Abstract
In recent years, it has become widely recognized that histone modification plays a pivotal role in controlling gene expression, and is involved in a wide spectrum of disease regulation. Histone acetylation is a major modification that affects gene transcription and is controlled by histone acetyltransferases (HATs) and histone deacetylases (HDAC). HATs acetylate lysines of histone proteins, resulting in relaxation of chromatin structure, and they also facilitate gene activation. Conversely, HDACs remove acetyl groups from hyperacetylated histones and suppress general gene transcription. In addition to histones, numerous non-histone proteins can be acetylated and deacetylated, and they are also involved in a wide range of disease regulation. To date, there are 18 HDACs in mammals classified into four classes based on homology to yeast HDACs. Accumulating evidence has revealed that HDACs play crucial roles in a variety of biological processes including inflammation, cell proliferation, apoptosis, and carcinogenesis. In this review, we summarize the current state of knowledge of HDACs in carcinogenesis and describe the involvement of HDACs in cancer-associated molecular processes. It is hoped than our understanding of the role of HDACs in cancer will lead to the design of more potent and specific drugs targeting selective HDAC proteins for the treatment of the disease.
Keywords: Histone deacetylase, Histone acetyation, Cancer, drug target
I. INTRODUCTION
In the eukaryotic cell, genomic DNA is wrapped around highly conserved histone proteins H2A, H2B, H3 and H4 into nucleosomes, which form the fundamental units of chromatin. Chromatin organization plays a key role in the control of gene expression. Epigenetic modifications can regulate heritable or reversible gene expression without altering the DNA sequence, and can also modify chromatin architecture and its accessibility.1,2 DNA methylation and post-translational acetylation of histones constitute two major mechanisms that are responsible for epigenetic regulation of gene expression.3,4 Acetylation opens the condensed chromatin structure by reducing the affinity of DNA for histones and releasing the histone tails from the linker DNA, thereby allowing transcription factors, co-factors, and RNA polymerase II-complexes to access the DNA. Acetylation levels are the result of the balance of the activities of histone acetyltransferase (HAT) and histone deacetylase (HDAC). HATs acetylate lysines within lysine-rich amino-terminal tails of histone proteins, resulting in charge neutralization and a more relaxed, open, and transcriptionally active chromatin structure. In contrast, HDACs are enzymes that remove acetyl groups from 1-N-acetyl lysine amino acids on histones, counteracting the effects of HATs by returning the histone to its basal state, with the concomitant suppressing gene expression in most cases.
HDACs can also regulate gene repression via non-histone substrates. For example, the tumor suppressor p53 can be directly deacetylated by HDAC1 and SIRT1.5 The p65 subunit of NF-κB can be deacetylated by HDAC3, which promotes its association with IκBa and leads to NF-κB nuclear exportation.6 Moreover, two members of the Class III HDACs: SIRT1 and SIRT3 deacetylate the transcription factors p73 and Ku70, respectively.7,8 Deacetylation of nonhistone proteins allows HDACs to exert direct effects on multiple physiologic processes, including differentiation, apoptosis, autophagy, inflammation, and metabolism.
Generally, cancer is considered to originate from a wide variety of genetic and genomic alterations, such as rearrangements, mutations, deletions, and amplifications, leading to aberrant expression of tumor suppressor genes and oncogenes. As we now know, cancer is not only restricted to the genetic changes, but may also involve epigenetic modifications. Accumulating evidence indicates that cancer is associated with abnormal cell functions that include autophagy, apoptosis, cell motility, and DNA repair. These cell functions are regulated at least in part by HDACs.
In this review, we summarized the current state of knowledge of HDACs in carcinogenesis and described the involvement of HDACs in cancer-associated molecular mechanisms. Moreover, we also discussed the clinical trials of specific drugs that target individual HDAC proteins for the treatment of cancer. We hope that this will provide novel clues to develop a potent and specific drug to treat different types of human malignancies.
II. HISTONE DEACETYLASES
HDACs are typically present within larger, co-repressor multiprotein complexes, where they mediate the removal of an acetyl group from lysines on histone tails. To date, 18 HDACs have been identified in mammals.9 These HDACs can be subdivided into four classes based upon their homology with their yeast counterparts.10 Class I includes HDAC1, -2, -3, and -8, which show homology with Rpd3 in yeast, and they are usually detected in the nucleus. Class I HDACS are ubiquitously expressed in various mammalian cell lines and tissues.11 Class II comprises HDAC4, -5, -6, -7, -9 and -10, which have a high degree of homology with the Hda1 protein and can be subdivided into two subclasses: classes IIa (HDAC4, -5, -7, and -9) and IIb (HDAC6 and -10). Class II exhibits tissue specific expression and can shuttle between the nucleus and cytoplasm in a phosphorylation-regulated manner, which suggests that Class II HDACs may be involved in the acetylation of non-histone proteins. Class III consists of Sirt1, -2, -3, -4, -5, -6 and -7, which are homologous to the yeast transcriptional repressor silent information regulator-2 (Sir2) and sirtuin (SIRT1-7). The structure and function of Class III HDACs are distinct from all the other HDACs.12 HDAC11 is the sole member of the class IV HDACs. Classes I, II, and IV HDACs share common features such as their dependence on zinc2+ for their enzymatic activity and inhibition by broad spectrum HDAC inhibitors, such as trichostatin A (TSA), suberoylanilide hydroxamic acid (SAHA) and LB589, whereas class III enzymes are NAD+-dependent deacetylases, which are insensitive to TSA.13 HDACs have an increased expression in many kind of cancers, including ovarian, breast, bladder, and other cancers14–16 and are believed to promote carcinogenesis through the acetylation and interaction with key transcriptional regulators. Thus, HDAC enzymes are identified as attractive targets for cancer therapy. Table 1 provides an overview of HDACs with particular reference to their role in prognosis.
Table 1.
Cancer related HDACs (Partial list)
| Cancer type | Up-regulated HDACs | Effect on prognosis | Reference | |
|---|---|---|---|---|
| Hematological malignancies | HL | HDAC1-3 | Poor | 29 |
| ALL | HDAC1-2, HDAC4, HDAC8 | Poor | 30 | |
| DLBCL | HDAC1-2, HDAC6 | Poor | 31 | |
| APL | HDAC1 | ? | 33 | |
| Prostate cancer | HDAC1-3 | Poor | 36 | |
| HDAC1-5 | Poor | 37 | ||
| HDAC11 | Poor | 39 | ||
| Ovarian cancer | HDAC1-3 | Poor | 16, 43–45 | |
| HDAC4 | Poor | 46 | ||
| Bladder cancer | HDAC1-3 | Poor | 16 | |
| Hepatocellular carcinoma | HDAC1-3, 7 | Poor | 53 | |
| HDAC3 | Poor | 54 | ||
| Colon cancer | HDAC3 | Poor | 58 | |
| HDAC1-4 | Poor | 62 | ||
Abbreviations: HL, Hodgkin’s lymphoma; ALL, Acute lymphoblastic leukemia; DLBCL, diffuse large B-cell lymphoma; APL, acute promyelocytic leukemia.
