Abstract
Quantitative trait loci conferring high grain yield under drought in rice are important genomic resources for climate resilient breeding. Major and consistent drought grain yield QTLs usually co-locate with flowering and/or plant height QTLs, which could be due to either linkage or pleiotropy. Five mapping populations used for the identification of major and consistent drought grain yield QTLs underwent multiple-trait, multiple-interval mapping test (MT-MIM) to estimate the significance of pleiotropy effects. Results indicated towards possible linkages between the drought grain yield QTLs with co-locating flowering and/or plant height QTLs. Linkages of days to flowering and plant height were eliminated through a marker-assisted breeding approach. Drought grain yield QTLs also showed interaction effects with flowering QTLs. Drought responsiveness of the flowering locus on chromosome 3 (qDTY3.2) has been revealed through allelic analysis. Considering linkage and interaction effects associated with drought QTLs, a comprehensive marker-assisted breeding strategy was followed to develop rice genotypes with improved grain yield under drought stress.
Introduction
Genetic linkages and interactions are the two most important aspects accounting for the complexity of quantitative traits such as drought tolerance in rice. Such issues could not be efficiently addressed through a classical Mendelian approach [1]. However, recent advances in molecular marker applications have enabled plant breeders to better understand the genetic basis of complex traits [2,3,4,5]. The genetic basis of drought tolerance in rice, a model crop species, has been examined through mapping and fine mapping of QTLs. Major-effect drought grain yield QTLs have now been identified, but the use of tightly linked markers is still a limitation because of their co-location with other loci that may be undesirable for a plant breeder. For example, the major and consistent drought grain yield (GY) QTLs that can significantly enhance grain yield under reproductive-stage drought stress (RS) usually coincide with QTLs for plant height (PH) and/or flowering (DTF). The most consistent drought GY QTL identified so far, qDTY1.1, co-located with QTLs related to PH and DTF under stress. Interestingly, qDTY1.1 harbours green revolution gene 'sd1', and it has long been debated whether linkages or pleiotropic effects are associated with this gene [6,7,8,9,10]. Positive allele of qDTY1.1 has been reported by upland adapted traditional cultivars N22 and Dhagaddesi [9,11]. Another major QTL, qDTY3.1, identified for lowland drought stress co-located with QTLs for DTF. This QTL was also contributed by an upland adapted variety ‘Apo’ [12]. The largest effect drought GY QTL reported to date, qDTY12.1, also co-located with QTLs for DTF, PH, and some other drought-related traits such as biomass, harvest index, panicle number, and drought response index [13]. qDTY12.1 was identified in Vandana/Way Rarem population, both parents are upland adapted cultivars, positive allele being contributed by Way Rarem. Similar to the drought GY loci, other drought QTLs also co-locate with the regions governing different drought-related traits. qDTY3.2 is one such example. This locus, known as ‘HD9,’ was first identified as a major flowering locus [14] later on DTF, PH and some other drought related traits positive allele being contributed from Vandana [13]. This QTL is recently reported to be associated with GY under drought [9] and markers underlying this QTL region were reported to interact with drought GY QTLs [15]. Characterization of qDTY3.2 is therefore important to understand its genetic and genomic basis for practical breeding applications.
From a plant breeder’s perspective transfer of a DTH/PH loci collocating with drought GY loci may not be a preferred phenomenon and is a common phenomenon due positive allele contribution from tall and early donor genotypes. As an example, for rain-fed lowland situation medium to long duration variety with semi-dwarf height is preferred apart from its tolerance to drought. Co-location of the drought-related QTLs could be for three possible reasons: genetic linkage, pleiotropic effects, and cause-and-effect association [16]. From among these three possibilities, linkages between genes for individual traits or pleiotropic effects of genes affecting multiple traits are more commonly supported [17,18]. Recently, efforts have been made to characterize these co-locations for some rice drought GY QTLs. Covariate analysis has been carried out to remove the confounding effect of DTF on GY under drought [9,12].
Main and interaction effects of QTLs are usually estimated through composite interval mapping (CIM) models that consider a single trait at a time, but this approach does not allow an evaluation of pleiotropic effects. Multiple-trait mapping methods offer a powerful alternative approach for testing pleiotropic effects of the QTLs and therefore help in increasing the accuracy of mapping correlated traits [19]. Distinguishing linkage from pleiotropy is considered to be a difficult task requiring a large population of segregating individuals, greater marker saturation in the QTL region, and suitable multiple-trait analysis methods [19]. Despite these challenges, testing genetic linkage versus pleiotropy is recommended prior to QTL application in marker-assisted breeding (MAB). Considering these challenges, a suitable MAB strategy for drought tolerance in rice needs to be defined. In view of this the objectives of our study were framed which include- (1) testing the significance of pleiotropism for three major drought GY QTLs (qDTY1.1, qDTY3.1, and qDTY12.1); (2) validation of the qDTY3.2 effect on DTF and GY under RS and analysis of its interaction effects with other QTLs for GY under drought (3) marker-assisted linkage elimination from the drought GY QTLs- qDTY1.1, qDTY3.1, and qDTY12.1.
Results
Test of pleiotropism of the drought GY QTLs through multiple-trait mapping
MT-MIM was carried out for three QTLs in five populations to determine the LOD of the pleiotropic effects of qDTY1.1, qDTY3.1, and qDTY12.1. GY QTLs were analyzed in pairs with DTF/PH QTLs for each of the three regions and LOD, and the significance of the pleiotropic effects was determined. The LOD of pleiotropic effects for the GY-PH QTL pair in qDTY1.1 ranged from 0.7 to 2.4 and from 1.3 to 2.1 for the GY-DTF QTL pair in the same region. None of the LOD values for GY-PH and GY-DTF for different QTLs were significant (Table 1).
Table 1. Multiple-trait, multiple-interval mapping (MT-MIM) to test the pleiotropic effect of drought grain yield QTLs on flowering and plant height.
| QTL name | Population | Marker interval | Trait 1 | Trait 2 | Season | LOD of pleiotropy effect | LOD threshold |
|---|---|---|---|---|---|---|---|
| qDTY1.1 | N22/Swarna | RM11943—RM12091 | GY | DTF | Year 1 | 1.4 | 2.4 |
| Year 2 | 1.5 | 2.1 | |||||
| GY | PHT | Year 1 | 1.7 | 2.3 | |||
| Year 2 | 0.7 | 2.1 | |||||
| N22/IR64 | RM11943—RM12091 | GY | DTF | Year 1 | 1.3 | 2.5 | |
| Year 2 | 2.1 | 2.4 | |||||
| GY | PHT | Year 1 | 2.4 | 2.7 | |||
| Year 2 | 0.7 | 2.2 | |||||
| qDTY3.1 | Apo/2*Swarna | RM520—RM16030 | GY | DTF | Year 1 | 1.3 | 2.7 |
| Year 2 | 1.9 | 3.1 | |||||
| GY | PHT | Year 1 | 1.1 | 2.6 | |||
| Year 2 | 0.7 | 2.5 | |||||
| qDTY12.1 | Vandana/Way Rarem | RM28048—RM511 | GY | DTF | Year 1 | 0.7 | 4.2 |
| Year 2 | 1.2 | 13.3 | |||||
| GY | PHT | Year 1 | 1.5 | 3.3 | |||
| Year 2 | 2.6 | 9.3 | |||||
| IR74371-46-1-1/2*Sabitri | RM28166—RM28199 | GY | DTF | Year 1 | 1.7 | 6.2 | |
| Year 2 | 0.6 | 13.2 | |||||
| GY | PHT | Year 1 | 0.4 | 3.9 | |||
| Year 2 | 0.6 | 12.6 |
LOD threshold was significant at 1%.
