Abstract
Background
A fascinating aspect of bile acid homeostasis is the coordination between bile acid uptake in intestine and hepatic bile acid synthesis. In response to bile acid uptake in enterocytes, FXR is activated and induces transcription of fibroblast growth factor (FGF) 15 in mice, or FGF19 in humans. FGF15/19 is secreted into the enterohepatic circulation, and through activation of hepatic receptors, leads to repression of Cyp7a1, a rate-limiting enzyme for bile acid synthesis. Using a genetic approach, we identified a novel protein, Diet1, as a control point for FGF15/19 production.
Key Messages
Mice with a Diet1 null mutation have reduced FGF15 secretion, causing impaired feedback repression of hepatic bile acid synthesis, and increased fecal bile acid excretion. As a result, Diet1-deficient mice constitutively convert cholesterol to bile acids and are resistant to diet-induced hypercholesterolemia and atherosclerosis. Diet1 affects FGF15/19 production at the post-transcriptional level, and the proteins appear to have overlapping subcellular localization in enterocytes. Diet1 appears to be a control point for the production of FGF15/19 in enterocytes, and thus a regulator of bile acid and lipid homeostasis. Studies to evaluate the role of common and rare DIET1 genetic variants in human health and disease are warranted.
Conclusions
Further elucidation of the Diet1–FGF15/19 interaction will provide new insights into the intricate regulatory mechanisms underlying bile acid metabolism.
Metabolic Syndrome is a constellation of metabolic conditions—visceral obesity, insulin resistance, hypertriglyceridemia, reduced high density lipoprotein (HDL) levels, and elevated blood pressure—that occur together and promote the development of diabetes and cardiovascular disease [1]. Recent studies indicate that bile acid levels are associated with several components of the Metabolic Syndrome. For example, bile acid levels are inversely correlated with triglyceride levels, bile acids promote insulin sensitivity, and specific bile acid species are implicated in the onset of obesity via modulation of the microbiome [2–7]. Despite the importance of bile acids in metabolic homeostasis, aspects of bile acid regulation remain to be elucidated.
Bile acid activities and homeostasis
Metabolic effects of bile acids range from roles in lipid absorption and metabolism to glucose homeostasis, inflammation, maintenence of intestinal microflora, hepatocarcinogenesis, and energy expenditure [8,9]. These activities may be exerted through the action of bile acids as ligands for multiple nuclear receptors: farnesoid X receptor (FXR), pregnane X receptor (PXR), vitamin D receptor, and the G protein-coupled receptor, TGR5 [8,10]. Bile acids activate these receptors to regulate gene expression in metabolic tissues such as intestine, liver, brown adipose tissue, macrophages, and brain.
Bile acid homeostasis requires transcriptional regulation of bile acid synthetic genes in liver, as well as coordination between hepatic bile acid synthesis and excretion from intestine [8,11]. The bulk of bile acid synthesis occurs through the action of cholesterol 7α-hydroxylase (CYP7A1). An alternate pathway, contributing 10–25% of bile acid synthesis, is catalyzed by sterol-27 hydroxylase (CYP27A1). Bile acids act to negatively regulate their own synthesis through the FXR-short heterodimeric partner (SHP), which represses the expression of CYP8B1 (encoding the enzyme responsible for cholic acid synthesis) and to a lesser extent CYP7A1.
Of the bile acids secreted into the intestine, approximately 5% traverse the length of the intestine and are excreted in the feces. The majority, however, are reabsorbed by enterocytes in the ileum via the apical sodium dependent bile acid transporter (ASBT), and returned to the liver through the enterohepatic circulation [9]. To prevent overproduction of bile acids by liver, the intestine communicates to the liver regarding the levels of bile acids that are reabsorbed. Through bile acid activation of FXR in enterocytes, transcription of the gene encoding fibroblast growth factor (FGF) 15 (in mouse) or FGF19 (in humans) is induced. FGF15/19 is secreted into the enterohepatic circulation, binds to receptors on hepatocytes, and activates kinase signaling cascades that ultimately repress CYP7A1 transcription [12,13]. Thus, the levels of FGF15/19 secreted from enterocytes play a role in regulating the levels of bile acid synthesis that must occur to maintain homeostasis.
