Abstract
Real-time probing of structural transitions of a photoactive protein is challenging owing to the lack of a universal time-resolved technique that can probe the changes in both global conformation and light-absorbing chromophore of the protein. In this work, we combine time-resolved X-ray solution scattering (TRXSS) and transient absorption (TA) spectroscopy to investigate how the global conformational changes involved in the photoinduced signal transduction of photoactive yellow protein is temporally and spatially related with the local structural change around the light-absorbing chromophore. In particular, we examine the role of internal proton transfer in developing a signaling state of photoactive yellow protein by employing its E46Q mutant, where the internal proton transfer is inhibited by the replacement of a proton donor. The comparison of TRXSS and TA spectroscopy data directly reveals that the global conformational change of the protein, which is probed by TRXSS, is temporally delayed by tens of microseconds from the local structural change of the chromophore, which is probed by TA spectroscopy. The molecular shape of the signaling state reconstructed from the TRXSS curves directly visualizes the three-dimensional conformations of protein intermeidates and reveals that the smaller structural change in E46Q-PYP than in wt-PYP suggested by previous studies is manifested in terms of much smaller protrusion, confirming that the signaling state of E46Q-PYP is only partially developed compared with that of wt-PYP. This finding provides a direct evidence of how the environmental change in the vicinity of the chromophore alters the conformational change of the entire protein matrix.
Introduction
Elucidating the molecular mechanisms of protein structural transitions is crucial to understanding protein function. For this, it is necessary to characterize the three-dimensional structures (or conformations) of transient intermediates formed during the structural transitions of the protein. To capture the transient structures of the protein in real time, both superb temporal resolution and high structural sensitivity is required because the dynamic evolution of the protein structure often occurs on time scales ranging from sub-picosecond to seconds.1-5 As a means of probing such fast protein structural transitions in solution, various optical spectroscopic methods have been employed.2,5-11 However, spectroscopic techniques are mostly selective to states of specific energies and local structure around a light-absorbing chromophore and cannot directly probe the global conformation of the protein. To complement time-resolved optical spectroscopies, time-resolved X-ray solution scattering (TRXSS), also known as time-resolved X-ray liquidography (TRXL),1,12-18 can be used to monitor the changes in global conformation of the protein, but it is not able to resolve small changes in the local structure around the chromophore. In this regard, an approach of combining time-resolved optical spectroscopy with time-resolved X-ray solution scattering may be the most effective way of obtaining a comprehensive view of protein structural transitions. In this study, we applied both TRXSS and transient absorption (TA) spectroscopy to a mutant of photoactive yellow protein (PYP), one of the most extensively studied model systems for protein structural dynamics.
PYP, a structural prototype of the PAS domain superfamily, is a biological photosensor involved in the negative phototaxis of a bacterium called Halorhodospira halophila.19-22 Upon absorption of blue-light by the p-coumaric acid (pCA) chromophore, PYP undergoes a photocycle that involves transitions among various spectrally red-shifted (pR or I1) and blue-shifted (pB or I2) intermediates through synchronous movements of pCA and surrounding hydrogen-bonding network.8,9,23-31 When the putative signaling state, pB (λmax = 355 nm), is formed, the hydrogen bonds between pCA and the surrounding residues (Tyr42 and Glu46) are broken and the protonation of pCA induced by internal proton transfer from Glu46 leads to significant blue shift of the absorption spectrum compared with that of the ground state, pG (λmax = 446 nm).8,20,26,27 Previous studies have shown that the internal proton transfer from Glu46 to pCA induces the regulated change in protein conformation and thus is a key step leading to the formation of the signaling state.14,20,26-28,32-36 However, the exact role of internal proton transfer in the formation of the signaling state has not been elucidated at the molecular level yet.
In this work, to examine how internal proton transfer influences the dynamics and the mechanism of the structural transition of PYP, we investigated the structural dynamics of the E46Q mutant of PYP using a combination of two complementary time-resolved techniques: TRXSS and TA spectroscopy. In the mutant PYP (E46Q-PYP), the 46th residue, glutamate (Glu or E), is replaced by glutamine (Gln or Q) as shown in Fig. 1 and the mutation makes the hydrogen bond between the 46th residue and pCA much weaker than in wt-PYP.37-41 In particular, we focus on how the local structural change around the chromophore, to which TA spectroscopy is sensitive, is temporally and spatially related to the global conformational change, which can be probed by TRXSS. By comparing the kinetic components obtained from measurements using the two time-resolved techniques, we found that the global conformational change involved in the transition to the signaling state is temporally delayed from the local structural change around the chromophore. This result demonstrates that each time-resolved technique gives only limited information on the protein dynamics and thus the combination of the two techniques is relevant for obtaining a comprehensive view of the protein structural dynamics. The molecular shape of the signaling state reconstructed from the TRXSS curves directly visualizes the three-dimensional conformations of protein intermediates and shows how the smaller structural change in E46Q-PYP than in wt-PYP, which was suggested by previous FTIR27 and NMR42 studies, is manifested in the three-dimensional conformation. Specifically, the comparison of the molecular shapes shows that the protrusion in a local region is much smaller in E46Q-PYP, suggesting that the signaling state of E46Q-PYP is only partially developed compared with that of wt-PYP. This finding provides a direct evidence of how the environmental change in the vicinity of the chromophore alters the conformational change of the entire protein matrix.
