Abstract
We adapted a mathematical model of post-hepatectomy liver regeneration using data from a subset of patients in the Adult-to-Adult Living Donor Liver Transplantation (A2ALL) cohort. The original model addressed changes in the number of quiescent, primed, and proliferating cells. Our adapted model takes into account hypertrophy of primed and replicating cells, and is better able to predict liver volume. In addition, by building off the hypothesis that cell cycle parameters are approximately the same across all mammals, we found that changing only a single parameter characterizing metabolic load could model liver regeneration in five species of mammals. In conclusion, we improved a mathematical model of liver regeneration, predicted mammalian liver regeneration based on metabolism, and found correlations between model parameters and physiological measurements from liver donors.
Keywords: Live donor liver transplants (LDLT), transplant, donor, theory, model
INTRODUCTION
Live donor liver transplants (LDLT) are increasingly used to treat end-stage liver diseases such as hepatocellular carcinomas, non-alcoholic fatty liver disease, primary sclerosing cholangitis, and others, due to shortages of cadaveric organs. LDLT have the advantage of proactive treatment before the recipient’s condition deteriorates, but rare complications have occurred in donors.
The Adult-to-Adult Living Donor Liver Transplantation Cohort Study (A2ALL) was undertaken to investigate the risks and benefits to LDLT donors and recipients. A subset of donors in the A2ALL study was recruited for a detailed 6-month study of hepatic function and regeneration known as the DQLFT (Donor Quantitative Liver Function Tests) study (1). Liver volume measurements and blood measurements were taken at 4 time points (0 days, 4 days, 3 months, 6 months post-surgery) from these donors.
The DQLFT study was distinct in that it quantified liver regeneration during the initial 2-week period when human livers regenerate most quickly (2–6). This detailed data led us to make improvements in a mathematical model of rat liver regeneration developed by Furchtgott et al. (7) and adapted with limited data for human liver regeneration by Periwal et al. (8)
Finally, liver regeneration has been studied in several other mammals besides humans and rats, such as mice, pigs, dogs, and rabbits. In this study, we tested the hypothesis that only varying the metabolic load parameter between species while keeping all other cell cycle and biochemical parameters constant would allow us to predict liver regeneration in five mammalian species: mouse, rat, dog, rabbit and human.
METHODS
The mathematical model
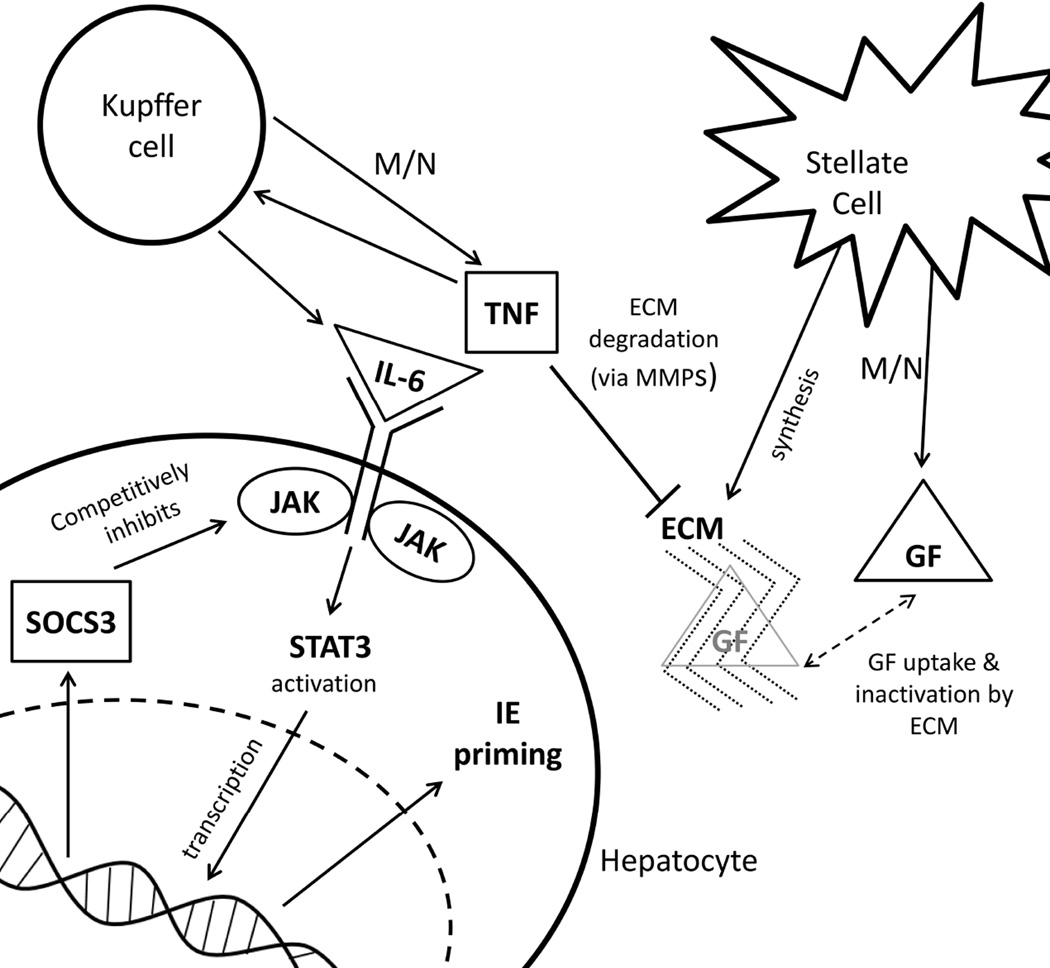
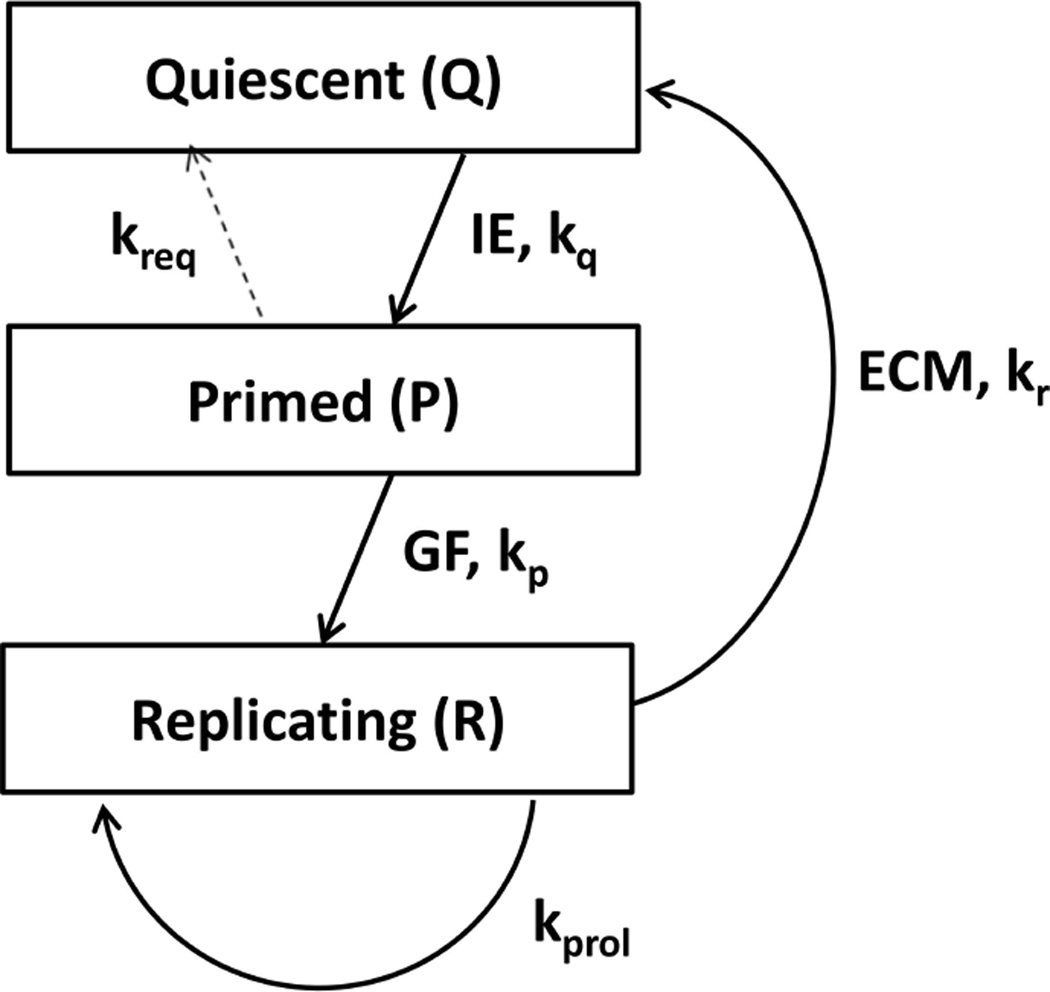
The model of liver regeneration is based on the work of Fausto and Riehle (9) and tracks three populations of cells – quiescent, primed, and replicating. Hepatocytes transition from a quiescent to a primed state shortly after liver resection, and then to a replicating state. After sufficient liver volume is achieved, the cells return to quiescence. The model replicates the biochemical changes that occur in liver regeneration (Figure 1). Hepatocytes shift from quiescent to primed due to changes in intermediate-early gene expression. Growth factors shift the cells from primed to replicating while extracellular matrix accumulation and decreased cytokine and growth factor secretion returns replicating cells to quiescence (10) (Figure 2).
