Highlight
Allantoin, a stress-related purine metabolite, can activate JA responses via ABA in Arabidopsis, suggesting its possible involvement in the homeostasis of these phytohormones and their interplay in stress signaling.
Key words: ABA, Arabidopsis, JA, purine metabolism, small metabolite, stress hormone, stress response, ureide.
Abstract
Allantoin is a metabolic intermediate of purine catabolism that often accumulates in stressed plants. Recently, we used Arabidopsis knockout mutants (aln) of ALLANTOINASE to show that this purine metabolite activates abscisic acid (ABA) production, thereby stimulating stress-related gene expression and enhancing seedling tolerance to abiotic stress. A detailed re-examination of the microarray data of an aln mutant (aln-1) confirmed the increased expression of ABA-related genes and also revealed altered expression of genes involved in jasmonic acid (JA) responses, probably under the control of MYC2, a master switch in the JA signaling pathway. Consistent with the transcriptome profiles, the aln-1 mutant displayed increased JA levels and enhanced responses to mechanical wounding and exogenous JA. Moreover, aln mutants demonstrated modestly increased susceptibility to Pseudomonas syringae and Pectobacterium carotovorum, probably reflecting the antagonistic action of MYC2 on the defense against these bacterial phytopathogens. Exogenously administered allantoin elicited the expression of JA-responsive genes, including MYC2, in wild-type plants, supporting the idea that allantoin might be responsible for the observed JA-related phenotypes of aln mutants. However, mutants deficient in bioactive JA (jar1-1), insensitive to JA (myc2-3), or deficient in ABA (aba2-1 and bglu18) suppressed the effect of exogenous allantoin. The suppression was further confirmed in aln-1 jar1-1 and aln-1 bglu18 double mutants. These results indicate that allantoin can activate the MYC2-regulated JA signaling pathway through ABA production. Overall, this study suggests a possible connection of purine catabolism with stress hormone homeostasis and signaling, and highlights the potential importance of allantoin in these interactions.
Introduction
Plants have evolved a complex diversity of metabolic pathways to maintain and improve their growth and survival in natural environments, including stressful conditions. Plants confronted with stressful conditions typically accumulate an amazing array of small organic compounds, including metabolic products and intermediates. Numerous in vitro and in vivo studies have shown that various small metabolites serve specialized functions in cellular protection, such as scavenging reactive oxygen species (ROS), osmoregulation, stabilization of protein structure, and maintenance of membrane integrity (Chen and Murata, 2002). Certain small metabolites also play signaling and regulatory roles in mediating plant responses and adaptation to stress (see, for example, Szabados and Savouré, 2010; Wind et al., 2010; Hussain et al., 2011; Lunn et al., 2014). However, the physiological function and significance of the accumulation of many stress-associated small compounds remain elusive.
Allantoin, a nitrogen-rich heterocyclic compound, occurs ubiquitously in plants as an intermediary metabolite of purine catabolism. Allantoin and its acyclic metabolite allantoate are collectively called ureides and, in tropical legumes, they serve as the vehicle for storage and xylem transport of symbiotically fixed nitrogen (Smith and Atkins, 2002). In non-legume plants, purine catabolism is believed to function as a fundamental route for nitrogen recycling and remobilization, since the sequential degradation of the purine ring, which initiates from oxidation of xanthine, proceeds through ureide hydrolysis and releases four molar equivalents of ammonia (Supplementary Fig. S1 at JXB online; Zrenner et al., 2006; Werner and Witte, 2011). However, conventional metabolite analyses and recent metabolomics studies revealed allantoin as a major purine metabolite in Arabidopsis (Arabidopsis thaliana), rice (Oryza sativa L.), and other species under various stress conditions such as drought (Oliver et al., 2011; Silvente et al., 2011; Yobi et al., 2013), high salinity (Kanani et al., 2010; Wu et al., 2012; Wang et al., 2016), cold (Kaplan et al., 2004), nutrient constraint (Nikiforova et al., 2005; Rose et al., 2012; Coneva et al., 2014), extended darkness (Brychkova et al., 2008), and pathogen invasion (Montalbini, 1991).
The possible involvement of allantoin in stress protection was initially suggested from studies on XANTHINE DEHYDROGENASE (XDH) knockout/knockdown mutants (xdh1 and RNAi lines) of Arabidopsis. In these mutants, defects in xanthine oxidation resulted in significantly impaired stress tolerance, as well as stress-induced symptoms such as early senescence (Nakagawa et al., 2007; Brychkova et al., 2008; Watanabe et al., 2010, 2014a). The stress-susceptible xdh phenotype might be at least in part attributable to purine metabolite deficiency, because supplementation with allantoin or its precursor uric acid, a product of XDH, reversed the mutant phenotype. Recent studies on knockout mutants (aln) of the Arabidopsis ALLANTOINASE gene (ALN) provided more direct evidence; the aln mutants showed constitutive accumulation of allantoin and enhanced seedling survival and growth under drought and osmotic stress (Watanabe et al., 2014b). Allantoin may function in stress protection by scavenging ROS; indeed, leaf disc assays showed that exogenous allantoin alleviated ROS accumulation and cell death (Brychkova et al., 2008). Allantoin may also activate stress responses, as its accumulation in aln mutants or supplementation of wild-type (WT) seedlings increased basal levels of the stress phytohormone abscisic acid (ABA), with concomitant activation of stress-related gene expression (Watanabe et al., 2014b).
ABA has pivotal roles in plant growth and adaptive responses to various environmental stresses, mostly through antagonistic, synergistic, and additive interactions with other phytohormones such as jasmonic acid (JA), salicylic acid (SA), and ethylene. ABA, in particular, has long been known to regulate JA responses positively. JA and its metabolites, such as the bioactive metabolite jasmonoyl-l-isoleucine (JA-Ile), are collectively known as jasmonates; the jasmonates regulate diverse aspects of plant development and defense responses to insect herbivores and microbial pathogens (Wasternack and Hause, 2013). The complex interplay of the ABA and JA pathways controls defense gene expression in response to biotic stress. Early studies in potato and tomato plants reported that ABA elicits wounding responses and wound-inducible JA production (Pēna-Cortés et al., 1989, 1995). Recent studies in Arabidopsis demonstrated that ABA can increase JA levels in disease conditions, and activate JA signaling for systemic induced resistance against herbivores (Adie et al., 2007; Fan et al., 2009; Vos et al., 2013).
In Arabidopsis, the JA signaling pathway consists of two antagonistically acting branches, the ABA-co-regulated MYC branch and the ethylene-co-regulated ETHYLENE RESPONSE FACTOR (ERF) branch. The MYC branch responds to herbivore feeding and mechanical wounding, and is controlled by the basic helix–loop–helix leucine zipper transcription factors MYC2/3/4 (Anderson et al., 2004; Fernández-Calvo et al., 2011; Niu et al., 2011). The ERF branch responds to infection with necrotrophic pathogens and is controlled by the APETALA2 (AP2)/ERF domain transcription factors ERF1 and OCTADECANOID-RESPONSIVE ARABIDOPSIS AP2/ERF 59 (ORA59) (Penninckx et al., 1998; Lorenzo et al., 2004; Pré et al., 2008). ABA acts synergistically on the MYC branch and antagonistically on the ERF branch pathways (Anderson et al., 2004).
The effects of ABA on these branches are mediated, at least in part, through transcriptional regulation by the ABA/JA-responsive MYC2 transcription factor [also known as JASMONATE INSENSITIVE1 (JAI1/JIN1)] (Anderson et al., 2004; Kazan and Manners, 2013). Originally identified as a positive regulator of ABA-dependent drought responses (Abe et al., 1997), MYC2 orchestrates the expression of early JA-responsive genes that play key roles in JA signaling and responses. While activating herbivore and wounding responses, MYC2 acts as a negative regulator of resistance to necrotrophic pathogens by repressing ERF1 and ORA59 (Lorenzo et al., 2004; Dombrecht et al., 2007). MYC2 also suppresses SA-dependent defenses against biotrophic pathogens and innate immune responses (Laurie-Berry et al., 2006; Millet et al., 2010). In addition to its antagonistic co-ordination of biotic stress responses to herbivores and pathogens, MYC2 also participates in regulation of a broad spectrum of JA-related responses, including the biosynthesis of JA and anthocyanin, JA-induced root growth inhibition, and oxidative stress tolerance (Lorenzo et al., 2004; Dombrecht et al., 2007; Kazan and Manners, 2013; Wasternack and Hause, 2013). Thus, MYC2 is considered a master regulator of most aspects of the JA signaling pathway as well as a hub that integrates ABA and JA signaling.
Given the critical role of JA and its intimate regulatory interaction with ABA in plant stress responses, this study investigated whether allantoin affects JA signaling and responsiveness in Arabidopsis. Detailed phenotypic and transcriptomic analyses of allantoin-accumulating aln mutants revealed activation of JA responses at the levels of gene expression, metabolism, physiology, and pathophysiology, in a MYC2-regulated manner. These JA-related mutant phenotypes involve allantoin because exogenous application of allantoin in WT Arabidopsis plants induced the expression of MYC2 and MYC2-regulated JA-responsive genes. However, disrupting JA signaling or blocking ABA production abrogated the effect of both the aln mutation and exogenous allantoin. The results presented here thus provide evidence that allantoin can activate JA responses via ABA, and suggest that purine catabolism may have the potential to affect the homeostasis of these phytohormones and their interplay in stress signaling.
