Abstract
Calcineurin is a calcium/calmodulin dependent protein phosphatase in eukaryotes that consists of a catalytic subunit A and a regulatory subunit B. Previous studies in the filamentous fungus Neurospora crassa had suggested that the catalytic subunit of calcineurin might be an essential protein. We generated N. crassa strains expressing the A (cna-1) and B (cnb-1) subunit genes under the regulation of Ptcu-1, a copper-responsive promoter. In these strains, addition of bathocuproinedisulfonic acid (BCS), a copper chelator, results in induction of cna-1 and cnb-1, while excess Cu2+ represses gene expression. Through analysis of these strains under repressing and inducing conditions, we found that the calcineurin is required for normal growth, asexual development and female fertility in N. crassa. Moreover, we isolated and analyzed cnb-1 mutant alleles generated by repeat-induced point mutation (RIP), with the results further supporting roles for calcineurin in growth and fertility in N. crassa. We demonstrated a direct interaction between the CNA-1 and CNB-1 proteins using an assay system developed to study protein-protein interactions in N. crassa.
Introduction
Calcineurin is the only serine/threonine protein phosphatase that depends on calcium/calmodulin (Ca2+/CaM) [1, 2]. The heterodimeric calcineurin protein consists of a catalytic subunit (A) that binds to calcium sensor CaM, and a regulatory subunit (B) that contains four Ca2+-binding domains [1–4]. CaM binds to the regulatory domain of the catalytic subunit in response to the cytosolic Ca2+concentration, and stimulates phosphatase activity that converts signals to various outputs by dephosphorylating target proteins [5, 6]. The functions of calcineurin have been studied in various organisms, including mammals, invertebrates, plants, parasites, and fungi [4]. The well-studied targets of calcineurin are the transcription factor Nuclear Factor of Activated T cells (NFAT) in mammals and the calcineurin responsive zinc-finger 1 (Crz1) in fungi, which upon dephosphorylation by activated calcineurin, are localized to the nucleus to regulate expression of target genes [7].
In mammals, the NFAT transcription factors are essential for several processes during organogenesis, including endocrine, immune, nervous, respiratory, skeletal and vascular systems [8, 9]. Defective calcineurin pathways have been linked to diseases such as cancer, cardiac hypertrophy, diabetes, Down’s syndrome and other degenerative brain diseases in humans [10]. Calcineurin is essential for the development of Drosophila melanogaster, in that flies with a mutation in the calcineurin B2 gene die at the late larval or early pupal stage [11]. In the worm Caenorhabidits elegans, calcineurin controls sensitivity to odorants, and may play a role in either motor neuron or muscle development [12]. In plants, the calcineurin B-like proteins (CBLs) are calcium sensors that regulate the activity of the CBL-interacting protein kinase (CIPK) and CBL-CIPK pathways function in signaling processes including environmental stress responses, nutrient sensing and adaptation [13]. In the malaria parasite Plasmodium falciparum, the chaperone heat shock protein 90 (Hsp90) associates with calcineurin, in a process that is necessary for erythrocytic replication of the parasite in the host [14].
In fungi, calcineurin has been mainly studied in the budding yeast Saccharomyces cerevisiae and certain fungal pathogens. In S. cerevisiae, the catalytic subunit is encoded by the CNA1/CMP1 and CNA2/CMP2 genes, while the regulatory subunit is encoded by the CNB1 gene; all three calcineurin subunits are individually or collectively non-essential for growth [15–17]. Calcineurin in S. cerevisiae is involved in the pheromone response and tolerance to high ion stress [15–18]. In Candida albicans, calcineurin subunits CNA and CNB are not essential, but play roles in cation homeostasis, and are essential for survival during membrane stress, in serum-containing media, and for virulence in a mouse model of systemic infection [19, 20]. In the opportunistic human fungal pathogen Cryptococcus neoformans, CNA1 and CNB1 are essential for fungal virulence and for growth at 37°C [21, 22]. In addition, calcineurin also regulates hyphal elongation and mating in C. neoformans [23]. In the rice blast fungus, Magnaporthe oryzae, the calcineurin catalytic subunit MCNA and the CNB-like genes play a role in infection-related differentiation and pathogenicity [24, 25]. In Mucor circinelloides, the causative zygomycete of human mucormycosis, there are three catalytic A subunit genes, cnaA, cnaB, and cnaC, and one regulatory gene, cnbR [26]. The calcineurin pathway plays a key role in dimorphic transitions and virulence in M. circinelloides [26]. In Ustilago maydis, a dimorphic basidiomycete that causes corn smut disease, the ucn1 catalytic subunit regulates morphogenesis and pathogenesis [27].
In the filamentous fungus Neurospora crassa, the calcineurin catalytic subunit cna-1 has been suggested to be an essential gene, and knock-down experiments using cna-1 antisense RNA revealed its requirement for normal hyphal branching, growth, and maintenance of the apical tip-high Ca2+ gradient [28]. In addition, analysis of insertional and repeat-induced point mutation (RIP) [29] mutants of the calcineurin regulatory subunit B, cnb-1, suggested requirements for normal vegetative growth in N. crassa [30]. Moreover, it has been demonstrated that CNB-1 binds to the calcineurin-dependent response element (CDRE) in the copper-induced metallothionein (CuMT) gene, suggesting a putative role for calcineurin in the regulation of CuMT in N. crassa [31]. Since homokaryotic knockout mutants of the essential calcineurin catalytic subunit A are not available, and no experiments have investigated its function using temperature-sensitive alleles or regulatable promoters, information regarding detailed functions of calcineurin in N. crassa have remained elusive.
In N. crassa, the tcu-1 gene encodes a high affinity copper transporter and its expression is precisely controlled by copper availability [32]. In addition, the kinetics of induction and repression of heterologous genes under the tcu-1 promoter (Ptcu-1) are rapid and stable [33]. We therefore used this system to investigate functions of cna-1 and cnb-1 by generating N. crassa strains with these genes under the control of Ptcu-1. We found that calcineurin is required for normal growth, asexual development, and sexual fertility in N. crassa. These findings were supported by results from analysis of cnb-1 mutant alleles generated by RIP. In addition, we demonstrated a direct interaction between the CNA-1 and CNB-1 proteins using an assay system developed to study protein-protein interactions in N. crassa [34].
Materials and Methods
Media, Growth Conditions, and Transformation Procedure
Neurospora crassa wild type strains 74-OR23-IVA and ORS-SL6a and all other strains were either obtained from the Fungal Genetics Stock Center (FGSC, Manhattan, KS) or generated in our laboratory (Table 1). Strains were cultured on Vogel’s minimal medium (VM) [35] to support vegetative growth, while sexual development was induced using synthetic crossing medium (SCM) [36]. Conidia used for inoculating cultures were propagated in 13x100 mm glass tubes containing VM with 1% agar, and grown for 3 days at 30°C in the dark and for 4 days at 25°C in the light [37, 38]. The growth of N. crassa strains was measured essentially as described previously [39–41]. Sorbose-containing medium (FGS) was used for ascospore germination and isolation of transformant colonies [42]. VM with proline as the nitrogen source was used for selection with Ignite [43]. When indicated, the growth medium was supplemented with pantothenate (Product number P2250; Sigma-Aldrich, St. Louis, MO), hygromycin (Calbiochem; San Diego, CA), Ignite (extracted from Finale; Farnam Companies INC.; Phoenix, AZ) [44, 45], or Nourseothricin (clonNAT; WERNER BioAgents; Germany) at a concentration of 10, 220, 400, and 50 μg/ml, respectively. Stocks of pantothenate and inositol were prepared and used as described previously [46; http://www.fgsc.net/methods/stanford.html]. The standard VM contains 50 mM CuSO4; where indicated, bathocuproinedisulfonic acid (BCS; Catalog number 164060010, Acros Organics, Geel, Belgium) and copper II sulfate (CuSO4; Catalog number 830521, Chempure, Texas, USA) were added to VM at the indicated concentrations. All sequencing was performed at the Genomics Core, Institute for Integrative Genome Biology, UC Riverside. E. coli strain DH5α [47] was the recipient for plasmid transformations. Transformation of N. crassa strains was performed using an Eppendorf Electroporator 2510 at a setting of 2000 volts as described previously [48].
