Abstract
The mechanisms underlying the increase in the numbers of regulatory T (Treg) cells in chronic infection settings remain unclear. Here we have delineated the phenotype and transcriptional profiles of Treg cells from 18 filarial-infected (Fil+) and 19 filarial-uninfected (Fil-) subjects. We found that the frequencies of Foxp3+ Treg cells expressing CTLA-4, GITR, LAG-3 and IL-10 were significantly higher in Fil+ subjects compared with that in Fil- subjects. Foxp3-expressing Treg-cell populations in Fil+ subjects were also more heterogeneous and had higher expression of IL-10, CCL-4, IL-29, CTLA-4 and TGF-β than Fil- subjects, each of these cytokines having been implicated in immune suppression. Moreover, Foxp3-expressing Treg cells from Fil+ subjects had markedly upregulated expression of activation-induced apoptotic genes with concomitant downregulation of those involved in cell survival. To determine whether the expression of apoptotic genes was due to Treg-cell activation, we found that the expression of CTLA-4, CDk8, RAD50, TNFRSF1A, FOXO3 and RHOA were significantly upregulated in stimulated cells compared with unstimulated cells.
Taken together, our results suggest that in patent filarial infection, the expanded Treg-cell populations are heterogeneous, short-lived, activated and express higher levels of molecules known to modulate immune responsiveness, suggesting that filarial infection is associated with high Treg-cell turnover.
Keywords: chronic infection, Foxp3, regulatory molecules, regulatory T cells
Introduction
Regulatory T (Treg) cells play a crucial role in modulating immune responses, modulation that may have either beneficial or detrimental consequences. Their important beneficial role has been best demonstrated in maintaining immune homeostasis by preventing autoimmune and allergic diseases, and promoting tolerance after transplantation [1-4]. In contrast, Treg cells have a deleterious effect in chronic infections and cancer in which they increase in numbers and frequencies and where they suppress tumor-specific or antigen-specific immune responses [5-8]. There exist two major subsets of Treg cells that are largely dependent on the expression of the canonical transcription factor Foxp3 and their site of differentiation; these are the thymus-derived (tTreg) cells and peripherally-derived (pTreg) cells; however, Foxp3+ Treg cells can be induced in vitro and called induced Treg or iTreg cells [9]. In addition to the Foxp3-expressing Treg cells, other populations of Treg cells include the IL-10-producing type 1 (Tr1) and the TGF-β-producing type 3 (Th3) regulatory T cells [10, 11]. Although the evidence of de novo or in vitro induction of Foxp3+ Treg populations (pTreg cells and iTreg cells) is very clear, the molecular basis for the differentiation of tTreg cells and pTreg cells/iTreg cells populations is less so [12].
A number of studies have shown that the frequencies of both Foxp3-expressing tTreg cells and pTreg cells and other Treg-cell populations (e.g. Tr1 and Th3) are increased during chronic infections and surrounding/within solid tumors. Human lymphatic filarial infection (LF) is one such chronic infection. Caused by Wuchereria bancrofti, Brugia malayi and Brugia timori, LF is subclinical in most people, due in large part to the presence of a regulatory environment that not only suppresses filarial-specific T-cell responses but also diminishes, albeit less profoundly, the immune responses to bystander antigens [13] including those that are vaccine deliverable [14-16]. This downregulated immune responsiveness associated with chronic filarial infections is accompanied by the expansion of Foxp3-expressing Treg cells (tTreg cells and/or pTreg cells) [17-19]. Although the expansion of CD4+CD25+Foxp3+-expressing Treg cells has been demonstrated in W. bancrofti infections [18, 20-23], relatively little is known about their phenotype and the activation status. Thus, we sought to investigate the nature of Foxp-3+ Treg cells in the context of chronic filarial infection through transcriptional profiling and flow cytometry. Our data suggest that in LF the expansion of Foxp3-expressing Treg-cell populations reflects transcriptional heterogeneity related to high turnover and increased expression of inhibitory cell surface molecules known to play important roles in immune regulation.
Results
Study Population
Subjects were enrolled from two neighboring villages in Mali. Filarial-infected subjects were gender-, location- and age-matched to mininize variation in sampling. Thirty-seven subjects participated from, were enrolled in the study with 18 Fil+ and 19 Fil- subjects as described in Table 1. Apart from their infection status and their levels of BMA-specific IgG4, which were significantly higher in the Fil+ compared with that of the Fil- groups (p = 0.04, Table 1), there were no other demographic or clinically significant differences between the 2 groups.
Table 1. Study Population.
| Filarial Status | |||
|---|---|---|---|
|
| |||
| Negative (n = 19) | Positive (n = 18) | p value | |
| Gender (Male/Female) | 14/5 | 13/5 | NSa |
| Wb Circulating Ag Positive GM (U/ml) [95% CI]) | < 32 | 124.1[22.52 – 683.5] | NDa |
| Mp/Wb microfilaremia median (mf/ml) [range] | 0 | 17[0.0 – 136.0] | ND |
| BmA-specific Ab IgG (ug/ml) | 14.7[3.5 – 68.2] | 31.75[5.5 – 344.0] | NSb |
| Median [range] IgG4 (ng/ml) | 0.0[0.0 – 19395.0] | 824.0[0.0 – 7514] | 0.04 |
Not determined
Non significant
Treg cells from Fil+ subjects have higher frequencies of CTLA-4+, GITR+, LAG-3+ and IL-10+ cells
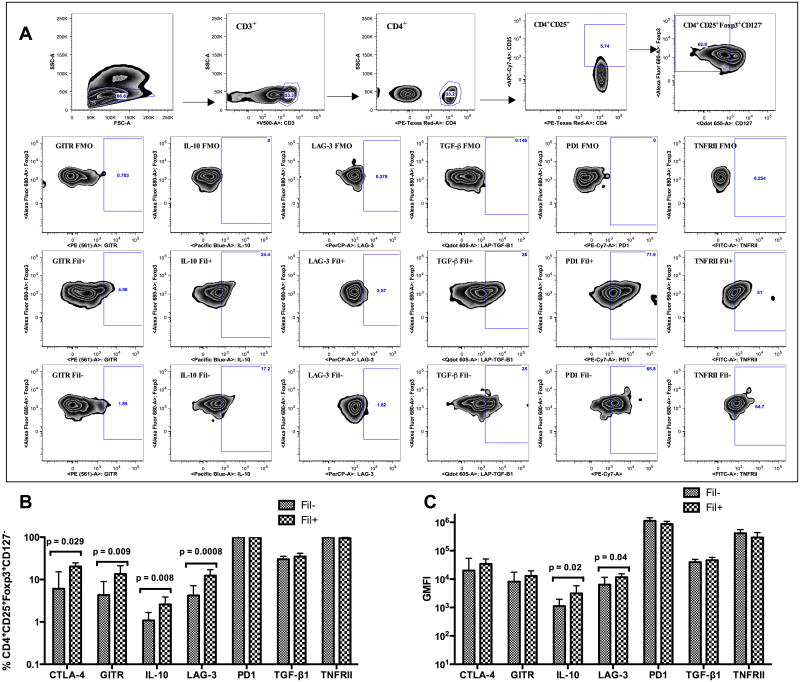
Multiparameter flow cytometry was used to compare the surface expression of the regulatory molecules (CTLA-4, GITR, LAG-3, PD1, LAP-TGF-β, TNFRII) and the expression of intracellular IL-10 on Treg cells in Fil+ and Fil- subjects (gating strategy shown in Figure 1A). As shown in Figure 1B, the frequencies of CD3+CD4+CD25+Foxp3+CD127low Treg cells expressing CTLA-4, GITR, LAG-3 or intracellular IL-10 were significantly increased in the Fil+ compared with that of the Fil- subjects (p = 0.029, 0.009, 0.0008 and 0.008 respectively). When the integrated geometric mean fluorescence intensity (iGMFI) was assessed (Figure 1C), the relative per-cell production of IL-10 and per-cell expression level of LAG-3 by Treg cells were also significantly higher (p = 0.02 and p = 0.04 respectively) in the Fil+ group compared to the Fil- group. However there were no differences in the surface expression of PD1, TGF-β and TNFRII by Treg cells from Fil+ and Fil-.
Figure 1. Treg cells from Fil+ subjects have higher frequencies of CTLA-4+, GITR+, LAG-3+ and IL-10+ cells.