Materials and methods
Plant materials and growth conditions
Arabidopsis thaliana (L.) Heynh., accession Columbia-0, was used for all experiments. Seeds of the following mutants and T-DNA insertion lines were obtained from the Arabidopsis Biological Resource Center (Ohio State University, Columbus, OH, USA): aah (SALK_112631; Todd and Polacco, 2006), aba2-1 (CS156; Léon-Kloosterziel et al., 1996), aln-1 (SALK_000325; Yang and Han, 2004; Watanabe et al., 2014b; also known as aln), aln-2 (SALK_146783; Watanabe et al., 2014b), bglu18 (SALK_075731C; Ogasawara et al., 2009), jar1-1 (CS8072; Staswick et al., 1992), myc2-3 (SALK_061267; identical to jin1-8 reported in Lorenzo et al., 2004), and xdh1 (SALK_148366; Watanabe et al., 2014b). The double mutants aln-1 bglu18 and aln-1 jar1-1 were constructed by manual cross-pollination. A homozygous complemented line of aln-1 (aln-1 35S:ALN) was described in our previous study (Watanabe et al., 2014b). Under standard conditions, surface-sterilized seeds were sown on solid medium containing half-strength Murashige and Skoog (1/2MS) salts, 1.0% (w/v) sucrose, and 0.3% (w/v) gellan gum. After 2 d at 4 °C to break seed dormancy, plates were placed in a growth chamber maintained at 23 °C under long-day conditions (16h light/8h dark) with 70 µmol photons m−2 s−1.
Gene expression analyses
For transcriptome analysis, the previously deposited raw data from the microarray data sets (NCBI Gene Expression Omnibus GSE44922) were parametrically normalized by the three-parameter lognormal distribution method (Konishi, 2004), using the SuperNORM data service (Skylight Biotech Inc., Akita, Japan). The renormalized data have been deposited under accession number GSE73841. The significance of differentially expressed genes showing ≥3-fold changes was statistically tested by a two-way ANOVA with a significance threshold set at 0.001 (Konishi, 2011). Functional enrichment analyses of the Biological Process Gene Ontology (GO) terms were carried out using the BioMaps tool of VirtualPlant version 1.3 (Katari et al., 2010; http://virtualplant.bio.nyu.edu/cgi-bin/vpweb/, accessed 22 February 2016) in the default-setting mode (Fisher’s exact test with false discovery rate correction, P<0.01), with the Arabidopsis genome annotation (TAIR release 10; https://www.arabidopsis.org/, accessed 22 February 2016) as the background.
For quantitative transcript analysis, samples of aerial parts were collected from 2-week-old seedlings that were grown aseptically on solid 1/2MS medium with or without added allantoin (Wako Pure Chemical Industries, Ltd, Osaka, Japan) or allantoic acid (Carbosynth Limited, Berkshire, UK). RNA extraction and real-time reverse transcription quantitative PCR (RT-qPCR) were performed as described previously (Watanabe et al., 2014b). Relative gene expression levels were calculated by the 2−∆∆Ct method (Livak and Schmittgen, 2001) after normalization to ACTIN2 expression levels. Primer sequences for target genes are listed in Supplementary Table S1.
Quantification of JA and JA-Ile
JA and JA-Ile were quantified following the method of Preston et al. (2009) with minor modifications, by using the stable isotope-labeled compounds [2H2]JA and [13C6]JA-Ile as internal standards for quantitative LC-electrospray ionization-tandem MS (LC-ESI-MS/MS), as detailed in Supplementary Methods S1; Supplementary Table S2.
Metabolite analyses
Anthocyanin was extracted from the aerial parts of 8-day-old seedlings with methanol acidified with 1% (v/v) hydrochloric acid, and the anthocyanin level was calculated based on the absorbance at 530nm and 657nm according to Teng et al. (2005). Allantoin was determined in 2-week-old seedlings as described previously (Watanabe et al., 2014b).
Root growth inhibition assay
Seeds were surface sterilized and sown onto square plates of solid 1/2MS medium containing methyl jasmonate (MeJA; Sigma-Aldrich, St Louis, MO, USA). Square plates were set vertically in a growth chamber and, 8 d after germination, the length of each primary root was measured using ImageJ version 1.45 (http://rsbweb.nih.gov/ij/, accessed 22 February 2016).
Wounding treatment
All rosette leaves of 2-week-old aseptically grown seedlings were wounded mechanically once by crushing with tweezers. Aerial parts of the plants were collected at the indicated time after wounding and then quick-frozen in liquid nitrogen for RNA extraction.
Pathogen inoculation
Surface-sterilized seeds were germinated and grown on 0.3% (w/v) Phytagel-solidified 1/2MS medium at 24 °C under a photoperiod of 12h light/12h dark with a light intensity of 150–200 µmol photons m−2 s−1. Two weeks after germination, seedlings were used for evaluation of resistance to Pseudomonas syringae pv. tomato (Pst) strain DC3000 according to Ishiga et al. (2011) and to Pectobacterium carotovorum subsp. carotovorum (Pcc) EC1 as described in Higashi et al. (2008). For Pst DC3000, seedlings were inoculated by flooding with a bacterial suspension [5×106 colony-forming units (CFU) ml−1]. At 48h and 96h post-inoculation (hpi), aerial parts were collected. After being surface sterilized with 5% (v/v) H2O2, samples were homogenized in sterile water to recover internal bacteria and, upon appropriate dilution, bacterial populations were determined by colony formation on MG agar medium containing rifampicin. For infection with Pcc EC1, intact leaves were drop-inoculated with 5 μl of a bacterial suspension in 0.9% (w/v) NaCl; this suspension was pre-adjusted to a 600nm optical density of 0.01. The levels of disease symptoms were categorized as: level 0, no symptoms; level 1, symptoms restricted within the inoculated region; level 2, symptoms extended outside of the inoculated region.
Statistical analyses
All data are shown as means ± SE. Means of two groups were compared by Student’s t-test at a 5% level of significance with Microsoft Excel. Means of three or more groups were compared by Tukey’s multiple comparison test, which was performed with IBM SPSS statistic, version 21.0 (IBM, New York, NY, USA). Statistical difference of Pcc EC1 resistance was tested with Fisher’s exact test using RStudio (version 0.98; https://www.rstudio.com/, accessed 22 February 2016).
Accession numbers
Arabidopsis Genome Initiative numbers for the genes mentioned in this article are summarized in Supplementary Methods S1.
Results
The aln mutants show increased expression of ABA- and JA-responsive genes and decreased expression of SA-related genes
Previous work reported that normally grown 2-week-old aln mutant (aln-1) seedlings exhibited genome-wide alterations in the levels of transcripts associated with stress responses, with increased expression of ABA-responsive genes. Although not conspicuous in the GO analysis (Table 1 of Watanabe et al., 2014b ), the aln mutants also showed increased expression of JA response signatures including genes involved in JA metabolism and signaling, such as 13-LIPOXYGENASE (LOX) and JASMONATE ZIM-DOMAIN (JAZ) PROTEINS (Supplementary Table S2 of Watanabe et al., 2014b). For this reason, and to improve the interpretation of the transcriptome profiles, the first step in this study was to renormalize these microarray data according to a three-parameter lognormal distribution model (Konishi, 2004). This refinement of the data resulted in identification of 324 genes (211 increased, Supplementary Table S3; 113 reduced, Supplementary Table S4) with a significant change of at least 3.0-fold in their transcript levels (P<0.001). The physiological and functional aspects of these differentially expressed genes were assessed by GO enrichment analysis for biological process terms using VirtualPlant software (P<0.01). As shown in Supplementary Fig. S2 as a hierarchical tree graph, 21 out of 47 of the significantly enriched GO terms for genes with increased transcript levels were relevant to the ‘response to stimulus’ category, which included genes associated with ABA and abiotic stress responses (‘response to abscisic acid stimulus’ and ‘response to abiotic stimulus’ along with its eight descendant GO terms), and also genes associated with JA-related stress responses (‘response to jasmonic acid stimulus’, ‘response to wounding’, ‘defense response to fungus’, and ‘response to biotic stimulus’). Moreover, ‘jasmonic acid metabolic process’ was identified as significantly enriched in the ‘cellular process’ category. For genes with reduced transcript levels, more than half (20 out of 33) of the significantly enriched GO terms belonged to three related categories (‘response to stimulus’, ‘multi-organism process’, and ‘immune system process’), which emphasized biotic defense responses mainly involving SA, such as innate and acquired immunity (Supplementary Fig. S3). This downward shift in basal expression levels of SA-responsive genes was confirmed by RT-qPCR of the canonical SA-marker PATHOGENESIS-RELATED PROTEIN 1 (PR-1) (Supplementary Fig. S4). Taken together, the results of re-examining the transcriptome profiling suggested that the aln mutation caused increased transcript levels of ABA- and JA-responsive genes but decreased transcript levels of SA-related genes at the seedling stage under normal growth conditions.
The aln mutants show altered basal expression of MYC2 and MYC2-regulated genes in JA metabolism, signaling, and responses
Next, mapping of the relevant microarray-derived expression data onto the current model of the MYC2-modulated JA signaling pathways was conducted to obtain a more detailed picture of JA-associated gene expression in the aln-1 mutant (Kazan and Manners, 2013), with emphasis on the crosstalk with ABA and SA (Fig. 1A). In the ABA-co-regulated MYC branch, this mapping revealed a significant increase in basal expression levels of two of the three genes encoding Arabidopsis NAM/ATAF/CUC (ANAC) transcription factors, ANAC019, ANAC055, and ANAC072, immediate target genes positively regulated by MYC2 (Bu et al., 2008). Transcript levels of downstream MYC branch marker genes that these ANAC transcription factors regulate, positively or negatively, also showed major changes. Genes with increased transcript levels included the marker VEGETATIVE STORAGE PROTEIN 1 (VSP1) along with SABATH METHYLTRANSFERASE (also known as BSMT1) and SALICYLIC ACID GLUCOSYLTRANSFERASE 1 (SAGT1), both of which encode SA-inactivating enzymes. Genes with decreased transcript levels included ISOCHORISMATE SYNTHASE 1 (ICS1), which encodes an SA biosynthetic enzyme. For ERF branch genes, markedly reduced basal expression was observed for the key transcription factor gene ORA59 whose expression is negatively regulated by MYC2 (Dombrecht et al., 2007), and the ORA59 downstream target and representative ERF branch marker PLANT DEFENSIN 1.2 (PDF1.2a and PDF1.2b). Overall, the altered expression of these JA and SA markers was in accordance with the known modes of MYC2 action on its direct and downstream target genes.