Table 1. Neurospora crassa strains used in this study.
| Strain | Genotype | Reference |
|---|---|---|
| 74-OR23-IVA | Wild type; mat A | FGSC 2489 |
| ORS-SL6 a | Wild type; mat a | FGSC 4200 |
| 2074 | Δcnb-1::hph; mat a (heterokaryon) | FGSC 2074 |
| 2173 | Δcna-1::hph; mat a (heterokaryon) | FGSC 2173 |
| 51-4-1 | Δrid-1::nat; Δmus-51::nat; mat a | aOuyang et al., submitted |
| T-51-4-1 (28) | Δrid-1::nat; Δmus-51::nat; ∆pan-2::Ptcu-1::cnb-1::v5::gfp; mat a (heterokaryon) | This study |
| 52-4-9 | Δmus-52::nat; mat A | FGSC 2479 |
| T-52-4-9 (41) | Δmus-52::nat; ∆cna-1::hph; ∆pan-2::Ptcu-1::cna-1::V5::gfp; mat A (heterokaryon) | This study |
| T-52-4-9 (632) | Δmus-52::nat; ∆cna-1::hph; ∆inl:: Pccg-1::cna-1::S-tag::rfp; mat A (heterokaryon) | This study |
| T-52-4-9 (679) | Δmus-52::nat; ∆cnb-1::hph; ∆pan-2::Pccg-1::cnb-1::V5::gfp; mat A (heterokaryon) | This study |
| 2994 | ∆pan-2::Pccg-1::V5::gfp; mat a | aOuyang et al., submitted |
| 2995 | ∆inl:: Pccg-1::S-tag::rfp; mat a | aOuyang et al., submitted |
| 540 | ∆cna-1::hph; ∆pan-2::Ptcu-1::cna-1::V5::gfp; mat a | This study |
| 554 | ∆pan-2::Ptcu-1::cnb-1::V5::gfp; mat A | This study |
| 555 | ∆pan-2::Ptcu-1::cnb-1::V5::gfp; mat A | This study |
| 597 | ∆cnb-1::hph; ∆pan-2::Ptcu-1::cnb-1::V5::gfp; mat a | This study |
| 599 | ∆cnb-1::hph; ∆pan-2::Ptcu-1::cnb-1RIP::V5::gfp; mat A | This study |
| 600 | ∆cnb-1::hph; ∆pan-2:: Ptcu-1::cnb-1 RIP::V5::gfp; mat A | This study |
| 602 | ∆cnb-1::hph; ∆pan-2::Ptcu-1::cnb-1RIP::V5::gfp; mat A | This study |
| 727 | ∆cnb-1::hph; ∆pan-2::Pccg-1::cnb-1::V5::gfp; mat a | This study |
| 767 | ∆cna-1::hph; ∆inl::Pccg-1::cna-1::S-tag::rfp; mat a | This study |
| CNB-1GFP+CNA-1RFP#5 | Heterokaryon of strain 727 + strain 767 | This study |
| GFP+RFP#7 | Heterokaryon of strain 2994 + strain 2995 | This study |
| CNB-1GFP+RFP#18 | Heterokaryon of strain 727 + strain 2995 | This study |
a Shouqiang Ouyang, Ilva E. Cabrera, Asharie J. Campbell and Katherine A. Borkovich, submitted.
Construction of Strains Expressing Tagged Versions of cna-1 or cnb-1
∆cna-1::hph; ∆pan-2::Ptcu-1::cna-1::V5::gfp; mat a (strain 540)
The 2018 bp open reading frame (ORF) of calcineurin catalytic subunit A (NCU03804) was PCR-amplified using the primer pairs CNA-1-FOR and CNA-1-REV-GFP (Table 2), and gel purified. The pan-2 (NCU10048.7) locus targeting vector pRS426PVGTCU1.5 (Ptcu-1::5xGly::V5::gfp) [34] was linearized by restriction digestion with PacI and SmaI (New England Biolabs, Ipswich, MA) and gel-purified. Approximately 100 ng each of the gel-purified linearized vector and the PCR product were joined in the yeast strain FY834 [49] using yeast recombinational cloning [50]. DNA from the transformed yeast was isolated using the yeast smash and grab method [51] and transformed into Escherichia coli DH5α ultra-competent cells [47]. One clone, pRTCNA-1(18), was confirmed by sequencing. The pRTCNA-1(18) vector was transformed into N. crassa strain 52-4-9 (Table 1). The initial transformants were selected on FGS plates supplemented with Ignite and pantothenate. A transformant strain, T-52-4-9 (41) (Table 1), was isolated and crossed with a heterokaryotic Δcna-1::hph; mat a strain (2173; Table 1). Ascospores from this cross were germinated on FGS medium supplemented with pantothenate and BCS and then screened for resistance to hygromycin and Ignite, and pantothenate auxotrophy.
Table 2. Primers used in this study.
| Primer | Sequence (5’→3’) |
|---|---|
| aCNA-1-FOR | CGCACACACATCCCCAACCAACCATGGAAAGCAACAATGGTACCGGCGC |
| aCNA-1-REV-GFP | GTTAGGGATAGGCTTTCCGCCGCCTCCGCCCGATCGCTTGCGGTCACTCAAC |
| aPAN2-PTCU1-0.5-FW | CCTTGCGTATATTCTGGACCGGTACACGGAACATCTCGTGAACAAGAAGG |
| CNA-1-5F | CTCTGAAAGAGGGCCTTGCC |
| b5HPHR | ATCCACTTAACGTTACTGAAATC |
| aCNB-1-FOR | CGCACACACATCCCCAACCAACCATGGGCAACACCACCAGCTCCGTCC |
| aCNB-1-REV-GFP | GTTAGGGATAGGCTTTCCGCCGCCTCCGCCGAATTGATCTGTTAAAGCGTCGACTGTC |
| CNB-1-5F | CACCACTTCCTCTCCATGTC |
| aCNA-1-pCCG-RFP-FW | CCACTTTCACAACCCCTCACATCAACCAAAATGGAAAGCAACAATGGTACCGGCGC |
| aCNA-1-pCCG-RFP-RV | AGCAGCGGTTTCTTTTCCGCCGCCTCCGCCCGATCGCTTGCGGTCACTCAAC |
| aCNB-1-pCCG-GFP-FW | CCACTTTCACAACCCCTCACATCAACCAAAATGGGCAACACCACCAGCTCCGTCC |
| PCCG-1-SEQ-FW | CCATCATCAGCCAACAAAGC |
| RFP-RV | TAGGGAGGTCGCAGTATCTG |
| GFP-RV | AACTCCAGCAGGACCATGTG |
a Sequences are from Ouyang, Cabrera, Campbell and Borkovich (submitted). See Materials and Methods for details.
b Ref. [39]
We isolated a progeny with genotype ∆cna-1::hph; ∆pan-2::Ptcu-1::cna-1::v5::gfp; mat a (540; Table 1) that showed resistance to hygromycin, Ignite, and requirements for pantothenate and BCS. We verified the presence of the cna-1 allele at the pan-2 locus by PCR amplification with primers PAN2-PTCU1-0.5-FW and CNA-1-REV-GFP (Table 2), and further confirmed by sequencing. In addition, the ∆cna-1::hph allele in strain 540 was verified by PCR using primers CNA-5F and 5HPHR (Table 2).
∆cnb-1::hph; ∆pan-2::Ptcu-1::cnb-1::V5::gfp; mat a (strains 597, 599, 600, and 602)
The 1178 bp fragment of calcineurin regulatory subunit B (NCU03833) was PCR-amplified using the primer pairs CNB-1-FOR and CNB-1-REV-GFP (Table 2) and assembled with the pan-2 locus targeting vector pRS426PVGTCU1.5 in yeast as described above. One clone, pRTCNB-1 (28), was confirmed by sequencing. The pRTCNB-1 (28) construct was transformed into the N. crassa strain 51-4-1 (Table 1). One transformant, T-51-4-1 (28), was first crossed with wild type mat A strain (FGSC 2489) and two homokaryotic progeny with the genotype ∆pan-2::Ptcu-1::cnb-1::v5::gfp; mat A (strains 554 and 555; Table 1) were isolated.
In order to isolate strains with the cnb-1 gene under control of the tcu-1 promoter in an otherwise Δcnb-1::hph background, the 554 and 555 strains were crossed with the Δcnb-1::hph; mat a strain (2074; Table 1). Ascospores from these crosses were germinated on medium supplemented with pantothenate and BCS and screened for resistance to hygromycin and Ignite, and pantothenate auxotrophy. We isolated three progeny (597, 599 and 600) from the cross of strain 554 to strain 2074 and one progeny (602) from the cross of strain 555 to strain 2074 of genotype ∆cnb-1::hph; ∆pan-2::Ptcu-1::cnb-1::v5::gfp; mat a (Table 1). The presence of the cna-1 allele at the pan-2 locus was verified by PCR amplification and further confirmed by sequencing (Genomics core, UCR). In addition, the ∆cnb-1::hph allele in strain 597 was verified by PCR using primers CNB-1-5F and 5HPHR (Table 2).