Multiparameter flow cytometry was used to determine the frequencies of Treg cells expressing CTLA-4, GITR, LAG-3, PD1, TNFRII, LAP-TGF-β, or IL-10. (A) The gating strategy for the identification of Treg cells, the FMO gating for positive events and representative graphs of each marker on Treg cells from a Fil+ and Fil- subject are shown. (B) The frequencies of Treg cells (CD4+CD25+Foxp3+CD127low) expressing CTLA-4, GITR, LAG-3, PD1, TNFRII, LAP-TGF-β, or IL-10 were determined by flow cytometry. (C) The integrated geometric mean (GM) fluorescence intensity (iGMFI) for each subset is also shown. Data are shown as geometric mean + 95% confidence interval of 18 Fil+ or 19 Fil− subjects. Statistical significance determined by Mann-Whitney U test.
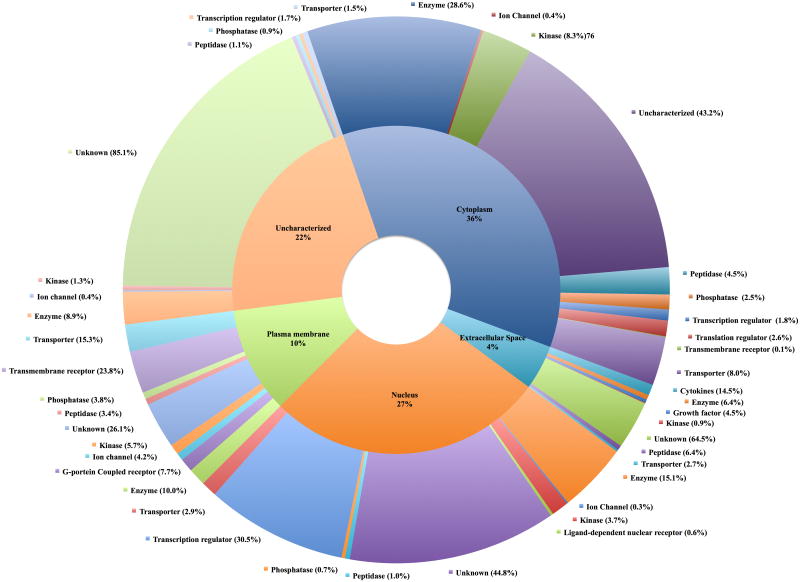
Differentially regulated genes in Treg cells from Fil+ subjects have no known functional category
Highly purified Treg cells from Fil+ and Fil- subjects were used for transcriptional profiling using microarray analysis. The purity of the purified Treg cells was comparable to that of sorted CD4+CD25+Foxp+CD127- as assessed by flow cytometry and was estimated to be more than 95% (Supporting Information figure 1 and figure 2). RNA was extracted from purified Treg cells from Fil+ and Fil- subjects and used for microarray analysis; the fold change of differentially regulated genes of Treg cells from Fil+ over those from Fil- were calculated. The two-fold up- or downregulated genes were analyzed using IPA™ to determine their cellular location and functional category (Figure 2). As can be seen, most (89%) genes could be assigned a cellular location. However, when looking at the genes known to code for proteins predicted to be found in the extracellular space a number with known immunoregulatory functions were found to be upregulated in Treg cells from Fil+ compared with those from Fil- subjects (Table 2). In fact among the top 10% of genes differentially regulated, molecules that have been shown to possess regulatory or suppressive function such as VEGFA, EREG, IL-10, TNFSF15 and PDGFA were respectively 28.7-, 17.4-, 11.2-, 8.7- and 6.21-fold upregulated in Treg cells from Fil+ compared with those from Fil-. Furthermore, CCL-4 [24] and CXCL-16 [25, 26] that have been shown to recruit Treg cells to the site of inflammation [25-27] and IL-29 [28-30] that not only is suppressive but also potentiates dendritic cells to induce Treg cells were 6.1-, 5.5- and 3.1-fold increased in Treg cells from Fil+ compared with those from Fil- (Table 2).
Figure 2. Differentially regulated genes in Treg cells from Fil+ subjects have no known functional category.
Differentially expressed genes were imported into the IPA software to determine the cellular location and functional categories. These data were used to plot the doughnut graph. The inner pie chart represents the cellular location of the differentially expressed gene and the outer pie chart represents the different functional category for each cellular location. For functional category, the percentage shown represents the relative abundance of that category in a specific location.
Table 2.
List of immunoregulatorymolecules in the top 10% and 2-fold up-regulated in Tregs from filarial-infected subjects.
| Symbol | GenBank ID | Fold change | Biological functions |
|---|---|---|---|
| VEGFA | 7422 | 28.681 | May promote tumor growth and possess immunosuppressive properties |
| EREG | 2069 | 17.44 | May stimulate cell proliferation and/or angiogenesis |
| IL10 | 3586 | 11.167 | May function as an angiogenesis inhibitor |
| TNFSF15 | 9966 | 8.739 | May function as an angiogenesis inhibitor |
| PDGFA | 5154 | 6.225 | May promote tumor growth and possess immunosuppressive properties |
| CCL4 | 6351 | 6.134 | Recruit regulatory T cells to inflammatory sites |
| TNFSF4 | 7292 | 5.704 | Potential inhibitor of hypersensitivity |
| CXCL16 | 58191 | 5.574 | Recruit Tregs to tumor sites |
| IL29 | 100620067 | 3.094 | Contribute to Treg expansion by APCs |
Heterogeneous populations of Treg cells with upregulated expression of regulatory markers
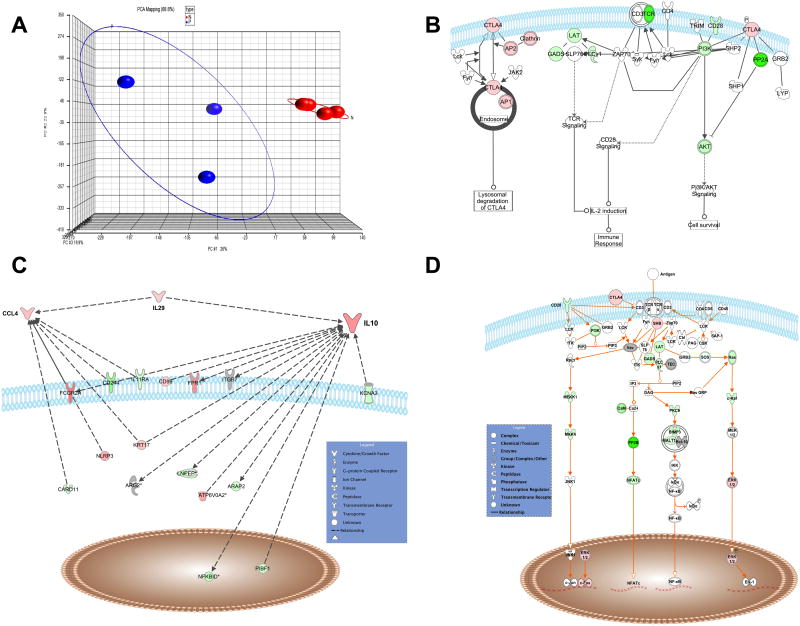
Gene expression profiles of Treg cells from Fil+ and Fil- were compared using Principal Component Analysis (PCA). In figure 3A, PCA demonstrates the distinct clustering of Treg-cell transcription profiles from Fil- and Fil+ patients. Moreover, Treg cells from Fil- patients form a tight cluster indicating the homogeneity in their transcriptional profile whereas Treg cells from Fil+ subjects are relatively scattered reflecting greater Treg-cell heterogeneity in this group. The data were ranked, and genes that were both within the top 10% and significantly two-fold differentially expressed between Fil- and Fil+ were used for pathway analysis (see Materials and methods and supporting Table S2). Based on previously described information [21, 31-33] and flow cytometry data demonstrating that the frequency of Treg cells expressing CTLA-4 and IL-10 were significantly higher in Fil+ subjects, we focused on the pathways related to CTLA-4 and IL-10. As seen in figure 3B, the CTLA-4 signaling pathway was clearly upregulated in the Fil+ compared to Fil- subjects as demonstrated by not only the upregulation of CTLA-4 expression on Treg cells from Fil+ subjects, but also by those molecules involved in the trafficking of CTLA-4 from the endosome to the cell surface. Moreover, IL-10 and the related member of the IL-10 family, IL-29, along with CCL4 (molecules that have been shown to be associated with the modulation of the immune response [28, 34-37]) were upregulated in Treg cells from Fil+ compared with those from Fil- (Figure 3C). In addition, those molecules known to function in opposition to CTLA4 in the TCR signaling pathways such as CD28, Linker for activation of T cell (LAT), phosphatidylinositol-3 kinase (PI3K), phosphoinositide phospholipase C gamma1 (PLCγ1) were downregulated in Treg cells from Fil+ (Figure 3D). The downregulation of T-cell receptor (TCR) signaling processes reflected by the downregulation of CD28 and sets of signaling molecules associated with TCR-mediated activation, in parallel with the coincident upregulation of CTLA-4 is reminiscent of the consequences of effector T-cell activation. Thus, the comparative pathways analysis of Treg cells from Fil+ over Fil- reveals that in chronic filarial infection the increased number of Treg cells is associated with down regulation of TCR signaling, a process that occurs subsequent to T-cell activation.