Characterization of the flowering locus for co-localized drought related traits
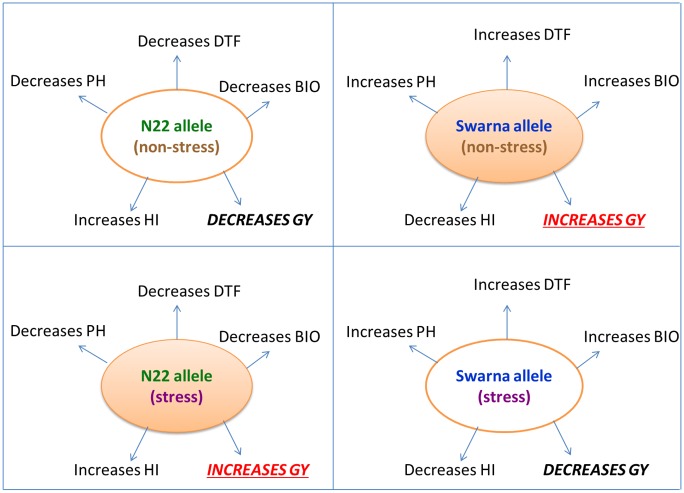
The flowering locus qDTY3.2 showed significant effects for GY and DTF in the N22/Swarna BIL population under reproductive-stage drought stress (RS) drought and non-stress (NS) situations of two seasons, DS2011 and DS2012. QTL effects have been presented as additive effects percentage, a measure which is more practical compared to other measures. This was estimated as the percentage of population mean contributed by QTL [Additive effect/population mean × 100]. Additive effects were contributed by N22 and Swarna (denoted as ‘-’ sign in Table 2). Additive effects, F-values and allele contributions have been presented in a separate column in Table 2. Increases in DTF, PH, and BIO under RS and NS were contributed by Swarna, whereas N22 contributed the HI-increasing allele under RS as well as NS for qDTY3.2. Interestingly, enhancement of GY under RS came from N22, whereas it came from Swarna under NS, indicating the drought responsiveness of qDTY3.2. The effect of the N22 and Swarna alleles in the qDTY3.2 region was similar in the N22/Swarna BIL and N22/Swarna RIL populations (Table 2, Fig 1).
Table 2. Effect of qDTY3.2 for high grain yield under drought in N22/Swarna BIL and RIL populations.
| F-value | Additive effect % | Allele contribution | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Season | Season1 | Season2 | Season1 | Season2 | Season1 | Season2 | ||||||
| Environment | Non-stress | Stress | Non-stress | Stress | Non-stress | Stress | Non-stress | Stress | Non-stress | Stress | Non-stress | Stress |
| N22/Swarna BIL population | ||||||||||||
| GY | 13.7 | 27.1 | 29.8 | 42.6 | -17.1 | 23.9 | -22.8 | 49.2 | Swarna | N22 | Swarna | N22 |
| DTF | 60.7 | 85.1 | 62.8 | 52.6 | -14.8 | -15.6 | -8.9 | -10.1 | Swarna | Swarna | Swarna | Swarna |
| PH | 26.9 | 46.7 | 14.5 | 5.2 | -5.3 | -5.8 | -4.3 | -2.9 | Swarna | Swarna | Swarna | Swarna |
| BIO | 18.9 | 4.6 | NA | NA | -14.1 | -9.1 | NA | NA | Swarna | Swarna | NA | NA |
| HI | 5.7 | 42.8 | NA | NA | 7.7 | 35.3 | NA | NA | N22 | N22 | NA | NA |
| N22/Swarna RIL population | ||||||||||||
| GY | 3.70 NS | 3.16 NS | 7.8 | 17.1 | -3.4 | 3.4 | -6.8 | 13.6 | Swarna | N22 | Swarna | N22 |
| DTF | 125.5 | 135.5 | 113.2 | 79.6 | -5.3 | -8 | -5.6 | -4 | Swarna | Swarna | Swarna | Swarna |
| PH | 12.5 | 6.7 NS | 10.9 | 0.5NS | -3.9 | -1.1 | -3.5 | -1.9 | Swarna | Swarna | Swarna | Swarna |
| BIO | 58.7 | 24.6 | 39.3 | 0.8 NS | -20.5 | -10.2 | -12 | -0.9 | Swarna | Swarna | Swarna | Swarna |
| HI | 36.4 | 13.5 | 8.2 | 73.6 | 10.7 | 11.1 | 3.8 | 19.9 | N22 | N22 | N22 | N22 |
NA: not available, BIO and HI were recorded in only one season in N22/Swarna BIL population.
NS: non-significant. F-value: represent the significance of QTLs and their effects. Additive effect (%): Additive effect / Population mean × 100. Allele contribution: represents the parents contributing positive allele for effect of QTL.
Fig 1. Figure representing the effect of N22 and Swarna alleles at qDTY3.2 locus.
GY increase under RS due to N22 allele and under NS due to Swarna allele confirms the drought responsiveness of the qDTY3.2 locus.
Interaction analysis of drought related grain yield and flowering loci
qDTY1.1 showed a significant epistatic interaction with qDTY3.2 for DTF in two seasons (DS2009 and DS2010) under RS in the N22/RIL population. Both loci had a main effect on DTF and there was an additive-additive interaction between the two loci (Table 3, S1 Fig). Also in a N22/Swarna BIL population additive-additive interaction between qDTY1.1 and qDTY3.2 was observed under RS in the DS2012 experiment (data not presented). Consistent results were therefore obtained in N22 derived RIL and BIL populations. qDTY3.2 also showed an additive interaction with another drought GY QTL qDTY12.1 [15]. Therefore to better understand the effect of flowering locus qDTY3.2 on GY loci qDTY1.1 and qDTY12.1 a class analysis was carried out. QTL classes, including individual QTLs (qDTY1.1, qDTY12.1, and qDTY3.2) as well as QTL combinations (qDTY1.1 + qDTY3.2 and qDTY12.1 + qDTY3.2), were analyzed for their effect on GY and DTF under RS and NS. Results have been presented in S2 and S3 Figs. Though significant differences between different classes were not observed, a common trend was seen among three classes. Trait values (DTF or GY) of QTL combination classes showed that qDTY3.2 interact with qDTY1.1 and qDTY12.1 to reduce DTF and enhance GY under RS in different populations. Similarly, under NS, lines with qDTY1.1 / qDTY12.1 and qDTY3.2 (qDTY1.1 / qDTY12.1 + qDTY3.2) had lower yield as compared to the lines with only qDTY1.1 / qDTY12.1.
Table 3. qDTY3.2 main and interaction effect analysis for flowering in N22/IR64 RIL population.
| Season | QTL interval 1 | QTL interval 2 | AA | AE% | P-value |
|---|---|---|---|---|---|
| DS2009 | RM11943-RM431 (qDTY1.1) | RM22-RM231 (qDTY3.2) | -0.8 | -1.2 | 0.003 |
| DS2010 | RM11943-RM431 (qDTY1.1) | RM22-RM231 (qDTY3.2) | -1.3 | -1.6 | 0.005 |
DS2009: dry season of 2009; DS2010: dry season of 2010. AA: additive-additive interaction effect of two QTLs (qDTY1.1 and qDTY3.2). AE%: additive effect as the percent of trial mean.
