Abstract
TNF is a pleiotropic cytokine with important functions in homeostasis and disease pathogenesis. Recent discoveries have provided insights into TNF biology that introduce new concepts for the development of therapeutics for TNF-mediated diseases. The model of TNF receptor signalling has been extended to include linear ubiquitination and the formation of distinct signalling complexes that are linked with different functional outcomes, such as inflammation, apoptosis and necroptosis. Our understanding of TNF-induced gene expression has been enriched by the discovery of epigenetic mechanisms and concepts related to cellular priming, tolerization and induction of ‘short-term transcriptional memory’. Identification of distinct homeostatic or pathogenic TNF-induced signalling pathways has introduced the concept of selectively inhibiting the deleterious effects of TNF while preserving its homeostatic bioactivities for therapeutic purposes. In this Review, we present molecular mechanisms underlying the roles of TNF in homeostasis and inflammatory disease pathogenesis, and discuss novel strategies to advance therapeutic paradigms for the treatment of TNF-mediated diseases.
Forty years have passed since the description of a serum factor inducing tumour necrosis, 30 years since the cloning and purification of TNF, and almost 20 years since the approval of the first drug that targets TNF1. The initial concept of TNF as a potential drug for the treatment of cancer was followed by the opposite concept of TNF as a drug-target for inflammatory diseases2,3. Currently, five biologic agents targeting TNF are approved for the treatment of rheumatoid arthritis (RA), inflammatory bowel disease (IBD; for example, Crohn disease and ulcerative colitis), psoriasis, psoriatic arthritis, ankylosing spondylitis, juvenile idiopathic arthritis (JIA) and, most recently, hidradenitis suppurativa4,5 (TABLE 1). Notably, lower-cost biosimilar TNF-inhibitors have already been developed and introduced in the clinic6. In addition to the approved indications, TNF-blockade is also used, off-label, in Behçet disease, non-infectious ocular inflammation, and pyoderma gangrenosum, as well as in patients with TNF-receptor associated periodic fever syndrome (TRAPS), adult-onset Still disease and systemic-onset JIA7. In this Review, we focus on the latest discoveries about the biology of TNF, and outline new concepts that have been introduced in therapeutics for TNF-mediated diseases.
Table 1.
TNF inhibitors and approved indications
| Drug | Description | Indications |
|---|---|---|
| Monoclonal antibodies | ||
| Infliximab | Chimeric (mouse and human) whole mAb against TNF | Crohn disease (adult and paediatric), ulcerative colitis (adult and paediatric), RA, PsA, plaque psoriasis, AS |
| Adalimumab | Human whole mAb against TNF | RA, ulcerative colitis, Crohn disease, plaque psoriasis, PsA, AS, JIA (paediatric), hidradenitis suppurativa |
| Certolizumab pegol | Humanized PEGylated Fab fragment of a mAb against TNF | RA, PsA, AS, Crohn's disease |
| Golimumab | Human whole mAb against TNF | RA, PsA, AS, ulcerative colitis |
| Soluble TNFR | ||
| Etanercept | TNFR2 fused to IgG1 Fc | RA, plaque psoriasis, PsA, AS, JIA |
Indications are for adult patients unless noted otherwise. AS, ankylosing spondylitis; JIA, juvenile idiopathic arthritis; mAb, monoclonal antibody; PsA, psoriatic arthritis; RA, rheumatoid arthritis; TNFR, TNF receptor.
TNF-induced signal transduction
TNF receptors
Newly synthesized TNF is expressed initially as a transmembrane protein, which requires proteolytic cleavage by TNFα-converting enzyme (TACE, also named ADAM17) to release soluble TNF8. TACE-dependent release of soluble TNF has been implicated in TNF-mediated inflammatory pathology in disease models9. TNF exerts versatile bioactivities via binding to, and activation of, two distinct receptors: TNF receptor 1 (TNFR1) and TNFR2 (REF. 10). TNFR1 is expressed ubiquitously, bears conserved death-domain motifs, and is activated by both soluble and transmembrane TNF. Expression of TNFR2 is restricted to specific cell types, such as neurons, immune cells and endothelial cells. TNFR2 lacks a death domain and thus is unable to induce programmed cell death directly; this receptor has been proposed to be activated primarily by transmembrane TNF11. One model of TNFR signalling proposes that TNFR1 primarily promotes inflammation and tissue degeneration, whereas TNFR2 mediates local homeostatic effects, such as cell survival and tissue regeneration12 (FIG. 1). This model suggests that selective therapeutic blockade of TNFR1 would keep homeostatic TNFR2 signalling intact; indeed, such a strategy is under development for the treatment of TNF-mediated diseases12.
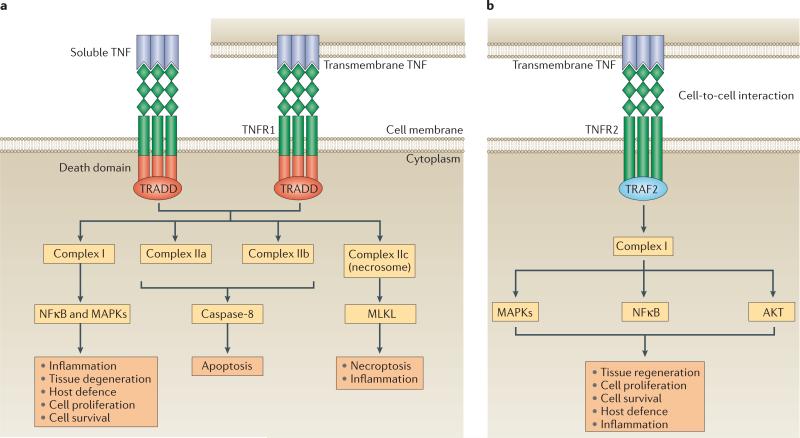
Figure 1. Signalling modalities and bioactivities downstream of TNF receptors.
a | TNF receptor 1 (TNFR1) signalling is activated by both soluble and transmembrane TNF. TNFR1 bears a death domain that recruits the adaptor protein TNFR1-associated death domain protein (TRADD). Ligation of TNFR1 by soluble TNF or transmembrane TNF leads initially to the assembly of complex I, which activates nuclear factor κB (NFκB) and mitogen-activated protein kinases (MAPKs). TNFR1–complex I signalling induces inflammation, tissue degeneration, cell survival and proliferation, and orchestrates the immune defence against pathogens. Alternative signalling modalities, associated with programmed cell death, can also be activated downstream of TNFR1. The formation of the complexes IIa and IIb (also known as ripoptosome) results in apoptosis, whereas complex IIc (necrosome) induces necroptosis and inflammation. b | TNFR2 is proposed to be fully activated primarily by transmembrane TNF, in the context of cell-to-cell interactions. TNFR2 recruits TNFR-associated factor 2 (TRAF2) via its TRAF domain, triggering the formation of complex I and the downstream activation of NFκB, MAPKs and AKT. TNFR2 mediates primarily homeostatic bioactivities including tissue regeneration, cell proliferation and cell survival. This pathway can also initiate inflammatory effects and host defence against pathogens. MLKL, mixed lineage kinase domain-like protein.
Homotrimers of TNF bind to homotrimeric TNFRs to induce signalling10. Ligand-binding to TNFR1 results in the recruitment of the adaptor molecule TNFR1-associated death domain protein (TRADD) and the assembly of distinct signalling complexes, termed complexes I, IIa, IIb and IIc, which lead to distinct functional outcomes13,14 (FIG. 1a).
TNF receptor signalling
TNFR signalling via complex I
Upon TNF binding to TNFR, complex I is assembled at TNFR cyto plasmic domains at the plasma membrane and comprises TRADD, receptor-interacting serine/threonine-protein kinase 1 (RIPK1), TNFR-associated factor 2 (TRAF2), cellular inhibitor of apoptosis protein 1 (cIAP1) or cIAP2, and linear ubiquitin chain assembly complex (LUBAC)14 (FIG. 2). LUBAC consists of haem-oxidized IRP2 ubiquitin ligase-1 (HOIL-1), HOIP (HOIL-1 interacting protein, also known as E3 ubiquitin-protein ligase RNF31) and Sharpin. The current TNFR signalling model postulates the building of a scaffolding ubiquitin network in a step-wise process15. Initially, TRAF2 and cIAP1 or cIAP2 mediate Lys63-linked ubiquitination of complex I components. Subsequently, the LUBAC complex adds Met1-linked linear ubiquitin chains that strengthen the ubiquitin network, resulting in the stabilization of complex I and amplification of signalling16,17. Additional ‘atypical’ ubiquitin chains, such as Lys11-linked chains, can also facilitate signal transduction through TNFRs18.
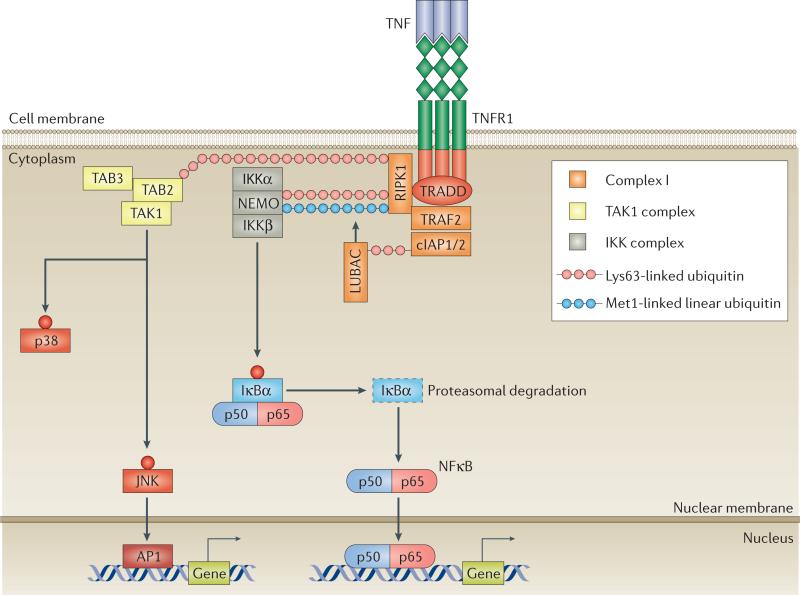
Figure 2. A model of TNFR–complex I signalling.
The binding of homotrimeric TNF to homotrimeric TNF receptors (TNFRs) induces the formation of complex I, comprising TNFR1-associated death domain protein (TRADD), receptor-interacting serine/threonine-protein kinase 1 (RIPK1), TNFR-associated factor 2 (TRAF2), cellular inhibitor of apoptosis protein 1 (cIAP1) or cIAP2, and linear ubiquitin chain assembly complex (LUBAC). cIAPs and LUBAC decorate RIPK1 with scaffolding Lys63-linked and Met1-linked polyubiquitin chains, inducing the recruitment of transforming growth factor (TGF)-β-activated kinase 1 (TAK1) and inhibitor of κB (IκB) kinase (IKK) complexes. TAK1 activates p38 and JUN N-terminal kinase (JNK), leading to the transcription of AP1-target genes. IKKβ phosphorylates IκB, inducing its proteasomal degradation and the release of nuclear factor κB (NFκB). Free NFκB translocates to the nucleus, where it induces the expression of target genes. NEMO, NFκB essential modulator; TAB, TAK1-binding protein.
