaz insertion lines, none of these lines exhibited altered disease phenotypes compared to wild-type plants (data not shown).
Fig. 4.
A null T-DNA insertional inactivation line of JAZ7 does not affect resistance to F. oxysporum. (A) Schematic representation of the jaz7-1 (WiscDsLox7H11) T-DNA insertion line. 5′ and 3′ UTR are shaded in gray, exons in black and the only intron as a removed segment. (B) JAZ7 expression was examined in leaves of wild-type (WT), jaz7-1D and jaz7-1 plants before or 4 d after F. oxysporum inoculation. Values are averages ±SE of three biological replicates comprising 5–10 plants. Gene expression levels are relative to the internal control β-actin genes. (C-D) WT, jaz7-1D and jaz7-1 were inoculated with F. oxysporum and disease symptoms recorded with (C) necrotic leaves per plant at 10 d and (D) survival rates at 14 d post-inoculation. Values are averages ±SE (n=60). Asterisks indicate values that are significantly different (**, P<0.01; Student’s t-test) from WT. Similar results were obtained in independent experiments. (E) F. oxysporum culture filtrate was applied to detached WT, jaz7-1D and jaz7-1 leaves. Representative leaves are shown from three replicates 6 d post-treatment.
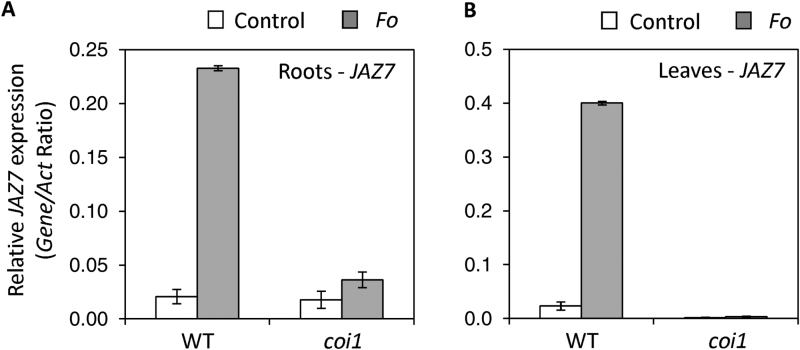
Given that increased JAZ7 expression in the jaz7-1D mutant correlated with increased susceptibility to F. oxysporum and JAZ proteins act as repressors in JA-signaling, we asked whether the Fusarium inducibility of JAZ7 requires COI1. As shown in Fig. 5, F. oxysporum inducibility of JAZ7 was abolished in both roots and leaves of the coi1 mutant, suggesting that COI1 (or JA-sensing) is required for pathogen inducible JAZ7 expression.
Fig. 5.
Fusarium induced JAZ7 expression is COI1-dependent. JAZ7 expression was monitored in (A) roots and (B) leaves of control or F. oxysporum (Fo)-challenged wild-type (WT) or coi1 plants at 4 d post-infection. Values are averages ±SE of three biological replicates consisting of pools of 10–30 plants. Gene expression levels are relative to the internal control β-actin genes. Similar results were obtained in an independent experiment.
jaz7-1D shows differential resistance to other pathogens and an early flowering phenotype
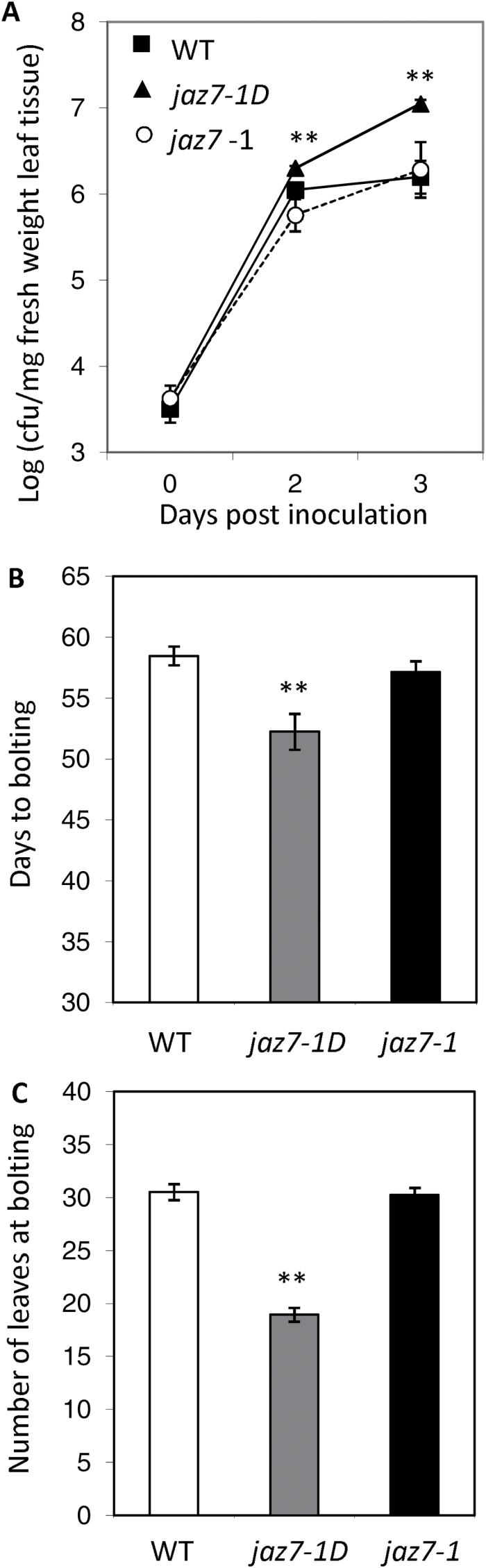
JA-signaling in Arabidopsis is also known to affect resistance to pathogens other than F. oxysporum. For instance, as with F. oxysporum, JA-signaling promotes susceptibility to the bacterial pathogen Pst DC3000 (Kloek et al., 2001) whereas intact JA-signaling is required for resistance to the leaf-infecting necrotrophic pathogen Alternaria brassicicola (Thomma et al., 1998). We therefore tested jaz7-1D and jaz7-1 mutants against both of these pathogens. Similar to its response to F. oxysporum, the jaz7-1D mutant showed significantly increased susceptibility to Pst (Fig. 6A) while, consistent with de Torres et al. (2015) no effect of the jaz7-1 mutation on resistance was evident. In contrast, jaz7-1D and jaz7-1 showed no significant difference in resistance or susceptibility to A. brassicicola relative to wild-type plants. Combined, these results implicate JAZ7 in resistance against specific pathogens.
Fig. 6.
jaz7-1D is highly susceptible to Pseudomonas syringae pv tomato and exhibits an early flowering phenotype. (A) Pathogen infection of wild-type (WT), jaz7-1D and jaz7-1. Log P. syringae counts from leaf tissue after Pst DC3000 infection over 3 d. Values are averages ±SE of four biological replicates consisting of pools of four leaves. Flowering time as noted by (B) days to bolting and (C) number of rosette leaves at bolting. Values are averages ±SE of two biological replicates consisting of pools of 10 plants. Asterisks indicate values that are significantly different (**, P<0.01; Student’s t-test) from WT. Similar results were obtained in independent experiments.
In addition to compromised disease resistance, we noted that the jaz7-1D mutant flowered earlier than jaz7-1 and wild-type plants under short-day conditions (Fig. 6B, C).
The jaz7-1D mutant shows increased JA-sensitivity and JA-responsive gene expression
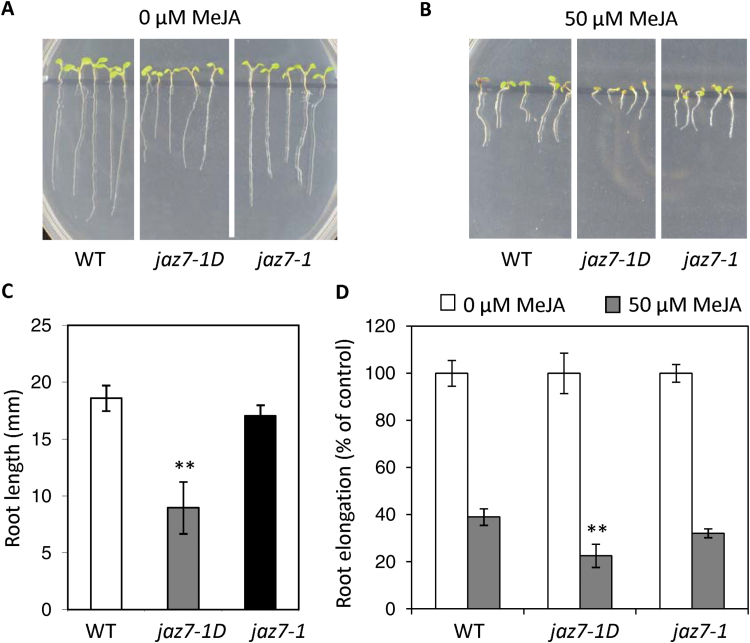
As JAZ proteins act as repressors of JA signaling, we hypothesized that JA-dependent plant responses such as JA-mediated inhibition of primary root elongation and JA-responsive gene expression may be altered in the jaz7-1D mutant due to constitutive JAZ7 expression. We first tested the JA-mediated root growth inhibition phenotype of jaz7-1D and jaz7-1 mutants. In the absence of MeJA, jaz7-1D roots were shorter than those of wild-type and jaz7-1 (Fig. 7A, C). The percent inhibition of root elongation by MeJA was also greater in jaz7-1D than in wild-type and jaz7-1 (Fig. 7B, D), suggesting activated JAZ7 expression in the jaz7-1D mutant confers increased JA sensitivity rather than the decreased sensitivity expected from a repressor.
