Abstract
3,3′-Diindolylmethane (DIM), a natural indole found in cruciferous vegetables, has significant anti-cancer and anti-inflammatory properties. In this current study, we investigated the effects of DIM on acute lung injury (ALI) induced by exposure to staphylococcal enterotoxin B (SEB). We found that pretreatment of mice with DIM led to attenuation of SEB-induced inflammation in the lungs, vascular leak, and IFN-γ secretion. Additionally, DIM could induce cell-cycle arrest and cell death in SEB-activated T cells in a concentration-dependent manner. Interestingly, microRNA (miRNA) microarray analysis uncovered an altered miRNA profile in lung-infiltrating mononuclear cells after DIM treatment of SEB-exposed mice. Moreover, computational analysis of miRNA gene targets and regulation networks indicated that DIM alters miRNA in the cell death and cell-cycle progression pathways. Specifically, DIM treatment significantly downregulated several miRNA and a correlative increase associated gene targets. Furthermore, overexpression and inhibition studies demonstrated that DIM-induced cell death, at least in part, used miR-222. Collectively, these studies demonstrate for the first time that DIM treatment attenuates SEB-induced ALI and may do so through the induction of microRNAs that promote apoptosis and cell-cycle arrest in SEB-activated T cells.
Introduction
Staphylococcus aureus, a Gram-positive opportunistic pathogen, has become a major cause of nosocomial infections and secretes staphylococcal enterotoxin (SEB) among other virulence factors (Lowy, 1998; Foster, 2004). Moreover, the Centers for Disease Control and Prevention lists SEB as a category B select agent. Thus, SEB poses a significant public health threat if it is used as a biologic weapon. The consequences of SEB exposure can range from mild food poisoning to potentially fatal toxic shock (Pinchuk et al., 2010) and stem from its ability to induce the expansion of a significant proportion of T cells (Herman et al., 1991). SEB binds directly to class II major histocompatibility complex expressed on antigen-presenting cells and consecutively binds the T-cell receptor (TCR) via the variable region on the β-chain of the TCR (Seth et al., 1994). Inhalation of SEB leads to recruitment of immune cells to the lungs, secretion of cytokines (IL-2, IL-4, tumor necrosis factor-α, IFN-γ, etc.), and damage of the endothelial-epithelial interface in lungs, creating increased vascular permeability and ultimately respiratory failure (Krakauer, 2005; Liu et al., 2009; Saeed et al., 2012).
3,3′-Diindolylmethane (DIM) is a natural indole compound derived from its precursor indole-3-carbinol, found in cruciferous vegetables, such as kale, cabbage, broccoli, and cauliflower (Ali et al., 2013). Indigoid compounds have been extensively studied for their anti-cancer properties (Ahmad et al., 2010; Banerjee et al., 2011). The immunosuppressive role for DIM has been studied in a number of inflammatory diseases, including experimental arthritis (Dong et al., 2010), colitis (Huang et al., 2013), and autoimmune encephalomyelitis (Rouse et al., 2013).
MicroRNAs (miRNAs), a class of small (∼22 nt) noncoding RNAs, have recently been implicated as evolutionarily conserved gene regulators (Zhang and Su, 2009). Moreover, the role of microRNA in the regulation of inflammation has become apparent (Baltimore et al., 2008; Hoefig and Heissmeyer, 2008). Furthermore, our laboratory has recently proposed a role for microRNA in SEB-mediated toxicity (Rao et al., 2015). MicroRNAs perform their regulatory role by binding to the 3′ untranslated region (UTR) of target mRNA, ultimately causing translational repression via sequestration or degradation of mRNA (Plank et al., 2013). Target mRNAs are determined by the microRNA seed sequence, and microRNA-target gene pairings can be predicted using computational algorithms (miRANDA, PicTar, TargetScan, etc.) (Watanabe et al., 2007). Recently, our laboratory has also discovered a role for DIM-mediated miRNA regulation in inflammatory disease (Rouse et al., 2014; Tomar et al., 2014).
In the current study, we investigated the therapeutic potential of DIM in the attenuation of SEB-induced acute lung injury (ALI) and the role microRNA might play. We demonstrate that DIM ameliorates the aberrant inflammation via suppression of cellular proliferation, thereby protecting mice against SEB-induced ALI. Moreover, our data suggest that DIM may mitigate lung injury through miRNAs that regulate the cell-cycle and cell death pathways.
Materials and Methods
Animal Use and Care.
Female C57BL/6 mice (6–8 weeks of age) were procured from the National Institutes of Health National Cancer Institute (Frederick, MD). Animals were maintained under typical pathogen-free conditions, and all animal care and experimental procedures were approved by the University of South Carolina Institutional Animal Care and Use Committee, 2012.
Induction of SEB-Induced Lung Inflammation and DIM Treatment Regimen.
Groups of four to five mice were randomized and exposed to SEB (BT202; Toxin Technologies Inc., Sarasota, FL), 50 μg/mouse in 25 μl of sterile phosphate-buffered saline (PBS) intranasally, as described (Saeed et al., 2012). DIM (D9568; Sigma-Aldrich, St. Louis, MO) was prepared in sterile corn oil and administered via oral gavage on days −1, 0, and 1 at a 100 mg/kg body weight. Anderton et al. (2004) showed that mice given 250 mg/kg DIM via oral gavage had wide bioavailability, including both plasma and lung tissues, and no associated toxicity. Control groups received intranasal administration of sterile PBS and sterile corn oil (gavage). Mice were euthanized 48 hours after SEB exposure.
Measurement of Capillary Leak.
Capillary leak in the lungs was measured via Evans blue assay as previously described (Rieder et al., 2012). SEB and DIM were similarly administered, and 48 hours after SEB exposure, mice were injected with 1% Evans blue dye in PBS i.v.; 2 hours after dye injection, the mice were sacrificed under anesthesia. After perfusion with heparinized PBS, the lungs were removed and placed in formamide at 37°C for 24 hours. The amount of dye in the lungs was calculated by measuring the absorbance of the supernatants at 620 nm. The following equation was used to calculate percent increase in capillary: (ODsample − ODcontrol)/ODcontrol) × 100.
Histopathology.
SEB and DIM were similarly administered as described, and 48 hours after SEB exposure, lungs were collected and placed in 10% formalin overnight. Tissue samples were embedded in paraffin and processed for H&E staining. Briefly, tissues were deparaffinzed using xylene and alcohol dehydration series (90%, 95%, and 100%). Sections were then cut and mounted on slides. Slides were stained with H&E and assessed using a Nikon E600 light microscope.
Cytokine Detection of Serum and Bronchoalveolar Lavage Fluid.
Mice were euthanized 48 hours after SEB exposure. Intact lungs were removed after suture of the trachea. Cold sterile PBS (1 ml) was used to collect the lavage fluid from lungs of vehicle- or DIM-treated mice. Mice were bled befor sacrifice and blood spun down for isolation of serum. Cytokine levels for IFN-γ was measured in bronchoalveolar lavage (BAL) fluid and serum. All cytokines were measured using Biolegend ELISA Max kits (430801; Biolegend, San Diego, CA) as described (Rao et al., 2014).
Antibodies and Flow Cytometry.
Cells were stained with fluorescent conjugated antibodies and analyzed using the Beckman Coulter FC500 (Indianapolis, IN) to determine phenotypes of infiltrating lung mononuclear cells. The following antibodies were used: allophycocyanin (APC)-conjugated anti-CD3 (clone: 145.2 C11), fluorescein isothiocyanate–conjugated anti-CD8 (clone: 53-6.7), anti-CD3 (clone: 145.2 C11) anti-vβ8 (clone: KJ16-133.18), phycoerthyrin (PE)-conjugated anti-CD4 (clone: GK 1.5), and anti-NK1.1 (clone: PK136) from Biolegend.
Isolation of Lung-Infiltrating Cells.
SEB and DIM were administered as described, and 48 hours after SEB exposure, mice were euthanized. Lungs were harvested and homogenized using Stomacher 80 Biomaster in 10 ml of sterile PBS. Cell suspensions were washed and isolated using Ficoll gradient separation. Briefly, cells resuspended in sterile PBS were carefully layered onto Ficoll (Histopaque-1077, no. 10771; Sigma-Aldrich) and centrifuged at 500g for 30 minutes at room temperature (25°C) with brake off. The mononuclear cells were collected at the interface. Live cells were then enumerated using a hemocytometer and trypan blue exclusion.
MicroRNA and mRNA Isolation
Using miRNeasy Kit (217004; Qiagen, Valencia, CA), total RNAs (including small RNAs) were isolated from lung-infiltrating mononuclear cells or in vitro splenocytes cultures. Purity and concentration of RNA were determined using the NanoDrop 2000 spectrophotometer from Thermo Scientific (Wilmington, DE).