∆cna-1::hph; ∆inl:: Pccg-1::cna-1::S-tag::rfp; mat a (767) and ∆cnb-1::hph;∆pan-2::Pccg-1::cnb-1::V5::gfp; mat a (727) strains
The 2018 bp ORF of calcineurin subunit A (NCU03804) and the 1178 bp ORF of calcineurin subunit B (NCU03833) were PCR-amplified using the primer pairs CNA-1-pCCG-RFP-FW and CNA-1-pCCG-RFP-RV, and CNB-1-pCCG-GFP-FW and CNB-1-REV-GFP, respectively (Table 2). PCR products for the calcineurin subunits A and B were assembled, respectively, in the inl locus (NCU06666.7) targeting vector pRS426ISR (Pccg-1::5xGly::S-tag::rfp) and pan-2 locus targeting vector pRS426PVG (Pccg-1::5xGly::V5::gfp) [34] that were linearized by restriction digestion with PacI and SmaI (NEB). These fragments were assembled in yeast as described above, and pRTCNA-1 (6) and pRTCNB-1 (3) were isolated and confirmed by sequencing. The pRTCNA-1(6) and pRTCNB-1(3) constructs were transformed into N. crassa strain 52-4-9 (Table 1). The initial transformants were selected on VM supplemented with Ignite and pantothenate. Strains T-52-4-9 (632) and T-52-4-9 (679), transformed with the pRTCNA-1(6) and pRTCNB-1(3), respectively, were isolated and crossed with the heterokaryotic strains ∆cna-1::hph; mat a (2173) and ∆cnb-1::hph; mat a (2074), respectively. From these crosses, we isolated homokaryotic strains ∆cna-1::hph; ∆pan-2::Pccg-1::cna-1::S-tag::rfp; mat a (767) and ∆cnb-1::hph; ∆pan-2::Pccg-1::cnb-1::V5::gfp; mat a (727) (Table 1). The cna-1 and cnb-1 transgenes in the two strains were verified by PCR amplification with primer PCCG-1-SEQ-FW in combination with RFP-RV and GFP-RV, respectively (Table 2). In addition, the two strains (767 and 727) were resistant to hygromycin B and the presence of the ∆cna-1::hph and the ∆cnb-1::hph allele in the respective strains was confirmed, as described above for strains 540 and 597, respectively.
Western Blot and Co-Immunoprecipitation Analysis
Strains were grown in liquid VM for 16 h as described in the figure legends and whole cell extracts were isolated essentially as described previously [52, 53]. Briefly, mycelia from these cultures were collected on a filter using vacuum, and then pulverized to a powder using a mortar and pestle in liquid nitrogen. Extraction buffer [50 mM HEPES (pH 7.7), 2 mM EGTA (pH 8.0), 2 mM EDTA (pH 8.0), 1% SDS, 10% glycerol, 100 mM NaCl, 1 mM Na3VO4 and 1 mM NaF] was added to the mycelial powder, and the samples were heated at 85°C for 5 min, after which 10 μl of 100 mM PMSF and 1 μl of fungal protease inhibitor cocktail (Product number P8215; Sigma-Aldrich, St. Louis, MO) were added. The crude protein extracts from these samples were isolated by centrifugation at 4000xg for 15 min at 4°C, and protein quantification was performed on the supernatants using the Bradford method (Protein Assay Dye Reagent Concentrate, Catalog number 500–0006; Bio-Rad, Hercules, CA). Samples containing 50 μg of protein were separated using a 10% SDS-PAGE gel and transferred onto a nitrocellulose membrane [54]. Membranes were probed using anti-GFP antibody at a dilution of (1:5000) (Catalog number A6455; Life Technologies; Carlsbad, CA) or anti S-tag antibody at a dilution of 1:3000 (Catalog number A190-135A; Bethyl Laboratories, Montgomery, TX) antibodies [54]. A horseradish peroxidase secondary antibody was used at a dilution of 1:2000 and bands were visualized by chemiluminescent detection as previously described [54]. In addition, after detecting via Western blotting, membranes were stained with an amido black solution (0.1% amido black from the Sigma-Aldrich Product number N3393, 10% acetic acid, 25% isopropanol) to show all proteins as an indication of protein transfer.
For co-immunoprecipitation analysis, forced heterokaryons (Table 1) were made by co-inoculating the individual strains in flasks containing VM agar and incubating at 30°C in the dark for 3 days and at room-temperature for four days under constant light. Conidia were isolated as described above and then inoculated in flasks containing 500 ml liquid VM at a concentration of 1x106 conidia/ml. Flasks were incubated at 30°C for 16 h with shaking at 200 rpm. Cells were isolated by filtration through sterile shop towels and then ground using a mortar and pestle with liquid nitrogen. The powdered mycelia were transferred to a bead-beater (Biospec Products, Bartlesville, OK) containing glass beads and extraction buffer [50 mM Tris-Cl (pH 7.5), 1 mM EDTA, 6 mM MgCl2, 2.5 mM PMSF and 0.1% (v/v) of fungal protease inhibitor cocktail (Product number P8215; Sigma-Aldrich, St. Louis, MO)]. Cells were broken using three pulses of one minute each, with one minute intervals in between. The mixture was centrifuged at 16,000xg for 15 minutes at 4°C. The supernatant containing the whole cell extract was carefully transferred into a fresh tube and protein quantified using the Bradford assay (Bio-Rad, Hercules, CA). Then, an aliquot of 3 μl GFP-Trap agarose beads (Product numbergta-20; ChromoTek; GmbH, Germany) was taken in a microcentrifuge tube and beads were equilibrated by washing once with 1 ml of ice-cold solutions of 1X Tris-buffered saline with Tween-20 (TBST) [51] and twice with 1 ml of ice-cold extraction buffer by centrifugation at 500xg for 1 min at 4°C. An aliquot of the sample containing 10 mg of protein extract was then added tothe microcentrifuge tube containing the equilibrated GFP-Trap agarose beads and incubated on a rotating shaker at 4°C overnight. The beads with bound protein were collected by centrifugation at 500xg for 1 min at 4°C and then resuspended in 500 μl of ice-cold extraction buffer, followed by centrifugation at 500xg for 1 min at 4°C. This step was repeated once more. The bound protein was liberated by adding 30 μl of 5x Laemmli SDS-PAGE sample buffer [54] and heating at 95°C for 5 minutes, followed by centrifugation at 500xg for 5 min at 4°C. The supernatants were removed, separated using 10% SDS-PAGE and analyzed by immunoblot using anti-GFP or anti-S tag antibodies as described above.
Results
Expression of cna-1 and cnb-1 under Control of the tcu-1 Promoter
Previous work in N. crassa has suggested that the catalytic subunit of calcineurin, cna-1, is an essential gene [28], while the regulatory subunit, cnb-1, is not [30]. In order to address the putative essential nature of cna-1, we produced strains that express cna-1 under the control of a regulatable promoter. We included cnb-1 in these studies as a nonessential gene that would serve as a control for the method. We took advantage of a recently developed system using the copper-regulated tcu-1 promoter [33] in N. crassa, inserting the constructs at the pan-2 locus [34]. The tcu-1 promoter is expressed in limiting copper II, which can be imposed through addition of a copper chelator such as BCS to the growth medium [33].
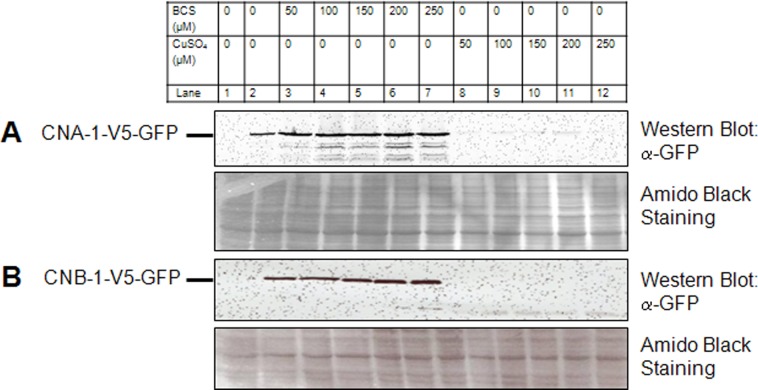
To verify that expression of the CNA-1 and CNB-1 proteins was regulated by copper availability, the strains were grown in minimal medium (VM) supplemented with various amounts of BCS and/or copper sulfate and protein expression was examined using Western analysis. We included a wild type strain lacking the GFP-tagged constructs for CNA-1 and CNB-1 as a negative control for the presence of the two proteins (Fig 1). The CNA-1 and CNB-1 proteins were abundantly expressed in the medium supplemented with BCS, but not detectable in the presence of copper sulfate (Fig 1). These observations suggest that expression of the CNA-1 and CNB-1 proteins is controlled by copper availability in the medium, similar to results demonstrated for other heterologous proteins in the previous study [33].
Fig 1. Copper-regulated expression of CNA-1 and CNB-1 under control of the tcu-1 promoter.
The effect of BCS or copper sulfate on Ptcu-1 driven expression of CNA-1 from the ∆cna-1::hph; ∆pan-2::Ptcu-1::cna-1::v5::gfp; mat a (540) strain (A), and CNB-1 from the ∆cnb-1::hph; ∆pan-2::Ptcu-1::cnb-1::v5::gfp; mat a (597) strain (B) was determined (lanes 2–12) after growth of strains in liquid medium under the indicated conditions for 22 h. Protein isolated from the wild type strain (FGSC 2489) was used as control (lane 1). Protein extracts were prepared and samples containing 50 μg of total protein were analyzed by Western blot using rabbit anti-GFP antibody as indicated in the Materials and Methods. The solid lines indicate positions of CNA-1::V5::GFP (~93 kDa) and CNB-1::V5::GFP (~48 kDa) proteins. The amido black staining of the membrane, shown in the lower panel, was done to demonstrate equal protein loading.