Figure 3. Heterogeneous populations of Treg cells with upregulated expression of regulatory markers.
(A) The principal component analysis (PCA) in which each sphere represents the result from an individual sample (chip) with the plotted location based upon the grouping of each sample relative to the others is shown. The blue spheres represent Treg cells from Fil+ and those from Fil− are shown in red. (B-D) The list of the top 10% and two-fold differentially regulated genes of Treg cells from Fil+ compared with Treg cells from Fil− was then imported into IPA for pathway anlaysis. The pathways of (B) CTLA-4, (C) IL-10 and (D) TCR signaling with differentially upregulated (red) or downregulated (green) genes in Treg cells from Fil+ relative to Treg cells from Fil− are shown.
Chronic filarial infection is associated with Treg-cell apoptosis
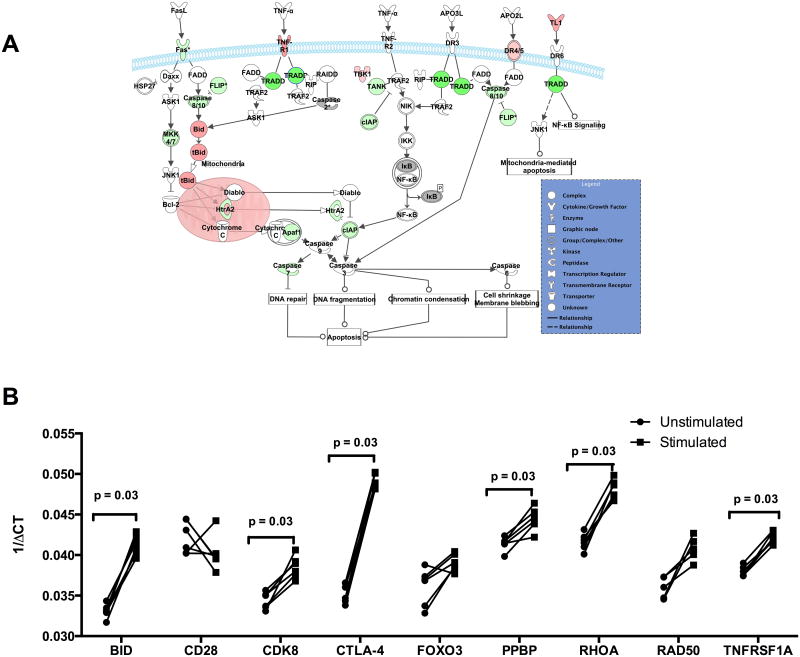
Among the top down- and upregulated genes were those involved in mediating apoptosis or cell death as demonstrated by an increase of the pro-apoptotic genes (Table 3) that include XAF1 (XIAP associated factor 1) and BID (BH3 interacting domain death agonist). Additionally there was an accompanying downregulation of the pro-inflammatory and anti-apoptotic or proliferative genes including CDK8 (cyclin-dependent kinase 8), TNFRSF1A (TNF receptor superfamily member 1A), TNFRSF5, MAP3K4 (mitogen-activated protein kinase kinase kinase 4) and API5 (apoptosis inhibitor 5) among many other genes (Table 3). In addition to the upregulation of pro-apoptotic genes and downregulation of anti-apoptotic genes, the death receptor-associated apoptotic genes including DR4/5 and the mitochondria-associated apoptotic genes including cytochrome c were upregulated in Treg cells from Fil+ subjects (Figure 4A). Furthermore, analysis of all known cell survival genes showed that 142 of the 259 presumed survival genes (Supporting table S3) were down regulated consistent with decreased cell survival (z-score -3.195, overlap p value 3.59 × 10-4).
Table 3. List of apoptotic genes.
| Symbol | Gene Name | Fold Change |
|---|---|---|
| BID | BH3 interacting domain death agonist | 9.28 |
| XAF1 | XIAP associated factor 1 | 8.19 |
| SIRPA | Signal-regulatory protein alpha | 6.90 |
| CTNNA1 | Catenin (cadherin-associated protein), alpha 1, 102kDa | 4.68 |
| MXD1 | MAX dimerization protein 1 | 3.26 |
| TGFBR1 | Transforming growth factor, beta receptor 1 | 2.94 |
| ITGAM | Integrin, alpha M (complement component 3 receptor 3 subunit) | 2.93 |
| FOXO3 | Forkhead box O3 | 2.51 |
| NBN | Nibrin | 2.46 |
| MAP3K8 | Mitogen-activated protein kinase kinasekinase 8 | 2.25 |
| PTEN | Phosphatase and tensin homolog | 2.07 |
| IL6ST | Interleukin 6 signal transducer (gp130, oncostatin M receptor) | -2.16 |
| FOXP1 | Forkhead box P1 | -2.30 |
| API5 | Apoptosis inhibitor 5 | -2.66 |
| VPRBP | Vpr (HIV-1) binding protein | -2.85 |
| TM2D1 | TM2 domain containing 1 | -2.86 |
| TAF9B | TAF9B RNA polymerase II, TATA box binding protein (TBP)-associated factor, 31kDa | -2.90 |
| ZNF10 | Zinc finger protein 10 | -2.99 |
| INPP4A | Inositol polyphosphate-4-phosphatase, type I, 107kDa | -3.21 |
| PPID | Peptidylprolylisomerase D | -3.38 |
| ITGA4 | Integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) | -4.09 |
| MAP3K4 | Mitogen-activated protein kinase kinasekinase 4 | -4.30 |
| POU2AF1 | POU class 2 associating factor 1 | -4.78 |
| HUWE1 | HECT, UBA and WWE domain containing 1, E3 ubiquitin protein ligase | -4.94 |
| ZNF148 | Zinc finger protein 148 | -5.72 |
| IFNAR1 | Interferon (alpha, beta and omega) receptor 1 | -5.85 |
| TNFRSF5 | TNF receptor superfamily member 5 | -6.51 |
| RHOA | Ras homolog family member A | -7.15 |
| PRMT2 | Protein arginine methyltransferase 2 | -7.90 |
| CDK8 | Cyclin-dependent kinase 8 | -17.06 |
Figure 4. Patent filarial infection is associated with upregulation of activation-induced apoptotic genes.
(A) The death receptor-associated apoptosis pathways of the top 10% and two-fold differentially regulated genes in Treg cells from Fil+ subjects, relative to Treg cells from Fil− subjects are shown. Upregulated genes are shaded red while downregulated genes are shaded green. (B) The expression levels of apoptosis and/or survival genes in Treg cells from healthy donors (n= 15) left unstimulated (circle) or stimulated (square) with anti-CD3/CD28 beads are shown. Statistical significance determined by Wilcoxon matched-pairs signed rank test.
To ascertain whether the upregulation of the apoptosis pathways could be a consequence of Treg activation, Treg cells from healthy donors recruited from the National Institutes of Health (NIH) Blood Bank were stimulated with anti-CD3/CD28 beads and assessed by qRT-PCR for the expression of those genes identified in the previous analyses. As can be seen, the activation of the Treg cells was associated with the significant transcriptional upregulation of the genes BID, CDK8, CTLA-4, PPBP, RHOA and TNFRSF1A, molecules shown to be associated with proliferation and apoptosis (Figure 4B).
