Abstract
Age-associated cognitive decline is an inevitable phenomenon that predisposes individuals for neurological and psychiatric disorders eventually affecting the quality of life. Scientists have endeavored to identify the key molecular switches that drive cognitive decline with advancing age. These newly identified molecules are then targeted as recovery of cognitive aging and related disorders. Cognitive decline during aging is multi-factorial and amongst several factors influencing this trajectory, gene expression changes are pivotal. Identifying these genes would elucidate the neurobiological underpinnings as well as offer clues that make certain individuals resilient to withstand the inevitable age-related deteriorations. Our laboratory has focused on this aspect and investigated a wide spectrum of genes involved in crucial brain functions that attribute to senescence induced cognitive deficits. We have recently identified master switches in the epigenome regulating gene expression alteration during brain aging. Interestingly, these factors when manipulated by chemical or genetic strategies successfully reverse the age-related cognitive impairments. In the present article, we review findings from our laboratory and others combined with supporting literary evidences on molecular switches of brain aging and their potential as recovery targets.
Keywords: cognitive decline, brain aging, gene expression, molecular mechanism, recovery
Cognitive decline is a major hurdle for the attainment of healthy aging. Advancing age impacts our cognitive capacities of perceptual speed, attention, reasoning, and the most crucial, learning and memory processes dependent on hippocampus and frontal cortex [1]. Mild impairment in these brain functions is manifested even in physically healthy aged individuals and its severity disposes to neurodegenerative disorders. The demographic survey worldwide estimates that 40% of population above 60 years is affected by varying degrees of cognitive decline, which is persistently increasing in prevalence and transition to disorders [2]. In this regard, major research efforts focus towards understanding the pathological aging such as Alzheimer’s and Parkinson’s disease. It is worth mentioning here that even if these disorders are devastating, all the aged people are not afflicted with dementia. However, majority of individuals suffer from subtle age-associated cognitive deficits which deteriorate quality of life. Therefore, unraveling the mechanisms of cognitive decline associated with normal aging is of utmost importance and would offer a hope for recovery targets of degenerative pathologies.
The molecular mechanisms underlying the onset of age-associated deteriorations in brain is still a mystery and one of the greatest health challenges of the decade. However, every cloud has a silver lining as certain examples of healthy aged brain raised immense hope among the researchers. The neuroscientists from University of Groningen, Netherlands assessed the cognitive abilities of 113-year old Dutch woman, Hendrikje van Andel-Schipper, and examined her brain immediately after the death at the age of 115. Her cognitive performances were better than healthy adults of 60-75 years. Her brain was without plaques, vascular changes and neuron number corresponded with brains of healthy people of 60-80 years [3].
These observations have made researchers to believe that there are no limits to healthy life of brain and challenged them to search the measures which maintain cognitively intact brain till late age. In this context, the major questions that strike the neuroscientists are why some aged individuals can preserve cognitive abilities while others can’t and which factors switch normal or non-pathological brain aging to disease conditions.
Gene expression changes in aging brain: Central to cognitive decline
The triggers and consequences of brain aging and associated cognitive decline are considered multidimensional, though accumulating studies implicate that gene expression changes are pivotal [4]. Alterations in gene expression robustly affect brain functions from cellular to behavioral level with critical influence on cognitive processes. Although major investigations have been conducted for gene expression changes in neurodegenerative disorders, little is known about their profiles in aged brain. Moreover, studies conducted till date reveal that genes related to stress, inflammation, immune response, mitochondrial functions, growth factors, neuronal survival and calcium homeostasis are altered during aging [5,6]. In general, genes that are stress responsive and related to inflammation and DNA repair are upregulated while genes involved in neuronal growth and survival and mitochondrial functions are downregulated with advancing age in several organisms [7]. Gene expression changes during brain aging also exhibit regional differences and sexual dimorphism [8]. The forebrain regions including superior frontal gyrus, entire cortex and hippocampal CA1 are more susceptible to aging and exhibit a large number of gene alterations [9].
Studies from our laboratory have primarily focused on specific categories of genes influencing cognitive functions during aging and also implicated in neurodegenerative disorders. As hormonal changes are also crucial during aging process, we have initially examined steroid hormones and their receptor encoding genes in brain aging. In parallel, we studied causative and susceptibility genes of neurodegenerative disorders in normal brain aging process and checked their regulation by the steroids. These studies revealed very promising results and prompted us to believe that cognitive impairment during aging also involves some other factors. Further literature evidences indicated the concept of synaptic plasticity decline in brain aging. In the past few years, age-associated cognitive decline has been primarily attributed to loss of synaptic connectivity and plasticity rather than complete degeneration of neurons. Therefore, we started investigating genes involved in different stages of synaptic plasticity and their influence on cognition, particularly learning and memory in aging brain.
Sex steroids and receptors
An important factor influencing cognitive functions of aging brain is hormone. In particular, alteration in the level of sex steroid hormones including testosterone, estrogen and progesterone [10] is considered a serious risk factor for accelerated brain aging. Amongst these steroids, estrogen stands out for its direct influence on cognitive decline. The estrogen deprivation in postmenopausal women is integral to aberrant changes in hippocampal morphology and ensuing cognitive impairment. Estrogen acts mainly through its intracellular receptors, estrogen receptor (ER) α and β. These receptors are implicated in several crucial brain functions including neuronal growth and differentiation, migration and synapse formation [11, 12].