Repression of CNA-1 and CNB-1 Results in Severe Defects in Hyphal Growth and Aerial Hyphae Development
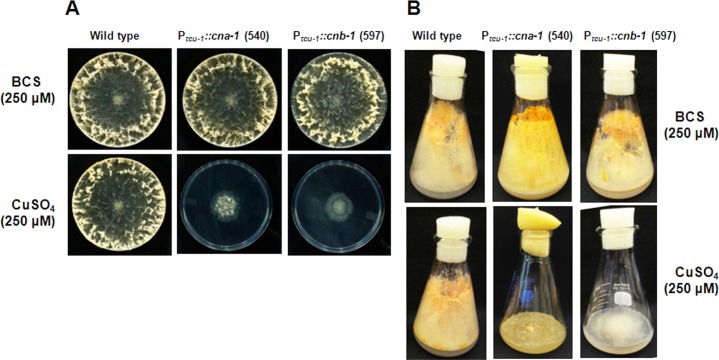
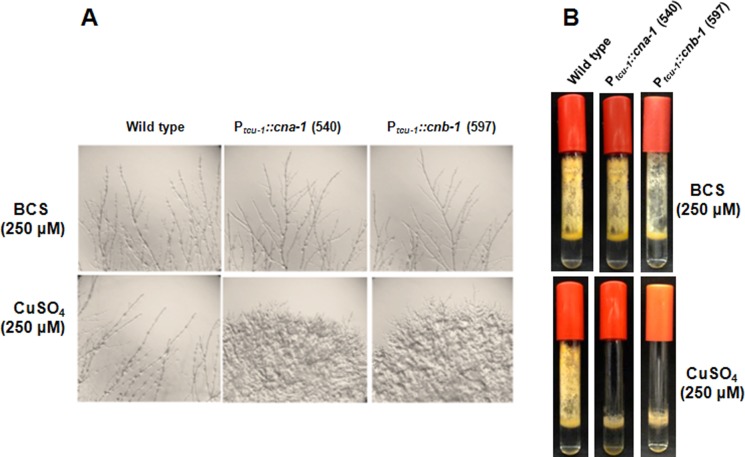
To test the requirement for CNA-1 and CNB-1 during growth and development of N. crassa, we determined the effect of copper availability on morphology of the strains with cna-1 or cnb-1 under the control of the tcu-1 promoter (Figs 2 and 3). The 540 and 597 strains exhibited normal growth on agar medium supplemented with BCS; however, severe impairment was observed on the medium supplemented with excess copper sulfate (Fig 2). The average colony diameter of the two strains was reduced ~74% in the medium supplemented with excess BCS vs. copper sulfate (Table 3). Furthermore, consecutive transfers of the strains on the repressing medium continued to show severe growth impairment, but growth was reverted back to normal after inoculation of the strains on medium containing BCS (data not shown). Therefore, the growth impairment of strains 540 and 597 by excess copper sulfate was reversible. In addition, the hyphal growth of 540 and 597 was abnormal, with bulged hyphal tips produced in the presence of excess copper sulfate (Fig 3A). Moreover, aerial hyphae production in the 540 and 597 strains was greatly reduced in the repressing high copper medium (Fig 3B). These results indicate thatCNA-1 and CNB-1 play an important role in regulating vegetative growth and hyphal development in N. crassa.
Fig 2. Effect of reduced expression of calcineurin on colony morphology.
(A) Colony morphology. Wild type, ∆cna-1::hph; ∆pan-2::Ptcu-1::cna-1::v5::gfp; mat a (540), and ∆cnb-1::hph; ∆pan-2::Ptcu-1::cnb-1::v5::gfp; mat a (597) strains were cultured on VM plates supplemented with 250 μM of BCS (upper panel) or CuSO4 (lower panel). Strains were grown for one day at 30°C in the dark, followed by two days under constant light at room temperature and photographed using a Canon G10 camera. (B) Morphology of flask cultures. Cultures of wild type, ∆cna-1::hph; ∆pan-2::Ptcu-1::cna-1::v5::gfp; mat a (540), and ∆cnb-1::hph; ∆pan-2::Ptcu-1::cnb-1::v5::gfp; mat a (597) strains were grown in flasks containing VM agar medium supplemented with 250 μM of BCS (upper panel) or CuSO4 (lower panel). Strains were grown for three days at 30°C in dark and four days under light at room temperature and photographed using a Canon G10 camera.
Fig 3. Effect of reduced expression of calcineurin on hyphal growth.
(A) Hyphal morphology. The wild type, ∆cna-1::hph; ∆pan-2::Ptcu-1::cna-1::v5::gfp; mat a (540), and ∆cnb-1::hph; ∆pan-2:: Ptcu-1::cnb-1::v5::gfp; mat a (597) strains were cultured on VM medium supplemented with 250 μM of BCS (upper panel) or CuSO4 (lower panel) for 24 h at 30°C and then photographed using a Canon G10 camera with an Olympus SZX9 stereo microscope. (B) Aerial hyphae growth. Wild type, ∆cna-1::hph; ∆pan-2:: Ptcu-1::cna-1::v5::gfp; mat a (540), and ∆cnb-1::hph; ∆pan-2::Ptcu-1::cnb-1::v5::gfp; mat a (597) strains in VM liquid medium supplemented with 250 μM of BCS (upper panel) or CuSO4 (lower panel). Strains were grown for three days at 30°C in dark and four days under light at room temperature and photographed using a Canon G10 camera.
Table 3. Average growth rate.
| Strain | Average growth rate (cm h-1) | |
|---|---|---|
| VM+pantothenate+BCS | VM+pantothenate+CuSO4 | |
| Wild type: 74-OR23-IVA | 0.377 ±0.031 | 0.380 ±0.025 |
| Strain 540: ∆cna-1::hph; ∆pan-2::Ptcu-1::cna-1::V5::gfp; mat a | 0.391 ±0.006 | 0.103 ±0.005 |
| Strain 597: ∆cnb-1::hph; ∆pan-2::Ptcu-1::cnb-1::V5::gfp; mat a | 0.303 ±0.011 | 0.079 ±0.004 |
Reduced Expression of CNA-1 and CNB-1 Affects Female Sexual Fertility
To determine possible roles for CNA-1 and CNB-1 in sexual fertility, we analyzed crosses involving strains 540 and 597 on sexual crossing medium (SCM) supplemented with either BCS or excess copper. In the inducing BCS medium, the crosses involving strains 540 and 597 as female parents with a wild type male parent were fertile (produced thousands of ascospores; Table 4), with strain 540 slightly less fertile than the wild type control (Table 4). Under repressing conditions, crosses involving strain 540 as the female parent with a wild type male produced peithecia-like structures; however, when these were dissected, they were devoid of asci and ascospores. Thus strain 540 is female-sterile in excess copper sulfate, indicating a strict requirement for CNA-1 in female structures during the sexual cycle (Table 4). The need for CNB-1 appeared to be not as critical, as crosses with strain 597 as the female parent produced an intermediate number of ascospores relative to wild type (Table 4). Crosses involving wild type as the female parent and strain 540 or 597 as the male parent in the SCM supplemented with only pantothenate (Table 4) or minimal SCM were intermediate and fertile, respectively. Therefore, of the two subunits of calcineurin, CNA-1 seems to be essential for female fertility and may also influences male fertility.
Table 4. Sexual cycle phenotypes.