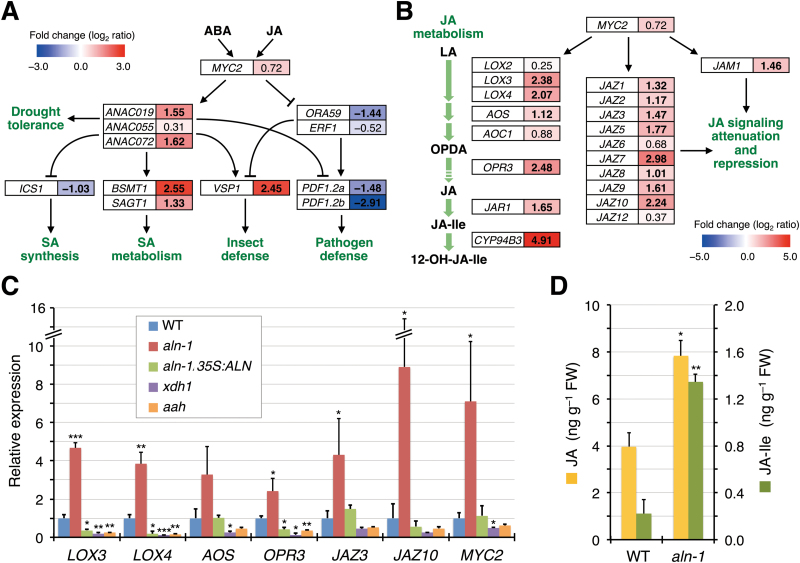
Fig. 1.
The aln-1 mutant shows altered levels of basal expression of MYC2-regulated JA-related genes and has enhanced JA and JA-Ile levels. (A and B) Simplified schemes of MYC2-mediated signaling processes, with emphasis on JA–ABA and JA–SA crosstalk in abiotic and biotic responses (A; mostly based on Kazan and Manners, 2013) and MYC2-mediated transcriptional activation of genes for JA metabolic enzymes and those involved in JA signaling (B). Genes are indicated in open boxes, and their positive and negative actions are denoted by lines with an arrow and a bar end, respectively. For each gene, the microarray-based relative expression ratio (aln-1 mutant versus the WT) is represented in log2 scale and by a boxed color on the blue-to-red gradient scale, with significant fold changes (|log2(aln-1/WT)| ≥ 1) in bold. Abbreviations not defined in the text are: LA, linolenic acid; OPDA, 12-oxo-phytodienoic acid; 12-OH-JA-Ile, 12-hydroxyjasmonoyl-l-isoleucine. (C) RT-qPCR for expression of the selected genes in the WT, three purine catabolic mutants (aln-1, xdh1, and aah), and the complemented mutant (aln-1 35S:ALN). RNA was extracted from the aerial parts of 2-week-old seedlings grown under normal aseptic conditions. Relative mRNA levels for each gene were determined using ACTIN2 expression as reference and are presented as values relative to those of the WT. (D) Endogenous jasmonate levels of WT and aln-1 mutants were quantified by LC-ESI-MS/MS in 2-week-old seedlings grown under normal aseptic conditions. In (C) and (D), data are means ±SE from three independent experiments, and asterisks denote significant differences between WT and mutant plants (*P<0.05; **P<0.01; ***P<0.001 by Student’s t-test).
MYC2 also acts as a transcriptional activator of JA-responsive genes encoding JA metabolic enzymes such as LOX, allene oxide synthase (AOS), allene oxide cyclase (AOC), 12-oxophytodienoate reductase (OPR), jasmonate-amido synthetase (also known as JASMONATE RESISTANCE1; JAR1), and cytochrome P450 monooxygenase 94B3 (CYP94B3) (Lorenzo et al., 2004; Sasaki-Sekimoto et al., 2013). MYC2 also activates genes encoding JA signaling components including JAZ and JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1) repressor proteins (Chini et al., 2007; Nakata et al., 2013; Sasaki-Sekimoto et al., 2013). As shown by microarray analysis (Fig. 1B), the basal expression levels of most of these genes significantly increased in the aln-1 mutant. This was confirmed by RT-qPCR for selected genes (LOX3, LOX4, AOS, OPR3, JAZ3, JAZ10, and MYC2) using a separate set of RNA samples from those used for microarray analysis (Fig. 1C). The RT-qPCR clearly demonstrated the significant activation of MYC2 expression, although the microarray analysis only showed a modest increase in the MYC2 transcript level (Fig. 1A, B). Genetic complementation of the aln-1 mutant with the WT ALN cDNA (aln-1 35S:ALN) resulted in the reversion of the transcript levels of all these genes to the normal WT levels, corroborating the effect of the mutation.
The increased expression of these JA-responsive genes could be caused by general inhibition of the purine catabolic pathway, rather than by the specific blockage of allantoin degradation. To exclude this possibility, two other knockout mutations, xdh1 and aah (allantoate amidohydrolase), were examined which impair the metabolic steps upstream and downstream of allantoin degradation, respectively (Supplementary Fig. S1). The basal expression levels of JA-responsive genes in xdh1 and aah mutants were comparable with or lower than those of the WT (Fig. 1C), supporting the idea that the aln mutation preventing allantoin degradation was indeed responsible for the observed gene expression phenotype.
The aln mutants have increased levels of endogenous JA and JA-Ile
Based on the observation that expression of genes involved in JA metabolism was significantly increased in the aln-1 mutant (Fig. 1B, C), endogenous JA and JA-Ile levels were measured in the WT and the mutant by LC-ESI-MS/MS (Fig. 1D). The concentrations of JA and JA-Ile in WT seedlings (4.0±0.6ng g−1 FW and 0.22±0.12ng g−1 FW, respectively) were similar to those reported previously (Pan et al., 2010). Compared with WT seedlings, the aln-1 mutant showed a 2-fold increase in JA levels and a 6-fold increase in JA-Ile levels. This increase in jasmonates correlated with the observed expression profiles (Fig. 1B, C), suggesting that the aln mutation promotes jasmonate accumulation in normally grown Arabidopsis seedlings, probably through the transcriptional activation of JA biosynthetic genes.
The aln mutants show enhanced sensitivity to exogenous jasmonate
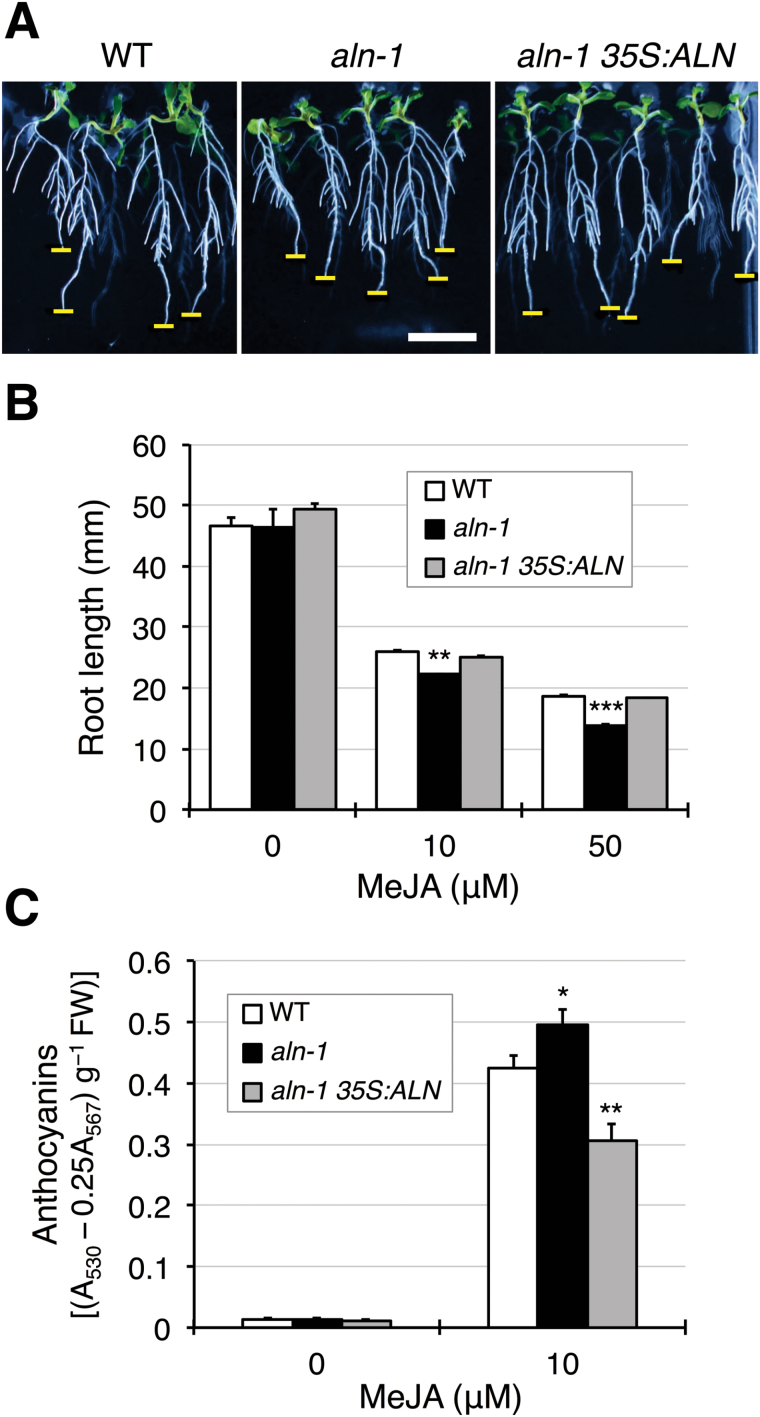
In addition to JA biosynthesis, MYC2 also positively regulates jasmonate-induced root growth inhibition and anthocyanin production (Dombrecht et al., 2007). To determine whether the aln mutants show increased activation of these MYC2-modulated JA responses, the sensitivity of the WT, aln-1 mutants, and the complemented line to exogenous MeJA was tested. When these genotypes were grown for 8 d on medium containing MeJA, they seemed to develop lateral roots well (Fig. 2A), probably reflecting the stimulatory effect of MeJA (Raya-González et al. 2012). Simultaneously, the aln-1 seedlings showed a slight but statistically significant reduction in primary root length, compared with the WT and the complemented mutant (Fig. 2B). However, when the aln mutation was introduced into the JA-insensitive jar1-1 mutant, which is defective in JA-Ile production (Staswick et al., 1992), the resultant aln-1 jar1-1 double mutant displayed no change in root growth in the presence of MeJA (Supplementary Fig. S5). Similar to the results of root growth inhibition, the aln-1 mutant responded more strongly than the WT to exogenous MeJA in anthocyanin production, further increasing its anthocyanin accumulation in response to MeJA treatment (Fig. 2C). Conversely, the complemented line was the least responsive to MeJA treatment as it had lower levels of anthocyanin than the WT and the aln-1 mutant. The aah mutant also compromised the metabolic response to MeJA (Supplementary Fig. S6). These results support the idea that the aln mutation enhances the MYC2-mediated physiological and metabolic responses to JA.