Linkage elimination from drought GY QTLs
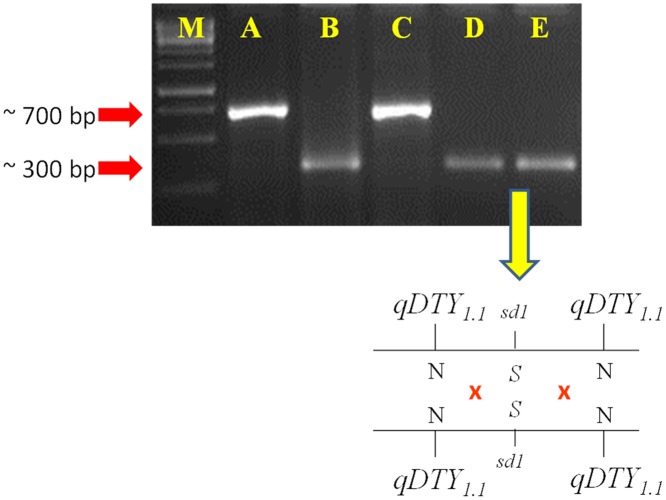
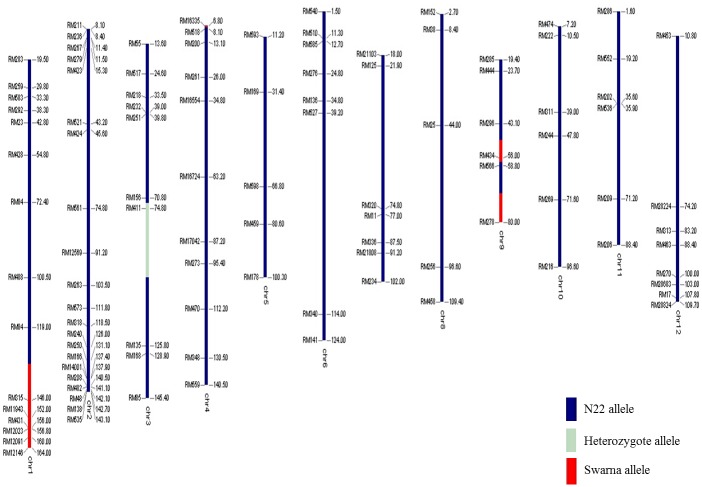
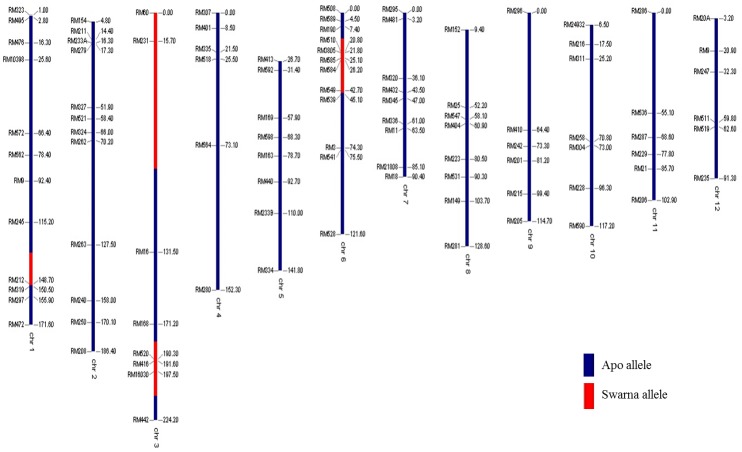
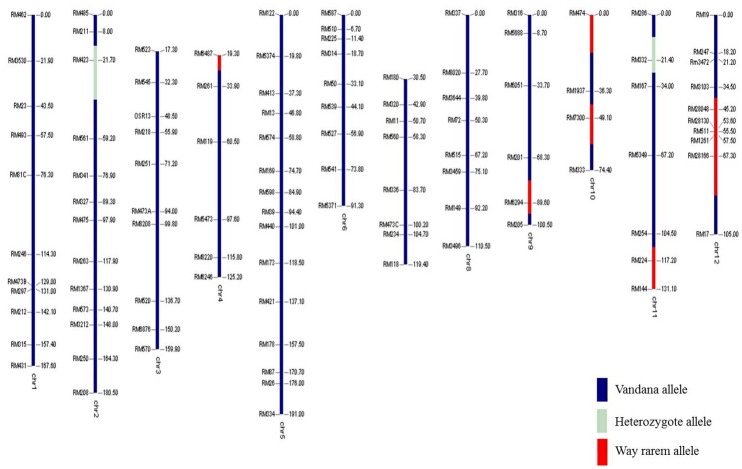
The performance of linkage-eliminated lines under RS is provided in Table 4. The qDTY3.1 Apo/Swarna BILs (IR88287-524-1-B-B and IR88287-63-1-B-B) showed significant GY increases under RS in two seasons of study. IR88287-524-1-B-B and IR88287-63-1-B-B, respectively, showed 344% and 386% increases over Swarna in the first season and 100% and 164% increases over Swarna in the second season under RS (Table 4). The qDTY12.1 lines IR84984-83-15-332-B and IR84984-83-15-817-B, respectively, showed GY enhancement of 349% and 109% over Vandana in the first experiment and 383% and 194% over Vandana in the second experiment under RS (Table 4). The dwarf qDTY1.1 N22/Swarna lines IR91659-41-59-B and IR91659-54-17-B showed a double cross-over event around sd1 gene and significant GY increases of more than 300% in two experiments conducted in two different environments (Fig 2, Table 4). Genotypes developed through MAB recovered over 85% of the recipient parent genome (Figs 3, 4 and 5).
Table 4. Means (±SED) of QTL lines developed through marker-assisted breeding for high grain yield under drought.
| QTL | Entry | Stress 1 | Stress 2 | Non-stress | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| DTF | PHT | GY | DTF | PHT | GY | DTF | PHT | GY | ||
| qDTY3.1 | IR88287-524-1-B-B | 108± 4* | 61± 4* | 1511± 360* | 104± 1* | 64± 3* | 772± 202* | 102± 4* | 81± 4* | 5358± 873NS |
| qDTY3.1 | IR88287-63-1-B-B | 104± 4* | 62± 4* | 1654± 360* | 100± 1* | 66± 3* | 1018± 202* | 96± 4* | 73± 4* | 4334± 873NS |
| Swarna | 109± 4* | 62± 4* | 340± 360* | 109± 1* | 62± 3* | 385± 202* | 103± 4* | 82± 4* | 4121± 873NS | |
| qDTY12.1 | IR84984-83-15-332-B | 66± 3* | 76± 4* | 691± 175* | 70± 3* | 70± 4* | 348± 73* | 57± 1.4* | 126± 4.6NS | 3747± 744* |
| qDTY12.1 | IR84984-83-15-817-B | 64± 3* | 71± 4* | 322± 175* | 72± 3* | 61± 4* | 212± 73* | 56± 1.4* | 118± 4.6NS | 4130± 744* |
| Vandana | 69± 3* | 66± 4* | 154± 175* | 70± 3* | 64± 4* | 72± 73* | 54± 1.4* | 120± 4.6NS | 3556± 744* | |
| qDTY1.1 | IR91659:41-59-B | 100± 3* | 70± 5NS | 2475± 645* | 89± 3* | 63± 3* | 386± 28* | 89± 11NS | 97± 6NS | 4428± 113NS |
| qDTY1.1 | IR91659:54-17-B | 93± 3* | 75± 5NS | 2440± 645* | 90± 3* | 71± 3* | 383± 28* | 82± 11NS | 98± 6NS | 4222± 113NS |
| Swarna | 98± 3* | 75± 5NS | 553± 645* | N | N | 0 | 101± 11NS | 91± 6NS | 4778± 113NS | |
Stress 1 and Stress 2 correspond to the two different experiments. qDTY3.1 lines were screened in two different seasons whereas qDTY1.1 and qDTY12.1 lines were screened in only one season. qDTY1.1 lines were screened in two different environments (stress 1 = field conditions and stress 2 = rainout shelter). qDTY12.1 lines were screened on two different dates at the same location (stress 1 = first date and stress 2 = second date). Improved genotypes can be compared with their respective parents for each QTL (qDTY3.1, qDTY12.1 and qDTY1.1). DTF: days; PHT: cm, GY: kg ha–1. N: Swarna did not flower under a rainout shelter (severe stress).