MicroRNA Array, Pathway Analysis, and Quantitative Real-Time Polymerase Chain Reaction.
MiRNA expression profile of lung infiltrating mononuclear cells was determined using Affymetrix GeneChip miRNA 1.0 array platform (Affymetrix Inc, Santa Clara, CA) as described previously (Hegde et al., 2013). Ingenuity Pathway Analysis (IPA) software (Qiagen) was used to analyze the microarray data set. Top miRNA molecules were determined using a log2 1.5-fold change between treatment groups. Pathways and mRNA targets were selected using IPA; either strongly predicted or experimentally validated interactions in published literature were pursued. This method was used to create miRNA-mRNA pathways as previously described (Hegde et al., 2013). Select miRNAs were validated by real-time quantitative polymerase chain reaction (qPCR). Using miScript II RT kit (218160; Qiagen), cDNA was prepared from total RNA samples. Mature miRNA detection was performed using QuantiTect SYBR Green PCR kit (218073; Qiagen) per the manufacturer’s instructions and the Viia7 PCR thermal cycler for the following: mmu-miR-222, mmu-miR-34a, mmu-miR-494, mmu-miR-706, mmu-miR-125b, mmu-miR-155, and control RNU_1a or Snord96a primers obtained from Qiagen. Relative quantification by 2(ΔΔCq) method and expressed relative to endogenous control.
Cell-Cycle Analysis.
Splenocytes were isolated from naïve C57BL/6 mice and cultured with SEB (1 μg/ml) in the absence or presence of vehicle or DIM (10–50 μM) for 6, 12, 18, and 24 hours. In vitro concentrations of DIM were based on previous work from our laboratory and well within attainable concentrations found after oral administration of DIM in mice (Anderton et al., 2004; Rouse et al., 2013; Busbee et al., 2014). Cells were harvested and fixed with 2% paraformaldehyde for 1 hour at 4°C. After washing with staining buffer, cells were incubated in a solution containing 0.2% Tween 20, 100 U/ml RNase, and 50 μg/ml propidium iodide for 20 minutes at 37°C. Cell-cycle analysis was performed using flow cytometry in which samples were gated on live cells. Listmode (LMD) files were further analyzed using ModFit LT (Verity Software House, Topsham, ME).
Transfection of miRNA.
Transfection was performed as described (Busbee et al., 2015). Splenocytes were cultured in complete (10% FBS, 10 mM l-glutamine, 10 mM Hepes, 50 μM β-mercaptoethanol, and 100 μg/ml penicillin) RPMI 1640 medium (11875-093; Life Technologies Corp., Waltham, MA). Cells were seeded at 2.5 × 105 cells/well in a 24-well plate and transfected for 24 hours with mock control, 40 nM synthetic mmu-miR-222 (MSY0000670), and 100 nM mmu-mmiR-222 (MIN0000670) using HiPerFect transfection reagent (301704; Qiagen), according to manufacturer’s instructions. Cells were then cultured in the absence or presence of 25 μM DIM for 6 hours. Cells were collected and stained using AnnexinV/PI kit to ascertain extent of cell death. Additionally, total RNA and protein were extracted for analysis.
Western Blotting.
Total protein was isolated from lung mononuclear cells and in vitro transfection reactions using RIPA buffer (sc-24948; Santa Cruz Biotechnology, Santa Cruz, CA) and then quantified using Pierce BCA Protein Assay Kit (23225; Thermo Fisher Scientific, Somerset, NJ). For each sample, 10–20 μg of protein was loaded for each sample. Protein extracts were separated by 12% polyacrylamide gel electrophoresis and transferred onto nitrocellulose membrane. Membranes were then probed with antibodies against: p27kip1, BIM, and/or controls β-actin and γ-tubulin as shown (Santa Cruz). Blots were developed using WesternSure PREMIUM Chemiluminescent Substrate and scanned using the C-Digit Blot Scanner from Licor (Lincoln, NE).
Statistics.
Statistical analysis was performed using GraphPad Prism 5.0 (San Diego, CA). The data using mice were generated using groups of four or five and depicted as mean ± S.E.M. The in vitro assays were performed at least three times to confirm the reproducibility. Statistical difference was calculated using analysis of variance and Student’s t test, and where stated post hoc analysis was performed via Tukey’s method. P value < 0.05 was considered statistically significant.
Results
DIM Attenuates SEB-Induced ALI.
Inhalation exposure of SEB leads to acute pulmonary damage comprising inflammation, cytokine secretion, capillary leak, and edema. In this study, we investigated the ameliorative potential of DIM in this model of inflammation. Previously, our laboratory has shown that 40 mg/kg body weight of DIM was able to attenuate other inflammatory disorders (Busbee et al., 2014; Rouse et al., 2014). To account for lower bioavailability of DIM in lung tissue, we used a dose of 100 mg/kg body weight of DIM in this study, described further in Materials and Methods. Moreover, previous studies showed that DIM could attenuate brain inflammation in a lipopolysaccharide model at similar dosage via oral gavage (Kim et al., 2014). Mice were pretreated with DIM or vehicle (corn oil) 24 hours and 1 hour before SEB exposure. Twenty-four hours after SEB exposure, mice were given the final dose of DIM or vehicle. A group of mice were also given DIM only, receiving the same three treatments as stated herein.
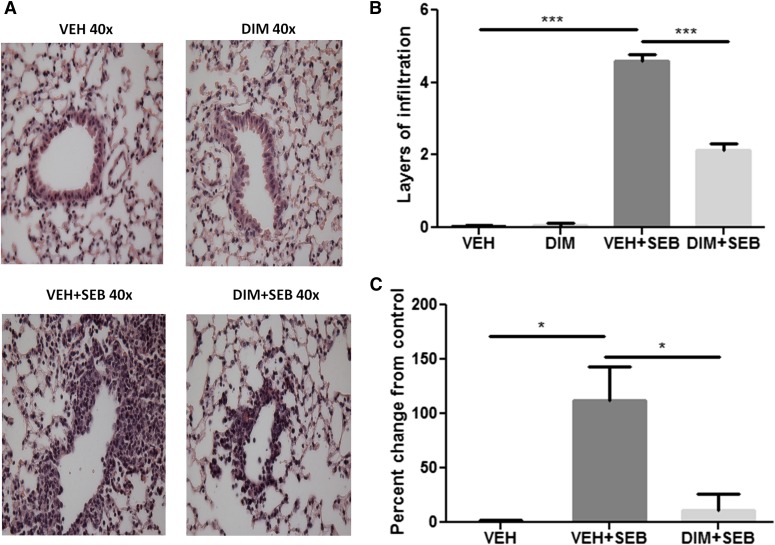
Forty-eight hours after SEB exposure, histopathological analysis of H&E-stained lung sections showed extensive infiltration of cells in VEH + SEB group compared with control (Fig. 1, A and B). The DIM + SEB group showed significantly reduced infiltration, and DIM alone treatment showed no infiltration (Fig. 1, A and B). We analyzed the extent of vascular leak using Evans blue dye as previously described (Saeed et al., 2012). Indeed, SEB exposure led to a significant increase in vascular leak compared with vehicle alone; however, DIM treatment could significantly reduce vascular leak (Fig. 1C).
Fig. 1.
DIM treatment reduces SEB-induced inflammation and capillary leak in the lungs. (A) H&E staining of lung sections from different treatment groups. The photographs were taken at 40× magnification. (B) Quantification of the histopathology data. Layers of infiltrating cells were counted around 15 different capillaries of the same size, and the data represent the mean ± S.E.M. for each individual group. (C) Measurement of capillary leak in the lungs using Evans blue dye extravasation. Percent increase in vascular leak was calculated compared with control. Vertical bars represent data collected from four or five mice per group expressed as means ± S.E.M. Analysis of variance, ***P < 0.001; *P < 0.05 with Tukey’s test.
DIM Lessens Lung Infiltration and IFN-γ Secretion.