Figure 1. Biochemistry of liver regeneration for the model.
After partial hepatectomy, hepatocyte number is reduced which increases metabolic load (M/N). Increased metabolic load drives hepatocyte growth factor (GF) and tumor necrosis factor (TNF) expression. TNF degrades the ECM through matrix metalloproteases (MMPs). TNF also leads to the expression of IL-6, which initiates Janus kinase (JAK) signaling by activating STAT3. STAT3 leads to transcription of suppressor of cytokine signaling 3 (SOCS3), and intermediate-early genes (IE) that cause the hepatocyte to shift into a primed state. As regeneration progresses and hepatocyte numbers increase, TNF levels decrease and the ECM reforms and takes up GF. This stops the primed to replicating cell transition, and drives the replicating to quiescence cell transition.
Figure 2. Cell cycle of liver regeneration.
The total number of cells is Q + P + R, but the total volume of the liver is larger due to hypertrophy of primed and replicating cells. We estimated total volume to be Q + 2P + 1.5 R.
Detailed explanations of the differential equations and parameters can be found in Furchtgott et al. (7). We kept virtually all cell cycle and biochemical parameters the same as in this reference. The only parameter we allowed to vary is the metabolic load per hepatocyte, M. M is a theoretical measure of metabolic demand on the liver, normalized to the original mass of the donor liver. Thus, when a fraction f of the liver remains after surgery, the regeneration process is driven by M/f. This leads to a one-parameter family of models, with cell cycle duration and transitions that vary when M is changed, and stronger regenerative impulse for smaller remnant fractions, as demonstrated extensively in Furchtgott et al. (7). Given the sparse sampling of time points available for most species, this avoids over-fitting the data. Because M is species-dependent, the model shows differences in cell cycle duration between species. We used the adjusted value for kapop (the apoptosis rate parameter) used in Periwal et al. (8), which was 1/24th the rat kapop parameter.
Volume multipliers
The existing model describes cell number changes, but CT scans (or other imaging techniques) provide measurements of liver volume. Hypertrophy has also been observed in replicating hepatocytes (11). While this hypertrophy is not a concern if the data only covers the beginning and close to the end of the liver regeneration process, the availability of the DQLFT data implies the model must take hypertrophy of cells into account in order to accurately compare cell number predictions with volume measurements. We adapted the model predictions to DQLFT data by including volume multipliers for primed and replicating cells. The model still describes the number of cells in the quiescent (Q), primed (P) or replicating (R) states, but the volume multipliers allow the model to account for hypertrophy approximately.
The total number of hepatocytes is Q+P+R. To translate cell number to liver volume measurements, we included two volume multipliers for primed and replicating cells (2.0 and 1.5, respectively). The total volume of liver is therefore modeled to be Q + 2.0*P + 1.5*R.