Fig. 1.
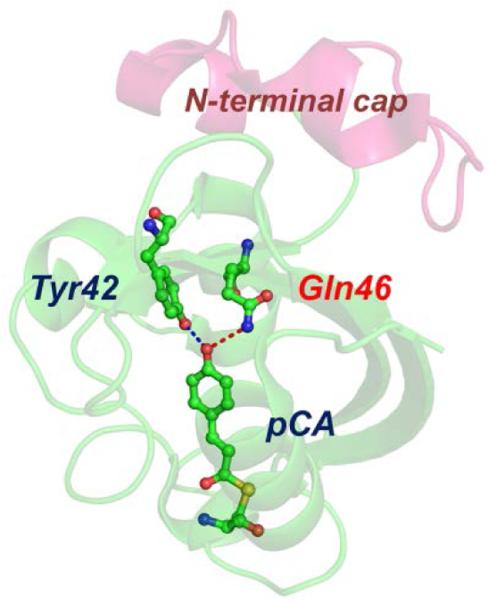
Structure of E46Q-PYP. Structures of the protein backbone (ribbon) and the active site around the chromophore (ball-and-stick) in E46Q-PYP (PDB: 1OTA). In E46Q-PYP, the glutamic acid (Glu46) in wild-type PYP was replaced by a glutamine (Gln46). The dashed lines indicate the hydrogen bonds between p-coumaric acid (pCA) and the neighbouring residues in the ground state of E46Q. Due to the effect of mutation, the hydrogen bond between pCA and Gln46 is longer than the one between pCA and Glu46 in wild-type PYP.19
Results
Time-resolved X-ray solution scattering
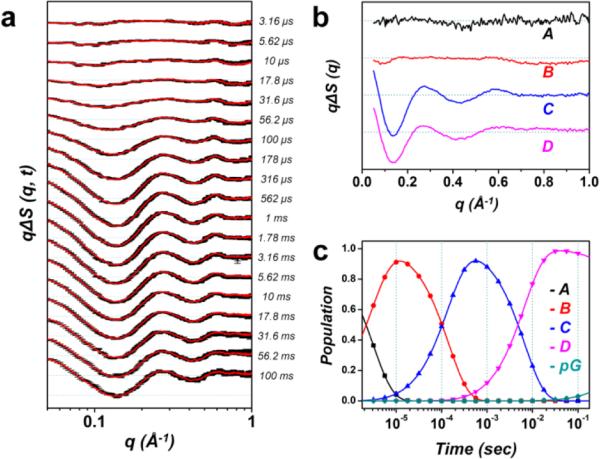
Time-resolved X-ray solution scattering (TRXSS) enables real-time monitoring of structural changes of proteins with high structural sensitivity over a broad range of time scales.1,12-18,43-47 We performed a TRXSS experiment to investigate the structural dynamics of the E46Q-PYP protein in a wide time range from 3.16 μs to 100 ms. The details of the TRXSS measurement are described in the experimental section. Fig. 2a shows time-resolved difference scattering curves, qΔS(q, t), measured at various time delays. The difference scattering curves show not only significant amplitude in the small-angle region (q < 0.3 Å−1), which is related with global structural change, but also oscillatory features in the wide-angle region (0.3 < q < 1.0 Å−1), which is attributed to subtle structural change such as the rearrangement of the secondary structure. Here we note that the solvent heating induced by the pump laser excitation also contributes to the scattering in the wide-angle region as shown in Fig. S1 in the ESI.† Therefore, to extract only the photoinduced signal of pure protein from the measured difference scattering curves, the contribution of solvent heating was removed by subtracting the signal that was obtained from a separate experiment on a solution of laser dye (see the ESI†).
Fig. 2.
Time-resolved X-ray solution scattering data of the E46Q-PYP photocycle and global kinetic analysis. (a) Experimental (black) and theoretical (red) difference scattering curves. (b) Species-associated difference scattering curves extracted from the PCA analysis. (c) Time-dependent population change of the intermediates following the proposed kinetic model determined by the PCA analysis.