Fig. 2.
The aln-1 mutant exhibits increased sensitivity to exogenous MeJA. Sterile seedlings of the WT, aln-1 mutant, and the complemented mutant (aln-1 35S:ALN) were grown for 8 d on standard medium supplemented with MeJA and examined for root growth and anthocyanin accumulation. (A) Typical root growth in the presence of 10 µM MeJA. Horizontal bars indicate a scale of 10mm in length (thick) and the position of a primary root tip (thin). (B) Primary root length in the presence of 10 µM and 50 µM MeJA. Data are means ±SE (n≥16). (C) Anthocyanin levels in the presence of 10 µM MeJA. Data are means ±SE (n=8). Asterisks denote significant differences between WT and mutant plants (*P<0.05; **P<0.01; ***P<0.001 by Student’s t-test). (This figure is available in colour at JXB online.)
The aln mutants show enhanced wounding-inducible expression of JA-responsive genes
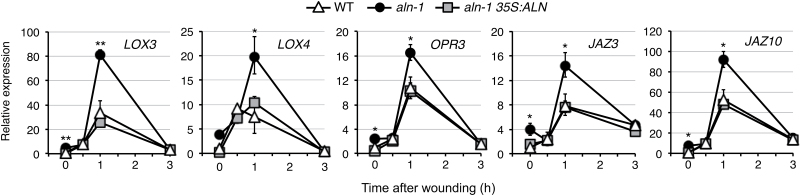
Next, this study investigated whether the aln mutation has a stimulatory effect on MYC2-mediated JA responses under stress conditions known to activate the JA signaling pathway. Wounding stress strongly enhances the transcript levels of genes involved in JA biosynthesis and signaling in a MYC2-dependent manner (Reymond et al., 2000; Lorenzo et al., 2004; Chung et al., 2008). Thus, the expression of these JA-responsive genes (LOX3, LOX4, and OPR3 for JA biosynthesis and JAZ3 and JAZ10 for JA signaling) was monitored in wounded leaves of 2-week-old seedlings (Fig. 3). Although the transcripts for these genes accumulated in response to wounding, in all cases the maximum levels were significantly higher in the mutant than in the WT, possibly reflecting the increased basal expression levels. The complemented mutant showed essentially the same transcript expression profiles as those observed in the WT. These results suggest that the aln mutation activates the MYC2-mediated JA response in normal growth conditions and under JA-associated stress conditions.
Fig. 3.
The aln-1 mutant enhances wounding-inducible expression of genes involved in JA biosynthesis and signaling. RNA was extracted from aerial parts of 2-week-old seedlings of the WT, aln-1 mutant, and the complemented mutant (aln-1 35S:ALN) at the indicated time points after wounding of rosette leaves. Relative mRNA levels were determined as described in Fig. 1C and are presented as values relative to those of the WT at zero time. Data are means ±SE from three independent experiments, and asterisks denote significant differences between WT and mutant plants (*P<0.05; **P<0.01 by Student’s t-test).
The aln mutation affects basal resistance to Pseudomonas syringae DC3000
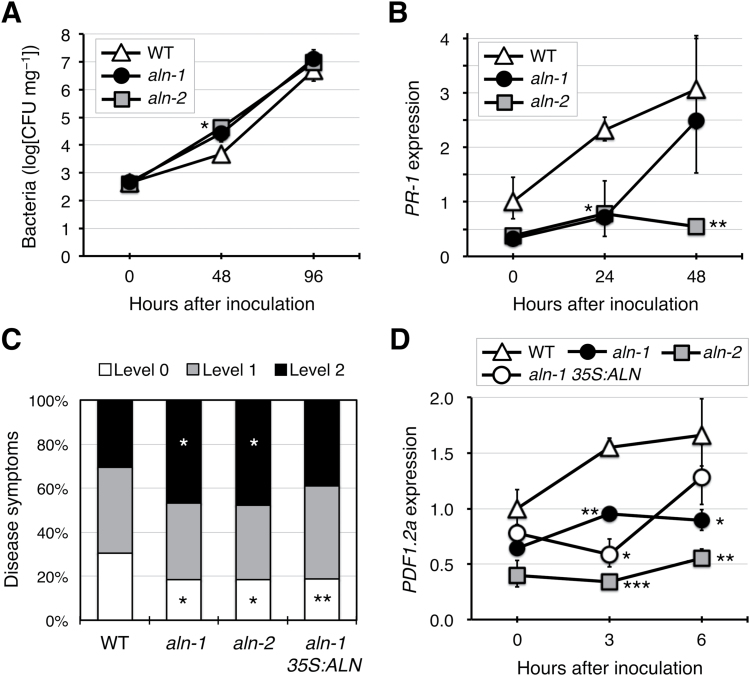
MYC2 negatively regulates the SA signaling pathway and suppresses SA-mediated host defenses against the bacterial pathogen Pst DC3000 (Nickstadt et al., 2004; Laurie-Berry et al., 2006). Because the gene expression data described above suggested that the aln mutation led to attenuation of SA signaling through activation of MYC2 (Fig. 1A; Supplementary Figs S3, S4), the resistance of two aln mutant alleles, aln-1 and aln-2, to Pst DC3000 was examined by monitoring bacterial growth in inoculated seedlings (Fig. 4A). At 48 hpi, bacterial growth was evident in both the WT and the mutants, but the aln-2 mutants had significantly more bacteria (11-fold, P<0.05) than the WT. Although not statistically significant, the same tendency was observed in the aln-1 mutant (a 9-fold higher population; P=0.068). At 96 hpi, bacterial populations showed further increases, but no longer differed between the WT and the aln mutants. Simultaneous monitoring of expression of the SA marker PR-1 during the course of infection with Pst DC3000 gave overall results consistent with those of the infection experiments, as induction of PR-1 expression was compromised in the aln-1 and aln-2 mutants (Fig. 4B). These results suggest that the aln mutation reduced the resistance to Pst DC3000 at early stages of pathogen infection, probably due to constitutive activation of the MYC branch of JA signaling.
Fig. 4.
Effect of aln mutations on resistance to bacterial pathogens. (A) Resistance to Pst DC3000 was evaluated for 2-week-old seedlings of the WT and two allelic aln mutants (aln-1 and aln-2). At 48 and 96 hpi, internal bacterial populations were determined as mg-1 FW of whole rosettes (n=4). (B) Time-course expression of the SA marker PR-1 in leaves inoculated with Pst DC3000. Relative mRNA levels were determined as described in Fig. 1C and are presented as values relative to those of the WT at zero time. Data are means ±SE from three independent experiments. (C) Resistance to Pcc EC1 was evaluated for leaves from 2-week-old seedlings of the WT, two allelic aln mutants, and the complemented mutant (aln-1 35S:ALN). Disease symptoms were scored at 24 hpi according to three criteria: level 0, no symptoms; level 1, symptoms restricted within the inoculated region; level 2, symptoms extended over the inoculated region (n≥169). (D) Time-course expression of the ERF branch marker PDF1.2a in leaves inoculated with Pcc EC1 was determined as described in (B). Asterisks denote significant differences between WT and mutant plants [*P<0.05; **P<0.01; ***P<0.001 by Student’s t-test, except in (C), where Fisher’s exact test was used].
The aln mutation suppresses basal resistance against Pectobacterium carotovorum
MYC2 down-regulates the ERF branch of JA signaling, which is critical for Arabidopsis resistance to phytopathogens such as Pcc EC1 (Norman-Setterblad et al., 2000; Hiruma et al., 2011). To test if the aln mutation altered resistance to this bacterium, Pcc EC1 was drop-inoculated onto intact leaves of 2-week-old seedlings and disease symptoms were evaluated at 24 hpi. As shown in Fig. 4C, the aln-1 and aln-2 mutants had compromised resistance to Pcc EC1, showing a significantly higher proportion of level 2 (most serious) symptoms and a lower proportion of level 0 (no detectable) symptoms, compared with the WT. The complemented aln-1 mutant plants showed substantially recovered disease resistance, with a proportion of level 2 symptoms non-significantly different from the WT. The expression levels of the disease resistance gene PDF1.2a, a marker of the ERF branch, were also examined at early stages of infection (0, 3, and 6 hpi). As expected from the enhanced susceptibility to Pcc EC1, the aln mutants showed consistently lower levels of PDF1.2a expression compared with the WT (Fig. 4D). For unknown reasons, complementation of the aln-1 mutant failed to restore PDF1.2a expression to the WT levels, which might explain why the resistance of the complemented mutant to Pcc EC1 was not fully recovered. Nevertheless, these results showed the compromised resistance of the aln mutants to Pcc EC1, providing more evidence for aln-mediated activation of MYC2.