The identification of Diet1
As described above, it is established that FGF15/19 expression in enterocytes is regulated by bile acid activation of FXR. However, little is known about the post-translational itinerary of FGF15/19 in enterocytes. A recently identified protein, Diet1, appears to influence the levels of FGF15/19 that are secreted from enterocytes, and therefore may serve as an additional regulatory component in the FGF15/19 axis [14]. Herein, we describe the identification of the Diet1 gene and protein and the current understanding of its function in enterohepatic bile acid homeostasis. The groundwork for the identification of Diet1 was laid several years ago, with the characterization of a mouse strain that was protected from the development of hypercholesterolemia and atherosclerosis when fed an atherogenic diet [15]. The mouse strain, C57BL/6ByJ, is a close relative of the widely studied and atherosclerosis-susceptible C57BL6/J strain. Further characterization revealed that the atherosclerosis resistance in C57BL/6ByJ mice could not be attributed to alterations in food intake, dietary cholesterol absorption, or endogenous cholesterol synthesis [15,16]. However, the two strains differed in bile acid metabolism: C57BL/6ByJ mice had enhanced bile acid excretion into the urine and feces, and elevated serum bile acid levels [16]. Gene expression profiling indicated that bile acid synthetic gene expression was elevated. Together, these findings led to the hypothesis that increased conversion of cholesterol to bile acids, and enhanced bile acid excretion, prevented the accumulation of high circulating cholesterol levels [16]. Genetic mapping studies showed that the responsible mutation was located on mouse chromosome 2 in a region containing hundreds of genes (49). The map location ruled out known players in cholesterol and bile acid metabolism as candidate genes, and suggested involvement of a novel gene, which was given the name Diet1.
We set out to identify the Diet1 gene and the mutation responsible for altered bile acid metabolism in C57BL/6ByJ mice using a positional cloning approach. This approach identifies the locus responsible for a phenotypic trait based strictly on its location in the genome, without knowledge or assumptions regarding the function of the responsible gene [17]. We first established that serum bile acid levels co-segregate with cholesterol levels and markers on chromosome 2, indicating that the same locus is responsible for both the resistance to hypercholesterolemia and elevated bile acid levels [16]. We reasoned that elevated bile acid levels represent the phenotype that is most directly related to the gene mutation in C57BL/6ByJ mice, with cholesterol levels being affected secondarily. To genetically position the mutant gene, we analyzed ~800 offspring of a cross between C57BL/6ByJ and an unrelated strain for bile acid levels, and mapped the responsible locus to a small region of chromosome 2 that harbored only a handful of genes. Extensive sequencing and mRNA transcript analysis in C57BL/6ByJ compared to C57BL/6J mice led to the identification of a gene that spanned 700 kb and overlapped with several small predicted open reading frames [14]. We defined the gene and mRNA structure of Diet1, and determined that genomic DNA from C57BL6ByJ mice contains a rearrangement in this gene. Specifically, the Diet1 region in these mice has a partial gene duplication leading to a frame-shift and premature stop codon. Diet1 mRNA cannot be detected in tissues of C57BL/6ByJ mice, most likely because the premature stop codon induces nonsense mediated mRNA decay. The Diet1 gene in C57BL/6ByJ mice therefore produces no functional mRNA or protein.