The expression of these receptors encoding genes is significantly reduced in aging brain that is influenced by estrogen, growth, thyroid and other hormones [13]. Cumulative evidences link the decrease of ERs with enhanced vulnerability for cognitive deficits in aging as well as neurodegenerative disorders. A linear relationship was found between cognitive function and level of ERα in the frontal cortex of Alzheimer's disease (AD) patients [14]. In rodent models, aged females showed lower ER α and β immunoreactivity consistent with reduction in the number of hippocampal synapses [15].
ER α and β following ligand binding recruit a set of coactivators and corepressor proteins termed as coregulators which regulate expression of specific genes. Interestingly, the expression of these coregulators and their interaction with ERs also alter in aging brain. As compared to young, old mice exhibited significant decrease in the level ERα coregulators such as PELP1, RIP140, PGC1a and BAF60. PELP1 and RIP140 are key players of estrogen-induced neurogenesis and learning and memory, whereas p68 and PGC1α have been implicated in formation and maintenance of dendritic spine and neuronal growth. These data provided evidence that age dependent decline in ERα coregulators might be responsible for cognitive dysfunctions [16]. ER β coregulators AIB1, ERAP 140, TrkA, Src, pCREB and CREB also decreased in aging brain and eventually influenced estrogen signaling. CREB is central to learning and memory regulating a wide variety of intracellular signaling cascades responsible for synaptic efficacy and long-lasting changes in synaptic plasticity. CREB is phosphorylated (pCREB) in the nucleus by various protein kinases via secondary messengers such as cAMP and/or Ca2+ for regulating specific genes. Alterations in CREB signaling lead to cognitive deficits in aging and degenerative pathologies [17]. These compelling evidences have put forward the use of estrogen replacement therapy in reversal of cognitive deficits. However, estrogen replacement therapy in post-menopausal women may be a risk factor for development of breast cancer. To overcome this problem, selective estrogen receptor modulators (SERM) may play a promising role due to its selective action as an agonist and antagonist in tissue dependent manner. Recently, Kindler et al [18] reported that raloxifene, a benzothiopene derivative SERM improved hippocampal activity and associated learning in schizophrenia patients. Earlier reports have shown that raloxifene treatment prevented osteoporosis and reduced the risk of breast cancer in post-menopausal women. Thus, selective modulation of estrogen receptors and coregulators may be identified as potential therapeutics in aging [19, 20, 21].
AD associated genes
The severity in aging associated cognitive decline predisposes to neurodegenerative diseases such as AD. Although majority of patients develop late-onset AD at an age above 65 years, 2-10% of patients show clinical symptoms much earlier referred to as early-onset AD. Therefore, cognitive aging is not causative rather predisposing factor making individuals vulnerable for AD. Age related structural and functional deteriorations in brain are considered to be the greatest risk factor for late onset AD. Mutations in different genes including amyloid precursor protein (APP), presenilin (PS) 1 and 2 are considered to cause familial forms of AD whereas apolipoprotein E (ApoE) 4 allele increases the susceptibility. We hypothesized that these candidate genes that pose risk for AD might also alter during normal aging. Expression analysis of these AD related genes during aging in vulnerable brain regions of cerebral cortex and hippocampus confirmed our speculation. Moreover, we also noted pronounced influence of estradiol and testosterone on the expression of these genes in aging brain.
AD is characterized by accumulation of amyloid beta peptides produced by proteolytic cleavage of APP. APP gene expression showed significant upregulation in the cerebral cortex of old male and female mice. Further, estradiol modulated the expression APP mRNA as well its alternative splicing in old mouse cerebral cortex. APP has various isoforms (APP770, APP751 and APP695) amongst which APP695 is predominantly found in the brain and remains elevated in non-pathological conditions. APP695 was significantly upregulated by estrogen in aged female mouse brain [22].
Other genes associated with early onset of AD are PS1 and PS2, which are transmembrane aspartyl proteases. They constitute the catalytic core of the gamma-secretase enzyme which cleaves βAPP to generate Aβ peptides. We observed reduction in PS1 expression but increase in PS2 in the cerebral cortex of old mice of both sexes. Such expression pattern was modulated by 17β-estradiol and testosterone [23, 24].
Interestingly, all the AD associated genes were not affected during aging as we noted no significant change in the expression of ApoE associated with late-onset sporadic and familial AD. ApoE is a lipid transport protein involved in membrane repair and synaptic plasticity and implicated in atherosclerosis and neurodegeneration [25, 26, 27], but its overall impact during normal aging remains to be determined. We reported that ApoE expression decreased from young to adult and then remained unchanged in old mouse cerebral cortex. Certainly, such expression profile suggested the involvement of other factors in maintaining ApoE level and dependent brain functions in old [28].
Synaptic plasticity genes
Over the past decade, enormous evidences from human and animal models suggest that brain aging accompanied cognitive decline is primarily due to changes in neuronal morphology and synaptic plasticity. Synaptic plasticity events form the core of learning and memory processes that are affected almost in every species during aging. Aging rodents show reduced hippocampal dependent recognition memory [29], spatial memory, fear memory [30] and associative memory. Similarly, primates and non-primate mammals show decline in working memory [31].
Memory processes dependent on hippocampus (declarative memory) and dorsolateral prefrontal cortex (working memory) show maximum decline with advancing age [32, 33, 34]. In the light of this information, we were intrigued to investigate the genes regulating synaptic plasticity in aging brain. Neuronal activity induced genes such as brain derived neurotrophic factor (BDNF) and activity regulated cytoskeleton associated protein (Arc) have been well reported in synaptic plasticity changes of aged brain [35]. We explored a new class of molecule, activity dependent neural proteases in brain aging. Upon neuronal stimulation, these proteases are secreted in the synaptic cleft and cleave extracellular matrix proteins and/or cell adhesion molecules. This proteolytic cleavage immediately causes synaptic remodeling or the cleaved products activate certain signaling cascades, eventually affecting synaptic connectivity and plasticity.