Discussion
The frequencies of Treg cells increase during chronic infection and at the site of many solid tumors, but the mechanisms underlying their increased numbers and their mode of action remains unclear [1, 3, 4, 38]. In the context of human lymphatic filarial infection, populations of Foxp3-expressing Treg cells and Foxp3-negative Tr1 and Th3 cells expand significantly during the chronic phase of the infection and are associated with a wide range of immune regulatory processes postulated to allow the parasite to persist in the human host [39]. In the current studies we used different approaches to delineate the phenotype and function of Foxp3-expressing Treg cells in the context of a chronic tissue-invasive helminth infection.
In the present study we found that chronic filarial infection was associated with the expansion of multiple and heterogeneous populations of Foxp3-expressing Treg cells. Although the population had received two round of treatment under the mass drug administration program (MDA), filarial infection was still prevalent as shown by the detection of microfilaria and circulating filarial antigen in the population. In fact, the goal of MDA is to eliminate lymphatic filarial infection through disruption of transmission; the results have been mixed despite multiple annual rounds of treatment [40-47]. Furthermore, we found that these Treg cells from Fil+ subjects were associated with upregulated pro-apoptotic genes and the concomitant down regulation of anti-apoptotic and cell survival genes. Although the implication of the complex mechanism of apoptosis and cell survival genes was not investigated in the current study, our data show that in the chronic filarial infection, the expanded population of Foxp3-expressing Treg cells is diverse, highly activated and expresses higher levels of regulatory surface molecules and cytokines. The increased heterogeneity suggests that in the chronic settings Treg cells are being constantly activated, expanding, undergoing apoptosis and repopulating. The increased expression of pro-apoptotic genes combined with down regulation of anti-apoptotic and survival genes observed in our studies corroborates previous data that not only demonstrated high turnover and increased apoptosis as a mechanism underlying Treg-cell accumulation and suppressive activity [48-50] but also demonstrated that filarial infection induced apoptosis of CD4+ T cells [51].
The heterogeneity of Foxp3-expressing Treg cells in Fil+ may also reflect the expansion of multiple subsets of Treg cells or the activation status of Treg cells in peripheral circulation where high levels of filarial antigens are persistent and T cell activation perpetual. In fact, based on the differentiation site, there exist two major subsets of Treg cells in circulation, the thymus-derived (tTreg cells) or peripherally differentiated (pTreg cells) [9]. In addition, it has been shown that Treg cells purified from different organs or generated in varied cytokine environments have distinct transcriptional profiles [52]. Thus, in chronic filarial infection where antigen persistence leads to continual T cell activation, T cells in the peripheral circulation are henceforth comprised of both activated and naïve cells. Furthermore, in addition to the high antigenemia and cytokine environment that clearly contributes to the differentiation of varying Treg subsets [53-57], parasite products may be contributing to the heterogeneity of Foxp3-expressing Treg cells as has been shown for the helminth parasites Heligmosomoides polygyrus and Teladorsagia circumcincta [58]. In fact, it has been shown that helminth parasite (H. polygyrus and T. circumcincta)-derived TGF-β induces de novo Treg differentiation [58]. Whether secreted human filarial parasites products have the capability to induce regulatory T cells remains to be determined though all human filarial parasites for example, secrete a homolog of human TGF-β [59-62].
Because filarial-associated T-cell hyporesponsiveness has been associated with an IL-10-mediated diminution of T-cell proliferation and IFN-γ and IL-2 production [63-66] the assumption has been made that this reflects an underproduction of APC-derived IL-12 [67-70]. In the current study we have been able to show that filarial infection is not only associated with increased frequencies of Treg cells but also that those Treg cells express demonstrable levels of IL-10 as well as CTLA-4, LAG-3, IL-29 and CCL-4 each known to play a role in regulating T-cell activation. Although, we did not assess the suppressive function of these regulatory T cells, a recent study in Indonesia found that depletion of CD25hi cells significantly upregulated cytokine production and the proliferation of B and T cells in patent filarial infection, a finding suggesting that Treg cells from chronically infected LF patients were functionally more suppressive [22].
Despite not being the major source of IL-10 in chronic human infections [18, 19, 71-73] our data demonstrate that Treg cells in chronic filarial infection produce a variety of other regulatory molecules. Among these, CCL-4 plays an important role in Treg-cell-associated suppression of tumor-specific and inflammatory responses in mice [37, 74] and humans [75]. Our data also demonstrate that Treg cells in Fil+ subjects produce IL-29, a member of the IL-10 cytokine family with anti-tumor and anti-viral activity [34, 76, 77] that can program naïve DCs to induce Treg-cell differentiation [30, 78].
In conclusion, our results suggest that chronic filarial infection is associated with the expansion of short-lived heterogeneous populations of Treg cells that express high levels regulatory molecules (e.g. CTLA-4, IL-10, GITR LAG-3, IL-29, and CCL4) known to mediate immune unresponsiveness in T effector cells. Taken together, our results suggest a complex mechanism involving activation, upregulation of suppressive and chemotactic factors that underlies the increased frequencies and numbers of Treg cells associated with chronic infection.
Materials and Methods
Study site and patients
The study was conducted in two villages, Bougoudiana and Tienebougou situated northwest of Bamako in the Kolokani district of Mali. The study was approved by the Institutional Review Board (IRB, NCT00471666) of the National Institutes of Allergy and Infectious Diseases and the Ethics committee of the University of Mali. The study was explained to all participants in French or in Bambara, the local language, and written informed consent was obtained from all participants. This area is endemic for W. bancrofti and Mansonella perstans as has been described previously [79]. Unlike our previous studies when patients had not taken, anti-filaria drug, [18, 79, 80], the region at the time of the current study had received 2 annual doses of ivermectin/albendazole as part of the mass drug administration (MDA) activities of the Malian National Program for the Elimination of Lymphatic Filariasis. Filarial infected subjects were gender-, location- and age-matched to minimize variation in sampling.
Eighteen Fil+ subjects and 19 Fil- subjects were studied (see Table 1 for details). Fil+ was defined by a positive test for circulating filarial Ag (FilAg) (TropBio ELISA) and/or detectable microfilariae (Wb or Mp) in peripheral blood samples drawn between 10 p.m. and 2 a.m. Microfilaremia was assessed by calibrated thick smear (total blood volume = 60 μl). Fil− had a negative test for circulating filarial antigen (FilAg) and no microfilariae seen on thick smear. Blood, serum, and plasma were collected for processing. Filaria-specific antibodies (IgG and IgG4) were determined by ELISA as described previously [81]. PBMCs were purified from whole blood using lymphocyte separation media (LSM) (MP Biomedicals, Solon, OH).
Cell purification
Treg cells defined as CD3+CD4+CD127-CD49d-CD25+ and hereafter called Treg cells, were purified using the EasySep™ Human CD4+CD127lowCD49d- Regulatory T Cell Enrichment Kit (StemCell Technologies, Vancouver, Canada) following the manufacturer's protocol. Briefly CD3+CD4+CD127-CD49d- cells were enriched in a rosetting step and CD25+ cells were selected using magnetic beads. The purity of enriched cells was verified by flow cytometry (Supporting figure S1). After purification, Treg cells from Fil+ and Fil- were directly suspended in 350 μl of Buffer RLT (Qiagen Valencia, CA, USA) containing 1% β-Mercaptoethanol (Sigma Aldrich, St. Louis, MO, USA) and stored at -80°C until processed for microarray analysis.
