Abstract
Our recent studies of microRNA (miRNA) expression signatures in human cancers revealed that microRNA-26a (miRNA-26a) and microRNA-26b (miRNA-26b) were significantly reduced in cancer tissues. To date, few reports have provided functional analyses of miR-26a or miR-26b in renal cell carcinoma (RCC). The aim of the present study was to investigate the functional significance of miR-26a and miR-26b in RCC and to identify novel miR-26a/b-mediated cancer pathways and target genes involved in RCC oncogenesis and metastasis. Downregulation of miR-26a or miR-26b was confirmed in RCC clinical specimens. Restoration of miR-26a or miR-26b in RCC cell lines (786-O and A498) revealed that these miRNAs significantly inhibited cancer cell migration and invasion. Our in silico analysis and luciferase reporter assays showed that lysyl oxidase-like 2 (LOXL2) and procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 (PLOD2) were directly regulated by these miRNAs. Moreover, downregulating the PLOD2 gene significantly inhibited cell migration and invasion in RCC cells. Thus, our data showed that two genes promoting metastasis, LOXL2 and PLOD2, were epigenetically regulated by tumor-suppressive microRNAs, miR-26a and miR-26b, providing important insights into the molecular mechanisms of RCC metastasis.
Keywords: LOXL2, microRNA, miR-26a, miR-26b, PLOD2, renal cell carcinoma
Introduction
Renal cell carcinoma (RCC) is a disease in which cells in the kidney tubules undergo oncogenic transformation. RCC has multiple subtypes and may occur in hereditary (2–3% of RCC) or sporadic forms (1,2). RCC is the third most common urological cancer and accounts for 3% of all adult neoplasias. Clear cell RCC (ccRCC) is the most common subtype of sporadic RCC (~80%) (1). The standard curative treatment for localized diseases remains surgical excision with total nephrectomy. In contrast, at diagnosis, ~30% of RCCs have already metastasized. The 5-year survival rate in patients with advanced stage RCC is poor (5–10%) due to recurrence or distant metastasis (3,4). Recent molecularly targeted therapy has improved the survival rate of patients with advanced RCC (5,6). However, almost all patients eventually relapse or show distant metastasis due to acquired resistance to molecularly targeted therapy. Identifying molecular pathways responsible for RCC metastasis could provide novel approaches for the development of therapies that block the RCC metastatic pathways.
The discovery of microRNA (miRNA) in the human genome provided new directions in cancer research. The miRNAs are endogenous small RNA molecules (19–22 bases long) that regulate protein coding gene expression by repressing translation or cleaving RNA transcripts in a sequence-specific manner (7,8). Numerous studies have shown that miRNAs are aberrantly expressed in many human cancers, and they have significant roles in the initiation, development and metastasis of those cancers (9–11). Moreover, normal regulatory mechanisms can be disrupted by the aberrant expression of tumor-suppressive or oncogenic miRNAs in cancer cells. Therefore, identifying aberrantly expressed miRNAs is an important first step toward elucidating miRNA-mediated oncogenic pathways.
Using miRNA expression signatures, we have identified molecular pathways in RCC that are mediated by aberrantly expressed miRNAs (12–15). For example, downregulation of tumor-suppressive miR-218 promoted cancer cell migration and invasion through dysregulation of the focal adhesion pathway. In this regard, caveolin-2 has an oncogenic function in RCC cells (13). The epithelial-mesenchymal transition (EMT)-related miR-200 family (miR-200a/b/c, miR-141 and miR-429) is significantly downregulated in RCC where they act as tumor suppressors that target the focal adhesion and ErbB signaling pathways (14). The miR-143/145 cluster was frequently reduced in RCC tissues; restoration of these miRNAs significantly inhibited RCC cell proliferation and invasion through targeting of hexokinase-2 (16). More recently, expression of the miR-23b/27b cluster was significantly decreased in ccRCC tissues and associated with pathological grade and stage of the disease (17).
Our miRNA expression signatures of human cancers revealed that miR-26a and miR-26b were frequently downregulated in various types of cancer tissues (10,18,19), suggesting that these miRNAs act as tumor suppressors targeting several oncogenic pathways. Database searches revealed that there were few reports of functional analyses of miR-26a or miR-26b in RCC. The aim of the present study was to investigate the functional significance of miR-26a and miR-26b and to identify molecular targets and pathways contributing to metastasis in RCC cells by miR-26a or miR-26b regulation. We expect that this analysis will provide important insights into the potential molecular mechanisms of RCC oncogenesis and metastasis and will facilitate the development of therapeutic strategies for the treatment of the disease.
Materials and methods
RCC clinical specimens and cell culture
A total of 15 pairs of ccRCC specimens and corresponding non-cancerous specimens were collected from patients who had undergone radical nephrectomy at Chiba University Hospital (Chiba, Japan) from 2012 to 2015. These specimens were staged according to the General Rule for Clinical and Pathological Studies on Renal Cell Carcinoma based on the American Joint Committee on Cancer (AJCC)-UICC TNM classification. The clinicopathological characteristics of the patients are summarized in Table I. Before tissue collection, written informed consent of tissue donation for research purposes was obtained from all the patients.
Table I.
Characteristics of ccRCC clinical specimens.
| No. | Pathology | Grade | pT | INF | v | ly | eg or ig | fc | im | rc | rp | s |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Clear cell | G2 | T1a | a | 0 | 0 | eg | 1 | 0 | 0 | 0 | 0 |
| 2 | Clear cell | G1>G2 | T1a | a | 0 | 0 | eg | 1 | 0 | 0 | 0 | 0 |
| 3 | Clear cell | G3>G2 | T1b | a | 0 | 0 | eg | 1 | 0 | 0 | 0 | 0 |
| 4 | Clear cell | G2>G3>G1 | T1a | a | 0 | 0 | eg | 1 | 0 | 0 | 0 | 0 |
| 5 | Clear cell | G2>G3 | T1b | a | 0 | 0 | eg | 1 | 1 | 0 | 0 | 0 |
| 6 | Clear cell | G2>G3 | T3a | a | 1 | 0 | eg | 1 | 0 | 0 | 0 | 0 |
| 7 | Clear cell | G2>G3>G1 | T3a | b | 1 | 0 | ig | 0 | 1 | 1 | 0 | 0 |
| 8 | Clear cell | G2>G3>G1 | T3a | b | 1 | 0 | ig | 1 | 0 | 0 | 0 | 0 |
| 9 | Clear cell | G3 | T3a | b | 1 | 0 | ig | 0 | 0 | 0 | 0 | 0 |
| 10 | Clear cell | G1>G2 | T1b | a | 0 | 0 | eg | 1 | 0 | 0 | 0 | 0 |
| 11 | Clear cell | G2>G1>G3 | T3a | b | 1 | 0 | ig | 0 | 0 | 0 | 0 | 0 |
| 12 | Clear cell | G2 | T1a | a | 0 | 0 | eg | 0 | 0 | 0 | 0 | 0 |
| 13 | Clear cell | G2>G1>>G3 | T1b | b | 0 | 0 | eg | 1 | 0 | 0 | 0 | 0 |
| 14 | Clear cell | G2>G1 | T1a | b | 0 | 0 | eg | 1 | 0 | 0 | 0 | 0 |
| 15 | Clear cell | G2 | T1b | a | 0 | 0 | eg | 0 | 0 | 0 | 0 | 0 |
INF, infiltration; v, vein; ly, lymph node; eg, expansive growth; ig, infiltrative growth; fc, capsular formation; im, intrarenal metastasis; rc, renal capsule invasion; rp, pelvis invasion; s, sinus invasion.