Fig. 7.
jaz7-1D shows increased JA-sensitivity. Sensitivity of wild-type (WT), jaz7-1D and jaz7-1 seedlings to JA was determined by MeJA inhibition of root growth on control media versus media containing MeJA at 7 d post-germination. Representative images of seedlings on (A) control (0 µM MeJA) or (B) MeJA media (50 µM). jaz7-1D mutants have shorter roots under basal conditions (C) and their root elongation (D) shows increased sensitivity to MeJA. Root elongation of each line when grown on control media or media containing MeJA was calculated as a percentage relative to control treatment. Values are averages ±SE of three biological replicates consisting of pools of 10–15 seedlings. Values that differed significantly from the WT were identified by the one-way ANOVA and Dunnet’s post-hoc test (**, P<0.01). Similar results were obtained in independent experiments.
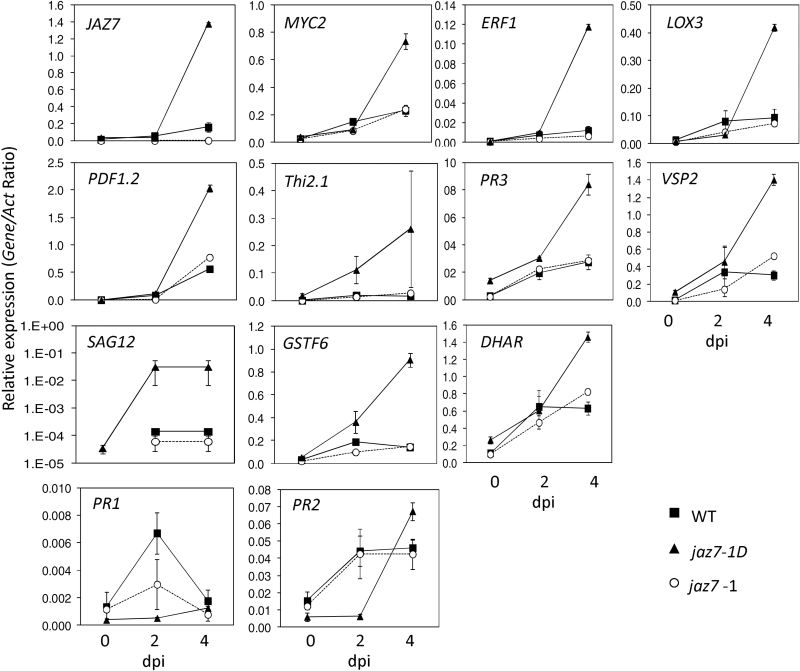
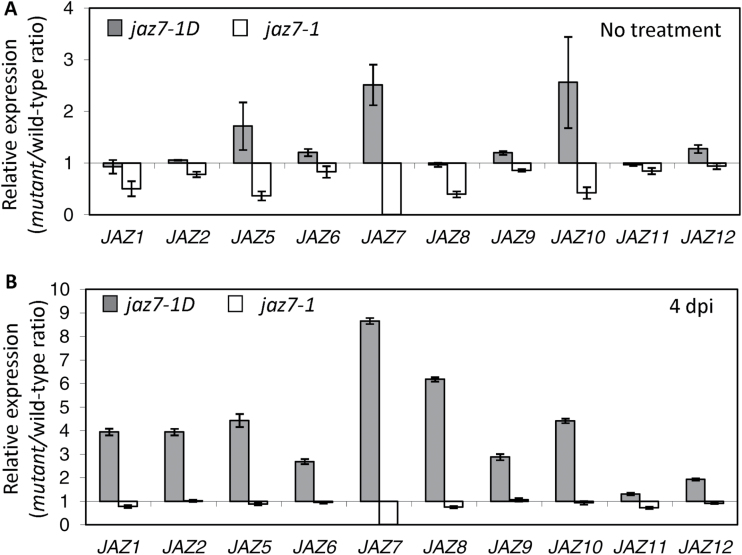
We next analyzed the F. oxysporum-induced expression of JA-responsive genes in the two jaz7 mutants after inoculations with F. oxysporum (Fig. 8). Genes encoding JA-responsive transcription factors (e.g. MYC2 and ERF1), a JA-biosynthesis enzyme (e.g. LOX3) and JA-related defense proteins (e.g. PDF1.2, Thi2.1, PR3 and VSP2) were induced more strongly in the leaves of inoculated jaz7-1D plants than in jaz7-1 and wild-type plants at 4 dpi. Expression of senescence or oxidative stress associated transcripts (e.g. SAG12, GSTF6, DHAR) were also up-regulated in jaz7-1D. Furthermore, analysis of JAZ gene expression after F. oxysporum inoculations revealed that transcript levels of almost all JAZ genes were up-regulated in jaz7-1D while in jaz7-1 levels were either reduced or did not differ from wild-type levels (Fig. 9). Overall, this indicates JA-regulated gene expression is up-regulated in jaz7-1D plants.
Fig. 8.
jaz7-1D shows increased JA-responsive gene expression under Fusarium infection. Gene expression was monitored in leaf tissue of untreated or F. oxysporum-challenged wild-type (WT), jaz7-1D and jaz7-1 plants at 2 and 4 d post-infection (dpi). Values are averages ±SE of three biological replicates consisting of pools of five plants. Gene expression levels are relative to the internal control β-actin genes.
Fig. 9.
JAZ transcripts are up-regulated in jaz7-1D. JAZ expression was monitored in leaf tissue of (A) untreated or (B) F. oxysporum-challenged wild-type (WT), jaz7-1D and jaz7-1 plants at 4 d post-infection (dpi). Values are expressed relative to WT values at that time-point. Values are averages ±SE of three biological replicates consisting of pools of five plants. Gene expression levels are relative to the internal control β-actin genes. JAZ3 and JAZ4 expression was not examined due to lack of F. oxysporum inducibility (Fig. 1).
In parallel to the overall increases observed in JA-responsive gene expression, the SA marker genes PR1 and PR2 showed reduced or delayed induction in response to F. oxysporum inoculations (Fig. 8). These gene expression studies together with JA root inhibition data suggest that jaz7-1D plants exhibit altered regulation of the JA-pathway in response to F. oxysporum infection of Arabidopsis.
Genome-wide identification of differentially expressed genes in jaz7-1D
To further dissect the effect of the jaz7-1D mutant on JA-responsive gene expression, we conducted genome-wide identification of genes differentially regulated in the jaz7-1D mutant following a control or MeJA treatment. This involved microarray analysis of jaz7-1D and wild-type plants from four independent replicates using the Arabidopsis Affymetrix ATH1 Genome Array. Stringent analysis of the expression data was performed using two-way ANOVA (P<0.05) on the entire dataset with the inclusion of the Benjamini and Hochberg FDR. A comparison of differentially regulated genes by genotype identified 113 up-regulated and 25 down-regulated genes showing ≥2-fold in mock-treated jaz7-1D relative to mock-treated wild-type plants (Supplementary Tables S4–6). To gain insight into the functions of these genes, we performed GO term enrichment analysis. Significantly enriched biological processes from genes up-regulated in jaz7-1D were those involved in defense responses, multi-organism processes, and responses to stress, fungi, other organisms, biotic and abiotic stimulus, and organic substances, while those from the down-regulated dataset were enriched for genes involved in response secondary metabolic processes and response to stimulus (FDR<0.05). The majority of genes up-regulated in jaz7-1D are also associated with JA-signaling, plant defense and/or senescence including Thi2.1 which we previously identified as being up-regulated (Fig. 8). For example, three of the up-regulated genes in jaz7-1D, encoding a protease 1-like protein (AT2G38860), an alpha-beta hydrolyase superfamily/lipase protein (AT2G42690) and a cyclic nucleotide-regulated ion channel (AT2G46450), respectively, have been identified as senescence markers in Arabidopsis (Yoshida et al., 2001; Gepstein et al., 2003). Notably, the most highly up-regulated gene in jaz7-1D encoding a N-acetyltransferase (AT2G39030/NATA1) is highly JA-inducible and linked to Pst susceptibility (Adio et al., 2011). In addition, expression of genes (e.g. DET2/DWF6) known to promote flowering (Chory et al., 1991; Li et al., 2010) are up-regulated while those (e.g. MADS Box type II protein) associated with negative regulation of flowering (Ratcliffe et al., 2003) are down-regulated in the mutant, thus correlating with the jaz7-1D early flowering phenotype. Previously, the det2 mutant was shown to display a lack of leaf senescence and also delayed flowering time (Chory et al., 1991). The increased DET2/DWF6 expression in the jaz7-1D mutant is therefore consistent with the role of this gene as a positive regulator of senescence and flowering time. Importantly, two genes encoding pectin methylesterase inhibitors were down-regulated in the jaz7-1D mutant (Supplementary Table S6). Again, this is consistent with the increased susceptibility phenotype of the jaz7-1D mutant as overexpression of methylesterase inhibitors in Arabidopsis provides increased pathogen resistance (Lionetti et al., 2007).