Allantoin activates expression of MYC2 and MYC2-modulated JA-responsive genes
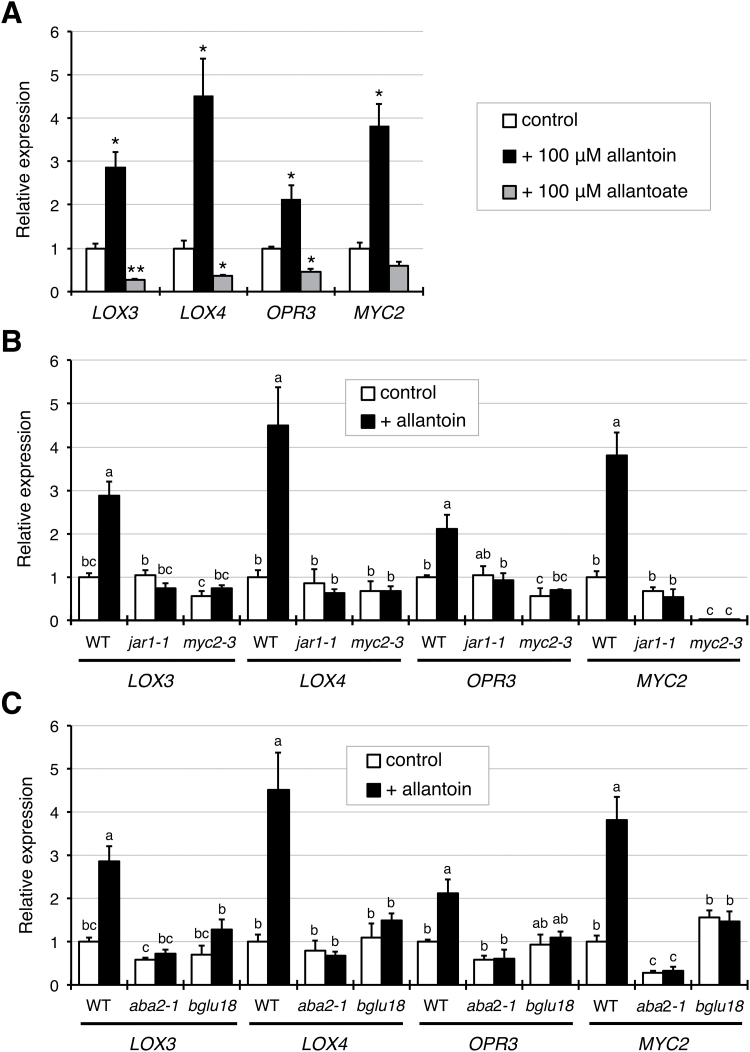
To address whether the observed JA-related aln phenotype was attributable to the accumulation of allantoin as a result of the aln mutation, the effects of exogenous allantoin on expression of MYC2 and the MYC2-modulated JA-responsive genes LOX3, LOX4, and OPR3 were examined. When seedlings of the WT were grown aseptically for 2 weeks on standard medium containing 100 µM allantoin, the four genes showed significantly increased transcript levels, compared with the non-treated control (Fig. 5A). Allantoate, which is similar in structure to allantoin as the immediate downstream metabolite, was also tested. When exogenously supplied at 100 µM, allantoate did not affect transcript levels, or caused a decrease in transcript levels (Fig. 5A), further supporting the specific efficacy of allantoin in inducing these JA-responsive genes. These results supported the idea that the JA-related aln mutant phenotypes result from accumulation of allantoin.
Fig. 5.
Exogenous allantoin induces JA-responsive gene expression in an ABA-dependent manner. (A) Effects of exogenous allantoin and allantoate on JA-responsive gene expression in the WT. RNA was extracted from aerial parts of sterile seedlings that had been grown for 2 weeks on standard medium containing allantoin or allantoate at 100 µM, and relative mRNA levels for each gene were determined as described in Fig. 1C. Data are means ±SE from three independent experiments, and asterisks denote significant differences between control and treated seedlings (*P<0.01; **P<0.001 by Student’s t-test). (B and C) Effect of exogenous allantoin on JA-responsive gene expression in bioactive JA-deficient jar1-1 and JA-insensitive myc2-3 mutants (B), and in ABA-deficient aba2-1 and bglu18 mutants (C). WT data are the same as presented in (A) and are shown for comparison. Data are means ±SE from three independent experiments, and different letters indicate significant differences (P<0.05 by Tukey’s test).
Allantoin requires ABA to activate JA-responsive genes
To gain insight into how allantoin activates JA-dependent responses, the next step was to investigate the effects of exogenous allantoin on JA-responsive gene expression in the two mutants, JA-insensitive jar1-1 and MYC2-deficient myc2-3. In marked contrast to the WT, both jar1-1 and myc2-3 mutants showed no responses to allantoin (Fig. 5B), demonstrating that the action of allantoin relies on an intact JA signaling pathway and requires the formation of bioactive JA-Ile. Because MYC2 is responsive to ABA (Abe et al., 1997), and because ABA levels increase in the aln mutants and in response to exogenous allantoin in WT plants (Watanabe et al., 2014b), expression of JA-responsive genes was also examined in the ABA-deficient mutants aba2-1 (abscisic acid deficient 2-1) and bglu18 (ß-glucosidase 18), which impair de novo ABA synthesis and regeneration of ABA from inactive ABA glucosides, respectively (Léon-Kloosterziel et al., 1996; Lee et al., 2006; Ogasawara et al., 2009). Similar to the JA-insensitive mutants, neither aba2-1 nor bglu18 mutants showed increases in the transcript levels of JA-responsive genes in response to exogenous allantoin (Fig. 5C).
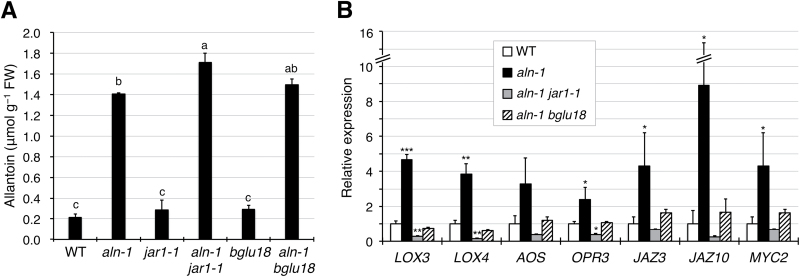
To obtain further evidence supporting this, the next experiments tested if these JA- and ABA-related mutations also suppress the effect of allantoin accumulation in the aln-1 background. For this, the aln-1 mutant was crossed with the bglu18 mutant to establish the aln-1 bglu18 double mutant (Supplementary Fig. S7), and the transcript levels of seven JA-responsive genes (the same set as reported in Fig. 1C) were measured in the aln-1 jar1-1 and aln-1 bglu18 double mutants. Although these double mutants exhibited allantoin levels comparable with the aln-1 mutant (Fig. 6A), the expression levels of seven JA-responsive genes no longer increased significantly (Fig. 6B). Thus, ABA may be required for activation of JA responses by allantoin.
Fig. 6.
The aln mutation activates JA-responsive gene expression in an ABA-dependent manner. (A) Endogenous allantoin levels in aseptically grown 2-week-old seedlings of JA- and ABA-related single and double mutants. Data are means ±SE from three independent experiments, and different letters indicate significant differences (P<0.05 by Tukey’s test). (B) RT-qPCR was performed for RNA samples extracted from the aerial parts of 2-week-old sterile seedlings of aln-1 jar1-1 or aln-1 bglu18 double mutants. Relative mRNA levels for each gene were determined and are presented as values relative to WT data that are taken from Fig. 1C, to simplify comparison. Data are means ±SE from three independent experiments, and asterisks denote significant differences between WT and mutant plants (*P<0.05; **P<0.01; ***P<0.001 by Student’s t-test).
Discussion
Evidence for the involvement of purine catabolism in stress protection in plants has only recently emerged (Brychkova et al., 2008; Watanabe et al., 2010), and the mechanism behind this remains unclear. Previous studies using Arabidopsis mutants defective in purine catabolism showed that the intermediary metabolite allantoin stimulated ABA production and enhanced abiotic stress tolerance (Watanabe et al., 2010, 2014a, b ). To gain more insight into the relationship between this purine metabolite and stress hormones, the present study investigated the effects of allantoin on JA signaling and responses, because the interplay between ABA and JA constitutes a fundamental regulatory mechanism in plant defense reactions (Kazan and Manners, 2013; Wasternack and Hause, 2013). The current results show that allantoin can activate JA metabolism, signaling, and responses via mechanisms involving ABA, which is supported by several lines of evidence. First, with the improved microarray data using a superior renormalization technique and a stringent cut-off threshold (Konishi, 2004, 2011), GO-based over-representation analysis of the aln-1 mutant, which accumulates allantoin, revealed a strong effect on JA responses (Supplementary Fig. S2). This was supported by the significantly increased levels of JA and JA-Ile in the aln-1 mutants (Fig. 1D). This analysis also reproduced the enhanced responses to ABA and abiotic stress conditions that were previously observed with the aln-1 mutant (Watanabe et al., 2014b), indicating that the aln mutation could cause the activation of both JA and ABA responses. Such transcriptome profiles provided a plausible explanation for the simultaneous suppression of SA responses (Fig. 1A; Supplementary Figs S3, S4), since antagonistic action of JA and ABA on SA signaling has been reported (Laurie-Berry et al., 2006; Yasuda et al., 2008; Millet et al., 2010). Secondly, the detailed transcriptome profiling and RT-qPCR analyses emphasized the massive influence of MYC2, a master regulator of JA signaling, on global gene expression in this mutant (Fig. 1A−C). As discussed below, this finding was supported by significant alterations in metabolic, physiological, and pathophysiological responses that are mainly controlled by MYC2 (Figs 2−4). Most of these alterations were reversed upon genetic complementation of the aln mutation (Figs 1C, 2, 3, 4C, D), while the other mutations that disrupt different steps in purine degradation had little effect on activating JA-responsive genes (Fig. 1C). These results confirmed the aln mutation as the primary cause behind the observed phenotype, and ruled out non-specific effects of inhibiting purine catabolism. Thirdly, exogenously supplied allantoin, but not its immediate metabolite allantoate, activated expression of JA-responsive genes in the WT plants (Fig. 5A). The observation is consistent with the expression data for aln-1 and aah mutants, which endogenously accumulate allantoin and allantoate, respectively (Fig. 1C). It thus appears that the accumulation of allantoin as a result of the aln mutation indeed triggers JA responses. Finally, not only JA-insensitive mutants, but also ABA-deficient mutants, showed no effect of allantoin that was either exogenously supplied or endogenously accumulated due to the aln-1 mutation (Figs 5B, C, 6). These findings revealed the requirement for ABA in the action of allantoin to elicit JA responses, suggesting that allantoin may exert its effects through the interplay between the two phytohormones.