*: Significance at 1%;
NS: Non-significant difference;
Fig 2. Figure representing the double cross-over event within qDTY1.1 QTL region in two N22/Swarna BILs.
The semi-dwarfing gene, ‘sd1’ lies within qDTY1.1 region. BILs had dwarfing allele of the sd1 gene whereas N22 alleles on its both sides. M = 1Kb Ladder, A = N22, B = Swarna, C = IR 91659:25-50-B (Tall but drought susceptible N22/Swarna BIL), D = IR91659:41-59-B and E = IR91659:54-17-B (Dwarf drought tolerant qDTY1.1 N22/Swarna BILs).
Fig 3. Genotypic background of qDTY1.1 BILs.
Fig 4. Genotypic background of qDTY3.1 BILs.
Fig 5. Genotypic background of qDTY12.1 BIL.
Discussion
Molecular markers associated with drought resistance in plants offer a promise of increased selective gain to ensure sustainable targeted breeding progress [20]. Marker-assisted breeding using major drought GY QTLs in rice is a fast-track alternative approach to improve yield under drought [21,22]. Major drought GY QTLs that are potential candidates for an efficient drought molecular breeding program usually co-locate with other QTLs for traits such as DTF and PH, suggesting possible linkage or pleiotropy effects [9,12,13]. In addition, interactions of DTF and GY QTLs are highly likely because of the drought escape mechanism associated with DTF QTLs. Our study was conducted to test the pleiotropic effects of major drought GY QTLs, characterize interaction effects of drought GY and DTF QTLs, and finally define a molecular breeding strategy suitable for high GY under RS in rice.
The MT-MIM approach is being followed to test the significance of pleiotropic effects of the co-located QTLs in a particular chromosomal region [23,24]. The LOD values of pleiotropic effects obtained during the MT-MIM test with three drought GY QTLs were not significant (Table 1), clearly suggesting that there is no significant pleiotropic effect. Hence, there could be possible linkage between DTF/PH QTLs (or genes) and drought GY QTLs (or genes) co-locating in the same genomic region. Since the three drought GY QTLs (qDTY1.1, qDTY3.1, and qDTY12.1) analysed in our study co-located with QTLs for DTF under RS, it is worth looking into their effect under NS conditions. A QTL is considered as a flowering locus if it significantly affects DTF under both RS and NS conditions. Notably, qDTY1.1, qDTY3.1, and qDTY12.1 didn’t affect DTF under NS. Second, qDTY1.1 and qDTY3.1 significantly enhanced GY under RS even after covariate adjustment of DTF [9,12]. qDTY12.1 was identified in a subset of the population (242 lines out of 436) in which the effect of DTF was minimized [13]. Third, the MT-MIM test carried out in our study also supports possible linkage instead of pleiotropy.
Similar to DTF, the above-mentioned three drought GY QTLs co-located with PH QTLs under RS. Two of them (qDTY3.1 and qDTY12.1) were not associated with PH under NS, clearly indicating that the increase in PH was in response to drought. Second, these QTLs did not show a significant pleiotropic effect on GY and PH (Table 1). Unlike qDTY3.1 and qDTY12.1, qDTY1.1 was significantly associated with PH under NS. Contradictorily, in a CT9993/IR86626 population, qDTY1.1 showed a significant association with GY under RS but not with PH [7]. In three N22-derived RIL populations, Vikram et al. [25] have successfully proven that qDTY1.1, is linked with the sd1 gene. The qDTY1.1 BILs developed in present study (IR91659:41-59-B and IR91659:54-17-B) had PH comparable with that of Swarna under both RS and NS conditions, suggesting a double crossover event at this locus (Table 4). Further analysis of these two lines with the sd1 gene based marker confirmed the occurrence of double cross-over event (Fig 2).
In addition to linkages, genomic interactions play a crucial role in a drought MAB program. Digenic interactions have been reported to affect drought-related traits significantly [26]. Another recent example is the digenic interaction of a drought QTL on chromosome 3 (qDTY3.2) with another one on chromosome 12 (qDTY12.1) for GY under RS [15]. qDTY3.2 is a well-known flowering locus identified to show a digenic interaction with qDTY1.1 for DTF under RS in an N22/IR64 RIL population (Table 3). The digenic interaction of qDTY3.2 and qDTY1.1 was also observed in an N22/Swarna BIL population in DS2012. The N22 allele contributed to the reduction in DTF under RS in the two N22-derived populations. Interestingly, N22 alleles at qDTY3.2 and qDTY1.1 loci reduced DTF in the N22/Swarna RIL population, but no interaction was reported [9]. To better understand the effect of qDTY3.2 on two other drought GY QTLs, a QTL class analysis was carried out. The QTL class analysis of qDTY3.2, qDTY1.1, and qDTY12.1 revealed that qDTY3.2 interacts with other drought GY QTLs, resulting in decreased DTF under RS, consequently increasing GY under RS. On the other hand, GY under NS declines when DTY QTLs interact with qDTY3.2 (S2 and S3 Figs). To better understand the genetic effect of qDTY3.2 QTL, marker alleles at this locus was analyzed in an N22/Swarna BIL population and compared with results of a previous study on the N22/Swarna RIL population by Vikram et al. [9] where this QTL was first reported for high GY under RS. Reduction in DTF under RS and NS was due to the N22 allele. However, for GY under RS and NS, N22 and Swarna alleles had different effects. Under RS situation the N22 allele contributed for a GY enhancement whereas under NS situation Swarna allele contributed for the same. GY enhancement from the N22 allele under RS and from the Swarna allele under NS demonstrates that qDTY3.2 is a drought-responsive locus (Table 2, Fig 1). Another important note would be the effects of N22 and Swarna alleles on HI and BIO. The N22 allele increased HI and decreased BIO under both RS and NS situations. Similarly, Swarna allele decreased HI and increased BIO under RS as well as NS situations. GY increase under RS with the effect of N22 allele is likely to be due to mobilization of solutes from source to sink under RS. GY increase under NS with effect of the Swarna allele could be due the overall increase in BIO of plant. Drought-responsive and interacting loci such as qDTY3.2 should be pyramided with other desirable QTLs in a drought MAB program. Pyramiding can be followed for the development of relatively short-duration varieties (with minimum grain yield compromise), which are preferred for rainfed upland conditions.