III. HISTONE DEACETYLASE INHIBITORS
HDAC inhibitors (HDACi) are natural or synthetic chemical compounds that have broad functions in the cell. Various HDACi were designed to target the catalytic sites of HDACs. Thus, HDACi can change the balance between HATs and HDACs, leading to the accumulation of acetylated histones/non-histone proteins, which induces transcriptional and related molecular effects. According to their structure and specificity, HDACi can be grouped into several classes, including hydroxamates, cyclic peptides, aliphatic acids, and benzamides. TSA was found to inhibit HDACs in 1990, and this was the first natural hydroxamate discovered with the function of HDAC inhibition.17 Vorinostat is structurally related to TSA, and it is the first of the new HDACi to be approved by the Food and Drug Administration for the treatment of cutaneous T-cell lymphoma patients.18 Depsipeptide was approved in 2009. In addition, FK228, PXD101, PCI-24781, ITF2357, MGCD0103, MS-275, valproic acid (VPA), and LBH589 have also demonstrated therapeutic potential in monotherapy or in combination with other anti-tumor drugs in malignancies.19 HDACi represent exciting clinical drugs with broad therapeutic spectrum and are, therefore, currently being investigated for use in the treatment of cancer. As we know, classes I, II, and IV HDACs share zinc-dependent homologies. Therefore, many inhibitors are non-specific and can be used to inhibit multiple isoforms of HDACs. For example, TSA can inhibit both class I and II HDACs. The majority of currently available HDAC inhibitors are pan-HDAC inhibitors, which inhibit the activity of a broad spectrum of HDACs. However, specific inhibitors that target individual HDACs have also been explored. An example is that HDAC6 activity was selectively inhibited by tubacin.20
IV. HDACs AND INFLAMMATION
Accumulating evidence has revealed that chronic inflammation is predisposed to many kinds of cancer. Even in these cancers that do not develop in inflamed tissues, inflammatory mediators are usually present, and it is now known to be an essential part of the malignant microenvironment. Inflammation was considered to be the seventh hallmark of cancer. Cancer-related inflammation mediators include cytokines such as tumor necrosis factor (TNF), interleukin (IL)-1, IL-6, and chemokines such as CCL2, CXCL8, CXCL5.21 Recently, it was reported that HDACs participated in the regulation of cancer-related inflammation. Cytokine secretion by cancer cells contributes to cancer-induced symptoms and angiogenesis.
HDACs can act as both positive and negative regulators of inflammatory factors. T cell-specific loss of HDAC1 led to an increased inflammatory response in an in vivo allergic airway inflammation model. The study also shows that mice with HDAC1-deficient T cells displayed an increase in parenchymal lung inflammation in the Th2-type asthma model. These data provide genetic evidence that HDAC1 controls the magnitude of an inflammatory response by modulating cytokine expression in effector T cells.22 HDAC-1 can suppress CCL2 and CXCL10 in a model of chronic liver inflammation and fibrosis, and it is thought that the p50:p50:HDAC-1 complex is a master negative regulator of inflammation.23 Accordingly, HDAC2 interacts with the transcriptional activator metastatic tumor antigen (MTA)1 to decrease the expression of inflammatory cytokine genes in macrophages.24 By contrast, SIRT6 enhances cytokine IL8 and TNF secretions and cell motility in pancreatic cancer cells by activating Ca2+ signaling. The results suggest that SIRT6 is a target to modifying cancer cell pro-inflammatory phenotype and migratory propensity.25 HDAC3-deficient macrophages were unable to activate almost half of the inflammatory gene expression program when stimulated with LPS.26 Reducing expression of histone deacetylase genes (HDAC 2, 3 and 9) altered the global modification of histones and decreased expression of pro-inflammatory genes (RIPK2 and COX2).27 In addition, in the mice model of colitis, which is induced by azoxymethane and dextran sulphate sodium, treatment with the HDAC inhibitor SAHA or ITF2357 suppressed inflammation and inhibited tumorigenesis profoundly.28
V. HDAC AND HEMATOLOGICAL MALIGNANCIES
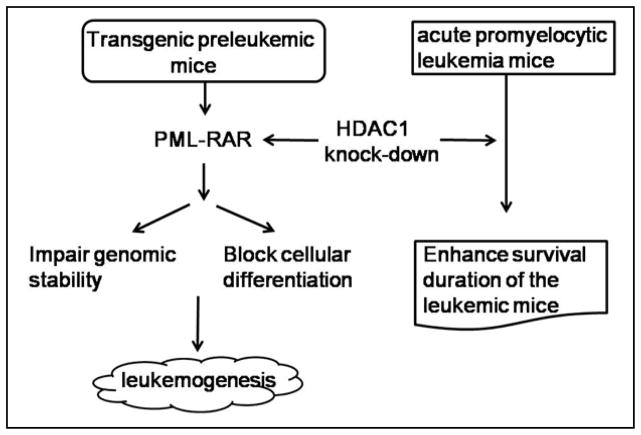
Several lines of evidence show that HDACs play a crucial role in hematological malignancies. The first HDACi, Vorinostat, to be approved by the Food and Drug Administration, was used to treat cutaneous T-cell lymphoma patients.18 In recent years, more roles and molecular mechanisms of HDACs in hematological malignancies were discovered. In hematological malignancies, the aberrant expression and activity of HDACs often occur. It was shown that Class I HDACs 1, 2 and 3 are highly expressed in classical Hodgkin’s lymphoma (HL), and a decreased HDAC1 expression is accompanied by a worse outcome in HL.29 Acute lymphoblastic leukemia (ALL) is the most common childhood malignancy. Furthermore, Gruhn and colleagues identified the relevance of HDACs for childhood ALL. In this experiment, HDAC1-11 expression was determined in 93 primary ALL and eight healthy donor samples. They found that HDAC1, HDAC2 and HDAC8 were significantly higher expressed in ALL samples. A high HDAC4 expression was associated with a high initial leukocyte count, T cell ALL, and prednisone poor-response. These data show that HDAC4 can act as a drug target in childhood ALL, especially in prednisone poor-responders.30 Accordingly, HDAC1, HDAC2, and HDAC6 are over-expressed in diffuse large B-cell lymphoma and peripheral T-cell lymphoma. Within these HDACs, HDAC6 may be an important prognostic marker associated with a favorable outcome in diffuse large B-cell lymphoma but a more aggressive course in peripheral T-cell lymphoma.31 Interestingly, HDAC1 performed dual roles in the regulation of acute promyelocytic leukemia: oncosuppressive in the early stages, and oncogenic in established tumor cells.32 Knock-down of HDAC1 dramatically accelerates leukemogenesis in transgenic preleukemic mice through counteracting the activity of PML-RAR, an oncoprotein, which blocks cellular differentiation and increases genomic instability. In contrast, knock-down of HDAC1 in transplanted PML-RAR–expressing leukemia cells prolonged the survival time of the recipient mice, supporting the view that HDAC1 has oncogenic activity in established tumor cells32 (Figure. 1).
Figure 1.