We used two human RCC cell lines (786-O and A498) obtained from the American Type Culture Collection (ATCC; Manassas, VA, USA) as previously described (12–14).
Quantitative real-time reverse transcription polymerase chain reaction (qRT-PCR)
The procedure for PCR quantification was previously described. TaqMan probes and primers for LOXL2 (P/N: Hs00158757_ml; Applied Biosystems, Foster City, CA, USA), PLOD2 (P/N: Hs01118190_ml; Applied Biosystems) and GUSB (the internal control; P/N: Hs00939627_ml; Applied Biosystems) were assay-on-demand gene expression products. The expression levels of miR-26a (assay ID: 000405; Applied Biosystems) and miR-26b (assay ID: 000407; Applied Biosystems) were analyzed by TaqMan quantitative real-time RT-PCR (TaqMan MicroRNA assay; Applied Biosystems) and normalized to the expression of RNU48 as previously described (12,20,21).
Transfection with mature miRNAs and siRNAs
The following mature miRNAs were used: Ambion Pre-miR miRNA precursor for hsa-miR-26a-5p (product ID: PM10249; Applied Biosystems) and for hsa-miR-26b-5p (product ID: PM12899; Applied Biosystems). The following siRNAs were used: Stealth Select RNAi si-RNA, si-PLOD2 (cat nos. HSS108124 and HSS182371; Invitrogen) and negative control miRNA/siRNA (P/N: AM17111; Applied Biosystems). RNAs were incubated with Opti-MEM (Invitrogen) and Lipofectamine RNAiMax transfection reagent (Invitrogen) as previously described (12,20,21).
Cell proliferation, migration and invasion assays
786-O and A498 cells were transfected with 10 nM miRNAs or si-RNAs by reverse transfection. Cell proliferation, migration and invasion assays were performed as previously described (12,20,21).
Western blotting
Cells were harvested 72 h after transfection, and lysates were prepared. Protein lysates (20 μg) were separated on Mini-PROTEAN TGX gels (Bio-Rad Laboratories, Hercules, CA, USA) and transferred to PVDF membranes. Immunoblotting was performed with rabbit anti-LOXL2 antibodies (1:1000; ab96233; Abcam, Cambridge, UK) and rabbit anti-PLOD2 antibodies (1:300; 21214-1-AP; Proteintech Group, Inc., Chicago, IL, USA). Anti-GAPDH antibodies (1:1,000; ab8245; Abcam) were used as an internal loading control. The membranes were washed and incubated with anti-rabbit IgG horseradish peroxidase (HRP)-linked antibodies (#7074; Cell Signaling Technology). Complexes were visualized with Clarity Western Substrate (Bio-Rad Laboratories).
Screening of miR-26a and miR-26b target genes using in silico analysis and gene expression data
To identify miR-26a/b target genes, we used in silico analysis and genome-wide gene expression analysis. First, we screened genes using TargetScan release 6.2 (http://www.targetscan.org/). Next, to identify upregulated genes in ccRCC clinical specimens, we analyzed publicly available gene expression profiles in the GEO database (accession nos. GSE22541 and GSE36895). Our strategies for miRNA target screening were previously described (12,20,21).
Plasmid construction and dual-luciferase reporter assay
Partial wild-type sequences of the LOXL2 and PLOD2 3′-untranslated region (UTR) or those with deleted miR-26a/b binding sites were inserted between the XhoI-PmeI restriction sites in the 3′-UTR of the hRluc gene in the psiCHECK-2 vector (C8021; Promega, Madison, WI, USA). The procedure for the dual-luciferase reporter assay was previously described (12,20,21).
Statistical analysis
The relationships between the two groups and the numerical values obtained by real-time RT-PCR were analyzed using the Mann-Whitney U-test. The relationships among the three variables and numerical values were analyzed using the Bonferroni-adjusted Mann-Whitney U test. Spearman's rank test was used to evaluate the correlations between the expression of (miR-26a and LOXL2), (miR-26a and PLOD2), (miR-26b and LOXL2) and (miR-26b and PLOD2). All analyses were performed using Expert StatView (version 5; SAS Institute, Inc., Cary, NC, USA).
Results
Expression levels of miR-26a and miR-26b in ccRCC clinical specimens and cell lines
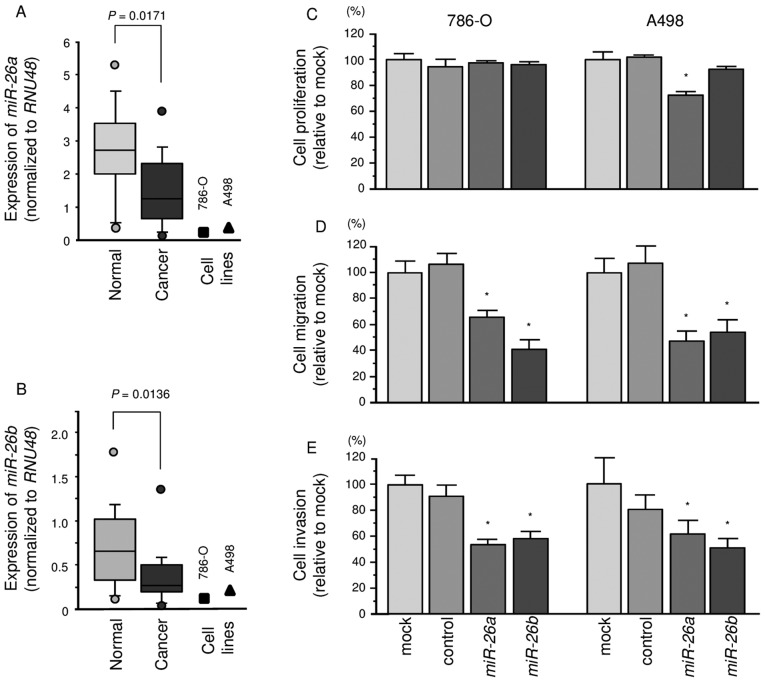
The expression levels of miR-26a and miR-26b were significantly lower in ccRCC specimens than in corresponding non-cancerous specimens (P=0.0171 and P=0.0136, respectively; Fig. 1A and B). In 786-O and A498 cells, the expression levels of miR-26a or miR-26b were lower than in non-cancerous specimens.