Flow cytomtery
To assess the expression of regulatory markers on Treg cells, peripheral blood mononuclear cells (PBMCs) from Fil+ and Fil- subjects were isolated, fixed as described previously [18, 70, 80] and transported in liquid nitrogen to the National Institutes of Health (NIH), USA. The fixed cells were thawed and stained with anti-human CD3-V500 (clone UCHT1), CD25- Allophycocyanin-Cy7 (clone M-A251), CD152 –Allophycocyanin (CTLA-4, clone BNI3), (all from BD Bioscience, San Jose, CA, USA), anti-human CD4-PE-Texas Red (clone S3.5) (Life Technologies, Carlsbad, CA, USA), TNFRII-FITC (clone 22235) (R&D Systems, Minneapolis, MN, USA), CD127-eFluor 650NC (clone eBioRDR5), Foxp3-Alexa Fluor 700 (clone PCH101), IL-10-Pacific blue (clone JES3-9D7), PD1-PE-Cy7 (clone eBioJ105),GITR-PE (clone eBioAITR) (all from eBioscience, San Diego, CA, USA), and LAP-TGF-β (clone 27235) was custom-conjugated to Qdot 605 (Life Technologies, Carlsbad, CA, USA). To assess the purity of isolated Treg cells, pre- and post-purification cells were stained with anti-human CD3-Alexa Fluor 700 (clone UCHT1), CD4-Allophycocyanin-Cy7 (clone RPA-T4), CD127-PE-Cy5 (clone eBioRDR5), Foxp3-Alexa Fluor 488 (clone PCH101) (all from eBioscience, San Diego, CA, USA), CD25-PE (Clone M-A251) (BD Bioscience, San Jose, CA, USA). Cells were stained using the routine flow cytometry protocol, acquired on a BD LSRFortessa (BD Bioscience, San Jose, CA, USA) and analyzed using FlowJo (Tree Star, Ashland, OR, USA).
RNA purification for microarray
Eight filaria-infected subjects with the same microfilaremia levels, were age-, sex- and location-matched with eight filaria-uninfected and were selected for microarray analysis. RNA was extracted from Treg cell pellets following the RNeasy 96-well protocol (Qiagen, Valencia, CA) with an additional on-column DNase I treatment. RNA concentrations were measured using Quant-iT Ribogreen RNA reagent (Life Technologies, Carlsbad, CA) on a VICTOR™ X3 Multilabel plate reader (Perkin-Elmer, Waltman, MA). Five nanograms of Treg RNA was subjected to DNA microarray target synthesis using WT-OvationTM Pico system RNA amplification kit and WT-Amp plus ST RNA amplification systems kit according to manufacturer's instructions (Nugen, San Carlos, CA). DNA microarray data was validated by quantitative Real Time PCR (qRT-PCR). qRT-PCR primer design and analysis was done as described previously [82]. Designed primers and probes are listed in the Supporting Information Table 1.
Microarray processing and analysis
Samples were processed as described previously [83] with several changes as follows: Hybridization, fluidics and scanning were performed according to standard Affymetrix protocols (http://www.affymetrix.com). Command Console (CC v3.1, http://www.Affymetrix.com) software was used to convert the image files to cell intensity data (cel files). All cel files, representing individual samples, were normalized using the scaling method within expression console (EC v1.1, http://www.Affymetrix.com) and a scaled target of 1250 to produce the analyzed cel files (chp files) along with the report files. The cel files were imported into Partek Genomics Suite software (Partek, inc. St. Louis, Mo., v6.6 6.12.0420) and quantile normalized to produce the principal component analysis (PCA) graph. An ANOVA was performed within Partek to obtain multiple test corrected p-values using the false discovery rate method [84]. Rankings were assigned from p-values, fold change, signal confidence, and call consistency. The rankings were then combined using custom Excel templates to view trends from the lowest p-value, highest signal confidence, fold change and call consistency performing probe sets. Using the top ranked 10% and a two fold cutoff to determine the differential gene expression list between Treg cells from Fil+ and those from Fil- (Supporting information table 2) were put in IPA (Ingenuity Systems, www.ingenuity.com) for network and functional analyses. Microarray results were confirmed by qRT-PCR with a Pearson Product Moment Correlation coefficient of 0.81. The raw data from the microarrays can be accessed at (http://www.ncbi.nlm.nih.gov/geo/).
Real time quantitative PCR
To ascertain that T cell receptor stimulation in Treg cells are associated with up regulation of pro-apoptotic genes, Treg cells purified from buffy coats obtained from healthy volunteers under a protocol approved by the NIH Clinical Center's Institutional Review Board National Institutes of Health (IRB# 99-CC-0168) were left unstimulated or stimulated with anti-CD3/CD28 beads (Invitrogen, Carlsbad, CA, USA) for 24 hours. RNA was purified using the RNeasy kit (Qiagen, Valencia, CA, USA) according to the manufacturer's instructions and was subsequently converted into cDNA using TaqMan reverse transcription reagents (ABI, Carlsbad, CA, USA) according to the manufacturer's protocol. Real-time quantitative RT-PCR was performed in an ABI 7900HT sequence detection system (ABI, Carlsbad, CA, USA) using TaqMan Assays for CDK8 (Assay ID: Hs00176209_m1), CD28 (Assay ID: Hs01007422_m1), RAD50 (Assay ID: Hs00990023_m1), RUNX1T1 (Assay ID: Hs00231702_m1), PPBP (Assay ID: Hs00234077_m1), BID (Assay ID: Hs00609632_m1), FOXO3 (Assay ID: Hs00818121_m1), RHOA (Assay ID: Hs00357608_m1), TNFRSF1A (Assay ID: Hs00236902_m1) and CTLA-4 (Assay ID: Hs03044418_m1). An endogenous 18s ribosomal (Assay ID: Hs99999901_s1) was used as an RNA control. Relative transcripts were determined by the formula: ΔCT = CTsample − CT18S control and graphed as 1/ΔCT.
Statistical analyses
Data analyses for everything but the microarray data were performed using GraphPad PRISM (GraphPad Software version 6, Inc., San Diego, CA, USA). Statistically significant differences between two groups were analyzed using the non-parametric Mann-Whitney U test for unpaired data and the Wilcoxon matched-pairs signed rank test for paired data. The p values were adjusted for multiple comparisons using the Holm's correction.
Supplementary Material
1.
This work was supported by the Intramural Research Program of the Division of Intramural Research, National Institute of Allergy and Infectious Diseases, National Institutes of Health.
Footnotes
Conflict of Interest Disclosure: Because S. Metenou, D. Sturdevant, S. F. Porcella, A. Klion, and T. B. Nutman are government employees and this is a government work, the work is in the public domain in the United States. Notwithstanding any other agreements, the NIH reserves the right to provide the work to PubMedCentral for display and use by the public, and PubMedCentral may tag or modify the work consistent with its customary practices. You can establish rights outside of the U.S. subject to a government use license.