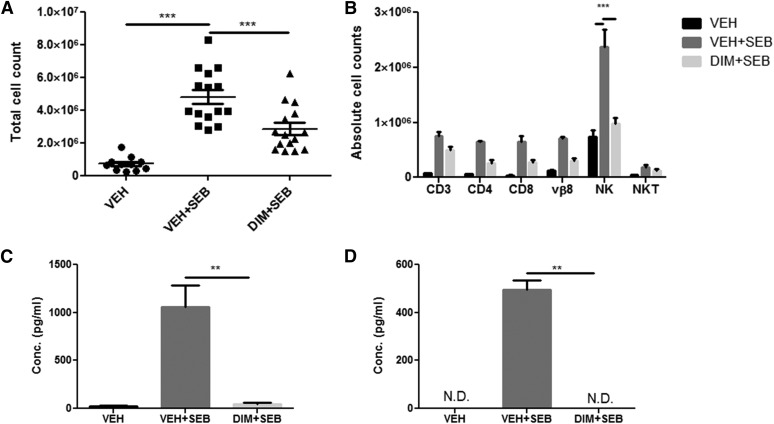
Substantial cellular infiltration and subsequent cytokine secretion characterize ALI induced by SEB. Overall inflammatory burden was next assessed by enumerating the total mononuclear cells from the lungs of each group of mice. SEB-treated mice had a significant increase in lung mononuclear cells, which was decreased after DIM treatment (Fig. 2A). Lung-infiltrating cells were further analyzed to identify different immune subsets. We found that SEB exposure produced an increase in the total number of cells expressing CD3+, CD4+, CD8+, Vβ8+, NK1.1+ (NK cells), and NK1.1+CD3+ (NKT cells). Furthermore, DIM reduced these cell subsets infiltrating into the lungs (Fig. 2B). Our laboratory has recently shown the importance of γ interferon (IFN-γ) in the inflammatory cascade and subsequent cell damage in ALI (Rao et al., 2014). We therefore examined the expression of IFN-γ in the serum and BAL fluid. The IFN-γ levels in the serum and BAL fluid were significantly elevated in SEB-treated mice compared with vehicle controls. DIM treatment was able to significantly reduce to IFN-γ expression in serum and BAL fluid (Fig. 2, C and D).
Fig. 2.
DIM decreases immune cell infiltration and IFN-γ expression in the lungs. All experiments were performed 48 hours after SEB exposure. (A) Total number of mononuclear cells from lungs of mice is expressed as per mouse. (B) Immune cells were further stained with monoclonal antibodies to determine the following subsets: T cells (CD3), T-helper cells (CD4), T-cytotoxic cells (CD8), natural killer cells (NK), and natural killer T-cells (NKT). The percentages were multiplied by the total cell number to yield the absolute cell counts shown. (C) IFN-γ levels in BAL fluid. (D) IFN-γ expression in serum. All cytokines were determined using enzyme linked immunosorbent assay (ELISA) from Biolegend. Vertical bars in (B–D) represent mean ± S.E.M. from groups of four or five mice. Analysis of variance, ***P < 0.001; **P < 0.01 with Tukey’s post hoc test.
DIM Induces Apoptosis and Cell-Cycle Arrest.
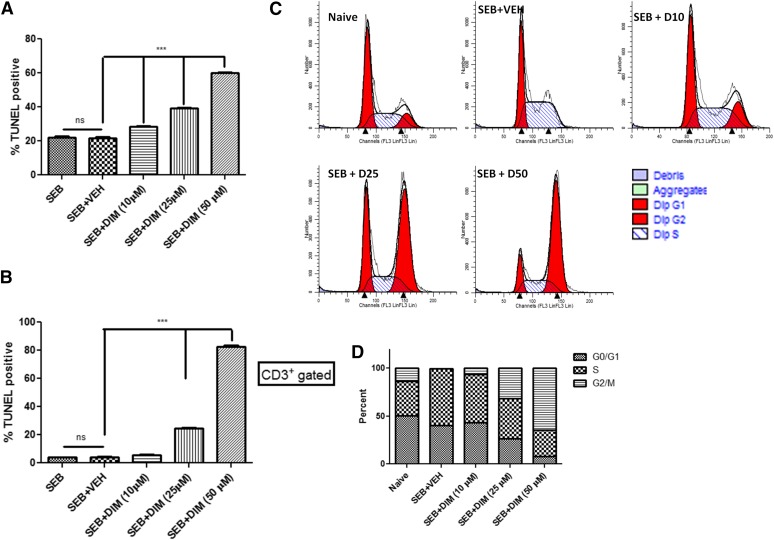
DIM has been shown to have potent antiproliferative properties in a multitude of cancer cells lines, inducing apoptosis and/or cell-cycle disruption (Shorey et al., 2012; Weng et al., 2012a; Jin et al., 2015). In this current study, we determined the possibility that DIM may induce cell death in SEB-activated T cells. To this end, we activated splenocytes with SEB (1 μg/ml) for 24 hours in the presence of a DIM concentration range (10, 25, 50 μM). Cells were then collected and stained with anti-CD3 and terminal deoxynucleotidyl transferase–mediated digoxigenin-deoxyuridine nick-end labeling (Roche) and analyzed by flow cytometry. After 24 hours, DIM showed a significant increase in apoptosis in SEB-activated T cells compared with vehicle. This response could be seen in a concentration dependent manner. It should be noted that SEB alone led to apoptosis (∼20% overall and ∼4% T cells), which can be attributed to activation induced cell death (Fig. 3, A and B).
Fig. 3.
DIM induces apoptosis and cell-cycle arrest in SEB-activated cells. (A) 3,3′-diindolylmethane induces cell death in a concentration-dependent manner. Splenocytes from C57Bl/6 mice were activated with SEB (1 μg/ml) in the presence of either vehicle or DIM (10, 25, 50 μM) for 24 hours. Cells were stained for CD3 analyzed for apoptosis based on terminal deoxynucleotidyl transferase–mediated digoxigenin-deoxyuridine nick-end labeling (TUNEL). (B) TUNEL-positive cells from above gated on CD3. (C) Cells were treated similarly and collected at 6, 12, and 18 hours. Cells were analyzed with povidone iodine staining, flow cytometry, and ModFit software. (D) Quantification of cell-cycle analysis. Vertical bars in panels represent mean ± S.E.M. from groups of four or five mice. Analysis of variance, ***P < 0.001; **P < 0.01 with Tukey’s post hoc test.
Additionally, we tested DIM’s potential to arrest the cell cycle in SEB-activated T cells. Splenocytes were treated similarly as described herein; however, cells were collected at 6, 12, and 18 hours and analyzed for cell cycle using propidium iodide staining, flow cytometry and ModFit LT software. Activation with SEB for 6 hours led to an increase in cells found in S phase as compared with naïve control, which were found to be in G0/G1 phase, with a minor percentage in S and G2/M phases. Interestingly, DIM treatment caused most of the cells to be found in G2/M phase; this was seen in a concentration-dependent manner (Fig. 3, C and D). Similar results were seen at 12 and 18 hours (data not shown). These results substantiate the ability of DIM to alter the cellular proliferation induced by SEB activation via G2/M arrest and apoptosis.
DIM Treatment of SEB-Exposed Mice Significantly Alters the MicroRNA Profile of Lung Mononuclear Cells.
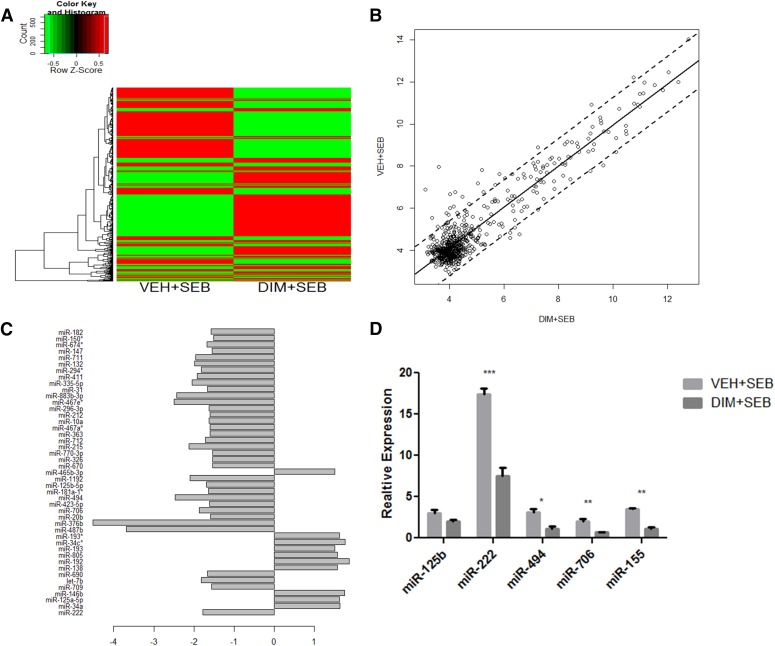
MicroRNAs have recently been implicated as having a regulatory role in cellular proliferation, cell-cycle progression, and inflammation. Furthermore, miRNAs have been shown to play a key role in lung inflammation (Foster et al., 2013) and particularly in SEB-induced lung inflammation (Rao et al., 2015). Therefore, we investigated the role of microRNA in the DIM-mediated suppression of inflammation, specifically in the lungs of SEB exposed mice. Forty-eight hours after administration of SEB, we isolated lung mononuclear infiltrating cells from SEB exposed mice treated with DIM or vehicle. Total RNA was collected and analyzed using Affymetrix GeneChip miRNA 1.0 array platform. Differentially expressed miRNAs were represented in a heat map. Mice treated with DIM and SEB showed a significant change in the microRNA expression profile compared with the VEH + SEB group (Fig. 4A). On further analysis, we were able to identify 10 miRNAs that were significantly upregulated and 25 miRNAs that were significantly downregulated in DIM + SEB groups compared with VEH + SEB treatment (Fig. 4, B and C). After the miRNA array, we used Ingenuity Systems IPA analysis to further fine tune select miRNAs. These miRNAs are compiled in (Table 1), along with their seed sequence and respective fold change. Six microRNAs were randomly selected for further validation. DIM treatment led to a significant downregulation of four of the selected miRNAs in SEB exposed mice (Fig. 4D). Collectively, these data suggested that microRNA may have an integral role in the antiproliferative effect of DIM in SEB-induced ALI.