Neuropsin (NP) is one such activity induced serine protease predominantly expressed in hippocampus CA1-CA3 subregions, amygdala and moderately in prefrontal cortex. NP gene ablation impaired long-term potentiation (LTP) and memory acquisition process [36, 37, 38]. NP cleaves L1CAM, an immunoglobulin superfamily neural cell adhesion molecule, and is implicated in hippocampal plasticity [39]. L1CAM is also known to regulate neurite growth of hippocampal neurons by inducing the expression of microtubule associated protein (MAP)2c [40]. With this background, we investigated NP and its effectors L1CAM and MAP2c in aging mouse brain. We observed age and region dependent variation in NP expression. NP mRNA and protein expression was drastically decreased in the olfactory bulb, cerebral cortex and hippocampus of old mice as compared to young. In cerebral cortex, the decline was gradual with age while in olfactory bulb and hippocampus there was a robust increase in adult and decrease in old. As these brain regions are pivotal for memory processes, it indicated that NP might be a critical determinant of regional differences in memory decline during aging. Analysis of L1CAM cleavage and MAP2c protein level revealed regional and age dependent strong positive correlation with NP expression pattern. These findings suggested that NP dependent LICAM cleavage might regulate neurite growth marker MAP2c attributing to alterations in synaptic plasticity and memory during aging [41].
Neurite growth inhibitory genes implicated in synaptic plasticity were also altered during aging. Nogo-A is a myelin-associated neurite growth inhibitory protein, that binds to its receptor Nogo-66 receptor1 (NgR1) and regulates filamentous actin dynamics and involved in structural plasticity of the synapses. We demonstrated that Nogo-A protein level decreased in male and female forebrain of old mice while NgR1 remained unaltered [42]. In recent years, another set of synaptic proteins neurexins (Nrxns) and neuroligins (Nlgns) have emerged as potential candidates for alterations in structural synaptic plasticity. Presynaptic Nrxns form complexes with postsynaptic Nlgns [43] and facilitate the differentiation, maturation, and stabilization of synapses [44, 45]. We analyzed expression pattern of these genes in aging mouse brain and observed a significant decrease of both Nrxn1 and Nlgn3 level in cerebral cortex of old mice. Interestingly, this pattern strongly correlated with a crucial marker of functional synapse, synaptophysin during aging [46].
Master regulators of gene expression: Epigenetic switch?
The molecular switches driving these wide spectrums of gene expression alteration in aged brain deserve utmost attention. Research endeavors over past few years hypothesize that epigenetic mechanisms of DNA methylation and post translational modifications of histones modulate gene transcription resulting in age-related cognitive deficits. In particular, methylation of DNA and histone acetylation is considered pivotal for memory-linked gene transcription [47].
DNA methylation occurs at 5′C of cytosine in CpG dinucleotide and is distributed as CpG islands at the promoter region of gene and on repetitive sequences in the genome. DNA methylation directly interferes with the binding of transcription factors or recruits methyl binding proteins (MBDs and MeCP2) forming repressor complex at the gene promoter and thereby down regulates gene expression. Global methyl cytosine level is decreased while promoter specific methylation either gets decreased or increased in the brain during aging. Such alteration in cytosine methylation patterns affects synaptic plasticity gene expression and function, particularly learning and memory during aging. Further, alteration in DNA methylation is one of the key factors leading to neurodegeneration and cognitive dysfunction in AD and other neurological disorders. AD human subjects showed decline in DNA methylation level in CA1, CA3 and dentate gyrus region of hippocampus as compared to age matched controls [48, 49, 50, 51].
Although there is reduction in global level of DNA methylation during aging or AD, methylation at gene promoter level is different. It is observed that DNA methylation at the promoter region of Arc is upregulated in the hippocampus of old mice as compared to adult [52]. This upregulation in DNA methylation is associated with decline in Arc expression and memory consolidation in old mice. BDNF, a well known synaptic plasticity gene and marker for learning and memory, plays an important role in neuronal survival and synaptic activity. DNA methylation at the promoter region of BDNF is upregulated in the peripheral blood sample of AD patients as compared to age matched control [53]. Moreover, APP gene upregulation causing Aβ deposition in brain of AD patients has been attributed to hypomethylation at gene promoter [54, 55]. DNA methylation is catalyzed by DNA methyl transferases (DNMTs) categorized into DNMT1, 3a and 3b. DNMT3a and 3b are de novo methyl transferases that methylate previously unmethylated sites while DNMT1 is maintenance methyltransferase methylating hemi methylated sites on DNA strands. Earlier reports have shown that DNMTs play a crucial role in brain development, neuronal differentiation, apoptosis and cognition. We reported that DNMT1 is down regulated in the cerebral cortex and hippocampus and positively correlated with decline in recognition memory consolidation during aging [56]. In consistent with our findings, cumulative studies on DNMT1 established its crucial role in memory processes. Homozygous DNMT1 mutation caused lethality at embryonic stages [57] while conditional DNMT1 mutation in rodents led to cortical and hippocampal neurodegeneration, and reduced their volume and memory consolidation [58, 59, 60] DNMT1 mutation in human caused hereditary sensory neuropathy characterized by reduced cytosine DNA methylation level, hearing loss, neurodegeneration and dementia [61].