Figure 1.
Expression levels of miR-26a and miR-26b in ccRCC clinical specimens and cell lines 786-O and A498. (A and B) Quantitative real-time RT-PCR showed that the expression levels of miR-26a and miR-26b were significantly lower in ccRCC tissues and RCC cell lines than in normal kidney tissues. RNU48 was used as an internal control. (C–E) Effects of miR-26a or miR-26b transfection on RCC cell lines 786-O and A498. (C) Cell proliferation was determined by XTT assays 72 h after transfection with miR-26a or miR-26b (10 nM). (D) Cell migration activity was determined by wound-healing assays 48 h after transfection with miR-26a or miR-26b (10 nM). (E) Cell invasion activity was determined by Matrigel invasion assays 48 h after transfection with miR-26a or miR-26b (10 nM). *P<0.001.
Effects of miR-26a and miR-26b restoration on cell proliferation, migration and invasion activities in ccRCC cells
To investigate the functional effects of miR-26a or miR-26b, we performed gain-of-function studies using mature miRNA transfection of 786-O and A498 cells.
The XTT assays demonstrated that cell proliferation was not inhibited in miR-26a or miR-26b transfectants in comparison with the mock or miR-control transfectants (Fig. 1C).
In contrast, the migration assays demonstrated that cell migration activity was significantly inhibited in miR-26a or miR-26b transfectant cells in comparison with the mock or miR-control transfectants (Fig. 1D). The Matrigel invasion assays demonstrated that cell invasion activity was significantly inhibited in miR-26a or miR-26b transfectant cells in comparison with the mock or miR-control transfectants (Fig. 1E).
Identification of candidate target genes of miR-26a and miR-26b in ccRCC cells
To identify target genes of miR-26a and miR-26b (the seed sequences of the two miRNAs are identical), we used in silico analysis and genome-wide gene expression data. First, we searched the TargetScan database (release 6.2: http://www.targetscan.org/) and identified 3,419 genes that had putative target sites for miR-26a and miR-26b in their 3′-UTRs. Next, we pared down the list of putative candidate genes based on upregulated genes determined by the gene expression data set of RCC clinical specimens in the GEO (Gene Expression Omnibus) database (accession numbers: GSE36895, GSE22541). The flow chart outlining our strategy for identification of candidate target genes of miR-26a and miR-26b is shown in Fig. 2.
Figure 2.
Strategy for selecting target genes regulated by miR-26a and miR-26b in RCC cells.
From this selection, 39 candidate genes were identified as targets of miR-26a and miR-26b (Table II). Among these candidate genes, we focused on LOXL2 and PLOD2 genes because these genes have two conserved target sites for miR-26a and miR-26b in their 3′-UTRs, and function as collagen cross-linking enzymes associated with extracellular matrix (ECM) stiffness. Recent studies showed that aberrantly expressed ECM contributes to cancer cell metastasis (22,23). Therefore, these two genes were chosen for further analysis.
Table II.
Putative candidate target genes regulated by miR-26a and miR-26b in RCC cells.
| Entrez gene ID | Symbol | Gene name | Location | No. of conserved target sites | No. of poorly conserved target sites | GEO (GSE36895, GSE22541 average fold-change |
|---|---|---|---|---|---|---|
| 5352 | PLOD2 | Procollagen-lysine, 2-oxoglutarate 5-dioxygenase 2 | 3q24 | 2 | 0 | 2.2220507 |
| 4017 | LOXL2 | Lysyl oxidase-like 2 | 8p21.3 | 2 | 0 | 2.7719142 |
| 2146 | EZH2 | Enhancer of zeste homolog 2 (Drosophila) | 7q35-q36 | 1 | 0 | 2.0032272 |
| 3625 | INHBB | Inhibin, β B | 2cen-q13 | 1 | 0 | 3.7558112 |
| 3678 | ITGA5 | Integrin, α 5 (fibronectin receptor, α polypeptide) | 12q11-q13 | 1 | 0 | 2.8391342 |
| 23023 | TMCC1 | Transmembrane and coiled-coil domain family 1 | 3q22.1 | 1 | 1 | 2.226072 |
| 1404 | HAPLN1 | Hyaluronan and proteoglycan link protein 1 | 5q14.3 | 1 | 1 | 2.7813237 |
| 7903 | ST8SIA4 | ST8 α-N-acetyl-neuraminide α-2,8-sialyltransferase 4 |
5q21 | 1 | 0 | 3.1741676 |
| 1846 | DUSP4 | Dual specificity phosphatase 4 | 8p12-p11 | 1 | 0 | 2.1518986 |
| 6890 | TAP1 | Transporter 1, ATP-binding cassette, sub-family B (MDR/TAP) | 6p21.3 | 0 | 1 | 2.0403051 |
| 7272 | TTK | TTK protein kinase | 6q14.1 | 0 | 1 | 2.3837836 |
| 170384 | FUT11 | Fucosyltransferase 11 (α (1,3) fucosyltransferase) | 10q22.2 | 0 | 1 | 2.0443428 |
| 22974 | TPX2 | TPX2, microtubule-associated, homolog (Xenopus laevis) | 20q11.2 | 0 | 1 | 2.662108 |
| 2210 | FCGR1B | Fc fragment of IgG, high affinity Ib, receptor (CD64) | 1p11.2 | 0 | 1 | 2.294377 |
| 4747 | NEFL | Neurofilament, light polypeptide | 8p21 | 0 | 1 | 2.1319628 |
| 5836 | PYGL | Phosphorylase, glycogen, liver | 14q21-q22 | 0 | 1 | 2.0643747 |
| 1234 | CCR5 | Chemokine (C-C motif) receptor 5 | 3p21.31 | 0 | 1 | 3.3846455 |
| 55165 | CEP55 | Centrosomal protein 55 kDa | 10q23.33 | 0 | 1 | 2.0711598 |
| 10288 | LILRB2 | Leukocyte immunoglobulin-like receptor, subfamily B (with TM and ITIM domains), member 2 | 19q13.4 | 0 | 1 | 2.454539 |