We chose the value of 2.0 for the primed cell volume multiplier, because of the assumption that a primed hepatocyte about to undergo cell division would be twice as large as a quiescent cell. For replicating cells, we assumed half the cells were about to replicate and therefore twice the size of a quiescent cell, and that half the cells had finished replication and were the size of a quiescent cell. We averaged the two values together to get 1.5 for the value of the replicating cell volume multiplier. We note that this is a crude assumption based on the two extremes: a cell about to replicate, and a daughter cell that has just been formed. In reality, cell sizes in any state should be modeled with a cell-size distribution, with probabilities for replication dependent on the cell size. Furthermore, the assumption that these volume multipliers are the same between species is a simplifying assumption due to the sparse time sampling of data available, and is independent of the changes in cell cycle phases and duration that result from each species having a specific metabolic load parameter.
Mammalian data sets for model fitting
Human
We used data from the 12 donors in the DQLFT study (1). Two patients were excluded because they were missing all but the initial measurement due to post-operative complications. Two of the 10 patients were missing day 7 measurements, but were included. The 8 remaining patients had complete liver fraction measurements taken at 0 days, 7 days, 3 months, and 6 months post-surgery. Resections ranged from 62.61% to 72.78%, with a mean of 68.77% and standard deviation of 3.07%.
Rat
Data points were extrapolated from studies by Tanoue et al. (12) 7-week old male Sprague-Dawley rats underwent a 70% hepatectomy, and regeneration was measured in 4–6 rats at each time point 0, 1, 3, and 7 days post-surgery.
Mouse
Data points were extrapolated from studies by Shu et al. (13) 8–12 week old mice underwent a 68% hepatectomy, and regeneration was measured in at least 4 mice at each time point 0, 12, 24, 36, 48, 60, 72, and 96 hours post-surgery.
Dog
Data points were obtained from studies by Child et al. (14) Six mongrel dogs underwent a 70% hepatectomy, and regeneration was measured 35–57 days post-surgery. Due to small sample sizes, liver regeneration was only measured at one time point.
Rabbit