For the heating-free experimental data, we performed the kinetic analysis based on singular value decomposition (SVD)13,48 to extract the kinetics of structural transitions of the protein (see the ESI†). First, we applied the SVD analysis in the q-range of 0.05 – 1.0 Å−1 and the time range from 3.16 μs to 100 ms and identified four structurally distinct intermediates and four time constants: 3.1 (± 2.2) μs, 143 (± 4) μs, 6.7 (± 1.0) ms, and ~1 s. Based on the results of the SVD analysis, we applied a sequential kinetic model as follows:
where A, B, C, and D are photoinduced protein intermediates and pG is the ground state. For the transitions among the intermediates, the time constants determined from the SVD analysis are shown together. To extract the structural features of the protein intermediates included in this kinetic model, we constructed scattering curves that directly reflect the structures of the intermediates (termed species-associated difference scattering curves) by performing the principal component analysis (PCA). In the PCA, the theoretical time-resolved difference scattering curve at each time delay is generated by a sum of species-associated difference scattering curves obeying the kinetics determined by the sequential kinetic model shown above. By globally minimizing the differences between the experimental and theoretical difference scattering curves at all time delays, we extracted the species-associated difference scattering curves (Fig. 2b) for the four intermediates. Also, time-dependent populations of the species-associated difference scattering curves were obtained as shown in Fig. 2c. As shown in Fig. 2a, the final theoretical difference scattering curves show excellent agreement with the experimental curves, confirming that the experimental data are fit well by the sequential kinetic model.
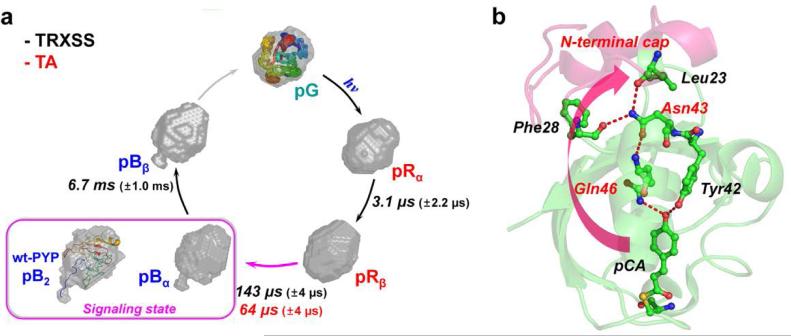
In principle, each species-associated difference scattering curve contains the information on the structure of a protein intermediate. To construct the three-dimensional molecular shape of individual intermediates, we converted the species-associated “difference” scattering curves into species-associated “static” scattering curves by adding the static scattering curve of pG multiplied by an appropriate scaling factor (see the ESI†). Subsequently, we applied ab-initio shape reconstruction algorithm to each of the species-associated static scattering curves for the four intermediates. Specifically, using the GNOM program,49 pair distribution functions (PDFs) were determined from the experimental species-associated static scattering curves. Then, the configuration of randomly oriented dummy atoms was varied until the discrepancy between the experimental and the theoretical scattering curves becomes minimized. For each species-associated static scattering curve, the above procedure of shape reconstruction was repeated about 50 times and the resulting structures were filtered to yield the final molecular shape of the intermediate (see the ESI†). Figure 4a shows the reconstructed molecular shapes of the intermediates together with the associated time constants for the transitions in the sequential kinetic model.
Fig. 4.
Structural dynamics of E46Q-PYP photocycle and hydrogen-bonding network from the chromophore binding pocket to the N-terminal cap. (a) Global kinetics of E46Q-PYP photocycle. The reconstructed molecular shapes of the intermediates were extracted from the structural analysis of the scattering data. The photocycle of E46Q-PYP includes four intermediates (pRα, pRβ, pBα, and pBβ) and the transition rates among the intermediates were determined from time-resolved X-ray solution scattering (TRXSS) and transient absorption spectroscopy (TA). The global conformational change of the protein is maximal in the pRβ → pBα transition occurring on the time scale of 143 μs as supported by the reconstructed shape from TRXSS. The inset (magenta box) shows the reconstructed molecular shape of the signaling state (pB2) of wt-PYP14 with the atomistic structure determined from the previous study combining DEER, NMR, and SAXS/WAXS experiment.51 (b) The local structural change in the vicinity of chromophore (pCA) can be propagated up to the N-terminal cap region through the hydrogen-bonding network (red dashed lines). The weak hydrogen bond between pCA and Gln46 in E46Q PYP may result in the smaller global conformational change compared with the change in wild-type PYP.