| 1356 | CP | Ceruloplasmin (ferroxidase) | 3q23-q25 | 0 | 1 | 3.9467278 |
| 3910 | LAMA4 | Laminin, α 4 | 6q21 | 0 | 1 | 2.2182174 |
| 163404 | LPPR5 | Lipid phosphate phosphatase-related protein type 5 | 1p21.3 | 0 | 1 | 2.450066 |
| 5027 | P2RX7 | Purinergic receptor P2X, ligand-gated ion channel, 7 | 12q24 | 0 | 3 | 3.0084689 |
| 330 | BIRC3 | Baculoviral IAP repeat containing 3 | 11q22 | 0 | 1 | 2.2927191 |
| 6507 | SLC1A3 | Solute carrier family 1 (glial high affinity glutamate transporter), member 3 | 5p13 | 0 | 1 | 2.1052346 |
| 2335 | FN1 | Fibronectin 1 | 2q34 | 0 | 1 | 2.4469628 |
| 8701 | DNAH11 | Dynein, axonemal, heavy chain 11 | 7p21 | 0 | 1 | 2.2785249 |
| 79850 | FAM57A | Family with sequence similarity 57, member A | 17p13.3 | 0 | 1 | 2.2900116 |
| 1462 | VCAN | Versican | 5q14.3 | 0 | 1 | 2.524361 |
| 128346 | C1orf162 | Chromosome 1 open reading frame 162 | 1p13.2 | 0 | 1 | 2.2255776 |
| 4015 | LOX | Lysyl oxidase | 5q23.2 | 0 | 1 | 3.3194032 |
| 115761 | ARL11 | ADP-ribosylation factor-like 11 | 13q14.2 | 0 | 1 | 2.4013827 |
| 286336 | FAM78A | Family with sequence similarity 78, member A | 9q34 | 0 | 1 | 2.1942985 |
| 6664 | SOX11 | SRY (sex determining region Y)-box 11 | 2p25 | 0 | 1 | 2.577679 |
| 9770 | RASSF2 | Ras association (RalGDS/AF-6) domain family member 2 | 20p13 | 0 | 1 | 2.619857 |
| 57823 | SLAMF7 | SLAM family member 7 | 1q23.1-q24.1 | 0 | 1 | 2.063896 |
| 58475 | MS4A7 | Membrane-spanning 4-domains, subfamily A, member 7 | 11q12 | 0 | 1 | 2.0315962 |
| 79742 | CXorf36 | Chromosome X open reading frame 36 | Xp11.3 | 0 | 1 | 2.3148956 |
| 146857 | SLFN13 | Schlafen family member 13 | 17q12 | 0 | 1 | 2.6972997 |
Direct regulation of LOXL2 and PLOD2 by miR-26a and miR-26b in ccRCC cells
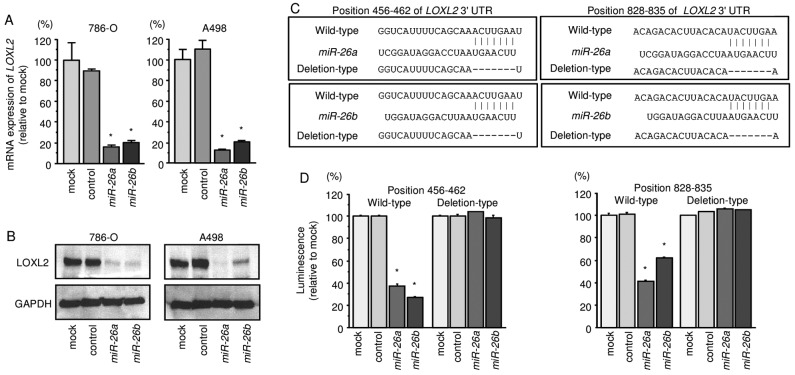
We first performed qRT-PCR and Western blotting to investigate whether expression of the LOXL2 gene and protein were reduced by restoration of miR-26a or miR-26b in 786-O and A498 cells. We found that the mRNA and protein expression levels of LOXL2/LOXL2 were significantly repressed in miR-26a or miR-26b transfectant cells in comparison with mock or miR-control transfectants (Fig. 3A and B).
Figure 3.
Expression of the gene encoding LOXL2 is suppressed by transfection of RCC cell lines 786-O and A498 with miR-26a or miR-26b. (A) LOXL2 mRNA expression was evaluated by quantitative RT-PCR 72 h after transfection with miR-26a or miR-26b (10 nM). GUSB was used as an internal control. *P<0.01. (B) LOXL2 protein expression was evaluated by western blotting 72 h after transfection with miR-26a or miR-26b (10 nM). GAPDH was used as a loading control. (C) miR-26a and miR-26b binding sites in the 3′-UTR of LOXL2 mRNA. (D) Luciferase reporter assays in A498 cells using vectors encoding putative miR-26a and miR-26b target sites at position 456-462 and 828-835 of the LOXL2 3′-UTR. Renilla luciferase values were normalized to firefly luciferase values. *P<0.0001.
Next, to investigate whether LOXL2 mRNA had target sites for miR-26a or miR-26b, we performed luciferase reporter assays in 786-O cells. We used vectors encoding either the partial wild-type sequence of the 3′-UTR of LOXL2, including the predicted miR-26a/b target sites, or deletion vectors lacking the miR-26a/b target sites. We found that the luminescence intensities were significantly reduced by transfection with miR-26a or miR-26b and vectors carrying the wild-type 3′-UTR of LOXL2, whereas transfection with deletion vectors blocked the decrease in luminescence. These data suggested that miR-26a or miR-26b bound directly to specific sites in the 3′-UTR of LOXL2 (Fig. 3C and D).
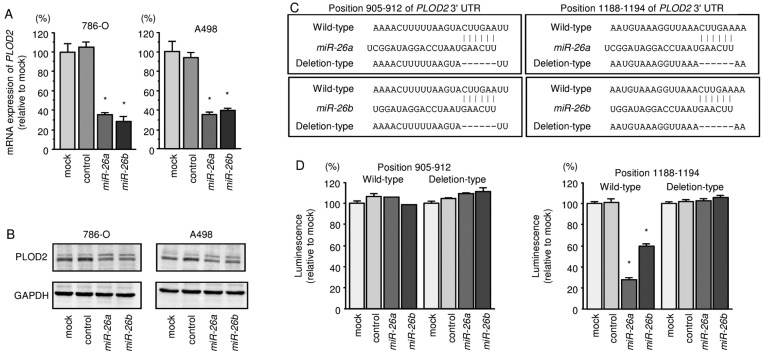
We also found that the mRNA and protein expression levels of PLOD2/PLOD2 were significantly repressed in miR-26a or miR-26b transfectant cells in comparison with mock or miR-control transfectants (Fig. 4A and B). We also observed that the luminescence intensities were significantly reduced by transfection with miR-26a or miR-26b and vectors carrying the wild-type 3′-UTR of PLOD2, whereas transfection with deletion vectors blocked the decrease in luminescence. These data suggested that miR-26a or miR-26b bound directly to specific sites in the 3′-UTR of PLOD2 (Fig. 4C and D).
Figure 4.
Expression of the gene encoding PLOD2 is suppressed by transfection of RCC cell lines 786-O and A498 with miR-26a or miR-26b. (A) PLOD2 mRNA expression was evaluated by quantitative RT-PCR 72 h after transfection with miR-26a or miR-26b (10 nM). GUSB was used as an internal control. *P<0.01. (B) PLOD2 protein expression was evaluated by western blotting 72 h after transfection with miR-26a or miR-26b (10 nM). GAPDH was used as a loading control. (C) miR-26a and miR-26b binding site in the 3′-UTR of PLOD2 mRNA. (D) Luciferase reporter assays in A498 cells using a vector encoding a putative miR-26a and miR-26b target sites at position 905-912 and 1188-1194 of the PLOD2 3′-UTR. Renilla luciferase values were normalized to firefly luciferase values. *P<0.0001.
