Abstract
Disruption of steroidogenesis by environmental chemicals can result in altered hormone levels causing adverse reproductive and developmental effects. A high-throughput assay using H295R human adrenocortical carcinoma cells was used to evaluate the effect of 2060 chemical samples on steroidogenesis via high-performance liquid chromatography followed by tandem mass spectrometry quantification of 10 steroid hormones, including progestagens, glucocorticoids, androgens, and estrogens. The study employed a 3 stage screening strategy. The first stage established the maximum tolerated concentration (MTC; ≥ 70% viability) per sample. The second stage quantified changes in hormone levels at the MTC whereas the third stage performed concentration-response (CR) on a subset of samples. At all stages, cells were prestimulated with 10 µM forskolin for 48 h to induce steroidogenesis followed by chemical treatment for 48 h. Of the 2060 chemical samples evaluated, 524 samples were selected for 6-point CR screening, based in part on significantly altering at least 4 hormones at the MTC. CR screening identified 232 chemical samples with concentration-dependent effects on 17β-estradiol and/or testosterone, with 411 chemical samples showing an effect on at least one hormone across the steroidogenesis pathway. Clustering of the concentration-dependent chemical-mediated steroid hormone effects grouped chemical samples into 5 distinct profiles generally representing putative mechanisms of action, including CYP17A1 and HSD3B inhibition. A distinct pattern was observed between imidazole and triazole fungicides suggesting potentially distinct mechanisms of action. From a chemical testing and prioritization perspective, this assay platform provides a robust model for high-throughput screening of chemicals for effects on steroidogenesis.
Keywords: steroidogenesis, H295R cells, ToxCast, high-throughput screening.
Identifying potential endocrine disrupting compounds (EDCs) and understanding their mode of action is of high priority for chemical safety and regulatory agencies. The World Health Organization defines an endocrine disruptor as, “an exogenous substance or mixture that alters function(s) of the endocrine system and consequently causes adverse health effects in an intact organism or its progeny or (sub)populations” (WHO/UNEP, 2013). EDCs have been associated with adverse reproductive and developmental effects as well as increased risk of hormone-dependent cancers (Yeung et al., 2011).
Most EDC research has largely focused on chemical interaction with hormone nuclear receptors, mainly estrogen and androgen receptors, and cellular signaling pathways for these receptors. For example, the ToxCast high-throughput screening program includes 18 unique assays that evaluate chemical interactions across the estrogen signaling pathway focusing on estrogen receptor binding and activation as well as downstream effects on cell proliferation (Judson et al., 2015) . However, independent of receptor activity, dysregulation, or interference with steroid hormone biosynthesis and metabolism (ie, steroidogenesis) can alter steroid hormone levels contributing to endocrine disruption and ultimately result in impaired reproductive and sexual development (Sanderson, 2006; Whitehead and Rice, 2006; Ye et al., 2014). Altered hormone levels contributing to endocrine disruption is not unique to steroid hormones; for example, mechanisms other than alterations in receptor occupancy have been known in thyroid hormone-related endocrine disruption (Capen and Martin, 1989).
Given their key role in the formation of endogenous steroid hormones, enzymes involved in steroidogenesis are considered important targets for endocrine disrupting chemicals. However, the development of in vitro steroidogenesis assays has been a challenge because only adrenal and gonadal cells express appreciable cytochrome P450 side chain cleavage enzyme (CYP11A1), the initial enzyme in the steroid metabolism pathway required to commit cholesterol to a steroid hormone fate by forming pregnenolone (PREG) from cholesterol (Payne and Hales, 2004). CYP11A1 is expressed in the zona glomerulosa of the adrenal gland, Leydig cells of the testes, and theca cells of the ovary. Subsequent enzymes involved in steroidogenesis, including CYP17A1 and CYP19A1, are expressed in a variety of tissues including the liver, skin, and the nervous system (Payne and Hales, 2004). To date, no steroidogenically competent in vitro platform has been developed from ovarian cells. Several cell lines have been established from rodent Leydig cells, including R2C, mLTC, and BLTK1 cells for the evaluation of progestagen and androgen levels (Ascoli, 1981; Forgacs et al., 2012; Rebois, 1982; Shin et al., 1968). The only human-derived, steroidogenically competent cell line is the human adrenocortical H295R cells (Gracia et al., 2006). The EPA’s Endocrine Disruptor Screening Program (EDSP) has incorporated the OECD validated H295R-based in vitro steroidogenesis assay (Test Guideline 456, OECD, 2011) as part of the Tier 1 screening battery (USEPA, 2006). The current design of the assay measures concentration-dependent changes in 17β-estradiol and testosterone levels upon chemical treatment in a 24-well plate format (OECD, 2011). Given the need to screen large numbers of chemicals for potential endocrine disruption, and specifically for effects on steroidogenesis, the development of a high-throughput steroidogenesis assay to complement the high-throughput nuclear receptor assays is required.
In this study, we modified the existing OECD H295R in vitro steroidogenesis assay to perform high-throughput measurement of 13 hormones in 96-well format using a novel high-throughput HPLC-MS/MS (high-performance liquid chromatography followed by tandem mass spectrometry) method. This approach provides the ability to quantitatively assess changes in 4 hormone classes including glucocorticoids, progestagens, androgens, and estrogens. The assay was used to screen 2060 chemical samples in single concentration format. Those samples having significant effects on at least 4 hormones were subsequently evaluated in concentration-response (CR). Comprehensively evaluating a panel of hormones across the steroidogenesis pathway allowed for the identification of different putative mechanisms of action for chemicals. The data not only offered unique insights into the effects of a diverse library of chemicals on hormone biosynthesis and metabolism thereby complementing current EDC screening assays, but also demonstrate a novel use for H295R cells as a high-throughput and multiplexed screening platform for the disruption of steroidogenesis.
MATERIALS AND METHODS
Chemical library
The chemical library used for this study contained 2060 samples representing 1998 unique test chemicals (Supplementary Table 1). Chemicals were selected from multiple ToxCast chemical lists established based on solubility in DMSO, commercial availability, and affordability of the compound. The chemical inventory for this study included the ToxCast Phase I, Phase II, as well as a selection of Phase III chemicals. Phase I was largely composed of conventional pesticide actives whereas Phases II and III generally encompassed environmental chemicals of interest. In addition, the E1K chemical library, a set of roughly 800 chemicals enriched for endocrine active chemicals, was included. Information on the complete ToxCast chemical library is publicly available for download (http://www.epa.gov/ncct/dsstox/). A top nominal stock concentration of 100 mM was attempted, solubility permitting, for the entire library.