Data points were obtained from studies by Fleig et al. (15) Rabbits underwent a 60% hepatectomy. Regeneration was measured in 3–6 rabbits at each time point 0, 12, 15, 18, 21, 24, 30, 36, 40, 48, 72, 96, and 168 hours post-surgery.
Mathematical and statistical analysis was performed in Matlab version 7.12.0 (Mathworks, Natick, MA).
Data points from published figures were obtained using the open source DigitizeIt software.
RESULTS
Improved fit to DQLFT data
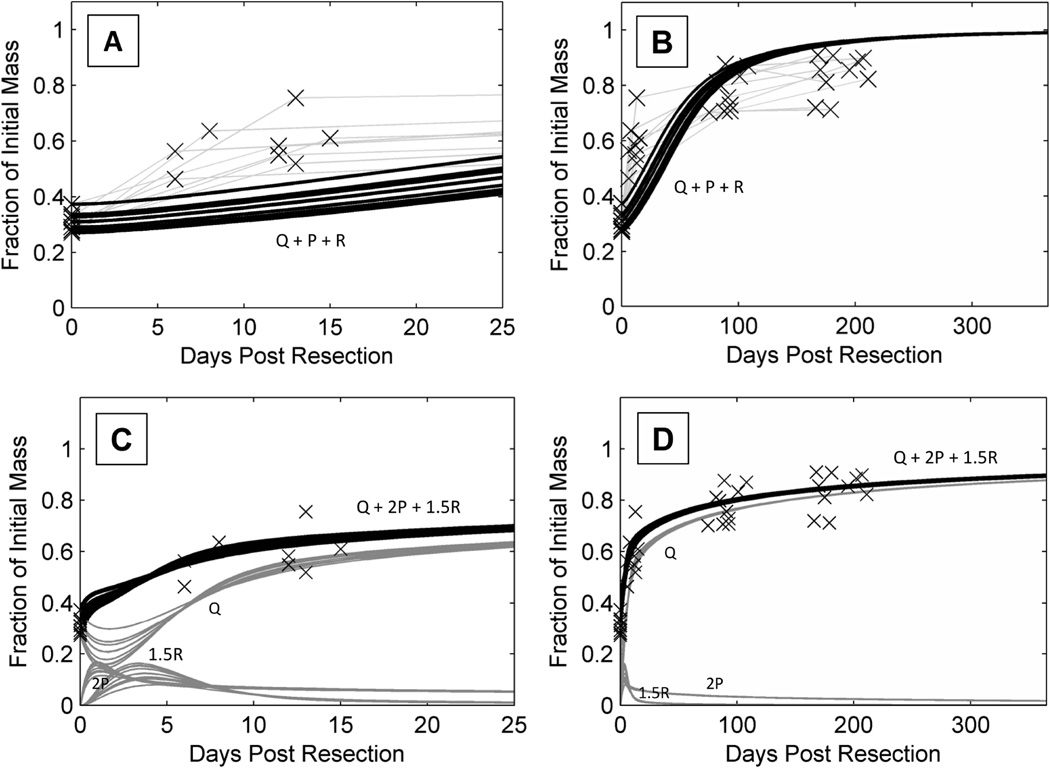
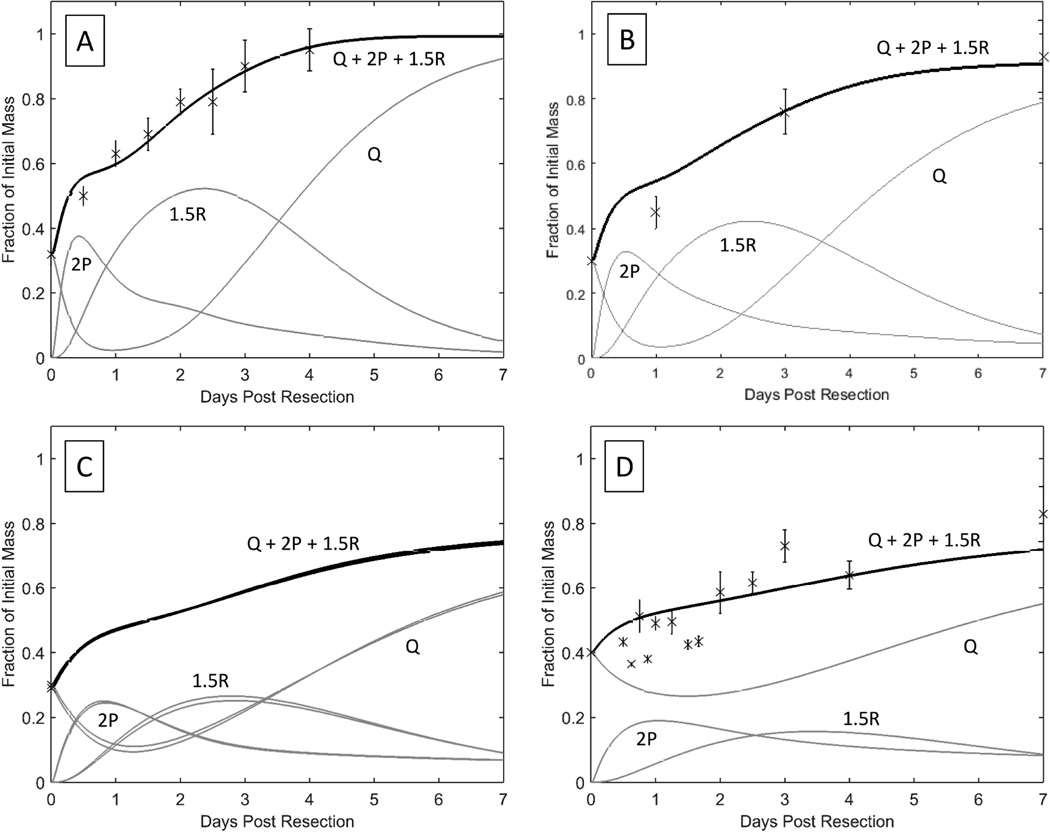
Reverting to using rat cell cycle parameters from Furchgott et al. (7), and optimizing the value of the metabolic load parameter, M, led to a remarkably improved fit (Figure 3A-D).
Figure 3. Improved mathematical model of human liver regeneration.
A and B show the original model applied to DQLFT data with the same cell cycle parameters used in Periwal et al (2014). C and D show the updated model with the original rat cell cycle parameters and optimized metabolic load parameter, including individual populations of Q, P, R cells.
A and C show liver regeneration over the span of 25 days (n = 8), B and D show liver regeneration over the span of 1 year (n = 10).
Multi-species model of mammalian liver regeneration
We hypothesized that because cell cycles are virtually the same among all mammals (16), rat, mouse, dog, rabbit, and human liver regeneration should have the same cell cycle and biochemical parameters. This is in contrast to the naïve scaling employed by Periwal et al. (8) Thus, the only thing that needs to be changed in a multi-species model of liver regeneration is the metabolic load parameter, M. It should be noted that this parameter is fixed for each species independently and does not depend on an individual subject or animal.
We applied the model to four non-human species, and used an optimized M value for each. Results are shown in Figure 4 and 5, and the optimized M values are shown in Table 1.
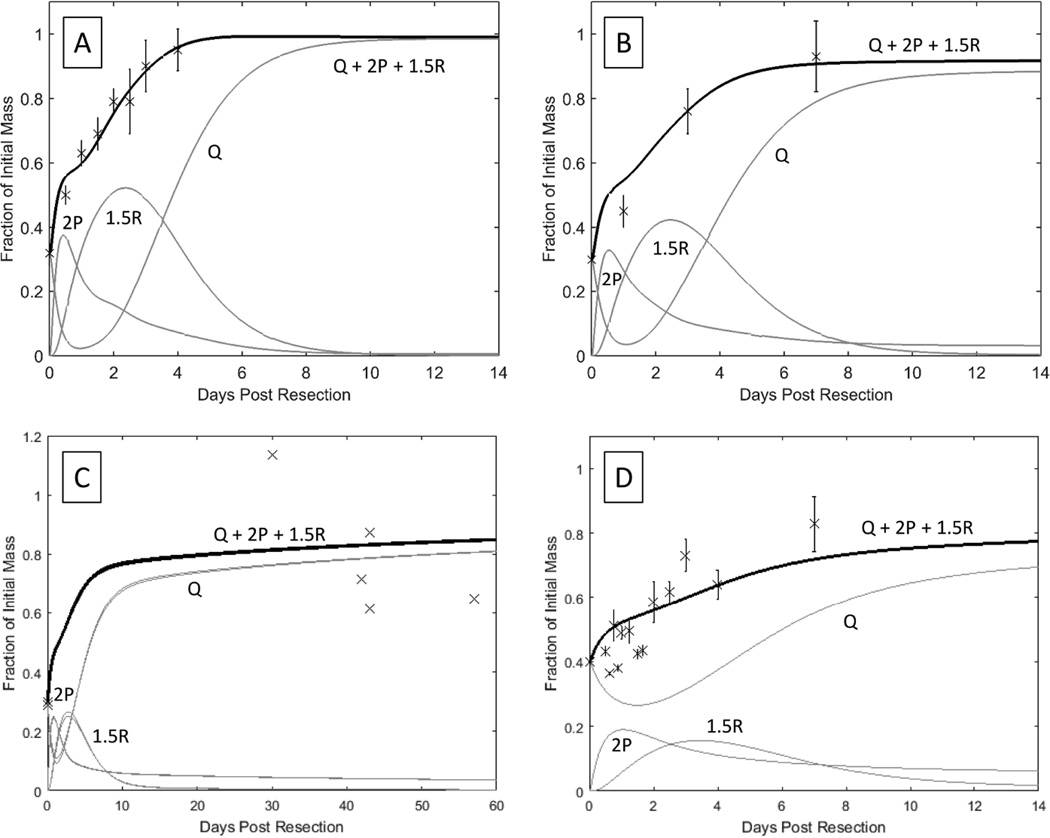
Figure 4. Models of liver regeneration in four mammalian species until complete liver regeneration.
A, B, C, D models liver regeneration in mice (n ≥ 4 at each time point), rats (n = 4–6 at each time point), dogs (n = 6), and rabbits (n = 3–6 at each time point) respectively. The limitations of the dog and rabbit data resulted in a less good fit with our model.