Fig. 4.
miRNA expression profiling. miRNA was isolated from lung mononuclear cells from mice exposed to SEB or SEB and DIM as described in Fig 1. (A) Differential expression heat map of 609 miRNAs between VEH + SEB and DIM + SEB groups. (B) Pearson correlation scatter plot of mi99RNA log2 expression values for VEH + SEB and DIM + SEB groups. (C) Bar plot of fold changes of differentially expressed miRNA in DIM + SEB showing >1.5-fold difference in expression compared with VEH + SEB. (D) Quantitative real-time PCR validation of select dysregulated miRNA. RNU1a and Snord96a were used as endogenous control, and expression level was normalized to vehicle. Vertical bars in (D) represent mean ± S.E.M. from groups of four or five mice. **P < 0.01; *P < 0.05 compared with vehicle.
TABLE 1.
Highly upregulated and downregulated miRNAs upon DIM treatment in SEB-exposed mice
| miRNA Identifier | MiRBase Accession No. | Chromosome | Sequencea | FC |
|---|---|---|---|---|
| mmu-miR-142b | MIMAT0031402 | Chr11 | uccauaaaguaggaaacacu | 1.7↑ |
| mmu-miR-34a-5p | MIMAT0000542 | Chr4 | uggcagugucuuagcugguugu | 1.6↑ |
| mmu-miR-465b-3p | MIMAT0004872 | ChrX | gaucagggccuuucuaaguaga | 1.5↑ |
| mmu-miR-125a-5p | MIMAT0031402 | Chr17 | ucccugagacccuuuaaccuguga | 1.6↑ |
| mmu-miR-222-3p | MIMAT0000670 | ChrX | agcuacaucuggcuacuggu | 1.8↓ |
| mmu-miR-494-3p | MIMAT0003182 | Chr12 | ugaaacauacacgggaaaccuc | 2.5↓ |
| mmu-miR-376b | MIMAT0001092 | Chr12 | aucauagaggaacauccacuu | 4.5↓ |
| mmu-miR-706 | MIMAT0003496 | Chr6 | agagaaacccugucucaaaaaa | 1.9↓ |
Fold change (FC),
7-mer seed sequence in bold.
DIM Treatment Decreases the Expression of miR-222 and miR-494 to Regulate p27kip1 and BIM Expression.
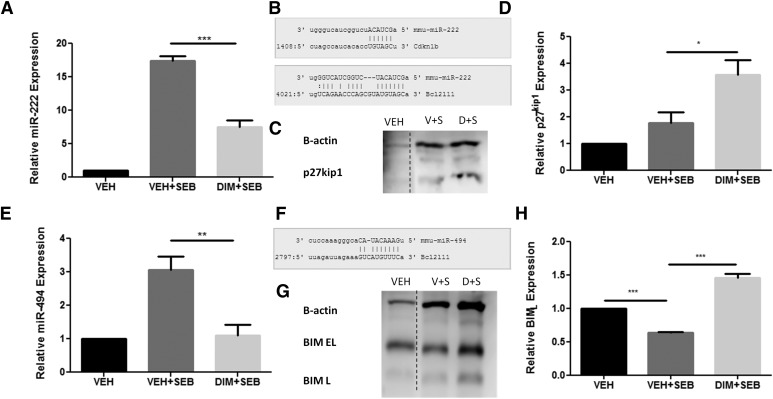
Among the miRs screened, miR-222 and miR-494 were significantly increased upon SEB exposure compared with vehicle control mice and DIM treatment led to a significant decrease (Fig. 5, A and E). The miR-222 was predicted to target cyclin-dependent kinase inhibitor (CDKN1b) (p27kip1), and miR-494 was predicted to target bcl2l11 (BIM), both having a miRWalk score of 3 and putative 3′ UTR targeting was seen for each miRNA-mRNA pairing using microRNA.org (www.microRNA.org) alignment tool (Fig. 5, B and F). A complete list of mRNA targets for miR-222 and miR-494 can be found in Table 2. Furthermore, p27kip1 and BIM expression levels were significantly increased in DIM + SEB compared with VEH + SEB group, respectively (Fig. 5, C, D, G, and H). Together, these results suggested that DIM treatment in mice exposed to SEB might alter the miRNA expression profile, which may lead to the halt of cell cycle progression and induction of apoptosis, therefore attenuating the aberrant lung inflammatory response.
Fig. 5.
In vivo miRNA and target gene expression for miR-222 and miR-494. Mice were exposed to SEB or SEB and DIM as described in Fig. 1. Total RNA and protein were isolated from lung mononuclear cells. (A) miR-222 expression level by qPCR. (B) Schematic of miRNA-mRNA pairing of miR-222 with CCKN1B (p27kip1) and BCL2L11 (BIM). (C) Western blot analysis of p27kip1. (D) Densitometry analysis of p27kip1 relative expression normalized to β-actin using ImageJ software. (E) miR-494 expression level by qPCR. (F) Schematic of miRNA-mRNA pairing of miR-494 with BCL2L11 (BIM). (G) Western blot analysis of BIM. (H) Densitometry analysis of BIML relative expression normalized to β-actin using ImageJ software. Data presented as means ± S.E.M. (n = 3 per group). *, **, and *** represent significant difference with P values < 0.05, < 0.01, and < 0.001, respectively.
TABLE 2.
MicroRNAs -222, -494, and experimentally proven target genes
| miRNA | Experimentally Observed Targets |
|---|---|
| mmu-miR-222-3p | p27kip1, p57, BIM, PUMA, PTEN |
| mmu-miR-494-3p | BIM, CCNB1, CDC2, PTEN, CDK6 |
Mir-222 Downregulation May Be Integral to DIM-Mediated Cell Death.
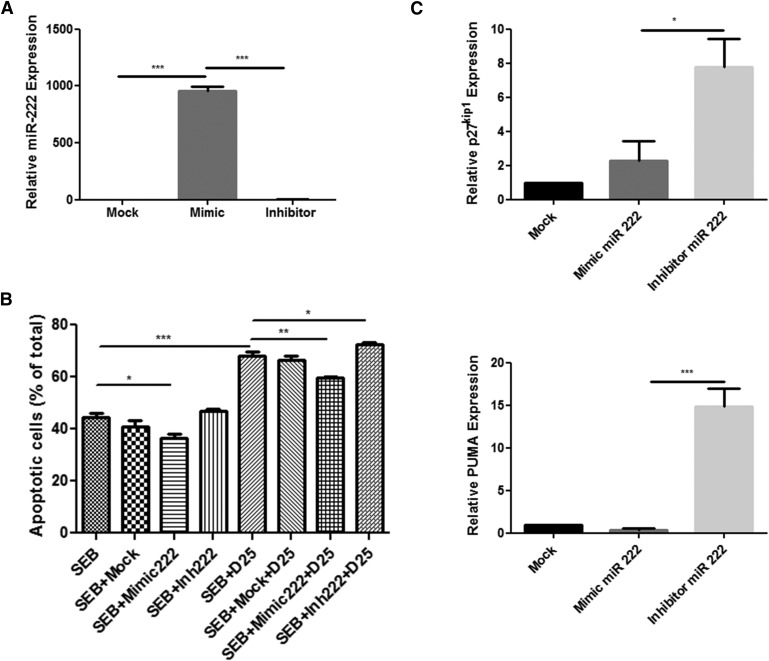
To further confirm the role of DIM-induced miRNAs in cell death, we transfected splenocytes with miR-222 mimic and inhibitor. After 18 hours, cells were treated with DIM (25 μM) or vehicle (dimethyl sufoxide) for 6 hours. MiR-222 expression after mimic transfection increased 960-fold compared with mock control and inhibitor saw no significant change (Fig. 6A). Furthermore, miR-222 mimic significantly decreased cell death compared with mock control and inhibitor saw no change in the absence of DIM. DIM treatment led to a significant increase in cell death compared with SEB. Mimic transfection could rescue cells from DIM-mediated apoptosis significantly. Moreover, inhibitor led to a significant increase in cell death in the presence of DIM (Fig. 6B). We then investigated the effect of miR-222 mimic and inhibitor in these SEB activated splenocytes on the level of expression of p27kip1 and PUMA and found that miR-222 mimic could decrease expression of PUMA but not p27kip1. Moreover, miR-222 inhibitor transfection led a significant increase in both p27kip1 and PUMA expression (Fig. 6C). Together, these data suggested that DIM may induce apoptosis through the downregulation of miR-222 and subsequent increase in gene targets p27kip1 and PUMA.