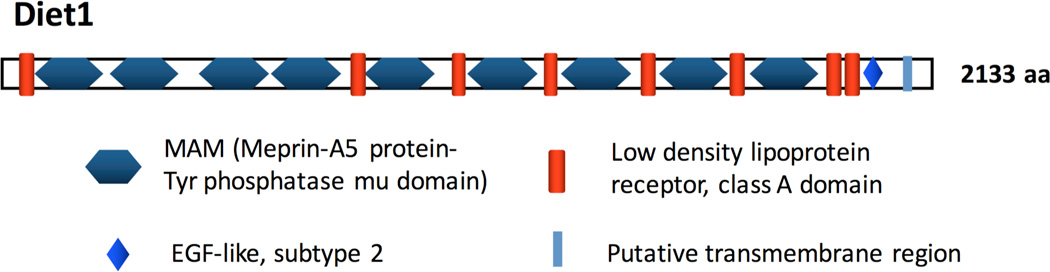
Diet1 encodes a protein with predicted molecular weight of 236 kDa, and the primary amino acid sequence is highly conserved between mouse and human (~70% identity), as well as other mammals, birds, amphibians, and fish [14]. Diet1 contains amino acid domains that have been identified in other proteins (Fig. 1). These include nine copies of the MAM (meprin-A5-tyrosine phosphatase μ) domain, which is present in other proteins of diverse functions and is thought to promote protein-protein interactions. The MAM domains are irregularly interspersed with copies of the low density lipoprotein (LDLR) receptor class A domain, which is present in multiple members of the LDLR family. Both the MAM and LDLR domains contain multiple cysteine residues that are suspected to form intra-domain disulfide bridges. Diet1 also contains a predicted membrane-spanning domain at the C-terminus. Based on primary amino acid sequence, Diet1 is most closely related to endotubin, a 140 kDa membrane glycoprotein that contains multiple copies of MAM and LDLR domains and is thought to have a role in vesicular trafficking in epithelial cells [18]. Based on its structural features, the official gene name for the Diet1 gene has been changed to MAM and LDL Receptor Class A Domain containing 1 (MALRD1 in humans, and Malrd1 in mice). However, we will continue to use the name Diet1 in this article, as this is the name used in all published work to date.
Figure 1.
Domain structure of mouse Diet1 protein.
Diet1 is present in intestinal epithelial cells
Detailed analysis of Diet1/DIET1 mRNA tissue distribution was performed using panels of tissue RNA samples, as well as in situ hybridization of whole mouse sections [14]. These studies revealed that expression occurs primarily in intestinal epithelial cells and specific regions of kidney cortex, with low levels also detected in testes. This is quite different from the ubiquitous expression pattern of the structurally related protein, endotubin [18]. During mouse development, Diet1 mRNA is first detectable in the intestinal primordium during late gestation, and levels increase through postnatal development to reach a maximum in adult intestine. DIET1 mRNA expression is induced more than 100-fold during differentiation of the human Caco-2 intestinal cell line, suggesting a role for Diet1 primarily in mature intestinal epithelial cells [14].
In both mouse intestinal epithelium and cultured intestinal cells, Diet1 protein is localized to puncta within the cytosol [14]. Despite the presence of a predicted membrane-spanning domain, Diet1 has not been detected at enterocyte plasma membranes. This suggests that Diet1 is associated with membranes in an intracellular compartment, although it remains possible that Diet1 traffics to the cell membrane under specific conditions.
Diet1 is a control point in enterohepatic bile acid homeostasis
As described above, the C57BL/6ByJ mice that are Diet1-deficient have elevated serum bile acid levels and enhanced fecal bile acid excretion. Detailed analysis of bile acid levels and composition in the blood, liver and gastrointestinal tract revealed that the bile acid pool size is increased in all of these compartments [14]. Diet1-deficient mice exhibit enhanced expression of key bile acid synthetic enzyme genes including Cyp7a1 and Cyp27. The levels and localization of ASBT, which mediates bile acid uptake in the ileum, were normal in Diet1-deficient mice, indicating that the increased bile acid excretion does not likely result from defective function of this transporter.
In searching for the mechanism by which reduced Diet1 activity in intestine leads to increased hepatic bile acid synthesis, we noted that the expression of genes encoding many proteins with roles in intestinal bile acid regulation and transport were normal, including FXR and SHP, bile acid transporters on apical and basolateral enterocyte membranes, and bile acid binding proteins. However, levels of FGF15 protein was reduced in the ileum of Diet1-deficient mice [14] (Fig. 2A). We hypothesized that Diet1 in the small intestine influences hepatic regulation of bile acid synthesis through effects on FGF15 levels. A number of experiments showed that this is indeed the case, as outlined below.