| Female Parent | Male Parent | Supplement | Phenotype |
|---|---|---|---|
| Wild type mat A | Wild type mat a | Pantothenate+BCS | Fertile, tens of thousands of ascospores |
| Wild type mat a | Wild type mat A | Pantothenate+BCS | Fertile, tens of thousands of ascospores |
| Strain 540 mat a | Wild type mat A | Pantothenate+BCS | Intermediate, few thousands of ascospores |
| Strain 597 mat a | Wild type mat A | Pantothenate+BCS | Fertile, tens of thousands of ascospores |
| Wild type mat A | Wild type mat a | Pantothenate+CuSO4 | Fertile, tens of thousands of ascospores |
| Wild type mat a | Wild type mat A | Pantothenate+CuSO4 | Fertile, tens of thousands of ascospores |
| Strain 540 mat a | Wild type mat A | Pantothenate+CuSO4 | Sterile, no ascospores |
| Strain 597 mat a | Wild type mat A | Pantothenate+CuSO4 | Intermediate, few hundreds of ascospores |
| Wild type mat A | Wild type mat a | Pantothenate | Fertile, tens of thousands of ascospores |
| Wild type mat a | Wild type mat A | Pantothenate | Fertile, tens of thousands of ascospores |
| Wild type mat A | Strain 540 mat a | Pantothenate | Intermediate, few thousands of ascospores |
| Wild type mat A | Strain 540 mat a | Pantothenate+BCS | Intermediate, few thousands of ascospores |
| Wild type mat A | Strain 597 mat a | Pantothenate | Fertile, tens of thousands of ascospores |
cnb-1 RIP-Mutants Possess Defects in Growth and Fertility
Due to the lack of a ∆cnb-1::hph mat A strain, it was necessary to perform two crosses using the mat a transformants with the tcu-1 promoted version of cnb-1 in order to create the final ∆cnb-1::hph; ∆pan-2:: Ptcu-1::cnb-1::v5::gfp strains (see Materials and Methods). This meant that the cnb-1 ORF in these strains might be subjected to RIP during the second cross. For this reason, it was necessary to sequence the cnb-1 ORF in the final cross progeny. We determined that strains 599, 600 and 602 had accrued RIP-induced mutations spread throughout the cnb-1 gene, including the Ca2+-binding domains (Materials and Methods; S1 Fig). Several nonsynonymous substitutions were also identified in the cnb-1RIP allele of these strains (S1 Fig). Strain 599 contains a M31I (ATG →ATA codon change) mutation in the EF1 domain. Strain 600 contains M31I (ATG →ATA codon change) mutation in the EF1 domain, D103N (GAC→AAC codon change), D107N (GAC→AAC codon change) and M121I (ATG→ATA codon change) mutations in the EF hand 3. Strain 602 contains M31I (ATG →ATA codon change) mutation in the EF1 domain, and D105N (GAC→AAC codon change) mutation in the EF hand loop 3, respectively.
Using Western analysis, we determined that the CNB-1 protein was expressed in the three RIP mutant strains under inducing conditions (S2 Fig). This result indicated that the RIP mutant proteins were translated and stable. However, phenotypic analysis demonstrated that these strains exhibited defects in colony morphology, growth rate, and aerial hyphae development in medium supplemented with excess copper (S3 Fig; S1 Table). The colony morphology of these strains was defective even in the inducing medium, and the cnb-1RIP (602) mutant showed a severe growth defect (S3 Fig; S1 Table). Furthermore, aerial hyphae development in the cnb-1RIP (600) and cnb-1RIP (602) mutants was defective even in inducing medium (S3 Fig). In addition, in the inducing medium, when used as a female parent with the wild type as male parent, strain cnb-1RIP (599) exhibited an intermediate phenotype, strain cnb-1RIP (600) was fertile, and strain cnb-1RIP (602) was sterile (S2 Table). In the repressing medium, when used as a female parent with the wild type as male parent, the cnb-1RIP (599) and cnb-1RIP (600) strains were fertile; while strain cnb-1RIP (602) was sterile (S2 Table). All three cnb-1RIP-mutants were fertile when used as males with a wild type female parent in SCM supplemented with pantothenate or minimal SCM (S2 Table). Therefore, cnb-1RIP (599) and cnb-1RIP (600) mutants were fully fertile either as a male or female parent; however, cnb-1RIP (602) mutant was sterile as a female parent in both inducing and repressing medium. Thus, the different RIP-induced alterations in the cnb-1 gene of cnb-1RIP mutants (599, 600, and 602) result in phenotypic differences.
CNA-1 and CNB-1 Form a Complex
To test whether CNA-1 and CNB-1 form a complex in vivo, we produced N. crassa strains that expressed RFP and GFP-tagged versions of these proteins from the inl and pan-2 loci, respectively (see Materials and Methods). We isolated homokaryotic stains expressing CNA-1::S-tag::RFP (strain 767; requires inositol) and CNB-1::V5::GFP (strain 727; requires pantothenate) tagged proteins.The CNA-1 and CNB-1 proteins contain 562 and 174 amino acid residues having predicted molecular weights of 64.5 and 19.8 kDa, respectively. The combined predicted molecular weights of the tagged proteins, CNA-1::S-tag::RFP and CNB-1::V5::GFP, are, respectively, 92.6 and 48.4 kDa. Expression of CNA-1 and CNB-1 tagged proteins of the correct size was verified using Western analysis (data not shown).
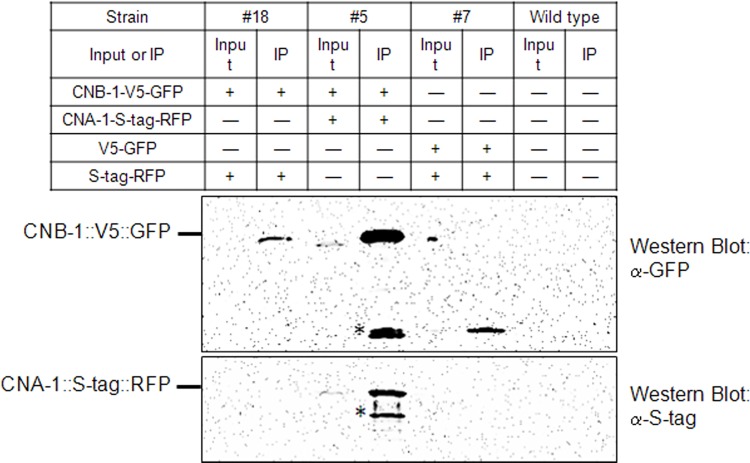
The tagged CNA-1 and CNB-1 proteins were brought together in the same cytoplasm through formation of forced heterokaryons between strains 767 and 727 (Table 1). The two strains were co-inoculated on Vogel’s minimal agar medium lacking inositol and pantothenate, conditions under which only the heterokaryon will grow. Control heterokaryons contained the CNB-1-GFP strain and a strain only expressing RFP (Fig 4; Table 1). Heterokaryons were cultivated in liquid medium and protein extracts were prepared. Extracts were treated with anti-GFP antibody coupled to agarose beads, and the immunoprecipitated proteins were analyzed by immunoblot using anti-GFP and anti-S-tag (detects S-tag-RFP-tagged protein) antibodies (Fig 4). The results demonstrated that CNB-1-V5-GFP was immunoprecipitated in all strains where it was present using the anti-GFP beads (Fig 4). Importantly, the ~93 kDa CNA-1-S-tag-RFP protein was visible in the input from the CNB-1-GFP+CNA-1-RFP strain and also co-immunoprecipitated with CNB-1-GFP (Fig 4). In control reactions, CNA-1-S-tag-RFP was not co-immunoprecipitated in strains expressing only V5-GFP, and S-tag-RFP was not co-immunoprecipitated using CNB-1-V5-GFP (Fig 4), showing that the fusion proteins do not interact with the fluorescent protein tags. In addition, microscopy analysis revealed co-localization of the tagged CNA-1 and CNB-1 proteins, which supports in vivo interaction between two calcineurin subunits (S4 Fig). These results indicate that CNA-1 and CNB-1 form a complex in N. crassa.
Fig 4. Co-immunoprecipitation of CNA-1 and CNB-1 proteins.
The indicated heterokaryons (#5, 7, and 18), and the wild type strain were grown for 16 h in submerged culture as described in the Materials and Methods. The soluble fraction was isolated and immunoprecipitated using anti-GFP antibody coupled to agarose beads. Samples of whole cell lysates (Input) and immunoprecipitated proteins (IP) were subjected to Western blot analysis using GFP (top), and S-tag (bottom) antibodies as described in Materials and Methods. The line indicates position for bands of the CNB-1::V5::GFP (~48 kDa; Top Blot) and CNA-1::S-tag::RFP (~ 93 kDa; Bottom blot) proteins. The asterisk is pointing to a smaller molecular weight molecule possibly resulted from proteolytic cleavage of CNB-1::V5::GFP and CNA-1::S-tag::RFP.
Discussion
Calcineurin is the only protein phosphatase that depends on both Ca2+ and CaM for its activity [1, 2]. Calcineurin consists of two subunits, a catalytic A and a regulatory B that are, respectively, encoded by the cna-1 and the cnb-1 genes in N. crassa. We studied here the cellular roles of calcineurin via its heterologous expression under the conditional tcu-1 promoter. In this system, expression of the heterologous gene cloned under the Ptcu-1 is controlled by copper availability in a rapid and stable manner [32, 33]. Using this system, we found that both CNA-1 and CNB-1 were required for normal vegetative and hyphal growth, and sexual development of N. crassa. Previous work using cna-1 antisense RNA also showed its essential role in normal hyphal growth, morphology, and maintenance of the apical tip-high Ca2+ gradient [28]. Moreover, immunosuppressive fungicidal drugs cyclosporin A (CsA) and FK506 were shown to inhibit hyphal growth of N. crassa by inactivating calcineurin. In addition, studies using the insertional and RIP mutants of the cnb-1 suggested its requirement for normal vegetative growth and a possible role in repressing the asexual developmental program in N. crassa [30]. Calcineurin is also required for normal hyphal growth in fungal pathogens in C. neoformans, A. fumigatus, M. oryzae, and U. maydis, suggesting a general role of calcineurin in governing hyphal growth [21, 22, 24, 25, 27, 55, 56].