Fig. 6.
miR-222 regulates p27kip1 and PUMA expression to induce cell death in SEB-activated splenocytes. Splenocytes were transfected with control (mock) or miRNA mimic and inhibitor for miRNA-222 in the presence of SEB (1 μg/ml). Eighteen hours later, cells were treated with DIM (25 μM) or vehicle for 6 hours. (A) Total RNA was isolated from cells and analyzed for miR-222 expression by qPCR. (B) Cells were collected and stained with AnnexinV/PI staining to determine apoptosis and then quantified for cell death. (C) Relative expression level of gene targets p27kip1 and PUMA by quantitative PCR. Data are presented as means ± S.E.M. (n = 3 per group). *, **, and *** represent significant difference with P values < 0.05, < 0.01, and < 0.001, respectively.
Discussion
ALI and its more severe form acute respiratory distress syndrome are clinical disorders with characteristic pulmonary bilateral infiltrates, edema, and hypoxemia and can have a wide range of causes: sepsis, reperfusion, pneumonia, and smoke or toxic chemical inhalation (Ware and Matthay, 2000). Mortality rates still remain relatively high (∼30%–50%), depending on causation, and treatment modalities remain restricted with limited efficacy (Dushianthan et al., 2011). Therefore, there still remains the need for novel treatment modalities with improved safety and efficacy. Although animal models do not perfectly resemble clinical manifestations of ALI, they do provide invaluable insight into facets of human pathophysiology (Matute-Bello et al., 2008). For example, inhalation of SEB can lead to dramatic respiratory inflammation, which mimics the infiltration and edema seen in clinical ALI (Neumann et al., 1997). In this study, we used intranasal administration of SEB to induce lung inflammation and for the first time demonstrated the clinical benefits of DIM in this model, which may result from the ability of naturally derived indole to suppress inflammation by modulating miRNA expression, which targets integral parts of the cell-cycle and cell death pathways. In the current study, we demonstrated that DIM can protect from SEB-induced ALI by decreasing pulmonary infiltration, attenuating IFN-γ secretion, and limiting vascular leak. Additionally, our data suggest that the changes in miRNA expression profile by DIM in vivo could be involved in the prophylactic role. DIM treatment significantly decreased the expression of miR-222 and miR-494 in the lung mononuclear cells, with a coordinate increase in the expression of p27kip1 and BIM. Moreover, using miR-222 synthetic mimic and inhibitor, we performed gain and loss of function analysis to demonstrate the potential role of miR-222 in DIM-mediated suppression of proliferation caused by SEB. In the current study, we used 100 mg/kg of body weight of DIM to attenuate SEB-induced ALI in the mouse model. These doses are clinically relevant because in human studies, doses up to 600 mg daily of DIM have been used (Heath et al., 2010). The dose of 100 mg/kg in mice, when converted into human equivalent dose, works out to 480 mg, considering an adult to weigh 60 kg. Thus, the dose used in this study is translationally relevant.
SEB exposure drives extensive release of chemokines and cytokines, T-cell expansion, and both localized and systemic tissue damage (Krakauer, 1999; Saeed et al., 2012; Busbee et al., 2014). This atypical immune response can be attributed to the unique mechanism of T-cell activation that SEB uses, expanding a considerable proportion of T cells by binding sequentially the vβ8 region of the TCR on T cells and major histocompatibility class II molecule on antigen presenting cells (Herman et al., 1991; Xia et al., 2014). Our results are consistent with these findings, as we observed a dramatic increase in infiltrating mononuclear cells and significant endothelial-epithelial barrier destruction in the lungs. Moreover, systemic and bronchoalveolar IFN-γ expression was significantly increased, which has been shown to be integral to SEB-mediated toxic shock syndrome and lung injury (Tilahun et al., 2011; Rao et al., 2014).
The molecular mechanisms have been well described for SEB-mediated inflammation. Upon TCR activation and costimulation, canonical and alternative signaling pathways are activated simultaneously (Deane and Fruman, 2004; Bueno et al., 2006; Park et al., 2009). These conditions synergize to promote proliferative and proinflammatory gene expression in T cells (Krakauer, 2013); however, recently a new regulatory factor has been discovered: miRNA. Extensive studies of miRNA have provided evidence to show its critical role in fine-tuning biologic processes. MicroRNAs exert their regulatory efforts by targeting the 3′ UTR of mRNA, thereby inhibiting gene translation via sequestration or degradation of mRNA (Ranganathan and Sivasankar, 2014). The importance of miRNAs in immune cell development and response has also been elucidated (Baltimore et al., 2008). Moreover, a study from our laboratory has expounded the role of miRNAs in SEB-induced ALI and mortality (Rao et al., 2015). The importance of individual (miR-155) and clustered miRNAs (miR-17-92) in SEB-induced ALI has also been studied (Rao et al., 2014, 2015).
The roles of miR-222 and miR-494 have predominantly been studied in the context of cancer; however, miR-222 was elevated in activated human T cells and lung mononuclear cells of SEB-exposed mice, and miR-494 was dysregulated in T cells in multiple sclerosis patients (Grigoryev et al., 2011; Jernås et al., 2013; Rao et al., 2015). Given the limited knowledge of how these function in lymphocytes, we sought to determine their regulatory role. MicroRNA-222 is encoded in tandem with homologous miR-221 on the X chromosome and has been implicated as a tumor suppressor or oncomiR targeting p27kip1, p57, PUMA, PTEN, and BIM in varying cancer cell lines (Garofalo et al., 2012). Interestingly, c-Jun and NFκB bind to the promoter of miR-221/222 to induce their expression (Galardi et al., 2011), and activation of ERK1/2 induces miR-221/222 (Terasawa et al., 2009). Similarly, miR-494 (Chr12) is also a known oncomiR targeting CCNB1, CDC2, PTEN, BCL2L11 (BIM), and CDK6 (Olaru et al., 2011; Yamanaka et al., 2012; Li et al., 2015). Romano et al. (2012) found that the ability of miR-494 to suppress the proapoptotic protein BIM was inhibited upon extracellular signal–regulated kinase (ERK)1/2 inactivation. Overexpression and loss of function studies with both miR-222 and -494 have shown regulatory roles in cell-cycle progression and proliferation in cell lines (Zhang et al., 2010; G. Sun et al., 2013; H. G. Sun et al., 2014). Similar to these reports, SEB exposure led a significant increase in the expression of miR-222 and -494 in lung mononuclear cells, suggesting that these may have functions consistent with those seen in cancer cells.
CDKN1B, or p27kip1, is member of the cyclin-dependent kinase inhibitor family and has been reported as a direct target of miR-222 (C. Sun et al., 2013). p27kip1 inhibits the activation of cyclin E-CDK2 or cyclin D-CDK4, thereby controlling cell-cycle progression at G1; however, this result was not replicated in another study performed by Wurz et al. (2010) in ovarian carcinoma, which showed a correlation between CDKN1C (p57) but not p27kip1 and miR-221/222. The timing, cell status, and cell type may be integral to finding this correlation as suggested as a feedback loop mechanism (Frenquelli et al., 2010). These findings may explain the lack of p27kip1 repression in SEB-exposed mice and miR-222 mimic studies conducted. Moreover, PUMA, another direct target of miR-222 (Zhang et al., 2010), was significantly reduced after induction of miR-222 mimic. Cell environment and timing are critical factors in miRNA regulation as changes can occur within hours of activation (Bronevetsky et al., 2013). Another confounding issue is that miRNAs can act synergistically and antagonistic with each other; both miR-222 and miR-494 directly target the proapoptotic BH3-only protein BCL2L11, or BIM (Terasawa et al., 2009; Romano et al., 2012). SEB-exposed mice showed decreased BIM protein in lung mononuclear cells correlating with the increased miR-222 and -494 levels. Concurrent with these findings, miR-222 mimic could induce cell survival, in part by modulating mRNA gene targets. Inhibition of miR-222 led to a significant increase in p27kip1 and PUMA expression but could not significantly increase apoptosis.