The post translational modifications of histones remodeling structure of chromatin are also considered significant for gene expression changes in aging brain. Early in eighties, we reported for the first time that histone acetylation [62], phosphorylation [63] and methylation [64] levels declined in the brain of aging rats. The decline in histone acetylation level was positively correlated with in vitro mRNA synthesis during aging. Several studies thereafter highlighted the role of histone acetylation in cognitive decline during aging and disorders [30, 65, 66]. Further it is reported that H4K12 histone acetylation level is reduced in the hippocampus of old mice leading to decline in memory consolidation. H4K12 regulates the expression of several synaptic plasticity genes such as Fmn2, Myst4, Prkca etc during brain aging.
Histone acetylation and deacetylation are regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs) enzymes. We demonstrated that HDAC2 is upregulated in the hippocampus of old mice during aging and this is negatively correlated with decline in recognition memory during aging [29]. We also identified that HDAC2 mediated decrease in H3K9 and H3K14 acetylation level at the promoter region of Arc and BDNF results in deficits of recognition memory consolidation in aging as well as chemically induced amnesia [67]. Other gene manipulation studies revealed that HDAC2 negatively regulates learning and memory as its over-expression decreased dendritic spine density and spatial memory consolidation [68]. HDAC2 upregulation by H2O2 mediated oxidative stress and β amyloid peptide is also attributed to AD pathology and cognitive deficits [69].
Histone methylation has also gained importance in memory decline during aging. It is well reported that hippocampal neurogenesis decreases with age and contributes to memory deficits [70]. Doublecortin (DCX) is a cytoskeletal protein expressed by neuronal precursor cells and used as a marker for neurogenesis. It is reported that decrease in H3K4 trimethylation and increase in H3K27 trimethylation at the promoter region of DCX gene reduces its expression, eventually leading to impaired neurogenesis and memory decline in old mice [71]. These evidences clearly indicate that modifications of histones are crucial and warrant further investigations in relation to aging and neurodegeneration.
Recovery avenues
Multiple approaches are being adopted to retard brain aging associated cognitive decline and vulnerability of disorders. These approaches including herbal interventions, lifestyle modulation, hormonal replacement therapy and stem cell based regenerative medicine eventually endeavor to modulate the molecular switches of cognitive deficits. In this regard, our laboratory has contributed on natural remedies of brain aging highlighting nootropic potential of Ashwagandha and Brahmi extracts. We reported that in animal models mimicking age related cognitive deficits, these extracts exhibit remarkable potential to enhance the expression of plasticity genes (BDNF, Arc, Neuropsin), promote neurite growth and reverse memory decline [72].
Genes altered during brain aging are also targeted specifically as recovery strategies. In AD-transgenic mice, BDNF gene delivery reversed synapse loss, normalized aberrant gene expression, improved cell signaling and restored learning and memory. Several protein kinases including protein kinase A and PI3K/AKT/mTOR pathway have been identified as crucial molecular targets in reversal of age-associated deficits in memory consolidation. Another molecular switch that is integral to brain aging related cognitive decline is the CREB dependent transcriptional machinery of memory linked genes and is considered to be a potent therapeutic target in disorders of the CNS [73].
Although these diverse avenues have proven to be beneficial, their effects are targeted and partial, and necessitate a holistic approach driving global gene expression changes. Such presumptions highlighted epigenetic modifiers as key therapeutic avenues in aging and neurodegenerative diseases. In particular, administration of different HDAC inhibitors, sodium butyrate (NaB) and Suberoylanilide hydroxamic acid (SAHA), rescued age-associated decline in H4K12 Ac and memory consolidation [74, 75]. HDAC2 selective inhibition by antisense oligonucleotides altered the histone acetylation status of synaptic plasticity genes promoter and enhanced memory in scopolamine induced amnesic mice [67]. NaB treatment increased the expression of pCREB and learning and memory in D-galactose induced aging mouse model [76]. NaB treatment also rescued cognitive impairment in APPswe/PS1dE9 transgeneic mice model of AD [77]. HDAC inhibitors have been used in different neurodegenerative and psychiatric disorders like Parkinson’s disease, Huntington’s disease, ischemic stroke, anxiety, depression, Schizophrenia etc [78]. However, the currently used HDAC inhibitors are non-selective inhibiting the activity of several HDAC enzymes and causing side effects including gastrointestinal, hematological, cardiac and metabolic dysfunctions [79]. Therefore, targeted therapy using selective HDAC inhibitors that alleviate neurological dysfunctions as well as maintain physiological balance is necessary [80]. These specific HDAC inhibitors are still to be elucidated and might come up as novel therapeutic agents in treatment of age associated neurodegenerative disorders. Besides histone acetylation, compounds altering DNA methylation have also proven beneficial in brain aging. Treatment with S-adenosylmethionine (SAM) in human neuroblastoma SK-N-SH cell cultures as well as AD transgenic mouse models silenced the expression of PS1 and Beta-secretase (BACE), reduced oxidative damage and improved cognitive performance [81,82,83,84].