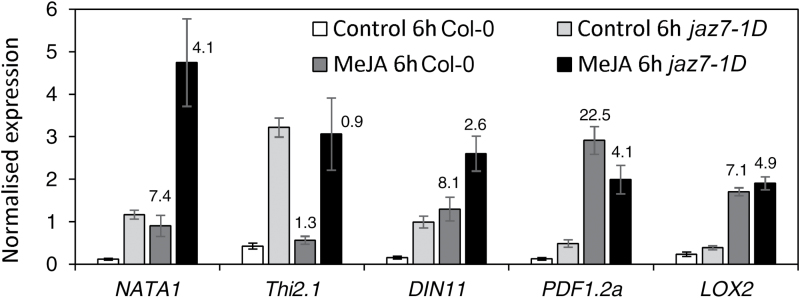
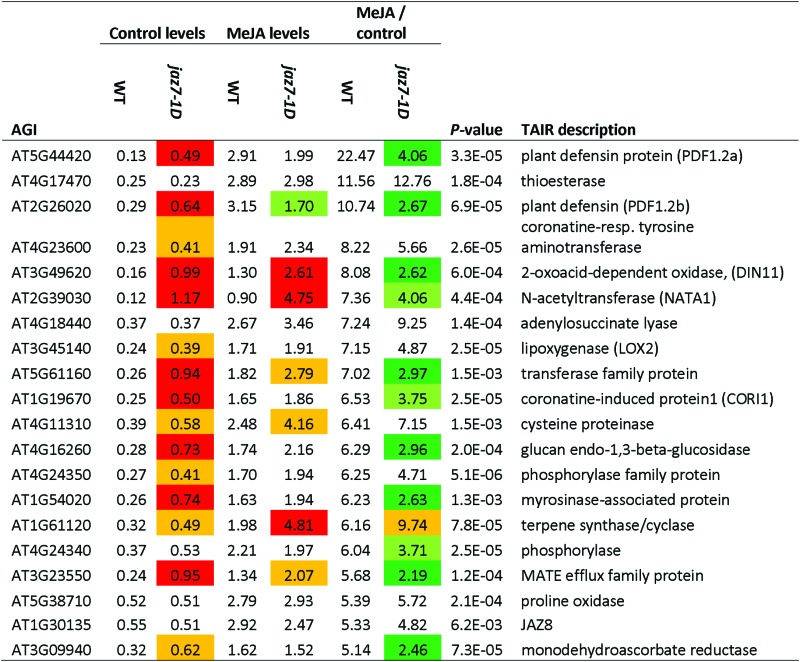
We next analyzed the two-way ANOVA data for genes differentially regulated by MeJA treatment and identified 56 up-regulated and 21 down-regulated ≥2-fold in MeJA-treated jaz7-1D relative to MeJA-treated wild-type plants (Supplementary Tables S7–9). Genes in the up-regulated dataset were enriched for lipid biosynthetic and metabolic processes, response to external stimulus, localization and transport, while the down-regulated dataset were enriched for response to stimulus, stress, chemicals and organic substances. NATA1 was the highest up-regulated gene, as was it under control treatment. However, we noticed the MeJA inducibility of this gene and others in jaz7-1D over its control levels was nearly 2-fold less than its inducibility in wild-type plants, suggesting the primed expression of these genes prevents the same level of induction observed in wild-type plants (Fig. 10). To dissect this phenomenon further, we took the ANOVA data and examined MeJA-inducible expression of genes in wild-type and jaz7-1D relative to their levels in control samples. Highly MeJA-inducible genes in wild-type were generally not as inducible in jaz7-1D (Table 1, Supplementary Table S10). This included genes involved in JA-responses, defense and senescence such as the two defensins PDF1.2a (AT5G544420) and PDF1.2b (AT2G26020), LOX2 (lipoxygenase 2/AT3G45140), COR1 (coronatine-responsive protein/AT1G19670), a glucan endo-1,3-beta-glucosidase (AT4G16260) and DIN11 (DARK INDUCIBLE 11/AT3G49620). Overall, these results suggest the primed JA-response in jaz7-1D may limit further JA-mediated fold-induction and/or that JAZ7 may have a role in inhibition of JA-regulated responses.
Table 1.
Subset of genes differentially regulated by MeJA treatment from the microarray
Shown are the top 20 wild-type MeJA/control-induced genes (data obtained from Supplementary Table S10). Colour coding: change in jaz7-1D over wild-type (WT) under each analysis; >2-fold, red; >1.5-fold, orange; <2-fold, green; <1.5-fold, lime.
|
Transgenic over-expression of JAZ7 does not reproduce jaz7-1D phenotypes
The finding that JAZ7 and JA-regulated gene expression is up-regulated in the jaz7-1D mutant prompted us to generate JAZ7 overexpression lines. We generated three independent lines overexpressing JAZ7 under the control of the constitutive 35S promoter (JAZ7-OX) with expression ranging from 9-fold to 1800-fold over wild-type levels (Supplementary Fig. S4A). Interestingly, the JAZ7-OX lines did not exhibit the small rosette size or reduced root length phenotypes of jaz7-1D under normal growing conditions, but did exhibit, although not significantly, increased basal expression of some but not all JA-marker genes tested (Supplementary Fig. S4B–D). We also examined JA-sensitivity and Fusarium susceptibility in the overexpression lines and found only the lowest JAZ7 expression line JAZ7-OX1 (with JAZ7 levels comparable to jaz7-1D) displayed increased JA-sensitivity and increased Fusarium susceptibility, but only at early stages of infection (Supplementary Fig. S4E–G).
Possibilities for the JAZ7-OX lines not phenocopying jaz7-1D may be jaz7-1D producing altered JAZ7 transcripts such as those harboring mutations, or formed as a result of altered splicing or altered transcription start sites (TSSs), or the presence of additional undetected T-DNA insertions in jaz7-1D. Therefore, we sequenced JAZ7 transcripts from Col-0, jaz7-1D and JAZ7-OX, but found no sequence variation. Further, inspection of RNA-seq data from Yan et al. (2014), who used SALK_040835C in their studies, revealed no differences in JAZ7 transcripts (SNPs, truncations, mis-splicing or altered TSSs) compared to wild-type Col-0. Next, to consider the possibility of additional insertions (not collated by SALK) in jaz7-1D affecting its phenotypes, we produced a backcrossed (to Col-0) line. The F2 progeny segregated 2:1 heterozygous jaz7-1D:Col-0 (confirmed via PCR) as suggestive of a dominant mutation, reiterating our previous results showing that homozygous lines of this insertion mutant may be lethal. The heterozygous progeny also conferred jaz7-1D phenotypes of short roots (this study; Yan et al., 2014) and JA-hypersensitivity (Supplementary Fig. S5). If the JA-hypersensitive phenotypes in jaz7-1D were due to an additional T-DNA insertion we would expect to see this phenotype segregate, unless the insertion is closely linked. Therefore, combined with our JAZ7-OX results, it is possible that jaz7-1D JA-related phenotypes are a result of ectopic cell or tissue-specific JAZ7 expression as a consequence of the T-DNA insertion in the JAZ7 promoter and/or high levels of JAZ7 in jaz7-1D plants interfering within COI1-JAZ-TPL-TF multiprotein complexes.
JAZ7 contains a functional EAR-repression motif
Increased JA-sensitivity and JA-mediated gene expression in jaz7-1D suggests a role for JAZ7 in activation of JA responses. However, JAZ proteins repress JA-responses through direct (EAR repressor motif) or indirect (NINJA-mediated) recruitment of co-repressor(s) and the EAR motif associated with gene repression is present in JAZ7 (Kazan, 2006; Kagale et al., 2010). JAZ5, JAZ6, JAZ8 and JAZ13 also contain EAR motifs required for interactions with the co-repressor protein TOPLESS (TPL) (Kagale et al., 2010; Causier et al., 2012; Shyu et al. 2012; Thireault et al., 2015) and the transcriptional repression function of this motif in JAZ8 has been confirmed (Shyu et al., 2012). The functionality of the predicted EAR motif found in JAZ7 has, however, not been experimentally tested. To determine whether the JAZ7 EAR motif acts as a functional repressor, we fused an extended version of this motif to the carboxy terminus of the CUC1 (CUP-SHAPED COTYLEDON1) transcription factor which should convert this transcriptional activator to an active repressor and produce plants with a cup-shaped cotyledon phenotype (Hiratsu et al., 2003). As shown in Fig. 11A, B, Arabidopsis plants transformed with 35S:CUC1-JAZ7 EAR showed a typical cup-shaped cotyledon formation, indicating that the JAZ7 EAR motif can act as a repression domain. Of the 13 selected T1 seedlings, severity of fused cotyledons varied, with eight displaying the cup-shaped cotyledon phenotype.
Fig. 11.
JAZ7 contains an active EAR-repression motif mediating its interaction with TPL, and interacts in vitro with MYC3, MYC4 and JAM1. (A) JAZ7 repression domain sequence used to generate a Cup-Shaped Cotyledon (CUC) repression construct. The EAR motif is red underlined. (B) The chimeric protein in which CUC1 was fused to the JAZ7EAR domain was expressed in transgenic plants (35S:CUC1-JAZ7 EAR). Representative wild-type (WT) and 35S:CUC1-JAZ7 EAR seedlings are shown. (C) Y2H assays between Topless (TPL), MYC2, MYC3, MYC4 or JAM1 and JAZ7, a version of JAZ7 containing a mutated EAR-motif (JAZ7mEAR), JAZ5 or JAZ8. Y2Hs were performed on non-selective SD-Leu-Trp media as well as selective SD-Leu-Trp-His or SD-Leu-Trp-His-Ade media. No interactions were seen for the binding domain (BD) only or when an empty activation domain (AD) was used on selective media. JAZ8 and JAZ5 were used as positive controls for TPL interactions. Results were replicated in several independent experiments.