Calcineurin also plays a role in fertility and stress response pathways in various organisms. In the model nematode C. elegans, calcineurin regulates fertility [57]. In addition, calcineurin is also reported to play a role in stress and pheromone response pathways in in other fungi, such as S. cerevisiae, Schizosaccharomyces pombe, C. albicans, and C. neoformans [15–18, 23, 58]. We have demonstrated that calcineurin subunit A plays an essential role during sexual development in N. crassa. In N. crassa, the sexual phase starts with fertilization between two opposite mating type nuclei in limited nitrogen and nutrient starvation, a stress condition. In addition, sexual development in N. crassa normally proceeds with development of protoperithecia and then perithecia, but the perithecia were empty under repressing conditions for the CNA-1 strain (540) as the female parent, suggesting that this protein must be important for some aspect of meiosis or ascospore development. Moreover, CNA-1 strain as the male parent was less productive, which could indicate a possible involvement of this protein in initial steps of mating. In addition, the CNB-1 appeared to be not as critical, and thus, that the phenotypes of the strains CNA-1 (540) and CNB-1 (597) in sexual development differ, suggesting that CNA-1 may have novel functions independent of CNB-1. In this study, we also isolated RIP mutated alleles of cnb-1. Among the non-synonymous substitutions, the cnb-1RIP (599) allele contains M31I alteration in the EF1. Similarly, the cnb-1RIP (600) allele contains M31I in the EF1, and D103N, D107N, and M121I alterations in the EF3. In addition, the cnb-1RIP (602) allele contains M31I and D105N alterations in the EF1 and EF3, respectively. The EF hand contains a helix-loop-helix structural unit known as EF hand or Ca2+ binding loop that contains characteristic 12 amino acid residues involved in the Ca2+ binding [59]. Ca2+ is coordinated directly by oxygen atoms provided by the side chains of residues at positions 1(+x), 3(+y), 5(+z) and 12 (-z) [59]. The residues D103 and D107 constitute the 1(+x) and 5(+z) position of the EF hand loop 3, respectively [59]. The Ca2+ coordinating residues D103 and D107 are also conserved among the CNB1 from different fungi, Drosophila, and mammals (data not shown). Therefore, the D103N and D107N alterations in the cnb-1RIP (600) allele are expected to cause a defect in Ca2+ co-ordination in this mutant, and this could explain the growth defect of the ∆cnb-1::hph; ∆pan-2::Ptcu-1::cnb-1 RIP::v5::gfp; mat a (600) strain even under inducing conditions (S3 Fig). EF3 of the cnb-1RIP (600) allele contains RIP mutations both inside and outside of the EF hand loop. The major alteration found in the cnb-1RIP (602) allele was D105N in the EF3. The D105 residue is the coordinate 3(+y) involved in Ca2+ binding through the oxygen atoms of aspartate [59]. Therefore, the cnb-1RIP (602) allele contains an EF3 domain with defective Ca2+ binding ability and this could explain the growth and fertility defects of the ∆cnb-1::hph; ∆pan-2::Ptcu-1::cnb-1 RIP::v5::gfp; mat a (602) strain in both inducing and repressing medium (S3 Fig; S2 Table). The cnb-1RIP (602) was sterile under both inducing and repressing medium, while the wild type allele was fertile. This suggests that very little wild type CNB-1 protein is necessary to support female fertility, while the cnb-1RIP (602) mutation impairs function so that even high levels of protein do not support female fertility. Thus, this analysis has identified D103, D105, and D107 as critical amino acid residues in the EF3 domain of cnb-1.
Interaction between the recombinant forms of the bovine A and rat B subunits of calcineurin, expressed in E. coli, was demonstrated in vitro [60]. The interaction between the calcineurin A and B subunits were shown in S. cerevisiae using the yeast two-hybrid system [61]. We demonstrated in vivo interaction between tagged versions of CNA-1 and CNB-1 by co-immunoprecipitation and co-localization studies which indicates formation of a complex of these two proteins in N. crassa (Fig 4; S4 Fig).
In this study, we showed the cellular roles of calcineurin in growth, development, and fertility in N. crassa. Moreover, we demonstrated in vivo interaction between the calcineurin subunits, CNA-1 and CNB-1, and identified three critical amino acid residues in CNB-1. Future work will identify the target transcription factors of calcineurin in N. crassa.
Supporting Information
A. Alignment of the wild type and cnb-1RIP alleles. Mutations in the intronic regions, and synonymous and non-synonymous substitutionsare shown as tick, arrow head and solid arrow marks, respectively, above the alignment. B. Alignment of the protein sequences of the CNB-1 and CNB-1RIP proteins. The positions of the EF-hand loops, EF1-EF4 (solid line) and the calcium binding regions (dotted line) are indicated above the sequence, as revealed by Uniprot analysis (http://www.uniprot.org/uniprot/P87072). The arrows indicate the altered amino acid residues. Conserved residues are indicated in black (100%), dark gray (>80%) and light gray (>60%).
(TIF)
Effects of BCS and copper sulfate on Ptcu-1 driven expression of CNB-1 from the cnb-1RIP strains. Samples containing 50 μg of total protein were analyzed by Western blot using rabbit anti-GFP antibody. An extract from untransformed wild type, grown in minimal VM, was used as a control. The expression level of CNB-1::V5::GFP in extracts from ∆cnb-1::hph; Ptcu-1::cnb-1::v5::gfp::∆pan-2; mat a strains (599, 600, and 602) treated with 250 μM of BCS (B) or copper (C) for 22 h, is indicated. The solid lines indicate positions of the CNB-1::V5::GFP protein of molecular weight (MW) ~ 48 in the strains 602, 600, and another band (possibly truncated) of ~25 kDa appeared in the lane for the strain 599 on the blot. The membrane was stained with amido black as a protein loading control (lower panel).
(TIF)
A. Hyphal morphology. Wild type and cnb-1RIP mutants were grown for 24 h at 30°C on VM medium supplemented with 250 μM of BCS (upper panel) or CuSO4 (lower panel). B. Colony morphology. Wild type and the cnb-1RIP mutants were cultured for 24 h at 30°C in dark and 48 h under light at room temperature on VM plates supplemented with 250 μM of BCS (upper panel) or CuSO4 (lower panel). C. Growth of aerial hyphae. Aerial hyphae of the wild type and the cnb-1RIP mutants grown for 72 h at 30°C in dark and four days under light at room-temperature in VM liquid medium supplemented with 250 μM of BCS (upper panel) or CuSO4 (lower panel). All the strains were photographed using a Canon G10 camera.
(TIF)
A heterokaryon (CNB-1GFP+CNA-1RFP#5, Table 1) expressing two fluorescent proteins, CNA-1::S-tag::RFP and CNB-1::V5::GFP, was analyzed using a confocal microscope to investigate the localization and in vivo interaction of the two caclineurin subunits. Forced heterokaryons were made same as described for co-immunoprecipitation analysis (see Materials and Methods), conidia were then isolated and inoculated in 5 ml liquid VM and incubated at 30°C for 6 h with shaking at 200 rpm. Germlings were analyzed using a Leica TCS SP8 (DMi8) confocal microscope with a 63x oil objective, 4x zoom, 1024x1024 pixels resolution, and scan speed of 400 Hz (Leica Microsystems CMS, GmbH, Germany). The CNA-1::S-tag::RFP and CNB-1::V5::GFP heterokaryons were visualized with the Hybrid Detection system (HyD) laser. Images were captured sequentially; RFP images were obtained with excitation at 543 nm and emission from 555–700 nm, and GFP images were obtained by excitation at 488 nm, with emission collected from 500–535 nm. DIC, RFP and GFP fluorescent, and merged images are shown in the columns from left to right, respectively. The arrowhead, asterisk, and solid arrow indicate CNA-1::S-tag::RFP, CNB-1::V5::GFP, and co-localization of the tagged calcineurin subunits, respectively. Scale bar = 5 μm.