The scaffolding ubiquitin network enables the recruitment and activation of two signalling complexes: the transforming growth factor (TGF)-β-activated kinase 1 (TAK1) complex, comprising TAK1, TAK1-binding protein 2 (TAB2) and TAB3, and the inhibitor of κB (IκB) kinase (IKK) complex, comprising NFκB essential modulator (NEMO, also known as IKKγ), IKK subunit-α (IKKα) and IKKβ14 (FIG. 2). TAK1 triggers mitogen-activated kinase (MAPK) signalling cascades that lead to activation of downstream JUN N-terminal kinase (JNK) and p38, as well as AP1 transcription factors, whereas IKKβ activates the canonical nuclear factor κB (NFκB) pathway (FIG. 2). Thus, induction of signalling complex I leads to expression of NFκB and AP1 target genes that are important in inflammation, host defence, and cell proliferation and survival1,14.
TNFR signalling via complexes IIa, IIb and IIc
In contrast to complex I, complexes IIa, IIb and IIc are assembled in the cytoplasm and have distinct signalling and functional outcomes13,14,19–21 (FIG. 1a). Complexes IIa and IIb lead to activation of a caspase cascade that results in TNF-induced cell death via apoptosis21,22. A highly controlled process, apoptosis is implicated in the turnover of cells during development and organogenesis, epi thelial homeostasis, inflammation, immunity, and disease pathogenesis. Apoptotic cells remain intact and are rapidly phagocytized by macrophages, a process which has suppressive effects and diminishes inflammatory cytokine production14.
Complex IIc activates the necroptosis effector mixed lineage kinase domain-like protein (MLKL) by a RIPK3-dependent mechanism13,19,23. In contrast to apoptosis, necroptosis results in plasma-membrane rupture, which releases intracellular contents and triggers local inflammation13. TNF-induced necroptosis of cells at barrier surfaces such as the skin and intestinal mucosa can compromise barrier function and thereby contribute to inflammation24–26. The implications of TNF-induced necroptosis in the pathogenesis of inflammatory diseases are the focus of active investigations19, and the RIPK1 and RIPK3 kinases that control these processes are considered potential therapeutic targets25–27.
Fine-tuning of TNFR-signalling
Fine-tuning the magnitude and kinetics of complex I signalling
TNF-induced complex I signalling requires precise control to prevent the development of deleterious excessive or chronic inflammation. Several studies support the concept that negative regulators are used to fine-tune the magnitude, amplitude and kinetics of TNFR signalling. Additionally, it has been suggested that specific negative modulators might set cell-type-specific thresholds for TNFR activation. These ‘signalling brakes’ operate by targeting receptor-proximal signalling events, by inhibiting IKK-complex activity, or by regulating downstream checkpoints. The mech anisms utilized by these negative regulators include cleavage or internalization of TNFRs10, destabilization of signalling complexes28, displacement of signalling molecules from complexes by antagonists29–34, as well as degradation (proteasomal or lysosomal)31,35–37, inactivation (for example, dephosphory lation)38–41, or seques tration of signalling factors42–44. Although the expression and activity of negative regulators can be constitutive, induction by TNF is consistent with a model of ‘applying the brakes’ as part of inducible inhibitory-feedback loops42. The three major classes of negative regulators of TNF-induced complex I signalling include regulators of ubiquitination, phosphatases, and IκBs (summarized in TABLE 2).
Table 2.
Inhibitors of TNFR signalling
| Inhibitor | Description | Mechanisms |
|---|---|---|
| Regulators of ubiquitination | ||
| A20 | • OTU-domain DUB • Inducible by TNF, via a GSK-3β-dependent mechanism76 • Interacts with partners, such as Itch, TAX1BP1, RNF11, protein Ymer and ABINs, to form ubiquitin-editing complexes36,131-133 • Polymorphisms or mutations of the gene encoding A20 (TNFAIP3) are associated with inflammatory diseases such as RA, SLE, psoriasis, and IBD134 |
• DUB: cleaves Lys63-linked ubiquitin chains from RIPK1 and NEMO135 • E3 ubiquitin ligase: conjugates Lys48-linked ubiquitin chains to RIPK1, UbcH5c and Ubc13, inducing their proteasomal degradation135 • Antagonistic ubiquitin binder: antagonizes the binding of LUBAC, NEMO, and E2 ubiquitin-conjugating enzymes32,33 • Targets signalling effectors for lysosomal degradation37 |
| Cezanne | • OTU-domain DUB • Inducible by TNF |
DUB: cleaves primarily Lys11-linked ubiquitin chains from targets such as RIPK1 (REF. 136) |
| Ubiquitin thioesterase otulin | • OTU-domain DUB • Not inducible by TNF • Has high specificity for inhibiting events related to linear ubiquitination |
• DUB: cleaves linear ubiquitin chains from RIPK1 and NEMO, destabilizing complex I137 • Antagonistic ubiquitin binder: antagonizes the binding of NEMO to Met1-linked ubiquitin chains34 • Regulator: interacts directly with HOIP, regulating its ligase-activity138,139 |
| Ubiquitin carboxyl-terminal hydrolase CYLD | • USP-family DUB • Interacts with partners, such as Itch, p62, HOIP and optineurin35 • Individuals with mutations of CYLD are predisposed to cylindromatosis140 |
Cleaves primarily Lys63-linked ubiquitin chains and secondarily linear chains from NEMO, TRAF2 and TAK1 (REF. 141) |
| USP4 | USP-family DUB | • Cleaves Lys63-linked ubiquitin chains from TRAF2, RIPK1 and TAK1 (REF. 142) |
| USP11 | USP-family DUB | • Cleaves Lys48-linked ubiquitin chains from IκBs, preventing their proteasomal degradation and the release of NFκB28 |
| USP15 | USP-family DUB | • Cleaves Lys48-linked ubiquitin chains from IκBs, preventing their proteasomal degradation and the release of NFκB28 |
| USP21 | USP-family DUB | • Cleaves Lys63-linked ubiquitin chains from TRAF2, RIPK1 and TAK1 (REF. 28) |
| USP31 | USP-family DUB | • Upstream: cleaves Lys63-linked chains from TRAF2 (REF. 28) • Downstream: cleaves ubiquitin chains from p65 modifying its transcriptional activity28 |
| MCPIP1 | • Unclassified DUB • Inducible by TNF • Interaction with diverse adaptor proteins can instruct MCPIP1 to function as either RNase or DUB |
• Upstream DUB: cleaves Lys63-linked chains from TRAF2 and RIPK1 (REF. 143) • Downstream DUB: cleaves Lys48-linked chains from IκBα143 • RNase: promotes degradation of inflammatory mRNAs144 |
| Itch | • E3 ubiquitin ligase • Interacts with A20 or CYLD to form ubiquitin-editing complexes35,36 |
Conjugates Lys48-linked ubiquitin chains to substrates, inducing their proteasomal degradation35,36 |
| Optineurin | • Antagonistic ubiquitin binder inducible by TNF • Mutations in OPTN have been identified in patients with POAG and ALS29, including ALS-associated mutations that disrupt the ubiquitin-binding capacity of optineurin and abolish its inhibitory functions in TNFR signalling29 |
Antagonizes the binding of NEMO to polyubiquitinated RIPK1 (REFS 29,30) |
| p47 | Antagonistic ubiquitin binder | Binds to polyubiquitinated NEMO directing its lysosomal degradation31 |
| Phosphatases | ||
| PP1 | Phosphatase | • Targets IKK-complex39 • Binding to IKK-complex is directed by the adaptor protein CUEDC2 (REF. 39) |
| PPP2CA-PPP2R1A-PPP2R5C (PP2A holoenzyme) | Phosphatase | Targets TRAF2 |
| PPP2CB-PPP2R1A (PP2A core enzyme) | Phosphatase | Targets IKK-complex |
| PPP2CA-PPP2R1B (PP2A core enzyme) | Phosphatase | Removes Ser536-phosphorylation from p65, modifying its transcriptional activity |
| PP2Cα (PP1A) | Phosphatase | Targets IKK-complex |
| PP2CP (PP1B) | Phosphatase | • Targets TAK1; binding to TAK1 is directed by the adaptor protein 14-3-3E40 • Targets IKK-complex |
| WIP1 (PP1D) | Phosphatase | Removes Ser536-phosphorylation from RelA/p65, modifying its transcriptional activity41 |
| Inhibitors of NFκB | ||
| IκBα | Rapidly inducible by TNF43 | Newly synthesized IκBα enters the nucleus, dissociates NFκB dimers from chromatin, and exports NFκB to the cytoplasm42 |
| IκBε | Inducible by TNF with slower kinetics (compared with IκBα)43,44 | Functions as a back-up or fail-safe mechanism43,44 |
| IκBδ (p100) | Inducible by TNF with delayed and sustained kinetics43 | Sequesters NFκB subunits in cytoplasm |
ABIN, A20-binding inhibitor of NFκB activation; ALS, amyotropic lateral sclerosis; CUEDC2, CUE domain-containing protein 2; DUB, deubiquitinating enzyme; GSK-3β, glycogen synthase kinase 3β; HOIP, HOIL-1-interacting protein (E3 ubiquitin-protein ligase RNF31); IBD, inflammatory bowel disease; IκB, inhibitor of κB; IKK, IκB kinase; LUBAC, linear ubiquitin chain assembly complex; MCPIP1, MCP-induced protein 1 (ribonuclease ZC3H12A); NEMO, NFκB essential modulator; NFκB, nuclear factor κB; OTU, ovarian tumour; POAG, primary open-angle glaucoma; RA, rheumatoid arthritis; RIPK1, receptor-interacting serine/threonine-protein kinase 1; RNF11, RING finger protein 11; SLE, systemic lupus erythematosus; TAK, transforming growth factor (TGF)-β-activated kinase 1; TAX1BP1, Tax1-binding protein 1; TNFR, TNF receptor; TRAF, TNFR-associated factor; USP, ubiquitin specific peptidase.
Reports from several studies of complex I signalling kinetics have shown that TNF induces oscillations of NFκB activity that dampen over time42,45–47. This observation suggests that the coordinated function of signalling brakes does not simply terminate TNF-induced signalling, but progressively diminishes the amplitude of waves of NFκB dimers that enter and exit the cell nucleus. The fraction of responding cells and the number of waves are related to the concentration of TNF47. However, the oscillatory behaviour of TNFR signalling can depend on cell type and context. For example, a single pulse with a saturating dose of TNF induces sustained activation of NFκB in fibroblast-like synoviocytes (FLS) from patients with RA48. Although the concentration of TNF gradually diminished over time in culture, the activity of IKK complexes, as well as the nuclear localization and binding of target genes by NFκB subunit p65 (encoded by RELA), were maintained for several days after the single TNF-pulse48. Further studies are necessary to investigate whether the sustained NFκB activity in RA FLS is attributable to functional incompetence of signalling brakes.
Fine-tuning the direction of TNFR-signalling: ‘live or die?’
TNF and TNFR1 are at the crossroads of inflammation, survival, apoptosis and necroptosis13,14. The factors that determine whether TNFR1 engagement induces complex I, leading to inflammatory responses, or complexes IIa, IIb and IIc, leading to cell death, are not well understood. The current model of this ‘live or die’ decision-making suggests that the expression levels of antiapoptotic molecules such as the long form of cellular FLICE-inhibitory protein (c-FLIPL), and the degree of RIPK1 ubiquitination are critical elements in determining which pathway is activated. c-FLIPL binds to caspase-8 and blocks specifically its proapoptotic effector functions14. Regarding the ubiquitination status of RIPK1, the model proposes that ubiquitinated RIPK1 is tethered to complex I, whereas stripping of RIPK1 from the ubiquitin cloak results in its cytoplasmic trafficking and interaction with programmed-cell-death-inducing machinery49. Studies published in the past 5 years have revealed the roles of Sharpin50–54, A20 (REF. 55) and ubiquitin carboxyl-terminal hydrolase CYLD49 in the decision to undergo cell death. Other factors related to cell differentiation and activation state, as well as additional environmental signals, are likely to affect this decision; thus, the outcome of TNFR1 signalling is context-dependent. Similarly, regulation of the balance of complex IIa/IIb versus complex IIc formation is incompletely understood, but RIPK1 and RIPK3 seem to have an important role in this process27,56–58.