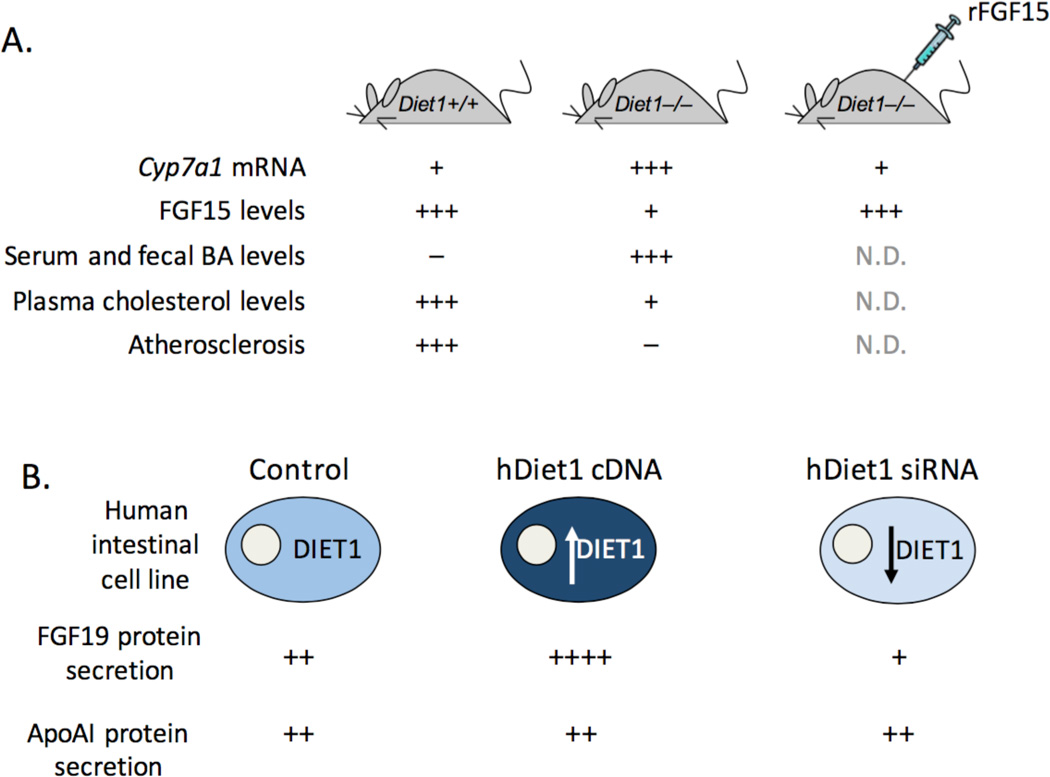
Figure 2.
Modulation of Diet1 levels in mouse and in human intestinal cells alters FGF15/19 protein levels. A. The relative levels of hepatic Cyp7a1 mRNA, ileal FGF15 protein, serum and fecal bile acids (BA), plasma cholesterol levels, and aortic lesion formation in mice fed an atherogenic diet. rFGF15, recombinant adenovirus FGF15 expression vector administered for 6 days. N.D., not determined. B. Effects of human DIET1 expression changes on secretion of FGF19 protein from HT-29 human intestinal cell line. ApoAI, apolipoprotein AI, an irrelevant secreted protein.
To demonstrate that Diet1 impacts hepatic bile acid levels through effects on FGF15, we manipulated Diet1 levels genetically and assessed effects on Fgf15 and Cyp7a1 expression levels [14]. The expression of a Diet1 transgene at low levels in C57BL/6ByJ mice revealed that levels of Diet1 mRNA in intestine are significantly correlated with intestinal Fgf15 mRNA levels, and inversely correlated with hepatic Cyp7a1 levels. We also complemented Diet1-deficient mice with FGF15 by adenovirus expression. After 6 days, the normally elevated Cyp7a1 mRNA levels were dramatically repressed, indicating that the impaired feedback regulation of bile acid synthesis in Diet1-deficient mice can be reversed with FGF15 (Fig. 2A).
The in vivo studies described above suggest that Diet1 has a role in the regulation of enterohepatic bile acid regulation through effects on FGF15. Consistent with this interpretation, a comparison of mouse models with genetic ablation of various components of the enterohepatic signaling axis indicates that Diet1 deficiency has similarities to those with direct disruption of FGF15 or FGF15 hepatic receptors (FGFR4, β-klotho) [reviewed in [19]; see also [12,20]]. All of these models have increased hepatic Cyp7a1 mRNA levels and increased fecal bile acid excretion. Cyp7a1 transgenic mice also have these characteristics, as well as reduced plasma cholesterol levels and resistance to atherosclerosis, as observed in Diet1–deficient mice [21,22].