Silencing PLOD2 affected cell proliferation, migration and invasion activities in ccRCC cells
We recently presented a loss-of-function study of LOXL2 in RCC cells (786-O and A498) by using two siRNAs (786-O and A498) (12). Those data showed that the silencing of LOXL2 significantly suppressed cancer cell migration and invasion activities in RCC cells.
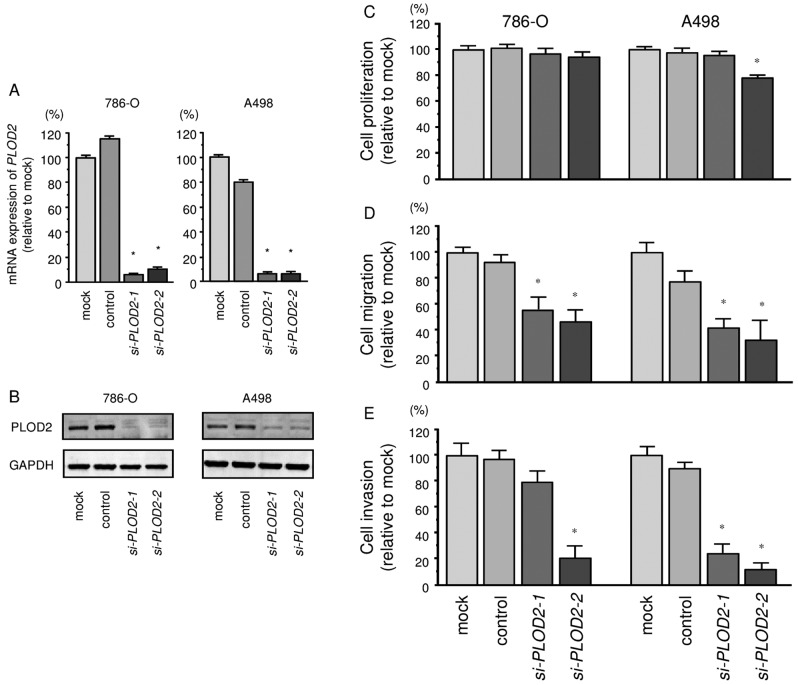
To investigate the functional role of PLOD2 in ccRCC cells, we performed a loss-of-function study using si-PLOD2 transfected cells. First, we evaluated the knockdown efficiency of si-PLOD2 transfection in 786-O and A498 cells. qRT-PCR and western blotting indicated that si-PLOD2 transfection effectively downregulated PLOD2 expression in both cell lines (786-O, P<0.0001; A498, P<0.0001; Fig. 5A and B).
Figure 5.
Effects on RCC cell proliferation, migration and invasion after silencing of PLOD2 mRNA and protein expression with si-PLOD2 transfection. (A) PLOD2 mRNA expression levels were evaluated by quantitative RT-PCR 72 h after transfection with si-PLOD2 (10 nM). GUSB was used as an internal control. *P<0.0001. (B) PLOD2 protein expression levels were evaluated by western blotting 72 h after transfection with si-PLOD2 (10 nM). GAPDH was used as a loading control. (C) Cell proliferation was determined by XTT assays. (D) Cell migration activity was determined by wound-healing assays. (E) Cell invasion activity was determined by Matrigel invasion assays. *P<0.0001.
The XTT assay demonstrated that cell proliferation was not inhibited significantly in si-PLOD2 transfectant cells in comparison with the mock or negative control transfectants (Fig. 5C).
In contrast, the migration assay demonstrated that cell migration activity was significantly inhibited in si-PLOD2 transfectants in comparison with the mock or negative control transfectants (Fig. 5D). The Matrigel invasion assay demonstrated that invasive activity was significantly inhibited in si-PLOD2 transfectants in comparison with the mock or negative control transfectants (Fig. 5E).
Expression of LOXL2 and PLOD2 in ccRCC clinical specimens
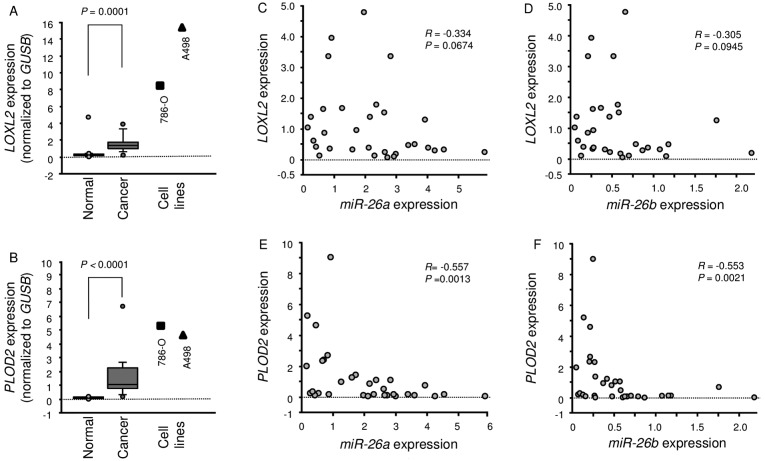
A total of 15 pairs of ccRCC specimens and corresponding non-cancerous specimens were used for expression studies of LOXL2 and PLOD2 using RT-PCR. We showed that LOXL2 and PLOD2 were significantly upregulated in cancer tissues compared with normal tissues (P=0.0001 and P<0.0001, respectively; Fig. 6A and B). Furthermore, Spearman's rank test showed a negative correlation between the expression of miR-26a/PLOD2 and miR-26b/PLOD2 (Fig. 6E and F).
Figure 6.
Expression levels of LOXL2 and PLOD2 in ccRCC clinical specimens and cell lines 786-O and A498. (A and B) Quantitative real-time RT-PCR showed that the expression levels of LOXL2 and PLOD2 were significantly higher in ccRCC tissues and RCC cell lines than in normal kidney tissues. GUSB was used as an internal control. (C and D) Correlations between LOXL2-miR-26a expression or LOXL2-miR-26b expression were determined in RCC clinical specimens. (E and F) Correlations between PLOD2-miR-26a expression or PLOD2-miR-26b expression were determined in RCC clinical specimens.