Cell culture and treatment
All cell culture and treatment procedures were carried out at CeeTox, Inc (Kalamazoo, Michigan). Cell culture conditions and media preparation were conducted in accordance with the OECD Test No. 456 guidelines (OECD, 2011), with minor modifications. Briefly, H295R cells (ATCC CRL-2128) were expanded for 5 passages and frozen in batches in liquid nitrogen. Prior to conducting steroidogenesis evaluation, batches of H295R cells were thawed and passed at least 4 times. The maximum passage number used for steroidogenesis evaluation was 10. Cells were maintained in a 1:1 mixture of Dulbecco’s Modified Eagle’s Medium with Ham’s F-12 Nutrient mixture (DMEM/F12) supplemented with 5 ml/l ITS+ Premix (BD Bioscience) and 12.5 ml/L Nu-Serum (BD Bioscience). After seeding cells at 50%–60% confluency into 96-well plates, an overnight acclimation period was allowed for cell adherence. Subsequently culture medium was replaced with 175 µl media containing 10 µM forskolin (FOR) to stimulate steroidogenesis for 48 h. Finally, the FOR stimulus media was replaced, this time test chemical was added to a final concentration of 0.1% DMSO. Duplicates were included for all chemical treatments as well as controls [10 µM FOR and 3 µM prochloraz (PRO)] in addition to 4 DMSO solvent controls on each 96-well plate. Following 48 h of chemical treatment, media was removed, split into 2 vials of approximately 75 µl media each, and stored at −80 °C prior to steroid hormone quantification.
MTT cytotoxicity assay
Cell viability was evaluated by MTT cytotoxicity assay after chemical treatment in all studies. Briefly, after chemical exposure and removal of media, 500 µl of 0.5 mg/ml 3-[4,5-dimethylthiazol-2-y]2,5-diphenyltetrazoliumbromide (MTT) solution was added to the cells remaining in the 96-well treatment plates. Following a 4 h incubation at 37 °C and 5% CO2 to allow formazan-MTT crystal formation, the MTT solution was removed and blue formazan salt crystals were solubilized using 500 µl anhydrous isopropanol with shaking for 20 min. Absorbance at 570 and 650 nm were measured using a Packard Fusion plate reader. Background correction of absorbance units was used to determine percent change relative to controls. All plates contained multiple control wells including 10 µM FOR (n = 4; control for stimulation of steroidogenesis), 3 µM PRO (n = 4; control for the inhibition of steroidogenesis), and digitonin (n = 4; control for cell death).
Initially, cytotoxicity was used to establish a maximum tolerated concentration (MTC) per chemical sample whereby the ToxCast chemicals were evaluated at a maximum nominal concentration of 100 µM, where possible. A target cell viability ≥ 70% was sought. Chemicals resulting in H295R cell viability of 20%–70% were diluted 10-fold, whereas those with < 20% viability were diluted 100-fold and re-evaluated. Dilutions were made until ≥ 70% viability was achieved for all chemicals establishing the MTC. The 70% cutoff criteria was established based on 5 times the baseline median absolute deviation (5 × BMAD), which uses the estimated baseline noise level of the assay to inform on significant changes in cell viability. For the MTT data analysis, BMAD was defined as the median absolute deviation (MAD) of all chemical samples evaluated during MTC determination, resulting in a BMAD of 6%; hence with a 5 × BMAD cutoff the allowable viability loss was set as 30%. MTT cytotoxicity evaluation was also conducted for the duplicates of all concentrations for chemicals tested in the CR studies (CR; 6-point CR established by 3-fold serial dilutions from the MTC). MTC and percent viability are summarized in Supplementary Table 1.
Quantification of steroid hormones
Media samples from treated cells were transported on dry ice to OpAns, LLC (Durham, North Carolina) where all subsequent extractions and quantification were performed. Briefly, samples were thawed to room temperature prior to liquid-liquid extraction. Steroid hormones were extracted from media samples using methyl tert-butyl ether (MTBE). An extra derivatization with dansyl chloride was included for estrogen (estrone and estradiol) detection only. Steroid hormones were separated and quantified using high-performance liquid chromatography followed by tandem mass spectrometry (HPLC-MS/MS). More specifically, reverse phase C18 gradient elution with electrospray positive ionization was used followed by MS/MS detection. All data were acquired using MassHunter Workstation Acquisition version B03.01 (Agilent Technologies, Inc), and processed using MassHunter Quantitative Analysis for QQQ.
To support accurate quantification of samples, the linear dynamic range was determined for each hormone analyte from 3 standards (Table 1). The lower limit of quantification (LLOQ) and upper limit of quantification (ULOQ) were established using a 7-point standard curve. Furthermore, precision and accuracy of the extraction and quantification methods were calculated as the percent relative standard deviation (%RSD) of the spiked standards and percent spiked standard recovered, respectively. The goal was to achieve 100% accuracy (ie, recover all spiked-in standard at quantification with minimal loss during run time) and good precision (ie, have %RSD < 15% to ensure reproducibility [Abdel-Khalik et al., 2013; USFDA, 2001]). The data collected in the study were within the specified margins, confirming high quality.
TABLE 1.
Steroid Hormones Evaluated: Limits of Quantification, Precision, and Accuracy of HPLC-MS/MS Measurement
| Steroid Hormone | Short Name | LLOQ (ng/ml) | ULOQ (ng/ml) | Standard | Standards (ng/ml) | Precisiona (%) | Accuracyb (%) | |
|---|---|---|---|---|---|---|---|---|
| Glucocorticoid | 11-Deoxycortisol | 11DCORT | 5 | 1000 | [2H11]-11DCORT | 20, 50, 800 | 5.0 | 101.7 |
| Deoxycorticosterone | DOC | 0.5 | 100 | [2H8]-DOC | 4, 10, 160 | 3.7 | 99.9 | |
| Cortisol | CORTISOL | 0.5 | 100 | [2H4]-CORTISOL | 20, 50, 800 | 3.3 | 99.7 | |
| Corticosterone | CORTICO | 0.5 | 100 | [2H8]-CORTICO | 20, 50, 800 | 4.7 | 100.5 | |
| Progestagen | 17α-Hydroxyprogesterone | OHPROG | 0.2 | 40 | [2H8]-OHPROG | 4, 10, 160 | 4.0 | 99.6 |
| 17α-Hydroxypregnenolone | OHPREG | 5 | 1000 | [2H3]-OHPREG | 20, 50, 800 | 6.7 | 100 | |
| Progesterone | PROG | 0.2 | 40 | [2H9]-PROG | 4, 10, 160 | 3.3 | 98.1 | |
| Pregnenolone | PREG | 2 | 400 | [2H4]-PREG | 20, 50, 800 | 10.0 | 100.9 | |
| Androgen | Dehydroepiandrosterone | DHEA | 3 | 600 | [2H5]-DHEA | 20, 50, 800 | 4.0 | 100.1 |
| Androstenedione | ANDR | 1 | 200 | [2H5]-ANDRO | 4, 10, 160 | 4.7 | 99.9 | |
| Testosterone | TESTO | 0.1 | 20 | [2H5]-TESTO | 0.4, 1, 16 | 5.7 | 100.7 | |
| Estrogen | Estrone | ESTRONE | 0.03 | 6 | [2H4]-ESTRO | 0.4, 1, 16 | 5.0 | 100.4 |
| Estradiol | ESTRADIOL | 0.03 | 6 | [2H5]-ESTRA | 0.4, 1, 16 | 6.3 | 101.4 |
ULOQ, upper limit of quantification
aPrecision was calculated as the %RSD of the standards.
bAccuracy was calculated as the percent standard recovered.