To complement the in vivo mouse studies, we investigated the effects of Diet1 levels on FGF19 expression and secretion in human intestinal cell lines. We determined that the HT-29 cell line robustly expresses and secretes FGF19. We modulated the levels of human Diet1 by transfection with a human DIET1 expression vector or by inhibition of endogenous DIET1 expression using siRNA. The acute modulation of DIET1 mRNA levels produced a corresponding alteration in FGF19 secretion into the culture medium (Fig. 2B). Thus, increased DIET1 expression caused a 3-fold increase, and partial DIET1 knockdown caused a 40% reduction, in secreted FGF19; no effects were observed on other secreted proteins, such as apolipoprotein AI.
The finding that altering Diet1 expression levels had modest effects on FGF19 mRNA levels, but substantial effects on levels of secreted FGF19 protein, suggested that Diet1 influences FGF15/19 (at least in part) through a post-transcriptional mechanism. This was investigated by modulating the Diet1 levels in cultured cells and quantifying the cellular and secreted FGF19 produced from a heterologous promoter [14]. The use of a heterologous promoter to drive FGF19 expression at a constitutive level allowed the detection of changes in FGF19 levels that occurred in a transcription-independent manner. The results revealed that increased Diet1 levels led to increased levels of both cellular and secreted FGF19 protein. These results are consistent with a role for Diet1 as a determinant of FGF19 protein production/secretion downstream of effects on gene transcription.
Diet1 and FGF15/19 proteins co-localize and interact
Little is known about the itinerary of FGF15/19 protein in enterocytes prior to secretion. The studies described above suggest that Diet1 may influence FGF15/19 protein trafficking and/or secretion from enterocytes. To assess whether Diet1 and FGF15/19 physically interact, we examined the subcellular localization of these proteins in a cultured intestinal cell line. Rat IEC-6 cells were used because they afford a large cytoplasmic volume in which to visualize protein subcellular localization. Diet1 and FGF15 each appeared as punctate structures in the cytoplasm of IEC-6 cells, and a proportion of these puncta co-localized [14]. Furthermore, Diet1 and FGF15 appeared to reside in vesicle-like structures, such as might be found in the endosomal/lysosomal trafficking compartment. However, these structures did not contain markers typical of early or late endosomes (e.g., early endosomal antigen 1, Rab 5, Rab7), nor lysosomes.
Additional evidence of Diet1 and FGF15/19 protein interaction was provided by coimmunoprecipitation of mouse Diet1 and FGF15, as well as human Diet1 and FGF19 [14]. The conservation of the Diet1–FGF15/19 interaction across species is notable because the primary amino acid sequence of FGF15 and FGF19 diverges by about 50% [23]. Taken together, the results available thus far suggest that Diet1 and FGF15 may associate in a unique vesicular compartment in enterocytes, and additional analysis is warranted. Candidates for potential protein components of Diet1-FGF15 vesicles include known players in protein secretion from the basolateral membranes of epithelial cells [24,25].
Does Diet1 impact glucose homeostasis?
A link between bile acid metabolism and type 2 diabetes has been noted by numerous investigators, although the results have not always been consistent [5]. In diabetic patients that do not respond to typical antidiabetic therapeutics (including insulin), treatment with bile acid sequestrants tends to improve glucose levels and whole-body insulin sensitivity [26,27]. Various mechanisms may contribute to the effects of bile acids on glucose homeostasis. Part of the effect may occur though FGF15/19, as FGF15/19 represses gluconeogenesis and stimulates glycogen synthesis in liver [28,29]. As a result, FGF15-deficient mice are glucose intolerant and store only about half the levels of glycogen as wild-type mice [28]. Interestingly, Diet1-deficient mice have somewhat elevated fasting glucose levels, which are reduced by ~25% in response to adenoviral FGF15 complementation [14]. Some evidence suggests that bile acid composition also impacts glucose homeostasis, with an elevated 12α-hydroxylated/non-12α-hydroxylated bile acid ratio associated with insulin resistance [30]. Diet1-deficient mice have a 3-fold elevation in this ratio [14], and therefore may be useful to test whether elevations in 12α-hydroxylated bile acids can drive changes in insulin resistance without confounding factors found in other models, such as obesity and hyperlipidemia. FGF15/19 also stimulates hepatic protein synthesis through a kinase signaling pathway that induces the phosphorylation of ribosomal protein S6 [29]. It will be interesting to determine whether the reduced FGF15 levels in Diet1-deficient mice also influence hepatic protein synthesis.