DIM has been shown to attenuate a wide array of inflammatory diseases and uses complementary functions. Recent reports purport the effective use of DIM in murine models of arthritis and steatosis (Huang et al., 2013; Liu et al., 2014). In mouse models of LPS-induced acute liver failure and experimental autoimmune encephalomyelitis, we have displayed its ability to attenuate disease progression, alleviating liver damage and halting central nervous system demyelination by altering microRNA expression profiles in macrophages and CD4+ T cells, respectively (Rouse et al., 2014; Tomar et al., 2014) inasmuch as our data demonstrate significant reduction in pulmonary infiltrates, local and systemic IFN-γ expression, and edema. We previously reported that DIM’s effects can be attributed, in part, to promotion of apoptosis and G1/S arrest in ConA-activated T cells (Busbee et al., 2014; Rouse et al., 2014). In the current study, DIM treatment led an increase in cell death in vitro; however, we also found that DIM halted SEB-activated cells in G2/M phase. Interestingly, DIM has been shown to halt cell-cycle progression in both the G1/S and G2/M phases, relying on suppression of ERK and Akt pathways and NFκB-Akt, MAPK and p53 signaling pathways, respectively, based on studies performed in different cancer cell lines and concentrations of DIM (Rajoria et al., 2011; Weng et al., 2012b). Such studies further emphasize the importance of cell-type specificity and the concentrations of DIM used. Furthermore, recent studies have demonstrated that DIM increases p27kip1 expression and a synthetic derivative 2,2′-diphenyl-3,3′-diindolylmethane can increase the expression of BIM (Wang et al., 2008; Bhowmik et al., 2013). These data are consistent with our findings that DIM significantly increased expression of both p27kip1 and BIM.
In the current study, we noted that DIM treatment altered the miRNA expression profile in lung mononuclear cells significantly, specifically downregulating the expression of miR-222 and -494 in lung mononuclear cells, which could explain the concurrent increase in p27kip1 and BIM protein levels observed in the same cells. Although the mechanism is unknown, DIM decreases NFκB nuclear translocation and ERK1/2 phosphorylation, both of which are proven to promote the transcription of these miRNAs (Rahman et al., 2007; Rajoria et al., 2011). In another study, formulated DIM (BR-DIM) could increase expression of miR-221 and consequently decrease p27kip1 and PUMA expression in a pancreatic cancer cell line (panc-1) (Sarkar et al., 2013). In addition, we noted that DIM-induced increased cell death in SEB-activated splenocytes, and transfection of miR-222 mimic could rescue cells from apoptosis. Inhibition of miR-222 significantly increased DIM-mediated cell death, and increased expression of miR-222 gene targets p27kip1 and PUMA in the presence of DIM could be one explanation for this effect.
It is not clear whether DIM would exert in vivo a direct effect on SEB biotransformation or affect the growth of S. aureus. A recent study indicated that DIM and synthesized DIM derivatives have antibacterial properties, including inhibition of S. aureus in vitro, at high concentrations (Roy et al., 2013). If DIM were to be effective in vivo to inhibit the bacteria, there could be a potential synergistic effect of DIM, acting on both the host immune response to SEB and the pathogen. It is currently unclear whether DIM has any direct effects on the biotransformation of SEB. Previous studies (Vabulas et al., 1996) have shown that administration of SEB into mice leads to its accumulation in the lymph nodes in 5 minutes, followed by clearance primarily through the kidneys within 10–24 hours. Thus, because SEB can come in contact with immune cells within 5 minutes, and its excretion through kidneys occurs much later, by which time, the T cells are already activated, we believe that DIM-mediated effects are less likely to result from its effect on SEB biotransformation.
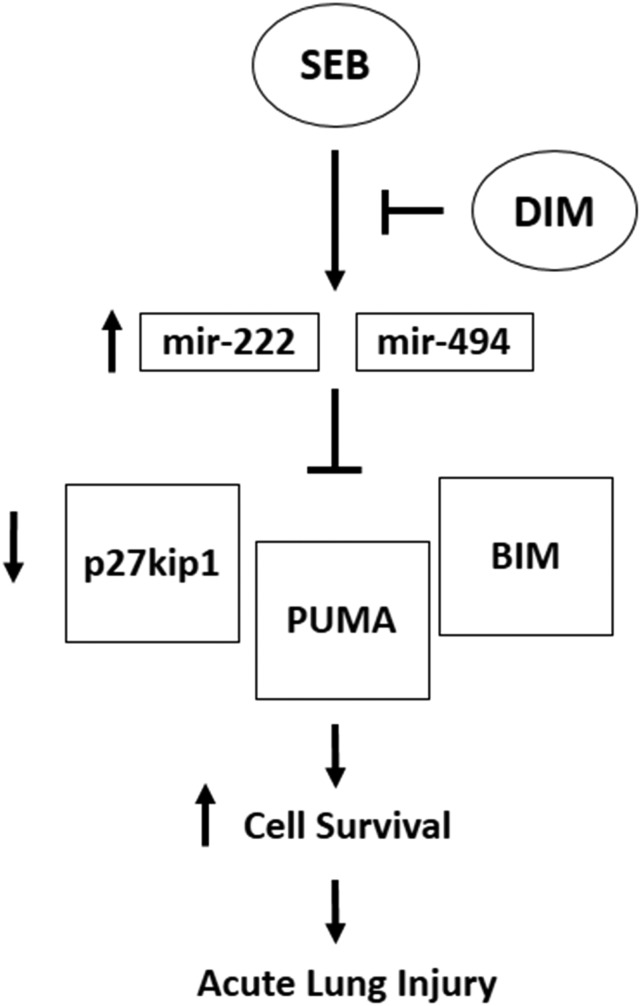
The current study demonstrates the use of DIM as a potential therapy against SEB-induced ALI and related pulmonary inflammatory diseases. This beneficial effect of DIM is mediated, at least in part, by the downregulation of microRNAs-222 and -494, leading to an increased expression of p27kip1, PUMA, and BIM to halt cell-cycle progression and promote cell death in SEB-activated cells (Fig. 7). These findings add to the growing evidence supporting the anti-inflammatory properties of DIM.
Fig. 7.
Schematic mechanism of DIM on SEB activated T cells. Exposure to SEB leads to massive activation and proliferation of T cells through canonical pathways, with those governing apoptotic status playing a critical role. Given our findings, microRNAs also have a key role in the regulation of this response. In particular, SEB alters expression of certain microRNAs, such as miR-222 and miR-494. These can act to inhibit the expression the proapoptotic factors PUMA and BIM, as well as a key regulator of cell-cycle progression, p27kip1, thereby promoting cell survival and proliferative response in T cells and causing ALI. Our studies suggest a new mechanism whereby DIM treatment leads to a decrease in the aforementioned microRNA, shifting the balance to an antiproliferative state, induction cell-cycle arrest, and cell death, ultimately leading to increased apoptosis of SEB-activated T cells and attenuation of ALI.
Abbreviations
- ALI
acute lung injury
- BAL
bronchoalveolar lavage
- CDK
cyclin-dependent kinase
- DIM 3
3′-diindolylmethane
- ERK
extracellular signal regulated kinase
- miRNA
microRNA
- PBS
phosphate-buffered saline
- SEB
staphylococcal enterotoxin B
- TCR
T-cell receptor
- UTR
untranslated region
Authorship Contributions
Participated in research design: Elliott, M. Nagarkatti, P. S. Nagarkatti.
Conducted experiments: Elliott.
Contributed new reagents or analytic tools: M. Nagarkatti, P. S. Nagarkatti.
Performed data analysis: Elliott, M. Nagarkatti, P. S. Nagarkatti.
Wrote or contributed to the writing of the manuscript: Elliott, M. Nagarkatti, P. S. Nagarkatti.