HDAC1 performs dual roles in the regulation of acute promyelocytic leukemia: oncosuppressive in the early stages and oncogenic in established tumor cells
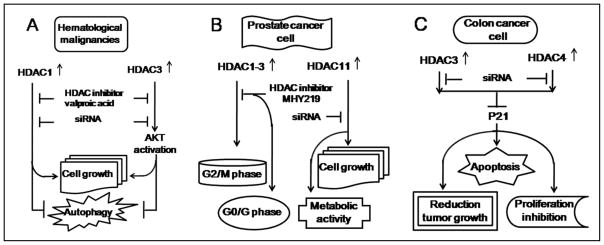
BARD1 (BRCA1-associated RING domain 1) is involved in the pathogenesis of different cancers, including breast, uterine, ovarian, colon, and lung. Specific BARD1 isoforms might act as tumor diagnostic and prognostic markers. In human acute myeloid leukemias (AML), Vorinostat reduces BARD1 mRNA levels through increasing miR-19a and miR-19b expression levels that directly target BARD1.33 These results reveal the novel molecular mechanism of Vorinostat in the treatment of AML. VPA, a class I HDAC inhibitor, suppresses tumor cell growth and induces autophagy in Burkitt leukemia/lymphoma though two distinct pathways (Figure 2A): VPA promotes temsirolimus to induce autophagy through inhibiting HDAC1. The specific targeting of HDAC1 using small interfering RNA (siRNA) attenuated VPA-mediated regulation of CDKN1A, CDKN1B and LC3-I/II, regression of tumor cell growth, and induction of autophagy. Meanwhile, VPA counteracted temsirolimus-induced AKT activation via HDAC3 inhibition. HDAC3 siRNA abrogated the ability of VPA to modulate AKT phosphorylation, to suppress tumor cell growth, and to induce autophagy.34 The tumor suppressor E-cadherin gene is frequently silenced in chronic lymphocytic leukemia cells and results in wnt-pathway activation, which promotes cancer development. The E-cadherin gene is epigenetically modified and hypoacetylated in lymphocytic leukemia leukemic cells. The treatment of lymphocytic leukemia cells from patients with HDACi MS-275 activates transcription from this silent gene with expression of more correctly spliced E-cadherin transcripts as compared to the aberrant exon11 skipped transcripts that in turn inhibits the wnt signaling pathway.35 These data reveal the novel molecular mechanism of HDACs in hematological malignancies and provide an insight of clinical application in treating the disease.
Figure 2.
The roles of HDACs in the regulation of cancer processing. (A) HDAC1 and HDACs regulate hematological malignancies. (B) HDAC1-3 and HDAC11 regulate prostate cancer. (C) HDAC3 and HDAC4 regulate colon cancer.
VI. HDACs AND PROSTATE CANCER
HDAC expressions in 192 prostate carcinomas (Pca) were detected by immunohistochemistry. The results show that HDACs 1, 2, and 3 are highly expressed in the majority of cases. HDACs were accompanied by enhanced tumor cell proliferation. This study pointed out that HDAC2 is an important prognostic marker of prostate cancer.36 Wang and his colleagues analyzed the expression levels of HDACs in benign and malignant human prostate tissue and various PCa cell lines. The results indicated that HDAC1-5 increased in these specimens. Moreover, the HDAC inhibitor SAHA suppressed, in particular, prostate cancer cell growth and invasion.37 miR-449a is frequently downregulated in prostate cancer tissue, relative to patient matched control tissues. An introduction of miR-449a into PC-3 prostate cancer cells resulted in cell cycle arrest, apoptosis, and a senescent-like phenotype concomitantly with the suppressing of the expression of HDAC1. The data confirmed that miR-449a inhibition of prostate cancer is involved in the mechanism of the direct target HDAC1.38 In addition, HDAC11 was strongly expressed in many cancer cell lines, including the PC-3 prostate cancer cell line. The specific targeting of HDAC11 using siRNA is sufficient to cause cell death and to inhibit metabolic activity in PC-3.39
The above findings suggest that HDAC specific targeting could serve as a potential therapeutic agent in prostate cancer. The effects of HDAC inhibitors VPA and TSA on ERG-positive prostate cancer cells were tested. It indicated that VPA and TSA could induce apoptosis, up-regulate p21/Waf1/CIP1, repress TMPRSS2-ERG expression, and affect the acetylation status of p53 in ERG-positive prostate cancer cells.40 Recently, several novel HDACis have been shown to treat prostate cancer. The anticancer effect of MHY219, a novel HDACi, was evaluated in the prostate cancer cell lines DU145, LNCaP, and PC3. The results indicate that MHY219 increased histone H3 hyperacetylation and reduced the expression of HDAC1-3 in prostate cancer cells. MHY219 significantly induced G2/M phase arrest in DU145 and PC3 cells and arrested the cell cycle at G0/G1 phase in LNCaP cells. Furthermore, MHY219 effectively increased apoptosis in DU145 and LNCaP cells41 (Figure 2B). A248, a novel synthetic HDAC inhibitor, decreased the expression of survivin and Mcl-1 through suppressing specificity protein 1 (Sp1) expression. Thus, this leads to inhibiting the growth of DU145 and PC3 cells while inducing apoptosis.42
VII. HDACs AND OVARIAN CANCER
The expression levels of Class I HDACs (HDAC1-3), but not Class II HDACs, are significantly higher in ovarian cancers in comparison to normal ovarian tissues. The HDAC inhibitor romidepsin (FK228) can selectively inhibit the class I HDACs. The knocking down of the gene expression of HDACs 1, 2, and 3 by siRNA suppresses ovarian cancer cell growth.43,44 A high-level expression of HDAC1-3 is associated with a poor prognosis in ovarian endometrioid carcinomas, which suggests that these targets should be explored as predictive factors in ovarian and endometrial carcinomas, prospectively.45 Ovarian cancer cells frequently acquire resistance to platinum chemotherapy, representing a major challenge for improving patient survival. HDAC4 is increased in platinum resistant ovarian tumors.46 A clinical selection of HDAC4 over-expressing tumor cells, upon exposure to chemotherapy, promoted STAT1 deacetylation and cancer cell survival. These findings suggest HDAC4 as a novel, therapeutically tractable target to countering platinum resistance in ovarian cancer.46 The roles of class I HDAC inhibitors thailandepsin A and thailandepsin B on five ovarian cancer cell lines were investigated. Thailandepsins decreased cell viability of four of the five ovarian cancer cell lines at nanonomolar concentrations. Thailandepsin B had both greater inhibitory effects on cell viability and ability on promoting cell apoptosis than thailandepsin A.47 Notably, thailandepsin B had greater cytotoxic effects than thailandepsin A in vitro. VPA performs anti-proliferative and pro-apoptotic activities, and the anticancer effects of VPA in ovarian cancer cells were at least partly mediated through HDAC inhibition and selective reduction of expression of HDAC7 and HDAC2 at both the transcriptional and translational levels.48
VIII. HDACs AND BLADDER CANCER
The studies of HDACs in bladder cancer are emerging in recent years. The expression levels of HDAC1-3 are significantly increased in bladder cancer.16 High-grade noninvasive papillary bladder tumors are associated with high expression levels of HDAC1 and HDAC2.16 In addition, the HDAC4 expression level is significantly higher in urinary bladder transitional cell carcinomas in comparison to normal bladder tissue. In both HTB4 and HTB9 bladder cancer, HDAC inhibitor VPN potently increased transcription of the XPC gene; deficiency or attenuation of this gene has been strongly associated with a high incidence of cancers.49 Treatment of 5637, T24, J82, and RT4 urothelial lines with the HDAC inhibitor elinostat caused a significant linear dose-dependent growth inhibition. Within these, 5637 cell line, which was derived from a superficial papillary tumor, was the most sensitive to treatment. An intraperitoneal injection of belinostat into the superficial bladder cancer mice (established by Ha-ras transgenic mice) caused lower bladder weight, reduced cell proliferation, and higher expression of p21WAF1.50 In addition, Both MS-275 and TSA blocked T24 human bladder cancer cell cycling in the G0/G1 phase and induced a significant increase in cell apoptosis in a concentration and time-dependent manner. The molecular mechanism is associated with increased level of histone acetylation, down-regulation of p21WAF1/CIP1 expression, and up-regulation of cyclin A expression.51 Moreover, the influence of the VPA on TCCSUP and RT-112 bladder cancer cell adhesion in vitro was investigated. VPA (0.5mM and 1mM) significantly prevented binding of both RT-112 and TCCSUP cells to collagen, as compared with the untreated controls.52