Figure 5. Models of liver regeneration in four mammalian species over 7 days.
A, B, C, D model liver regeneration in mice (n ≥ 4 at each time point), rats (n = 4–6 at each time point), dogs (n = 6), and rabbits (n = 3–6 at each time point) respectively. The figure highlights the differences in rates of liver regeneration due to species-dependent factors such as metabolic load.
Table 1.
Optimized M values for 5 mammalian species
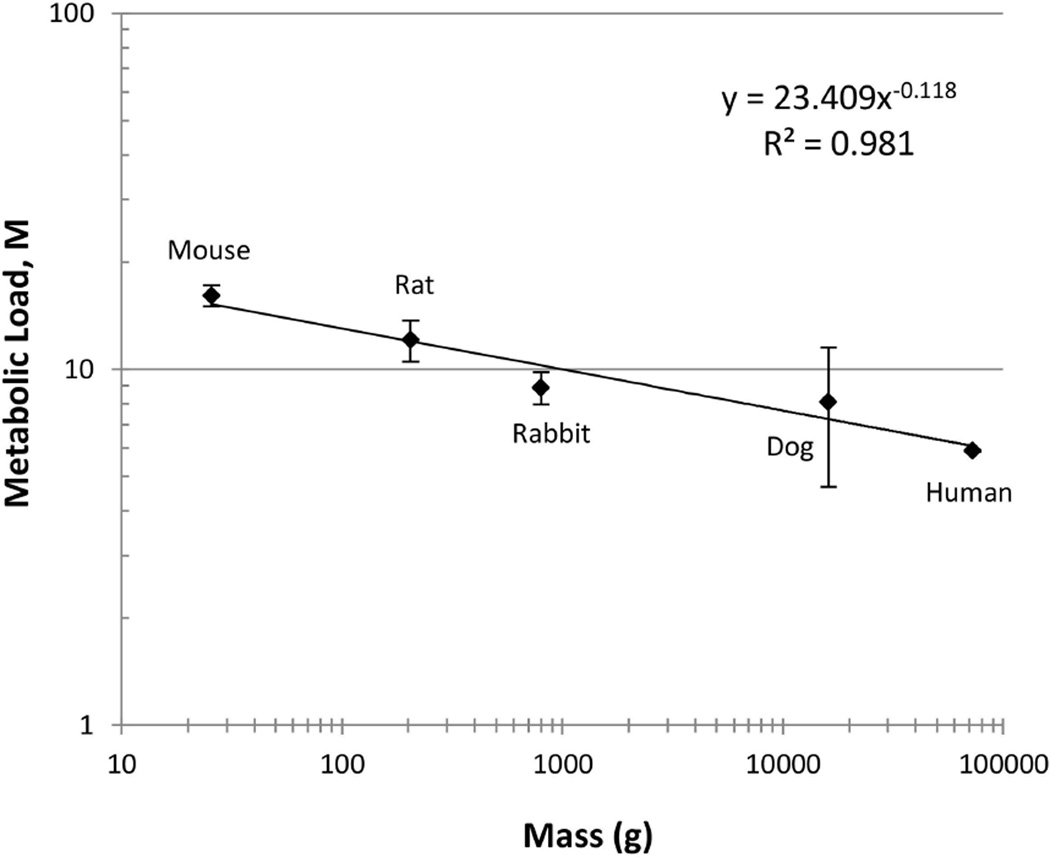
Because metabolic scaling is of great interest, we plotted the metabolic load as a function of the organism’s mass (Figure 6). The plot indicates a simple power law relationship between the metabolic load and mass (n = 5, p < 0.002).
Figure 6. Metabolic scaling for a model parameter of liver regeneration.
A log-log graph of the metabolic load parameter, M, versus the organism’s mass suggests a simple power law relationship between metabolism and mass.
To quantify the improved fit from adding the volume multipliers to the model, we calculated the Bayesian Information Criterion (BIC) for several species (Table 2).
Table 2.
Comparison of models with and without volume multipliers
| BIC | With volume multipliers | Without volume multipliers |
|---|---|---|
| Human | 135.93 | 135.05 |
| Mouse | 325.11 | 447.55 |
| Rat | 73.23 | 97.87 |
| Total | 534.27 | 680.48 |
Lower BIC denotes a better fit. We were unable to calculate BIC for dog and rabbit due to limitations in data.
DISCUSSION
This study adapted a mathematical model of liver regeneration to translate hepatocyte number to liver mass, and found a simple power law scaling in a metabolic load parameter of liver regeneration among mammalian species. This power law scaling in the metabolic load parameter is cell number independent as it is defined as a metabolic load per hepatocyte, and differences in this parameter’s values in distinct species lead to cell cycle differences independent of the fact that the lobes resected, and hence the remnant volume, differs from species to species. Thus the metabolic scaling we find is independent of the morphology of liver in distinct species, which dictates achievable remnant fractions.