Footnotes
The research was funded in part by National Institutes of Health [Grants R01AT006888, P01AT003961, P20RR032684, R01ES019313, R01MH094755, and VA Merit Award 1I01BX001357]. The funding agency had no role in experimental design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Ahmad A, Sakr WA, Rahman KMW. (2010) Anticancer properties of indole compounds: mechanism of apoptosis induction and role in chemotherapy. Curr Drug Targets 11:652–666. [DOI] [PubMed] [Google Scholar]
- Ali NAS, Dar BA, Pradhan V, Farooqui M. (2013) Chemistry and biology of indoles and indazoles: a mini-review. Mini Rev Med Chem 13:1792–1800. [DOI] [PubMed] [Google Scholar]
- Anderton MJ, Manson MM, Verschoyle R, Gescher A, Steward WP, Williams ML, Mager DE. (2004) Physiological modeling of formulated and crystalline 3,3′-diindolylmethane pharmacokinetics following oral administration in mice. Drug Metab Dispos 32:632–638. [DOI] [PubMed] [Google Scholar]
- Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. (2008) MicroRNAs: new regulators of immune cell development and function. Nat Immunol 9:839–845. [DOI] [PubMed] [Google Scholar]
- Banerjee S, Kong D, Wang Z, Bao B, Hillman GG, Sarkar FH. (2011) Attenuation of multi-targeted proliferation-linked signaling by 3,3′-diindolylmethane (DIM): from bench to clinic. Mutat Res 728:47–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhowmik A, Das N, Pal U, Mandal M, Bhattacharya S, Sarkar M, Jaisankar P, Maiti NC, Ghosh MK. (2013) 2,2′-diphenyl-3,3′-diindolylmethane: a potent compound induces apoptosis in breast cancer cells by inhibiting EGFR pathway. PLoS One 8:e59798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronevetsky Y, Villarino AV, Eisley CJ, Barbeau R, Barczak AJ, Heinz GA, Kremmer E, Heissmeyer V, McManus MT, Erle DJ, et al. (2013) T cell activation induces proteasomal degradation of Argonaute and rapid remodeling of the microRNA repertoire. J Exp Med 210:417–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno C, Lemke CD, Criado G, Baroja ML, Ferguson SSG, Rahman AKMN-U, Tsoukas CD, McCormick JK, Madrenas J. (2006) Bacterial superantigens bypass Lck-dependent T cell receptor signaling by activating a Galpha11-dependent, PLC-beta-mediated pathway. Immunity 25:67–78. [DOI] [PubMed] [Google Scholar]
- Busbee PB, Nagarkatti M, Nagarkatti PS. (2014) Natural indoles, indole-3-carbinol and 3,3′-diindolymethane, inhibit T cell activation by staphylococcal enterotoxin B through epigenetic regulation involving HDAC expression. Toxicol Appl Pharmacol 274:7–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busbee PB, Nagarkatti M, Nagarkatti PS. (2015) Natural indoles, indole-3-carbinol (I3C) and 3,3′-diindolylmethane (DIM), attenuate staphylococcal enterotoxin B-mediated liver injury by downregulating miR-31 expression and promoting caspase-2-mediated apoptosis. PLoS One 10:e0118506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane JA, Fruman DA. (2004) Phosphoinositide 3-kinase: diverse roles in immune cell activation. Annu Rev Immunol 22:563–598. [DOI] [PubMed] [Google Scholar]
- Dong L, Xia S, Gao F, Zhang D, Chen J, Zhang J. (2010) 3,3′-Diindolylmethane attenuates experimental arthritis and osteoclastogenesis. Biochem Pharmacol 79:715–721. [DOI] [PubMed] [Google Scholar]
- Dushianthan A, Grocott MPW, Postle AD, Cusack R. (2011) Acute respiratory distress syndrome and acute lung injury. Postgrad Med J 87:612–622. [DOI] [PubMed] [Google Scholar]
- Foster PS, Plank M, Collison A, Tay HL, Kaiko GE, Li J, Johnston SL, Hansbro PM, Kumar RK, Yang M, et al. (2013) The emerging role of microRNAs in regulating immune and inflammatory responses in the lung. Immunol Rev 253:198–215. [DOI] [PubMed] [Google Scholar]
- Foster TJ. (2004) The Staphylococcus aureus “superbug”. J Clin Invest 114:1693–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenquelli M, Muzio M, Scielzo C, Fazi C, Scarfò L, Rossi C, Ferrari G, Ghia P, Caligaris-Cappio F. (2010) MicroRNA and proliferation control in chronic lymphocytic leukemia: functional relationship between miR-221/222 cluster and p27. Blood 115:3949–3959. [DOI] [PubMed] [Google Scholar]
- Galardi S, Mercatelli N, Farace MG, Ciafrè SA. (2011) NF-kB and c-Jun induce the expression of the oncogenic miR-221 and miR-222 in prostate carcinoma and glioblastoma cells. Nucleic Acids Res 39:3892–3902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G. (2012) miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med 12:27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoryev YA, Kurian SM, Hart T, Nakorchevsky AA, Chen C, Campbell D, Head SR, Yates JR, 3rd, Salomon DR. (2011) MicroRNA regulation of molecular networks mapped by global microRNA, mRNA, and protein expression in activated T lymphocytes. J Immunol 187:2233–2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath EI, Heilbrun LK, Li J, Vaishampayan U, Harper F, Pemberton P, Sarkar FH. (2010) A phase I dose-escalation study of oral BR-DIM (BioResponse 3,3′- Diindolylmethane) in castrate-resistant, non-metastatic prostate cancer. Am J Transl Res 2:402–411. [PMC free article] [PubMed] [Google Scholar]
- Hegde VL, Tomar S, Jackson A, Rao R, Yang X, Singh UP, Singh NP, Nagarkatti PS, Nagarkatti M. (2013) Distinct microRNA expression profile and targeted biological pathways in functional myeloid-derived suppressor cells induced by Δ9-tetrahydrocannabinol in vivo: regulation of CCAAT/enhancer-binding protein α by microRNA-690. J Biol Chem 288:36810–36826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman A, Kappler JW, Marrack P, Pullen AM. (1991) Superantigens: mechanism of T-cell stimulation and role in immune responses. Annu Rev Immunol 9:745–772. [DOI] [PubMed] [Google Scholar]
- Hoefig KP, Heissmeyer V. (2008) MicroRNAs grow up in the immune system. Curr Opin Immunol 20:281–287. [DOI] [PubMed] [Google Scholar]
- Huang Z, Jiang Y, Yang Y, Shao J, Sun X, Chen J, Dong L, Zhang J. (2013) 3,3′-Diindolylmethane alleviates oxazolone-induced colitis through Th2/Th17 suppression and Treg induction. Mol Immunol 53:335–344. [DOI] [PubMed] [Google Scholar]
- Jernås M, Malmeström C, Axelsson M, Nookaew I, Wadenvik H, Lycke J, Olsson B. (2013) MicroRNA regulate immune pathways in T-cells in multiple sclerosis (MS). BMC Immunol 14:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin H, Park MH, Kim SM. (2015) 3,3′-Diindolylmethane potentiates paclitaxel-induced antitumor effects on gastric cancer cells through the Akt/FOXM1 signaling cascade. Oncol Rep 33:2031–2036. [DOI] [PubMed] [Google Scholar]
- Kim HW, Kim J, Kim J, Lee S, Choi B-R, Han J-S, Lee KW, Lee HJ. (2014) 3,3′-Diindolylmethane inhibits lipopolysaccharide-induced microglial hyperactivation and attenuates brain inflammation. Toxicol Sci 137:158–167. [DOI] [PubMed] [Google Scholar]
- Krakauer T. (2005) Chemotherapeutics targeting immune activation by staphylococcal superantigens. Med Sci Monit 11:RA290–RA295. [PubMed] [Google Scholar]
- Krakauer T. (1999) Immune response to staphylococcal superantigens. Immunol Res 20:163–173. [DOI] [PubMed] [Google Scholar]
- Krakauer T. (2013) Update on staphylococcal superantigen-induced signaling pathways and therapeutic interventions. Toxins (Basel) 5:1629–1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X-T, Wang H-Z, Wu Z-W, Yang T-Q, Zhao Z-H, Chen G-L, Xie X-S, Li B, Wei Y-X, Huang Y-L, et al. (2015) miR-494-3p regulates cellular proliferation, invasion, migration, and apoptosis by PTEN/AKT signaling in human glioblastoma cells. Cell Mol Neurobiol 35:679–687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D, Zienkiewicz J, DiGiandomenico A, Hawiger J. (2009) Suppression of acute lung inflammation by intracellular peptide delivery of a nuclear import inhibitor. Mol Ther 17:796–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, She W, Wang F, Li J, Wang J, Jiang W. (2014) 3, 3′-Diindolylmethane alleviates steatosis and the progression of NASH partly through shifting the imbalance of Treg/Th17 cells to Treg dominance. Int Immunopharmacol 23:489–498. [DOI] [PubMed] [Google Scholar]
- Lowy FD. (1998) Staphylococcus aureus infections. N Engl J Med 339:520–532. [DOI] [PubMed] [Google Scholar]
- Matute-Bello G, Frevert CW, Martin TR. (2008) Animal models of acute lung injury. Am J Physiol Lung Cell Mol Physiol 295:L379–L399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann B, Engelhardt B, Wagner H, and Holzmann B (1997) Induction of acute inflammatory lung injury by staphylococcal enterotoxin B. J Immunol 158:1862–1871. [PubMed]