Nonstandard abbreviations used: CI, confidence interval; Fil-, filarial-uninfected; Fil+, filaria infected; GM, geometric mean; GMFI- geometric mean fluorescence intensity; iGMFI, integrated geometric mean fluorescence intensity; mf, microfilaria; Treg cells, natural regulatory T cells; Teffs, effector T cells
References
- 1.Alunno A, Nocentini G, Bistoni O, Petrillo MG, Bartoloni Bocci E, Ronchetti S, Lo Vaglio E, et al. Expansion of CD4+CD25-GITR+ regulatory T-cell subset in the peripheral blood of patients with primary Sjogren's syndrome: correlation with disease activity. Reumatismo. 2012;64:293–298. doi: 10.4081/reumatismo.2012.293. [DOI] [PubMed] [Google Scholar]
- 2.Thiruppathi M, Rowin J, Ganesh B, Sheng JR, Prabhakar BS, Meriggioli MN. Impaired regulatory function in circulating CD4(+)CD25(high)CD127(low/-) T cells in patients with myasthenia gravis. Clin Immunol. 2012;145:209–223. doi: 10.1016/j.clim.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boni-Schnetzler M, Boller S, Debray S, Bouzakri K, Meier DT, Prazak R, Kerr-Conte J, et al. Free fatty acids induce a proinflammatory response in islets via the abundantly expressed interleukin-1 receptor I. Endocrinology. 2009;150:5218–5229. doi: 10.1210/en.2009-0543. [DOI] [PubMed] [Google Scholar]
- 4.Prantner D, Darville T, Sikes JD, Andrews CW, Jr, Brade H, Rank RG, Nagarajan UM. Critical role for interleukin-1beta (IL-1beta) during Chlamydia muridarum genital infection and bacterial replication-independent secretion of IL-1beta in mouse macrophages. Infect Immun. 2009;77:5334–5346. doi: 10.1128/IAI.00883-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu Q, Darveau RP, Samaranayake LP, Wang CY, Jin L. Differential modulation of human {beta}-defensins expression in human gingival epithelia by Porphyromonas gingivalis lipopolysaccharide with tetra- and penta-acylated lipid A structures. Innate Immun. 2009;15:325–335. doi: 10.1177/1753425909104899. [DOI] [PubMed] [Google Scholar]
- 6.Baranova IN, Kurlander R, Bocharov AV, Vishnyakova TG, Chen Z, Remaley AT, Csako G, et al. Role of human CD36 in bacterial recognition, phagocytosis, and pathogen-induced JNK-mediated signaling. J Immunol. 2008;181:7147–7156. doi: 10.4049/jimmunol.181.10.7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brant KA, Fabisiak JP. Nickel alterations of TLR2-dependent chemokine profiles in lung fibroblasts are mediated by COX-2. Am J Respir Cell Mol Biol. 2008;38:591–599. doi: 10.1165/rcmb.2007-0314OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kubo-Murai M, Hazeki K, Nigorikawa K, Omoto T, Inoue N, Hazeki O. IRAK-4-dependent degradation of IRAK-1 is a negative feedback signal for TLR-mediated NF-kappaB activation. J Biochem. 2008;143:295–302. doi: 10.1093/jb/mvm234. [DOI] [PubMed] [Google Scholar]
- 9.Abbas AK, Benoist C, Bluestone JA, Campbell DJ, Ghosh S, Hori S, Jiang S, et al. Regulatory T cells: recommendations to simplify the nomenclature. Nat Immunol. 2013;14:307–308. doi: 10.1038/ni.2554. [DOI] [PubMed] [Google Scholar]
- 10.Roncarolo MG, Bacchetta R, Bordignon C, Narula S, Levings MK. Type 1 T regulatory cells. Immunol Rev. 2001;182:68–79. doi: 10.1034/j.1600-065x.2001.1820105.x. [DOI] [PubMed] [Google Scholar]
- 11.Weiner HL. Oral tolerance: immune mechanisms and the generation of Th3-type TGF-beta-secreting regulatory cells. Microbes Infect. 2001;3:947–954. doi: 10.1016/s1286-4579(01)01456-3. [DOI] [PubMed] [Google Scholar]
- 12.Himmel ME, MacDonald KG, Garcia RV, Steiner TS, Levings MK. Helios+ and Helios- cells coexist within the natural FOXP3+ T regulatory cell subset in humans. J Immunol. 2013;190:2001–2008. doi: 10.4049/jimmunol.1201379. [DOI] [PubMed] [Google Scholar]
- 13.Metenou S, Kovacs M, Dembele B, Coulibaly YI, Klion AD, Nutman TB. Interferon regulatory factor modulation underlies the bystander suppression of malaria antigen-driven IL-12 and IFN-gamma in filaria-malaria co-infection. European journal of immunology. 2011 doi: 10.1002/eji.201141991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cooper MA, Fehniger TA, Ponnappan A, Mehta V, Wewers MD, Caligiuri MA. Interleukin-1beta costimulates interferon-gamma production by human natural killer cells. Eur J Immunol. 2001;31:792–801. doi: 10.1002/1521-4141(200103)31:3<792::aid-immu792>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 15.Elias D, Britton S, Aseffa A, Engers H, Akuffo H. Poor immunogenicity of BCG in helminth infected population is associated with increased in vitro TGF-beta production. Vaccine. 2008;26:3897–3902. doi: 10.1016/j.vaccine.2008.04.083. [DOI] [PubMed] [Google Scholar]
- 16.Sabin EA, Araujo MI, Carvalho EM, Pearce EJ. Impairment of tetanus toxoid-specific Th1-like immune responses in humans infected with Schistosoma mansoni. J Infect Dis. 1996;173:269–272. doi: 10.1093/infdis/173.1.269. [DOI] [PubMed] [Google Scholar]
- 17.Babu S, Kumaraswami V, Nutman TB. Transcriptional control of impaired Th1 responses in patent lymphatic filariasis by T-box expressed in T cells and suppressor of cytokine signaling genes. Infection and immunity. 2005;73:3394–3401. doi: 10.1128/IAI.73.6.3394-3401.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metenou S, Dembele B, Konate S, Dolo H, Coulibaly SY, Coulibaly YI, Diallo AA, et al. At homeostasis filarial infections have expanded adaptive T regulatory but not classical Th2 cells. J Immunol. 2010;184:5375–5382. doi: 10.4049/jimmunol.0904067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mitre E, Chien D, Nutman TB. CD4(+) (and not CD25+) T cells are the predominant interleukin-10-producing cells in the circulation of filaria-infected patients. J Infect Dis. 2008;197:94–101. doi: 10.1086/524301. [DOI] [PubMed] [Google Scholar]
- 20.Korten S, Hoerauf A, Kaifi JT, Buttner DW. Low levels of transforming growth factor-beta (TGF-beta) and reduced suppression of Th2-mediated inflammation in hyperreactive human onchocerciasis. Parasitology. 2011;138:35–45. doi: 10.1017/S0031182010000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Taylor MD, LeGoff L, Harris A, Malone E, Allen JE, Maizels RM. Removal of regulatory T cell activity reverses hyporesponsiveness and leads to filarial parasite clearance in vivo. J Immunol. 2005;174:4924–4933. doi: 10.4049/jimmunol.174.8.4924. [DOI] [PubMed] [Google Scholar]
- 22.Wammes LJ, Hamid F, Wiria AE, Wibowo H, Sartono E, Maizels RM, Smits HH, et al. Regulatory T cells in human lymphatic filariasis: stronger functional activity in microfilaremics. PLoS Negl Trop Dis. 2012;6:e1655. doi: 10.1371/journal.pntd.0001655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Babu S, Blauvelt CP, Kumaraswami V, Nutman TB. Regulatory networks induced by live parasites impair both Th1 and Th2 pathways in patent lymphatic filariasis: implications for parasite persistence. J Immunol. 2006;176:3248–3256. doi: 10.4049/jimmunol.176.5.3248. [DOI] [PubMed] [Google Scholar]
- 24.Ablamunits V, Bisikirska BC, Herold KC. Human regulatory CD8 T cells. Ann N Y Acad Sci. 2008;1150:234–238. doi: 10.1196/annals.1447.000. [DOI] [PubMed] [Google Scholar]
- 25.Deng L, Chen N, Li Y, Zheng H, Lei Q. CXCR6/CXCL16 functions as a regulator in metastasis and progression of cancer. Biochim Biophys Acta. 2010;1806:42–49. doi: 10.1016/j.bbcan.2010.01.004. [DOI] [PubMed] [Google Scholar]
- 26.Darash-Yahana M, Gillespie JW, Hewitt SM, Chen YY, Maeda S, Stein I, Singh SP, et al. The chemokine CXCL16 and its receptor, CXCR6, as markers and promoters of inflammation-associated cancers. PLoS ONE. 2009;4:e6695. doi: 10.1371/journal.pone.0006695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y, Zhu XY, Du MR, Li DJ. Human trophoblasts recruited T lymphocytes and monocytes into decidua by secretion of chemokine CXCL16 and interaction with CXCR6 in the first-trimester pregnancy. J Immunol. 2008;180:2367–2375. doi: 10.4049/jimmunol.180.4.2367. [DOI] [PubMed] [Google Scholar]