IX. HDACs AND HEPATOCELLULAR CARCINOMA
HDAC2 could be an independent predictor of survival in hepatocellula carcinoma (HCC). HDACs 1, 2, 3, and 7 expression levels in 170 surgically resected primary HCC adjacent uninvolved tissues were detected, and their correlation with clinical data and patient survival were evaluated. HDACs 1, 2, and 3 were expressed significantly higher in cancer cells compared to normal tissue. The expression levels of HDACs 1, 2 and 3 are highly related with the growth of tumor grades. A high level of HDAC2 was also associated with poor survival in low-grade and early-stage tumors.53 Over-expression of HDAC3 was associated with a poor outcome in liver cancer. HDAC inhibitors could render liver cancer stem cells sensitive to a therapy of sorafenib. HDAC3 was selectively expressed in the liver for self-renewal of cancer stem cells and plays a critical role in regulating liver cancer stem cells.54 In HCC cells, up-regulation of HDAC1-3 reduces expression of miR-449. MiR-449 can promote apoptosis and reduce proliferation of liver cells through targeting c-MET mRNA that encodes the receptor tyrosine kinase for the hepatocyte growth factor.55
Suppression of HDAC activity by trichostatin A and sodium butyrate in human hepatocarcinoma cells led to the inhibition of metastasis and invasion through up-regulation of early growth response gene-1 and claudin-3.56 Through inhibition of HDAC4, sodium butyrate performs its anticancer role on HCC cells, SMMC-7721, and HepG2. Treatment with sodium butyrate at high concentrations significantly inhibited the growth of various HCC cells, such as inducing cell cycle arrest, apoptosis, and inhibition of cell migration/invasion. HDAC4 and matrix metalloproteinase 7 may be involved in these functions of sodium butyrate.57 Moreover, HDAC inhibition suppresses HCC cell growth by inducing autophagy. The human HCC cell lines Hep3B, HepG2, and Huh7 were treated with HDAC inhibitors OSU-HDAC42 and SAHA, which induced autophagy through down-regulation of Akt/mTOR signaling and induction of ER stress response. The data indicates that SAHA might be attractive for the treatment of HCC and pharmacological targeting of autophagy.58
X. HDACs AND COLON CANCER
Numerous evidences have shown that HDACs play a crucial role in the regulation of colon cancer. Elevated levels of several HDACs have been reported in colon cancer. The HDAC3 protein is increased in human colon tumors and in duodenal adenomas from Apc1638N/+mice. Silencing of HDAC3 expression in colon cancer cell lines resulted in growth inhibition, a decrease in cell survival, and increased apoptosis. Concurrently, over-expression of HDAC3 and other Class I HDACs inhibited basal and butyrate-induced p21 expressions.59 The other study also shows an increase in HDAC3 expression in colon cancer cell lines, such as SW480 and HT-29 cells. Interestingly, HDAC3 had a higher expression in SW480 compared to other colon cancer cell lines. P21 was poorly induced in SW480 cells relative to the lower HDAC3-expressing HT-29 cells. RNAi-induced reduction of HDAC3 in SW480 cells increased their constitutive, butyrate-, TSA-, and TNF-a-induced expression of p2160 (Fig. 2C). HDAC4 is expressed in a tissue-specific manner, and it represses differentiation of a specific cell type.61 Silencing HDAC4 expression by siRNA in HCT116 cells induced growth inhibition and apoptosis in vitro, reduced xenograft tumor growth, and increased p21 transcription. Conversely, over-expression of HDAC4 repressed p21 promoter activity (Fig. 2C). The studies imply HDAC4 as a regulator of colon cell proliferation through repression of p21.62 The data above show that p21 plays a critical role in mediating HDAC regulation in the colon cancer process.
Immunohistochemical staining showed that HDAC1-4 expression was significantly increased in colorectal adenocarcinoma specimens as compared to healthy control tissues. SAHA significantly induced tumor necrosis and inhibited the growth of colon tumors; SAHA inhibited the growth of colon tumors by decreasing HDACs and the expression of cyclin D1.63 In the rat colon carcinogenesis model induced by azoxymethane, Dietary Ohio State University HDAC42 (OSU-HDAC42), which is a selective HDAC2 inhibitor, produced a dose-dependent inhibition of colonic aberrant crypt foci formation. Accordingly, OSU-HDAC42 significantly inhibited small-intestinal polyp and colon tumor growths in APCmin/+ transgenic mice that spontaneously develop into intestinal tumors.64 However, TSA increases the activity and protein expression of matrix metalloproteinase 11, which is associated with tumor progression and a poor prognosis through ERK1/2-mitogen-activated protein kinase (MAPK) signaling in human epithelial colon adenocarcinoma cell lines BCS-TC2.65 It seems that TSA induces colon cancer progression.
Notably, nutrition-derived HDACis were discovered in colon cancer cell lines. Epigallocatechin 3-gallate, an active compound in green tea, contributes to the degradation of HDAC3 in HCT116 human colon cancer cells.66 Moreover, Kaempferol, a natural polyphenol belonging to the group of flavonoids, produces a distinct epigenetic activity by inhibition of HDACs. In vitro profiling of all conserved human HDACs of class I, II and IV showed that kaempferol inhibited all tested HDACs (HDAC1-11) in HCT116 colon cancer cells. Furthermore, kaempferol mediated prominent reductions in cell viability and proliferation rate.67 These findings provided a novel insight to treating colon cancer by inhibition of HDACs.
XI. CONCLUSIONS AND PERSPECTIVES
The specific interactions of various HDACs and their effects on many types of cancer were clarified. HDACs are often highly expressed in various types of cancer and promote cancer progress, so they are attractive anticancer targets. HDACi can interact with the catalytic domain of histone deacetylase. Thus, HDACi change the balance between the deacetylating activity of HDACs and the acetylating activity of HATs, which lead to increased histone acetylation and up-regulated gene expression. Currently, the majority of HDACi that are either in clinical trial or approved by the Food and Drug Administration are mostly non-selective (pan-HDAC inhibitors). For example, vorinostat, LAQ-824, LBH-589, and belinostat can inhibit class I and IIa HDACs.68,69 Non-specific targeting HDACi, which broadly inhibit many HDACs, have the potential risk of increasing side effects or cytotoxicity. Clinical trials show that treatment with oral vorinostat is associated with fatigue, diarrhea, anorexia, dehydration, and moderate thrombocytopenia.70 Chronic administration of MS-275 brings gastrointestinal side effects and fatigue.71 Deletion of both HDAC1 and HDAC2 in developing neurons leads to severe hippocampal abnormalities, absence of cerebellar foliation, disorganization of cortical neurons, and lethality by postnatal day 7.72 Accordingly, mice lacking HDAC1/2 in Schwann cells exhibit severe dysmyelination and cell cycle arrest at the immature stage due to NF-KB deacetylation.73 Global deletion of HDAC3 in embryo resulted in lethality by E9.5, and mice with cardiac-specific deletion of HDAC3 just survived about 4 months.74 In addition, HDAC3 deletion in the postnatal mouse liver leads to hepatocyte hypertrophy due to imbalances between carbohydrate and lipid metabolisms.75 Thus, when broad spectrum HDACi were used to treat cancer, some HDACs which have no relation to the cancer but are necessary for normal cell function will be blocked, leading to side-effects.
