Defects in enzymes involved in glycogen metabolism result in glycogen storage diseases (GSDs), which may affect the skeletal and sometimes also the cardiac muscle. The most frequent abnormality causing GSDs is glycogen storage, whereas other and uncommon forms of GSD are due to a perturbation of the branching structure of glycogen. These latter GSDs are characterized by an accumulation of polyglucosan (PG),1 an abnormal polysaccharide with few branched points and excessively long peripheral chains. PG is accumulated in PG bodies that can be easily identified in muscle by their typical features using histopathologic (strong periodic acid–Schiff [PAS] reaction, resistance to diastase digestion) and ultrastructural analyses.
Among the disorders in which PG storage is almost exclusively limited to striated muscle, those with an identified genetic cause include glycogenosis type IV (GBE1 gene), glycogenosis type VII (PFKM gene), and a new group of diseases termed polyglucosan body myopathies (PGBMs).1
PGBM type 1 (PGBM1) is due to mutations in the RBCK1 gene encoding a ubiquitin ligase, and it is clinically characterized by an early-onset myopathy and cardiomyopathy, with or without immunodeficiency.2,3 Recently, PGBM type 2 (PGBM2) has been recognized to be due to mutations in the GYG1 gene4 encoding glycogenin 1, the same defective enzyme that causes GSD type XV, in which cardiac involvement is also present.5
Glycogenin 1 is the muscle-specific isoform of a glycosyltransferase that catalyzes 2 autoglycosylation reactions using uridine diphosphoglucose as the donor substrate to start glycogen synthesis; the oligosaccharide chain so synthesized is then elongated by glycogen synthase and branched by branching enzyme.
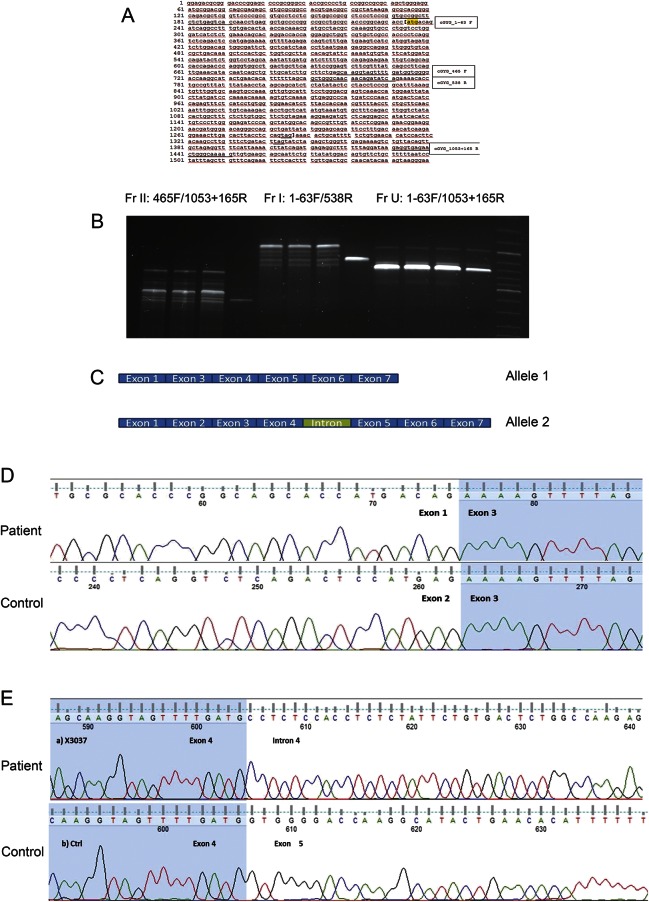
We have previously reported an Italian family6 including 3 affected relatives (2 siblings, 1 cousin) who presented a late-onset PGBM without cardiac involvement of unknown genetic origin (GBE1, PFKM, and RBCK1 gene sequences were normal). Following identification of the GYG1 gene as a possible cause of PGBM,4 we performed targeted reanalysis of whole-exome sequencing and MotorPlex7 data. This resulted in the identification of a single nucleotide substitution at the donor splice site in intron 2 of the GYG1 gene (c.143+3G>C, p.Asp3Glufs*4), either in homozygous (in the 2 siblings) or in compound heterozygous state (in the cousin), confirming the diagnosis of PGBM2. In the cousin, the second mutant allele was identified by cDNA analysis, which showed the inclusion of a cryptic exon, resulting from a deep intronic base change. Genomic sequencing revealed a novel variant (chr 3: g.148.717.967C>G) that enhances a cryptic 5′ splice site in intron 4 (figure).
Figure. Genetic characterization of the novel GYG1 gene mutation.