JAZ7 interacts with the co-repressor TPL
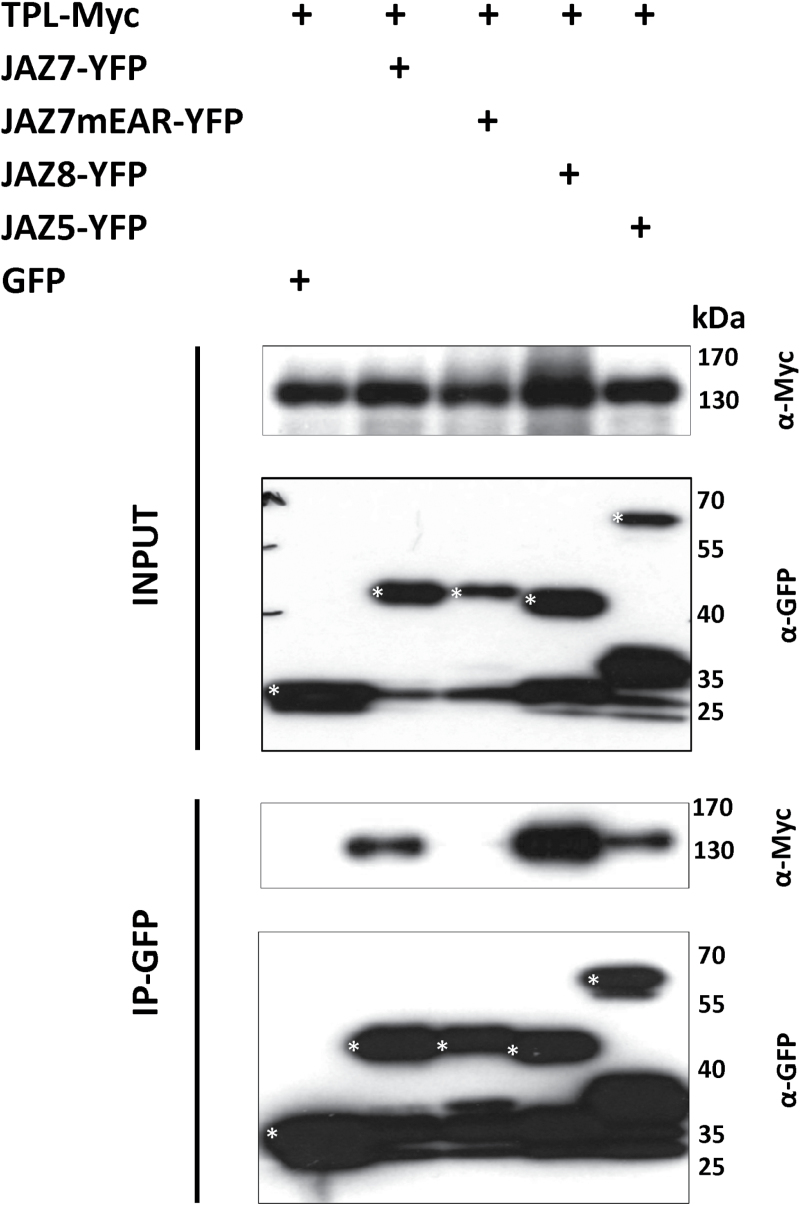
The finding that ectopic overexpression of the JAZ7 EAR motif has repressor activity prompted us to determine whether JAZ7 could interact with the co-repressor TPL, an interaction which had not yet been demonstrated. Using JAZ5-TPL and JAZ8-TPL interactions as positive controls (Causier et al., 2012; Shyu et al., 2012), an interaction between JAZ7 and TPL in Y2H studies was observed (Fig. 11C). To determine if the interaction is mediated through the EAR motif of JAZ7, we generated a JAZ7 construct containing a mutated version of the EAR motif (JAZ7mEAR). No interaction between JAZ7mEAR and TPL was observed (Fig. 11C). In follow up in vivo studies, we conducted co-immunoprecipitation experiments between TPL and JAZ7, JAZ7mEAR, JAZ8 and JAZ5 through transient expression in Nicotiana benthamiana. Correlating with the Y2H results, TPL interactions with JAZ7, JAZ5 and JAZ8 were observed, but when the EAR motif of JAZ7 was mutated, the TPL and JAZ7 interaction was strongly diminished (Fig. 12). These results strongly support an in planta interaction between TPL and JAZ7 mediated via its EAR-motif.
Fig. 12.
JAZ7 binds TPL in vivo. Co-immunoprecipitation analysis from transiently expressed proteins in N. benthamiana leaves showing association between TPL and JAZ7, JAZ8 or JAZ5 but not JAZ7mEAR. TPL was fused to C-terminal Myc tag while JAZ7, JAZ5, JAZ8 and JAZ7mEAR are fused to C-terminal YFP tag. Immunoblots show the presence of proteins in total extracts (input, top panels) and after immunoprecipitation with anti-GFP beads (IP-GFP, bottom panels). JAZ8 and JAZ5 were used as positive controls and GFP vector as a background control. Protein molecular mass ladder is shown to the right of each blot. Results were replicated in an independent experiment. Asterisks indicate the expected protein bands.
Fig. 13.
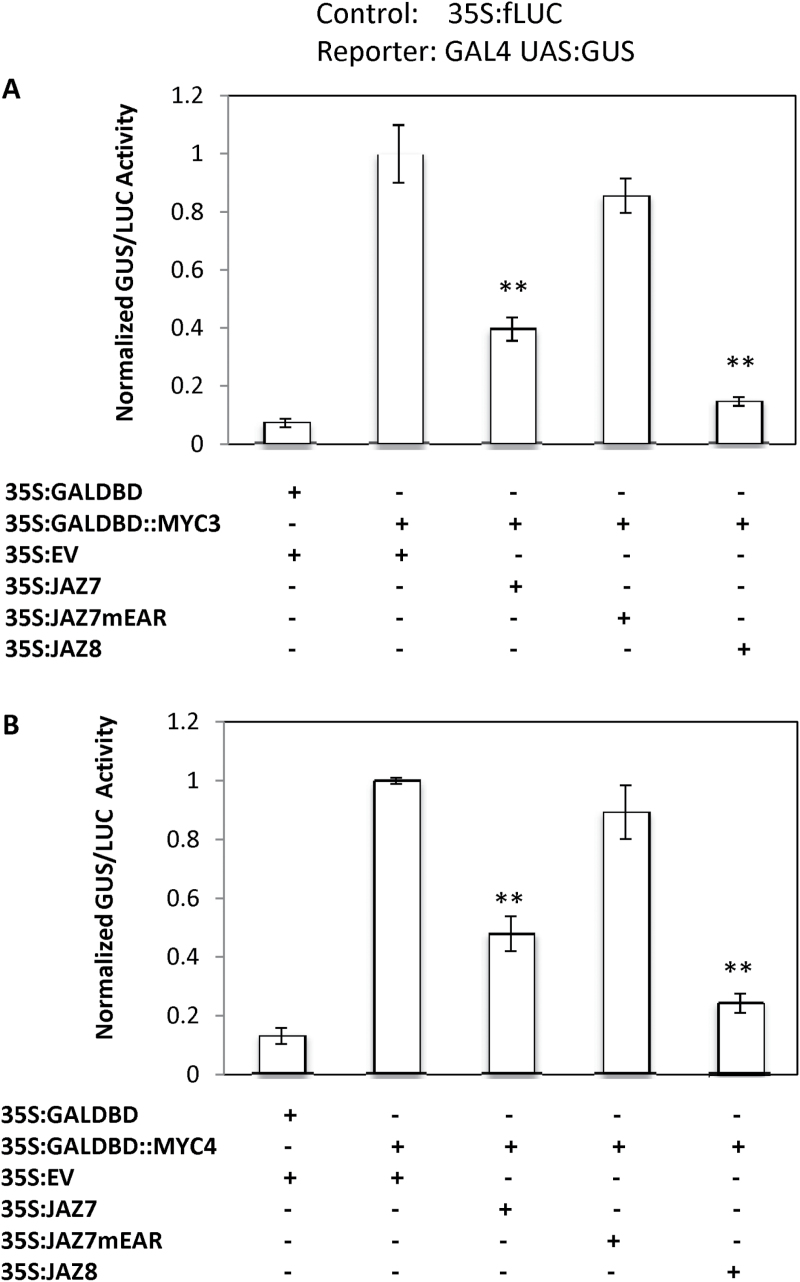
MYC3 and MYC4 transcription activities are repressed by JAZ7 and JAZ8 but not by JAZ7mutEAR in transient activation assays. Transient expression assays in Arabidopsis thaliana leaves show that JAZ7 and JAZ8 but not JAZ7mutEAR suppress (A) MYC3- and (B) MYC4-mediated transcription activation using the GAL4 binding domain (DBD) and upstream GAL4-binding sequences (GAL4-UAS) fused to the GUS gene. The activity of the reporter gene (GUS) was normalized to the activity of the firefly LUC gene. Data are means (±SD) of three biological replicates of two bombarded leaves. Statistical significance was assessed using the unpaired Student’s t-test (**, P<0.01). These experiments were carried out twice with similar results.
JAZ7 interacts with the transcriptional activators MYC3 and MYC4, and the transcriptional repressor JAM1
To dissect the potential mechanism of JAZ7 in JA-responses we tested for JAZ7 interactions with the transcriptional activators MYC2, MYC3 and MYC4 that can bind to most JAZ proteins (Chini et al., 2009; Cheng et al., 2011; Fernandez-Calvo et al., 2011; Niu et al., 2011). Using Y2H approaches, several groups have reported JAZ7 binding to MYC2, MYC3 and MYC4, while others have not detected these interactions (Chini et al., 2009; Arabidopsis Interactome Mapping Consortium, 2011; Cheng et al., 2011; Fernandez-Calvo et al., 2011; Qi et al., 2011). To address this, we conducted Y2H studies using JAZ5 and JAZ8 as positive controls; both interact with MYC2, MYC3 and MYC4 in all published studies to our knowledge (Cheng et al., 2011; Fernandez-Calvo et al., 2011). We found a strong interaction between JAZ7-MYC3 and JAZ7-MYC4, but failed to identify a JAZ7-MYC2 interaction (Fig. 11C).
To determine whether JAZ7 has the capacity to repress these transcriptional activators we conducted transcriptional activation assays with JAZ7 against MYC3 and MYC4. In these experiments, we co-bombarded a reporter gene construct containing the GAL4 upstream activation sequence (pGAL4UAS) linked to the GUS gene (pGAL4UAS-GUS), together with CaMV35S expression constructs of MYC3 or MYC4 fused to the GAL4 DNA binding domain (GAL4BD) or GAL4BD alone, as well as empty vector, JAZ7, JAZ7mEAR or JAZ8 under CaMV35S promoter (Fig. 13). In addition, an expression construct of the firefly luciferase (LUC) gene was co-bombarded as a normalization control. The addition of the vector constructs expressing either MYC3- or MYC4-GAL4BD produced significantly higher transcription activity of the GUS reporter gene compared to the control effector plasmid (GAL4BD only) when co-bombarded with the empty vector. However, transcription activation abilities of the MYC3 and MYC4-GAL4BD fusion proteins were significantly reduced when co-bombarded with JAZ7 or JAZ8, but not with JAZ7mEAR. These experiments demonstrate JAZ7 and JAZ8, but not JAZ7mEAR, repress MYC3 and MYC4-mediated transcriptional activities.