Transient absorption spectroscopy
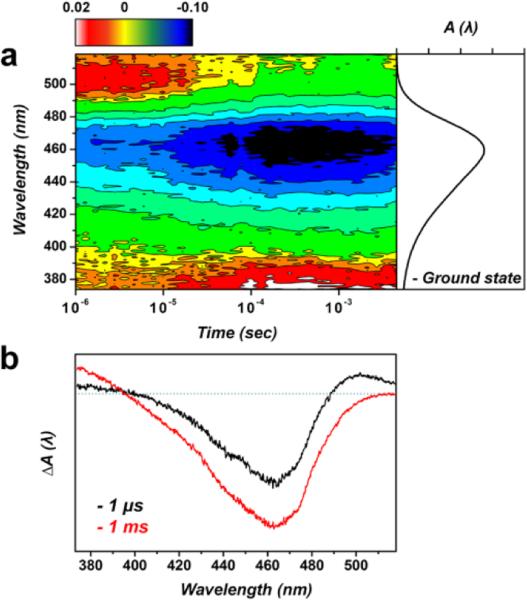
Transient absorption (TA) spectroscopy monitors time evolution of spectral features associated with various intermediates formed during the photocycle.8,37 We measured the transient change of the absorption spectrum of E46Q-PYP using TA spectroscopy to follow the transition dynamics among the protein intermediates, which were originally defined by the spectral features of their electronic transitions. Previously, the signaling process in the photocycle of E46Q-PYP was studied by nanosecond TA spectroscopy,8 but the TA spectra were recorded only at selected wavelengths with 10 nm wavelength step. In the present work, we comprehensively measured TA spectra in a wide spectral range (370 – 520 nm) and in the time range of 1 μs – 7 ms (Fig. 3). At the time delay of 1 μs, the TA spectrum shows a large negative feature around 460 nm and a small positive feature around 500 nm. The negative feature at 460 nm arises from the bleaching of the pG state, considering that its maximum matches with the absorption spectrum of the pG state of E46Q-PYP. The positive feature at 500 nm can be assigned to the absorption by a spectrally red-shifted intermediate (pR) in agreement with the previous TA study on E46Q-PYP.8 As the time progresses, the positive feature at 500 nm decreases gradually and disappears within 100 μs. In concurrence with the decrease of the 500 nm feature, another positive feature at 370 nm appears around 30 μs. This new positive feature can be assigned to the absorption by a spectrally blue-shifted intermediate (pB) of E46Q-PYP.
Fig. 3.
Transient absorption spectra of E46Q-PYP. (a) The contour shows the time evolution of transient absorption spectra. The ground state absorption spectrum of E46Q-PYP is shown together on the right side of the TA spectra. At ~64 μs, the decay of the positive feature at ~500 nm concurs with the growth of positive signal on ~380 nm, which is the signature of the spectrally blue-shifted intermediate called pB. (b) TA spectra at two representative time delays, 1 μs and 1 ms.
To determine the kinetics of the transitions between the protein intermediates, we performed the SVD analysis of the TA data and identified a kinetic component with the time constant of 64 (± 4) μs in the time range of our measurement (1 μs – 7 ms) as shown in Fig. S9 in the ESI.† In Fig. 3b, we compare the TA spectra before (1 μs) and after (1 ms) the time delay corresponding to the time constant of the kinetic component determined from the SVD analysis. When comparing the two TA spectra, we can see that the positive absorption around 380 nm increases and the positive absorption around 500 nm decreases between 1 μs and 1 ms, confirming that the 64 μs kinetic component corresponds to the transition from pR to pB as follows:
According to previous studies of wt-PYP,20,31 the pCA chromophore of pR has a negative charge and is stabilized by hydrogen bonds with surrounding residues, resulting in the spectral red shift of the absorption spectrum. In contrast, in pB, the pCA is protonated by the internal proton transfer from Glu46 and the hydrogen bonds are broken, leading to the spectral blue shift of the absorption spectrum. The dynamics of the pR-to-pB transition determined in the present work is slightly slower than the ones probed in the previous TA study of E46Q-PYP.8 Such difference in the dynamics can be attributed to the difference in the sample environment between the present (Na-phosphate buffer and pH 7.0) and the previous (MEG buffer and pH 6.5) works. Since we performed the TA spectroscopy and TRXSS measurements on the samples prepared under the same conditions (20 mM Na-phosphate buffer, 20 mM NaCl, and pH 7.0) in the present work, we note that our TA result is more relevant for comparison with the kinetics obtained from the TRXSS measurement.