Discussion
A growing body of evidence has shown that aberrantly expressed miRNAs can disrupt tightly regulated RNA networks in cancer cells and promote human oncogenesis and metastasis (7,9,24–26). Recently, our studies identified a variety of novel RCC molecular pathways regulated by tumor-suppressive miRNAs (12–15). In the present study, we focused on miR-26a and miR-26b because the expression levels of these miRNAs were reduced in the miRNA signatures of various types of cancers (10,18,19,27). Moreover, the functional roles of these miRNAs in RCC cells are not clear. Our present data showed that miR-26a and miR-26b act as tumor suppressors that modulate cancer cell migration and invasion in RCC cells. Our previous studies of oral cancer and prostate cancer demonstrated the tumor-suppressive roles of these miRNAs (19,20), and those findings support the present results obtained with RCC cells. Downregulation and tumor-suppressive roles of miR-26a or miR-26b have been reported in several types of cancer, such as bladder, breast, hepatocellular carcinoma and oral cancer (19,28–30).
In the human genome, the miR-26 family consists of three subtypes of miRNAs: miR-26a-1, miR-26a-2 and miR-26b. The mature sequences of miR-26a-1 and miR-26a-2 are identical, whereas the two nucleotides differ from that of miR-26b (miRBase release 21; http://www.mirbase.org/). The molecular mechanisms responsible for silencing the expression of the miR-26 family are still unclear. A recent study indicated that MYC oncogene directly bound to the promoter regions of miR-26a-1, miR-26a-2 and miR-26b and negatively regulated expression of these miRNAs in prostate cancer cells (31). Overexpression of MYC was observed in RCC clinical specimens (15,32), suggesting MYC might be a mediator for expression control of tumor-suppressive miRNAs in cancer cells.
A single miRNA may regulate multiple protein-coding genes; indeed, bioinformatics studies have shown that miRNAs regulate >30–60% of the protein-coding genes in the human genome (7,33). Reduced expression of tumor-suppressive miRNAs may cause overexpression of oncogenic genes in cancer cells. To better understand RCC oncogenesis and metastasis, we identified miR-26a and miR-26b target genes using in silico analysis. Recent miRNA studies in our laboratory have utilized this strategy to identify novel molecular targets and pathways regulated by tumor-suppressive miRNAs in several cancers, including RCC (12,20).
A total of 39 putative target genes of miR-26a and miR-26b were identified in the present study. Among these genes, we focused on LOXL2 and PLOD2 because they function as collagen cross-linking enzymes. Numerous studies have shown that aberrant expression of collagen cross-linking enzymes promotes extracellular matrix (ECM) stiffening, resulting in enhanced cancer cell migration and invasion (22,34–39). Overexpression of ECM components has been observed in several cancers (21,23,40). Recently, a number of studies indicated that several miRNAs regulated ECM component genes, and aberrantly expressed miRNAs have contributed to cancer cell progression by dysregulation of cell adhesion, polarity and ECM remodeling (21,23). Our past studies found that the tumor-suppressive miR-29-family (miR-29a, miR-29b and miR-29c) and miR-218 directly regulated laminins (LAMC2 and LAMB3) and integrins (ITGA6 and ITGB3), such that restoration of these miRNAs inhibited cancer cell migration and invasion (21,41,42).
Once collagen is secreted, collagen cross-linking occurs on lysine and hydroxylysine residues by the lysyl oxidase (LOX) family of enzymes (22,43). More recently, we showed that the miR-29s-family directly targeted LOXL2 in RCC and lung cancers (12). Overexpression of LOXL2 was observed in RCC clinical specimens and silencing of LOXL2 inhibited cancer cell migration and invasion in ccRCC cell lines (12). Other research groups found that increased expression of LOXL2 is correlated with disease progression, including RCC (34,44). The function of the LOX-family is covalent crosslinking of collagen and/or elastin in the ECM (35,36). Aberrant expression of LOX-family proteins has been reported in several diseases, including cancers (34–39). Interestingly, LOXL2 is a direct transcriptional target of HIF-1. Moreover, nuclear LOXL2 interacts with transcription factor SNAIL1 and represses E-cadherin as well as induces EMT (45,46). In this study, we demonstrated direct regulation of LOXL2 by miR-26a and miR-26b in RCC cells as observed with the miR-29s-family. These findings showed that tumor-suppressive miR-26a/b-LOXL2 is the pivotal pathway contributing to cancer cell migration and invasion in RCC.
In this study, we also focused on the PLOD2 (procollagen-lysine 2-oxyglutarate-dioxygenase) gene as a target of miR-26a and miR-26b and demonstrated the direct regulation of these miRNAs by luciferase reporter assays. PLOD2 encodes an enzyme that mediates collagen lysine hydroxylation. Collagen cross-linking that are derived from hydroxylated lysine residues have increased stability compared with non-hydroxylated lysine residues (22,47). Overexpression of PLOD2 in ccRCC clinical specimens and promoting migration and invasion in cancer cells were observed in the present study. In breast cancer, Kaplan-Meier curves of disease-specific survival stratified by PLOD2 expression revealed that high PLOD2 expression was significantly associated with decreased disease-specific survival (48). Moreover, PLOD2 expression promoted tumor stiffness and was required for metastasis to lymph nodes and lungs (22,48).
In conclusion, miR-26a and miR-26b were significantly downregulated in ccRCC clinical specimens and appeared to function as tumor suppressors through regulation of collagen cross-linking enzymes, LOXL2 and PLOD2, both of which function as oncogenes in this disease. The identification of novel molecular targets and pathways regulated by the tumor-suppressive miR-26a and miR-26b may lead to a better understanding of ccRCC and the development of new therapeutic strategies to treat this disease.
Acknowledgements
The present study was supported by the Japanese Society for the Promotion of Science (KAKENHI), grant numbers (C) 15K10801, (C) 15K20070, (C) 15K20071, and (B) 25293333.