Data processing and analysis
All data were analyzed in R (R 3.1.1; R Foundation for Statistical Computing). In the single concentration MTC screen, samples were deemed to have a significant effect on hormone levels using a cutoff of ≥ |1.5-fold change| over DMSO controls on a per plate basis. Analysis of the CR data was performed using the ToxCast pipeline package in R (tcpl; http://epa.gov/ncct/toxcast/data.html). Briefly, all data were analyzed on a per sample basis for each hormone endpoint independently. Data were not adjusted for cytotoxicity, all MTT cytotoxicity assay results are available in Supplementary Tables 1 and 2. CR data were fit using 3 models: constant, hill, and gain-loss, the winning model was chosen as that having the lowest Akaike Information Criterion. Curve parameters from the winning model such as logAC50 (the log10 concentration in micromolar at which 50% of the maximum fold change was achieved; modl_ga) and the modeled top of the curve (modl_tp) were used to quantify potency and efficacy, respectively. A sample was identified as having a significant effect on a hormone if it was fit with one of the CR models and if the efficacy (ie, change in hormone levels) exceeded a specified threshold defined by a cutoff of ≥ 6 times the BMAD (the MAD of all normalized response values at the lowest 2 tested concentrations across all samples). All data are available through the ToxCast data download website (http://epa.gov/ncct/toxcast/data.html) and ToxCast Dashboard (http://actor.epa.gov/dashboard/). Supplementary files for all data are also included herein: stage II MTT results including the determined MTC for each chemical (Supplementary Table 1), stage III MTT results (Supplementary Table 2), stage II MTC screening raw hormone data (Supplementary Table 3), stage II MTC screening hit calls (Supplementary Table 4), stage III CR evaluation raw hormone data (Supplementary Table 5), and stage III CR evaluation hormone data curve fitting and tcpl outputs (Supplementary Table 6). Finally, all data processing R scripts are also available as Supplementary File S7.
Correlation analyses between MTC screening and CR results were conducted using Pearson’s linear correlation per hormone analyte using the median log2-fold change of results from MTC treatment. This was possible due to the MTC having been evaluated in both the MTC screening and CR stages of the study. Robust Z-prime (Z′) and strictly standardized mean difference (SSMD) values were calculated per hormone using the median and MAD of the control wells (DMSO, PRO, or FOR, respectively) raw response values included on all plates. The median Z′ and SSMD values from across all treated plates (from all stages with hormone quantification), were reported for PRO and FOR independently to demonstrate the robustness of the assay under generally stimulatory and inhibitory conditions. Clustering of the CR hormone profiles was conducted at the chemical-level using the modeled top (modl_tp) multiplied by the signal direction (ie, 1 for upregulation and −1 for downregulation). Chemical-level analyses, aggregating the 524 samples run in CR down to the 514 unique tested chemicals, were performed using the “tcpl” package’s “tcplSubsetChid” function. Figure generation for K-means clustering, hierarchical clustering, and heatmap generation were all conducted using the “gplots” package in R. All scripts developed to perform the analyses in the study are available as Supplemental file S7.
RESULTS
Assay Workflow and Summary
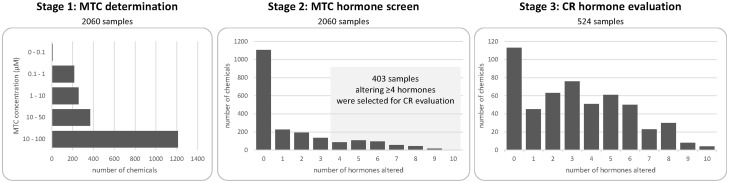
The high-throughput steroidogenesis assay involved 3 stages: I. establishing the MTC, II. Screening chemical samples at the MTC for effects on hormone levels, III. CR evaluation for select chemical samples (Figure 1). Initially, 2060 chemical samples were evaluated for cytotoxicity to establish an MTC (ie, a single concentration with > 70% cell viability). In total, 1203 chemical samples had an MTC of 100 µM, 812 chemical samples had MTC between 1 and 99 µM, and 45 chemical samples had MTC less than 0.9 µM. In the MTC screen, all chemical samples were tested at their MTC for effects on steroidogenesis by quantification of 13 hormones (Table 1). Three hormones, PREG, dehydroepiandrosterone, and corticosterone, consistently had data below the LLOQ and were not included in subsequent analyses. For MTC screening purposes, an absolute fold change cutoff of ≥ 1.5-fold (relative to DMSO controls on a per-plate basis) was used to identify chemical samples eliciting effects on hormone levels. In total, 955 chemical samples altered at least one hormone and 403 samples had effects on ≥ 4 hormones.
FIG. 1.
High-throughput H295R steroidogenesis assay workflow and summary. This study was conducted in 3 stages: I. Determination of an MTC (maximum concentration achieving ≥ 70% cell viability), II. Quantification of hormone levels upon MTC treatment, III. CR evaluation for selected chemicals. MTC concentrations were established for 2060 chemical samples, with the majority of samples having an MTC ≥ 10 µM, as shown in the stage 1 graph. All 2060 chemical samples were evaluated for MTC effects on hormone levels. Samples altering ≥ 4 hormones in the MTC screen (highlighted in the shaded region in the stage 2 graph), in addition to 121 randomly selected chemical samples that did not meet the selection criteria were included for CR evaluation. The final stage 3 graph illustrates sum of how many hormones showed a concentration-dependent response upon treatment among the 524 chemical samples included in the CR evaluation. CR, concentration-response; MTC, maximum tolerated concentration.
All 403 chemical samples identified using the criteria of ≥ 4 hormones altered at the MTC were included in the stage III CR evaluation. Additionally, 121 chemical samples not meeting this criteria were randomly chosen in order to evaluate the sensitivity of our selection criteria. As a result, 524 samples were evaluated in CR. Of the CR tested samples, a total of 411 chemical samples had concentration-dependent effects on at least 1 hormone analyte. From these 411, 347 chemical samples had been selected based on the CR selection criteria, resulting in a selection sensitivity or recall of 86% when using the CR selection criteria. In contrast, only 64 of the additional 121 chemical samples not meeting the CR selection criteria altered at least one hormone in stage III, a selection sensitivity of 53% (Supplementary Table 8 summarizes the selection sensitivity with different CR selection criteria). For each hormone, statistically derived cutoff criteria were calculated based on 6 times BMAD (BMAD values were in log2 fold-change) allowing for a data-driven cutoff that directly reflects the relative baseline noise level for each endpoint. The resulting cutoffs ranged from 1.5-fold (BMAD of 0.097) for 11DCORT to a cutoff of 2-fold (BMAD of 0.17) for both ESTRADIOL and TESTO (Table 2).