The significant association of drought-related traits GY, PH, and DTF under RS in three important genomic regions (qDTY1.1, qDTY3.1, and qDTY12.1) is therefore due to their linkage effect, and this linkage need to be broken prior to their application in MAB. Practically, elimination of these linkages of PH and DTF from DTY QTLs could be carried out at the BCnF2 (n = 2/3) stage, that is, during selection of QTL homozygotes. There are background effects at the BCnF2 stage in addition to these linkages because of unidentified introgressions and unknown genomic interactions. A MAB strategy was followed to remove the effect of unwanted linkages, interactions and other background effects at the BCnF2 (n = 2/3) stage (Fig 6). Following this strategy dwarf qDTY1.1 Swarna lines with plant type and grain type similar to those of Swarna were developed showing more than a 300% yield advantage over Swarna under drought stress. Similarly, Swarna and Vandana introgression lines with qDTY3.1 and qDTY12.1 were also developed (Table 4).
Fig 6. Figure presents the five important steps in marker-assisted breeding for high grain yield under drought in rice through major drought grain yield QTLs- qDTY1.1, qDTY12.1 and qDTY3.1. Figure also represents a separate strategy for elimination of linkage of the plant height gene from qDTY1.1 (sd1 gene locus).
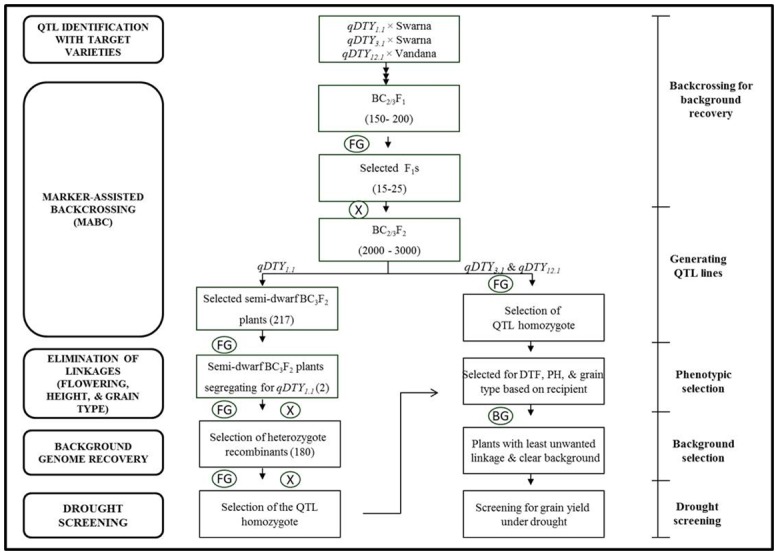
FG = Genotyping with fore ground markers; BG = Genotyping for recipient background recovery.
Varying QTL effects in different backgrounds, unwanted linkages, and genomic interactions are the three most probable impediments in the deployment of drought QTLs in MAB programs. Drought GY QTLs offer promise of a selective advantage in drought MAB. The three major drought GY QTLs analyzed in our study through MT-MIM indicated possible linkages among PH, DTF, and GY genes/QTLs, which were eliminated during MAB. Interaction effects of DTF QTLs such as qDTY3.2 with qDTY1.1 and with qDTY12.1 in a previous study by Dixit et al. [15] suggested that these interacting loci also play a strategic role in a MAB program. It is advisable to use this information when defining a suitable MAB strategy for high GY under RS. Considering the linkage-interaction analysis of DTY QTLs in this study a suitable MAB protocol can be formulated with following steps: (1) QTLs for GY under RS could be identified in populations with target varieties in multiple populations simultaneously to identify major drought GY QTLs showing a significant effect across multiple, elite, and preferred genetic backgrounds [9]; (2) Co-locating QTLs could be tested for linkage vs pleiotropy for co-locating QTLs associated with phenotypically correlated traits; (3) Linkages of DTF and PH could be removed at the BC2/3F2 stage using a large population. In our study, linkages from the three drought QTLs (qDTY1.1, qDTY3.1, and qDTY12.1) were broken and drought-tolerant lines were developed; (4) The background of QTL homozygote lines could be tested genotypically in order to select lines with a higher representation of the recurrent parent genome; (5) Finally, screening of selected lines for GY under RS could be carried out to take advantage of other unidentified QTLs, genomic interactions, and/or irreversible epigenetic mechanisms (Fig 6). Such a modified MAB strategy would be advantageous in developing drought-tolerant rice varieties with a desired duration and increased grain yield under drought.
Conclusion
Major and consistent drought GY QTLs qDTY1.1, qDTY3.1 and qDTY12.1 collocate with DTF and PH QTLs and this collocation is not due to pleiotropic effect, thereby indicating towards possible linkage. Allelic analysis of the qDTY3.2 revealed that it is a drought responsive locus. qDTY3.2 interacts qDTY1.1 and qDTY12.1 for reduction in flowering duration. A MAB strategy was followed for high grain yield under drought which involved elimination of the unwanted linkages and development of lines similar to recurrent parent. This MAB strategy could be followed for the development of drought tolerant lines with desired characters of recipient parents. Genetic improvement of the popular varieties which are susceptible to drought could be followed through this approach.
Materials and Methods
Our study included a multiple-trait multiple-interval mapping (MT-MIM) analysis using phenotypic and genotypic data of populations in which qDTY1.1, qDTY3.1, and qDTY12.1 were originally identified [9,12,13]. Validation of the interacting locus, qDTY3.2, was carried out in N22-derived RILs and BILs. Further, epistatic interaction of qDTY3.2 with qDTY1.1 and qDTY12.1 was evaluated through a QTL class analysis to better understand the interaction of DTF and GY QTLs under RS and NS conditions. Finally, marker-assisted linkage elimination for all three QTLs was carried out.
Mapping population data for testing pleiotropic effect of drought GY QTLs
Phenotypic and genotypic data of five mapping populations originally used for the QTL identification study were subjected to a test of pleiotropism. These populations were N22/Swarna, N22/IR64, Vandana/Way Rarem (RIL populations), and Apo/2*Swarna and IR74371-46-1-1/2*Sabitri (BIL populations). qDTY1.1 was identified in N22/Swarna and N22/IR64 [9], qDTY3.1 in Apo/2*Swarna [12], and qDTY12.1 in Vandana/Way Rarem [13] and IR74371-46-1-1/2*Sabitri [27].
MT-MIM analysis of DTY-QTLs
A multiple-trait analysis model was used to test the pleiotropic effect of drought GY QTLs qDTY1.1, qDTY3.1 and qDTY12.1. This analysis was carried out through a Java-based QTL analysis software Q Gene version 4.3.10 [28]. The multiple-trait, multiple-interval mapping (MT-MIM) model used for analysis was similar to the multiple-trait composite interval mapping (MT-CIM) model explained by Kao et al. [29]. The only difference between these two models was that MT-MIM tests more than one QTL (denoted as ‘q’ in the formula) at a time, unlike MT-CIM, in which only one QTL is tested at a time [30]. The model used for MT-MIM was
| (1) |
where 'q' is the number of QTLs being fitted simultaneously, t is the number of analyzed traits, 'n' is number of observation, 'p' is the number of non-genetic fixed factors, 'ai' and 'di' are additive and dominant effects of a QTL, 'X' refers to the non-genetic fixed effect, 'B' is incidence matrix that links observation of the data with fixed effects and 'E' is random error Eq (1). The hypotheses to be tested are H0: ai = 0, di = 0 and E = 0 vs. HA: a ≠ 0, d ≠ 0 and E ≠ 0 (i.e., testing if the QTL effects are zero in at least one trait). In this case, there are multiple null hypotheses.