Conclusion
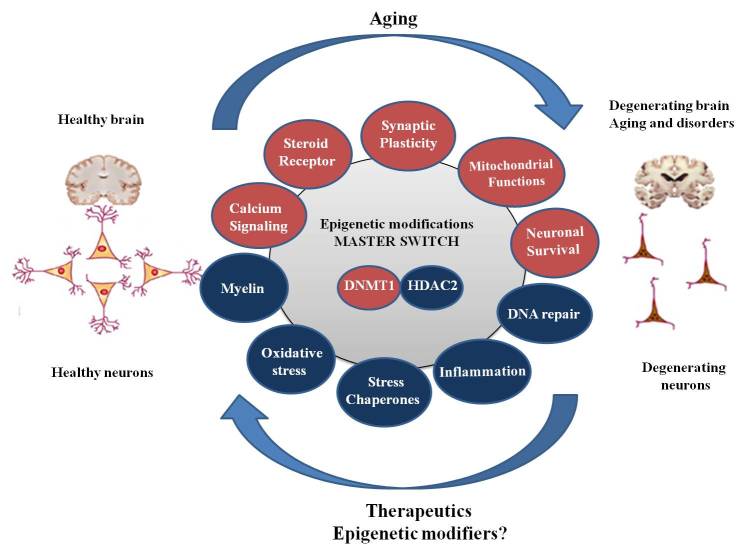
Age-related cognitive decline is an increasing biomedical concern. The exploration of underlying mechanisms highlighted the aberrant gene expression changes as pivotal for cognitive disabilities. However, these genes belong to multiple pathways making the recovery approach complex and arduous. Recent reports pointed towards a master molecular switch involving the epigenome driving global gene expression changes (Fig 1). In particular, chemicals or gene manipulation strategies that can promote epigenetic changes have been highlighted as holding promise for alleviating cognitive dysfunction. Further studies are warranted to investigate epigenetic marks in the regulation of cognition and reversal of age-associated cognitive decline.
Figure 1.
Gene expression changes and master switch driving age related cognitive decline Brain aging accompanies alteration in expression (red circles represent downregulation; blue circles represent upregulation) of genes belonging to multiple pathways. Epigenetic modifications particularly decrease in DNMT1 and increase in HDAC2 level might be the master regulators and accordingly epigenetic modifiers might prove ideal therapeutic targets.
Acknowledgement
The authors thank Department of Science and Technology (DST), Government of India for INSPIRE faculty award to AK, PS is a recipient of Senior Research Fellowship from the Council of Scientific and Industrial Research (CSIR) Government of India. The work cited in this article has been supported by grants from the DBT, DST, CSIR, ICMR, Government of India to MKT.
References
- [1].Hedden T,Gabrieli JDE (2004). Insights into the aging mind: a view from cognitive neuroscience. Nat Rev Neurosci, 5:87-97. [DOI] [PubMed] [Google Scholar]
- [2].Andrade C,Radhekrishan R (2009). The prevention and treatment of cognitive decline and dementia: An overview of recent research on experimental treatments. Indian J Psychiatry, 51:12-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].den Dunnen WF,Brouwer WH, et al. (2008). No disease in the brain of a 115-year-old woman. Neurobiol Aging, 29:1127-32. [DOI] [PubMed] [Google Scholar]
- [4].Nicolle MM,Gonzalez J,Sugaya K, et al. (2001). Signatures of hippocampal oxidative stress in aged spatial learning-impaired rodents. Neuroscience, 107:415-31 [DOI] [PubMed] [Google Scholar]
- [5].Lu T,Pan Y,Kao SY,Li C,Kohane I,Chan J,Yankner BA (2004). Gene regulation and DNA damage in the ageing human brain. Nature, 429:883-91. [DOI] [PubMed] [Google Scholar]
- [6].Loerch PM,Lu T,Dakin KA, et al. (2008). Evolution of the aging brain transcriptome and synaptic regulation. PLoS One, 3(10):e3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Yankner A,Lu T,Bruce LP (2008). The aging brain. Annu Rev Pathol Mech Dis, 3:41-66. [DOI] [PubMed] [Google Scholar]
- [8].Berchtold NC,Cribbs DH,Coleman PD, et al. (2008). Gene expression changes in the course of normal brain aging are sexually dimorphic. Proc Natl Acad Sci USA, 105:15605-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zeier Z,Madorsky I,Xu Y,Ogle WO,Notterpek L,Foster TC (2011). Gene expression in the hippocampus: regionally specific effects of aging and caloric restriction. Mech Ageing Dev, 132:8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Veiga S,Melcangib RC,DonCarlosc LL,Garcia-Seguraa LM,Azcoitiad I (2004). Sex hormones and brain aging. Exp Gerontol, 39:1623-31. [DOI] [PubMed] [Google Scholar]
- [11].Thakur MK,Sharma PK (2006). Aging of brain: role of estrogen. Neurochem Res, 31:1389-98. [DOI] [PubMed] [Google Scholar]
- [12].Mérot Y,Ferrière F,Debroas E,Flouriot G,Duval D,Saligaut C (2005). Estrogen receptor alpha mediates neuronal differentiation and neuroprotection in PC12 cells: critical role of the A/B domain of the receptor. J Mol Endocrinol, 35:257-67. [DOI] [PubMed] [Google Scholar]
- [13].Ghosh S,Thakur MK (2009). Interaction of estrogen receptor-alpha ligand binding domain with nuclear proteins of aging mouse brain. J Neurosci Res, 87: 2591-600. [DOI] [PubMed] [Google Scholar]
- [14].Kelly JF,Bienias JL,Shah A,Meeke KA,Schneider JA,Soriano E,Bennett DA (2008). Levels of estrogen receptors alpha and beta in frontal cortex of patients with Alzheimer's disease: relationship to Mini-Mental State Examination scores. Curr Alzheimer Res, 10:45-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Mehra RD,Sharma K,Nyakas C,Vij U (2005). Estrogen receptor alpha and beta immunoreactive neurons in normal adult and aged female rat hippocampus: a qualitative and quantitative study. Brain Res, 1056:22-35. [DOI] [PubMed] [Google Scholar]
- [16].Paramanik V,Thakur MK (2010). Interaction of estrogen receptor associated protein (ERAP) 140 with ER beta decreases but its expression increases in aging mouse cerebral cortex. Cell Mol Neurobiol, 30: 961-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Paramanik V,Thakur MK (2013). Role of CREB signaling in aging brain. Arch Ital Biol, 151:33-42. [DOI] [PubMed] [Google Scholar]