Discussion
Structural and spectroscopic kinetics of E46Q-PYP photocycle
Based on the results obtained by TRXSS and TA spectroscopy, we can establish the kinetics and structural mechanism of the E46Q-PYP photocycle. Simultaneously, such comparison also provides opportunities to compare the sensitivity of each technique to a specific aspect of structural dynamics and obtain more detailed information about structural dynamics than can be acquired by a single technique. The TRXSS measurement clearly identifies four intermediates that exhibit sequential transition kinetics. Meanwhile, only a single kinetic component with a 64 μs time constant was identified from the TA measurement and was assigned to the pR → pB transition. Based on the similarity of the time scales, the B → C transition observed by TRXSS must be closely related to the pR → pB transition but occurs on a slower time scale (~100 μs) than the pR → pB transition probed by the TA measurement. While the experimental conditions (for example, sample concentration and laser fluence) used for the TRXSS and TA measurements were different from each other, their effect on the measured dynamics should be minimal (see the ESI† for details) and cannot account for the difference in the dynamics of pR → pB transition obtained from the TRXSS and TA measurements. Previously, it was reported that the shift of the absorption maximum of the transient intermediates in the PYP photocycle is attributed to the change in the orientation of the pCA chromophore induced by the change of hydrogen-bonding interactions between pCA and surrounding amino acid residues.8,20,33,40 Therefore, the slower B → C transition (probed by TRXSS) than pR → pB transition (probed by TA) indicates that the conformational change of the protein matrix is temporally delayed from the local movement around the pCA chromophore. A previous study on wt-PYP using time-resolved optical rotary dispersion spectroscopy50 reported a similar observation that the change in the chromophore precedes the unfolding of secondary structure (protein backbone). Thus, the combined results of our TA (sensitive to chromophore) and TRXSS (sensitive to protein conformation) provide a direct evidence for the sequential occurrence of the local movement around the chromophore and the global conformational change in E46Q-PYP.
We note that the kinetic components with the time constants of 3.1 μs (A → B transition) and 6.7 ms (C → D transition) were detected by TRXSS, but not by TA spectroscopy. The lack of these kinetic components in the TA data indicates that the electronic absorption spectrum does not change appreciably by the transitions associated with those kinetic components. Accordingly, among the four intermediates (A, B, C, and D) in the kinetic model used for the analysis of the TRXSS data, the two early intermediates, A and B, can be assigned to the variants of the spectrally red-shifted intermediates, pRα and pRβ, respectively, and the two late intermediates, C and D, can be regarded as the variants of the spectrally blue-shifted intermediates, pBα and pBβ, respectively. Here we note that we termed the transient intermediates (pRα, pRβ, pBα, and pBβ) of E46Q-PYP differently from the ones involved in the photocycle of wt-PYP (pR1, pR2, pB1, and pB2)14 because the transient intermediates of the two proteins are not considered structurally identical to each other. Judging from the different shapes of species-associated difference scattering curves for these intermediates shown in Fig. 2b, pRα and pRβ (or pBα and pBβ) must have subtly different conformations from each other, as was the case for pR1 and pR2 (or pB1 and pB2) in wt-PYP.14 We combine the results of the TRXSS and TA measurements to propose the following sequential kinetic scheme for the photocycle of E46Q-PYP including spectrally red-shifted (pRα and pRβ) and blue-shifted (pBα and pBβ) intermediates.
As an overview of the kinetics obtained from the TRXSS and TA measurements, we summarized the time constants of the transitions among the protein intermediates obtained by the three different techniques in Fig. 4a.
Structure of the signaling state of E46Q-PYP
From the reconstructed molecular shapes of the protein intermediates shown in Fig. 4a and the Rg and Dmax values listed in Table 1, we can estimate the extent of conformational changes occurring in the photocycle of E46Q-PYP. The pRα → pRβ transition of E46Q-PYP involves relatively small changes in the global shape and the volume of the protein, as judged from Rg and Dmax values. In contrast, the pRβ → pBα transition leads to the significant increase of the protein volume. Specifically, the protein volume of pBα calculated from the Rg value is larger by 5 % than that of pG. The pBα → pBβ transition does not involve any notable change in the molecular shape (even in the protruded region) or the protein volume. However, the species-associated difference scattering curves of pBα and pBβ are clearly different from each other in the wide-angle region (0.5 – 1.0 Å−1) as can be seen in Fig. 2b, implying that the two intermediates have subtly different conformations from each other.
Table 1.
Structural parameters of the intermediates of E46Q-PYP photocycle compared with those of wt-PYP. The radius of gyration (Rg) and maximum distance (Dmax) were determined from the representative structure of each intermediate.
| E46Q | Rg (Å) | Dmax (Å) | Wild type§ | Rg (Å) | Dmax (Å) |
|---|---|---|---|---|---|
| pRα | 15.08 | 41.53 | pR1 | 15.4 | 57.47 |
| pRβ | 15.11 | 42.57 | pR2 | 14.88 | 48.09 |
| pBα | 15.36 | 49.84 | pB1 | 15.26 | 53.83 |
| pBβ | 15.32 | 48.09 | pB2 | 15.68 | 58.09 |
| pG | 15.10 | 41.68 | pG | 14.83 | 41.02 |
The structural parameters of wt-PYP were adapted from ref.14.