TABLE 2.
Assay Endpoint (hormone) Quality Statistics
| Hormone Short Name | Correlation of Maximum Fold Changea | Z′ (FOR)b | Z′ (PRO)c | SSMD (FOR)d | SSMD (PRO)e | BMADf | CR Cutoff (Fold-Change)g |
|---|---|---|---|---|---|---|---|
| 11DCORT | 0.82 | 0.76 | 0.88 | 15 | −27 | 0.097 | 1.50 |
| DOC | 0.45 | 0.65 | 0.51 | 10 | 7 | 0.154 | 1.90 |
| CORTISOL | 0.78 | 0.67 | 0.83 | 11 | −18 | 0.164 | 1.98 |
| PROG | 0.79 | 0 | 0.85 | 1 | 21 | 0.163 | 1.97 |
| OHPROG | 0.71 | 0.62 | 0 | 10 | −2 | 0.150 | 1.87 |
| OHPREG | 0.64 | 0.81 | 0.77 | 20 | 3 | 0.119 | 1.64 |
| ANDR | 0.81 | 0.68 | 0.84 | 11 | −19 | 0.127 | 1.70 |
| TESTO | 0.57 | 0.57 | 0.75 | 8 | −13 | 0.172 | 2.05 |
| ESTRONE | 0.65 | 0.79 | 0.76 | 18 | −14 | 0.135 | 1.75 |
| ESTRADIOL | 0.64 | 0.73 | 0.72 | 13 | −11 | 0.173 | 2.05 |
aCorrelation of log2 fold-change was calculated by comparing maximum response achieved by the MTC tested in the MTC screening versus CR evaluation for 524 samples. Values presented are r2 from Pearson correlations.
bMedian Z-prime across all tested plates using forskolin (FOR) as the control.
cMedian Z-prime across all tested plates using prochloraz (PRO) as the control.
dMedian strictly standardized median difference (SSMD) across all tested plates using FOR as the control.
eMedian SSMD across all tested plates using PRO as the control.
fBaseline median absolute deviation (BMAD; presented as log2 fold-change).
gCR cutoff shown as a fold-change equating to 6× BMAD.
Assay Performance Evaluation
Correlation analysis was conducted to evaluate the reproducibility of the log2 fold-change achieved at the MTC concentration between single concentration MTC screening and CR testing. Overall the squared Pearson’s correlation coefficient (r2) between the 2 phases of this study, which were conducted independently, was on average 0.70 across all measured hormones (Table 2) suggesting good reproducibility. As another measure of assay performance, Z′ and SSMD values were calculated using FOR and PRO as generally stimulatory and inhibitory controls, respectively (Table 2). Assay endpoints with Z′ values between 0.5 and 1 are considered to be excellent assays. Each measured hormone had a positive control with a Z′ ≥ 0.5. Furthermore, SSMD was used to demonstrate overall assay quality and directionality. FOR generally increased hormone quantities with excellent dynamic range based on SSMD values ≥ 7, whereas PRO generally inhibited hormone production with excellent dynamic range based on SSMD values ≤−7. Specifically, FOR stimulated androgen and estrogen hormone production while PRO inhibited the production of these hormones consistently and robustly.
Comparison to OECD Guideline Study
This study presents a high-throughput modification of an OECD-validated assay. Although high-throughput screening assays are not always amenable to traditional validation approaches, a performance-based approach was applied to help establish confidence in our identification of steroidogenesis disrupting chemicals. Among the 12 core chemicals included in the OECD H295R interlaboratory validation study (Hecker et al., 2011), the MTC screening dataset included 7 of the 12 core chemicals comprising FOR, PRO, atrazine, molinate, paraben, benomyl, and nonoxynol-9. The 5 core chemicals not included in our dataset were either not included in the ToxCast inventory or was a protein (human chorionic gonadotropin) and hence not in the scope of this study. MTC screening correctly identified the same effect as the OECD validation study with benomyl and nonoxynol-9 having no effect on any hormone and the others (FOR, PRO, atrazine, molinate, and paraben) eliciting increase/decrease in ESTRADIOL and/or TESTO. Furthermore, 4 of these chemicals (FOR, PRO, atrazine, and molinate) were also evaluated in the CR stage, confirming the desired concentration-dependent effects on ESTRADIOL and/or TESTO. In addition to correctly identifying the effects of the core chemicals on the 2 endpoints included in the OECD guideline assay, our study identified other hormones altered by these chemicals.
Profiling and Mechanism of Action Categorization
The nearly complete panel of hormones measured in this study provides a comprehensive evaluation of chemical effects on the steroid hormone biosynthesis and metabolism pathway. Distinguishing profiles of chemical-mediated changes in hormone levels can be useful for hypothesizing mechanisms of action for steroidogenesis disruption.
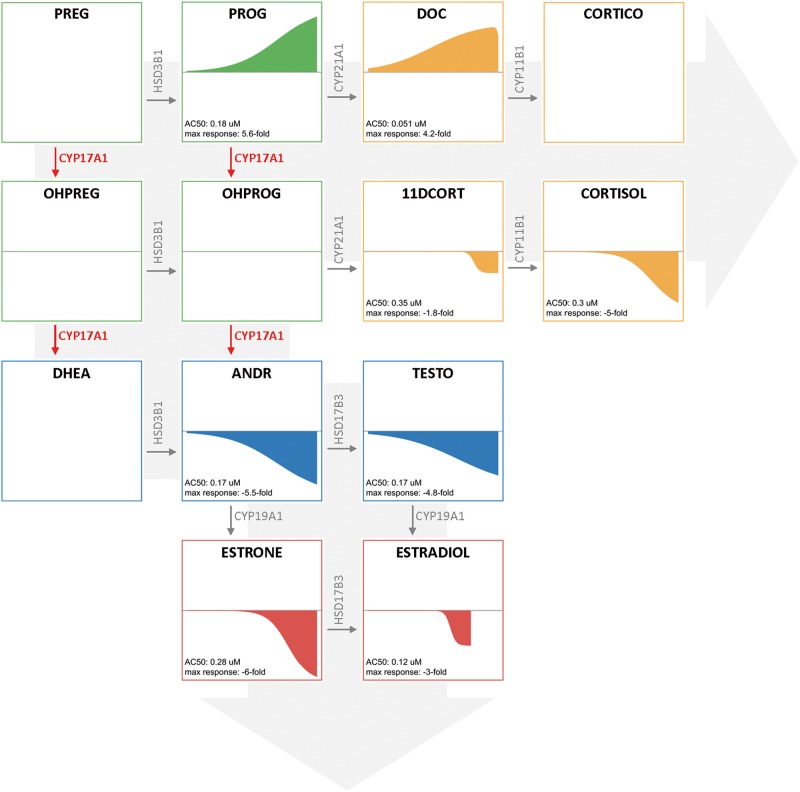
PRO, an imidazole fungicide, is the best characterized and most widely used reference chemical for steroidogenesis studies. PRO potently inhibits the hydroxylase activity of CYP17A1 resulting in the inhibition of androgen production (Blystone et al., 2007). The CYP17A1 enzyme is responsible for 2 steps in the steroidogenesis pathway catalyzing both a hydroxylation and 17, 20-lyase reaction. In our study, PRO significantly altered the levels of 8 of the 10 hormones evaluated (Figure 2). More specifically, the levels of PROG and DOC, which precede CYP17A1 hydroxylase activity were increased relative to DMSO controls, while all hormones produced as products of CYP17A1 activity were significantly decreased or unchanged. All 8 hormones that were altered by PRO had AC50 values over a tight range of 0.1–0.3 µM. The consistency of the potency and profile of altered hormone levels supports the reported CYP17A1 mechanism for PRO.