Due to side effects of non-specific target HDACs, it is necessary to discover the exact roles of individual HDACs in tissue specific cancer and the associated cellular and molecular mechanisms. Hopefully, the design of specific HDACi will improve the efficiency of treatment and optimize patient benefits. Although various HDACi were designed to target individual HDACs and were used in clinical trials for specific cancers in recent years, HDACi are rarely designed to target the crucial signaling of the cancer process that associates with specific HDAC. Furthermore, nutrition-derived HDACis that have low toxicity and side effects should be more carefully studied.
Acknowledgments
This study was supported by the National Natural Science Foundation of China (81260318) and the National Heart, Lung, and Blood Institute (R01 HL089405 and R01 HL115265).
ABBREVIATIONS
- ALL
Acute lymphoblastic leukemia
- HAT
Histone acetyltransferase
- HCC
Hepatocellula carcinoma
- HDAC
Histone deacetylases
- SAHA
Suberoylanilide hydroxamic acid
- VPA
Valproic acid
References
- 1.Arnsdorf EJ, Tummala P, Castillo AB, Zhang F, Jacobs CR. The Epigenetic Mechanism of Mechanically Induced Osteogenic Differentiation. Journal of Biomechanics. 2010;43:2881–2886. doi: 10.1016/j.jbiomech.2010.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pons D, de Vries FR, van den Elsen PJ, Heijmans BT, Quax PH, Jukema JW. Epigenetic Histone Acetylation Modifiers in Vascular Remodelling: New Targets for Therapy in Cardiovascular Disease. European Heart Journal. 2009;30:266–277. doi: 10.1093/eurheartj/ehn603. [DOI] [PubMed] [Google Scholar]
- 3.Nagase H, Ghosh S. Epigenetics: differential DNA methylation in mammalian somatic tissues. The FEBS Journal. 2008;275:1617–1623. doi: 10.1111/j.1742-4658.2008.06330.x. [DOI] [PubMed] [Google Scholar]
- 4.Vaissiere T, Sawan C, Herceg Z. Epigenetic Interplay Between Histone Modifications and DNA Methylation in Gene Silencing. Mutation Research. 2008;659:40–48. doi: 10.1016/j.mrrev.2008.02.004. [DOI] [PubMed] [Google Scholar]
- 5.Luo J, Su F, Chen D, Shiloh A, Gu W. Deacetylation of P53 Modulates Its Effect on Cell Growth and Apoptosis. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 6.Chen L, Fischle W, Verdin E, Greene WC. Duration of Nuclear NF-kappaB Action Regulated by Reversible Acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 7.Dai JM, Wang ZY, Sun DC, Lin RX, Wang SQ. SIRT1 Interacts with P73 and Suppresses P73-dependent Transcriptional Activity. Journal of Cellular Physiology. 2007;210:161–166. doi: 10.1002/jcp.20831. [DOI] [PubMed] [Google Scholar]
- 8.Sundaresan NR, Samant SA, Pillai VB, Rajamohan SB, Gupta MP. SIRT3 Is a Stress-responsive Deacetylase in Cardiomyocytes that Protects Cells from Stress-mediated Cell Death by Deacetylation of Ku70. Molecular and Cellular Biology. 2008;28:6384–6401. doi: 10.1128/MCB.00426-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verdin E, Dequiedt F, Kasler HG. Class II Histone Deacetylases: Versatile Regulators. Trends in Genetics. 2003;19:286–293. doi: 10.1016/S0168-9525(03)00073-8. [DOI] [PubMed] [Google Scholar]
- 10.Bolden JE, Peart MJ, Johnstone RW. Anticancer Activities of Histone Deacetylase Inhibitors. Nature Reviews Drug Discovery. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 11.Yang XJ, Seto E. The Rpd3/Hda1 Family of Lysine Deacetylases: from Bacteria and Yeast to Mice and Men. Nature Reviews Molecular Cell Biology. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily Conserved and Nonconserved Cellular Localizations and Functions of Human SIRT Proteins. Molecular Biology of The Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scroggins BT, Robzyk K, Wang D, Marcu MG, Tsutsumi S, Beebe K, Cotter RJ, Felts S, Toft D, Karnitz L, Rosen N, Neckers L. An Acetylation Site in the Middle Domain of Hsp90 Regulates Chaperone Function. Molecular Cell. 2007;25:151–159. doi: 10.1016/j.molcel.2006.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hayashi A, Horiuchi A, Kikuchi N, Hayashi T, Fuseya C, Suzuki A, Konishi I, Shiozawa T. Type-specific Roles of Histone Deacetylase (HDAC) Overexpression in Ovarian Carcinoma: HDAC1 Enhances Cell Proliferation and HDAC3 Stimulates Cell Migration with Downregulation of E-cadherin. International Journal of Cancer Journal International du Cancer. 2010;127:1332–1346. doi: 10.1002/ijc.25151. [DOI] [PubMed] [Google Scholar]
- 15.Muller BM, Jana L, Kasajima A, Lehmann A, Prinzler J, Budczies J, Winzer KJ, Dietel M, Weichert W, Denkert C. Differential Expression of Histone Deacetylases HDAC1, 2 and 3 in Human Breast Cancer-Overexpression of HDAC2 and HDAC3 is Associated with Clinicopathological Indicators of Disease Progression. BMC Cancer. 2013;13:215. doi: 10.1186/1471-2407-13-215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poyet C, Jentsch B, Hermanns T, Schweckendiek D, Seifert HH, Schmidtpeter M, Sulser T, Moch H, Wild PJ, Kristiansen G. Expression of Histone Deacetylases 1, 2 and 3 in Urothelial Bladder Cancer. BMC Clinical Pathology. 2014;14:10. doi: 10.1186/1472-6890-14-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoshida M, Kijima M, Akita M, Beppu T. Potent and Specific Inhibition of Mammalian Histone Deacetylase Both in Vivo and in Vitro by Trichostatin A. The Journal of Biological Chemistry. 1990;265:17174–17179. [PubMed] [Google Scholar]
- 18.Duvic M, Vu J. Vorinostat: a New Oral Histone Deacetylase Inhibitor Approved for Cutaneous T-cell Lymphoma. Expert Opinion on Investigational Drugs. 2007;16:1111–1120. doi: 10.1517/13543784.16.7.1111. [DOI] [PubMed] [Google Scholar]