In general, the signaling state of a protein often has a regulated, unfolded structure that is suitable for transferring the signal to its binding partner.51 According to previous studies on wt-PYP using TRXSS14 and a combination of structural probes (DEER, NMR, and SAXS/WAXS),52 the putative signaling state of wt-PYP is the second blue-shifted intermediate termed pB2, which is formed via the protrusion of N-terminus and the partial unfolding of the protein backbone. According to the results of our TRXSS measurement discussed above, the first blue-shifted intermediate, pBα, of E46Q-PYP has similar structural changes as pB2 of wt-PYP and thus can be regarded as at least an early form of the putative signaling state of E46Q-PYP. Here we note that the pBα intermediate is larger than pG by only 5 % in contrast to pB2 of wt-PYP, which is larger than pG by 18 %.14 The smaller volume increase of the signaling state in E46Q-PYP suggests that, compared with wt-PYP, E46Q-PYP undergoes restricted changes in global conformation in the transition to the signaling state.
Role of internal proton transfer in the signal transduction of PYP
The combined results of TRXSS and TA measurements provide a key evidence for the role of the Glu46 (E46) residue in the signal transduction of PYP. According to a previous time-resolved X-ray Laue crystallography (TR-Laue) study on a crystalline sample of E46Q-PYP,53 the E46Q mutation modifies the lengths and the strengths of hydrogen bonds between the pCA chromophore and the surrounding amino acid residues. In the same study, it was found that the spectrally blue-shifted intermediate is formed faster in E46Q-PYP than in wt-PYP, which was also reported by a previous TA study.8 Previous studies on E46Q-PYP using FTIR27 and NMR42 reported that, in the transition from pG to pB, the amide-I band absorption and chemical shift, respectively, change to the lesser extent in E46Q-PYP than in wt-PYP, implying smaller structural changes involved in the formation of the signaling state of E46Q-PYP. The global structure of the signaling state determined from our TRXSS measurement allows us to quantitatively connect the acceleration of the kinetics in E46Q-PYP with the degree of structural changes and to visualize the detailed structural difference accounting for the smaller changes in global conformation. As shown in Fig. 4a, the molecular shape of the signaling state reconstructed from the TRXSS curves directly visualizes how the smaller structural change in E46Q-PYP suggested by the previous studies is manifested in the three-dimensional conformation. Specifically, the signaling state of E46Q-PYP is formed with smaller degree of N-terminal protrusion and smaller volume increase than that of wt-PYP. Based on these observations, we can regard the signaling state of E46Q-PYP as a partially developed state compared with pB2 in wt-PYP. The restricted conformational change leading to the signaling state of E46Q-PYP will take less time than the structural change in wt-PYP, accounting for the accelerated formation of the signaling state in E46Q-PYP.8,39,53
We can attribute the origin of the partial structural development observed in the signaling state of E46Q-PYP to the weaker hydrogen bond between Gln46 and pCA in the E46Q mutant than in wt-PYP. As shown in Fig. 4b, the hydrogen-bonding network starting from pCA reaches up to the N-terminal cap through three hydrogen bonds (pCA → Gln46 → Asn43 → Phe28/Leu23) and can play an important role in triggering global conformational changes associated with the signal transduction.53-55 In the pG of E46Q-PYP, pCA forms two hydrogen bonds, one with the nitrogen atom of Gln46 and the other with the oxygen atom of Tyr42. The hydrogen bond between pCA and Gln46 is weaker than the one between pCA and Glu46 in wt-PYP because nitrogen has a smaller electron affinity than oxygen. As a result of the weaker hydrogen bond between Gln46 and pCA in E46Q-PYP, the structural perturbation towards the N-terminal cap is likely to be smaller in E46Q-PYP than in wt-PYP, resulting in the partially developed signaling state of E46Q-PYP.