FIG. 2.
Evaluation of prochloraz-mediated effects on steroidogenesis. CR evaluation of hormones across the steroidogenesis pathway identified 8 hormones significantly altered by prochloraz (AC50 range 0.1–0.3 µM). The illustrations of concentration-response curves for each hormone upon prochloraz treatment are laid out reflecting the steroidogenesis pathway. Hormones produced prior to CYP17A1 (highlighted in red text) in the pathway, namely PROG and DOC, were elevated while all hormones whose production requires CYP17A1 activity were decreased relative to DMSO controls. The intermediate hydroxylated progestagens were not detected. Hormone classes are highlighted in unique colors: progestagens (green), glucocorticoids (yellow), androgens (blue), and estrogens (red). Plots with a single flat line indicate no significant change (OHPREG and OHPROG). Blank plots (PREG, DHEA, and CORTICO) indicate a hormone that was omitted due to data below the LLOQ. LLOQ, lower limit of quantification
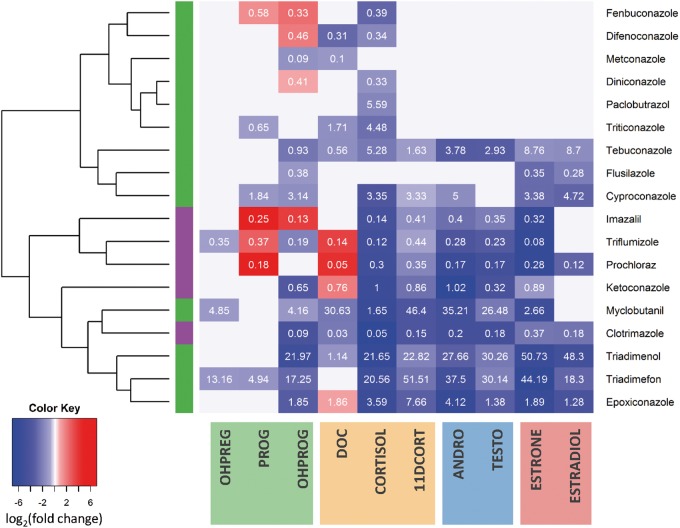
The current study included 18 conazoles evaluated in stage III CR, expanding upon the previous knowledge on the effects of conazoles on steroidogenesis. Previous studies have only investigated select conazoles for effects on steroidogenesis, namely PRO, ketoconazole, and imazalil (Ulleras et al., 2008). The evaluation of these conazole fungicides effects on steroidogenesis reveals that the 2 structurally distinguishable conazole classes, imidazole and triazole, can be approximately differentiated from one another based on their distinct profile of effects on steroidogenesis (Figure 3). With the exception of clotrimazole, the imidazoles cluster together due to increases in PROG and/or DOC in addition to strong decreases in COTRISOL, 11DCORT, ANDR, TESTO, ESTRONE, and ESTRADIOL. The triazole fungicides showed more diverse behavior with a generally conserved decrease in CORTISOL levels. Furthermore, most conazoles demonstrate consistent AC50 values across all hormones they concentration-dependently altered supporting the hypothesis that they have a single target mediating their primary effects on steroidogenesis.
FIG. 3.
Profiling conazole fungicides effects on steroidogenesis. Heatmap visualizing the effect of 18 conazoles across the 10 hormones. The heatmap was generated to visualize the maximum fold change (in log2) achieved from concentration-dependent increase (red) or decrease (blue) in hormone levels with hierarchical clustering conducted using Euclidean distance metric to sort the conazoles. The AC50 concentration (µM) for the effect is overlaid in white print. A bar on the left identifies imidazole (purple) versus triazole (green) conazoles. Hormones are grouped and highlighted based by progestagen (green), glucocorticoid (yellow), androgen (blue), and estrogen (red) across the bottom. In total, 20 conazoles were evaluated in CR, with the 18 depicted conazoles having concentration-dependent effects on at least one hormone.
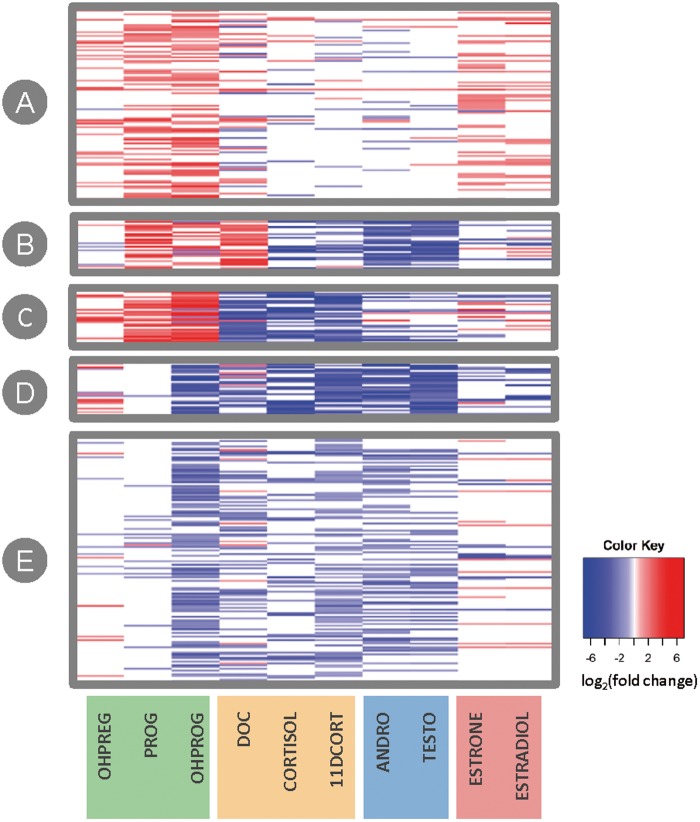
To investigate the profiles of steroidogenic disruption K-means clustering was used to identify 5 distinct profiles (Figure 4; Supplementary Table 9 summarizes the identity of chemicals per cluster). For this analysis, the 411 chemical samples that elicited concentration-dependent effects on ≥ 1 hormone in stage III CR evaluation were mapped to 401 unique chemicals which were used as input for clustering. Cluster A is composed of 137 chemicals eliciting overall increases in hormone levels, most strongly driven by increases in PROG, OHPROG, ESTRONE, and ESTRADIOL. This first profile may be a result of chemical-mediated stimulation of steroidogenesis or chemical-mediated disruption of hormone metabolism/turnover. For example, FOR, a chemical known to increase steroidogenesis across all hormones, can be found in Cluster A. Additionally, atrazine, a core chemical that the OECD validation study expected to weakly induce estrogen levels as a result of aromatase induction in vitro, was also in cluster A due to concentration-dependent increases in ESTRADIOL. Cluster B contains 35 chemicals with PRO-like profiles constituting marked increases in PROG and DOC levels and decreases among androgens, the remaining glucocorticoids, and estrogens suggesting putative CYP17A1 disruption. Cluster C is composed of 50 chemicals with increased progestagen levels (OHPREG, OHPROG, and PROG) with simultaneous decreases across the rest of the hormones evaluated, which may result from HSD3B1 disruption. Finally, clusters D and E showed overall decreases in hormone levels with both clusters having consistent decreases in glucocorticoid and androgen levels. The clusters are distinguished from one another by estrogen responses. Cluster D, composed of 36 chemicals, consistently decreased estrogens suggesting overall inhibition of the steroidogenic pathway (ie, putative CYP11A1 inhibition, or chemical-mediated effects upstream of steroidogenesis). In contrast, the 143 chemicals in cluster E have no change or increased estrogen levels overall.