(A) GYG1_cDNA sequence and position of primers used. (B) Product amplified by PCR on the GYG1 coding sequence using 3 different primers pairs (from left to right: Fr II [465F/1053+165R], Fr I [1-63F/538R], and Fr U [1-63F/1053+165R]) in the sample of a patient (the cousin) and control. FR II corresponds to the 3′ half of the gene (from exon 5 to exon 7), Fr I to the 5′ region of GYG1 cDNA (from exon 1 to exon 4), and Fr U to the whole cDNA (from exon 1 to exon 7). At 3 different annealing temperatures (60°C, 62°C, and 64°C), the patient's sample showed a number of bands of different sizes. These PCR products are all longer than the control band, suggesting the insertion of a cryptic exon. To verify this hypothesis, each fragment was isolated and cloned (TA cloning kit; Invitrogen Corporation, Carlsbad, CA). (C) Sequencing of each clone showed that there are 2 different cDNAs, encoded by either allele's cDNA: allele 1 shows the skipping of exon 2, while allele 2 shows the insertion of an intronic fragment. (D) Sequence analysis of the critical region corresponding to allele 1 shows that in the patient's sample, exon 2 is skipped. (E) Sequence analysis of the critical region corresponding to allele 2 shows that in the patient's sample, there is an insertion between exon 4 and exon 5. By BLAST search, we mapped this sequence in intron 4 (chr 3: 148.717.897–148.717.963). A set of primers was designed to amplify the intronic region on genomic DNA to characterize the sequence of this intron retention. The sequence analysis shows a single nucleotide variant (chr 3: g.148.717.967C>G) that enhances the 5′ splice site of this cryptic exon; the variant is novel and also absent in the EXAC database. To confirm that this cryptic exon is not a secondary extra product of splicing, we designed a PCR protocol to amplify allele 2 alone, with the forward primer mapped to the intronic sequence: no band was detected in the control sample, while a strong band was present in the patient's sample.
The c.143G>C homozygous mutation has been identified in other patients with PGBM2,4 in whom it was associated with some residual amount of glycogenin-1 protein. These data might explain the late-onset myopathic phenotype observed in such patients. Indeed, in our patient series, the onset of symptoms occurred later than 50 years of age, with weakness involving both limb-girdle muscles, and causing waddling gait and Gowers sign. The disease was invariably progressive, leading to inability to raise arms and walk unassisted approximately 10 years after onset in the 2 siblings.6
In this family with PGBM2 and GYG1 gene mutations, investigation of the pathogenetic events causing PG accumulation has suggested a role of ubiquitin-proteasomal and autophagic degradation pathways6: the accumulated PGs are likely to be insufficiently degraded by the ubiquitin-proteasomal system and may cause induction and impairment of the autophagic flux, as documented by increased LC3-II and p62/SQSTM1 accumulation.
Our report suggests that deficiency of glycogenin 1 should be considered and investigated as a possible genetic cause of PGBM.
Footnotes
Author contributions: Dr. Fanin designed the study, collected, analyzed, and interpreted the data, and drafted the manuscript. Drs. Torella and Savarese collected, analyzed, and interpreted the data. Drs. Nigro and Angelini revised the manuscript for intellectual content.
Study funding: The authors acknowledge the financial support of research grants from the Comitato Telethon Fondazione Onlus (grants GTB12001 and GUP13013 to C.A. and grants TGM11Z06, GUP10006, and GUP11006 to V.N.).
Disclosure: Dr. Fanin, Dr. Torella, Dr. Savarese, and Dr. Nigro report no disclosures relevant to this manuscript. Dr. Angelini has served on scientific advisory boards for and the editorial boards of Neuromuscular Disorders, Neurological Sciences, Current Opinion in Neurology, Neurology, and Therapeutical Advances in Neurological Disorders; has received funding for travel and/or speaker honoraria from Genzyme; and has received research support from Genzyme, Telethon, and the Association Française contre les Myopathies. Go to Neurology.org/ng for full disclosure forms. The Article Processing Charge was paid by the authors.
References
- 1.Oldfors A, DiMauro S. New insights in the field of muscle glycogenosis. Curr Opin Neurol 2013;26:544–553. [DOI] [PubMed] [Google Scholar]
- 2.Boisson B, Laplantine E, Prando C, et al. Immunodeficiency, autoinflammation and amylopectinosis in humans with inherited HOIL-1 and LUBAC deficiency. Nat Immunol 2012;13:1178–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nilsson J, Schoser B, Laforet P, et al. Polyglucosan body myopathy caused by defective ubiquitin ligase RBCK1. Ann Neurol 2013;74:914–919. [DOI] [PubMed] [Google Scholar]
- 4.Malfatti E, Nilsson J, Hedberg-Oldfors C, et al. A new muscle glycogen storage disease associated with glycogenin-1 deficiency. Ann Neurol 2014;76:891–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moslemi AR, Lindberg C, Nilsson J, Tajsharghi H, Andersson B, Oldfors A. Glycogenin-1 deficiency and inactivated priming of glycogen synthesis. N Engl J Med 2010;362:1203–1210. [DOI] [PubMed] [Google Scholar]
- 6.Fanin M, Nascimbeni AC, Savarese M, et al. Familial polyglucosan body myopathy with unusual phenotype. Neuropathol Appl Neurobiol 2015;41:385–390. [DOI] [PubMed] [Google Scholar]
- 7.Savarese M, Di Fruscio G, Mutarelli M, et al. MotorPlex provides accurate variant detection across large muscle genes both in single myopathic patients and in pools of DNA samples. Acta Neuropathol Commun 2014;2:100. [DOI] [PMC free article] [PubMed] [Google Scholar]