Conclusions
We combined time-resolved X-ray solution scattering (TRXSS) and transient absorption (TA) spectroscopy as probes of the dynamics of the protein conformation and the chromophore in the photocycle of E46Q-PYP, where the internal proton transfer is inhibited. We were able to establish the global kinetics of E46Q-PYP photocycle with the four structurally distinct intermediates by making detailed comparisons of the results obtained from the different time-resolved techniques. The spectral change related with the local structure of the chromophore (that is, pR → pB transition) probed by TA spectroscopy occurs in 64 μs and temporally precedes the change in the global conformation (143 μs) leading to the signaling state (that is, pRβ → pBα transition) probed by TRXSS. The delayed occurrence of the conformational change suggests that it takes time for the structural perturbation triggered by the chromophore to propagate through the hydrogen-bonding network. The pRα → pRβ and pBα → pBβ transitions occurring in 3.1 μs and 6.4 ms, respectively, accompany rather small changes in global conformation of the protein and were not detected by TA spectroscopy that lacks global structural sensitivity. The molecular shape of the signaling state reconstructed from the TRXSS curves reveals the smaller structural change of the signaling state of E46Q-PYP than that of wt-PYP and we revealed that the smaller change is associated with a smaller degree of protrusion, confirming that the suppression of the internal proton transfer induces the formation of the partially developed signaling state. In this work, we demonstrated that the approach of combining two time-resolved techniques allows us to obtain a comprehensive view of protein structural dynamics ranging from local structure to global conformation.
Experimental section
Preparation of photoactive yellow protein E46Q mutant
The procedure of protein preparation is based on a previously published protocol.56 The E46Q mutation was introduced into the pQE80L vector and was expressed in E. coli BL21 (DE3). The apo-E46Q-PYP was reconstituted with p-coumaric anhydride and purified by centrifugation and nickel affinity column chromatography. The protein solution was treated by enterokinase (EK) overnight after the buffer exchange and purified again by ion exchange column chromatography. The protein solution was dialyzed with the buffer for the time-resolved experiments (20 mM sodium phosphate buffer, 20 mM NaCl, and pH 7.0) and the purity index (Abs280 nm/Abs460 nm) of the solution was 0.43.
Time-resolved X-ray solution scattering
Time-resolved (pump-probe) X-ray solution scattering data were collected at ID14B beamline at Advanced Photon Source by following an established experimental protocol.13,14,17,46 Prior to the experiment for E46Q-PYP, the control experiment was performed with wt-PYP to check the experimental conditions. From the control experiment, the time constants determined from the previous experiment at ESRF were well reproduced. For the sample, a 4.4 mM E46Q-PYP solution dissolved in a 20 mM sodium phosphate buffer with 20 mM NaCl at pH 7.0 was enclosed in a quartz capillary (Hampton research). The capillary was mounted on a linear translational stage (Parker) to move the sealed capillary back and forth periodically. During the measurement, the temperature of the sample was kept at 293 K using a cryostream temperature controller (Oxford). The E46Q-PYP solution was irradiated by circularly polarized nanosecond (ns) laser pulses (1 mJ/mm2 fluence at 460 nm) and was probed by X-ray pulses incident at well-defined time delays. Polychromatic X-ray probe pulse (15 keV), which has a pulse duration of 1.5 μs, was generated from the 324-bunch mode operation at Advanced Photon Source. Time-dependent X-ray scattering patterns were recorded in a Mar165 CCD detector (Rayonix, USA), covered up to wide-angle region. The scattering pattern at a negative time-delay (−5 μs) was also collected and used as a reference for calculating time-resolved difference scattering curves. The scattering pattern at −5 μs time delay contains structural information of the pG ground state, while the data at positive time delays contain the contributions from a mixture of ground state and photoproducts. The time-resolved difference scattering curves, ΔS(q, t) = S(q, t) – S(q, −5 μs), were generated by subtracting the scattering curve at −5 μs from the curves at positive time delays. The ΔS(q, t) curves provide the information on the change caused by the laser photoexcitation. The data acquisition was implemented at a repetition rate of 2 Hz while translating the quartz capillary containing the sample. We measured the time-resolved X-ray solution scattering curves at a total of 19 time delays in a wide time range from 3.16 μs to 100 ms. At each time delay, >150 scattering curves were averaged to achieve a high signal-to-noise ratio.