In Arabidopsis, the MYC and ERF branches of the JA signaling pathway play critical roles in JA-mediated defense responses, and their antagonistic interaction is co-ordinated mainly by the MYC2 transcription factor (Kazan and Manners, 2013). Surprisingly, the aln mutation resulted in highly contrasting effects on these two major branches, activating the MYC branch while repressing the ERF branch (Fig. 1A). This finding suggested that MYC2 activity was augmented in the aln-1 mutant. Indeed, the aln-1 mutant showed significantly increased MYC2 transcript levels, which most probably accounted for transcriptional activation of MYC2 target genes (Fig. 1A−C). Consistent with these observations, aln-1 mutants also showed enhanced sensitivity to exogenous MeJA and mechanical wounding (Figs 2, 3), which are established MYC2-mediated responses. Modestly compromised resistance of aln mutants to bacterial pathogens (Fig. 4) also seems to result from high levels of MYC2 expression, perhaps due to suppression of SA signaling and the ERF branch of JA signaling (Fig. 1A). Some features of the aln mutant phenotype, such as enhanced sensitivity to exogenous JA, coincided with the phenotypes of transgenic Arabidopsis plants expressing MYC2 under a strong constitutive promoter (Lorenzo et al., 2004; Dombrecht et al., 2007). All together, these phenotypes of aln mutants can be interpreted as the consequences of MYC2 activation, suggesting that allantoin may activate JA responses by enhancing expression of this master regulator.
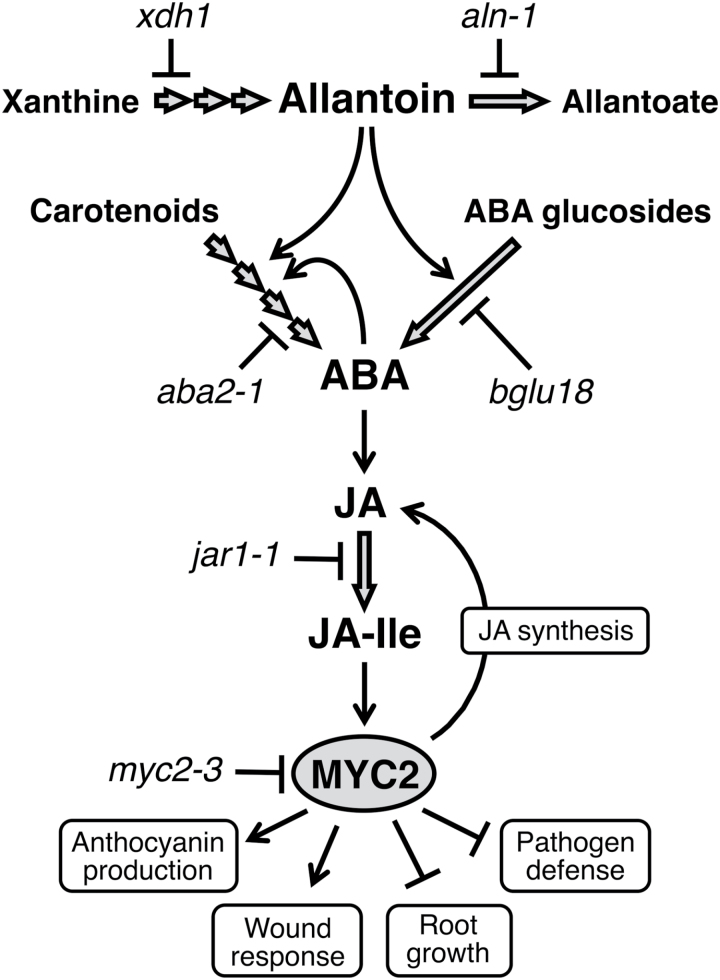
Arabidopsis MYC2 is rapidly induced by ABA (Abe et al., 1997), and it has been shown that ABA acts upstream of JA to regulate MYC2 expression (Lorenzo et al., 2004). Previous work showed that allantoin could increase ABA levels in Arabidopsis seedlings via two possible mechanisms: by stimulating the expression of a rate-limiting enzyme (9-cis-epoxycarotenoid dioxygenase 3) in de novo synthesis; and by post-translational activation of an enzyme (BGLU18 also known as BG1) responsible for hydrolytic conversion of inactive ABA glucosides (Watanabe et al., 2014b). Therefore, allantoin might activate JA responses through ABA-dependent MYC2 activation, although in Arabidopsis the aln mutation or exogenous allantoin increases ABA only at modest levels (Watanabe et al., 2014b) and increased ABA levels do not always result in significantly enhanced JA responses (García-Andrade et al., 2011). Here, this possibility was examined by quantifying expression of MYC2 and MYC2-regulated genes in ABA-deficient single mutants treated with exogenous allantoin, and in ABA-deficient aln-1 double mutants. The results revealed ABA to be an essential factor in allantoin-mediated MYC2 activation (Figs 5C, 6B). The similar expression analysis using the jar1-1 single and aln-1 jar1-1 double mutants gave the same results as those for the ABA mutants (Figs 5B, 6B), indicating that the effect of ABA depends on the formation of bioactive JA (JA-Ile). These results, together with those of a previous study (Watanabe et al., 2014b), support the idea that ABA might precede JA in the activation of MYC2 by allantoin, although how ABA enhances JA remains to be elucidated (Fig. 7).
Fig. 7.
Schematic model for how allantoin activates JA responses in Arabidopsis. Accumulation of allantoin causes enhanced ABA levels through de novo synthesis from carotenoids and/or hydrolytic conversion from inactive ABA glucosides, thereby activating ABA-dependent JA responses mediated by MYC2. The effect of allantoin on de novo synthesis may be indirect due to positive feedback of ABA released from the inactive pool (Watanabe et al., 2014b ). The open arrows indicate metabolic routes, and lines with arrows and bar ends represent positive and negative causal relationships, respectively.
Whether the activated JA response contributes to allantoin-mediated stress tolerance, as observed in the aln mutants (Watanabe et al., 2014b ), remains an important question regarding the mechanism of action of allantoin. Although ABA plays a central role in drought tolerance, JA participates in the regulation of ABA-mediated physiological responses to drought such as stomatal closure (Suhita et al., 2004). JA also affects expression of a significant fraction of drought-related genes that are responsive to ABA (Huang et al., 2008) and positively regulates freezing tolerance (Hu et al., 2013), which may share common protective mechanisms with drought tolerance. In fact, allantoin-accumulating aln mutants enhance ABA levels, activate stress-related genes, and increase drought and osmotic stress tolerance (Watanabe et al., 2014b ); the results described here show that the mutants also activate JA responses and JA-related gene expression. These observations may suggest a possible contribution of JA signaling to the abiotic stress tolerance mediated by allantoin, through interactions with ABA.
These results point to a previously unrecognized connection between allantoin and JA signaling, and imply its potential influence on the interplay between JA and SA (Figs 1A, 4; Supplementary Figs S3, S4). This suggests that purine catabolism may play a role in biotic stress responses. Indeed, purine catabolism has been implicated in plant–microbe interactions: in beans and wheat, changes in the enzyme activities and metabolites, including allantoin, occur as an early response to pathogens (Montalbini, 1991, 1992b ). Also, inhibition of the purine catabolic pathway by the XDH inhibitor allopurinol resulted in altered rust infection and compromised hypersensitive responses during incompatible interactions (Montalbini, 1992a), and the application of allopurinol enhanced virus susceptibility in tobacco (Silvestri et al., 2008). In addition, a recent transcriptome analysis of wheat revealed significantly increased expression of genes involved in ureide transport during fusarium infection (Lysøe et al., 2011). Although the above pathophysiological phenomena may be relevant to JA and SA signaling, further research is needed to obtain evidence that substantially supports the role of purine catabolism in biotic defense. In this context, it would be interesting to examine the effect of allantoin on herbivore resistance because the aln mutants activated the MYC branch pathway and enhanced wounding responses (Figs 1, 3).
An intriguing observation is that aah mutation and exogenous allantoate treatment, both causing allantoate accumulation, tend to suppress JA responses at the transcript and metabolite levels (Figs 1C, 5A; Supplementary Fig. S6). The aah mutation also tends to perturb the expression of certain ABA-related genes, although the ABA level in this mutant does not seem to be significantly different from that of the WT (Watanabe et al., 2014b ). These observations may imply that allantoate also affects stress-related gene expression, potentially with the opposite effect to that of allantoin reported here and previously (Watanabe et al., 2014b ). If such a possibility were likely, then allantoate could ensure the elimination of the effect of allantoin after its degradation. However, this remains to be investigated. A more extensive analysis, including further examination of the physiological effects of ureide accumulation and in-depth characterization of aln and aah mutants, would shed light on the possible relationship between allantoin and allantoate in the mechanism of stress response and adaptation.
Recent metabolomic studies showed that allantoin levels significantly increased in Arabidopsis, rice, and other species subjected to various stress conditions (see the Introduction). Some possibilities have been proposed to explain the physiological significance of stress-induced allantoin accumulation. The conventional view argues that this phenomenon results from a metabolic counter-balance to optimize carbon to nitrogen ratios, because, under stress conditions, reduced photosynthesis causes a shortage of carbon skeletons, which restricts assimilation of purine-derived ammonia and leads to accumulation of this toxic nitrogen compound. However, recent studies using Arabidopsis mutants indicated that allantoin might participate in stress protection and tolerance mechanisms. Brychkova et al. (2008) reported that this ureide compound could act as an antioxidant. Our previous and current studies showed that allantoin elicited phytohormone-mediated stress responses (Watanabe et al., 2014b; this study). In cultivated rice, allantoin also induced stress responses by significantly increasing the levels of osmoprotectants such as soluble sugars and free proline (Wang et al., 2012). Although none of these lines of evidence is as yet conclusive, it is worth mentioning that these possible actions of allantoin are not mutually exclusive, which suggests that this purine metabolite may have a role beyond its function as a metabolic intermediate or nitrogen reservoir under stress conditions. Multifunctionality of small metabolites, in particular their signaling and regulatory roles, has recently gained attention as part of the set of adaptive mechanisms that allow plants to survive and grow under changing environments. The importance of various effects on plant development and physiology has been documented for certain molecular species of sugars, amino acids, polyamines, and other kinds of small compounds that are not considered to be phytohormones (see the Introduction). Currently, the number of such small metabolites is limited, but is estimated to be much higher than that reported in the literature (Tholl and Aharoni, 2014). Thus, it seems possible that allantoin could serve in either nitrogen metabolism or stress protection, depending on the circumstances where plants live. For further substantiation of such a dual functionality of allantoin, its modes of action in stress responses and adaptation need to be firmly defined.