TNF-induced gene expression
Cell-type specificity and epigenetic regulation
At the cellular level, several hundred genes are regulated (induced or suppressed) by TNF in a cell-type-specific manner. For instance, in FLS, TNF induces copious amounts of IL-6, with rapid and sustained production48. In macrophages, lower levels of IL-6 are induced more transiently48, whereas neutrophils have limited capacity to produce IL-6 in response to TNF59. Cell-type specificity of gene expression results, at least in part, from the effects of cell differentiation and cell-state transitions on the chromatin landscape60.
The chromatin landscape refers to the genome-wide pattern of open chromatin sites that demarcate regulatory elements (promoters and enhancers) where transcription factors can readily bind to regulate gene expression60. During cell differentiation, lineage-specific transcription factors, termed ‘pioneer factors’ or ‘master transcription factors’, penetrate the barrier of histone-containing nucleosomes that maintain a ‘closed’ or inaccessible chromatin conformation to create lineage-specific topologies of open chromatin. According to this model, cellular patterns of gene expression in response to stimuli such as TNF are predetermined chiefly by the chromatin landscape. ‘Signalling transcription factors’, such as NFκB and AP1, which have limited ability to remodel chromatin, will predominantly bind to and activate genes that exhibit open chromatin at their promoters and enhancers60.
Studies in the past 3 years have modified this concept by showing that lineage-determining transcription factors can cooperate with various environmentally activated transcription factors to remodel the enhancer repertoire of a cell, and thus alter patterns of gene expression. For example, stimulation of macrophages with diverse agonists (including Toll-like receptor (TLR) ligands or cytokines) induced cooperative recruitment of master transcription factors and stimulus-specific transcription factors in new genomic topologies, generating novel enhancers termed ‘latent enhancers’ (REF. 61). In the same study, acquisition of latent enhancers was shown to alter responses to subsequent stimulation. Along these lines, another study showed that TLR8 agonists remodel the chromatin at the IL6 locus in neutro phils and uncover a latent enhancer, converting IL6 in neutrophils into a TNF-inducible gene59.
Evidence suggests that chronic pathological states are associated with disease-specific stable changes in gene expression that are consistent with epigenetic mechanisms. For instance, FLS derived from patients with RA have a DNA methylome that is different from that of osteoarthritis (OA) FLS62, and FLS derived from patients with early and longstanding RA have distinct DNA methylomes63. Notably, TNF alters chromatin states and induces higher levels of inflammatory cytokines and chemokines in RA FLS compared with OA FLS48. We hypothesize that, in the context of RA, unremitting inflammation induces disease-associated chromatin modifications, potentially altering cellular gene-induction responses to TNF. Consistently, inflammatory cytokines have been shown to modulate DNA hydroxyl-methylation in FLS and chondrocytes64,65. Overall, accumulating evidence suggests that epi genetic mechanisms, such as chromatin modifications or DNA methylation, established during differentiation or acquired in response to homeostatic or pathological environmental stimuli, contribute to the tissue-specific and disease-associated effects of TNF.
The microenvironment can also condition cellu lar responses to TNF independently of epigenetic mechanisms. Simultaneous engagement of TLRs and prostaglandin receptors cooperates with TNF to induce transcriptomes in monocytes, macrophages and dendritic cells that resemble chronic inflammatory states66. In addition, RA FLS modulate expression of approximately one-third of TNF-regulated genes in macrophages, including Myc-dependent, growth-factor-inducible and interferon (IFN)-inducible genes67. These findings suggest that signal integration and intercellular functional coupling will shape responses to TNF in complex inflammatory environments such as RA synovitis.
Expression kinetics of TNF-inducible genes
Three distinct patterns of induction kinetics for TNF-inducible genes have been identified: early, inter mediate and late68–70. The accessibility of chromatin is a critical factor in determining the expression kinetics of TNF-inducible genes. Genes with accessible chromatin are induced more rapidly by TNF (FIG. 3a), compared with genes that require chromatin remodelling (FIG. 3b). Genes requiring de novo protein synthesis for their induction are also usually expressed in a delayed manner. For instance, cell-type-specific clusters of genes exist that require the prior synthesis of IFN-β or the transcription factor IκBζ for optimal induction by TNF59,68 (FIG. 3c,3d). Regarding the IFN-β-dependent genes, TNF first induces NFκB activation and IFN-β synthesis68,71 (FIG. 3c). Subsequently, newly synthesized IFN-β operates in an autocrine manner, activating the Janus kinase (JAK) and signal transducer and activator of transcription (STAT) signalling pathway, which cooperates with NFκB and activates IFN-stimulated genes (ISGs) such as CXCL9 (encoding MIG) and CXCL10 (encoding IP-10)68. Regarding the IκBζ-dependent genes, TNF synergizes with TLR8 agonists or cytokines to induce IκBζ59, which binds to and promotes activation of a distinct subset of gene promoters by inducing an activating histone mark, histone H3 trimethylation at lysine 4 (H3K4me3). The need for prior histone modification results in delayed transcription of NFκB-dependent genes in a cell-type-specific manner59. These results suggest a model whereby TNF induces a cascade of transcription factors that sustains late-phase gene expression. This model is supported by evidence that TNF induces a cascade mediated by initial induction of AP1 that eventually leads to expression of NFATc1, which promotes osteoclastogenesis that can contribute to the bone-resorptive properties of TNF70. Discovery of additional transcriptional modules that mediate novel aspects of late-phase TNF-induced gene expression and function represents an important area for future research.
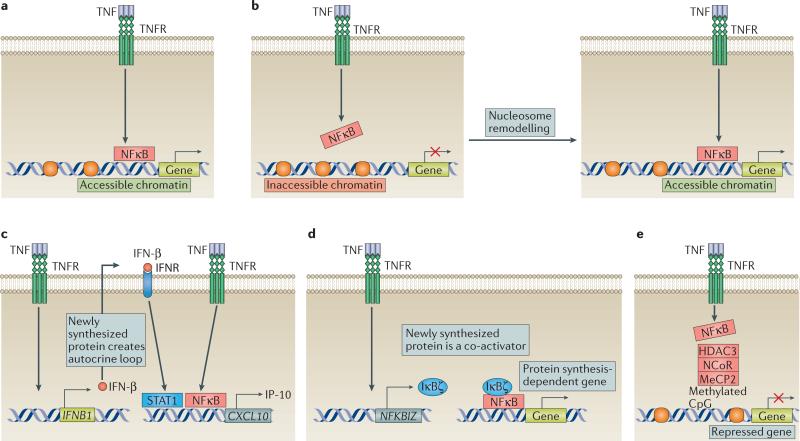
Figure 3. Molecular mechanisms of the differential induction kinetics of TNF-inducible genes.
a | Immediate early TNF-inducible genes have an accessible chromatin state, which enables the unopposed and rapid recruitment of nuclear factor κB (NFκB). b | Induction of genes that have inaccessible chromatin (due to the presence of a chromatin barrier, such as nucleosomes) by TNF is delayed, as it requires chromatin remodelling to remove the chromatin barrier and enable the recruitment of NFκB. c | For one class of de novo protein synthesis-dependent genes, TNF first induces the production of IFN-β, which subsequently functions in an autocrine manner to activate signal transducer and activator of transcription (STAT) signalling. Eventually, STATs cooperate with TNF-induced NFκB for the optimal induction of genes. d | For another class of de novo protein synthesis-dependent genes dependent on IκBζ production, TNF-induced IκBζ is recruited to the promoter by NFκB and operates as a co-activator to trigger gene-induction. e | The induction by TNF of transcription of repressed genes is prevented by the presence of a co-repressor complex. A well-known co-repressor is nuclear receptor co-repressor 1 (NCoR), which recruits histone deacetylase 3 (HDAC3) that removes acetyl groups from neighbouring histones, creating a chromatin environment that is inaccessible to NFκB. The NCoR complex is recruited to methylated CpG motifs via the adaptor protein methyl-CpG-binding protein 2 (MeCP2). TNFR, TNF receptor.
A currently accepted model suggests that chromatin barriers operate as rheostats to regulate the amplitude and timing of expression of TNF-inducible genes60. Genes induced with a delay before peak expression are typically associated with a nonpermissive chromatin environment, such as nucleosomes that occlude regulatory elements, repressive histone marks (for example, H3K27me3 and H3K9me3) and co-repressor complexes60 (FIG. 3). The nuclear receptor co-repressor 1 (NCoR) complex represses a cluster of TNF-inducible genes by recruiting the enzyme histone deacetylase 3 (HDAC3)72,73. Methyl-CpG-binding protein 2 (MeCP2), which binds to methylated CpGs, recruits the NCoR complex to gene promoters, suppressing their transcription74 (FIG. 3e). In macrophages, lack of MeCP2 resulted in increased expression of a cluster of inflammatory genes upon TNF-stimulation74. Overall, the need to overcome nonpermissive chromatin states before genes can be transcribed contributes to the delayed gene-induction observed after TNF stimulation.
TNF-inducible genes can be further subclassified as transient or sustained, on the basis of the duration of their expression48,69. TNF induces sustained expression of inflammatory mediators in RA FLS, potentially due to prolonged NFκB signalling and sustained chromatin remodelling48. For a subset of TNF-inducible genes, mRNA stability is an additional determinant of the duration of their expression69. The mechanisms that fine-tune the expression kinetics of TNF-inducible genes, including signalling brakes and rheostat control of chromatin, help to ensure a stepwise and sequential response to pathogens and inflammatory stimuli — optimal host defence relies on a rapid and transient inflammatory response, followed by a resolution phase to attain wound healing and tissue repair.
Altered cellular responses to other stimuli
One important function of cytokines is to change the physiological state of cells and the way they respond to environmental stimuli. This feature has been referred to as ‘memory’ or ‘training’, and includes enhanced responses to subsequent challenges (priming) or refractoriness to stimulation (desensitization or tolerance). Accumulating evidence suggests that TNF modifies cellular responses to other stimuli. For instance, TNF displays a priming effect on RA FLS, enhancing their subsequent inflammatory response to IFNs75. Notably, this priming was gene-specific and resulted in sensitization of RA FLS even to suboptimal concentrations of IFNs. In macrophages, by contrast, TNF desensitized the cells to the effects of lipopolysaccharide (LPS) and protected mice from LPS-induced lethality76.
Molecular mechanisms underlying TNF-induced priming and tolerization have been revealed (summarized in FIG. 4). A common theme for both phenomena is the induction of gene-specific chromatin modifications by TNF, which alter cell responses to subsequent stimulations. In RA FLS, TNF induces an accessible chromatin state in the promoter region of CXCL10, enabling the immediate and robust induction of IP-10 mRNA upon re-stimulation with IFNs75 (FIG. 4a). A genome-wide study revealed a similar capacity of TNF and other cytokines to induce faster or augmented responses to subsequent challenges, owing to a gene-specific induction of latent enhancers61, although TNF can also suppress remodelling of chromatin at genes such as IL6 (REF. 76).