References
- 1.Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373:1119–1132. doi: 10.1016/S0140-6736(09)60229-4. [DOI] [PubMed] [Google Scholar]
- 2.Randall JM, Millard F, Kurzrock R. Molecular aberrations, targeted therapy, and renal cell carcinoma: Current state-of-the-art. Cancer Metastasis Rev. 2014;33:1109–1124. doi: 10.1007/s10555-014-9533-1. [DOI] [PubMed] [Google Scholar]
- 3.Gupta K, Miller JD, Li JZ, Russell MW, Charbonneau C. Epidemiologic and socioeconomic burden of metastatic renal cell carcinoma (mRCC): A literature review. Cancer Treat Rev. 2008;34:193–205. doi: 10.1016/j.ctrv.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Motzer RJ, Russo P. Systemic therapy for renal cell carcinoma. J Urol. 2000;163:408–417. doi: 10.1016/S0022-5347(05)67889-5. [DOI] [PubMed] [Google Scholar]
- 5.Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Oudard S, Negrier S, Szczylik C, Pili R, Bjarnason GA, et al. Overall survival and updated results for sunitinib compared with interferon alfa in patients with meta-static renal cell carcinoma. J Clin Oncol. 2009;27:3584–3590. doi: 10.1200/JCO.2008.20.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Motzer RJ, Hutson TE, Tomczak P, Michaelson MD, Bukowski RM, Rixe O, Oudard S, Negrier S, Szczylik C, Kim ST, et al. Sunitinib versus interferon alfa in metastatic renal-cell carcinoma. N Engl J Med. 2007;356:115–124. doi: 10.1056/NEJMoa065044. [DOI] [PubMed] [Google Scholar]
- 7.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nat Rev Genet. 2008;9:102–114. doi: 10.1038/nrg2290. [DOI] [PubMed] [Google Scholar]
- 8.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Nelson KM, Weiss GJ. MicroRNAs and cancer: Past, present, and potential future. Mol Cancer Ther. 2008;7:3655–3660. doi: 10.1158/1535-7163.MCT-08-0586. [DOI] [PubMed] [Google Scholar]
- 10.Goto Y, Kurozumi A, Enokida H, Ichikawa T, Seki N. Functional significance of aberrantly expressed microRNAs in prostate cancer. Int J Urol. 2015;22:242–252. doi: 10.1111/iju.12700. [DOI] [PubMed] [Google Scholar]
- 11.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6:259–269. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 12.Nishikawa R, Chiyomaru T, Enokida H, Inoguchi S, Ishihara T, Matsushita R, Goto Y, Fukumoto I, Nakagawa M, Seki N. Tumour-suppressive microRNA-29s directly regulate LOXL2 expression and inhibit cancer cell migration and invasion in renal cell carcinoma. FEBS Lett. 2015;589:2136–2145. doi: 10.1016/j.febslet.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 13.Yamasaki T, Seki N, Yoshino H, Itesako T, Hidaka H, Yamada Y, Tatarano S, Yonezawa T, Kinoshita T, Nakagawa M, et al. MicroRNA-218 inhibits cell migration and invasion in renal cell carcinoma through targeting caveolin-2 involved in focal adhesion pathway. J Urol. 2013;190:1059–1068. doi: 10.1016/j.juro.2013.02.089. [DOI] [PubMed] [Google Scholar]
- 14.Yoshino H, Enokida H, Itesako T, Tatarano S, Kinoshita T, Fuse M, Kojima S, Nakagawa M, Seki N. Epithelial-mesenchymal transition-related microRNA-200s regulate molecular targets and pathways in renal cell carcinoma. J Hum Genet. 2013;58:508–516. doi: 10.1038/jhg.2013.31. [DOI] [PubMed] [Google Scholar]
- 15.Yamada Y, Hidaka H, Seki N, Yoshino H, Yamasaki T, Itesako T, Nakagawa M, Enokida H. Tumor-suppressive microRNA-135a inhibits cancer cell proliferation by targeting the c-MYC oncogene in renal cell carcinoma. Cancer Sci. 2013;104:304–312. doi: 10.1111/cas.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yoshino H, Enokida H, Itesako T, Kojima S, Kinoshita T, Tatarano S, Chiyomaru T, Nakagawa M, Seki N. Tumor-suppressive microRNA-143/145 cluster targets hexokinase-2 in renal cell carcinoma. Cancer Sci. 2013;104:1567–1574. doi: 10.1111/cas.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishihara T, Seki N, Inoguchi S, Yoshino H, Tatarano S, Yamada Y, Itesako T, Goto Y, Nishikawa R, Nakagawa M, et al. Expression of the tumor suppressive miRNA-23b/27b cluster is a good prognostic marker in clear cell renal cell carcinoma. J Urol. 2014;192:1822–1830. doi: 10.1016/j.juro.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 18.Kikkawa N, Hanazawa T, Fujimura L, Nohata N, Suzuki H, Chazono H, Sakurai D, Horiguchi S, Okamoto Y, Seki N. miR-489 is a tumour-suppressive miRNA target PTPN11 in hypopharyngeal squamous cell carcinoma (HSCC) Br J Cancer. 2010;103:877–884. doi: 10.1038/sj.bjc.6605811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fukumoto I, Hanazawa T, Kinoshita T, Kikkawa N, Koshizuka K, Goto Y, Nishikawa R, Chiyomaru T, Enokida H, Nakagawa M, et al. MicroRNA expression signature of oral squamous cell carcinoma: Functional role of microRNA-26a/b in the modulation of novel cancer pathways. Br J Cancer. 2015;112:891–900. doi: 10.1038/bjc.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kato M, Goto Y, Matsushita R, Kurozumi A, Fukumoto I, Nishikawa R, Sakamoto S, Enokida H, Nakagawa M, Ichikawa T, et al. MicroRNA-26a/b directly regulate La-related protein 1 and inhibit cancer cell invasion in prostate cancer. Int J Oncol. 2015;47:710–718. doi: 10.3892/ijo.2015.3043. [DOI] [PubMed] [Google Scholar]
- 21.Kinoshita T, Hanazawa T, Nohata N, Kikkawa N, Enokida H, Yoshino H, Yamasaki T, Hidaka H, Nakagawa M, Okamoto Y, et al. Tumor suppressive microRNA-218 inhibits cancer cell migration and invasion through targeting laminin-332 in head and neck squamous cell carcinoma. Oncotarget. 2012;3:1386–1400. doi: 10.18632/oncotarget.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilkes DM, Semenza GL, Wirtz D. Hypoxia and the extracellular matrix: Drivers of tumour metastasis. Nat Rev Cancer. 2014;14:430–439. doi: 10.1038/nrc3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kurozumi A, Goto Y, Matsushita R, Fukumoto I, Kato M, Nishikawa R, Sakamoto S, Enokida H, Nakagawa M, Ichikawa T, et al. Tumor-suppressive microRNA-223 inhibits cancer cell migration and invasion by targeting ITGA3/ITGB1 signaling in prostate cancer. Cancer Sci. 2015;107:84–94. doi: 10.1111/cas.12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wiemer EA. The role of microRNAs in cancer: No small matter. Eur J Cancer. 2007;43:1529–1544. doi: 10.1016/j.ejca.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 25.Singh SK, Pal Bhadra M, Girschick HJ, Bhadra U. MicroRNAs - micro in size but macro in function. FEBS J. 2008;275:4929–4944. doi: 10.1111/j.1742-4658.2008.06624.x. [DOI] [PubMed] [Google Scholar]