JAZ7 appears not to interact with the JA-ASSOCIATED MYC2-like transcriptional repressors (JAM) bHLH003/JAM3 or bHLH013/JAM2 (Song et al., 2013; Fonseca et al., 2014; Sasaki-Sekimoto et al., 2014), but one group has reported bHLH017/JAM1-JAZ7 binding (Fonseca et al., 2014). We therefore next tested for JAZ7-JAM1 binding, again using JAZ5 and JAZ8 as positive controls as both interact with JAM1 (Song et al., 2013; Fonseca et al., 2014), and confirmed that JAZ7 can bind to the transcriptional repressor JAM1 (Fig. 11C).
Combined, our results demonstrate through direct recruitment of TPL, in wild-type plants JAZ7 functions as a repressor within the JA-response network via its interaction with specific transcriptional regulators (e.g. MYC3, MYC4, JAM1). In jaz7-1D plants, we propose the misregulated expression of JAZ7 would obstruct the finely-tuned nature of the COI1-JAZ-TPL-TF multi-protein complex resulting in hyperactivation of JA-signaling.
Discussion
JA-signaling functions as a major determinant of disease outcome in Arabidopsis to the fungal pathogen F. oxysporum (Anderson et al., 2004; Berrocal-Lobo and Molina 2004; McGrath et al., 2005; Kidd et al., 2009; Thatcher et al., 2009, 2012a). In this study we analyzed the roles of JAZ proteins, repressors of JA-signaling, in F. oxysporum resistance or susceptibility. We identified a highly susceptible T-DNA insertion line (jaz7-1D) with a promoter insertion resulting in constitutive JAZ7 expression and enhanced susceptibility to F. oxysporum. The jaz7-1D line also conferred increased JA-sensitivity, up-regulation of defense and JA-mediated gene expression, and increased susceptibility to the bacterial pathogen Pst DC3000. Both F. oxysporum and Pst DC3000 appear to target host JA- signaling to elicit disease, the first to hyperactivate JA-signaling and senescence processes, and the second to antagonistically suppress defense responses mediated by salicylic acid signaling. Thus the jaz7-1D line interferes with defense responses that integrate signals downstream of pathogens with two different virulence strategies.
We found the majority of JAZ genes were induced following F. oxysporum inoculation, with the largest inductions observed in root tissues for JAZ5 and JAZ10 (Fig. 1). There were also differences in individual JAZ root and leaf temporal expression patterns suggesting that some JAZ proteins may play unique roles in different tissue types. The largest inductions were observed for JAZ5, JAZ7, JAZ8, JAZ9 and JAZ10 (Fig. 1). These genes are also highly induced by B. cinerea, Pst, and/or herbivory (Chung et al., 2008; data extracted from Genevestigator in Hruz et al., 2008; Demianski et al., 2012). JAZ7 and JAZ9 are also highly induced during senescence, which is promoted by F. oxysporum infection (data extracted from Genevestigator in Hruz et al., 2008).
The strong inducibility of several JAZ genes by F. oxysporum and other pathogens/pests led us to screen available T-DNA insertion lines in JAZ genes for altered F. oxysporum disease phenotypes. While most overexpression or knockout lines of individual JAZ genes lack observable JA-related phenotypes, suggesting functional redundancy amongst the JAZ proteins (reviewed in Wasternack and Hause, 2013), we identified the jaz7-1D T-DNA insertional activation mutant which conferred hyper-activation of JA-signaling including up-regulation of JA-regulated biosynthesis, defense and senescence-associated genes (Fig. 8), as well as up-regulation of most other JAZ genes (Fig. 9). In an unbiased approach to identify genes differentially regulated in jaz7-1D, our microarray analysis identified genes up-regulated ≥2-fold in jaz7-1D over wild-type to be significantly enriched for involvement in stress and defense responses. The most highly up-regulated gene (9.5-fold) NATA1 in the jaz7-1D mutant encodes a N-acetyltransferase, which acetylates ornithine to produce the defense-related metabolite Nδ-acetylornithine. Yan et al. (2014) also found this metabolite is more abundant in SALK_040835 (jaz7-1D) and its levels are highly up-regulated over wild-type following MeJA treatment. NATA1 expression is highly responsive to JA, Pst and herbivory (Adio et al., 2011) and a knockout mutant of NATA1 has increased resistance to Pst DC3000 (Adio et al., 2011), supporting our results for jaz7-1D. Adio et al. (2011) suggest that Pst DC3000 infection is promoted by coronatine/MeJA-induced expression of NATA1 and subsequent production of Nδ-acetylornithine. Although Thi2.1, the second most highly up-regulated gene in jaz7-1D, has been linked to increased F. oxysporum resistance (Epple et al., 1997; Chan et al., 2005; Thatcher et al., 2012a), Thi2.1 is not a single determinant of F. oxysporum resistance. Indeed, other mutants with constitutive Thi2.1 expression (e.g. cpr5) are highly susceptible while coi1 plants with severely compromised Thi2.1 expression are highly resistant (Bowling et al., 1997; Schenk et al., 2005; Thatcher et al., 2009). Another gene highly up-regulated in jaz7-1D was Histone1-3 (HIS1-3). HIS1-3 encodes a linker histone which functions as a stabilizer of chromatin structure and its expression is highly drought inducible, suggestive of a role in stress tolerance (Ascenzi and Gantt, 1999). Recently it was found that JAZ7 plays a role in negative regulation of dark-induced leaf senescence (Yu et al., 2015). Through analysis of the jaz7-1 (WiscDsLox7H11) knockout line, Yu and colleagues found senescence and H2O2-mediated responses and genes involved in these processes such as NATA1 and DIN11 were significantly up-regulated in jaz7-1 in darkness but not under light conditions. We found no alteration in Fusarium-induced senescence responses or oxidative stress responsive gene expression in jaz7-1 compared to wild-type plants (Figs 4, 8). Thus it appears JAZ7 plays contrasting roles in pathogen and dark-induced senescence responses.
In addition to hyperactivation of JA-responses, the jaz7-1D mutant displayed an early flowering phenotype (Fig. 6). Links between flowering time and altered JA-mediated pathogen resistance have been reported previously. For example, the pft1/med25 mutant is delayed in flowering, exhibits down-regulated JA-defense responses and increased resistance to F. oxysporum (Kidd et al., 2009). It has been shown COI1-dependent signaling delays flowering time via JAZ degradation and inhibiting the expression of FLOWERING LOCUS T (FT) (Zhai et al., 2015). Although increased JA-signaling and JAZ expression is evident in jaz7-1D plants, we did not detect altered expression of FT in our microarray analysis. However, other genes known to regulate flowering were altered (e.g. DET2/DWF6). The constitutive activation of JA-signaling in jaz7-1D may also be responsible for its small rosette phenotype and reduced root-length (Figs 2A, 7C). Many other mutants with constitutive JA-defense gene expression (e.g. cpr5, cev1, cet1, dnd1, dnd2) also show stunted growth (Bowling et al., 1997; Ellis and Turner, 2001; Hilpert et al., 2001; Genger et al., 2008). Without stringent regulation, constant activation of JA responses would place large demands on plant resources, repressing growth, and likely contribute to these dwarf phenotypes (Baldwin, 1998; Kazan and Manners, 2012; Pieterse et al., 2014). This is supported by the finding that defense and stress-related metabolites are increased in jaz7-1D/SALK_040835C which may limit resources available for growth (Yan et al., 2014). Basal expression of JA-marker genes in the JAZ7 overexpression lines (JAZ7-OX) that we generated was also increased, but not to the significantly high levels observed in jaz7-1D, and may account for why the JAZ7-OX lines did not exhibit the stunted jaz7-1D root and leaf phenotypes. To rule out the possibilities that altered JAZ7 transcripts (e.g. mutated, mis-spliced) or other T-DNA insertions in jaz7-1D are responsible for its JA-hyperactivation phenotypes, we conducted several additional analyses and backcrossed jaz7-1D to wild-type plants. Our results suggest the T-DNA insertion within the JAZ7 promoter is associated with the jaz7-1D phenotypes. However we cannot exclude the possibility that undetected secondary mutations or possible chromosomal rearrangements resulting from T-DNA transformation may contribute.