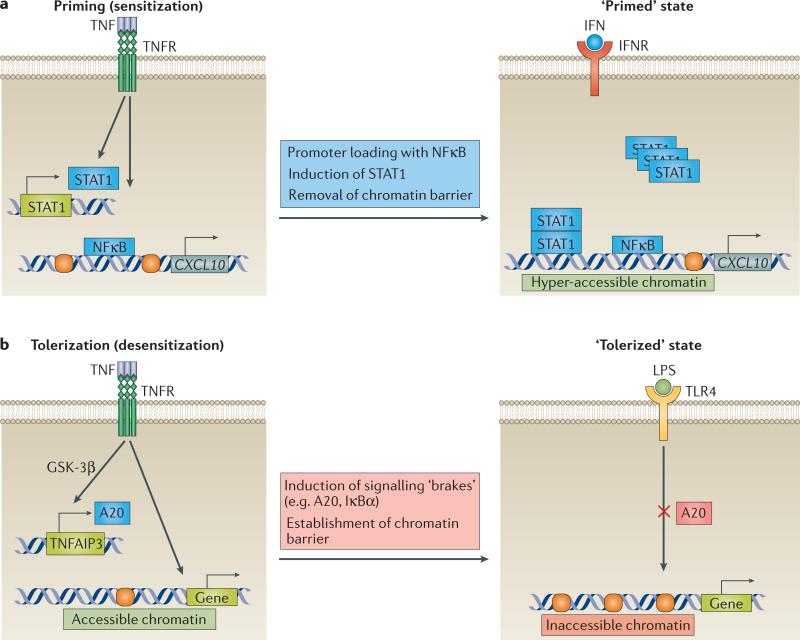
Figure 4. TNF modulates cellular responses to subsequent challenges and imposes short-term memory.
a | TNF-induced cell priming (sensitization). Prolonged exposure to TNF primes the chromatin of fibroblast-like synoviocytes (FLS) in a gene-specific manner, by histone eviction, acetylation of the remaining histones, and loading of nuclear factor κB (NFκB) to specific interferon (IFN)-target genes. In addition, TNF induces the expression of signal transducer and activator of transcription 1 (STAT1), increasing the intracellular STAT1 reservoir. This ‘primed’ state is maintained for several days, imposing a ‘short-term memory’ in cells. Primed FLS display hyperresponsiveness to saturating doses and sensitization to suboptimal doses of IFNs. b | TNF-induced cell tolerization (desensitization). TNF induces a ‘tolerized’ state in macrophages by triggering the expression of A20 via a glycogen synthase kinase 3β (GSK-3β)-dependent mechanism, and by establishing a gene-specific chromatin barrier. A20 attenuates the signalling input and the chromatin barrier prevents gene-expression upon subsequent stimulation with lipopolysaccharide (LPS). TLR, Toll-like receptor.
An additional mechanism that potentially contributes to TNF-induced cell priming or tolerization is the effect of TNF on signalling events upstream of chromatin. For instance, in RA FLS, prolonged exposure to TNF increases the intracellular reservoir of STAT1, amplifying the activation of STAT1 upon secondary challenge with interferons75 (FIG. 4a). Moreover, in macro phages, exposure to TNF induces the expression of signalling brakes, such as A20 and IκBα, which potentially restrict upstream signalling upon secondary stimulation with LPS76 (FIG. 4b).
An intriguing finding of these studies is the sustained effect of TNF on target cells, suggesting the establishment of ‘short-term transcriptional memory’. An attractive molecular explanation could be the induction by TNF of signalling molecules and chromatin modifications, which are not rapidly reversed77. Along these lines, TNF induces sustained expression of STAT1 (REF. 75), NFATc1 (REF. 70) and IκBδ43. Similarly, induction by TNF of the positive histone mark H3K4me1, which is associated with functional enhancers, is maintained for days in a large fraction of latent enhancers despite the absence of continuous stimulation61. Thus, short-term transcriptional memory is maintained by sequence-specific transcription factors and epigenetic mechanisms.
Role of TNF in health and disease
Homeostatic functions
Ablation of the Tnf gene in mice revealed that TNF has homeostatic functions in addition to its immune and inflammatory roles78–80. TNF is required for optimal defence against pathogens, proper lymphoid-organ architecture and germinal-centre formation, development of granulomas, resolution of inflammation and induction of tissue repair. Homeostatic functions of TNF are also supported by studies in mice lacking TNFRs12 and by cell-based functional assays76,81,82 (summarized in BOX 1). Several reports have identified molecular mech anisms that could explain some aspects of these unexpected homeostatic roles of TNF. One study showed that TNF desensitizes macrophages to the deleterious effects of secondary inflammatory challenges (tolerization)76. Others have described homeostatic effects of TNF in tissue regeneration, such as neuronal remyelination83, cardiac remodelling84 and cartilage regeneration85. In addition, TNF has suppressive effects on adaptive immune processes in models of autoimmunity such as autoimmune encephalitis12. The homeostatic potential of TNF has been suggested to be mediated, at least in part, by TNFR2 (REFS 12,86).
Pathogenic functions
Inflammatory and autoimmune diseases
TNF induces inflammation, activates vascular endothelium, orchestrates the tissue recruitment of immune cells and promotes tissue destruction2 (the pathogenic functions of TNF are summarized in BOX 1). Uncontrolled production or function of TNF has been linked to the development of inflammatory diseases such as RA, IBD, psoriasis, PsA, ankylosing spondylitis and specific types of JIA. In contrast to the well-defined role of TNF in these diseases, its role in the pathogenesis of multiple sclerosis is a conundrum12. Although initial studies suggested a pathogenic function for TNF in multiple sclerosis, clinical trials of a global inhibitor of TNF were discontinued owing to unexpected aggravation of the disease87. In addition, patients receiving TNF blockade for other diseases sporadically developed lesions with demyelination, such as optic neuritis88. These observations suggest that TNF can suppress autoimmune processes and/or exert essential homeostatic functions within the micro environment of the central nervous system (CNS). Several studies suggest that the homeostatic and pathogenic activities of TNF are mediated by distinct molecular and cellular pathways12. TNFR2, which is expressed on regulatory T (TREG) cells, oligodendro cytes and astrocytes, mediates immunoregulation, neuronal survival and re-meyelination; TNFR1, by contrast, induces CNS inflammation and neuronal demyelination12. Selective inhibitors of the neurotoxic TNFR1 pathway, which preserve the neuroprotective TNFR2 pathway, are being developed and could be a new avenue in therapeutics for patients with multiple sclerosis.
Hypernociception
TNF has been implicated in the hyperalgesia observed in the context of inflammation (inflammatory pain) and neuronal damage (neuropathic pain). In humans with RA and in several animal models of inflammation, neutralization of TNF induced rapid reduction of nociceptive neuronal activity in the afferent neurons, the spinal cord and the brain89–94. Notably, TNF blockade reverses hypernociception long before its effect on inflammation becomes obvious94. This observation suggests that the antinociceptive effects of TNF-blockade are distinct from their anti-inflammatory effects. At the cellular level, TNF seems to trigger hyperalgesia by inducing peripheral and central sensitization to mechanical stimulation93. Peripheral sensitization results from the direct action of TNF on C and Aδ nociceptive neurons, which innervate the site of inflammation91. Central sensitization is more complex, and potentially involves several cell types such as spinal cord nociceptive neurons, microglial cells and astrocytes93. At the molecular level, both peripheral and central sensitization are dependent on TNFR1-mediated signals90,93.
Cancer
TNF was discovered as a serum factor that induces necrosis of tumour cells, in part by acting on tumour vasculature to compromise blood supply95. The cytolytic potential of TNF has been explained by its capacity to induce programmed cell death. Subsequent studies revealed that TNF also displays cytostatic effects on specific tumour cell lines81. In a 2013 report, TNF was shown to cooperate with IFN-γ to induce tumour-cell senescence, a cell-state characterized by permanent growth arrest82. The widespread use of TNF as a drug for cancer has been prevented by the systemic toxicity of TNF96.
Accumulating evidence suggests that TNF also has the potential for opposite effects on cancer, either by inducing carcinogenesis or promoting the progression of established tumours97. TNF could be genotoxic by inducing de novo mutagenesis and suppressing the DNA-repair machinery. In the context of established tumours, TNF has the potential to promote the survival and proliferation of malignant cells, either directly by activating NFκB in tumour cells, or indirectly by inducing the production of tumour-promoting cytokines such as IL-6. In addition, TNF contributes to the escape of tumour cells from immunosurveillance by promoting the immunosuppressive capacity of myeloid-derived suppressor cells and TREG cells98,99. Finally, TNF facilitates tumour metastasis by promoting epithelial–mesenchymal transition and by inducing the synthesis of matrix metalloproteinases (MMPs).
Cardiovascular disease
Several studies suggest that TNF is cardiotoxic for the healthy myocardium, and potentially cardioprotective for the failing myocardium. Cardiotoxicity is primarily attributable to TNF-induced cardiomyocyte apoptosis100, whereas cardioprotection results from TNF-induced ectopic expression of keratin 8 and keratin 18 in cardiomyocytes84. In addition to its role in myocardial diseases, TNF is potentially implicated in the pathogenesis of atherosclerosis by affecting lipid metabolism, activating endothelial cells and inducing vascular inflammation101.
Fibrosis and Dupuytren disease
A study published in 2013 revealed a pathogenic role for TNF during the course of Dupuytren disease102. Dupuytren nodules are infiltrated by classically activated macrophages that secrete TNF. Fibroblasts and myofibroblasts from Dupuytren lesions had increased expression of both TNFR1 and TNFR2, suggesting that the disease micro environment sensitizes local stromal cells to the effects of TNF. Neutralization of TNF reduced the contractile activity of myofibroblasts, suggesting that the local administration of TNF blockers might be beneficial for patients with Dupuytren disease102.
Therapeutic targeting of TNF
Limitations of global inhibition
The approved anti-TNF agents have been a commercial success and a scientific breakthrough, improving the quality of life of millions of patients with TNF-mediated diseases. Despite these successes, the current therapeutic paradigm of global TNF blockade has several limitations, the three most important being low rates of disease remission, the development of adverse effects and the generation of antibodies against biologic TNF inhibitors103. Adverse effects include common and opportunistic infections, reactivation of latent tuberculosis, ‘paradoxical’ induction of autoantibodies, lupus-like symptoms, demyelination, psoriasis, sarcoidosis, as well as a potentially increased risk for specific malignancies, such as lymphomas. Development of fatal disseminated histoplasmosis and aggressive hepato splenic T-cell lymphomas in some patients undergoing treatment with TNF inhibitors resulted in the issue of ‘black box’ warnings for these drugs104,105. To overcome these limitations and improve efficacy and safety, alternative strategies are in development.
Improving TNF blockade
An attractive strategy to improve the rate of response to TNF blockade is to combine anti-TNF agents with other medications. Along these lines, combined inhibition of TNF and IL-17A with a bi-specific construct proved superior to single-cytokine blockade in inhibiting the production of inflammatory mediators in in vitro studies and in an arthritis model106. Notably, however, previous experience with combinations of biologic agents has shown that excessive immunosuppression can lead to an alarming rise in risk of infections107,108.
Combining TNF blockade with drugs that target pathogenic pathways or cells not implicated in host defence, to improve response rates without compromising safety, is a therapeutic approach worth considering. Several attractive candidate combinations have been suggested: targeting of TNF and angiogenesis (for example, with an antibody against vascular endothelial growth factor); targeting TNF and tissue destruction (for example, with anti-MMP14 or anti-RANKL (receptor activator of NFκB ligand) which suppresses osteoclastogenesis); and targeting TNF and FLS (for example, by inhibiting cadherin 11, a cell-surface molecule that regulates FLS adhesion and inflammatory functions)103.