Co-locating GY QTLs were tested in pairs with either DTF or PH in the same QTL region at a time for LOD of pleiotropy effects with an assumption that there is linkage between the two if the LOD of the pleiotropy effects is not significant and there is pleiotropy if the LOD is significant. LOD thresholds at 5% and 1% significance levels were determined using 1000 permutations.
Phenotyping
Phenotypic evaluation of two types of genetic material was performed in the study (1) N22/Swarna BIL population, and (2) QTL (qDTY1.1, qDTY3.1 and qDTY12.1) homozygote lines. Screening of the N22/Swarna BIL population of 297 lines was carried out under lowland RS and NS conditions in DS2011 and DS2012. Experiments were laid out in an alpha lattice design in two replications with a 5-meter (m) single-row plot and 0.2-m row spacing. Seeds were sown in a nursery and 21-day-old seedlings were transplanted with single seedlings per hill. Nitrogen, phosphorus, and potassium (NPK) were applied at 120:30:30 kg ha-1. Bayluscide (niclosamide, 0.25 kg a.i. ha-1) was sprayed immediately after transplanting for snail control. For weed control, Sofit (pretilachlor + safener, 0.3 kg a.i. ha-1), a post-emergence herbicide, was sprayed 4 days after transplanting (DAT). Furadan (carbofuran, 1 kg a.i. ha-1) and Cymbush (cypermethrin, 1 L ha-1) ± Dimotrin (cartap hydrochloride, 0.25 kg a.i. ha-1) were applied at 5 DAT and 16 DAT, respectively, for insect pest control.
A standing water level of 5 cm was maintained after transplanting throughout the crop season and drained before harvesting in the NS experiments. The RS experiments were maintained similar to the NS experiments until 30 DAT and water was drained for the imposition of stress at 30 DAT. The RS experiments were re-watered after 70% of the population lines showed severe leaf rolling symptoms. After 24 hours, the fields were drained again for a second RS cycle [31].
The phenotypic data for GY, DTF, and PH were recorded in DS2011. Further, two more parameters, biomass (BIO) and harvest index (HI), were added in the DS2012 experiment. When panicles of 50% of the plants in each plot were exerted, DTF was recorded. PH (cm) was measured as the height from the soil surface to the tip of the panicle on the main tiller before harvesting. BIO (g m-2 converted to kg ha-1) was taken from a 1-m2 area in each plot. Harvested samples were oven-dried, weighed, and threshed for grain weight. Grains were dried to 12% moisture before weighing [12, 13].
To calculate HI, the following formula Eq (2) was used:
| (2) |
QTL homozygote lines were screened in two different experiments (1) Stress 1 and Stress 2. qDTY3.1 lines were screened in two different seasons. qDTY1.1 lines were screened in two different environments (stress 1 = field conditions and stress 2 = rainout shelter). qDTY12.1 lines were screened on two different dates at the same location (stress 1 = first date and stress 2 = second date). Phenotypic data of an N22/IR64 population previously published by Vikram et al. (2011) were also used for the qDTY3.2 validation and interaction study.
Genotyping
Similar to the phenotyping, genotyping was also carried with (1) N22 derived BILs and RILs and (2) QTL homozygote lines. Genotyping of the N22/3*Swarna BIL population used for the qDTY3.2 validation in our study was carried out. In addition to this, an N22/IR64 RIL population was genotyped with markers for the qDTY3.2 QTL region to analyze its interaction effects with qDTY1.1. The N22/IR64 RIL population was originally used for the identification of qDTY1.1 [9].
Freeze-dried leaf samples were cut in eppendorf tubes, ground using a GENO grinder, and DNA was extracted following a modified CTAB method [32]. Quantification was done on a 0.8% agarose gel and concentrations adjusted to ~25 ng μL-1 for the PCR amplification. A 15-μL PCR mixture contained 50 ng DNA, 1 × PCR buffer, 100 μM dNTPs, 250 μM primers, and 1 unit Taq polymerase enzyme. PCR products were resolved in 8% non-denaturing polyacrylamide gel electrophoresis (PAGE). Genotyping of both N22/Swarna BIL and N22/IR64 RIL populations was done with markers for qDTY1.1 and qDTY3.2 (RM212, RM315, RM3825, RM11943, RM431, RM12023, RM12091, RM12146, RM22, and RM231).
QTL homozygote lines for three QTLs qDTY1.1, qDTY3.1 and qDTY12.1 were genotyped for their respective foreground markers reported by Vikram et al. [9], Venuprasad et al. [12] and Bernier et al. [13]. Marker analysis was also done for determining background recovery. A polymorphism survey was performed and markers spaced at equal distance throughout genome were selected for testing genetic background (Figs 3, 4 and 5).
Statistical and QTL analysis
Statistical analysis in the N22/Swarna BIL population was done using CROPSTAT v.4.2.3 (available at www.irri.org). Phenotypic means of entries were estimated using the following linear mixed model for the analysis of variance:
| (3) |
where Pijk is the measurement recorded on a plot, M is the mean over all plots, and R, B, L, and e are replications, blocks, lines, and error, respectively and i, j and k are number of replicates, blocks and lines respectively Eq (3). For estimating the entry means, replications and blocks within replicates were considered random whereas entries were fixed.
Genetic map distances between markers were estimated using their physical distances. Genetic distances between markers were estimated using Map Manager QTX software [33]. LOD threshold of 3.0, significance value of 1% (P ≤ 0.01) and Kosambi map function was used for the estimation of genetic distances. QTL analysis was with QTL Network software v2.1 using a mixed model-based composite interval mapping method [34] in which the significance level of the marker intervals was tested for their associations with traits of interest. Candidate marker intervals were selected and used as co-factors in a one-dimensional genome scan. Determination of the candidate intervals, detection of putative QTLs, and their additive effects were significant at P < 0.01. The F-value threshold was determined using 1000 permutation tests. Window size and walk speed used for the genome scan were 10 cM and 1 cM, respectively.
QTL class analysis for qDTY3.2 interaction analysis
Digenic interactions between DTF and PH QTLs in qDTY1.1 and qDTY3.2 regions were identified. To better understand the interaction of drought GY and DTF QTLs, QTL classes were formed. QTL classes were qDTY1.1, qDTY3.2, qDTY12.1, qDTY1.1 + qDTY3.2, and qDTY12.1 + qDTY3.2. Therefore there were total of four classes- (1) Class I with qDTY12.1 or qDTY1.1, (2) Class II with qDTY3.2, (3) Class III with qDTY3.2 + qDTY12.1 / qDTY1.1 and (4) Class IV with no QTL. Number of lines under class I, II, III and IV in N22/IR64 population were-17, 6, 12 and 4 respectively, whereas class I, II, III and IV had- 18, 7, 11 and 4 genotypes in N22/Swarna population respectively. In Vandana/ WayRarem population class I, III and IV had- 27, 2 and 30 genotypes respectively. None of genotype with only qDTY3.2 was found. However, one genotype with this QTL was taken in which background QTLs were in segregating condition. Phenotypic means of the lines with and without QTLs and QTL combinations were estimated. QTL class analysis was carried out with phenotypic data of RS as well as NS experiments.