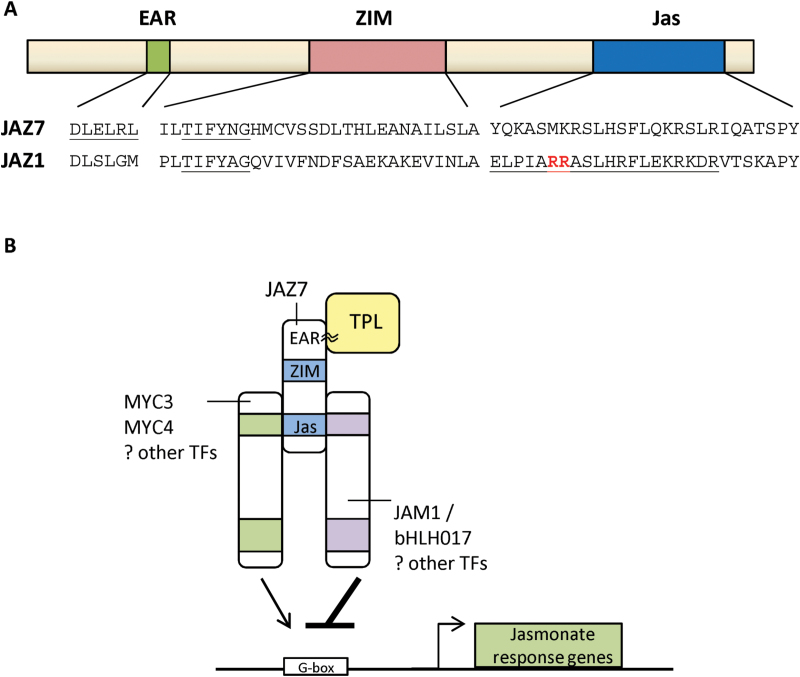
For other JAZ proteins characterized to date, JA-related phenotypes such as JA-insensitivity, sterility or altered tolerance to pathogens or pests have only been identified for JAZ8 and JAZ13 overexpressing lines (Shyu et al., 2012; Thireault et al., 2015), jaz10 T-DNA or RNAi knockdown lines (Cerrudo et al., 2012; Leone et al., 2014), or in modified JAZ proteins in which the conserved C-terminal Jas motif has been deleted or its critical amino acids modified. These alterations stabilize the JAZ protein by preventing its interaction with COI1 and subsequent ubiquitin-mediated degradation following JA-stimulation (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Chung et al., 2008; Chung and Howe, 2009). More specifically, the N-terminal domain of the Jas motif was identified as the COI1 and JA-Ile/COR binding site and termed the JAZ degron (Sheard et al., 2010). A LPIARR sequence in the JAZ degron binds JA-Ile and COI1 in a clamp (Sheard et al., 2010). This sequence is diverged in JAZ5, JAZ6, JAZ7 (Fig. 14A) and JAZ8, with both JAZ7 and JAZ8 lacking the RR or RK amino acid combination shown to be critical for COI1 binding (Melotto et al., 2008; Sheard et al., 2010; Shyu et al., 2012). Shyu et al. (2012) attributed the JAZ8 Jas motif’s divergence from the canonical JAZ degron to its very weak ability to associate with COI1. To our knowledge, the COI1-binding capacity of the JAZ7 Jas motif is unknown. However, with high similarity to the divergent JAZ8 Jas motif (Shyu et al. 2012), it is likely JAZ7 either does not or only weakly associates with COI1. JAZ13 was recently identified as a non-TIFY domain containing JAZ protein that represses JA-signaling and is most similar to JAZ8. It appears JAZ13 may also be resistant to JA-COI1-mediated degradation (Thireault et al., 2015). How poorly COI1-interacting JAZ proteins are degraded remains unknown, but Shyu et al. (2012) propose either they are eliminated via a COI1-independent mechanism, are degraded only under very high JA-Ile levels or through COI1-interactions mediated by molecules other than JA-Ile or COR. It is possible that JAZ7 escapes degradation through this mechanism and this allows JAZ7 to fine-tune the signaling pathway. Coupled with the high levels of JAZ7 expression in jaz7-1D, the low degradability of JAZ7 protein would produce a highly modified JA-signaling environment, as observed through our microarray data and jaz7-1D phenotypes.
Fig. 14.
JAZ7 domain structure and proposed mode of JA-hyper-activation in jaz7-1D plants. (A) JAZ7 domain structure highlighting the N-terminal EAR motif, ZIM and Jas domains, and a comparison against conserved JAZ interaction domains in JAZ1. The EAR motif, TIFY motif and JAZ degron for the ZIM and Jas domains respectively are underlined. Residues in the JAZ1 Jas motif shown in bold red are required for COI1-binding. In JAZ1, the ZIM domain mediates NINJA binding and JAZ homo- and heterodimerization, and the Jas domain mediates COI1 binding and interactions with several transcription factors. (B) Proposed model for JA-responses in jaz7-1D plants. Through its EAR domain, JAZ7 binds with the co-repressor TPL to facilitate transcriptional repression. High levels of JAZ7 are associated with hyper-activation of JA-signaling possibly through JAZ7 disturbing components of this network (e.g. TPL, JAM1).
Expected from JAZ repressors, knockdown of jaz1 or jaz10, and overexpression of JAZ8 or JAZ13 result in increased or reduced JA-sensitivity respectively (Yan et al., 2007; Grunewald et al., 2009; Demianski et al., 2012; Shyu et al., 2012; Thireault et al., 2015). The jaz7-1D mutant, however, displays increased JA-sensitivity coupled with up-regulated defense and JA-mediated gene expression. Our results, showing JAZ7 indeed acts as repressor and can bind to both transcriptional activators (e.g. MYC3) and repressors (e.g. JAM1) of JA-responses (Figs 10–12), suggest that JA-sensitivity in jaz7-1D may result from the high and/or ectopic levels of JAZ7 inhibiting normal COI1-JAZ-TPL-TF interactions and highlights one of the difficulties in dissecting the individual roles of proteins that act within multiprotein complexes. The MYC-like bHLH transcription factors bHLH017/JAM1, bHLH013/JAM2 and bHLH003/JAM3 are phylogenetically closely related to MYC2, MYC3 and MYC4, but they lack the conserved activation domain present in the later MYC transcription factors (Fonseca et al., 2014; Sasaki-Sekimoto et al., 2014). Interestingly, jaz7-1D and a loss-of-function jam1 mutant exhibit similar JA-related phenotypes, including small rosette size, increased JA-regulated gene expression (e.g. MYC2, VSP, JAZ10) and increased susceptibility to Pst DC3000 (Nakata et al., 2013; Fonseca et al., 2014). Other individual or triple loss-of-function jam mutants also display similar JA-related phenotypes (Nakata et al., 2013; Sasaki-Sekimoto et al., 2013; Song et al., 2013; Fonseca et al., 2014). The overlap in enhanced JA-responsiveness between jaz7-1D and these jam mutants (Sasaki-Sekimoto et al., 2013) suggests the jaz7-1D phenotypes we observed may be mediated by the high levels of JAZ7 titrating out transcriptional repressors such as JAM1.
Fig. 10.
Priming of JA-regulated gene expression in jaz7-1D. Highly MeJA inducible genes in wild-type were generally not as inducible in jaz7-1D. Shown is a subset of differentially regulated genes in the jaz7-1D mutant following a control or MeJA (6h) treatment as identified by microarray analysis. Col-0 control and MeJA: white and dark gray boxes, respectively. jaz7-1D control and MeJA: light gray and black boxes, respectively. The numbers above MeJA columns represent fold-induction over control treatment. Values are averages ±SE of four biological replicates consisting of pools of 20 plants.
JAM1, JAM2 and JAM3 bind the same DNA motif (G-box, CACGTG) as MYC2, MYC3 and MYC4 (Nakata et al., 2013; Fonseca et al., 2014), and through competitive binding for the same DNA-binding site, these transcriptional repressors and activators can fine-tune JA-mediated responses. An unbiased in silico search (TAIR motif analysis: Statistical Motif Analysis in Promoter or Upstream Gene Sequences, 1000bp) for G-box motifs (Dombrecht et al., 2007; Fernandez-Calvo et al., 2011) in the promoters of the up-regulated genes in jaz7-1D (Supplementary Table S5) identified 19 to contain the CACGTG G-box motif, and 43 and 38 to contain the MYC2 binding variants CACATG and CACGTT, respectively (Dombrecht et al., 2007). The promoters of down-regulated jaz7-1D (Supplementary Table S6) genes also contained these motifs (CACGTG: 7; CACATG: 8; CACGTT: 4). These findings suggest JAZ7 co-ordinates the expression of stress-responsive genes through its interaction with specific MYC or JAM transcription factors and their binding to G-box DNA motifs.
The ZIM domain of JAZ proteins mediates their homo- or heterodimerization (Chini et al., 2009; Chung and Howe, 2009; Chung et al., 2009), but JAZ7 appears to be the only JAZ protein incapable of homodimerizing or forming heterodimers with other JAZ proteins (Chini et al., 2009; Chung and Howe, 2009; reviewed by Pauwels and Goossens, 2011). Another TIFY-containing protein not capable of interacting with JAZ proteins is the non-JAZ protein TIFY8 (Cuéllar Pérez et al., 2014). Although TIFY8 has a functional ZIM domain that mediates transcriptional repression by recruiting TPL via NINJA, its ZIM domain does not confer interactions with JAZ proteins. The differences in JAZ7 protein-protein interactions suggest JAZ7 does not function like the other JAZ repressors. Further to this, although Jas and ZIM motifs in JAZ7 and JAZ8 are similar, suggestive of similar binding activity (Shyu et al., 2012; Wager and Browse, 2012), they regulate binding to different transcription factors. For example, we found JAZ7 and JAZ8 interacted with MYC3/4 and JAM1, but only JAZ8 interacted with MYC2. JAZ8 but not JAZ7, also interacts with JAM2 (Song et al., 2013; Fonseca et al., 2014), with two regulators of stamen development (MYB21 and MYB24) (Song et al., 2011) and with WD-repeat/bHLH/MYB complex members that regulate anthocyanin biosynthesis and trichome initiation (EGL3, GL3, TT8, MYB75, GL1, TTG1) (Qi et al., 2011). These differences in transcription factor binding may explain why JAZ8 overexpression confers reduced JA-sensitivity (Shyu et al., 2012) while high levels of JAZ7 in jaz7-1D plants confers increased JA-sensitivity (this work).
In summary, our results support a model in which F. oxysporum stimulates JA-signaling, resulting in increased JAZ7 expression and JAZ7-TPL-mediated repression contributing to the control of JA-responses and disease progression. Our characterization of the jaz7-1D mutant suggests the ectopic or non-wild-type high levels of JAZ7 in jaz7-1D is a major determinant of its phenotypes and that these abnormal levels may be detrimental to the normal COI1-JAZ-TPL-MYC/JAM regulatory network leading to hyperactivation of JA-signaling (Fig. 14B). Additionally, the unusual protein binding properties of JAZ7 compared to other JAZs may exacerbate this phenotype (e.g. lack of homo- or heterodimerization, divergent JAZ degron). Our results also exemplify the need to use caution when interpreting results from T-DNA insertion lines and proteins that act in multiprotein complexes. Nonetheless, identification of JA-hyperactivation in the jaz7-1D mutant has provided new insight into JA-signaling and why a plant needs many JAZ proteins to fine-tune JA-responses. Future research on JAZ7 expression (tissue/cell specificity) and its interacting partners should reveal mechanistic details on how JAZ7 functions in wild-type plants.
Supplementary data
Supplementary data are available at JXB online.
Fig. S1. Schematic representation of jaz T-DNA insertion lines.
Fig. S2. Screening of jaz T-DNA insertion lines in F. oxysporum disease assays.
Fig. S3. Detection of seed aborts in jaz7-1D and confirmation of jaz7-1.