- [18].Kindler J,Weickert CS,Skilleter AJ,Catts SV,Lenroot R,Weickert TW (2015). Selective estrogen receptor modulation increases hippocampal activity during probabilistic association learning in schizophrenia. Neuropsychopharmacology, 40:2388-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Diez-Perez A (2006). Selective estrogen receptor modulators (SERMS). Arq Bras Endocrinol Metab, 50: 720-34. [DOI] [PubMed] [Google Scholar]
- [20].Draper MW,Flowers DE,Huster WJ,Neild JA,Harper KD,Arnaud C (1996). A controlled trial of Raloxifene (LY 139481) HCl: impact on bone turnover and serum lipid profile in healthy postmenopausal women. J Bone Miner Res, 11:835-42. [DOI] [PubMed] [Google Scholar]
- [21].Maximov PY,Lee TM,Jordan VC (2013). The discovery and development of selective estrogen receptor modulators (SERMs) for clinical practice. Curr Clin Pharmacol, 8:135-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sivanandam TM,Thakur MK (2011). Amyloid precursor protein (APP) mRNA level is higher in the old mouse cerebral cortex and is regulated by sex steroids. J Mol Neurosci, 43:235-40. [DOI] [PubMed] [Google Scholar]
- [23].Ghosh S,Thakur MK (2008). PS1 expression is downregulated by gonadal steroids in adult mouse brain. Neurochem Res, 33:365-9. [DOI] [PubMed] [Google Scholar]
- [24].Ghosh S,Thakur MK (2008). PS2 protein expression is upregulated by sex steroids in the cerebral cortex of aging mice. Neurochem Int, 52:363-367. [DOI] [PubMed] [Google Scholar]
- [25].Mahley RW,Rall SC Jr (2000). Apolipoprotein E: far more than a lipid transport protein. Annu Rev Genomics Hum Genet, 1:507-37. [DOI] [PubMed] [Google Scholar]
- [26].Bales KR,Dodart JC,DeMattos RB,Holtzman DM,Paul SM (2002). Apolipoprotein E, amyloid, and Alzheimer disease. Mol Interv, 2:363-75. [DOI] [PubMed] [Google Scholar]
- [27].Mahley RW,Weisgraber KH,Huang Y (2006). Apolipoprotein E4: a causative factor and therapeutic target in neuropathology, including Alzheimer's disease. Proc Natl Acad Sci U S A, 103:5644-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Singh S,Thakur MK (2011). Age-dependent expression of apolipoprotein E in mouse cerebral cortex. J Mol Neurosci, 43(3):251-56. [DOI] [PubMed] [Google Scholar]
- [29].Singh P,Thakur MK (2014). Reduced recognition memory is correlated with decrease in DNA methyltransferase1 and increase in histone deacetylase2 protein expression in old male mice. Biogerontology, 15:339-46. [DOI] [PubMed] [Google Scholar]
- [30].Peleg S,Sananbenesi F,Zovoilis A, et al. (2010). Altered histone acetylation is associated with age-dependent memory impairment in mice. Science, 328:753-6. [DOI] [PubMed] [Google Scholar]
- [31].Smith DE,Rapp PR,McKay HM,Roberts JA,Tuszynski MH (2004). Memory impairment in aged primates is associated with focal death of cortical neurons and atrophy of subcortical neurons. J Neurosci, 24:4373-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Ystad MA,Lundervold AJ,Wehling E, et al. (2009). Hippocampal volumes are important predictors for memory function in elderly women. BMC Med Imaging, 9:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Brickman AM,Stern Y (2009). Aging and memory in humans. Vol. 1 In:Squire LR, editor. Encyclopedia of Neuroscience. Elsevier Ltd, 175-80. [Google Scholar]
- [34].Morrison JH,Baxter MG (2012). The ageing cortical synapse: hallmarks and implications for cognitive decline. Nat Rev Neurosci, 13:240-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Barrientos RM,Frank MG,Watkins LR,Maier SF (2012). Aging-related changes in neuroimmune-endocrine function: implications for hippocampal-dependent cognition. Horm Behav, 62:219-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Chen ZL,Yoshida S,Kato K,Momota Y, et al. (1995). Expression and activity-dependent changes of a novel limbic-serine protease gene in the hippocampus. J Neurosci, 75:5088-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Suzuki J,Yoshida S,Chen ZL,Momota Y,Kato K,Hirata A,Shiosaka S (1995). Ontogeny of neuropsin mRNA expression in the mouse brain. Neurosci Res, 23:345-51. [DOI] [PubMed] [Google Scholar]
- [38].Yousef GM,Diamandis EP (2001). The new human tissue kallikrein gene family: structure, function and association to disease. Endocr Rev, 22:184-204. [DOI] [PubMed] [Google Scholar]
- [39].Matsumoto-Miyai K,Ninomiya A,Yamasaki H,Tamura H,Nakamura Y,Shiosaka S (2003). NMDA-dependent proteolysis of presynaptic adhesion molecule L1 in the hippocampus by neuropsin. J Neurosci, 23:7727-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Poplawski GHD,Tranziska AK,Leshchyns'ka I, et al. (2012). L1CAM increases MAP2 expression via the MAPK pathway to promote neurite outgrowth. Mol Cell Neurosci, 50:169-78. [DOI] [PubMed] [Google Scholar]