- 28.Dolganiuc A, Kodys K, Marshall C, Saha B, Zhang S, Bala S, Szabo G. Type III interferons, IL-28 and IL-29, are increased in chronic HCV infection and induce myeloid dendritic cell-mediated FoxP3+ regulatory T cells. PLoS ONE. 2012;7:e44915. doi: 10.1371/journal.pone.0044915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He SH, Chen X, Song CH, Liu ZQ, Zhou LF, Ma WJ, Zhao LD, et al. Interferon-lambda mediates oral tolerance and inhibits antigen-specific, T-helper 2 cell-mediated inflammation in mouse intestine. Gastroenterology. 2011;141:249–258. 258 e241–242. doi: 10.1053/j.gastro.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 30.Mennechet FJ, Uze G. Interferon-lambda-treated dendritic cells specifically induce proliferation of FOXP3-expressing suppressor T cells. Blood. 2006;107:4417–4423. doi: 10.1182/blood-2005-10-4129. [DOI] [PubMed] [Google Scholar]
- 31.Joseph SK, Verma SK, Sahoo MK, Dixit S, Verma AK, Kushwaha V, Saxena K, et al. Sensitization with anti-inflammatory BmAFI of Brugia malayi allows L3 development in the hostile peritoneal cavity of Mastomys coucha. Acta Trop. 2011;120:191–205. doi: 10.1016/j.actatropica.2011.08.005. [DOI] [PubMed] [Google Scholar]
- 32.Satoguina J, Mempel M, Larbi J, Badusche M, Loliger C, Adjei O, Gachelin G, et al. Antigen-specific T regulatory-1 cells are associated with immunosuppression in a chronic helminth infection (onchocerciasis) Microbes Infect. 2002;4:1291–1300. doi: 10.1016/s1286-4579(02)00014-x. [DOI] [PubMed] [Google Scholar]
- 33.Satoguina JS, Adjobimey T, Arndts K, Hoch J, Oldenburg J, Layland LE, Hoerauf A. Tr1 and naturally occurring regulatory T cells induce IgG4 in B cells through GITR/GITR-L interaction, IL-10 and TGF-beta. Eur J Immunol. 2008;38:3101–3113. doi: 10.1002/eji.200838193. [DOI] [PubMed] [Google Scholar]
- 34.Tezuka Y, Endo S, Matsui A, Sato A, Saito K, Semba K, Takahashi M, et al. Potential anti-tumor effect of IFN-lambda2 (IL-28A) against human lung cancer cells. Lung Cancer. 2012;78:185–192. doi: 10.1016/j.lungcan.2012.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Suzuki M, Jagger AL, Konya C, Shimojima Y, Pryshchep S, Goronzy JJ, Weyand CM. CD8+CD45RA+CCR7+FOXP3+ T cells with immunosuppressive properties: a novel subset of inducible human regulatory T cells. J Immunol. 2012;189:2118–2130. doi: 10.4049/jimmunol.1200122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Allen C, Thornton P, Denes A, McColl BW, Pierozynski A, Monestier M, Pinteaux E, et al. Neutrophil cerebrovascular transmigration triggers rapid neurotoxicity through release of proteases associated with decondensed DNA. J Immunol. 2012;189:381–392. doi: 10.4049/jimmunol.1200409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobaczewski M, Xia Y, Bujak M, Gonzalez-Quesada C, Frangogiannis NG. CCR5 signaling suppresses inflammation and reduces adverse remodeling of the infarcted heart, mediating recruitment of regulatory T cells. Am J Pathol. 2010;176:2177–2187. doi: 10.2353/ajpath.2010.090759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Thiruppathi M, Rowin J, Li Jiang Q, Sheng JR, Prabhakar BS, Meriggioli MN. Functional defect in regulatory T cells in myasthenia gravis. Ann N Y Acad Sci. 2012;1274:68–76. doi: 10.1111/j.1749-6632.2012.06840.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hoerauf A, Satoguina J, Saeftel M, Specht S. Immunomodulation by filarial nematodes. Parasite Immunol. 2005;27:417–429. doi: 10.1111/j.1365-3024.2005.00792.x. [DOI] [PubMed] [Google Scholar]
- 40.Supali T, Djuardi Y, Bradley M, Noordin R, Ruckert P, Fischer PU. Impact of six rounds of mass drug administration on Brugian filariasis and soil-transmitted helminth infections in eastern Indonesia. PLoS Negl Trop Dis. 2013;7:e2586. doi: 10.1371/journal.pntd.0002586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sunish IP, Munirathinam A, Kalimuthu M, Ashok Kumar V, Tyagi BK. Persistence of Lymphatic Filarial Infection in the Paediatric Population of Rural Community, after Six Rounds of Annual Mass Drug Administrations. J Trop Pediatr. 2013 doi: 10.1093/tropej/fmt101. [DOI] [PubMed] [Google Scholar]
- 42.Ramaiah KD, Vanamail P. Surveillance of lymphatic filariasis after stopping ten years of mass drug administration in rural communities in south India. Trans R Soc Trop Med Hyg. 2013;107:293–300. doi: 10.1093/trstmh/trt011. [DOI] [PubMed] [Google Scholar]
- 43.Mitja O, Paru R, Hays R, Griffin L, Laban N, Samson M, Bassat Q. The impact of a filariasis control program on Lihir Island, Papua New Guinea. PLoS Negl Trop Dis. 2011;5:e1286. doi: 10.1371/journal.pntd.0001286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Centers for Disease, C. and Prevention. Progress toward elimination of lymphatic filariasis--Togo, 2000--2009. MMWR Morb Mortal Wkly Rep. 2011;60:989–991. [PubMed] [Google Scholar]
- 45.Liang JL, King JD, Ichimori K, Handzel T, Pa'au M, Lammie PJ. Impact of five annual rounds of mass drug administration with diethylcarbamazine and albendazole on Wuchereria bancrofti infection in American Samoa. Am J Trop Med Hyg. 2008;78:924–928. [PubMed] [Google Scholar]
- 46.W.H.O. Global programme to eliminate lymphatic filariasis. Wkly Epidemiol Rec. 2007;82:361–380. [PubMed] [Google Scholar]
- 47.Grady CA, de Rochars MB, Direny AN, Orelus JN, Wendt J, Radday J, Mathieu E, et al. Endpoints for lymphatic filariasis programs. Emerg Infect Dis. 2007;13:608–610. doi: 10.3201/eid1304.061063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banz A, Pontoux C, Papiernik M. Modulation of Fas-dependent apoptosis: a dynamic process controlling both the persistence and death of CD4 regulatory T cells and effector T cells. J Immunol. 2002;169:750–757. doi: 10.4049/jimmunol.169.2.750. [DOI] [PubMed] [Google Scholar]
- 49.Weiss EM, Schmidt A, Vobis D, Garbi N, Lahl K, Mayer CT, Sparwasser T, et al. Foxp3-mediated suppression of CD95L expression confers resistance to activation-induced cell death in regulatory T cells. J Immunol. 2011;187:1684–1691. doi: 10.4049/jimmunol.1002321. [DOI] [PubMed] [Google Scholar]
- 50.Yolcu ES, Ash S, Kaminitz A, Sagiv Y, Askenasy N, Yarkoni S. Apoptosis as a mechanism of T-regulatory cell homeostasis and suppression. Immunol Cell Biol. 2008;86:650–658. doi: 10.1038/icb.2008.62. [DOI] [PubMed] [Google Scholar]
- 51.Jenson JS, O'Connor R, Osborne J, Devaney E. Infection with Brugia microfilariae induces apoptosis of CD4(+) T lymphocytes: a mechanism of immune unresponsiveness in filariasis. Eur J Immunol. 2002;32:858–867. doi: 10.1002/1521-4141(200203)32:3<858::AID-IMMU858>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 52.Feuerer M, Hill JA, Kretschmer K, von Boehmer H, Mathis D, Benoist C. Genomic definition of multiple ex vivo regulatory T cell subphenotypes. Proc Natl Acad Sci U S A. 2010;107:5919–5924. doi: 10.1073/pnas.1002006107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li L, Boussiotis VA. Molecular and functional heterogeneity of T regulatory cells. Clin Immunol. 2011 doi: 10.1016/j.clim.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee BO, Jones JE, Peters CJ, Whitacre D, Frelin L, Hughes J, Kim WK, et al. Identification of a unique double-negative regulatory T-cell population. Immunology. 2011;134:434–447. doi: 10.1111/j.1365-2567.2011.03502.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hall JA, Grainger JR, Spencer SP, Belkaid Y. The role of retinoic acid in tolerance and immunity. Immunity. 2011;35:13–22. doi: 10.1016/j.immuni.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dominguez-Villar M, Baecher-Allan CM, Hafler DA. Identification of T helper type 1-like, Foxp3+ regulatory T cells in human autoimmune disease. Nat Med. 2011;17:673–675. doi: 10.1038/nm.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hori S. Developmental plasticity of Foxp3+ regulatory T cells. Curr Opin Immunol. 2010;22:575–582. doi: 10.1016/j.coi.2010.08.004. [DOI] [PubMed] [Google Scholar]