(TIF)
(DOCX)
(DOCX)
Acknowledgments
We thank the FGSC for some Neurospora strains. We thank Dr. Bula Choudhury from Guwahati Biotech Park, and Dr. Anand Tiwari and Dibakar Gohain from Indian Institute of Technology Guwahati for help in confocal microscopy.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Funded by Indo-US Science and Technology Forum (http://www.iusstf.org/) and National Institutes of Health (http://www.nih.gov/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Klee CB, Crouch TH, Krinks MH (1979) Calcineurin: a calcium- and calmodulin-binding protein of the nervous system. Proc Natl Acad Sci U S A 76: 6270–6273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Klee CB, Ren H, Wang X (1998) Regulation of the calmodulin-stimulated protein phosphatase, calcineurin. J Biol Chem 273: 13367–13370. [DOI] [PubMed] [Google Scholar]
- 3.Winkler MA, Merat DL, Tallant EA, Hawkins S, Cheung WY (1984) Catalytic site of calmodulin-dependent protein phosphatase from bovine brain resides in subunit A. Proc Natl Acad Sci U S A 81: 3054–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rusnak F, Mertz P (2000) Calcineurin: form and function. Physiol Rev 80: 1483–1521. [DOI] [PubMed] [Google Scholar]
- 5.Hogan PG, Chen L, Nardone J, Rao A (2003) Transcriptional regulation by calcium, calcineurin, and NFAT. Genes Dev 17: 2205–2232. [DOI] [PubMed] [Google Scholar]
- 6.Rumi-Masante J, Rusinga FI, Lester TE, Dunlap TB, Williams TD, Dunker AK, et al. (2012) Structural basis for activation of calcineurin by calmodulin. J Mol Biol 415: 307–317. 10.1016/j.jmb.2011.11.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Ying-Lien, Kozubowski L, Cardenas ME, Heitman J (2010) On the roles of calcineurin in fungal growth and pathogenesis. Curr Fungal Infect Rep 4: 244–255. [Google Scholar]
- 8.Rao A, Luo C, Hogan PG (1997) Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol 15: 707–747. [DOI] [PubMed] [Google Scholar]
- 9.Wu H, Peisley A, Graef IA, Crabtree GR (2007) NFAT signaling and the invention of vertebrates. Trends Cell Biol 17: 251–260. [DOI] [PubMed] [Google Scholar]
- 10.Musson RE, Cobbaert CM, Smit NP (2012) Molecular diagnostics of calcineurin-related pathologies. Clin Chem 58: 511–522. 10.1373/clinchem.2011.167296 [DOI] [PubMed] [Google Scholar]
- 11.Gajewski KM, Wang J, Schulz RA (2006) Calcineurin function is required for myofilament formation and troponin I isoform transition in Drosophila indirect flight muscle. Dev Biol 289: 17–29. [DOI] [PubMed] [Google Scholar]
- 12.Kuhara A, Inada H, Katsura I, Mori I (2002) Negative regulation and gain control of sensory neurons by the C. elegans calcineurin TAX-6. Neuron 33: 751–763. [DOI] [PubMed] [Google Scholar]
- 13.Luan S (2009) The CBL-CIPK network in plant calcium signaling. Trends Plant Sci 14: 37–42. 10.1016/j.tplants.2008.10.005 [DOI] [PubMed] [Google Scholar]
- 14.Kumar R, Musiyenko A, Barik S (2005) Plasmodium falciparum calcineurin and its association with heat shock protein 90: mechanisms for the antimalarial activity of cyclosporin A and synergism with geldanamycin. Mol Biochem Parasitol 141: 29–37. [DOI] [PubMed] [Google Scholar]
- 15.Cyert MS, Kunisawa R, Kaim D, Thorner J (1991). Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc Natl Acad Sci U S A 88: 7376–7380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cyert MS, Thorner J (1992) Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaptation to pheromone. Mol Cell Biol 12: 3460–3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Y, S. Ishii M, Tokai H, Tsutsumi O, Ohki R, Akada K, et al. (1991) The Saccharomyces cerevisiae genes (CMP1 and CMP2) encoding calmodulin-binding proteins homologous to the catalytic subunit of mammalian protein phosphatase 2B. Mol Gen Genet 227:52–59. [DOI] [PubMed] [Google Scholar]
- 18.Mendoza I, Quintero FJ, Bressan RA, Hasegawa PM, Pardo JM (1996) Activated calcineurin confers high tolerance to ion stress and alters the budding pattern and cell morphology of yeast cells. J Biol Chem 271: 23061–23067. [DOI] [PubMed] [Google Scholar]
- 19.Cruz MC, Goldstein AL, Blankenship JR, Del Poeta M, Davis D, Cardenas ME, et al. (2002) Calcineurin is essential for survival during membrane stress in Candida albicans. EMBO J 21:546–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sanglard D, Ischer F, Marchetti O, Entenza J, Bille J (2003) Calcineurin A of Candida albicans: involvement in antifungal tolerance, cell morphogenesis and virulence. Mol Microbiol 48: 959–976. [DOI] [PubMed] [Google Scholar]
- 21.Odom A, Muir S, Lim E, Toffaletti DL, Perfect J, Heitman J (1997) Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J 16:2576–2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fox DS, Cruz MC, Sia RA, Ke H, Cox GM, Cardenas ME, et al. (2001) Calcineurin regulatory subunit is essential for virulence and mediates interactions with FKBP12-FK506 in Cryptococcus neoformans. Mol Microbiol 39:835–849. [DOI] [PubMed] [Google Scholar]
- 23.Cruz MC, Fox DS, Heitman J (2001) Calcineurin is required for hyphal elongation during mating and haploid fruiting in Cryptococcus neoformans. EMBO J 20: 1020–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen QB, Kadotani N, Kasahara S, Tosa Y, Mayama S, Nakayashiki H (2008) Systematic functional analysis of calcium-signalling proteins in the genome of the rice-blast fungus, Magnaporthe oryzae, using a high-throughput RNA-silencing system. Mol Microbiol. 68:1348–1365. 10.1111/j.1365-2958.2008.06242.x [DOI] [PubMed] [Google Scholar]
- 25.Choi JH, Kim Y, Lee YH (2009) Functional analysis of MCNA, a gene encoding a catalytic subunit of calcineurin, in the rice blast fungus Magnaporthe oryzae. J Microbiol Biotechnol 19: 11–16. [PubMed] [Google Scholar]
- 26.Lee SC, Li A, Calo S, Heitman J (2013) Calcineurin plays key roles in the dimorphic transition and virulence of the human pathogenic zygomycete Mucor circinelloides. PLoS Pathog 9: e1003625 10.1371/journal.ppat.1003625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Egan JD, Garcia-Pedrajas MD, Andrews DL, Gold SE (2009) Calcineurin is an antagonist to PKA protein phosphorylation required for postmating filamentation and virulence, while PP2A is required for viability in Ustilago maydis. Mol Plant Microbe Interact 22: 1293–1301. 10.1094/MPMI-22-10-1293 [DOI] [PubMed] [Google Scholar]
- 28.Prokisch H, Yarden O, Dieminger M, Tropschug M, Barthelmess IB (1997) Impairment of calcineurin function in Neurospora crassa reveals its essential role in hyphal growth, morphology and maintenance of the apical Ca2+ gradient. Mol Gen Genet 256: 104–114. [DOI] [PubMed] [Google Scholar]
- 29.Cambareri EB, Jensen BC, Schabtach E, Selker EU (1989) Repeat-induced G-C to A-T mutations in Neurospora. Science 244: 1571–1575. [DOI] [PubMed] [Google Scholar]
- 30.Kothe GO, Free SJ (1998) Calcineurin subunit B is required for normal vegetative growth in Neurospora crassa. Fungal Genet Biol 23: 248–258. [DOI] [PubMed] [Google Scholar]
- 31.Kumar KS, Ravi Kumar B, Siddavattam D, Subramanyam C (2006) Characterization of calcineurin-dependent response element binding protein and its involvement in copper-metallothione in gene expression in Neurospora. Biochem Biophys Res Commun 345: 1010–1013. [DOI] [PubMed] [Google Scholar]
- 32.Korripally P, Tiwari A, Haritha A, Kiranmayi P, Bhanoori M (2010) Characterization of Ctr family genes and the elucidation of their role in the life cycle of Neurospora crassa. Fungal Genet Biol 47: 237–245. 10.1016/j.fgb.2009.12.006 [DOI] [PubMed] [Google Scholar]
- 33.Lamb TM, Vickery J, Bell-Pedersen D (2013) Regulation of gene expression in Neurospora crassa with a copper responsive promoter. G3 (Bethesda) 3: 2273–2280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ouyang S, Beecher CN, Wang K, Larive CK, Borkovich KA (2015) Metabolic impacts of using nitrogen and copper-regulated promoters to regulate gene expression in Neurospora crassa. G3 (Bethesda). 5(9):1899–908. 10.1534/g3.115.020073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vogel HJ (1956) A convenient growth medium for Neurospora (Medium N). Microbial Genet Bull 13: 42–43. [Google Scholar]
- 36.Westergaard M, Mitchell HK (1947) Neurospora. V. A synthetic medium favoring sexual reproduction. Am J Bot 34: 573–577. [Google Scholar]
- 37.Ivey FD, Kays AM, Borkovich KA (2002) Shared and independent roles for a Gαi protein and adenylyl cyclase in regulating development and stress responses in Neurospora crassa. Eukaryot Cell 1:634–642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barman A, Tamuli R (2015) Multiple cellular roles of Neurospora crassaplc-1, splA2, and cpe-1 in regulation of cytosolic free calcium, carotenoid accumulation, stress responses, and acquisition of thermotolerance. J Microbiol 53:226–235. 10.1007/s12275-015-4465-1 [DOI] [PubMed] [Google Scholar]