Fig. S4. Ectopic overexpression of JAZ7 in wild-type plants.
Fig. S5. Backcrossed F2 jaz7-1D seedlings have short roots and are JA-hypersensitive.
Table S1. jaz double and triple mutant lines screened in F. oxysporum disease assays.
Table S2. Primers used for the generation of transgenic plants and Y2-H and Co-IP constructs.
Table S3. Primers used for qRT-PCR.
Table S4. List of genes differentially regulated by genotype from the microarray.
Table S5. Genes differentially expressed ≥ 2-fold in the jaz7-1D line relative to wild-type.
Table S6. Genes differentially expressed ≤ 2-fold in the jaz7-1D line relative to wild-type.
Table S7. List of genes differentially regulated by MeJA treatment from the microarray.
Table S8. Genes differentially expressed ≥ 2-fold in the jaz7-1D line relative to wild-type under MeJA treatment.
Table S9. Genes differentially expressed ≤ 2-fold in the jaz7-1D line relative to wild-type under MeJA treatment.
Table S10. Differentially regulated by MeJA treatment genes sorted by MeJA inducibility in wild-type plants.
Acknowledgements
LFT was supported by a CSIRO OCE postdoctoral fellowship. We thank the AGRF and the support it receives from the Australian Government, the ABRC and NASC for the Arabidopsis T-DNA insertion lines (Alonso et al., 2003; Woody et al., 2007) and Roger Shivas (Queensland Department of Primary Industries and Fisheries, Australia) for the F. oxysporum. We also thank Shi Zhuge and Huan Zhao for technical assistance, Dr Laurence Tomlinson for Golden Gate cloning, and Drs Brendan Kidd and Jonathan Anderson for critical reading of the manuscript and useful discussions.
References
- Adio AM, Casteel CL, De Vos M, Kim JH, Joshi V, Li B, Juéry C, Daron J, Kliebenstein DJ, Jander G. 2011. Biosynthesis and defensive function of Nδ-acetylornithine, a jasmonate-induced Arabidopsis metabolite. The Plant Cell 23, 3303–3318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agrios GN. 2005. Plant Pathology 5th edn. Elsevier Academic Press. [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, et al. 2003. Genome-wide insertional mutagenesis of Arabidopsis thaliana . Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K. 2004. Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. The Plant Cell 16, 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium 2011. Evidence for network evolution in an Arabidopsis interactome map. Science 333, 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ascenzi R, Gantt JS. 1999. Molecular genetic analysis of the drought-inducible linker histone variant in Arabidopsis thaliana . Plant Molecular Biology 41, 159–169. [DOI] [PubMed] [Google Scholar]
- Baldwin I. 1998. Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proceedings of the National Academy of Sciences, USA 95, 8113–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A. 2004. Ethylene Response Factor 1 mediates Arabidopsis resistance to the soilborne fungus Fusarium oxysporum . Molecular Plant Microbe Interactions 17, 763–770. [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A. 2008. Arabidopsis defense response against Fusarium oxysporum . Trends in Plant Science 13, 145–150. [DOI] [PubMed] [Google Scholar]
- Berrocal-Lobo M, Molina A, Solano R. 2002. Constitutive expression of ETHYLENE-RESPONSE-FACTOR1 in Arabidopsis confers resistance to several necrotrophic fungi. The Plant Journal 29, 23–32 [DOI] [PubMed] [Google Scholar]
- Bowling SA, Clarke JD, Liu Y, Klessig DF, Dong X. 1997. The cpr5 mutant of Arabidopsis expresses both NPRl -dependent and NPRl -independent resistance. The Plant Cell 9, 1573–1584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Causier B, Ashworth M, Guo W, Davies B. 2012. The TOPLESS interactome: a framework for gene repression in Arabidopsis. Plant Physiology 158, 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerrudo I, Keller MM, Cargnel MD, Demkura PV, de Wit M, Patitucci MS, Pierik R, Pieterse CMJ, Ballaré CL. 2012. Low red/far-red ratios reduce Arabidopsis resistance to Botrytis cinerea and jasmonate responses via a COI1-JAZ10-dependent, salicylic acid-independent mechanism. Plant Physiology 158, 2042–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cevik V, Kazan K. 2013. Agroinfiltration of Nicotiana benthamiana leaves for co-localization of regulatory proteins involved in jasmonate signalling. Methods in Molecular Biology 1011, 199–208. [DOI] [PubMed] [Google Scholar]
- Cevik V, Kidd BN, Zhang P, et al. 2012. MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiology 160, 541–555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan YL, Prasad V, Chen SKH, Liu PC, Chan MT, Cheng CP. 2005. Transgenic tomato plants expressing an Arabidopsis thionin (Thi2.1) driven by a fruit-inactive promoter battle against phytopathogenic attack. Planta 221, 386–393. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Sun L, Qi T, Zhang B, Peng W, Liu Y, Xie D. 2011. The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Molecular Plant . 4, 279–288. [DOI] [PubMed] [Google Scholar]
- Chini A, Boter M, Solano R. 2009. Plant oxylipins: COI1/JAZs/MYC2 as the core jasmonic acid-signalling module. FEBS Journal 276, 4682–4692. [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernandez G, et al. 2007. The JAZ family of repressors is the missing link in jasmonate signaling. Nature 448, 666–671. [DOI] [PubMed] [Google Scholar]
- Chory J, Nagpal P, Peto CA. 1991. Phenotypic and genetic analysis of det2, a new mutant that affects light-regulated seedling development in Arabidopsis. The Plant Cell 3, 445–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Cooke TF, Depew CL, Patel LC, Ogawa N, Kobayashi Y, Howe GA. 2011. Alternative splicing expands the repertoire of dominant JAZ repressors of jasmonate signaling. The Plant Journal 63, 613–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Howe GA. 2009. A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. The Plant Cell 21, 131–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Koo AJK, Gao X, Jayanty S, Thines B, Jones D, Howe GA. 2008. Regulation and function of Arabidopsis JASMONATE ZIM-domain genes in response to wounding and herbivory. Plant Physiology 146, 952–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Niu Y, Browse J, Howe GA. 2009. Top hits in contemporary JAZ: an update on jasmonate signaling. Phytochemistry 70, 1547–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuéllar Pérez A, Nagels Durand A, Vanden Bossche R, et al. 2014. The non-JAZ TIFY protein TIFY8 from Arabidopsis thaliana is a transcriptional repressor. PLoS ONE 9, e84891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Torres Zabala M, Zhai B, Jayaraman S, Eleftheriadou G, Winsbury R, Yang R, Truman W, Tang S, Smirnoff N, Grant M. 2015. Novel JAZ co-operativity and unexpected JA dynamics underpin Arabidopsis defence responses to Pseudomonas syringae infection. New Phytologist doi: 10.1111/nph.13683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demianski AJ, Chung KM, Kunkel BN. 2012. Analysis of Arabidopsis JAZ gene expression during Pseudomonas syringae pathogenesis. Molecular Plant Pathology 13, 46–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pietro A, Madrid MP, Caracuel Z, Delgado-Jarana J, Roncero MIG. 2003. Fusarium oxysporum: exploring the molecular arsenal of a vascular wilt fungus. Molecular Plant Pathology 4, 315–325. [DOI] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, Kirkegaard JA, Ross JJ, Reid JB, Fitt GP, Sewelam N, Schenk PM, Manners JM, Kazan K. 2007. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. The Plant Cell 19, 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Z, Zhou X, Ling Y, Zhang Z, Su Z. 2010. agriGO: a GO analysis toolkit for the agricultural community. Nucleic Acids Research 38, W64–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earley K, Haag JR, Pontes O, Opper K, Juehne T, Song K, Pikaard CS. 2006. Gateway-compatible vectors for plant functional genomics and proteomics. Plant Journal 45, 616–629. [DOI] [PubMed] [Google Scholar]
- Ellis C, Turner JG. 2001. The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. The Plant Cell 13, 1025–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engler C, Kandzia R, Marillonnet S. 2008. A one pot, one step, precision cloning method with high throughput capability. PLoS One , 3, e3647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epple P, Apel K, Bohlmann H. 1997. Overexpression of an endogenous thionin enhances resistance of Arabidopsis against Fusarium oxysporum . The Plant Cell 9, 509–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, et al. 2011. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. The Plant Cell 23, 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca S, Fernández-Calvo P, Fernández GM, et al. 2014. bHLH003, bHLH013 and bHLH017 are new targets of JAZ repressors negatively regulating JA responses. PLoS One 9, e86182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genger RK, Jurkowski GI, McDowell JM, Lu H, Jung HW, Greenberg JT, Bent AF. 2008. Signaling pathways that regulate the enhanced disease resistance of Arabidopsis “defense, no death” mutants. Molecular Plant Microbe Interactions 21, 1285–1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepstein S, Sabehi G, Carp MJ, Hajouj T, Nesher MF, Yariv I, Dor C, Bassani M. 2003. Large-scale identification of leaf senescence-associated genes. The Plant Journal 36, 629–642. [DOI] [PubMed] [Google Scholar]
- Gleason C, Huang S, Thatcher LF, Foley RC, Anderson CR, Carroll AJ, Millar AH, Singh KB. 2011. Mitochondrial complex II has a key role in mitochondrial-derived reactive oxygen species influence on plant stress gene regulation and defense. Proceedings of the National Academy of Sciences, USA 108, 10768–10773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald W, Vanholme B, Pauwels L, Plovie E, Inzé D, Gheysen G, Goossens A. 2009. Expression of the Arabidopsis jasmonate signalling repressor JAZ1/TIFY10A is stimulated by auxin. EMBO Reports 10, 923–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilpert B, Bohlmann H, op den Camp RO, Przybyla D, Miersch O, Buchala A, Apel K. 2001. Isolation and characterization of signal transduction mutants of Arabidopsis thaliana that constitutively activate the octadecanoid pathway and form necrotic microlesions. The Plant Journal 26, 435–446. [DOI] [PubMed] [Google Scholar]