- 58.Grainger JR, Smith KA, Hewitson JP, McSorley HJ, Harcus Y, Filbey KJ, Finney CA, et al. Helminth secretions induce de novo T cell Foxp3 expression and regulatory function through the TGF-beta pathway. J Exp Med. 2010;207:2331–2341. doi: 10.1084/jem.20101074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bennuru S, Meng Z, Ribeiro JM, Semnani RT, Ghedin E, Chan K, Lucas DA, et al. Stage-specific proteomic expression patterns of the human filarial parasite Brugia malayi and its endosymbiont Wolbachia. Proc Natl Acad Sci U S A. 2011;108:9649–9654. doi: 10.1073/pnas.1011481108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bennuru S, Semnani R, Meng Z, Ribeiro JM, Veenstra TD, Nutman TB. Brugia malayi excreted/secreted proteins at the host/parasite interface: stage- and gender-specific proteomic profiling. PLoS Negl Trop Dis. 2009;3:e410. doi: 10.1371/journal.pntd.0000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gomez-Escobar N, Gregory WF, Maizels RM. Identification of tgh-2, a filarial nematode homolog of Caenorhabditis elegans daf-7 and human transforming growth factor beta, expressed in microfilarial and adult stages of Brugia malayi. Infect Immun. 2000;68:6402–6410. doi: 10.1128/iai.68.11.6402-6410.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Korten S, Buttner DW, Schmetz C, Hoerauf A, Mand S, Brattig N. The nematode parasite Onchocerca volvulus generates the transforming growth factor-beta (TGF-beta) Parasitol Res. 2009;105:731–741. doi: 10.1007/s00436-009-1450-9. [DOI] [PubMed] [Google Scholar]
- 63.Mahanty S, Mollis SN, Ravichandran M, Abrams JS, Kumaraswami V, Jayaraman K, Ottesen EA, et al. High levels of spontaneous and parasite antigen-driven interleukin-10 production are associated with antigen-specific hyporesponsiveness in human lymphatic filariasis. J Infect Dis. 1996;173:769–773. doi: 10.1093/infdis/173.3.769. [DOI] [PubMed] [Google Scholar]
- 64.Mahanty S, Luke HE, Kumaraswami V, Narayanan PR, Vijayshekaran V, Nutman TB. Stage-specific induction of cytokines regulates the immune response in lymphatic filariasis. Exp Parasitol. 1996;84:282–290. doi: 10.1006/expr.1996.0114. [DOI] [PubMed] [Google Scholar]
- 65.Mahanty S, Nutman TB. Immunoregulation in human lymphatic filariasis: the role of interleukin 10. Parasite Immunol. 1995;17:385–392. doi: 10.1111/j.1365-3024.1995.tb00906.x. [DOI] [PubMed] [Google Scholar]
- 66.Schonemeyer A, Lucius R, Sonnenburg B, Brattig N, Sabat R, Schilling K, Bradley J, et al. Modulation of human T cell responses and macrophage functions by onchocystatin, a secreted protein of the filarial nematode Onchocerca volvulus. J Immunol. 2001;167:3207–3215. doi: 10.4049/jimmunol.167.6.3207. [DOI] [PubMed] [Google Scholar]
- 67.De Becker G, Moulin V, Tielemans F, De Mattia F, Urbain J, Leo O, Moser M. Regulation of T helper cell differentiation in vivo by soluble and membrane proteins provided by antigen-presenting cells. Eur J Immunol. 1998;28:3161–3171. doi: 10.1002/(SICI)1521-4141(199810)28:10<3161::AID-IMMU3161>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 68.Taki S, Sato T, Ogasawara K, Fukuda T, Sato M, Hida S, Suzuki G, et al. Multistage regulation of Th1-type immune responses by the transcription factor IRF-1. Immunity. 1997;6:673–679. doi: 10.1016/s1074-7613(00)80443-4. [DOI] [PubMed] [Google Scholar]
- 69.Trinchieri G. Interleukin-12: a cytokine produced by antigen-presenting cells with immunoregulatory functions in the generation of T-helper cells type 1 and cytotoxic lymphocytes. Blood. 1994;84:4008–4027. [PubMed] [Google Scholar]
- 70.Metenou S, Kovacs M, Dembele B, Coulibaly YI, Klion AD, Nutman TB. Interferon regulatory factor modulation underlies the bystander suppression of malaria antigen-driven IL-12 and IFN-gamma in filaria-malaria co-infection. Eur J Immunol. 2012;42:641–650. doi: 10.1002/eji.201141991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Puntambekar SS, Bergmann CC, Savarin C, Karp CL, Phares TW, Parra GI, Hinton DR, et al. Shifting hierarchies of interleukin-10-producing T cell populations in the central nervous system during acute and persistent viral encephalomyelitis. J Virol. 2011;85:6702–6713. doi: 10.1128/JVI.00200-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nylen S, Maurya R, Eidsmo L, Manandhar KD, Sundar S, Sacks D. Splenic accumulation of IL-10 mRNA in T cells distinct from CD4+CD25+ (Foxp3) regulatory T cells in human visceral leishmaniasis. J Exp Med. 2007;204:805–817. doi: 10.1084/jem.20061141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Anderson CF, Oukka M, Kuchroo VJ, Sacks D. CD4(+)CD25(-)Foxp3(-) Th1 cells are the source of IL-10-mediated immune suppression in chronic cutaneous leishmaniasis. J Exp Med. 2007;204:285–297. doi: 10.1084/jem.20061886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Morlacchi S, Soldani C, Viola A, Sarukhan A. Self-antigen presentation by mouse B cells results in regulatory T-cell induction rather than anergy or clonal deletion. Blood. 2011;118:984–991. doi: 10.1182/blood-2011-02-336115. [DOI] [PubMed] [Google Scholar]
- 75.Joosten SA, van Meijgaarden KE, Savage ND, de Boer T, Triebel F, van der Wal A, de Heer E, et al. Identification of a human CD8+ regulatory T cell subset that mediates suppression through the chemokine CC chemokine ligand 4. Proc Natl Acad Sci U S A. 2007;104:8029–8034. doi: 10.1073/pnas.0702257104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu BS, Janssen HL, Boonstra A. Type I and III interferons enhance IL-10R expression on human monocytes and macrophages, resulting in IL-10-mediated suppression of TLR-induced IL-12. Eur J Immunol. 2012;42:2431–2440. doi: 10.1002/eji.201142360. [DOI] [PubMed] [Google Scholar]
- 77.Commins S, Steinke JW, Borish L. The extended IL-10 superfamily: IL-10, IL-19, IL-20, IL-22, IL-24, IL-26, IL-28, and IL-29. J Allergy Clin Immunol. 2008;121:1108–1111. doi: 10.1016/j.jaci.2008.02.026. [DOI] [PubMed] [Google Scholar]
- 78.Witte K, Witte E, Sabat R, Wolk K. IL-28A, IL-28B, and IL-29: promising cytokines with type I interferon-like properties. Cytokine Growth Factor Rev. 2010;21:237–251. doi: 10.1016/j.cytogfr.2010.04.002. [DOI] [PubMed] [Google Scholar]
- 79.Metenou S, Dembele B, Konate S, Dolo H, Coulibaly SY, Coulibaly YI, Diallo AA, et al. Patent filarial infection modulates malaria-specific type 1 cytokine responses in an IL-10-dependent manner in a filaria/malaria-coinfected population. J Immunol. 2009;183:916–924. doi: 10.4049/jimmunol.0900257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Metenou S, Dembele B, Konate S, Dolo H, Coulibaly YI, Diallo AA, Soumaoro L, et al. Filarial infection suppresses malaria-specific multifunctional Th1 and Th17 responses in malaria and filarial coinfections. J Immunol. 2011;186:4725–4733. doi: 10.4049/jimmunol.1003778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Lal RB, Ottesen EA. Enhanced diagnostic specificity in human filariasis by IgG4 antibody assessment. J Infect Dis. 1988;158:1034–1037. doi: 10.1093/infdis/158.5.1034. [DOI] [PubMed] [Google Scholar]
- 82.Virtaneva K, Porcella SF, Graham MR, Ireland RM, Johnson CA, Ricklefs SM, Babar I, et al. Longitudinal analysis of the group A Streptococcus transcriptome in experimental pharyngitis in cynomolgus macaques. Proc Natl Acad Sci U S A. 2005;102:9014–9019. doi: 10.1073/pnas.0503671102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Li M, Lai Y, Villaruz AE, Cha DJ, Sturdevant DE, Otto M. Gram-positive three-component antimicrobial peptide-sensing system. Proc Natl Acad Sci U S A. 2007;104:9469–9474. doi: 10.1073/pnas.0702159104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.