Another alternative could be to shift the current therapeutic paradigm of global TNF inhibition to selective targeting of the pathogenic bioactivities of TNF, keeping the homeostatic functions of this cytokine intact. The proposed functional dichotomy between the TNFR1 pathway (primarily pathogenic) and the TNFR2 pathway (primarily homeostatic) might provide an approach to selective inhibition12. Next-generation TNF-blockers that selectively block the TNFR1 pathway have been developed and are described in TABLE 3 (REFS 109–117). Notably, the development of selective TNFR2 agonists provides an option for the future: to combine selective blockade of the TNFR1 axis with selective boosting of TNFR2 signalling118.
Table 3.
Next-generation selective inhibitors of soluble TNF-TNFR1 pathway
| Class | Agent | Description |
|---|---|---|
| TNFR1-specific antibodies | ATROSAB110,111 | Full-length IgG against a specific epitope of TNFR1 |
| Maintains the conformation of TNFR1 in an inactive state and obstructs the binding of TNF and LTα | ||
| The Fc-region has been mutated to abolish the capacity to bind Fcγ receptor and complement, avoiding immune system activation | ||
| MDS5541 (REFS 109,112) | Domain-antibody comprising a single variable region with specificity for TNFR1 linked to another single variable region with specificity for albumin | |
| TROS113 | Trivalent nanobody comprising two distinct domains with specificy for TNFR1 (resulting in high-affinity binding to TNFR1) and an anti-albumin domain | |
| TNF muteins114,115 | XENP345 (REFS 145,146) and XPro1595 (REF. 147) | Dominant-negative TNF muteins that interact with native soluble TNF to form inactive heterotrimers, which have markedly reduced receptor-binding and signalling capacities |
| R1antTNF116,117 | TNFR1-selective antagonistic TNF mutein, that is, selective binding capacity for TNFR1 without signalling capacity | |
| Also has dominant negative TNF function, by forming inactive mixed heterotrimers with native TNF |
Notes: TNF muteins are TNF molecules engineered to introduce amino acid changes; such modifications in the receptor interface domain can impair the receptor-binding and signalling capacity of these molecules. ATROSAB, antagonistic TNF receptor one-specific antibody; LT-α, lymphotoxin-α; R1antTNF, TNF receptor 1 antagonist; TNFR, TNF receptor; TROS, TNF receptor-one silencer.
In 2011, progranulin was found to operate as a natural antagonist of TNF by binding to both TNFR1 and TNFR2 (REF. 119). Administration of recombinant progranulin or of an engineered progranulin analogue (Atsttrin) was beneficial in several animal models of inflammatory arthritis119. Notably, progranulin has anabolic effects in cartilage via TNFR2-mediated activation of MAPK1 and MAPK3 (REF. 85). This observation suggests that progranulin might alleviate arthritis via dual effects on TNFR-mediated pathways, by blocking deleterious TNFR1 signalling and by engaging TNFR2 to trigger MAPK-mediated anabolic effects in chondrocytes85.
Reducing immunogenicity
The development of antibodies against TNF-blocking biologic agents results in allergic reactions and in progressive loss of drug effectiveness. This issue of drug immunogenicity has been only partially addressed with the development of humanized or fully human biologic agents. An alternative approach is to trigger the production of natural neutralizing anti-TNF antibodies via active immunization. This method seemed feasible in a limited 12-month phase II study in patients with RA or IBD120, as active immunization with TNF-kinoid (composed of recombinant human TNF conjugated to the carrier protein KLH) resulted in the generation of neutralizing polyclonal antibodies against TNF. The clinical efficacy of this approach remains to be determined, and needs to be weighed against the potentially catastrophic consequences of life-long ablation of TNF.
Controlling TNF synthesis
Downregulation of the inappropriate transcription of TNF could be effective in ameliorating TNF-driven pathologies. IκBβ has been shown to function as a gene-selective co-activator, driving prolonged transcription of TNF121. Mice lacking IκBβ have reduced transcription of Tnf and are protected from LPS-induced lethality and collagen-induced arthritis. Gene-selectivity was explained by the specific interaction of the newly synthesized IκBβ with the p65–c-Rel NFκB dimer, which binds to the κB2 site in the promoter of Tnf121. Hence, blocking IκBβ might be a promising strategy to selectively inhibit the chronic phase of TNF production and treat chronic inflammatory diseases, such as RA.
As mentioned in this Review, precursor transmembrane TNF is cleaved by TACE, releasing soluble TNF. As TACE has important homeostatic functions by regulating EGFR-signalling in epithelial barriers, direct targeting of TACE raises safety concerns. However, inactive rhomboid protein 2 (iRhom2) has been identified as a myeloid-specific regulator of TACE that facilitates the shedding of membrane-bound TNF8. This observation suggests that therapeutic targeting of iRhom2 is a potentially attractive strategy to block the activity of TACE, specifically in macrophages, to prevent the release of pathogenic soluble TNF. This therapeutic strategy has the advantage of leaving intact the proposed homeostatic transmembrane TNF–TNFR2 pathway.
Inhibiting intracellular responses
Small molecules targeting chromatin regulators have the capacity to block specific pathogenic pathways downstream of TNFRs. In a 2014 study, pharmacologic inhibition of bromodomain and extra-terminal domain (BET) proteins suppressed TNF-induced endothelial cell activation, attenuating leukocyte rolling, adhesion and transmigration122. Notably, at the transcriptional level, inflammatory genes associated with TNF-induced super-enhancers were preferentially downregulated122. This observation suggests that BET inhibitors selectively block branches of the TNF-induced transcriptional programs.
Kinase inhibitors have also been described to suppress intracellular responses to TNF. For instance, tofacitinib, a JAK inhibitor approved for use in the treatment of RA, suppressed the late phase of TNF-induced STAT activation and cytokine production in macrophages123.
Conclusions
Important advances in the understanding of TNF biology and pathogenic functions are summarized in BOX 2. These advances have important therapeutic implications as they identify therapeutic targets — such as LUBAC components, mediators of necroptosis, epigenetic regulators (for example, BET proteins124 and HDACs125) and novel transcription factors — whose inhibition might have more-selective effects on disease-specific pathogenic mechanisms,possibly achieving comparable efficacy without the toxic effects associated with global TNF blockade. Recognition of the homeostatic functions of TNF has led to new concepts for the treatment of rheumatic diseases, presenting the possibility of strengthening mechanisms of tolerization and tissue repair as a complement to suppression of inflammatory pathways.
Despite decades of research, however, many important challenges and gaps remain in our understanding of the basic science of TNF and in how to translate this knowledge into better treatments. Important directions for future basic research include a deeper understanding of the molecular underpinnings and functional consequences of the alternative outcomes of ‘life’ versus ‘death’ signalling, of the epigenetic regulation of TNF responses, of mechanisms that restrain TNF responses, and of homeostatic functions that could be manipulated to alter the pathogenic profile of TNF. An intriguing research direction related to epigenetic mechanisms is the effect of TNF on the expression of non-coding RNAs, including microRNAs126,127 and long non-coding RNAs128. Another future research priority is the harnessing of basic science and the power of new high-throughput sequencing technologies and genome-wide approaches to study patients before and after TNF blockade therapy to identify markers predictive of clinical response and mech anisms associated with resistance to TNF blockade129. The identification of such markers and mechanisms is a major unmet need that will enable replacement of the current trial-and-error therapeutic approach with a stratified and increasingly precise strategy, in which the decision to introduce TNF blockade will be based on validated biomarkers that predict response and not on empirical criteria130. Such translational studies also offer hope for the discovery of alternative pathogenic pathways that can be targeted in patients who are resistant to therapy with TNF inhibitors.
Key points.
TNF is a pleiotropic cytokine that exerts homeostatic and pathogenic bioactivities
A new concept in therapeutics of TNF-mediated diseases is the selective inhibition of the pathogenic effects of TNF with preservation of its homeostatic functions
TNF-induced necroptosis is a new pathway potentially implicated in TNF-mediated pathologies
TNF induces cellular priming, tolerization, and short-term transcriptional memory in a context-dependent manner
Combining TNF-blockade with drugs that target pathogenic pathways or cells not implicated in host defence is an attractive approach to improve effectiveness without compromising safety
Box 1 | Functions of TNF.
Homeostatic functions
Defence against pathogens
Organogenesis and development: lymphoid organ architecture
Tissue regeneration: neuronal remyelination; cardiac remodelling; cartilage regeneration
Immunoregulation: tolerization (desensitization) of macrophages, apoptosis of inflammatory cells
Inhibition of tumorigenesis: necrosis, senescence (cytostatic effect)
Pathogenic functions
Inflammation: induction of inflammatory mediators (cytokines, lipid mediators); recruitment of inflammatory cells (induction of chemokines and adhesion molecules, endothelial cell activation); survival of inflammatory cells; necroptosis
Autoimmunity: inhibition of TREG cells
Tissue degeneration: induction of tissue-destructive enzymes; apoptosis; osteoclastogenesis
Hypernociception: peripheral neuronal sensitization (effects on C and Aδ neurons); central neuronal sensitization (effects on spinal cord nociceptive neurons, microglial cells and astrocytes)
Tumorigenesis: genotoxic effects (induction of de novo mutagenesis, impairment of DNA repair); induction of tumour-cell survival and proliferation; facilitation of tumour escape from immunosurveillance (effects on MDSCs and TREG cells); facilitation of tumour metastasis (induction of EMT and MMPs)
Atherogenesis: inflammation; endothelial cell activation; effect on lipid metabolism
EMT, epithelial–mesenchymal transition; MDSC, myeloid-derived suppressor cell; MMP, matrix metalloproteinase; TREG cell, regulatory T cell.
Box 2 | Recent advances in TNF biology and function.
Implication of the LUBAC complex and linear ubiquitination in TNF signalling, inflammatory responses and human diseases
Appreciation of the induction of complex IIc and the functions of RIPK3 and MLKL in TNF-induced necroptosis, which could have an important role in rheumatic disease pathogenesis
Recognition of the ability of TNF to induce pathogenic epigenetic modifications, which promote heightened inflammatory responses even to minor environmental challenges
Discovery of mechanisms, including induction of transcription modules mediated by novel TNF-induced transcription factors, that sustain cellular responses to TNF; these unremitting responses to TNF are probably important in sustaining chronic inflammation
Improved understanding of the importance of homeostatic functions of TNF, which include tolerization of macrophages, promotion of tissue repair possibly related to TNFR2 signalling, and suppressive effects on adaptive immunity
LUBAC, linear ubiquitin chain assembly complex; MLKL, mixed lineage kinase domain-like protein; RIPK3, receptor-interacting serine/threonine-protein kinase 3; TNFR2, TNF receptor 2.
Acknowledgements
This work was supported by grants from the NIH (to L.B.I.) and the Feldstein Medical Foundation (to G.D.K.).
Footnotes
Author contributions
Both authors researched data for article, made substantial contributions to discussions of the content, wrote the article and reviewed and/or edited the manuscript before submission.
Competing interests statement
The authors declare no competing interests.