- 19.Tan J, Cang S, Ma Y, Petrillo RL, Liu D. Novel Histone Deacetylase Inhibitors in Clinical Trials as Anti-cancer Agents. Journal of Hematology & Oncology. 2010;3:5. doi: 10.1186/1756-8722-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Haggarty SJ, Koeller KM, Wong JC, Grozinger CM, Schreiber SL. Domain-selective Small-molecule Inhibitor of Histone Deacetylase 6 (HDAC6)-mediated Tubulin Deacetylation. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4389–4394. doi: 10.1073/pnas.0430973100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related Inflammation, the Seventh Hallmark of Cancer: Links to Genetic Instability. Carcinogenesis. 2009;30:1073–1081. doi: 10.1093/carcin/bgp127. [DOI] [PubMed] [Google Scholar]
- 22.Grausenburger R, Bilic I, Boucheron N, Zupkovitz G, El-Housseiny L, Tschismarov R, Zhang Y, Rembold M, Gaisberger M, Hartl A, Epstein MM, Matthias P, Seiser C, Ellmeier W. Conditional Deletion of Histone Deacetylase 1 in T Cells Leads to Enhanced Airway Inflammation and Increased Th2 Cytokine Production. Journal of Immunology. 2010;185:3489–3497. doi: 10.4049/jimmunol.0903610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elsharkawy AM, Oakley F, Lin F, Packham G, Mann DA, Mann J. The NF-kappaB p50:p50:HDAC-1 Repressor Complex Orchestrates Transcriptional Inhibition of Multiple Pro-inflammatory Genes. Journal of Hepatology. 2010;53:519–527. doi: 10.1016/j.jhep.2010.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pakala SB, Bui-Nguyen TM, Reddy SD, Li DQ, Peng S, Rayala SK, Behringer RR, Kumar R. Regulation of NF-kappaB Circuitry by a Component of the Nucleosome Remodeling and Deacetylase Complex Controls Inflammatory Response Homeostasis. The Journal of Biological Chemistry. 2010;285:23590–23597. doi: 10.1074/jbc.M110.139469. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 25.Bauer I, Grozio A, Lasiglie D, Basile G, Sturla L, Magnone M, Sociali G, Soncini D, Caffa I, Poggi A, Zoppoli G, Cea M, Feldmann G, Mostoslavsky R, Ballestrero A, Patrone F, Bruzzone S, Nencioni A. The NAD+-dependent Histone Deacetylase SIRT6 Promotes Cytokine Production and Migration in Pancreatic Cancer Cells by Regulating Ca2+ Responses. The Journal of Biological Chemistry. 2012;287:40924–40937. doi: 10.1074/jbc.M112.405837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen X, Barozzi I, Termanini A, Prosperini E, Recchiuti A, Dalli J, Mietton F, Matteoli G, Hiebert S, Natoli G. Requirement for The Histone Deacetylase Hdac3 for the Inflammatory Gene Expression Program in Macrophages. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2865–E2874. doi: 10.1073/pnas.1121131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaliman P, Alvarez-Lopez MJ, Cosin-Tomas M, Rosenkranz MA, Lutz A, Davidson RJ. Rapid Changes in Histone Deacetylases and Inflammatory Gene Expression in Expert Meditators. Psychoneuroendocrinology. 2014;40:96–107. doi: 10.1016/j.psyneuen.2013.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Glauben R, Batra A, Stroh T, Erben U, Fedke I, Lehr HA, Leoni F, Mascagni P, Dinarello CA, Zeitz M, Siegmund B. Histone Deacetylases: Novel Targets for Prevention of Colitis-associated Cancer in Mice. Gut. 2008;57:613–622. doi: 10.1136/gut.2007.134650. [DOI] [PubMed] [Google Scholar]
- 29.Adams H, Fritzsche FR, Dirnhofer S, Kristiansen G, Tzankov A. Class I Histone Deacetylases 1, 2 and 3 Are Highly Expressed in Classical Hodgkin’s Lymphoma. Expert Opinion on Therapeutic Targets. 2010;14:577–584. doi: 10.1517/14728221003796609. [DOI] [PubMed] [Google Scholar]
- 30.Gruhn B, Naumann T, Gruner D, Walther M, Wittig S, Becker S, Beck JF, Sonnemann J. The Expression of Histone Deacetylase 4 is Associated with Prednisone Poor-response in Childhood Acute Lymphoblastic Leukemia. Leukemia Research. 2013;37:1200–1207. doi: 10.1016/j.leukres.2013.07.016. [DOI] [PubMed] [Google Scholar]
- 31.Marquard L, Poulsen CB, Gjerdrum LM, de Nully Brown P, Christensen IJ, Jensen PB, Sehested M, Johansen P, Ralfkiaer E. Histone Deacetylase 1, 2, 6 and Acetylated Histone H4 in B- and T-cell Lymphomas. Histopathology. 2009;54:688–698. doi: 10.1111/j.1365-2559.2009.03290.x. [DOI] [PubMed] [Google Scholar]
- 32.Santoro F, Botrugno OA, Dal Zuffo R, Pallavicini I, Matthews GM, Cluse L, Barozzi I, Senese S, Fornasari L, Moretti S, Altucci L, Pelicci PG, Chiocca S, Johnstone RW, Minucci S. A Dual Role for Hdac1: Oncosuppressor in Tumorigenesis, Oncogene in Tumor Maintenance. Blood. 2013;121:3459–3468. doi: 10.1182/blood-2012-10-461988. [DOI] [PubMed] [Google Scholar]
- 33.Lepore I, Dell’Aversana C, Pilyugin M, Conte M, Nebbioso A, De Bellis F, Tambaro FP, Izzo T, Garcia-Manero G, Ferrara F, Irminger-Finger I, Altucci L. HDAC Inhibitors Repress BARD1 Isoform Expression in Acute myeloid Leukemia Cells via Activation of MiR-19a and/or b. PloS One. 2013;8:e83018. doi: 10.1371/journal.pone.0083018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dong LH, Cheng S, Zheng Z, Wang L, Shen Y, Shen ZX, Chen SJ, Zhao WL. Histone Deacetylase Inhibitor Potentiated the Ability of MTOR Inhibitor to Induce Autophagic Cell Death in Burkitt Leukemia/Lymphoma. Journal of Hematology & Oncology. 2013;6:53. doi: 10.1186/1756-8722-6-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jordaan G, Liao W, Sharma S. E-cadherin Gene Re-expression in Chronic Lymphocytic Leukemia Cells by HDAC Inhibitors. BMC Cancer. 2013;13:88. doi: 10.1186/1471-2407-13-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weichert W, Roske A, Gekeler V, Beckers T, Stephan C, Jung K, Fritzsche FR, Niesporek S, Denkert C, Dietel M, Kristiansen G. Histone Deacetylases 1, 2 and 3 Are Highly Expressed in Prostate Cancer and HDAC2 Expression is Associated with Shorter PSA Relapse Time After Radical Prostatectomy. British Journal of Cancer. 2008;98:604–610. doi: 10.1038/sj.bjc.6604199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang L, Zou X, Berger AD, Twiss C, Peng Y, Li Y, Chiu J, Guo H, Satagopan J, Wilton A, Gerald W, Basch R, Wang Z, Osman I, Lee P. Increased Expression of Histone Deacetylaces (HDACs) and Inhibition of Prostate Cancer Growth and Invasion by HDAC Inhibitor SAHA. American Journal of Translational Research. 2009;1:62–71. [PMC free article] [PubMed] [Google Scholar]