- 39.Deka R, Kumar R, Tamuli R (2011) Neurospora crassa homologue of Neuronal Calcium Sensor-1 has a role in growth, calcium stress tolerance, and ultraviolet survival. Genetica139: 885–894. [DOI] [PubMed] [Google Scholar]
- 40.Deka, Tamuli R (2013) Neurospora crassa ncs-1, mid-1 and nca-2 double-mutant phenotypes suggest diverse interaction among three Ca2+-regulating gene products. J Gene 92: 559–563. [DOI] [PubMed] [Google Scholar]
- 41.Kumar R, Tamuli R (2014) Calcium/calmodulin-dependent kinases are involved in growth, thermotolerance, oxidative stress survival, and fertility in Neurospora crassa. Arch Microbiol196: 295–305. [DOI] [PubMed] [Google Scholar]
- 42.Davis RH, De Serres FJ (1970) Genetic and microbiological research techniques for Neurospora crassa. Methods Enzymol 17:79–143. [Google Scholar]
- 43.Hays S, Selker EU (2000) Making the selectable marker bar tighter and more economical. Fungal Genet. Newslett 47: 107. [Google Scholar]
- 44.Avalos J, Geever RF, Case ME (1989) Bialaphos resistance as a dominant selectable marker in Neurospora crassa. Curr Genet 16: 369–372. [DOI] [PubMed] [Google Scholar]
- 45.Pall ML (1993) The use of Ignite (Basta; glufosinate; phosphinothricin) to select transformants of bar-containing plasmids in Neurospora crassa. Fungal Genet News l40:58. [Google Scholar]
- 46.Murray NE, Perkins DD (1963) Stanford Neurospora methods. Neurospora News l4: 21–25. [Google Scholar]
- 47.Hanahan D (1985) Techniques for transformation of E. coli In:Glover DM ed. DNA cloning: a practical approach. 1st ed. Oxford, United Kingdom: IRL Press, pp1:109–135. [Google Scholar]
- 48.Ivey FD, Hodge PN, Turner GE, Borkovich KA (1996) The G alpha i homologue gna-1 controls multiple differentiation pathways in Neurospora crassa. Mol Biol Cell 7: 1283–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Winston F, Dollard C, Ricupero-Hovasse SL (1995) Construction of a set of convenient Saccharomyces cerevisiae strains that are isogenic to S288C. Yeast 11: 53–55. [DOI] [PubMed] [Google Scholar]
- 50.Colot HV, Park G, Turner GE, Ringelberg C, Crew CM, Litvinkova L, et al. (2006) A high-throughput gene knockout procedure for Neurospora reveals functions for multiple transcription factors. Proc Natl Acad Sci U S A.103:10352–10357. Erratum in: Proc Natl Acad Sci U S A. 2006 103:16614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoffman CS (2001) Preparation of yeast DNA. Curr Protoc Mol Biol Chapter 13: Unit13 11. [DOI] [PubMed] [Google Scholar]
- 52.Jones CA, Borkovich KA (2010) Analysis of mitogen-activated protein kinase phosphorylation in response to stimulation of histidine kinase signaling pathways in Neurospora. Methods Enzymol 471: 319–334. 10.1016/S0076-6879(10)71017-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghosh A, Servin JA, Park G, Borkovich KA (2014) Global analysis of serine/threonine and tyrosine protein phosphatase catalytic subunit genes in Neurospora crassa reveals interplay between phosphatases and the p38 mitogen-activated protein kinase. G3 (Bethesda) 4: 349–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krystofova S, Borkovich KA (2005) The heterotrimeric G-protein subunits GNG-1 and GNB-1 form a Gβγ dimer required for normal female fertility, asexual development, and Gα protein levels in Neurospora crassa. Eukaryot Cell 4: 365–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Steinbach WJ, Cramer RA Jr, Perfect BZ, Asfaw YG, Sauer TC, Najvar LK, et al. (2006) Calcineurin controls growth, morphology, and pathogenicity in Aspergillus fumigatus. Eukaryot Cell 5:1091–1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.da Silva Ferreira ME, Heinekamp T, Härtl A, Brakhage AA, Semighini CP, Harris SD, Savoldi M, et al. (2007) Functional characterization of the Aspergillus fumigatus calcineurin. Fungal Genet Biol 44:219–230. [DOI] [PubMed] [Google Scholar]
- 57.Bandyopadhyay J, Lee J, Lee J, Lee JI, Yu JR, Jee C, et al. (2002) Calcineurin, a calcium/calmodulin-dependent protein phosphatase, is involved in movement,fertility, egglaying, and growth in Caenorhabditis elegans. Mol Biol Cell 13:3281–3293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kraus PR, Heitman J (2003) Coping with stress: calmodulin and calcineurin in model and pathogenic fungi. Biochem Biophys Res Commun 311: 1151–1157. [DOI] [PubMed] [Google Scholar]
- 59.Gifford JL, Walsh MP, Vogel HJ (2007) Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem J 405: 199–221. [DOI] [PubMed] [Google Scholar]
- 60.Sikkink R, Haddy A, MacKelvie S, Mertz P, Litwiller R, Rusnak F (1995) Calcineurin subunit interactions: mapping the calcineurin B binding domain on calcineurin A. Biochemistry 34:8348–8356. [DOI] [PubMed] [Google Scholar]
- 61.Chaudhuri B, Hammerle M, Furst P (1995) The interaction between the catalytic A subunit of calcineurin and its autoinhibitory domain, in the yeast two-hybrid system, is disrupted by cyclosporin A and FK506. FEBS Lett 357: 221–226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A. Alignment of the wild type and cnb-1RIP alleles. Mutations in the intronic regions, and synonymous and non-synonymous substitutionsare shown as tick, arrow head and solid arrow marks, respectively, above the alignment. B. Alignment of the protein sequences of the CNB-1 and CNB-1RIP proteins. The positions of the EF-hand loops, EF1-EF4 (solid line) and the calcium binding regions (dotted line) are indicated above the sequence, as revealed by Uniprot analysis (http://www.uniprot.org/uniprot/P87072). The arrows indicate the altered amino acid residues. Conserved residues are indicated in black (100%), dark gray (>80%) and light gray (>60%).
(TIF)
Effects of BCS and copper sulfate on Ptcu-1 driven expression of CNB-1 from the cnb-1RIP strains. Samples containing 50 μg of total protein were analyzed by Western blot using rabbit anti-GFP antibody. An extract from untransformed wild type, grown in minimal VM, was used as a control. The expression level of CNB-1::V5::GFP in extracts from ∆cnb-1::hph; Ptcu-1::cnb-1::v5::gfp::∆pan-2; mat a strains (599, 600, and 602) treated with 250 μM of BCS (B) or copper (C) for 22 h, is indicated. The solid lines indicate positions of the CNB-1::V5::GFP protein of molecular weight (MW) ~ 48 in the strains 602, 600, and another band (possibly truncated) of ~25 kDa appeared in the lane for the strain 599 on the blot. The membrane was stained with amido black as a protein loading control (lower panel).
(TIF)
A. Hyphal morphology. Wild type and cnb-1RIP mutants were grown for 24 h at 30°C on VM medium supplemented with 250 μM of BCS (upper panel) or CuSO4 (lower panel). B. Colony morphology. Wild type and the cnb-1RIP mutants were cultured for 24 h at 30°C in dark and 48 h under light at room temperature on VM plates supplemented with 250 μM of BCS (upper panel) or CuSO4 (lower panel). C. Growth of aerial hyphae. Aerial hyphae of the wild type and the cnb-1RIP mutants grown for 72 h at 30°C in dark and four days under light at room-temperature in VM liquid medium supplemented with 250 μM of BCS (upper panel) or CuSO4 (lower panel). All the strains were photographed using a Canon G10 camera.
(TIF)
A heterokaryon (CNB-1GFP+CNA-1RFP#5, Table 1) expressing two fluorescent proteins, CNA-1::S-tag::RFP and CNB-1::V5::GFP, was analyzed using a confocal microscope to investigate the localization and in vivo interaction of the two caclineurin subunits. Forced heterokaryons were made same as described for co-immunoprecipitation analysis (see Materials and Methods), conidia were then isolated and inoculated in 5 ml liquid VM and incubated at 30°C for 6 h with shaking at 200 rpm. Germlings were analyzed using a Leica TCS SP8 (DMi8) confocal microscope with a 63x oil objective, 4x zoom, 1024x1024 pixels resolution, and scan speed of 400 Hz (Leica Microsystems CMS, GmbH, Germany). The CNA-1::S-tag::RFP and CNB-1::V5::GFP heterokaryons were visualized with the Hybrid Detection system (HyD) laser. Images were captured sequentially; RFP images were obtained with excitation at 543 nm and emission from 555–700 nm, and GFP images were obtained by excitation at 488 nm, with emission collected from 500–535 nm. DIC, RFP and GFP fluorescent, and merged images are shown in the columns from left to right, respectively. The arrowhead, asterisk, and solid arrow indicate CNA-1::S-tag::RFP, CNB-1::V5::GFP, and co-localization of the tagged calcineurin subunits, respectively. Scale bar = 5 μm.
(TIF)
(DOCX)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.