References
- 1.Aggarwal BB, Gupta SC, Kim JH. Historical perspectives on tumor necrosis factor and its superfamily: 25 years later, a golden journey. Blood. 2012;119:651–665. doi: 10.1182/blood-2011-04-325225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Feldmann M. Translating molecular insights in autoimmunity into effective therapy. Annu. Rev. Immunol. 2009;27:1–27. doi: 10.1146/annurev-immunol-082708-100732. [DOI] [PubMed] [Google Scholar]
- 3.Cerami A. The value of failure: the discovery of TNF and its natural inhibitor erythropoietin. J. Intern. Med. 2011;269:8–15. doi: 10.1111/j.1365-2796.2010.02319.x. [DOI] [PubMed] [Google Scholar]
- 4.Monaco C, Nanchahal J, Taylor P, Feldmann M. Anti-TNF therapy: past, present and future. Int. Immunol. 2015;27:55–62. doi: 10.1093/intimm/dxu102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee RA, Eisen DB. Treatment of hidradenitis suppurativa with biologic medications. J. Am. Acad. Dermatol. 2015;73:S82–88. doi: 10.1016/j.jaad.2015.07.053. [DOI] [PubMed] [Google Scholar]
- 6.Dorner T, Kay J. Biosimilars in rheumatology: current perspectives and lessons learnt. Nat. Rev. Rheumatol. 2015 doi: 10.1038/nrrheum.2015.110. http://dx.doi.org/10.1038/nrrheum.2015.110. [DOI] [PubMed]
- 7.Karampetsou MP, Liossis SN, Sfikakis PP. TNF-α antagonists beyond approved indications: stories of success and prospects for the future. QJM. 2010;103:917–928. doi: 10.1093/qjmed/hcq152. [DOI] [PubMed] [Google Scholar]
- 8.Issuree PD, et al. iRHOM2 is a critical pathogenic mediator of inflammatory arthritis. J. Clin. Invest. 2013;123:928–932. doi: 10.1172/JCI66168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horiuchi K, et al. Cutting edge: TNF-α-converting enzyme (TACE/ADAM17) inactivation in mouse myeloid cells prevents lethality from endotoxin shock. J. Immunol. 2007;179:2686–2689. doi: 10.4049/jimmunol.179.5.2686. [DOI] [PubMed] [Google Scholar]
- 10.Locksley RM, Killeen N, Lenardo MJ. The TNF and TNF receptor superfamilies: integrating mammalian biology. Cell. 2001;104:487–501. doi: 10.1016/s0092-8674(01)00237-9. [DOI] [PubMed] [Google Scholar]
- 11.Grell M, et al. The transmembrane form of tumor necrosis factor is the prime activating ligand of the 80 kDa tumor necrosis factor receptor. Cell. 1995;83:793–802. doi: 10.1016/0092-8674(95)90192-2. [DOI] [PubMed] [Google Scholar]
- 12.Probert L. TNF and its receptors in the CNS: the essential, the desirable and the deleterious effects. Neuroscience. 2015;302:2–22. doi: 10.1016/j.neuroscience.2015.06.038. [DOI] [PubMed] [Google Scholar]
- 13.Pasparakis M, Vandenabeele P. Necroptosis and its role in inflammation. Nature. 2015;517:311–320. doi: 10.1038/nature14191. [DOI] [PubMed] [Google Scholar]
- 14.Brenner D, Blaser H, Mak TW. Regulation of tumour necrosis factor signalling: live or let die. Nat. Rev. Immunol. 2015;15:362–374. doi: 10.1038/nri3834. [DOI] [PubMed] [Google Scholar]
- 15.Bianchi K, Meier P. A tangled web of ubiquitin chains: breaking news in TNF-R1 signaling. Mol. Cell. 2009;36:736–742. doi: 10.1016/j.molcel.2009.11.029. [DOI] [PubMed] [Google Scholar]
- 16.Haas TL, et al. Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol. Cell. 2009;36:831–844. doi: 10.1016/j.molcel.2009.10.013. [DOI] [PubMed] [Google Scholar]
- 17.Rahighi S, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-κB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 18.Dynek JN, et al. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 2010;29:4198–4209. doi: 10.1038/emboj.2010.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Linkermann A, Green DR. Necroptosis. N. Engl. J. Med. 2014;370:455–465. doi: 10.1056/NEJMra1310050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chan FK, Luz NF, Moriwaki K. Programmed necrosis in the cross talk of cell death and inflammation. Annu. Rev. Immunol. 2015;33:79–106. doi: 10.1146/annurev-immunol-032414-112248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Micheau O, Tschopp J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell. 2003;114:181–190. doi: 10.1016/s0092-8674(03)00521-x. [DOI] [PubMed] [Google Scholar]
- 22.Wang L, Du F, Wang X. TNF-α induces two distinct caspase-8 activation pathways. Cell. 2008;133:693–703. doi: 10.1016/j.cell.2008.03.036. [DOI] [PubMed] [Google Scholar]
- 23.Sun L, et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell. 2012;148:213–227. doi: 10.1016/j.cell.2011.11.031. [DOI] [PubMed] [Google Scholar]
- 24.Kumari S, Pasparakis M. Epithelial cell death and inflammation in skin. Curr. Top. Microbiol. Immunol. 2015 doi: 10.1007/82_2015_466. http://dx.doi.org/10.1007/82_2015_466. [DOI] [PubMed]
- 25.Dannappel M, et al. RIPK1 maintains epithelial homeostasis by inhibiting apoptosis and necroptosis. Nature. 2014;513:90–94. doi: 10.1038/nature13608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Takahashi N, et al. RIPK1 ensures intestinal homeostasis by protecting the epithelium against apoptosis. Nature. 2014;513:95–99. doi: 10.1038/nature13706. [DOI] [PubMed] [Google Scholar]
- 27.Silke J, Rickard JA, Gerlic M. The diverse role of RIP kinases in necroptosis and inflammation. Nat. Immunol. 2015;16:689–697. doi: 10.1038/ni.3206. [DOI] [PubMed] [Google Scholar]
- 28.Harhaj EW, Dixit VM. Regulation of NF-κB by deubiquitinases. Immunol. Rev. 2012;246:107–124. doi: 10.1111/j.1600-065X.2012.01100.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Maruyama H, et al. Mutations of optineurin in amyotrophic lateral sclerosis. Nature. 2010;465:223–226. doi: 10.1038/nature08971. [DOI] [PubMed] [Google Scholar]
- 30.Zhu G, Wu CJ, Zhao Y, Ashwell JD. Optineurin negatively regulates TNFα-induced NF-κB activation by competing with NEMO for ubiquitinated RIP. Curr. Biol. 2007;17:1438–1443. doi: 10.1016/j.cub.2007.07.041. [DOI] [PubMed] [Google Scholar]
- 31.Shibata Y, et al. p47 negatively regulates IKK activation by inducing the lysosomal degradation of polyubiquitinated NEMO. Nat. Commun. 2012;3:1061. doi: 10.1038/ncomms2068. [DOI] [PubMed] [Google Scholar]
- 32.Verhelst K, et al. A20 inhibits LUBAC-mediated NF-κB activation by binding linear polyubiquitin chains via its zinc finger 7. EMBO J. 2012;31:3845–3855. doi: 10.1038/emboj.2012.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tokunaga F, et al. Specific recognition of linear polyubiquitin by A20 zinc finger 7 is involved in NF-κB regulation. EMBO J. 2012;31:3856–3870. doi: 10.1038/emboj.2012.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Keusekotten K, et al. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell. 2013;153:1312–1326. doi: 10.1016/j.cell.2013.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ahmed N, et al. The E3 ligase Itch and deubiquitinase Cyld act together to regulate Tak1 and inflammation. Nat. Immunol. 2011;12:1176–1183. doi: 10.1038/ni.2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shembade N, et al. The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nat. Immunol. 2008;9:254–262. doi: 10.1038/ni1563. [DOI] [PubMed] [Google Scholar]
- 37.Li L, et al. Localization of A20 to a lysosome-associated compartment and its role in NFκB signaling. Biochim. Biophys. Acta. 2008;1783:1140–1149. doi: 10.1016/j.bbamcr.2008.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li S, Wang L, Berman MA, Zhang Y, Dorf ME. RNAi screen in mouse astrocytes identifies phosphatases that regulate NF-κB signaling. Mol. Cell. 2006;24:497–509. doi: 10.1016/j.molcel.2006.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li HY, et al. Deactivation of the kinase IKK by CUEDC2 through recruitment of the phosphatase PP1. Nat. Immunol. 2008;9:533–541. doi: 10.1038/ni.1600. [DOI] [PubMed] [Google Scholar]
- 40.Zuo S, et al. 14-3-3 epsilon dynamically interacts with key components of mitogen-activated protein kinase signal module for selective modulation of the TNF-α-induced time course-dependent NF-κB activity. J. Proteome Res. 2010;9:3465–3478. doi: 10.1021/pr9011377. [DOI] [PubMed] [Google Scholar]
- 41.Chew J, et al. WIP1 phosphatase is a negative regulator of NF-κB signalling. Nat. Cell Biol. 2009;11:659–666. doi: 10.1038/ncb1873. [DOI] [PubMed] [Google Scholar]
- 42.Hoffmann A, Levchenko A, Scott ML, Baltimore D. The IκB–NF-κB signaling module: temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- 43.Shih VF, et al. Kinetic control of negative feedback regulators of NF-κB/RelA determines their pathogen-and cytokine-receptor signaling specificity. Proc. Natl Acad. Sci. USA. 2009;106:9619–9624. doi: 10.1073/pnas.0812367106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kearns JD, Basak S, Werner SL, Huang CS, Hoffmann A. IκBε provides negative feedback to control NF-κB oscillations, signaling dynamics, and inflammatory gene expression. J. Cell Biol. 2006;173:659–664. doi: 10.1083/jcb.200510155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nelson DE, et al. Oscillations in NF-κB signaling control the dynamics of gene expression. Science. 2004;306:704–708. doi: 10.1126/science.1099962. [DOI] [PubMed] [Google Scholar]
- 46.Ashall L, et al. Pulsatile stimulation determines timing and specificity of NF-κB-dependent transcription. Science. 2009;324:242–246. doi: 10.1126/science.1164860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tay S, et al. Single-cell NF-κB dynamics reveal digital activation and analogue information processing. Nature. 2010;466:267–271. doi: 10.1038/nature09145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee A, et al. Tumor necrosis factor α induces sustained signaling and a prolonged and unremitting inflammatory response in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 2013;65:928–938. doi: 10.1002/art.37853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Justus SJ, Ting AT. Cloaked in ubiquitin, a killer hides in plain sight: the molecular regulation of RIPK1. Immunol. Rev. 2015;266:145–160. doi: 10.1111/imr.12304. [DOI] [PubMed] [Google Scholar]
- 50.Tokunaga F, et al. SHARPIN is a component of the NF-κB-activating linear ubiquitin chain assembly complex. Nature. 2011;471:633–636. doi: 10.1038/nature09815. [DOI] [PubMed] [Google Scholar]
- 51.Ikeda F, et al. SHARPIN forms a linear ubiquitin ligase complex regulating NF-κB activity and apoptosis. Nature. 2011;471:637–641. doi: 10.1038/nature09814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gerlach B, et al. Linear ubiquitination prevents inflammation and regulates immune signalling. Nature. 2011;471:591–596. doi: 10.1038/nature09816. [DOI] [PubMed] [Google Scholar]
- 53.Kumari S, et al. Sharpin prevents skin inflammation by inhibiting TNFR1-induced keratinocyte apoptosis. eLIFE. 2014;3:e03422. doi: 10.7554/eLife.03422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rickard JA, et al. TNFR1-dependent cell death drives inflammation in Sharpin-deficient mice. eLIFE. 2014;3:e03464. doi: 10.7554/eLife.03464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Onizawa M, et al. The ubiquitin-modifying enzyme A20 restricts ubiquitination of the kinase RIPK3 and protects cells from necroptosis. Nat. Immunol. 2015;16:618–627. doi: 10.1038/ni.3172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Newton K, et al. Activity of protein kinase RIPK3 determines whether cells die by necroptosis or apoptosis. Science. 2014;343:1357–1360. doi: 10.1126/science.1249361. [DOI] [PubMed] [Google Scholar]