- 38.Noonan EJ, Place RF, Pookot D, Basak S, Whitson JM, Hirata H, Giardina C, Dahiya R. MiR-449a Targets HDAC-1 and Induces Growth Arrest in Prostate Cancer. Oncogene. 2009;28:1714–1724. doi: 10.1038/onc.2009.19. [DOI] [PubMed] [Google Scholar]
- 39.Deubzer HE, Schier MC, Oehme I, Lodrini M, Haendler B, Sommer A, Sommer A, Witt O. HDAC11 Is a Novel Drug Target in Carcinomas. International Journal of Cancer Journal International du Cancer. 2013;132:2200–2208. doi: 10.1002/ijc.27876. [DOI] [PubMed] [Google Scholar]
- 40.Fortson WS, Kayarthodi S, Fujimura Y, Xu H, Matthews R, Grizzle WE, Rao VN, Bhat GK, Reddy ES. Histone Deacetylase Inhibitors, Valproic Acid and Trichostatin-A Induce Apoptosis and Affect Acetylation Status of P53 in ERG-positive Prostate Cancer Cells. International Journal of Oncology. 2011;39:111–1119. doi: 10.3892/ijo.2011.1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patra N, De U, Kim TH, Lee YJ, Ahn MY, Kim ND, Yoon JH, Choi WS, Moon HR, Lee BM, Kim HS. A Novel Histone Deacetylase (HDAC) Inhibitor MHY219 Induces Apoptosis Via Pp-regulation of Androgen Receptor Expression in Human Prostate Cancer Cells. Biomed Pharmacother. 2013;67:407–415. doi: 10.1016/j.biopha.2013.01.006. [DOI] [PubMed] [Google Scholar]
- 42.Choi ES, Han G, Park SK, Lee K, Kim HJ, Cho SD, Kim HM. A248, a Novel Synthetic HDAC Inhibitor, Induces Apoptosis Through the Inhibition of Specificity Protein 1 and Tts dDownstream Proteins in Human Prostate Cancer Cells. Molecular Medicine Reports. 2013;8:195–200. doi: 10.3892/mmr.2013.1481. [DOI] [PubMed] [Google Scholar]
- 43.Khabele D, Son DS, Parl AK, Goldberg GL, Augenlicht LH, Mariadason JM, Rice VM. Drug-induced Inactivation or Gene Silencing of Class I Histone Deacetylases Suppresses Ovarian Cancer Cell Growth: Implications for Therapy. Cancer Biology & Therapy. 2007;6:795–801. doi: 10.4161/cbt.6.5.4007. [DOI] [PubMed] [Google Scholar]
- 44.Jin KL, Pak JH, Park JY, Choi WH, Lee JY, Kim JH, Nam JH. Expression Profile of Histone Deacetylases 1, 2 and 3 in Ovarian Cancer Tissues. Journal of Gynecologic Oncology. 2008;19:185–190. doi: 10.3802/jgo.2008.19.3.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weichert W, Denkert C, Noske A, Darb-Esfahani S, Dietel M, Kalloger SE, Huntsman DG, Kobel M. Expression of Class I Histone Deacetylases Indicates Poor Prognosis in Endometrioid Subtypes of Ovarian and Endometrial Carcinomas. Neoplasia. 2008;10:1021–1027. doi: 10.1593/neo.08474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stronach EA, Alfraidi A, Rama N, Datler C, Studd JB, Agarwal R, Guney TG, Gourley C, Hennessy BT, Mills GB, Mai A, Brown R, Dina R, Gabra H. HDAC4-regulated STAT1 Activation Mediates Platinum Resistance in Ovarian Cancer. Cancer Research. 2011;7:4412–4422. doi: 10.1158/0008-5472.CAN-10-4111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wilson AJ, Cheng YQ, Khabele D. Thailandepsins Are New Small Molecule Class I HDAC Inhibitors With Potent Cytotoxic Activity in Ovarian Cancer Cells: a Preclinical Study of Epigenetic Ovarian Cancer Therapy. Journal of Ovarian Research. 2012;5:12. doi: 10.1186/1757-2215-5-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kwiecinska P, Wrobel A, Tauboll E, Gregoraszczuk EL. Valproic Acid, But Not Levetiracetam, Selectively Decreases HDAC7 and HDAC2 Expression in Human Ovarian Cancer Cells. Toxicology Letters. 2014;224:225–232. doi: 10.1016/j.toxlet.2013.10.035. [DOI] [PubMed] [Google Scholar]
- 49.Xu XS, Wang L, Abrams J, Wang G. Histone Deacetylases (HDACs) in XPC Gene Silencing and Bladder Cancer. Journal of Hematology & Oncology. 2011;4:17. doi: 10.1186/1756-8722-4-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Buckley MT, Yoon J, Yee H, Chiriboga L, Liebes L, Ara G, Qian X, Bajorin DF, Sun TT, Wu XR, Osman I. The Histone Deacetylase Inhibitor Belinostat (PXD101) Suppresses Bladder Cancer Cell Growth in Vitro and In Vivo. Journal of Translational Medicine. 2007;5:49. doi: 10.1186/1479-5876-5-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qu W, Kang YD, Zhou MS, Fu LL, Hua ZH, Wang LM. Experimental Study on Inhibitory Effects of Histone Deacetylase Inhibitor MS-275 and TSA on Bladder Cancer Cells. Urologic Oncology. 2010;28:648–654. doi: 10.1016/j.urolonc.2008.11.018. [DOI] [PubMed] [Google Scholar]
- 52.Juengel E, Meyer dos Santos S, Schneider T, Makarevic J, Hudak L, Bartsch G, Haferkamp A, Wiesner C, Blaheta RA. HDAC Inhibition Suppresses Bladder Cancer Cell Adhesion to Collagen under Flow Conditions. Experimental Biology and Medicine. 2013;238:1297–1304. doi: 10.1177/1535370213498975. [DOI] [PubMed] [Google Scholar]
- 53.Quint K, Agaimy A, Di Fazio P, Montalbano R, Steindorf C, Jung R, Hellerbrand C, Hartmann A, Sitter H, Neureiter D, Ocker M. Clinical Significance of Histone Deacetylases 1, 2, 3, and 7: HDAC2 Is an Independent Predictor of Survival in HCC. Virchows Archiv. 2011;459:129–139. doi: 10.1007/s00428-011-1103-0. [DOI] [PubMed] [Google Scholar]
- 54.Liu C, Liu L, Shan J, Shen J, Xu Y, Zhang Q, Yang Z, Wu L, Xia F, Bie P, Cui Y, Zhang X, Bian X, Qian C. Histone Deacetylase 3 Participates in Self-renewal of Liver Cancer Stem Cells Through Histone Modification. Cancer Letters. 2013;339:60–69. doi: 10.1016/j.canlet.2013.07.022. [DOI] [PubMed] [Google Scholar]
- 55.Buurman R, Gurlevik E, Schaffer V, Eilers M, Sandbothe M, Kreipe H, Wilkens L, Schlegelberger B, Kuhnel F, Skawran B. Histone Deacetylases Activate Hepatocyte Growth Factor Signaling by Repressing MicroRNA-449 in Hepatocellular Carcinoma Cells. Gastroenterology. 2012;143:811–820. doi: 10.1053/j.gastro.2012.05.033. [DOI] [PubMed] [Google Scholar]
- 56.Kim SO, Choi BT, Choi IW, Cheong J, Kim GY, Kwon TK, Kim ND, Choi YH. Anti-invasive Activity of Histone Deacetylase Inhibitors via the Induction of Egr-1 and the Modulation of Tight junction-related Proteins in Human Hepatocarcinoma Cells. BMB Reports. 2009;42:655–660. doi: 10.5483/bmbrep.2009.42.10.655. [DOI] [PubMed] [Google Scholar]