Potential role of DIET1 polymorphisms or mutations in human disease
A key unanswered question is whether common or rare genetic variants in DIET1 are associated with altered lipid or bile acid homeostasis in humans. As evidenced by data from the 1000 Genomes Project, dozens of potentially damaging missense Diet1 protein variants are present in “healthy” humans. In recent work, we have detected effects of some of these common amino acid variants on the ability of Diet1 to promote FGF19 secretion from cultured cells (our unpublished results). However, the assessment of the significance of such variants in vivo awaits the development of large study samples in which traits such as bile acid levels and FGF19 levels are available. Suggestive genome-wide genetic association signals for the region encompassing the DIET1 gene have been detected for plasma lipid levels. For example, in a Hispanic population, a polymorphism within a DIET1 intron was associated with the ratio of plasma triglyceride to high density lipoprotein cholesterol levels (p = 7.26 E-06) [31].
Aside from common population genetic variants, it is possible that rare DIET1 mutations are associated with disease conditions. It is interesting to speculate, for example, that the case of extreme resistance to hypercholesterolemia in an elderly man who consumed two dozen eggs daily, and had features in common with Diet1-deficient mice, could have been related to DIET1 dysfunction [32]. Unfortunately, DNA samples from the affected individual are not available, and this question will likely never be answered. Diet1-deficient mice also resemble individuals with primary bile acid malabsorption syndrome. Most strikingly, both exhibit reduced FGF15/19 levels, evidence of increased bile acid synthesis, and enhanced fecal bile acid excretion, despite normal intestinal bile acid absorption [33,34]. Efforts are underway to evaluate the potential role of DIET1 mutations in this disorder. Finally, case-control studies of Alzheimer’s disease identified a single nucleotide polymorphism that locates to an intron of the DIET1 gene with lateonset Alzheimer disease [35,36]. It is unclear how Diet1 might influence Alzheimer’s disease pathogenesis, but this potential relationship warrants further exploration.
Acknowledgments
This work was supported by Public Health Service grant P01 HL028481 (K.R.).
References
- 1.Lusis AJ, Attie AD, Reue K. Metabolic syndrome: From epidemiology to systems biology. Nat Rev Genet. 2008;9:819–830. doi: 10.1038/nrg2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charach G, Grosskopf I, Rabinovich A, Shochat M, Weintrabu M, Rabinovich P. The association of bile acid excretion and atherosclerotic coronary artery disease. Therap Adv Gastroenterol. 2011;4:95–101. doi: 10.1177/1756283X10388682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galman C, Angelin B, Rudling M. Pronounced variation in bile acid synthesis in humans is related to gender, hypertriglyceridaemia and circulating levels of fibroblast growth factor 19. J Intern Med. 2011;270:580–588. doi: 10.1111/j.1365-2796.2011.02466.x. [DOI] [PubMed] [Google Scholar]
- 4.Islam KB, Fukiya S, Hagio M, Fujii N, Ishizuka S, Ooka T, Ogura Y, Hayashi T, Yokota A. Bile acid is a host factor that regulates the composition of the cecal microbiota in rats. Gastroenterology. 2011;141:1773–1781. doi: 10.1053/j.gastro.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 5.Prawitt J, Caron S, Staels B. Bile acid metabolism and the pathogenesis of type 2 diabetes. Curr Diab Rep. 2011;11:160–166. doi: 10.1007/s11892-011-0187-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, Angelin B, Hyotylainen T, Oresic M, Backhed F. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring fxr antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 7.Steiner C, Othman A, Saely CH, Rein P, Drexel H, von Eckardstein A, Rentsch KM. Bile acid metabolites in serum: Intraindividual variation and associations with coronary heart disease, metabolic syndrome and diabetes mellitus. PLoS One. 2011;6:e25006. doi: 10.1371/journal.pone.0025006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Aguiar Vallim TQ, Tarling EJ, Edwards PA. Pleiotropic roles of bile acids in metabolism. Cell Metab. 2013;17:657–669. doi: 10.1016/j.cmet.2013.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hofmann AF. The enterohepatic circulation of bile acids in mammals: Form and functions. Front Biosci. 2009;14:2584–2598. doi: 10.2741/3399. [DOI] [PubMed] [Google Scholar]