- 57.Dillon CP, et al. RIPK1 blocks early postnatal lethality mediated by caspase-8 and RIPK3. Cell. 2014;157:1189–1202. doi: 10.1016/j.cell.2014.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Rickard JA, et al. RIPK1 regulates RIPK3-MLKL-driven systemic inflammation and emergency hematopoiesis. Cell. 2014;157:1175–1188. doi: 10.1016/j.cell.2014.04.019. [DOI] [PubMed] [Google Scholar]
- 59.Zimmermann M, et al. Chromatin remodelling and autocrine TNFα are required for optimal interleukin-6 expression in activated human neutrophils. Nat. Commun. 2015;6:6061. doi: 10.1038/ncomms7061. [DOI] [PubMed] [Google Scholar]
- 60.Smale ST, Tarakhovsky A, Natoli G. Chromatin contributions to the regulation of innate immunity. Annu. Rev. Immunol. 2014;32:489–511. doi: 10.1146/annurev-immunol-031210-101303. [DOI] [PubMed] [Google Scholar]
- 61.Ostuni R, et al. Latent enhancers activated by stimulation in differentiated cells. Cell. 2013;152:157–171. doi: 10.1016/j.cell.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 62.Nakano K, Whitaker JW, Boyle DL, Wang W, Firestein GS. DNA methylome signature in rheumatoid arthritis. Ann. Rheum. Dis. 2013;72:110–117. doi: 10.1136/annrheumdis-2012-201526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ai R, et al. DNA methylome signature in synoviocytes from patients with early rheumatoid arthritis compared to synoviocytes from patients with longstanding rheumatoid arthritis. Arthritis Rheumatol. 2015;67:1978–1980. doi: 10.1002/art.39123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nakano K, Boyle DL, Firestein GS. Regulation of DNA methylation in rheumatoid arthritis synoviocytes. J. Immunol. 2013;190:1297–1303. doi: 10.4049/jimmunol.1202572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Haseeb A, Makki MS, Haqqi TM. Modulation of ten-eleven translocation 1 (TET1), isocitrate dehydrogenase (IDH) expression, α-ketoglutarate (α-KG), and DNA hydroxymethylation levels by interleukin-1β in primary human chondrocytes. J. Biol. Chem. 2014;289:6877–6885. doi: 10.1074/jbc.M113.512269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xue J, et al. Transcriptome-based network analysis reveals a spectrum model of human macrophage activation. Immunity. 2014;40:274–288. doi: 10.1016/j.immuni.2014.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Donlin LT, Jayatilleke A, Giannopoulou EG, Kalliolias GD, Ivashkiv LB. Modulation of TNF-induced macrophage polarization by synovial fibroblasts. J. Immunol. 2014;193:2373–2383. doi: 10.4049/jimmunol.1400486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yarilina A, Park-Min KH, Antoniv T, Hu X, Ivashkiv LB. TNF activates an IRF1-dependent autocrine loop leading to sustained expression of chemokines and STAT1-dependent type I interferon-response genes. Nat. Immunol. 2008;9:378–387. doi: 10.1038/ni1576. [DOI] [PubMed] [Google Scholar]
- 69.Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat. Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yarilina A, Xu K, Chen J, Ivashkiv LB. TNF activates calcium-nuclear factor of activated T cells (NFAT)c1 signaling pathways in human macrophages. Proc. Natl Acad. Sci. USA. 2011;108:1573–1578. doi: 10.1073/pnas.1010030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rosengren S, Corr M, Firestein GS, Boyle DL. The JAK inhibitor CP-690,550 (tofacitinib) inhibits TNF-induced chemokine expression in fibroblast-like synoviocytes: autocrine role of type I interferon. Ann. Rheum. Dis. 2012;71:440–447. doi: 10.1136/ard.2011.150284. [DOI] [PubMed] [Google Scholar]
- 72.Ghisletti S, et al. Cooperative NCoR/SMRT interactions establish a corepressor-based strategy for integration of inflammatory and anti-inflammatory signaling pathways. Genes Dev. 2009;23:681–693. doi: 10.1101/gad.1773109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hargreaves DC, Horng T, Medzhitov R. Control of inducible gene expression by signal-dependent transcriptional elongation. Cell. 2009;138:129–145. doi: 10.1016/j.cell.2009.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cronk JC, et al. Methyl-CpG binding protein 2 regulates microglia and macrophage gene expression in response to inflammatory stimuli. Immunity. 2015;42:679–691. doi: 10.1016/j.immuni.2015.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sohn C, et al. Prolonged tumor necrosis factor α primes fibroblast-like synoviocytes in a gene-specific manner by altering chromatin. Arthritis Rheumatol. 2015;67:86–95. doi: 10.1002/art.38871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Park SH, Park-Min KH, Chen J, Hu X, Ivashkiv LB. Tumor necrosis factor induces GSK3 kinase-mediated cross-tolerance to endotoxin in macrophages. Nat. Immunol. 2011;12:607–615. doi: 10.1038/ni.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Monticelli S, Natoli G. Short-term memory of danger signals and environmental stimuli in immune cells. Nat. Immunol. 2013;14:777–784. doi: 10.1038/ni.2636. [DOI] [PubMed] [Google Scholar]
- 78.Pasparakis M, Alexopoulou L, Episkopou V, Kollias G. Immune and inflammatory responses in TNFα-deficient mice: a critical requirement for TNFα in the formation of primary B cell follicles, follicular dendritic cell networks and germinal centers, and in the maturation of the humoral immune response. J. Exp. Med. 1996;184:1397–1411. doi: 10.1084/jem.184.4.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Marino MW, et al. Characterization of tumor necrosis factor-deficient mice. Proc. Natl Acad. Sci. USA. 1997;94:8093–8098. doi: 10.1073/pnas.94.15.8093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu J, et al. TNF is a potent anti-inflammatory cytokine in autoimmune-mediated demyelination. Nat. Med. 1998;4:78–83. doi: 10.1038/nm0198-078. [DOI] [PubMed] [Google Scholar]
- 81.Sugarman BJ, et al. Recombinant human tumor necrosis factor-alpha: effects on proliferation of normal and transformed cells in vitro. Science. 1985;230:943–945. doi: 10.1126/science.3933111. [DOI] [PubMed] [Google Scholar]
- 82.Braumuller H, et al. T-helper-1-cell cytokines drive cancer into senescence. Nature. 2013;494:361–365. doi: 10.1038/nature11824. [DOI] [PubMed] [Google Scholar]
- 83.Arnett HA, et al. TNFα promotes proliferation of oligodendrocyte progenitors and remyelination. Nat. Neurosci. 2001;4:1116–1122. doi: 10.1038/nn738. [DOI] [PubMed] [Google Scholar]
- 84.Papathanasiou S, et al. Tumor necrosis factor-α confers cardioprotection through ectopic expression of keratins K8 and K18. Nat. Med. 2015;21:1076–1084. doi: 10.1038/nm.3925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zhao YP, et al. Progranulin protects against osteoarthritis through interacting with TNF-α and β-catenin signalling. Ann. Rheum. Dis. 2014;74:2244–2253. doi: 10.1136/annrheumdis-2014-205779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bluml S, et al. Antiinflammatory effects of tumor necrosis factor on hematopoietic cells in a murine model of erosive arthritis. Arthritis Rheum. 2010;62:1608–1619. doi: 10.1002/art.27399. [DOI] [PubMed] [Google Scholar]
- 87.The Lenercept Multiple Sclerosis Study Group and The University of British Columbia MS/MRI Analysis Group TNF neutralization in MS: results of a randomized, placebo-controlled multicenter study. Neurology. 1999;53:457–465. [PubMed] [Google Scholar]
- 88.Robinson WH, Genovese MC, Moreland LW. Demyelinating and neurologic events reported in association with tumor necrosis factor α antagonism: by what mechanisms could tumor necrosis factor α antagonists improve rheumatoid arthritis but exacerbate multiple sclerosis? Arthritis Rheum. 2001;44:1977–1983. doi: 10.1002/1529-0131(200109)44:9<1977::AID-ART345>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 89.Inglis JJ, et al. Collagen-induced arthritis as a model of hyperalgesia: functional and cellular analysis of the analgesic actions of tumor necrosis factor blockade. Arthritis Rheum. 2007;56:4015–4023. doi: 10.1002/art.23063. [DOI] [PubMed] [Google Scholar]
- 90.Boettger MK, et al. Antinociceptive effects of tumor necrosis factor α neutralization in a rat model of antigen-induced arthritis: evidence of a neuronal target. Arthritis Rheum. 2008;58:2368–2378. doi: 10.1002/art.23608. [DOI] [PubMed] [Google Scholar]
- 91.Richter F, et al. Tumor necrosis factor causes persistent sensitization of joint nociceptors to mechanical stimuli in rats. Arthritis Rheum. 2010;62:3806–3814. doi: 10.1002/art.27715. [DOI] [PubMed] [Google Scholar]
- 92.Boettger MK, et al. Spinal tumor necrosis factor alpha neutralization reduces peripheral inflammation and hyperalgesia and suppresses autonomic responses in experimental arthritis: a role for spinal tumor necrosis factor alpha during induction and maintenance of peripheral inflammation. Arthritis Rheum. 2010;62:1308–1318. doi: 10.1002/art.27380. [DOI] [PubMed] [Google Scholar]
- 93.Konig C, Zharsky M, Moller C, Schaible HG, Ebersberger A. Involvement of peripheral and spinal tumor necrosis factor alpha in spinal cord hyperexcitability during knee joint inflammation in rats. Arthritis Rheumatol. 2014;66:599–609. doi: 10.1002/art.38271. [DOI] [PubMed] [Google Scholar]
- 94.Hess A, et al. Blockade of TNF-α rapidly inhibits pain responses in the central nervous system. Proc. Natl Acad. Sci. USA. 2011;108:3731–3736. doi: 10.1073/pnas.1011774108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Carswell EA, et al. An endotoxin-induced serum factor that causes necrosis of tumors. Proc. Natl Acad. Sci. USA. 1975;72:3666–3670. doi: 10.1073/pnas.72.9.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Beutler B, Cerami A. Cachectin and tumour necrosis factor as two sides of the same biological coin. Nature. 1986;320:584–588. doi: 10.1038/320584a0. [DOI] [PubMed] [Google Scholar]
- 97.Elinav E, et al. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat. Rev. Cancer. 2013;13:759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 98.Zhao X, et al. TNF signaling drives myeloid-derived suppressor cell accumulation. J. Clin. Invest. 2012;122:4094–4104. doi: 10.1172/JCI64115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hu X, et al. Transmembrane TNF-α promotes suppressive activities of myeloid-derived suppressor cells via TNFR2. J. Immunol. 2014;192:1320–1331. doi: 10.4049/jimmunol.1203195. [DOI] [PubMed] [Google Scholar]
- 100.Haudek SB, Taffet GE, Schneider MD, Mann DL. TNF provokes cardiomyocyte apoptosis and cardiac remodeling through activation of multiple cell death pathways. J. Clin. Invest. 2007;117:2692–2701. doi: 10.1172/JCI29134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Skeoch S, Bruce IN. Atherosclerosis in rheumatoid arthritis: is it all about inflammation? Nat. Rev. Rheumatol. 2015;11:390–400. doi: 10.1038/nrrheum.2015.40. [DOI] [PubMed] [Google Scholar]