- Hiratsu K, Matsui K, Koyama T, Ohme-Takagi M. 2003. Dominant repression of target genes by chimeric repressors that include the EAR motif, a repression domain, in Arabidopsis. The Plant Journal 34, 733–739. [DOI] [PubMed] [Google Scholar]
- Hruz T, Laule O, Szabo G, Wessendorp F, Bleuler S, Oertle L, Widmayer P, Gruissem W, Zimmermann P. 2008. Genevestigator v3: a reference expression database for the meta-analysis of transcriptomes. Advances in Bioinformatics 2008, 420747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagale S, Links MG, Rozwadowski K. 2010. Genome-wide analysis of ethylene-responsive element binding factor-associated amphiphilic repression motif-containing transcriptional regulators in Arabidopsis. Plant Physiology 152, 1109–1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L, Schilmiller AL, Staswick PE, He SY, Howe GA. 2008. COI1 is a critical component of a receptor for jasmonate and the bacterial virulence factor coronatine. Proceedings of the National Academy of Sciences, USA 105, 7100–7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K. 2006. Negative regulation of defence and stress genes by EAR-motif-containing repressors. Trends in Plant Science 11, 109–112. [DOI] [PubMed] [Google Scholar]
- Kazan K, Manners JM. 2012. JAZ repressors and the orchestration of phytohormone crosstalk. Trends in Plant Science 17, 22–31. [DOI] [PubMed] [Google Scholar]
- Kidd BN, Edgar CI, Kumar KK, Aitken EA, Schenk PM, Manners JM, Kazan K. 2009. The mediator complex subunit PFT1 is a key regulator of jasmonate-dependent defense in Arabidopsis. Plant Cell 21, 2237–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kloek AP, Verbsky ML, Sharma SB, Schoelz JE, Vogel J, Klessig DF, Kunkel BN. 2001. Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. The Plant Journal 26, 509–522. [DOI] [PubMed] [Google Scholar]
- Leone M, Keller MM, Cerrudo I, Ballaré CL. 2014. To grow or defend? Low red: far-red ratios reduce jasmonate sensitivity in Arabidopsis seedlings by promoting DELLA degradation and increasing JAZ10 stability. New Phytologist 204, 355–367. [DOI] [PubMed] [Google Scholar]
- Li J, Li Y, Chen S, An L. 2010. Involvement of brassinosteroid signals in the floral-induction network of Arabidopsis. Journal of Experimental Botany 61, 4221–4230. [DOI] [PubMed] [Google Scholar]
- Lionetti V, Raiola A, Camardella L, Giovane A, Obel N, Pauly M, Favaron F, Cervone F, Bellincampi D. 2007. Overexpression of pectin methylesterase inhibitors in Arabidopsis restricts fungal infection by Botrytis cinerea . Plant Physiology 2143, 1871–1880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath KC, Dombrecht B, Manners JM, Schenk PM, Edgar CI, Maclean DJ, Scheible WR, Udvardi MK, Kazan K. 2005. Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiology 139, 949–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melotto M Mecey C Niu Y, et al. 2008. A critical role of two positively charged amino acids in the Jas motif of Arabidopsis JAZ proteins in mediating coronatine- and jasmonoyl isoleucine-dependent interactions with the COI1 F-box protein. The Plant Journal 55, 979–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M, Mitsuda N, Herde M, Koo AJ, Moreno JE, Suzuki K, Howe GA, Ohme-Takagi M. 2013. A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis. The Plant Cell 25, 1641–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata M, Ohme-Takagi M. 2013. Two bHLH-type transcription factors, JA-ASSOCIATED MYC2-LIKE2 and JAM3, are transcriptional repressors and affect male fertility. Plant Signaling and Behavior 8, e26473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Figueroa P, Browse J. 2011. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. Journal of Experimental Botany 62, 2143–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L Barbero GF Geerinck J, et al. 2010. NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauwels L, Goossens A. 2011. The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23, 3089–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieterse CM, Ronald Pierik R, Van Wees SC. 2014. Different shades of JAZ during plant growth and defense. New Phytologist 204, 261–264. [DOI] [PubMed] [Google Scholar]
- Qi T, Song S, Ren Q, Wu D, Huang H, Chen Y, Fan M, Peng W, Ren C, Xie D. 2011. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana . The Plant Cell 23, 1795–1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL. 2003. Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. The Plant Cell 15, 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y, Jikumaru Y, Obayashi T, Saito H, Masuda S, Kamiya Y, Ohta H, Shirasu K. 2013. Basic helix-loop-helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiology 163, 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y, Saito H, Masuda S, Shirasu K, Ohta H. 2014. Comprehensive analysis of protein interactions between JAZ proteins and bHLH transcription factors that negatively regulate jasmonate signaling. Plant Signaling and Behavior 9, e27639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Rusu AG, Manners JM, Maclean DJ. 2005. The SEN1 gene of Arabidopsis is regulated by signals that link plant defense responses and senescence. Plant Physiology and Biochemistry 43, 997–1005 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW. 2012. NIH Image to ImageJ: 25 years of image analysis. Nature Methods 9, 671–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheard LB, Tan X, Mao H, et al. 2010. Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shyu C, Figueroa P, Depew CL, Cooke TF, Sheard LB, Moreno JE, Katsir L, Zheng N, Browse J, Howe GA. 2012. JAZ8 lacks a canonical degron and has an EAR motif that mediates transcriptional repression of jasmonate responses in Arabidopsis. The Plant Cell 24, 536–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sohn KH, Hughes RK, Piquerez SJ, Jones JD, Banfield MJ. 2012. Distinct regions of the Pseudomonas syringae coiled-coil effector AvrRps4 are required for activation of immunity. Proceedings of the National Academy of Sciences, USA 109, 16371–16376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Fan M, Zhang X, Gao H, Huang H, Wu D, Guo H, Xie D. 2013. The bHLH subgroup IIId factors negatively regulate jasmonate-mediated plant defense and development. PLoS Genetics 9, e1003653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S, Qi T, Huang H, Ren Q, Wu D, Chang C, Peng W, Liu Y, Peng J, Xie D. 2011. The jasmonate-ZIM domain proteins interact with the R2R3-MYB transcription factors MYB21 and MYB24 to affect jasmonate-regulated stamen development in Arabidopsis. The Plant Cell 23, 1000–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher LF, Manners JM, Kazan K. 2009. Fusarium oxysporum hijacks COI1-mediated jasmonate signaling to promote disease development in Arabidopsis. The Plant Journal 58, 927–939. [DOI] [PubMed] [Google Scholar]
- Thatcher LF, Powell JJ, Aitken EAB, Kazan K, Manners JM. 2012. a The Lateral Organ Boundaries Domain transcription factor LBD20 functions in Fusarium wilt susceptibility and jasmonate signaling in Arabidopsis. Plant Physiology 160, 407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thatcher LF, Gardiner DM, Kazan K, Manners JM. 2012b. A highly conserved effector in Fusarium oxysporum is required for full virulence on Arabidopsis. Molecular Plant Microbe Interactions 25,180–190. [DOI] [PubMed] [Google Scholar]
- Thatcher LF, Kamphuis LG, Hane JK, Onate-Sanchez L, Singh KB. 2015. The Arabidopsis KH-domain RNA-binding protein ESR1 functions in components of jasmonate signalling, unlinking growth restraint and resistance to stress. PLoS ONE 10, . [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thines B, Katsir L, Melotto M, Niu Y, Mandaokar A, Liu G, Nomura K, He SY, Howe GA, Browse J. 2007. JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signaling. Nature 448, 661–665. [DOI] [PubMed] [Google Scholar]
- Thireault C, Shyu C, Yoshida Y, St Aubin B, Campos ML, Howe GA. 2015. Repression of jasmonate signaling by a non-TIFY JAZ protein in Arabidopsis. Plant Journal 82, 669–679. [DOI] [PubMed] [Google Scholar]
- Thomma BPHJ, Eggermont K, Penninckx IAMA, Mauch-Mani B, Vogelsang R, Cammue BPA, Broekaert WF. 1998. Separate jasmonate-dependent and salicylate-dependent defense-response pathways in Arabidopsis are essential for resistance to distinct microbial pathogens. Proceedings of the National Academy of Sciences, USA 95, 15107–15111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Yoo MJ, Koh J, Liu L, Chen Y, Acikgoz D, Wang Q, Chen S. 2014. Molecular reprogramming of Arabidopsis in response to perturbation of jasmonate signaling. Journal of Proteome Research 13, 5751–5766. [DOI] [PubMed] [Google Scholar]
- Yan J, Zhang C, Gu M, et al. 2009. The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. The Plant Cell 21, 2220–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y, Stolz S, Chetelat A, Reymond P, Pagni M, Dubugnon L, Farmer EE. 2007. A downstream mediator in the growth repression limb of the jasmonate pathway. The Plant Cell 19, 2470–2483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida S, Ito M, Nishida I, Watanabe A. 2001. Isolation and RNA gel blot analysis of genes that could serve as potential molecular markers for leaf senescence in Arabidopsis thaliana . Plant and Cell Physiology 242, 170–178. [DOI] [PubMed] [Google Scholar]
- Yu J, Zhang Y, Di C, et al. 2015. JAZ7 negatively regulates dark-induced leaf senescence in Arabidopsis. Journal of Experimental Botany pii: erv487 [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wager A, Browse J. 2012. Social network: JAZ protein interactions expand our knowledge of jasmonate signaling. Frontiers in Plant Science 3, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B. 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woody ST, Austin-Phillips S, Amasino RM, Krysan PJ. 2007. The WiscDsLox T-DNA collection: an Arabidopsis community resource generated by using an improved high-throughput T-DNA sequencing pipeline. Journal of Plant Research 120, 157–165. [DOI] [PubMed] [Google Scholar]
- Zhai Q, Zhang X, Wu F, Feng H, Deng L, Xu L, Zhang M, Wang Q, Li C. 2015. Transcriptional mechanism of jasmonate receptor COI1-mediated delay of flowering time in Arabidopsis. The Plant Cell 27, 2814–2828. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.