Marker-assisted linkage elimination
A marker-assisted breeding strategy was followed for the elimination of linkage of PH from DTY QTL qDTY1.1. The marker-assisted linkage elimination was done to remove the linkage of PH and DTF QTLs from qDTY1.1, qDTY3.1, and qDTY12.1. QTL lines for the three QTLs (qDTY1.1, qDTY3.1, and qDTY12.1) were backcrossed with their respective recipient parents to recover the recipient genotype background. qDTY1.1 and qDTY3.1 lines were backcrossed thrice whereas qDTY12.1 lines were backcrossed twice. The total number of F1s produced ranged from 150 to 200 in the three crosses. These F1s were genotyped with foreground markers and 15–25 lines segregating for QTLs were selected. Selected BC2F1s were selfed to generate 2000–3000 BCnF2 (BCnF2 refers to BC2F2 or BC3F2) plants that were further genotyped for foreground markers of the respective QTLs, and finally QTL homozygote lines were selected. This strategy was followed for qDTY3.1 and qDTY12.1 to remove PH and DTF QTL linkages. A slightly different strategy was followed for qDTY1.1 as described by Vikram et al [25]. Semi-dwarf plants were selected among BC3F2 plants and genotyped for qDTY1.1 foreground markers. Plants segregating for qDTY1.1 were allowed to segregate twice more to generate the QTL homozygote. Further selection among QTL homozygote lines was made for DTF and PH. Background genotyping of the QTL homozygote was carried out (Fig 3). Similarly, qDTY3.1 and qDTY12.1 BILs were genotyped to clear the background (Figs 4 and 5). Finally, lines with cleared backgrounds and recurrent parent phenology were screened for GY under RS. qDTY1.1 BIL lines were screened in lowland RS conditions in DS2012 at two locations: RS in the field (stress 1) and in a rainout shelter (stress 2). The qDTY3.1 BIL lines were screened under lowland RS in DS2011 (stress 1) and DS2012 (stress 2). The screening protocol used for phenotyping of the N22/Swarna BILs was followed. The qDTY12.1 BILs were screened under upland RS in DS2010 in two different experiments sown at two different dates (first date: stress 1 and second date: stress 2) using a protocol described by Bernier et al. [13].
Supporting Information
The additive interaction of qDTY3.2 with qDTY1.1 reduced days to 50% flowering under stress as well as non-stress situations.
(TIF)
Trait values (DTF or GY) and QTLs/ QTL combinations have been plotted on ‘Y’ and ‘X’ axis respectively. Trait values (DTF or GY) of QTL combination class circled with orange colour to show that how qDTY3.2 interacts with qDTY1.1 and qDTY12.1 to reduce flowering duration and enhance grain yield under drought in different populations. Standard errors of difference for different classes of three populations were (1) N22/IR64 RIL population: DTF = 1.589, GY = 232.1, (2) N22/Swarna: DTF = 2.409, GY = 256.3; (3) Vandana/Way Rarem: DTF = 3.415, GY = 128.2.
(TIF)
Trait values (DTF or GY) and QTLs/ QTL combinations have been plotted on ‘Y’ and ‘X’ axis respectively. Trait values (DTF or GY) of QTL combination class (qDTY3.2 and qDTY1.1 / qDTY12.1) circled with orange colour. Lines with qDTY1.1 and qDTY3.2 had lower yield as compared to the lines with only qDTY1.1. On the other hand qDTY12.1 and qDTY3.2 had lower grain yields compared to the lines with either qDTY12.1 or qDTY3.2. Standard errors of difference for different classes of three populations were (1) N22/IR64 RIL population: DTF = 2.010, GY = 502.7, (2) N22/Swarna: DTF = 1.754, GY = 497.8; (3) Vandana/Way Rarem: DTF = 3.913, GY = 304.9.
(TIF)
Acknowledgments
The authors are thankful to Dr. Amelia Henry for testing the lines in a rainout shelter at IRRI. Authors duly acknowledge the contribution of IRRI drought breeding group. We are also thankful to NRCPB, India for their support in genotyping for the sd1 gene. Experiments conducted in our study comply with the current laws of the Philippines.
Abbreviations
- BIL
Backcross inbred line
- BIO
Biomass
- CTAB
Cetyl trimethyl ammonium bromide
- DNA
Deoxyribonucleic acid
- DTF
Days to 50% flowering
- GY
Grain yield
- HI
Harvest index
- LOD
Logarithm of odds
- MAB
Marker-assisted breeding
- NS
Non-stress
- PCR
Polymerase chain reaction
- PAGE
Polyacrylamide gel electrophoresis
- PHT
Plant height
- QTL
Quantitative trait loci
- RS
Reproductive-stage drought stress
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Bill & Mellinda Gates Foundation (BMGF) funded Project: Stress Tolerant Rice for Poor Farmers of South Asia and Africa (STRASA), http://strasa.irri.org/.
References
- 1.Comstock RE (1978) Quantitative genetics in maize breeding In ‘Maize breeding and genetics’. Wiley-Interscience: New York, pp. 191–206, 1978. [Google Scholar]
- 2.Lander ES, Botstein D (1989) Mapping Mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng ZB (1994) Precision mapping of quantitative trait loci. Genetics 136:1457–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng ZB, Weir BS (1996) Statistical methods for mapping quantitative trait loci. Acta Agron Sin 22:535–549. [Google Scholar]
- 5.Yang J, Zhu J, Williams RW (2007) Mapping the genetic architecture of complex traits in experimental populations. Bioinformatics 23:1527–1536. [DOI] [PubMed] [Google Scholar]
- 6.Babu RC, Nguyen BD, Chamarerk V, Shanmugasundaram P, Chezhian P, Jeyaprakash P et al. (2003) Genetic analysis of drought resistance in rice by molecular markers: association between secondary traits and field performance. Crop Sci 43:1457–1469. [Google Scholar]
- 7.Kumar R, Venuprasad R, Atlin GN (2007) Genetic analysis of rainfed lowland rice drought tolerance under naturally-occurring stress in eastern India: heritability and QTL effects. Field Crops Res 103:42–52. [Google Scholar]
- 8.Gomez SM, Boopathi NM, Kumar SS, Ramasubramanian T, Chengsong Z, Jeyaprakash P, et al. (2010) Molecular mapping and location of QTLs for drought-resistance traits in indica rice (Oryza sativa L.) lines adapted to target environments. Acta Physiol Plant 32:355–364. [Google Scholar]
- 9.Vikram P, Swamy BPM, Dixit S, Sta Cruz MT, Ahmed HU, Singh AK, et al. (2011) qDTY1.1, a major QTL for rice GY under reproductive-stage drought stress with a consistent effect in multiple elite genetic backgrounds. BMC Genet 12:89 10.1186/1471-2156-12-89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Venuprasad R, Bool ME, Quiatchon L, Sta Cruz MT, Amante M, Atlin GN (2012) A large-effect QTL for rice GY under upland drought stress on chromosome 1. Mol. Breed 30 (1):533–547. [Google Scholar]
- 11.Ghimire KH, Quiatchon LA, Vikram P, Swamy BPM, Dixit S, Ahmed H, et al. (2012) Identification and mapping of a QTL (qDTY1.1) with a consistent effect on GY under drought. Field Crops Res 131:88–96. [Google Scholar]