- [41].Konar A,Thakur MK (2015). Neuropsin expression correlates with dendritic marker MAP2c level in different brain regions of aging mice. Mol Neurobiol, 51:1130-38. [DOI] [PubMed] [Google Scholar]
- [42].Kumari A,Thakur MK (2014). Age-dependent decline of nogo-a protein in the mouse cerebrum. Cell Mol Neurobiol, 34:1131-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Yamagata M,Sanes JR,Weiner JA (2003). Synaptic adhesion molecules. Curr Opin Cell Biol, 15:621-32. [DOI] [PubMed] [Google Scholar]
- [44].Budreck EC,Scheiffele P (2007). Neuroligin-3 is a neuronal adhesion protein at GABAergic and glutamatergic synapses. Eur J Neurosci, 26:1738-48. [DOI] [PubMed] [Google Scholar]
- [45].Krueger DD,Tuffy LP,Papadopoulos T,Brose N (2012). The role of neurexins and neuroligins in the formation, maturation, and function of vertebrate synapses. Curr Opin Neurobiol, 22:412-22. [DOI] [PubMed] [Google Scholar]
- [46].Kumar D,Thakur MK (2015). Age-related expression of Neurexin1 and Neuroligin3 is correlated with presynaptic density in the cerebral cortex and hippocampus of male mice. Age, 37:17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lubin FD,Roth TL,Sweatt JD (2008). Epigenetic regulation of bdnf gene transcription in the consolidation of fear memory. J Neurosci, 28: 10576-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chouliaras L,Mastroeni D,Delvaux E, et al. (2013). Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer's disease patients. Neurobiol Aging, 34:2091-99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Richardson B (2003). Impact of aging on DNA methylation. Ageing Res Rev. 2: 245-61. [DOI] [PubMed] [Google Scholar]
- [50].Mastroeni D,Grover A,Delvaux E,Whiteside C,Coleman PD,Rogers J (2011). Epigenetic mechanisms in Alzheimer's disease. Neurobiol Aging, 32:1161-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Mastroeni D,Chouliaras L,Grover A,Liang WS,Hauns K,Rogers J,Coleman PD (2013). Reduced RAN expression and disrupted transport between cytoplasm and nucleus; a key event in Alzheimer's disease pathophysiology. PLoS One, 8(1):e53349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Penner MR,Roth TL,Chawla MK,Hoang LT,Roth ED,Lubin FD,Sweatt JD,Worley PF,Barnes CA (2011). Age-related changes in Arc transcription and DNA methylation within the hippocampus. Neurobiol Aging, 32:2198-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Nagata T,Kobayashi N,Ishii J, et al. (2015). Association between DNA methylation of the BDNF promoter region and clinical presentation in Alzheimer's disease. Dement Geriatr Cogn Dis Extra, 5:64-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Zhang Y,Qian YP,Jiang KD,Yu SY,Wang DX,Zhang MY,Jiang SD (2006). Expression levels of APP and PS1 genes in patients with Alzheimer's Disease. Yi Chuan, 28:525-8. [PubMed] [Google Scholar]
- [55].Tohgi H,Utsugisawa K,Nagane Y,Yoshimura M,Genda Y,Ukitsu M (1999). Reduction with age in methylcytosine in the promoter region -224 approximately -101 of the amyloid precursor protein gene in autopsy human cortex. Brain Res Mol Brain Res, 70:288-92. [DOI] [PubMed] [Google Scholar]
- [56].Li E,Bestor TH,Jaenisch R (1992). Targeted mutation of the DNA methyltransferase gene results in embryonic lethality. Cell, 69:915-26. [DOI] [PubMed] [Google Scholar]
- [57].Golshani P,Hutnick L,Schweizer F,Fan G (2005). Conditional Dnmt1 deletion in dorsal forebrain disrupts development of somatosensory barrel cortex and thalamocortical long-term potentiation. Thalamus Relat Syst, 3:227-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Hutnick LK,Golshani P,Namihira M, et al. (2009). DNA hypomethylation restricted to the murine forebrain induces cortical degeneration and impairs postnatal neuronal maturation. Hum Mol Genet, 18:2875-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Feng J,Zhou Y,Campbell SL, et al. (2010). Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat Neurosci, 13: 423-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Liu L,Groen TV,Kadish I, et al. (2011). Insufficient DNA methylation affects healthy aging and promotes age-related health problems. Clin Epigenetics, 2: 349-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Klein CJ,Botuyan MV,Wu Y, et al. (2011). Mutations in DNMT1 cause hereditary sensory neuropathy with dementia and hearing loss. Nat Genet, 43:595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Kanungo MS,Thakur MK (1979). Modulation of acetylation of histones and transcription of chromatin by butyric acid and 17β- estradiol in the brain of rats of various ages. Biochem Biophys Res Commun, 87:266-71. [DOI] [PubMed] [Google Scholar]
- [63].Kanungo MS &Thakur MK (1977) Phosphorylation of chromosomal proteins as a function of age and its modulation by calcium. Biochem Biophys Res Commun 79: 1031-6. [DOI] [PubMed] [Google Scholar]