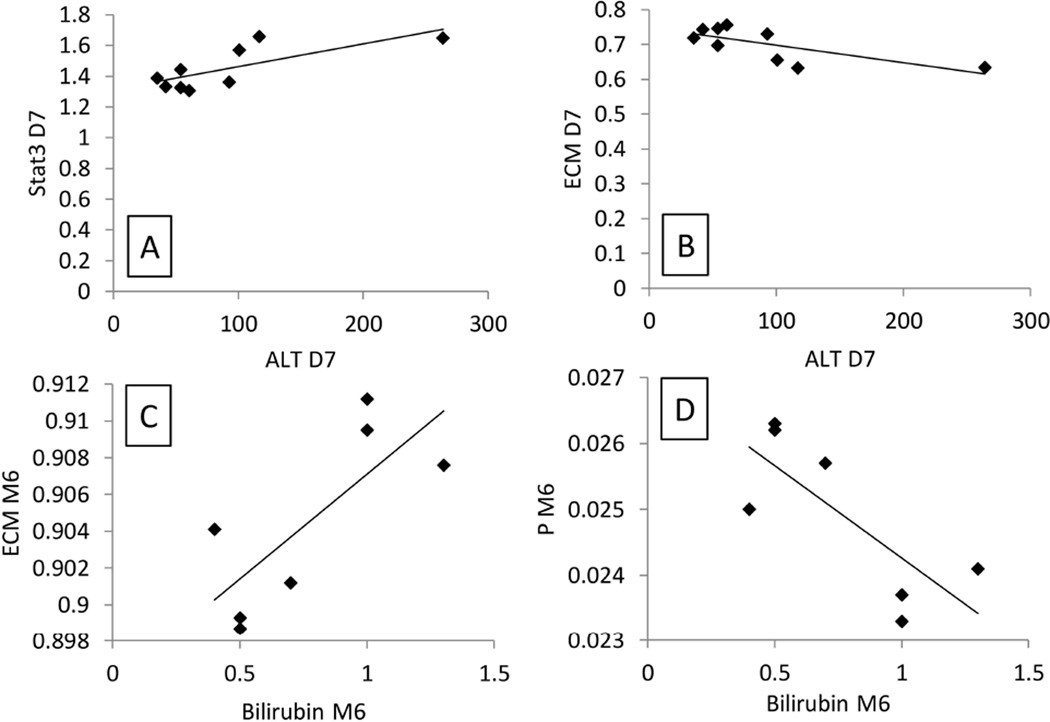
We calculated the correlation between the patient blood measurements from the DQLFT study with our model by using a correlation matrix (Table 3, Table 4, and Figure 7). Only alanine transaminase (ALT) at day 7 and bilirubin at month 6 had correlations with model molecular parameters. As a point of interest, a study of rhesus macaque liver regeneration by Gaglio et al. (17) found serum ALT was highest 7 days post-surgery.
Table 3.
(p < 0.05) correlations between model parameters and DQLFT patient blood measurements.
| Model parameters |
Day 7 ALT Correlation Coefficient n = 9 |
P | Month 6 Bilirubin Correlation Coefficient n = 7 |
p |
|---|---|---|---|---|
| TNF | 0.7326 | 0.02 | −0.7598 | 0.048 |
| Jak | 0.7339 | 0.02 | −0.7601 | 0.047 |
| Stat3 | 0.7439 | 0.02 | −0.7615 | 0.047 |
| SOC | 0.7288 | 0.03 | −0.7601 | 0.047 |
| ECM | −0.7195 | 0.03 | 0.7608 | 0.047 |
| IE | 0.7430 | 0.02 | −0.7592 | 0.048 |
| GF | 0.7356 | 0.02 | −0.7604 | 0.047 |
| Q | −0.7200 | 0.03 | 0.7594 | 0.048 |
| P | 0.6903 | 0.04 | −0.7698 | 0.043 |
| R | 0.7324 | 0.02 | ||
| Q+P+R | −0.7040 | 0.03 | 0.7612 | 0.047 |
TNF, tumor necrosis factor; JAK, Janus kinase; STAT3, signal transducer and activator of transcription 3; SOC, suppressor of cytokine signaling; ECM, extracellular matrix; IE, intermediate-early genes; GF, growth factor; Q, quiescent hepatocytes; P, primed hepatocytes; R, replicating hepatocytes.
Table 4.
Mean DQLFT patient blood measurements for model correlations
| Mean Day 7 ALT (units/L) n = 9 |
σ | Mean month 6 Bilirubin (mg/dL) n = 7 |
σ |
|---|---|---|---|
| 91 | 67 | 0.7 | 0.3 |
Figure 7. Correlations between model parameters and physiological measurements in human liver regeneration.
A and B depict positive and negative correlations for model parameters and blood measurements at 7 days post-surgery. C and D depict positive and negative correlations for model parameters and blood measurements at 6 months. The pairings with the largest correlation and lowest p values were chosen for graphical depiction. ECM, extracellular matrix parameter. P, primed hepatocytes. Stat3, Signal transducer and activator of transcription 3.
Though our model reasonably predicted liver regeneration in rats and mice, there was a discrepancy between our predictions and experimentally determined timing of DNA synthesis. Weglarz et al. (18) examined BrdU-labeled rat and mouse livers after two-thirds partial hepatectomy, and found that rat hepatocytes begin DNA synthesis 20 hours post-surgery, while mouse hepatocytes lag behind, beginning DNA synthesis 32 to 46 hours after surgery.
While our model did not directly predict initiation of DNA synthesis, it did model primed cells. Our model predicted that mouse and rat hepatocytes shift into the primed state at about the same time and mouse primed hepatocytes shift to the replicating state earlier than the rat’s. This discrepancy could be due to slight differences in cell cycle kinetics between species. Moreover, we had no data on the ploidy distribution of cells in any of the three states in our model. Given such data, it would be possible to include DNA synthesis explicitly in the model dynamics. With more data, our assumption that biochemical and cell cycle parameter differences between species depend only on the metabolic load parameter, while providing a reasonably good fit to the available data, could be improved upon as well.
We are aware that the accuracy of the metabolic load parameter we generated for each species was limited by the data sets available. We recognize there were only 5 species used, the data on mouse and rat liver regeneration was averaged, and there were only six dogs and one time point in the study by Child et al. (14). Nevertheless, the power-law scaling of body mass to the liver metabolic load parameter is statistically significant, and is in contrast to the curvature reported by Kolokotrones et al. (19) for metabolism as a whole.
Another limitation of our model is that we posited rough estimates of volume multipliers based on Miyaoka et al. (11). A model with variable volume multipliers could not be optimized as available data did not constrain the parameters adequately due to the sparsity of time points (data not shown). With data from more frequent time sampling, the model could be improved to determine these volume multipliers precisely. There may also be species dependence in the volume multipliers that we obviously cannot rule out. As noted, our model takes cell size in the proliferative stage to be a binary variable, with cells either about to replicate or daughter cells that have just been formed. Data on hepatocyte cell size distributions at different time points in the liver regeneration process does not appear to exist. Such data would be required to constrain a model taking continuous cell size distributions into account, and would be an experimental tour-de-force involving a large number of animals.
The BIC values in Table 2 show that the model with volume multipliers is very strongly preferred for mouse and rat, but not as strongly for human. This may be because we used the 22% difference between CT/MRI vs. SPECT estimates of liver volume found by Everson et al. (1) as our estimate of the variance in computing the BIC for human, while the uncertainty in the animal data is considerably less.