- 26.Iorio MV, Croce CM. MicroRNAs in cancer: Small molecules with a huge impact. J Clin Oncol. 2009;27:5848–5856. doi: 10.1200/JCO.2009.24.0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fuse M, Kojima S, Enokida H, Chiyomaru T, Yoshino H, Nohata N, Kinoshita T, Sakamoto S, Naya Y, Nakagawa M, et al. Tumor suppressive microRNAs (miR-222 and miR-31) regulate molecular pathways based on microRNA expression signature in prostate cancer. J Hum Genet. 2012;57:691–699. doi: 10.1038/jhg.2012.95. [DOI] [PubMed] [Google Scholar]
- 28.Lin Y, Chen H, Hu Z, Mao Y, Xu X, Zhu Y, Xu X, Wu J, Li S, Mao Q, et al. miR-26a inhibits proliferation and motility in bladder cancer by targeting HMGA1. FEBS Lett. 2013;587:2467–2473. doi: 10.1016/j.febslet.2013.06.021. [DOI] [PubMed] [Google Scholar]
- 29.Zhang B, Liu XX, He JR, Zhou CX, Guo M, He M, Li MF, Chen GQ, Zhao Q. Pathologically decreased miR-26a antagonizes apoptosis and facilitates carcinogenesis by targeting MTDH and EZH2 in breast cancer. Carcinogenesis. 2011;32:2–9. doi: 10.1093/carcin/bgq209. [DOI] [PubMed] [Google Scholar]
- 30.Zhu Y, Lu Y, Zhang Q, Liu JJ, Li TJ, Yang JR, Zeng C, Zhuang SM. MicroRNA-26a/b and their host genes cooperate to inhibit the G1/S transition by activating the pRb protein. Nucleic Acids Res. 2012;40:4615–4625. doi: 10.1093/nar/gkr1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Koh CM, Iwata T, Zheng Q, Bethel C, Yegnasubramanian S, De Marzo AM. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget. 2011;2:669–683. doi: 10.18632/oncotarget.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tang SW, Chang WH, Su YC, Chen YC, Lai YH, Wu PT, Hsu CI, Lin WC, Lai MK, Lin JY. MYC pathway is activated in clear cell renal cell carcinoma and essential for proliferation of clear cell renal cell carcinoma cells. Cancer Lett. 2009;273:35–43. doi: 10.1016/j.canlet.2008.07.038. [DOI] [PubMed] [Google Scholar]
- 33.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barker HE, Cox TR, Erler JT. The rationale for targeting the LOX family in cancer. Nat Rev Cancer. 2012;12:540–552. doi: 10.1038/nrc3319. [DOI] [PubMed] [Google Scholar]
- 35.Kagan HM, Li W. Lysyl oxidase: Properties, specificity, and biological roles inside and outside of the cell. J Cell Biochem. 2003;88:660–672. doi: 10.1002/jcb.10413. [DOI] [PubMed] [Google Scholar]
- 36.Vadasz Z, Kessler O, Akiri G, Gengrinovitch S, Kagan HM, Baruch Y, Izhak OB, Neufeld G. Abnormal deposition of collagen around hepatocytes in Wilson's disease is associated with hepatocyte specific expression of lysyl oxidase and lysyl oxidase like protein-2. J Hepatol. 2005;43:499–507. doi: 10.1016/j.jhep.2005.02.052. [DOI] [PubMed] [Google Scholar]
- 37.Kim YM, Kim EC, Kim Y. The human lysyl oxidase-like 2 protein functions as an amine oxidase toward collagen and elastin. Mol Biol Rep. 2011;38:145–149. doi: 10.1007/s11033-010-0088-0. [DOI] [PubMed] [Google Scholar]
- 38.Fong SF, Dietzsch E, Fong KS, Hollosi P, Asuncion L, He Q, Parker MI, Csiszar K. Lysyl oxidase-like 2 expression is increased in colon and esophageal tumors and associated with less differentiated colon tumors. Genes Chromosomes Cancer. 2007;46:644–655. doi: 10.1002/gcc.20444. [DOI] [PubMed] [Google Scholar]
- 39.Peinado H, Moreno-Bueno G, Hardisson D, Pérez-Gómez E, Santos V, Mendiola M, de Diego JI, Nistal M, Quintanilla M, Portillo F, et al. Lysyl oxidase-like 2 as a new poor prognosis marker of squamous cell carcinomas. Cancer Res. 2008;68:4541–4550. doi: 10.1158/0008-5472.CAN-07-6345. [DOI] [PubMed] [Google Scholar]
- 40.Liu LX, Jiang HC, Liu ZH, Zhou J, Zhang WH, Zhu AL, Wang XQ, Wu M. Integrin gene expression profiles of human hepatocellular carcinoma. World J Gastroenterol. 2002;8:631–637. doi: 10.3748/wjg.v8.i4.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kinoshita T, Nohata N, Hanazawa T, Kikkawa N, Yamamoto N, Yoshino H, Itesako T, Enokida H, Nakagawa M, Okamoto Y, et al. Tumour-suppressive microRNA-29s inhibit cancer cell migration and invasion by targeting laminin-integrin signalling in head and neck squamous cell carcinoma. Br J Cancer. 2013;109:2636–2645. doi: 10.1038/bjc.2013.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yamamoto N, Kinoshita T, Nohata N, Itesako T, Yoshino H, Enokida H, Nakagawa M, Shozu M, Seki N. Tumor suppressive microRNA-218 inhibits cancer cell migration and invasion by targeting focal adhesion pathways in cervical squamous cell carcinoma. Int J Oncol. 2013;42:1523–1532. doi: 10.3892/ijo.2013.1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gordon MK, Hahn RA. Collagens. Cell Tissue Res. 2010;339:247–257. doi: 10.1007/s00441-009-0844-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hase H, Jingushi K, Ueda Y, Kitae K, Egawa H, Ohshio I, Kawakami R, Kashiwagi Y, Tsukada Y, Kobayashi T, et al. LOXL2 status correlates with tumor stage and regulates integrin levels to promote tumor progression in ccRCC. Mol Cancer Res. 2014;12:1807–1817. doi: 10.1158/1541-7786.MCR-14-0233. [DOI] [PubMed] [Google Scholar]
- 45.Schietke R, Warnecke C, Wacker I, Schödel J, Mole DR, Campean V, Amann K, Goppelt-Struebe M, Behrens J, Eckardt KU, et al. The lysyl oxidases LOX and LOXL2 are necessary and sufficient to repress E-cadherin in hypoxia: Insights into cellular transformation processes mediated by HIF-1. J Biol Chem. 2010;285:6658–6669. doi: 10.1074/jbc.M109.042424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moon HJ, Finney J, Xu L, Moore D, Welch DR, Mure M. MCF-7 cells expressing nuclear associated lysyl oxidase-like 2 (LOXL2) exhibit an epithelial-to-mesenchymal transition (EMT) phenotype and are highly invasive in vitro. J Biol Chem. 2013;288:30000–30008. doi: 10.1074/jbc.C113.502310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Slot AJ, Zuurmond AM, van den Bogaerdt AJ, Ulrich MM, Middelkoop E, Boers W, Karel Ronday H, DeGroot J, Huizinga TW, Bank RA. Increased formation of pyridinoline cross-links due to higher telopeptide lysyl hydroxylase levels is a general fibrotic phenomenon. Matrix Biol. 2004;23:251–257. doi: 10.1016/j.matbio.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 48.Gilkes DM, Bajpai S, Wong CC, Chaturvedi P, Hubbi ME, Wirtz D, Semenza GL. Procollagen lysyl hydroxylase 2 is essential for hypoxia-induced breast cancer metastasis. Mol Cancer Res. 2013;11:456–466. doi: 10.1158/1541-7786.MCR-12-0629. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]