- 10.Gonzalez FJ. Nuclear receptor control of enterohepatic circulation. Compr Physiol. 2012;2:2811–2828. doi: 10.1002/cphy.c120007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Russell DW. Fifty years of advances in bile acid synthesis and metabolism. J Lipid Res. 2009;50(Suppl):S120–S125. doi: 10.1194/jlr.R800026-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Inagaki T, Choi M, Moschetta A, Peng L, Cummins CL, McDonald JG, Luo G, Jones SA, Goodwin B, Richardson JA, Gerard RD, Repa JJ, Mangelsdorf DJ, Kliewer SA. Fibroblast growth factor 15 functions as an enterohepatic signal to regulate bile acid homeostasis. Cell Metab. 2005;2:217–225. doi: 10.1016/j.cmet.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 13.Kong B, Wang L, Chiang JY, Zhang Y, Klaassen CD, Guo GL. Mechanism of tissue-specific farnesoid x receptor in suppressing the expression of genes in bileacid synthesis in mice. Hepatology. 2012;56:1034–1043. doi: 10.1002/hep.25740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vergnes L, Lee JM, Chin RG, Auwerx J, Reue K. Diet1 functions in the fgf15/19 enterohepatic signaling axis to modulate bile acid and lipid levels. Cell Metab. 2013;17:916–928. doi: 10.1016/j.cmet.2013.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mouzeyan A, Choi J, Allayee H, Wang X, Sinsheimer J, Phan J, Castellani LW, Reue K, Lusis AJ, Davis RC. A locus conferring resistance to diet-induced hypercholesterolemia and atherosclerosis on mouse chromosome 2. J Lipid Res. 2000;41:573–582. [PubMed] [Google Scholar]
- 16.Phan J, Pesaran T, Davis RC, Reue K. The diet1 locus confers protection against hypercholesterolemia through enhanced bile acid metabolism. J Biol Chem. 2002;277:469–477. doi: 10.1074/jbc.M107107200. [DOI] [PubMed] [Google Scholar]
- 17.Reue K, Vergnes L. Approaches to lipid metabolism gene identification and characterization in the postgenomic era. J Lipid Res. 2006;47:1891–1907. doi: 10.1194/jlr.R600020-JLR200. [DOI] [PubMed] [Google Scholar]
- 18.McCarter SD, Johnson DL, Kitt KN, Donohue C, Adams A, Wilson JM. Regulation of tight junction assembly and epithelial polarity by a resident protein of apical endosomes. Traffic. 2010;11:856–866. doi: 10.1111/j.1600-0854.2010.01052.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reue K, Lee JM, Vergnes L. Regulation of bile acid homeostasis by the intestinal diet1-fgf15/19 axis. Curr Opin Lipidol. 2014;25:140–147. doi: 10.1097/MOL.0000000000000060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu C, Wang F, Kan M, Jin C, Jones RB, Weinstein M, Deng CX, McKeehan WL. Elevated cholesterol metabolism and bile acid synthesis in mice lacking membrane tyrosine kinase receptor fgfr4. J Biol Chem. 2000;275:15482–15489. doi: 10.1074/jbc.275.20.15482. [DOI] [PubMed] [Google Scholar]
- 21.Li T, Matozel M, Boehme S, Kong B, Nilsson LM, Guo G, Ellis E, Chiang JY. Overexpression of cholesterol 7alpha-hydroxylase promotes hepatic bile acid synthesis and secretion and maintains cholesterol homeostasis. Hepatology. 2011;53:996–1006. doi: 10.1002/hep.24107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miyake JH, Duong-Polk XT, Taylor JM, Du EZ, Castellani LW, Lusis AJ, Davis RA. Transgenic expression of cholesterol-7-alpha-hydroxylase prevents atherosclerosis in c57bl/6j mice. Arterioscler Thromb Vasc Biol. 2002;22:121–126. doi: 10.1161/hq0102.102588. [DOI] [PubMed] [Google Scholar]