- 57.Wang HG, Huang XD, Shen P, Li LR, Xue HT, Ji GZ. Anticancer Effects of Sodium Butyrate on Hepatocellular Carcinoma Cells in Vitro. International Journal of Molecular Medicine. 2013;31:967–974. doi: 10.3892/ijmm.2013.1285. [DOI] [PubMed] [Google Scholar]
- 58.Liu YL, Yang PM, Shun CT, Wu MS, Weng JR, Chen CC. Autophagy Potentiates the Anti-cancer Effects of the Histone Deacetylase Inhibitors in Hepatocellular Carcinoma. Autophagy. 2010;6:1057–1065. doi: 10.4161/auto.6.8.13365. [DOI] [PubMed] [Google Scholar]
- 59.Wilson AJ, Byun DS, Popova N, Murray LB, L’Italien K, Sowa Y, Arango D, Velcich A, Augenlicht LH, Mariadason JM. Histone Deacetylase 3 (HDAC3) and Other Class I HDACs Regulate Colon Cell Caturation and P21 Expression and Are Deregulated in Human Colon Cancer. The Journal of Biological Chemistry. 2006;281:13548–13558. doi: 10.1074/jbc.M510023200. [DOI] [PubMed] [Google Scholar]
- 60.Spurling CC, Godman CA, Noonan EJ, Rasmussen TP, Rosenberg DW, Giardina C. HDAC3 Overexpression and Colon Cancer Cell Proliferation and Differentiation. Molecular Carcinogenesis. 2008;47:137–147. doi: 10.1002/mc.20373. [DOI] [PubMed] [Google Scholar]
- 61.Chen HP, Denicola M, Qin X, Zhao Y, Zhang L, Long XL, Zhuang S, Liu PY, Zhao TC. HDAC Inhibition Promotes Cardiogenesis and The survival of Embryonic Stem Cells Through Proteasome-dependent Pathway. Journal of Cellular Biochemistry. 2011;112:3246–3255. doi: 10.1002/jcb.23251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wilson AJ, Byun DS, Nasser S, Murray LB, Ayyanar K, Arango D, Figueroa M, Melnick A, Kao GD, Augenlicht LH, Mariadason JM. HDAC4 Promotes Growth of Colon Cancer Cells via Repression of P21. Molecular Biology of the Cell. 2008;19:4062–4075. doi: 10.1091/mbc.E08-02-0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jin JS, Tsao TY, Sun PC, Yu CP, Tzao C. SAHA Inhibits the Growth of Colon Tumors by Decreasing Histone Deacetylase and the Expression of Cyclin D1 and Survivin. Pathology Oncology Research. 2012;18:713–720. doi: 10.1007/s12253-012-9499-7. [DOI] [PubMed] [Google Scholar]
- 64.Ravillah D, Mohammed A, Qian L, Brewer M, Zhang Y, Biddick L, Steele VE, Rao CV. Chemopreventive Effects of an HDAC2-selective Inhibitor on Rat Colon Carcinogenesis and APCmin/+ Mouse Intestinal Tumorigenesis. The Journal of Pharmacology and Experimental Therapeutics. 2014;348:59–68. doi: 10.1124/jpet.113.208645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Barrasa JI, Olmo N, Santiago-Gomez A, Lecona E, Anglard P, Turnay J, Lizarbe MA. Histone Deacetylase Inhibitors Upregulate MMP11 Gene Expression Through Sp1/Smad Complexes in Human Colon Adenocarcinoma Cells. Biochimica et Biophysica Acta. 2012;1823:570–581. doi: 10.1016/j.bbamcr.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 66.Moseley VR, Morris J, Knackstedt RW, Wargovich MJ. Green Tea Polyphenol Epigallocatechin 3-gallate, Contributes to the Degradation of DNMT3A and HDAC3 in HCT 116 Human Colon Cancer Cells. Anticancer Research. 2013;33:5325–5333. [PMC free article] [PubMed] [Google Scholar]
- 67.Berger A, Venturelli S, Kallnischkies M, Bocker A, Busch C, Weiland T, Noor S, Leischner C, Weiss TS, Lauer UM, Bischoff SC, Bitzer M. Kaempferol, a New Nutrition-derived Pan-inhibitor of Human Histone Deacetylases. The Journal of Nutritional Biochemistry. 2013;24:977–985. doi: 10.1016/j.jnutbio.2012.07.001. [DOI] [PubMed] [Google Scholar]
- 68.Marks PA, Breslow R. Dimethyl Sulfoxide to Vorinostat: Development of this Histone Deacetylase Inhibitor as an Anticancer Drug. Nature Biotechnology. 2007;25:84–90. doi: 10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 69.Rasheed WK, Johnstone RW, Prince HM. Histone Deacetylase Inhibitors in Cancer Therapy. Expert Opinion on Investigational Drugs. 2007;16:659–678. doi: 10.1517/13543784.16.5.659. [DOI] [PubMed] [Google Scholar]
- 70.Kelly WK, O’Connor OA, Krug LM, Chiao JH, Heaney M, Curley T, MacGregore-Cortelli B, Tong W, Secrist JP, Schwartz L, Richardson S, Chu E, Olgac S, Marks PA, Scher H, Richon VM. Phase I Study of an Oral Histone Deacetylase Inhibitor, Suberoylanilide Hydroxamic Acid, in Patients with Advanced Cancer. Journal of Clinical Oncology. 2005;23:3923–3931. doi: 10.1200/JCO.2005.14.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ryan QC, Headlee D, Acharya M, Sparreboom A, Trepel JB, Ye J, Figg WD, Hwang K, Chung EJ, Murgo A, Melillo G, Elsayed Y, Monga M, Kalnitskiy M, Zwiebel J, Sausville EA. Phase I and Pharmacokinetic Study of MS-275, a Histone Deacetylase Inhibitor, in Patients with Advanced and Refractory Solid Tumors or Lymphoma. Journal of Clinical Oncology. 2005;23:3912–3922. doi: 10.1200/JCO.2005.02.188. [DOI] [PubMed] [Google Scholar]
- 72.Montgomery RL, Hsieh J, Barbosa AC, Richardson JA, Olson EN. Histone Deacetylases 1 and 2 Control the Progression of Neural Precursors to Neurons During Brain Development. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:7876–7881. doi: 10.1073/pnas.0902750106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chen Y, Wang H, Yoon SO, Xu X, Hottiger MO, Svaren J, Nave KA, Kim HA, Olson EN, Lu QR. HDAC-mediated Deacetylation of NF-kappaB is Critical for Schwann Cell Myelination. Nature Neuroscience. 2011;14:437–441. doi: 10.1038/nn.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Montgomery RL, Potthoff MJ, Haberland M, Qi X, Matsuzaki S, Humphries KM, Richardson JA, Bassel-Duby R, Olson EN. Maintenance of Cardiac Energy Metabolism by Histone Deacetylase 3 in Mice. The Journal of Clinical Investigation. 2008;118:3588–3597. doi: 10.1172/JCI35847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Knutson SK, Chyla BJ, Amann JM, Bhaskara S, Huppert SS, Hiebert SW. Liver-specific Deletion of Histone Deacetylase 3 Disrupts Metabolic Transcriptional Networks. The EMBO Journal. 2008;27:1017–1028. doi: 10.1038/emboj.2008.51. [DOI] [PMC free article] [PubMed] [Google Scholar]