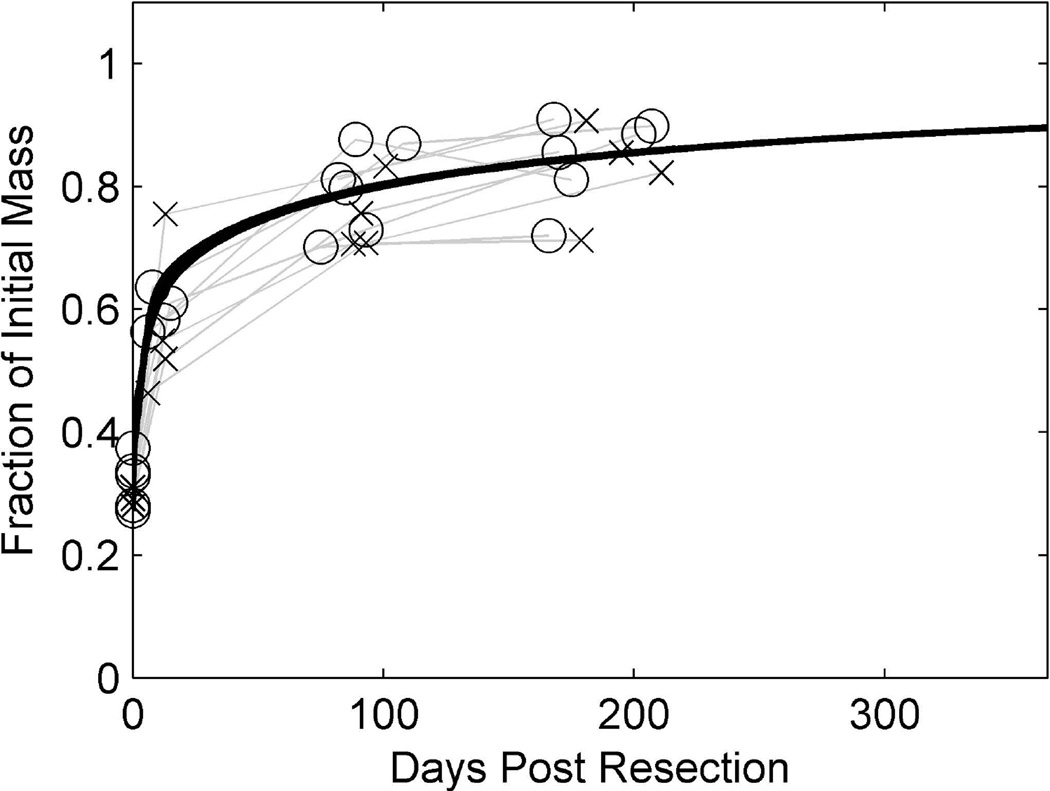
Finally, we found that in contrast to results reported by Pomfret et al. (2), liver regeneration does not differ significantly between males and females. Pomfret’s study of 27 male and 16 female live liver donors found that males had a statistically significant, larger percentage of regeneration. However, the DQLFT data and our model showed no difference between liver regeneration among the 4 men and 8 women in the DQLFT study (Figure 8). As well, the data shown in Marcos et al. (4) does not exhibit a separation between subjects based on gender. It could be that the remnant livers for males were smaller than for females in Pomfret et al.(2), leading to a more rapid regeneration driven by the larger resection rather than sex.
Figure 8. Liver regeneration among human male and female DQLFT donors.
Crosses denote male patients (n = 4), circles denote female patients (n = 6). There seems to be no difference in the rate of liver regeneration among male and female donors in the DQLFT study.
Future studies could focus on modeling liver regeneration in recipients, sorted by end-stage liver diseases leading the recipient to require a transplant. Recipient liver regeneration occurs at a noticeably faster rate than that of donor liver regeneration (20, 21) and genes for cellular proliferation are up-regulated (22). This could be due to recipients receiving certain immunosuppressive treatments (25, 26).
In conclusion, we were able to significantly improve an existing mathematical model of human liver regeneration with a detailed data set (1), and simplify the application of this model to different mammalian species.
Acknowledgments
Grants and financial support
This study was supported by the Intramural Research Program of the National Institutes of Health, NIDDK, project number Z01-DK075059-04.
Data from the A2ALL study was the basis of the analysis in this paper.
Abbreviations
- A2ALL
Adult-to-Adult Living Donor Liver Transplantation
- BIC
Bayesian Information Criterion
- ECM
extracellular matrix
- DQLFT
Donor Quantitative Liver Function Tests study
- GF
growth factor
- IE
intermediate-early genes
- JAK
Janus kinase
- LDLT
Live donor liver transplants
- M
Metabolic load parameter
- P
primed hepatocytes
- Q
quiescent hepatocytes
- R
replicating hepatocytes
- SOCS3
suppressor of cytokine signaling
- STAT3
signal transducer and activator of transcription 3
Footnotes
Conflicts of Interest
There are no conflicts of interest to report.