- 12.Venuprasad R, Dalid CO, Del Valle M, Zhao D, Espiritu M, Sta Cruz MT, et al. (2009) Identification and characterization of large-effect quantitative trait loci for GY under lowland drought stress in rice using bulk-segregant analysis. Theor Appl Genet 120:177–190. 10.1007/s00122-009-1168-1 [DOI] [PubMed] [Google Scholar]
- 13.Bernier J, Kumar A, Venuprasad R, Spaner D, Atlin GN (2007) A large-effect QTL for GY under reproductive-stage drought stress in upland rice. Crop Sci 47:507–516. [Google Scholar]
- 14.Lin H, Ashihikari M, Yamanouchi U, Sasaki T, Yano M (2002) Identification and characterization of a quantitative trait locus, HD9, controlling heading date in rice. Breed Sci 52:35–41. [Google Scholar]
- 15.Dixit S, Swamy BPM, Vikram P, Bernier J, Sta Cruz MT, Amante M et al. (2012) Increased drought tolerance and wider adaptability of qDTY12.1 conferred by its interaction with qDTY2.3 and qDTY3.2. Mol. Breed 30:1767–1779. [Google Scholar]
- 16.Lebreton C, Lazic-Jancic V, Steel A, Pekic S, Quarrie SA (1995) Identification of QTL for drought responses in maize and their use in testing causal relationships between traits. J Exp Bot 46:853–865. [Google Scholar]
- 17.Shashidhar HE, Hemamalini GS, Hittalmani S (1999) Molecular marker-assisted tagging of morphological and physiological traits at the peak vegetative stage: two contrasting moisture regimes In: Ito O., O'Toole J., Hardy B. (eds.). Genetic improvement of rice for water-limited environments. International Rice Research Institute, Manila, Philippines: pp 239–256. [Google Scholar]
- 18.Hittalmani S, Shashidhar HE, Bagali PG, Ning Huang, Sidhu JS, Singh VP, et al. (2002) Molecular mapping of quantitative trait loci for plant growth, yield and yield related traits across three diverse locations in a doubled haploid rice population. Euphytica 125:207–214. [Google Scholar]
- 19.Jiang C, Zeng Z-B (1995) Multiple trait analysis of genetic mapping for quantitative trait loci. Genetics 140:1111–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tuberosa R, Salvi S (2006) Genomics-based approaches to improve drought tolerance of crops. Trends Plant Sci 11(8):405–412. [DOI] [PubMed] [Google Scholar]
- 21.Swamy BPM, Kumar A (2011) Sustainable rice yield in water short drought prone environments: conventional and molecular approaches In: Teang Shui Lee (ed.), Irrigation Systems and Practices in Challenging Environments. InTech, Croatia: pp 149–168. [Google Scholar]
- 22.Vikram P, Swamy BPM, Dixit S, Sta Cruz MT, Ahmed HU, Singh AK (2012) Bulk segregant analysis: an effective approach for mapping consistent effect drought GY QTLs in rice. Field Crops Res 134:185–192. [Google Scholar]
- 23.Deng S, Wu X, Wu Y, Zhou R, Wang H (2011) Characterization and precise mapping of a QTL increasing spike number with pleiotropic effects in wheat. Theor Appl Genet 122:281–289. 10.1007/s00122-010-1443-1 [DOI] [PubMed] [Google Scholar]
- 24.Kumawat G, Raje RS, Bhutani S, Pal JK, Mithra ASVCR, Gaikwad K (2012) Molecular mapping of QTLs for plant type and earliness traits in pigeonpea (Cajanus cajan L. Millsp.). BMC Genet 13:84 10.1186/1471-2156-13-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vikram P, Swamy BPM, Dixit S, Singh R, Singh BP, Miro B, et al. (2015) Drought susceptibility of modern rice varieties: an effect of linkage of drought tolerance with undesirable traits. Sci Rep 5:14799 10.1038/srep14799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lanceras JC, Pantuwan G, Jongdee B, Toojinda T (2004) Quantitative trait loci associated with drought tolerance at reproductive stage in rice. Plant Physiol 135:384–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mishra KKM, Vikram P, Yadaw RB, Swamy BPM, Dixit S, Sta Cruz MT et al. (2013) qDTY 12.1: a locus with a consistent effect on grain yield under drought in rice. BMC Genet 14:12 10.1186/1471-2156-14-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joehanes R, Nelson JC (2008) QGene 4.0, an extensible Java QTL analysis platform. Bioinformatics 24:2788–2789. 10.1093/bioinformatics/btn523 [DOI] [PubMed] [Google Scholar]
- 29.Kao C-H, Zeng Z-B, Teasdale RD (1999) Multiple trait mapping for quantitative trait loci. Genetics 152:1203–1216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Joehanes R (2009) Multiple-trait multiple-interval mapping of quantitative-trait loci. Thesis report (master of science) from Department of Statistics, Gary L. Gadbury laboratory, Kansas State University, Manhattan, Kansas, USA. Available: http://hdl.handle.net/2097/1605
- 31.Venuprasad R, Lafitte HR, Atlin GN (2007) Response to direct selection for GY under drought stress in rice. Crop Sci 47:285–293. [Google Scholar]
- 32.Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Manly KF, Olson JM (1999) Overview of QTL mapping software and introduction to Map Manager QT. Mamm Genome 10:327–334. [DOI] [PubMed] [Google Scholar]
- 34.Yang J, Hu C, Hu H, Yu R, Xia Z, Ye X (2008) QTL Network: mapping and visualizing genetic architecture of complex traits in experimental populations. Bioinformatics 24:721–723. 10.1093/bioinformatics/btm494 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The additive interaction of qDTY3.2 with qDTY1.1 reduced days to 50% flowering under stress as well as non-stress situations.
(TIF)
Trait values (DTF or GY) and QTLs/ QTL combinations have been plotted on ‘Y’ and ‘X’ axis respectively. Trait values (DTF or GY) of QTL combination class circled with orange colour to show that how qDTY3.2 interacts with qDTY1.1 and qDTY12.1 to reduce flowering duration and enhance grain yield under drought in different populations. Standard errors of difference for different classes of three populations were (1) N22/IR64 RIL population: DTF = 1.589, GY = 232.1, (2) N22/Swarna: DTF = 2.409, GY = 256.3; (3) Vandana/Way Rarem: DTF = 3.415, GY = 128.2.
(TIF)
Trait values (DTF or GY) and QTLs/ QTL combinations have been plotted on ‘Y’ and ‘X’ axis respectively. Trait values (DTF or GY) of QTL combination class (qDTY3.2 and qDTY1.1 / qDTY12.1) circled with orange colour. Lines with qDTY1.1 and qDTY3.2 had lower yield as compared to the lines with only qDTY1.1. On the other hand qDTY12.1 and qDTY3.2 had lower grain yields compared to the lines with either qDTY12.1 or qDTY3.2. Standard errors of difference for different classes of three populations were (1) N22/IR64 RIL population: DTF = 2.010, GY = 502.7, (2) N22/Swarna: DTF = 1.754, GY = 497.8; (3) Vandana/Way Rarem: DTF = 3.913, GY = 304.9.
(TIF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.