Supplementary data
Supplementary data are available at JXB online.
Methods S1. Quantification of JA and JA-Ile; accession numbers.
Figure S1. The purine catabolism pathway and metabolites derived therefrom.
Figure S2. Hierarchical tree graph of over-represented GO terms for genes with significantly increased expression in the aln-1 mutant.
Figure S3. Hierarchical tree graph of over-represented GO terms for genes with significantly reduced expression in the aln-1 mutant.
Figure S4. Basal level expression of PR-1 as a canonical SA marker.
Figure S5. Characterization of the aln-1 jar1-1 double mutant.
Figure S6. Reduced response to MeJA of anthocyanin accumulation in the aah mutant.
Figure S7. Characterization of the aln-1 bglu18 double mutant.
Table S1. Primers used in this study.
Table S2. LC-ESI-MS/MS parameters for jasmonate determination.
Table S3. Genes with significantly increased expression in the aln-1 mutant.
Table S4. Genes with significantly reduced expression in the aln-1 mutant.
Acknowledgments
We thank Dr Masaru Nakata (NARO Agricultural Research Center, Niigata, Japan) for kind advice on the experimental procedures for exogenous MeJA treatments. This work was supported in part by the Ministry of Education, Culture, Sports, Science and Technology, Japan (MEXT) [Grants-in-Aid for Scientific Research for Innovative Areas (nos 23119514 and 25119717 to AS)], the Japan Society for Promotion of Science (JSPS) [a Grant-in-Aid Scientific Research (no. 26440147 to AS), and a JSPS Fellowship for Young Scientists (DC, no. 26-5367 to HT)], Tottori University [Joint Research Program of Arid Land Research Center (to AS)], and Hiroshima University Alumni Association [Graduate Student Research Grant (to HT)]. The hormone analysis was supported by the Japan Advanced Plant Science Network.
References
- Abe H, Yamaguchi-Shinozaki K, Urao T, Iwasaki T, Hosokawa D, Shinozaki K. 1997. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. The Plant Cell 9, 1859–1868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adie BAT, Péréz-Péréz J, Péréz-Péréz MM, Godoy M, Sánchez-Serrano J-J, Schmelz EA, Solano R. 2007. ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. The Plant Cell 19, 1665–1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson JP, Badruzsaufari E, Schenk PM, Manners JM, Desmond OJ, Ehlert C, Maclean DJ, Ebert PR, Kazan K. 2004. Antagonistic interaction between abscisic acid and jasmonate–ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. The Plant Cell 16, 3460–3479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brychkova G, Alikulov Z, Fluhr R, Sagi M. 2008. A critical role for ureides in dark and senescence-induced purine remobilization is unmasked in the Atxdh1 Arabidopsis mutant. The Plant Journal 54, 496–509. [DOI] [PubMed] [Google Scholar]
- Bu Q, Jiang H, Li C-B, Zhai Q, Zhang J, Wu X, Sun J., Xie Q, Li C. 2008. Role of the Arabidopsis thaliana NAC transcription factors ANAC019 and ANAC055 in regulating jasmonic acid-signaled defense responses. Cell Research 18, 756–767. [DOI] [PubMed] [Google Scholar]
- Chen THH, Murata N. 2002. Enhancement of tolerance of abiotic stress by metabolic engineering of betaines and other compatible solutes. Current Opinion in Plant Biology 5, 250–257. [DOI] [PubMed] [Google Scholar]
- Chini A, Fonseca S, Fernández G, et al. 2007. The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. [DOI] [PubMed] [Google Scholar]
- Chung HS, Koo AJK, Gao X, Jayanty S, Thines B, Jones AD, Howe GA. 2008. Regulation and function of Arabidopsis JASMONATE ZIM-Domain genes in response to wounding and herbivory. Plant Physiology 146, 952–964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coneva V, Simopoulos C, Casaretto JA, et al. 2014. Metabolic and co-expression network-based analyses associated with nitrate response in rice. BMC Genomics 15, 1056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dombrecht B, Xue GP, Sprague SJ, et al. 2007. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis . The Plant Cell 19, 2225–2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan J, Hill L, Crooks C, Doerner P, Lamb C. 2009. Abscisic acid has a key role in modulating diverse plant–pathogen interactions. Plant Physiology 150, 1750–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-Calvo P, Chini A, Fernández-Barbero G, et al. 2011. The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. The Plant Cell 23, 701–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Andrade J, Ramírez V, Flors V, Vera P. 2011. Arabidopsis ocp3 mutant reveals a mechanism linking ABA and JA to pathogen-induced callose deposition. The Plant Journal 67, 783–794. [DOI] [PubMed] [Google Scholar]
- Higashi K, Ishiga Y, Inagaki Y, Toyoda K, Shiraishi T, Ichinose Y. 2008. Modulation of defense signal transduction by flagellin-induced WRKY41 transcription factor in Arabidopsis thaliana . Molecular Genetics and Genomics 279, 303–312. [DOI] [PubMed] [Google Scholar]
- Hiruma K, Nishiuchi T, Kato T, Bednarek P, Okuno T, Schulze-Lefert P, Takano Y. 2011. Arabidopsis ENHANCED DISEASE RESISTANCE 1 is required for pathogen-induced expression of plant defensins in nonhost resistance, and acts through interference of MYC2-mediated repressor function. The Plant Journal 67, 980–992. [DOI] [PubMed] [Google Scholar]
- Hu Y, Jiang L, Wang F, Yu D. 2013. Jasmonate regulates the INDUCER OF CBF EXPRESSION-C-REPEAT BINDING FACTOR/DRE BINDING FACTOR1 cascade and freezing tolerance in Arabidopsis . The Plant Cell 25, 2907–2924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D, Wu W, Abrams SR, Cutler AJ. 2008. The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. Journal of Experimental Botany 59, 2991–3007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain SS, Ali M, Ahmad M, Siddique KHM. 2011. Polyamines: natural and engineered abiotic and biotic stress tolerance in plants. Biotechnology Advances 29, 300–311. [DOI] [PubMed] [Google Scholar]
- Ishiga Y, Ishiga T, Uppalapati SR, Mysore KS. 2011. Arabidopsis seedling flood-inoculation technique: a rapid and reliable assay for studying plant–bacterial interactions. Plant Methods 7, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanani H, Dutta B, Klapa MI. 2010. Individual vs. combinatorial effect of elevated CO2 conditions and salinity stress on Arabidopsis thaliana liquid cultures: comparing the early molecular response using time-series transcriptomic and metabolomic analyses. BMC Systems Biology 4, 177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaplan F, Kopka J, Haskell DW, Zhao W, Schiller KC, Gatzke N, Sung DY, Guy CL. 2004. Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiology 136, 4159–4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katari MS, Nowicki SD, Aceituno FF, et al. 2010. VirtualPlant: a software platform to support systems biology research. Plant Physiology 152, 500–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K, Manners JM. 2013. MYC2: the master in action. Molecular Plant 6, 686–703. [DOI] [PubMed] [Google Scholar]
- Konishi T. 2004. Three-parameter lognormal distribution ubiquitously found in cDNA microarray data and its application to parametric data treatment. BMC Bioinformatics 5, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konishi T. 2011. Microarray test results should not be compensated for multiplicity of gene contents. BMC Systems Biology 5, S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie-Berry N, Joardar V, Street IH, Kunkel BN. 2006. The Arabidopsis thaliana JASMONATE INSENSITIVE 1 gene is required for suppression of salicylic acid-dependent defenses during infection by Pseudomonas syringae . Molecular Plant-Microbe Interactions 19, 789–800. [DOI] [PubMed] [Google Scholar]
- Lee KH, Piao HL, Kim H-Y, Choi SM, Jiang F, Hartung W, Hwang I, Kwak JM, Lee I-J, Hwang I. 2006. Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126, 1109–1120. [DOI] [PubMed] [Google Scholar]
- Léon-Kloosterziel KM, Gil MA, Ruijs GJ, Jacobsen SE, Olszewski NE, Schwartz SH, Zeevaart JAD, Koornneef M. 1996. Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. The Plant Journal 10, 655–661. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2 − ∆∆Ct Method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Chico JM, Sánchez-Serrano JJ, Solano R. 2004. JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. The Plant Cell 16, 1938–1950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lunn JE, Delorge I, Figueroa CM, Van Dijck P, Stitt M. 2014. Trehalose metabolism in plants. The Plant Journal 79, 544–567. [DOI] [PubMed] [Google Scholar]
- Lysøe E, Seong K-Y, Kistler HC. 2011. The transcriptome of Fusarium graminearum during the infection of wheat. Molecular Plant-Microbe Interactions 24, 995–1000. [DOI] [PubMed] [Google Scholar]
- Millet YA, Danna CH, Clay NK, Songnuan W, Simon MD, Werck-Reichhart D, Ausubel FM. 2010. Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. The Plant Cell 22, 973–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montalbini P. 1991. Effect of rust infection on levels of uricase, allantoinase and ureides in susceptible and hypersensitive bean leaves. Physiological and Molecular Plant Pathology 39, 173–188. [Google Scholar]
- Montalbini P. 1992. a Inhibition of hypersensitive response by allopurinol applied to the host in the incompatible relationship between Phaseolus vulgaris and Uromyces phaseoli . Journal of Phytopathology 134, 218–228. [Google Scholar]
- Montalbini P. 1992. b Ureides and enzymes of ureide synthesis in wheat seeds and leaves and effect of allopurinol on Puccinia recondita f. sp. tritici infection. Plant Science 87, 225–231. [Google Scholar]