REFERENCES
- 1.Everson GT, Hoefs JC, Niemann CU, Olthoff KM, Dupuis R, Lauriski S, et al. Functional elements associated with hepatic regeneration in living donors after right hepatic lobectomy. Liver Transplantation. 2013;19:292–304. doi: 10.1002/lt.23592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pomfret EA, Pomposelli JJ, Gordon FD, Erbay N, Price LL, Lewis WD, et al. Liver regeneration and surgical outcome in donors of right-lobe liver grafts. Transplantation. 2003;76(1):5–10. doi: 10.1097/01.TP.0000079064.08263.8E. [DOI] [PubMed] [Google Scholar]
- 3.Kawasaki S, Makuuchi M, Matsunami H, Hashikura Y, Ikegami T, Nakazawa Y, et al. Living related liver transplantation in adults. Annals of Surgery. 1998;227(2):269–274. doi: 10.1097/00000658-199802000-00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marcos A, Fisher R, Ham J, Shiffman ML, Sanyal AJ, Luketic VA, et al. Liver regeneration and function in donor and recipient after right lobe adult to adult living donor liver transplantation. Transplantation. 2000;69(7):1375–1379. doi: 10.1097/00007890-200004150-00028. [DOI] [PubMed] [Google Scholar]
- 5.Nakagami M, Morimoto T, Itoh K, Arima Y, Yamamoto Y, Ikai I, et al. Patterns of restoration of remnant liver volume after graft harvesting in donors for living related liver transplantation. Transplantation Proceedings. 1998;30(1):195–199. doi: 10.1016/s0041-1345(97)01229-3. [DOI] [PubMed] [Google Scholar]
- 6.Yamanaka N, Okamoto E, Kawamura E, Kato T, Oriyama T, Fujimoto J, et al. Dynamics of normal and injured human liver regeneration after hepatectomy as assessed on the basis of computed tomography and liver function. Hepatology. 1993;18(1):79–85. [PubMed] [Google Scholar]
- 7.Furchtgott LA, Chow CC, Periwal V. A Model of Liver Regeneration. Biophysical Journal. 2009;96:3926–3935. doi: 10.1016/j.bpj.2009.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Periwal V, Gaillard JR, Needleman L, Doria C. Mathematical model of liver regeneration in human live donors. Journal of Cellular Physiology. 2014;229:599–606. doi: 10.1002/jcp.24482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fausto N, Riehle KJ. Mechanisms of liver regeneration and their clinical implications. Journal of Hepato-Biliary-Pancreatic Surgery. 2005;12(3):181–189. doi: 10.1007/s00534-005-0979-y. [DOI] [PubMed] [Google Scholar]
- 10.Taub R. Liver regeneration: from myth to mechanism. Nature Reviews Molecular Cell Biology. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 11.Miyaoka Y, Ebato K, Kato H, Arakawa S, Shimizu S, Miyajima A. Hypertrophy and unconventional cell division of hepatocytes underlie liver regeneration. Current Biology. 2012;22(13):1166–1175. doi: 10.1016/j.cub.2012.05.016. [DOI] [PubMed] [Google Scholar]
- 12.Tanoue S, Uto H, Kumamoto R, Arima S, Hashimoto S, Nasu Y, et al. Liver regeneration after partial hepatectomy in rat is more impaired in a steatotic liver induced by dietary fructose compared to dietary fat. Biochemical and Biophysical Research Communications. 2011;407:163–168. doi: 10.1016/j.bbrc.2011.02.131. [DOI] [PubMed] [Google Scholar]
- 13.Shu R-Z, Zhang F, Wang F, Feng D-C, Li X-H, Ren W-H, et al. Adiponectin deficiency impairs liver regeneration through attenuating STAT3 phosphorylation in mice. Laboratory Investigation. 2009;89:1043–1052. doi: 10.1038/labinvest.2009.63. [DOI] [PubMed] [Google Scholar]
- 14.Child CG, III, Barr D, Holswade GR, Harrison CS. Liver regeneration following portacaval transposition in dogs. Annals of Surgery. 1953;138(4):600–608. doi: 10.1097/00000658-195310000-00013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fleig WE, Lehmann H, Wagner H, Hoss G, Ditschuneit H. Hepatic regenerative stimulator substance in the rabbit: relation to liver regeneration after partial hepatectomy. Journal of Hepatology. 1986;3:19–26. doi: 10.1016/s0168-8278(86)80141-6. [DOI] [PubMed] [Google Scholar]
- 16.Nurse P. Universal control mechanism regulating onset of M-phase. Nature. 1990;344:503–508. doi: 10.1038/344503a0. [DOI] [PubMed] [Google Scholar]
- 17.Gaglio PJ, Baskin G, Bohm RJ, Blanchard J, Cheng S, Dunne B, et al. Partial hepatectomy and laparoscopic-guided liver biopsy in rhesus macaques (macaca mulatta): novel approach for study of liver regeneration. Comparative Medicine. 2000;50(4):363–368. [PubMed] [Google Scholar]
- 18.Weglarz TC, Sandgren EP. Timing of hepatocyte entry into DNA synthesis after partial hepatectomy is cell autonomous. Proceedings of the National Academy of Sciences in the United States of America. 2000;97(23):12595–12600. doi: 10.1073/pnas.220430497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolokotrones T, Savage V, Deeds EJ, Fontana W. Curvature in metabolic scaling. Nature. 2010;464(7289):753–756. doi: 10.1038/nature08920. [DOI] [PubMed] [Google Scholar]
- 20.Kamel IR, Erbay N, Warmbrand G, Kruskal JB, Pomfret EA, Raptopoulos V. Liver regeneration after living adult right lobe transplantation. Abdominal Imaging. 2003;28:53–57. doi: 10.1007/s00261-001-0192-7. [DOI] [PubMed] [Google Scholar]
- 21.Olthoff KM, Emond JC, Shearon TH, Everson G, Baker TB, Fisher RA, et al. Liver regeneration after living donor transplantation: adult-to-adult living donor liver transplantation cohort study. Liver Transplantation. 2015;21(1):79–88. doi: 10.1002/lt.23966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Jonge J, Kurian S, Shaked A, Reddy KR, Hancock W, Salomon DR, et al. Unique early gene expression patterns in human adult to adult living donor liver grafts compared to deceased donor grafts. American Journal of Transplantation. 2009;9(4):758–772. doi: 10.1111/j.1600-6143.2009.02557.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charles River. [Accessed July 2015];Sprague Dawley Rat. http://www.criver.com/products-services/basic-research/find-a-model/sprague-dawley-rat.
- 24.The Jackson Laboratory. [Accessed July 2015];Body weight information. http://jaxmice.jax.org/support/weight/000664.html.
- 25.Starzl TE, Porter KA, Mazzaferro V, Todo S, Fung J, Francavilla A. Hepatotrophic effects of FK506 in dogs. Transplantation. 1991;51:67–69. doi: 10.1097/00007890-199101000-00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mazzaferro V, Porter KA, Scotti-Foglieni CL, Venkataramanan R, Makowka L, Rossaro L, Francavilla A, Todo S, Van Thiel DH, Starzl TE. The hepatotropic influence of cyclosporine. Surgery. 1990;107:533–539. [PMC free article] [PubMed] [Google Scholar]