- [64].Thakur MK,Kanungo MS (1981). Methylation of chromosomal proteins and DNA of rat brain and its modulation by estradiol and calcium during aging. Exp Gerontol, 16:331-7. [DOI] [PubMed] [Google Scholar]
- [65].Fischer A,Sananbenesi F,Wang X,Dobbin M,Tsai LH (2007). Recovery of learning and memory is associated with chromatin remodelling. Nature, 447: 178-82. [DOI] [PubMed] [Google Scholar]
- [66].Pavlopoulos E,Jones S,Kosmidis S,Close M,Kim C,Kovalerchik O,Small SA,Kandel ER (2013). Molecular mechanism for age-related memory loss: the histone-binding protein RbAp48. Sci Transl Med, 5: 200ra115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Singh P,Konar A,Kumar A,Srivas S,Thakur MK (2015). Hippocampal chromatin modifying enzymes are pivotal for scopolamine- induced synaptic plasticity gene expression changes and memory impairment. J Neurochem, 134:642-51 [DOI] [PubMed] [Google Scholar]
- [68].Guan JS,Haggarty SJ,Giacometti E, et al. (2009). HDAC2 negatively regulates memory formation and synaptic plasticity. Nature, 459:55-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Gräff J,Rei D,Guan JS,Wang WY, et al. (2012). An epigenetic blockade of cognitive functions in the neurodegenerating brain. Nature, 483:222-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Kim SE,Ko IG,Kim BK, et al. (2010). Treadmill exercise prevents aging-induced failure of memory through an increase in neurogenesis and suppression of apoptosis in rat hippocampus. Exp Gerontol, 45:357-65. [DOI] [PubMed] [Google Scholar]
- [71].Kuzumaki N,Ikegami D,Tamura R, et al. (2010). Hippocampal epigenetic modification at the doublecortin gene is involved in the impairment of neurogenesis with aging. Synapse, 64:611-6. [DOI] [PubMed] [Google Scholar]
- [72].Thakur MK,Konar A (2014). Brain Aging and associated diseases-Text book on geriatric medicine. In:Sanchetee P, editor. Indian academy of geriatrics. Hyderabad: Paras Medical Publisher, 32-36 [Google Scholar]
- [73].Thakur MK,Konar A,Gautam A (2012). Brain Aging: A Critical Reappraisal. In:Thakur MK andRattan SIS, editors. Brain Aging and Therapeutic Interventions. Auckland: Springer, 1-18. [Google Scholar]
- [74].Intlekofer KA,Berchtold NC,Malvaez M, et al. (2013). Exercise and sodium butyrate transform a subthreshold learning event into long-term memory via a brain-derived neurotrophic factor-dependent mechanism. Neuropsychopharmacology, 38:2027-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Reolon GK,Maurmann N,Werenicz A, et al. (2011). Posttraining systemic administration of the histone deacetylase inhibitor sodium butyrate ameliorates aging-related memory decline in rats. Behav Brain Res, 221:329-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Yoo DY,Kim W,Lee CH,Shin BN,Nam SM,Choi JH,Won MH,Yoon YS,Hwang IK (2012). Melatonin improves D-galactose-induced aging effects on behavior, neurogenesis, and lipid peroxidation in the mouse dentate gyrus via increasing pCREB expression. J Pineal Res, 52:21-8. [DOI] [PubMed] [Google Scholar]
- [77].Kilgore M,Miller CA,Fass DM, et al. (2010). Inhibitors of class 1 histone deacetylases reverse contextual memory deficits in a mouse model of Alzheimer's disease. Neuropsychopharmacology. 35:870-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Abel T,Zukin RS (2008). Epigenetic targets of HDAC inhibition in neurodegenerative and psychiatric disorders. Curr Opin Pharmacol. 8:57-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Subramanian S,Bates SE,Wright JJ,Espinoza-Delgado I,Piekarz RL (2010) Clinical toxicities of histone deacetylase inhibitors, 3: 2751-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Kazantsev AG,Thompson LM (2008). Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nat Rev Drug Discov, 7:854-68. [DOI] [PubMed] [Google Scholar]
- [81].Sigfrido Scarpa,Andrea Fuso,Fabrizio D’Anselmi,Rosaria A Cavallaro (2003). Presenilin 1 gene silencing by S-adenosylmethionine: a treatment for Alzheimer disease? FEBS Letter, 541:145-8. [DOI] [PubMed] [Google Scholar]
- [82].Shea TB,Rogers E,Ashline D,Ortiz D,Sheu MS (2002). Apolipoprotein E deficiency promotes increased oxidative stress and compensatory increases in antioxidants in brain tissue. Free Radic Biol Med. 33:1115-20. [DOI] [PubMed] [Google Scholar]
- [83].Mihalick SM,Ortiz D,Kumar R,Rogers E,Shea TB (2003). Folate and vitamin E deficiency impair cognitive performance in mice subjected to oxidative stress: differential impact on normal mice and mice lacking apolipoprotein E. Neuromolecular Med, 4:197-202. [DOI] [PubMed] [Google Scholar]
- [84].Tchantchou F,Graves M,Ortiz D,Chan A,Rogers E,Shea TB (2006). S-adenosyl methionine: A connection between nutritional and genetic risk factors for neurodegeneration in Alzheimer's disease. J Nutr Health Aging, 10:541-4. [PubMed] [Google Scholar]