FIG. 4.
Profiling chemical effects on steroidogenesis. The 401 unique chemicals altering at least one hormone in CR (mapped from 411 chemical samples) were clustered using K-means to identify unique profiles of steroidogenesis disruption. The heatmap visualizes the maximum achieved log2 fold change (calculated using the top of the modeled response curve) from concentration-dependent increase (red) or decrease (blue) in hormone level ordered according to the 5 distinct K-means clusters. Each horizontal line within the heatmap (y-axis) represents one chemical. Hormone classes are highlighted in unique colors along the bottom x-axis: progestagens (green), glucocorticoids (yellow), androgens (blue), and estrogens (red).
DISCUSSION
In vitro evaluation of chemical-mediated disruption of estrogenic and androgenic activity has historically been focused on nuclear receptor interactions. However, it is paramount that effects on steroidogenesis are evaluated because changes in hormone levels can also result in adverse outcomes, independent of nuclear receptor binding. More specifically, disruption of steroidogenesis can result in reproductive and developmental toxicity including nipple retention, decreases in anogenital distance, and fertility (Sanderson, 2006; Whitehead and Rice, 2006; Ye et al., 2014). In males, chemical-mediated decreases in androgen levels as a result of HSD3B inhibition, CYP17A1 inhibition, or modulation of CYP19A1 activity have been associated with testicular dysgenesis syndrome and adverse reproductive effects (Edwards et al., 2006; Sharpe, 2006; Skakkebaek et al., 2001). For example, atrazine is an herbicide known to disrupt steroidogenesis and was one of the core chemicals in the OECD interlaboratory validation studies eliciting concentration-dependent effects on both TESTO and ESTRADIOL. Atrazine-mediated adverse reproductive effects in rodents have been attributed to the disruption of steroidogenesis in light of the fact that it does not interact with nuclear receptors (Connor et al., 1996; Forgacs et al., 2013; Roberge et al., 2004; Trentacoste et al., 2001), which was also confirmed in ToxCast assays where it does not result in any effects in AR or ER assays. Similarly, the fungicide PRO has in vivo anti-androgenic effects across species including fish, rodents, and frogs as a result of CYP17A1 inhibition which disrupts steroidogenesis leading to adverse effects on sexual development (Ankley and Gray, 2013; Blystone et al., 2007; Vinggaard et al., 2005).
Despite the well-characterized effects of a few prototype chemicals, very few others have been comprehensively evaluated for effects on steroidogenesis, much less characterized for mechanisms of action underlying these effects. The conazoles fungicides PRO, ketoconazole, and imazalil have been previously investigated and characterized as CYP17A1 inhibitors confirming results from the profiling approach herein (Blystone et al., 2007; Loose et al., 1983; Ohlsson et al., 2010; Ulleras et al., 2008); however, this study is the first to expand beyond small prototypical chemical sets and comprehensively evaluate the effects of a large library of structurally diverse chemicals across the steroid hormone biosynthetic pathway. The chemical inventory studied herein included multiple libraries from the ToxCast high-throughput screening program and was not a random selection of chemicals, but rather it was enriched for potentially endocrine-active chemicals. The OECD validated H295R steroidogenesis assay (OECD Test Guideline 456) was adapted to high-throughput and a novel HPLC-MS/MS approach was used to successfully quantify 10 hormone analytes. Efforts to apply metabolomics and/or quantify a larger panel of hormones using mass spectrometry in H295R cells have been attempted in the past (Abdel-Khalik et al., 2013; Jeanneret et al., 2015; Kang et al., 2013; Nielsen et al., 2012; Rijk et al., 2012; Ulleras et al., 2008); however, these studies also deviated from the OECD assay guideline and the current study is the first to apply this approach successfully on such a large scale. Our methodology was robust with high Z′ and SSMD values and good reproducibility across the MTC and CR stages of the study. The use of HPLC-MS/MS for quantification of the steroid hormones improved accuracy, throughput, and allowed lower detection limits compared to the traditional ELISA or RIA previously used in hormone studies. Furthermore, despite adaptation to high-throughput, inclusion of a FOR prestimulus, and use of a novel quantification method, our study design was able to correctly identify the chemical effects on steroidogenesis when benchmarking against the OECD guideline study. Notably, the high-throughput method herein not only correctly identified anticipated effects on ESTRADIOL and/or TESTO, but also provided insight on other hormone endpoints across the steroidogenesis pathway. It is important to note that the current study design was modified from the OECD guideline: all test samples were evaluated in duplicate rather than triplicate; 70% cell viability was accepted instead of 80%; FOR prestimulus was included to enable robust dynamic responses in both up- and downregulation of hormone levels; and concentration-dependent effects were evaluated using the ToxCast Data Analysis Pipeline rather than the OECD guideline recommended Dunnett’s Test to identify significant effects in consecutive chemical concentration groups. However, the added value of having quantified effects on 10 hormones rather than simply ESTRADIOL and/or TESTO allowed for the profiling of chemical-elicited disruption of steroidogenesis to help in hypothesis generation for putative mechanisms of action. Yet the capacity to use this assay for chemical safety decisions warrants further evaluation and study, including the detailed evaluation of selection criteria, in vivo relevance especially as it pertains to the profile of hormone changes, and integration of other relevant assays (eg, aromatase inhibition assays).