- 102.Verjee LS, et al. Unraveling the signaling pathways promoting fibrosis in Dupuytren's disease reveals TNF as a therapeutic target. Proc. Natl Acad. Sci. USA. 2013;110:E928–E937. doi: 10.1073/pnas.1301100110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Feldmann M, Maini RN. Perspectives from masters in rheumatology and autoimmunity: can we get closer to a cure for rheumatoid arthritis? Arthritis Rheumatol. 2015;67:2283–2291. doi: 10.1002/art.39269. [DOI] [PubMed] [Google Scholar]
- 104.Smith JA, Kauffman CA. Endemic fungal infections in patients receiving tumour necrosis factor-α inhibitor therapy. Drugs. 2009;69:1403–1415. doi: 10.2165/00003495-200969110-00002. [DOI] [PubMed] [Google Scholar]
- 105.Deepak P, et al. T-cell non-Hodgkin's lymphomas reported to the FDA AERS with tumor necrosis factor-alpha (TNF-α) inhibitors: results of the REFURBISH study. Am. J. Gastroenterol. 2013;108:99–105. doi: 10.1038/ajg.2012.334. [DOI] [PubMed] [Google Scholar]
- 106.Fischer JA, et al. Combined inhibition of tumor necrosis factor α and interleukin-17 as a therapeutic opportunity in rheumatoid arthritis: development and characterization of a novel bispecific antibody. Arthritis Rheumatol. 2015;67:51–62. doi: 10.1002/art.38896. [DOI] [PubMed] [Google Scholar]
- 107.Genovese MC, et al. Combination therapy with etanercept and anakinra in the treatment of patients with rheumatoid arthritis who have been treated unsuccessfully with methotrexate. Arthritis Rheum. 2004;50:1412–1419. doi: 10.1002/art.20221. [DOI] [PubMed] [Google Scholar]
- 108.Weinblatt M, et al. Selective costimulation modulation using abatacept in patients with active rheumatoid arthritis while receiving etanercept: a randomised clinical trial. Ann. Rheum. Dis. 2007;66:228–234. doi: 10.1136/ard.2006.055111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Schmidt EM, et al. Selective blockade of tumor necrosis factor receptor I inhibits proinflammatory cytokine and chemokine production in human rheumatoid arthritis synovial membrane cell cultures. Arthritis Rheum. 2013;65:2262–2273. doi: 10.1002/art.38055. [DOI] [PubMed] [Google Scholar]
- 110.Zettlitz KA, et al. ATROSAB, a humanized antagonistic anti-tumor necrosis factor receptor one-specific antibody. MAbs. 2010;2:639–647. doi: 10.4161/mabs.2.6.13583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Richter F, et al. Antagonistic TNF receptor one-specific antibody (ATROSAB): receptor binding and in vitro bioactivity. PLoS ONE. 2013;8:e72156. doi: 10.1371/journal.pone.0072156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.McCann FE, et al. Selective tumor necrosis factor receptor I blockade is antiinflammatory and reveals immunoregulatory role of tumor necrosis factor receptor II in collagen-induced arthritis. Arthritis Rheumatol. 2014;66:2728–2738. doi: 10.1002/art.38755. [DOI] [PubMed] [Google Scholar]
- 113.Steeland S, et al. Generation and characterization of small single domain antibodies inhibiting human tumor necrosis factor receptor 1. J. Biol. Chem. 2015;290:4022–4037. doi: 10.1074/jbc.M114.617787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Steed PM, et al. Inactivation of TNF signaling by rationally designed dominant-negative TNF variants. Science. 2003;301:1895–1898. doi: 10.1126/science.1081297. [DOI] [PubMed] [Google Scholar]
- 115.Zalevsky J, et al. Dominant-negative inhibitors of soluble TNF attenuate experimental arthritis without suppressing innate immunity to infection. J. Immunol. 2007;179:1872–1883. doi: 10.4049/jimmunol.179.3.1872. [DOI] [PubMed] [Google Scholar]
- 116.Shibata H, et al. Creation and X-ray structure analysis f the tumor necrosis factor receptor-1-selective mutant of a tumor necrosis factor-α antagonist. J. Biol. Chem. 2008;283:998–1007. doi: 10.1074/jbc.M707933200. [DOI] [PubMed] [Google Scholar]
- 117.Kitagaki M, et al. Novel TNF-α receptor 1 antagonist treatment attenuates arterial inflammation and intimal hyperplasia in mice. J. Atheroscler. Thromb. 2012;19:36–46. doi: 10.5551/jat.9746. [DOI] [PubMed] [Google Scholar]
- 118.Fischer R, et al. A TNF receptor 2 selective agonist rescues human neurons from oxidative stress-induced cell death. PLoS ONE. 2011;6:e27621. doi: 10.1371/journal.pone.0027621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Tang W, et al. The growth factor progranulin binds to TNF receptors and is therapeutic against inflammatory arthritis in mice. Science. 2011;332:478–484. doi: 10.1126/science.1199214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Durez P, et al. Therapeutic vaccination with TNF-Kinoid in TNF antagonist-resistant rheumatoid arthritis: a Phase II randomized, controlled clinical trial. PLoS ONE. 2014;9:e113465. doi: 10.1371/journal.pone.0113465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rao P, et al. IκBβ acts to inhibit and activate gene expression during the inflammatory response. Nature. 2010;466:1115–1119. doi: 10.1038/nature09283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Brown JD, et al. NF-κB directs dynamic super enhancer formation in inflammation and atherogenesis. Mol. Cell. 2014;56:219–231. doi: 10.1016/j.molcel.2014.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Yarilina A, Xu K, Chan C, Ivashkiv LB. Regulation of inflammatory responses in tumor necrosis factor-activated and rheumatoid arthritis synovial macrophages by JAK inhibitors. Arthritis Rheum. 2012;64:3856–3866. doi: 10.1002/art.37691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Klein K, Kabala PA, Grabiec AM. The bromodomain protein inhibitor I-BET151 suppresses expression of inflammatory genes and matrix degrading enzymes in rheumatoid arthritis synovial fibroblasts. Ann. Rheum. Dis. 2014 doi: 10.1136/annrheumdis-2014-205809. http://dx.doi.org/10.1136/annrheumdis-2014-205809. [DOI] [PubMed]
- 125.Grabiec AM, Korchynskyi O, Tak PP, Reedquist KA. Histone deacetylase inhibitors suppress rheumatoid arthritis fibroblast-like synoviocyte and macrophage IL-6 production by accelerating mRNA decay. Ann. Rheum. Dis. 2012;71:424–431. doi: 10.1136/ard.2011.154211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Pandis I, et al. Identification of microRNA-221/222 and microRNA-323-3p association with rheumatoid arthritis via predictions using the human tumour necrosis factor transgenic mouse model. Ann. Rheum. Dis. 2012;71:1716–1723. doi: 10.1136/annrheumdis-2011-200803. [DOI] [PubMed] [Google Scholar]
- 127.Trenkmann M, et al. Tumor necrosis factor α-induced microRNA-18a activates rheumatoid arthritis synovial fibroblasts through a feedback loop in NF-κB signaling. Arthritis Rheum. 2013;65:916–927. doi: 10.1002/art.37834. [DOI] [PubMed] [Google Scholar]
- 128.Rapicavoli NA, et al. A mammalian pseudogene lncRNA at the interface of inflammation and anti-inflammatory therapeutics. eLIFE. 2013;2:e00762. doi: 10.7554/eLife.00762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Ermann J, Rao DA, Teslovich NC, Brenner MB, Raychaudhuri S. Immune cell profiling to guide therapeutic decisions in rheumatic diseases. Nat. Rev. Rheumatol. 2015;11:541–551. doi: 10.1038/nrrheum.2015.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Plant D, Wilson AG, Barton A. Genetic and epigenetic predictors of responsiveness to treatment in RA. Nat. Rev. Rheumatol. 2014;10:329–337. doi: 10.1038/nrrheum.2014.16. [DOI] [PubMed] [Google Scholar]
- 131.Shembade N, Pujari R, Harhaj NS, Abbott DW, Harhaj EW. The kinase IKKα inhibits activation of the transcription factor NF-κB by phosphorylating the regulatory molecule TAX1BP1. Nat. Immunol. 2011;12:834–843. doi: 10.1038/ni.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Shembade N, Parvatiyar K, Harhaj NS, Harhaj EW. The ubiquitin-editing enzyme A20 requires RNF11 to downregulate NF-κB signalling. EMBO J. 2009;28:513–522. doi: 10.1038/emboj.2008.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Bohgaki M, et al. Involvement of Ymer in suppression of NF-κB activation by regulated interaction with lysine-63-linked polyubiquitin chain. Biochim. Biophys. Acta. 2008;1783:826–837. doi: 10.1016/j.bbamcr.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 134.Ma A, Malynn BA. A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nat. Rev. Immunol. 2012;12:774–785. doi: 10.1038/nri3313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Wertz IE, et al. De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-κB signalling. Nature. 2004;430:694–699. doi: 10.1038/nature02794. [DOI] [PubMed] [Google Scholar]
- 136.Bremm A, Freund SM, Komander D. Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat. Struct. Mol. Biol. 2010;17:939–947. doi: 10.1038/nsmb.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Fiil BK, et al. OTULIN restricts Met1-linked ubiquitination to control innate immune signaling. Mol. Cell. 2013;50:818–830. doi: 10.1016/j.molcel.2013.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Elliott PR, et al. Molecular basis and regulation of OTULIN–LUBAC interaction. Mol. Cell. 2014;54:335–348. doi: 10.1016/j.molcel.2014.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Schaeffer V, et al. Binding of OTULIN to the PUB domain of HOIP controls NF-κB signaling. Mol. Cell. 2014;54:349–361. doi: 10.1016/j.molcel.2014.03.016. [DOI] [PubMed] [Google Scholar]
- 140.Bignell GR, et al. Identification of the familial cylindromatosis tumour-suppressor gene. Nat. Genet. 2000;25:160–165. doi: 10.1038/76006. [DOI] [PubMed] [Google Scholar]
- 141.Sato Y, et al. Structures of CYLD USP with Met1- or Lys63-linked diubiquitin reveal mechanisms for dual specificity. Nat. Struct. Mol. Biol. 2015;22:222–229. doi: 10.1038/nsmb.2970. [DOI] [PubMed] [Google Scholar]
- 142.Xiao N, et al. Ubiquitin-specific protease 4 (USP4) targets TRAF2 and TRAF6 for deubiquitination and inhibits TNFα-induced cancer cell migration. Biochem. J. 2012;441:979–986. doi: 10.1042/BJ20111358. [DOI] [PubMed] [Google Scholar]
- 143.Liang J, et al. MCP-induced protein 1 deubiquitinates TRAF proteins and negatively regulates JNK and NF-κB signaling. J. Exp. Med. 2010;207:2959–2973. doi: 10.1084/jem.20092641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Matsushita K, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 145.McCoy MK, et al. Blocking soluble tumor necrosis factor signaling with dominant-negative tumor necrosis factor inhibitor attenuates loss of dopaminergic neurons in models of Parkinson's disease. J. Neurosci. 2006;26:9365–9375. doi: 10.1523/JNEUROSCI.1504-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.McAlpine FE, et al. Inhibition of soluble TNF signaling in a mouse model of Alzheimer's disease prevents pre-plaque amyloid-associated neuropathology. Neurobiol. Dis. 2009;34:163–177. doi: 10.1016/j.nbd.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Sama DM, et al. Inhibition of soluble tumor necrosis factor ameliorates synaptic alterations and Ca2+ dysregulation in aged rats. PLoS ONE. 2012;7:e38170. doi: 10.1371/journal.pone.0038170. [DOI] [PMC free article] [PubMed] [Google Scholar]