- Olaru AV, Ghiaur G, Yamanaka S, Luvsanjav D, An F, Popescu I, Alexandrescu S, Allen S, Pawlik TM, Torbenson M, et al. (2011) MicroRNA down-regulated in human cholangiocarcinoma control cell cycle through multiple targets involved in the G1/S checkpoint. Hepatology 54:2089–2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park S-G, Schulze-Luehrman J, Hayden MS, Hashimoto N, Ogawa W, Kasuga M, Ghosh S. (2009) The kinase PDK1 integrates T cell antigen receptor and CD28 coreceptor signaling to induce NF-kappaB and activate T cells. Nat Immunol 10:158–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinchuk IV, Beswick EJ, Reyes VE. (2010) Staphylococcal enterotoxins. Toxins (Basel) 2:2177–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plank M, Maltby S, Mattes J, Foster PS. (2013) Targeting translational control as a novel way to treat inflammatory disease: the emerging role of microRNAs. Clin Exp Allergy 43:981–999. [DOI] [PubMed] [Google Scholar]
- Rahman KMW, Ali S, Aboukameel A, Sarkar SH, Wang Z, Philip PA, Sakr WA, Raz A. (2007) Inactivation of NF-kappaB by 3,3′-diindolylmethane contributes to increased apoptosis induced by chemotherapeutic agent in breast cancer cells. Mol Cancer Ther 6:2757–2765. [DOI] [PubMed] [Google Scholar]
- Rajoria S, Suriano R, Wilson YL, Schantz SP, Moscatello A, Geliebter J, Tiwari RK. (2011) 3,3′-diindolylmethane inhibits migration and invasion of human cancer cells through combined suppression of ERK and AKT pathways. Oncol Rep 25:491–497. [DOI] [PubMed] [Google Scholar]
- Ranganathan K, Sivasankar V. (2014) MicroRNAs: biology and clinical applications. J Oral Maxillofac Pathol 18:229–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R, Nagarkatti P, Nagarkatti M. (2015) Role of miRNA in the regulation of inflammatory genes in staphylococcal enterotoxin B-induced acute inflammatory lung injury and mortality. Toxicol Sci Off J Soc Toxicol; DOI: 10.1093/toxsci/kfu315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao R, Rieder SA, Nagarkatti P, Nagarkatti M. (2014) Staphylococcal enterotoxin B-induced microRNA-155 targets SOCS1 to promote acute inflammatory lung injury. Infect Immun 82:2971–2979. Erratum in: Infect Immun (2014) Sep; 82(9): 3986. Author added. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder SA, Nagarkatti P, Nagarkatti M. (2012) Multiple anti-inflammatory pathways triggered by resveratrol lead to amelioration of staphylococcal enterotoxin B-induced lung injury. Br J Pharmacol 167:1244–1258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romano G, Acunzo M, Garofalo M, Di Leva G, Cascione L, Zanca C, Bolon B, Condorelli G, Croce CM. (2012) MiR-494 is regulated by ERK1/2 and modulates TRAIL-induced apoptosis in non-small-cell lung cancer through BIM down-regulation. Proc Natl Acad Sci USA 109:16570–16575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse M, Rao R, Nagarkatti M, Nagarkatti PS. (2014) 3,3′-diindolylmethane ameliorates experimental autoimmune encephalomyelitis by promoting cell cycle arrest and apoptosis in activated T cells through microRNA signaling pathways. J Pharmacol Exp Ther 350:341–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouse M, Singh NP, Nagarkatti PS, Nagarkatti M. (2013) Indoles mitigate the development of experimental autoimmune encephalomyelitis by induction of reciprocal differentiation of regulatory T cells and Th17 cells. Br J Pharmacol 169:1305–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Gajbhiye R, Mandal M, Pal C, Meyyapan A, Mukherjee J, Jaisankar P. (2013) Synthesis and antibacterial evaluation of 3,3′-diindolylmethane derivatives. Med Chem Res 23:1371–1377. [Google Scholar]
- Saeed AI, Rieder SA, Price RL, Barker J, Nagarkatti P, Nagarkatti M. (2012) Acute lung injury induced by Staphylococcal enterotoxin B: disruption of terminal vessels as a mechanism of induction of vascular leak. Microsc Microanal 18:445–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S, Dubaybo H, Ali S, Goncalves P, Kollepara SL, Sethi S, Philip PA, Li Y. (2013) Down-regulation of miR-221 inhibits proliferation of pancreatic cancer cells through up-regulation of PTEN, p27(kip1), p57(kip2), and PUMA. Am J Cancer Res 3:465–477. [PMC free article] [PubMed] [Google Scholar]
- Seth A, Stern LJ, Ottenhoff TH, Engel I, Owen MJ, Lamb JR, Klausner RD, Wiley DC. (1994) Binary and ternary complexes between T-cell receptor, class II MHC and superantigen in vitro. Nature 369:324–327. [DOI] [PubMed] [Google Scholar]
- Shorey LE, Hagman AM, Williams DE, Ho E, Dashwood RH, Benninghoff AD. (2012) 3,3′-Diindolylmethane induces G1 arrest and apoptosis in human acute T-cell lymphoblastic leukemia cells. PLoS One 7:e34975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Li N, Zhou B, Yang Z, Ding D, Weng D, Meng L, Wang S, Zhou J, Ma D, et al. (2013) miR-222 is upregulated in epithelial ovarian cancer and promotes cell proliferation by downregulating P27(kip1). Oncol Lett 6:507–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Zhou Y, Li H, Guo Y, Shan J, Xia M, Li Y, Li S, Long D, Feng L. (2013) Over-expression of microRNA-494 up-regulates hypoxia-inducible factor-1 alpha expression via PI3K/Akt pathway and protects against hypoxia-induced apoptosis. J Biomed Sci 20:100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H-B, Chen X, Ji H, Wu T, Lu H-W, Zhang Y, Li H, Li Y-M. (2014) miR‑494 is an independent prognostic factor and promotes cell migration and invasion in colorectal cancer by directly targeting PTEN. Int J Oncol 45:2486–2494. [DOI] [PubMed] [Google Scholar]
- Terasawa K, Ichimura A, Sato F, Shimizu K, Tsujimoto G. (2009) Sustained activation of ERK1/2 by NGF induces microRNA-221 and 222 in PC12 cells. FEBS J 276:3269–3276. [DOI] [PubMed] [Google Scholar]
- Tilahun AY, Holz M, Wu T-T, David CS, Rajagopalan G. (2011) Interferon gamma-dependent intestinal pathology contributes to the lethality in bacterial superantigen-induced toxic shock syndrome. PLoS One 6:e16764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar S, Nagarkatti M, Nagarkatti PS. (2014) 3,3′-Diindolylmethane attenuates LPS-mediated acute liver failure by upregulating miRNAs-106a and miRNA-20b that target IRAK4 to suppress toll-like receptor signaling. Br J Pharmacol 172:2133–2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vabulas R, Bittlingmaier R, Heeg K, Wagner H, Miethke T. (1996) Rapid clearance of the bacterial superantigen staphylococcal enterotoxin B in vivo. Infect Immun 64:4567–4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Yu BW, Rahman KMW, Ahmad F, Sarkar FH. (2008) Induction of growth arrest and apoptosis in human breast cancer cells by 3,3-diindolylmethane is associated with induction and nuclear localization of p27kip. Mol Cancer Ther 7:341–349. [DOI] [PubMed] [Google Scholar]
- Ware LB, Matthay MA. (2000) The acute respiratory distress syndrome. N Engl J Med 342:1334–1349. [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Tomita M, and Kanai A (2007) Computational methods for microRNA target prediction, in Methods in Enzymology (Hannon JJR and Jensen GJ eds) pp 65–86, Academic Press, New York. [DOI] [PubMed] [Google Scholar]
- Weng J-R, Bai L-Y, Chiu C-F, Wang Y-C, Tsai M-H. (2012) The dietary phytochemical 3,3′-diindolylmethane induces G2/M arrest and apoptosis in oral squamous cell carcinoma by modulating Akt-NF-κB, MAPK, and p53 signaling. Chem Biol Interact 195:224–230. [DOI] [PubMed] [Google Scholar]
- Wurz K, Garcia RL, Goff BA, Mitchell PS, Lee JH, Tewari M, Swisher EM. (2010) MiR-221 and MiR-222 alterations in sporadic ovarian carcinoma: Relationship to CDKN1B, CDKNIC and overall survival. Genes Chromosomes Cancer 49:577–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T, Liang S, Wang H, Hu S, Sun Y, Yu X, Han J, Li J, Guo S, Dai J, et al. (2014) Structural basis for the neutralization and specificity of Staphylococcal enterotoxin B against its MHC Class II binding site. MAbs 6:119–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamanaka S, Campbell NR, An F, Kuo SC, Potter JJ, Mezey E, Maitra A, Selaru FM. (2012) Coordinated effects of microRNA-494 induce G₂/M arrest in human cholangiocarcinoma. Cell Cycle 11:2729–2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C-Z, Zhang J-X, Zhang A-L, Shi Z-D, Han L, Jia Z-F, Yang W-D, Wang G-X, Jiang T, You Y-P, et al. (2010) MiR-221 and miR-222 target PUMA to induce cell survival in glioblastoma. Mol Cancer 9:229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R, Su B. (2009) Small but influential: the role of microRNAs on gene regulatory network and 3′UTR evolution. J Genet Genomics 36:1–6. [DOI] [PubMed] [Google Scholar]