- Nakagawa A, Sakamoto S, Takahashi T, Morikawa H, Sakamoto A. 2007. The RNAi-mediated silencing of xanthine dehydrogenase impairs growth and fertility and accelerates leaf senescence in transgenic Arabidopsis plants. Plant and Cell Physiology 48, 1484–1495. [DOI] [PubMed] [Google Scholar]
- Nakata M, Mitsuda N, Herde M, Koo AJK, Moreno JE, Suzuki K, Howe GA, Ohme-Takagi M. 2013. A bHLH-type transcription factor, ABA-INDUCIBLE BHLH-TYPE TRANSCRIPTION FACTOR/JA-ASSOCIATED MYC2-LIKE1, acts as a repressor to negatively regulate jasmonate signaling in Arabidopsis . The Plant Cell 25, 1641–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickstadt A, Thomma BPHJ, Feussner I, Kangasjárvi J, Zeier J, Loeffler C, Scheel D, Berger S. 2004. The jasmonate-insensitive mutant jin1 shows increased resistance to biotrophic as well as necrotrophic pathogens. Molecular Plant Pathology 5, 425–434. [DOI] [PubMed] [Google Scholar]
- Nikiforova VJ, Kopka J, Tolstikov V, Fiehn O, Hopkins L, Hawkesford MJ, Hesse H, Hoefgen R. 2005. Systems rebalancing of metabolism in response to sulfur deprivation, as revealed by metabolome analysis of Arabidopsis plants. Plant Physiology 138, 304–318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Figueroa P, Browse J. 2011. Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis . Journal of Experimental Botany 62, 2143–2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman-Setterblad C, Vidal S, Palva ET. 2000. Interacting signal pathways control defense gene expression in Arabidopsis in response to cell wall-degrading enzymes from Erwinia carotovora . Molecular Plant-Microbe Interactions 13, 430–438. [DOI] [PubMed] [Google Scholar]
- Ogasawara K, Yamada K, Christeller JT, Kondo M, Hatsugai N, Hara-Nishimura I, Nishimura M. 2009. Constitutive and inducible ER bodies of Arabidopsis thaliana accumulate distinct ß-glucosidases. Plant and Cell Physiology 50, 480–488. [DOI] [PubMed] [Google Scholar]
- Oliver MJ, Guo L, Alexander DC, Ryals JA, Wone BWM, Cushman JC. 2011. A sister group contrast using untargeted global metabolomic analysis delineates the biochemical regulation underlying desiccation tolerance in Sporobolus stapfianus . The Plant Cell 23, 1231–1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan X, Welti R, Wang X. 2010. Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography–mass spectrometry. Nature Protocols 5, 986–992. [DOI] [PubMed] [Google Scholar]
- Peña-Cortés H, Fisahn J, Willmitzer L. 1995. Signals involved in wound-induced proteinase inhibitor II gene expression in tomato and potato plants. Proceedings of the National Academy of Sciences, USA 92, 4106–4113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Cortés H, Sánchez-Serrano JJ, Mertens R, Willmitzer L, Prat S. 1989. Abscisic acid is involved in the wound-induced expression of the proteinase inhibitor II gene in potato and tomato. Proceedings of the National Academy of Sciences, USA 86, 9851–9855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx IAMA, Thomma BPHJ, Buchala A, Métraux J-P, Broekaert WF. 1998. Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. The Plant Cell 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pré M, Atallah M, Champion A, De Vos M, Pieterse CMJ, Memelink J. 2008. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiology 147, 1347–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston J, Tatematsu K, Kanno Y, Hobo T, Kimura M, Jikumaru Y, Yano R, Kamiya Y, Nambara E. 2009. Temporal expression patterns of hormone metabolism genes during imbibition of Arabidopsis thaliana seeds: a comparative study on dormant and non-dormant accessions. Plant and Cell Physiology 50, 1786–1800. [DOI] [PubMed] [Google Scholar]
- Raya-González J, Pelagio-Flores R, López-Bucio J. 2012. The jasmonate receptor COI1 plays a role in jasmonate-induced lateral root formation and lateral root positioning in Arabidopsis thaliana . Journal of Plant Physiology 169, 1348–1358. [DOI] [PubMed] [Google Scholar]
- Reymond P, Weber H, Damond M, Farmer EE. 2000. Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. The Plant Cell 12, 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MT, Rose TJ, Pariasca-Tanaka J, Yoshihashi T, Neuweger H, Goesmann A, Frei M, Wissuwa M. 2012. Root metabolic response of rice (Oryza sativa L.) genotypes with contrasting tolerance to zinc deficiency and bicarbonate excess. Planta 236, 959–973. [DOI] [PubMed] [Google Scholar]
- Sasaki-Sekimoto Y, Jikumaru Y, Obayashi T, Saito H, Masuda S, Kamiya Y, Ohta H, Shirasu K. 2013. Basic helix–loop–helix transcription factors JASMONATE-ASSOCIATED MYC2-LIKE1 (JAM1), JAM2, and JAM3 are negative regulators of jasmonate responses in Arabidopsis. Plant Physiology 163, 291–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvente S, Sobolev AP, Lara M. 2011. Metabolite adjustments in drought tolerant and sensitive soybean genotypes in response to water stress. PLoS One 7, e38554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silvestri S, Murphy AM, Buonaurio R, Carr JP. 2008. Allopurinol, an inhibitor of purine catabolism, enhances susceptibility of tobacco to Tobacco mosaic virus . Virus Research 137, 257–260. [DOI] [PubMed] [Google Scholar]
- Smith PMC, Atkins CA. 2002. Purine biosynthesis. Big in cell division, even bigger in nitrogen assimilation. Plant Physiology 128, 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE, Su W, Howell SH. 1992. Methyl jasmonate inhibition of root growth and induction of a leaf protein are decreased in an Arabidopsis thaliana mutant. Proceedings of the National Academy of Sciences, USA 89, 6837–6840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suhita D, Raghavendra AS, Kwak JM, Vavasseur A. 2004. Cytoplasmic alkalization precedes reactive oxygen species production during methyl jasmonate- and abscisic acid-induced stomatal closure. Plant Physiology 134, 1536–1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szabados L, Savouré A. 2010. Proline: a multifunctional amino acid. Trends in Plant Science 15, 89–97. [DOI] [PubMed] [Google Scholar]
- Teng S, Keurentjes J, Bentsink L, Koornneef M, Smeekens S. 2005. Sucrose-specific induction of anthocyanin biosynthesis in Arabidopsis requires the MYB75/PAP1 gene. Plant Physiology 139, 1840–1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tholl D, Aharoni A. 2014. Small molecules: from structural diversity to signalling and regulatory roles. The Plant Journal 79, 541–543. [DOI] [PubMed] [Google Scholar]
- Todd CD, Polacco JC. 2006. AtAAH encodes a protein with allantoate amidohydrolase activity from Arabidopsis thaliana . Planta 223, 1108–1113. [DOI] [PubMed] [Google Scholar]
- Vos IA, Verhage A, Schuurink RC, Watt LG, Pieterse CMJ, Van Wees SCM. 2013. Onset of herbivore-induced resistance in systemic tissue primed for jasmonate-dependent defenses is activated by abscisic acid. Frontiers in Plant Science 4, 539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Kong C-H, Sun B, Xu X-H. 2012. Distribution and function of allantoin (5-ureidohydantoin) in rice grains. Journal of Agricultural and Food Chemistry 60, 2793–2798. [DOI] [PubMed] [Google Scholar]
- Wang W-S, Zhao X-Q, Li M, Huang L-Y, Xu J-L, Zhang F, Cui Y-R, Fu B-Y, Li Z-K. 2016. Complex molecular mechanisms underlying seedling salt tolerance in rice revealed by comparative transcriptome and metabolomic profiling. Journal of Experimental Botany 67, 405–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasternack C, Hause B. 2013. Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Annals of Botany 111, 1021–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe S, Kounosu Y, Shimada H, Sakamoto A. 2014. a Arabidopsis xanthine dehydrogenase mutants defective in purine degradation show a compromised protective response to drought and oxidative stress. Plant Biotechnology 31, 173–178. [Google Scholar]
- Watanabe S, Matsumoto M, Hakomori Y, Takagi H, Shimada H, Sakamoto A. 2014. b The purine metabolite allantoin enhances abiotic stress tolerance through synergistic activation of abscisic acid metabolism. Plant, Cell & Environment 37, 1022–1036. [DOI] [PubMed] [Google Scholar]
- Watanabe S, Nakagawa A, Izumi S, Shimada H, Sakamoto A. 2010. RNA interference-mediated suppression of xanthine dehydrogenase reveals the role of purine metabolism in drought tolerance in Arabidopsis. FEBS Letters 584, 1181–1186. [DOI] [PubMed] [Google Scholar]
- Werner AK, Witte C-P. 2011. The biochemistry of nitrogen mobilization: purine ring catabolism. Trends in Plant Science 16, 381–387. [DOI] [PubMed] [Google Scholar]
- Wind J, Smeekens S, Hanson J. 2010. Sucrose: metabolite and signaling molecule. Phytochemistry 71, 1610–1614. [DOI] [PubMed] [Google Scholar]
- Wu H, Liu X, You L, Zhang L, Zhou D, Feng J, Zhao J, Yu J. 2012. Effects of salinity on metabolic profiles, gene expressions, and antioxidant enzymes in halophyte Suaeda salsa . Journal of Plant Growth Regulation 31, 332–341. [Google Scholar]
- Yang J, Han K-H. 2004. Functional characterization of allantoinase genes from Arabidopsis and a nonureide-type legume black locust. Plant Physiology 134, 1039–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda M, Ishikawa A, Jikumaru Y, . et al. 2008. Antagonistic interaction between systemic acquired resistance and the abscisic acid-mediated abiotic stress response in Arabidopsis . The Plant Cell 20, 1678–1692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yobi A, Wone BWM, Xu W, Alexander DC, Guo L, Ryals JA, Oliver MJ, Cushman JC. 2013. Metabolomic profiling in Selaginella lepidophylla at various hydration states provides new insights into the mechanistic basis of desiccation tolerance. Molecular Plant 6, 369–385. [DOI] [PubMed] [Google Scholar]
- Zrenner R, Stitt M, Sonnewald U, Boldt R. 2006. Pyrimidine and purine biosynthesis and degradation in plants. Annual Review of Plant Biology 57, 805–836. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.