- 23.Wright TJ, Ladher R, McWhirter J, Murre C, Schoenwolf GC, Mansour SL. Mouse fgf15 is the ortholog of human and chick fgf19, but is not uniquely required for otic induction. Dev Biol. 2004;269:264–275. doi: 10.1016/j.ydbio.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 24.Carmosino M, Valenti G, Caplan M, Svelto M. Polarized traffic towards the cell surface: How to find the route. Biol Cell. 2010;102:75–91. doi: 10.1042/BC20090134. [DOI] [PubMed] [Google Scholar]
- 25.He B, Guo W. The exocyst complex in polarized exocytosis. Curr Opin Cell Biol. 2009;21:537–542. doi: 10.1016/j.ceb.2009.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Beysen C, Murphy EJ, Deines K, Chan M, Tsang E, Glass A, Turner SM, Protasio J, Riiff T, Hellerstein MK. Effect of bile acid sequestrants on glucose metabolism, hepatic de novo lipogenesis, and cholesterol and bile acid kinetics in type 2 diabetes: A randomised controlled study. Diabetologia. 2012;55:432–442. doi: 10.1007/s00125-011-2382-3. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz SL, Lai YL, Xu J, Abby SL, Misir S, Jones MR, Nagendran S. The effect of colesevelam hydrochloride on insulin sensitivity and secretion in patients with type 2 diabetes: A pilot study. Metab Syndr Relat Disord. 2010;8:179–188. doi: 10.1089/met.2009.0049. [DOI] [PubMed] [Google Scholar]
- 28.Kir S, Beddow SA, Samuel VT, Miller P, Previs SF, Suino-Powell K, Xu HE, Shulman GI, Kliewer SA, Mangelsdorf DJ. Fgf19 as a postprandial, insulinindependent activator of hepatic protein and glycogen synthesis. Science. 2011;331:1621–1624. doi: 10.1126/science.1198363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Potthoff MJ, Boney-Montoya J, Choi M, He T, Sunny NE, Satapati S, Suino-Powell K, Xu HE, Gerard RD, Finck BN, Burgess SC, Mangelsdorf DJ, Kliewer SA. Fgf15/19 regulates hepatic glucose metabolism by inhibiting the creb-pgc-1alpha pathway. Cell Metab. 2011;13:729–738. doi: 10.1016/j.cmet.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haeusler RA, Astiarraga B, Camastra S, Accili D, Ferrannini E. Human insulin resistance is associated with increased plasma levels of 12alpha-hydroxylated bile acids. Diabetes. 2013;62:4184–4191. doi: 10.2337/db13-0639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Comuzzie AG, Cole SA, Laston SL, Voruganti S, Haack K, Gibbs RA, Butte NF. Novel genetic loci identified for the pathophysiogy of childhood obesity in the hispanic population. PLoS One. 2012;12:e51954. doi: 10.1371/journal.pone.0051954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kern F., Jr Normal plasma cholesterol in an 88-year-old man who eats 25 eggs a day. Mechanisms of adaptation. N Engl J Med. 1991;324:896–899. doi: 10.1056/NEJM199103283241306. [DOI] [PubMed] [Google Scholar]
- 33.Pattni SS, Brydon WG, Dew T, Walters JR. Fibroblast growth factor 19 and 7-alpha-hydroxy-4-cholesten-3-one in the diagnosis of patients with possible bile acid diarrhea. Clin Transl Gastroenterol. 2012;3:e18. doi: 10.1038/ctg.2012.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walters JR, Tasleem AM, Omer OS, Brydon WG, Dew T, le Roux CW. A new mechanism for bile acid diarrhea: Defective feedback inhibition of bile acid biosynthesis. Clin Gastroenterol Hepatol. 2009;7:1189–1194. doi: 10.1016/j.cgh.2009.04.024. [DOI] [PubMed] [Google Scholar]
- 35.Majores M, Bagli M, Papassotiropoulos A, Schwab SG, Jessen F, Rao ML, Maier W, Heun R. Allelic association between the d10s1423 marker and alzheimer's disease in a german population. Neurosci Lett. 2000;289:224–226. doi: 10.1016/s0304-3940(00)01283-0. [DOI] [PubMed] [Google Scholar]
- 36.Zubenco GS, Hughes HB, 3rd, Stiffler JS. D10s1423 identifies a susceptibility locus for alzheimer's disease in a prospective, longitudinal, double-blind study of asymptomatic individuals. Mol Psychiatry. 2001;6:413–419. doi: 10.1038/sj.mp.4000900. [DOI] [PubMed] [Google Scholar]