The 3 stage approach sought to establish an MTC to minimize any confounding effects of cytotoxicity, while selection of chemicals from stage II to stage III aimed to hone in on identifying chemical-mediated effects on steroidogenesis. The number of chemicals identified as having effects on hormone levels is not altogether surprising as the chemical inventory was selected in part based on potential endocrine-activity, and our selection criteria were intended to be inclusive for screening purposes to minimize false negatives. Additionally, the current screening effort profiles a larger portion of the steroidogenic pathway than ever before (ie. 10 hormones quantified rather than ESTRADIOL and TESTO only), inherently resulting in a larger number of chemicals having any effect on hormone levels as additional mechanisms of action can be detected. However, it is also important to note that chemicals not selected for CR evaluation should not be considered as inactive for disruption of steroidogenesis as the selection process was based on a combination of activity in MTC screen and overall testing efficiency. Given the number of hormones measured, the reproducibility across studies, and the distinct profiles observed, our results suggest that it is unlikely that chemicals not selected for the CR stage have a significant effect on steroidogenesis. Yet refining the chemical selection criteria, which herein simply required effects on ≥ 4 hormones in the MTC screening, may identify chemicals that should be tested in CR. Additional follow-up studies are required to investigate the potential for highly targeted hormone effects (eg, chemicals altering estrogen levels exclusively), which may have an impact on future chemical selection for CR follow-up.
The quantification of hormones across the steroidogenesis pathway in CR across a diversity of chemicals enabled a comprehensive profiling of chemical effects that can support hypothesis generation for putative mechanisms of steroidogenesis perturbation. More specifically, the evaluation of conazole fungicides revealed distinguishable profiles of hormone effects for triazole versus imidazole conazoles illustrating the influence of chemical structure on bioactivity. Furthermore, the profiling results highlight a multitude of possible targets that can result in different effects on hormone levels evident in 401 out of the 514 chemicals that elicited concentration-dependent effects on at least one hormone. For example, increased progestagen levels in coordination with decreased androgen and estrogen levels were observed with CYP17A1 inhibition by PRO. Other observed profiles included the decrease of androgen levels with concurrent increases in estrogens suggesting increased aromatase (CYP19A1) activity or decreased estrogen turnover by sulfotransferases. Future studies will seek to characterize the profiles identified, to screen additional chemicals in CR, and to develop in silico predictive models to identify chemicals targeting steroidogenesis.
The H295R high-throughput screening assay presented herein fills a critical gap in the ability to evaluate large chemical libraries for effects on steroidogenesis in vitro. This assay also complements the existing EPA EDSP Tier 1 testing battery which has included the evaluation of < 100 chemicals effects on steroidogenesis using 2 in vitro assays: the OECD validated H295R assay only evaluating TESTO and ESTRADIOL levels (ie, OCSPP 890.1550) and the CYP19A1 inhibition assay (ie, OCSPP 890.1200). The novel approach outlined in this study to quantify a majority of hormones in the steroidogenesis pathway in high-throughput provides insight into the unique profiles elicited by different mechanisms of action aiding hypothesis generation, and ultimately serving as the basis for models that can support chemical testing prioritization.
FUNDING
A.L.K. and D.L.F. were supported by appointments to the Research Participation Program of the U.S. Environmental Protection Agency, Office of Research and Development, administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. EPA.
Supplementary Material
ACKNOWLEDGMENTS
The authors would like to thank Dr Richard Judson, Sine Rosenberg, Dr Kevin Crofton, and Dr Russell Thomas for the critical reading of the manuscript, as well as the ToxCast scientists for many helpful discussions.
SUPPLEMENTARY DATA
Supplementary data are available online at http://toxsci.oxfordjournals.org/.
REFERENCES
- Abdel-Khalik J., Bjorklund E., Hansen M. (2013). Development of a solid phase extraction method for the simultaneous determination of steroid hormones in H295R cell line using liquid chromatography-tandem mass spectrometry. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 935, 61–69. [DOI] [PubMed] [Google Scholar]
- Ankley G. T., Gray L. E. (2013). Cross-species conservation of endocrine pathways: A critical analysis of tier 1 fish and rat screening assays with 12 model chemicals. Environ. Toxicol. Chem. 32, 1084–1087. [DOI] [PubMed] [Google Scholar]
- Ascoli M. (1981). Regulation of gonadotropin receptors and gonadotropin responses in a clonal strain of Leydig tumor cells by epidermal growth factor. J. Biol. Chem. 256, 179–183. [PubMed] [Google Scholar]
- Blystone C. R., Lambright C. S., Howdeshell K. L., Furr J., Sternberg R.M., Butterworth B. C., Durhan E. J., Makynenl E. A., Ankley G. T., Wilson V. S., et al. (2007). Sensitivity of fetal rat testicular steroidogenesis to maternal prochloraz exposure and the underlying mechanism of inhibition. Toxicol. Sci. 97, 512–519. [DOI] [PubMed] [Google Scholar]
- Capen C. C., Martin S. L. (1989). The effects of xenobiotics on the structure and function of thyroid follicular and C-cells. Toxicol. Pathol. 17, 266–293. [DOI] [PubMed] [Google Scholar]
- Connor K., Howell J., Chen I., Liu H., Berhane K., Sciarretta C., Safe S., Zacharewski T. (1996). Failure of chloro-S-triazine-derived compounds to induce estrogen receptor-mediated responses in vivo and in vitro. Fundam. Appl. Toxicol. 30, 93–101. [PubMed] [Google Scholar]
- Edwards T. M., Moore B. C., Guillette L. J., Jr. (2006). Reproductive dysgenesis in wildlife: A comparative view. Int. J. Androl. 29, 109–121. [DOI] [PubMed] [Google Scholar]
- Forgacs A. L., Ding Q., Jaremba R. G., Huhtaniemi I. T., Rahman N. A., Zacharewski T. R. (2012). BLTK1 murine Leydig cells: A novel steroidogenic model for evaluating the effects of reproductive and developmental toxicants. Toxicol. Sci. 127, 391–402. [DOI] [PubMed] [Google Scholar]
- Forgacs A. L., D'Souza M. L., Huhtaniemi I. T., Rahman N. A., Zacharewski T. R. (2013). Triazine herbicides and their chlorometabolites alter steroidogenesis in BLTK1 murine leydig cells. Toxicol. Sci.134155–167. [DOI] [PubMed] [Google Scholar]
- Gracia T., Hilscherova K., Jones P. D., et al. (2006). The H295R system for evaluation of endocrine-disrupting effects. Ecotoxicol. Environ. Saf. 65, 293–305. [DOI] [PubMed] [Google Scholar]
- Hecker M. Hollert H. Cooper R. Vinggaard A.M.,Akahori Y.,Murphy M.,Nellemann C.,Higley E.,Newsted J.,Laskey J.,. et al. , (2011). The OECD validation program of the H295R steroidogenesis assay: Phase 3. Final inter-laboratory validation study. Environ. Sci. Pollut. Res. Int. 18, 503–515. [DOI] [PubMed] [Google Scholar]
- Jeanneret F., Tonoli D., Rossier M. F., Saugy M., Boccard J., Rudaz S. (2015). Evaluation of steroidomics by liquid chromatography hyphenated to mass spectrometry as a powerful analytical strategy for measuring human steroid perturbations. J. Chromatogr. A 1430,97–112. [DOI] [PubMed] [Google Scholar]
- Judson R. S., Magpantay F. M., Chickarmane V., Haskell C., Tania N., Taylor J., Xia M., Huang R.,, Rotroff D. M., Filer D. L.et al. (2015).Integrated Model of Chemical Perturbations of a Biological Pathway Using 18 In Vitro High-Throughput Screening Assays for the Estrogen Receptor.Toxicol. Sci.148,137–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang S., Park S., Kim M. J., Oh S. M., Chung K. H., Lee S. (2013). A sensitive and selective LC-MS/MS analysis coupled with an online sample enrichment technique for H295R steroidogenesis assay and its application in the investigation of the effect of sildenafil on steroidogenesis. Anal. Bioanal. Chem. 405, 9489–9496. [DOI] [PubMed] [Google Scholar]
- Loose D. S., Kan P. B., Hirst M. A., Marcus R. A., Feldman D. (1983). Ketoconazole blocks adrenal steroidogenesis by inhibiting cytochrome P450-dependent enzymes. J. Clin. Invest. 71, 1495–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen F. K., Hansen C. H., Fey J. A., Hansen M., Jacobsen N. W., Halling-Sorensen B., Bjorklund E., Styrishave B., et al. (2012). H295R cells as a model for steroidogenic disruption: A broader perspective using simultaneous chemical analysis of 7 key steroid hormones. Toxicol. in vitro 26, 343–350. [DOI] [PubMed] [Google Scholar]
- OECD. (2011). Test No. 456: H295R Steroidogenesis Assay. OECD Publishing, Paris. [Google Scholar]
- Ohlsson A., Cedergreen N., Oskarsson A., Ulleras E. (2010). Mixture effects of imidazole fungicides on cortisol and aldosterone secretion in human adrenocortical H295R cells. Toxicology 275, 21–28. [DOI] [PubMed] [Google Scholar]
- Payne A. H., Hales D. B. (2004). Overview of steroidogenic enzymes in the pathway from cholesterol to active steroid hormones. Endocr. Rev. 25, 947–970. [DOI] [PubMed] [Google Scholar]
- Rebois R. V. (1982). Establishment of gonadotropin-responsive murine leydig tumor cell line. J. Cell Biol. 94, 70–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rijk J. C., Peijnenburg A. A., Blokland M. H., Lommen A., Hoogenboom R. L., Bovee T. F. (2012). Screening for modulatory effects on steroidogenesis using the human H295R adrenocortical cell line: A metabolomics approach. Chem. Res. Toxicol. 25, 1720–1731. [DOI] [PubMed] [Google Scholar]
- Roberge M., Hakk H., Larsen G. (2004). Atrazine is a competitive inhibitor of phosphodiesterase but does not affect the estrogen receptor. Toxicol. Lett. 154, 61–68. [DOI] [PubMed] [Google Scholar]
- Sanderson J. T. (2006). The steroid hormone biosynthesis pathway as a target for endocrine-disrupting chemicals. Toxicol. Sci. 94, 3–21. [DOI] [PubMed] [Google Scholar]
- Sharpe R. M. (2006). Pathways of endocrine disruption during male sexual differentiation and masculinization. Best Pract. Res. Clin. Endocrinol. Metab. 20, 91–110. [DOI] [PubMed] [Google Scholar]
- Shin S. I., Yasumura Y., Sato G. H. (1968). Studies on interstitial cells in tissue culture. II. Steroid biosynthesis by a clonal line of rat testicular interstitial cells. Endocrinology 82, 614–616. [DOI] [PubMed] [Google Scholar]
- Skakkebaek N. E., Rajpert-De Meyts E., Main K. M. (2001). Testicular dysgenesis syndrome: An increasingly common developmental disorder with environmental aspects. Hum. Reprod. 16, 972–978. [DOI] [PubMed] [Google Scholar]
- Trentacoste S. V., Friedmann A. S., Youker R. T., Breckenridge C. B., Zirkin B. R. (2001). Atrazine effects on testosterone levels and androgen-dependent reproductive organs in peripubertal male rats. J. Androl. 22, 142–148. [PubMed] [Google Scholar]
- Ulleras E., Ohlsson A., Oskarsson A. (2008). Secretion of cortisol and aldosterone as a vulnerable target for adrenal endocrine disruption–screening of 30 selected chemicals in the human H295R cell model. J. Appl. Toxicol. 28, 1045–1053. [DOI] [PubMed] [Google Scholar]
- USEPA. (2006). Decision documents for atrazine: Finalization of interim registration eligibility decision and completion of tolerance reassessment and reregistration eligibility process. In: EPA US, ed. Office of Prevention, Pesticides and Toxic Substances, Washington D.C. EPA Report Number: EPA-HQ-OPP-2005-0481
- USFDA. (2013). Guidance for industry: Bioanalytical method validation. In: US FDA, ed. Department of Health and Human Services, Center for Drug Evaluation and Research (CDER) , Center for Veterinary Medicine (CVM). Silver Spring, MD., eds.
- Vinggaard A. M., Christiansen S., Laier P., et al. (2005). Perinatal exposure to the fungicide prochloraz feminizes the male rat offspring. Toxicol. Sci. 85, 886–897. [DOI] [PubMed] [Google Scholar]
- Whitehead S. A., Rice S. (2006). Endocrine-disrupting chemicals as modulators of sex steroid synthesis. Best Pract. Res. Clin. Endocrinol. Metab. 20, 45–61. [DOI] [PubMed] [Google Scholar]
- WHO/UNEP (2013). State of the science of endocrine disrupting chemicals. (A. Bergman, J. J. Heindel, S. Jobling, K. A. Kidd, R. T. Zoeller, Eds.). ISBN: 978 92 4 150503 1, WHO Press, Geneva, Switzerland.
- Ye L., Guo J., Ge R. S. (2014). Environmental pollutants and hydroxysteroid dehydrogenases. Vitam. Horm. 94, 349–390. [DOI] [PubMed] [Google Scholar]
- Yeung B. H., Wan H. T., Law A. Y., Wong C. K. (2011). Endocrine disrupting chemicals: Multiple effects on testicular signaling and spermatogenesis. Spermatogenesis 1, 231–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.