X-ray scattering patterns contain rich information on the global structure of a protein. Especially, small angle X-ray scattering (SAXS) signal is sensitive to the global protein conformation such as shape and size of the protein. In principle, X-ray scattering intensity is related to the pair distribution function, P(r), of intramolecular atomic distances by Fourier transform:
| (1) |
where Dmax is the maximum intramolecular distance. Because the pair distribution function is a real space representation of the scattering pattern, it provides direct information on protein structure and can be used to visualize the global shape of the protein though three-dimensional shape reconstruction.57,58 To reconstruct the structure of a transient reaction intermediate from time-resolved X-ray scattering curves, it is needed to extract a difference scattering curve characteristic of the intermediate species, so-called species-associated difference scattering curve. The species-associated scattering curve can be obtained by kinetic analysis based on singular value decomposition (SVD) and principal component analysis (PCA) of the time-resolved scattering data.1,13,48,59 From the SVD, the number of structurally distinct intermediates and the transition kinetics among the intermediates can be obtained. Subsequently, by PCA, the experimental difference scattering curves are decomposed into the species-associated scattering curves of the intermediates using the following equation:
| (2) |
where ΔStheory (qi, tj) is the theoretical difference scattering curve at given q and t values, ΔSCk (qi) is the species-associated difference scattering curve corresponding to the k-th intermediate species at a given q value, Ck (tj) is the instantaneous population of the k-th intermediate at a given time delay and can be easily calculated using the kinetics obtained from the SVD analysis. We employed ab-initio shape reconstruction to extract the molecular shape of the protein from the species-associated difference scattering curves. This approach consists of two steps: (i) Species-associated “static” scattering curves are obtained by adding the scattering curve of the ground state to the scaled species-associated difference scattering curves of the intermediates. (ii) For the species-associated static scattering curves, the three-dimensional shape reconstruction is implemented by DAMMIN package60 based on the dummy atom modelling. The details of the analysis is described in the ESI†.
Transient absorption spectroscopy
Transient absorption spectroscopy measurement was performed using a conventional setup.61 A 70 μM E46Q-PYP dissolved in a 20 mM sodium phosphate buffer with 20 mM NaCl at pH 7.0 was flowed through a 2 mm quartz cell (Hellma Analytics) by using a syringe pump (KD Scientific). A pump beam (460 nm, 1 Hz) was generated from an optical parametric oscillator (LOTIS). The fluence of the pump beam was 76 μJ/mm2 per pulse at the sample position. A continuous probe beam was generated by a Xe lamp (Newport, 66902). The pump beam was perpendicularly crossed with the probe beam. The probe light was detected by a combination of a spectrometer (Princeton Instruments) and an ICCD camera (Andor). The transient absorption measurements were implemented at 293 K.
Supplementary Material
Acknowledgements
This work was supported by IBS-R004-G2. This work was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (NRF-2014R1A1A1002511). Use of the BioCARS Sector 14 at the APS was supported by NIH National Institute of General Medical Sciences grant R24GM111072. The time-resolved set up at Sector 14 was funded in part through collaboration with P. Anfinrud (NIH/NIDDK) through the Intramural Research Program of the NIDDK. Use of the APS was supported by the US Department of Energy, Basic Energy Sciences, Office of Science, under Contract No. DE-AC02-06CH11357.
Footnotes
Electronic Supplementary Information (ESI) available: Materials of data analysis for the time-resolved X-ray scattering signal, figures, and table.
References
- 1.Kim JG, Kim TW, Kim J, Ihee H. Acc. Chem. Res. 2015;48:2200–2208. doi: 10.1021/acs.accounts.5b00198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bredenbeck J, Helbing J, Kumita JR, Woolley GA, Hamm P. Proc. Natl. Acad. Sci. U.S.A. 2005;102:2379–2384. doi: 10.1073/pnas.0406948102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schotte F, Lim MH, Jackson TA, Smirnov AV, Soman J, Olson JS, Phillips GN, Wulff M, Anfinrud PA. Science. 2003;300:1944–1947. doi: 10.1126/science.1078797. [DOI] [PubMed] [Google Scholar]
- 4.Chen EF, Kumita JR, Woolley GA, Kliger DS. J. Am. Chem. Soc. 2003;125:12443–12449. doi: 10.1021/ja030277+. [DOI] [PubMed] [Google Scholar]
- 5.Lewis JW, Goldbeck RA, Kliger DS, Xie XL, Dunn RC, Simon JD. J. Phys. Chem. 1992;96:5243–5254. [Google Scholar]
- 6.Mizuno M, Kamikubo H, Kataoka M, Mizutani Y. J. Phys. Chem. B. 2011;115:9306–9310. doi: 10.1021/jp2029399. [DOI] [PubMed] [Google Scholar]
- 7.Schuler B, Eaton WA. Curr. Opin. Struct. Biol. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yeremenko S, van Stokkum IHM, Moffat K, Hellingwerf KJ. Biophys. J. 2006;90:4224–4235. doi: 10.1529/biophysj.105.074765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.van Wilderen LJGW, Van der Horst MA, van Stokkum IHM, Hellingwerf KJ, van Grondelle R, Groot ML. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15050–15055. doi: 10.1073/pnas.0603476103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kennis JTM, Larsen DS, Ohta K, Facciotti MT, Glaeser RM, Fleming GR. J. Phys. Chem. B. 2002;106:6067–6080. [Google Scholar]
- 11.Kao YT, Saxena C, He TF, Guo LJ, Wang LJ, Sancar A, Zhong DP. J. Am. Chem. Soc. 2008;130:13132–13139. doi: 10.1021/ja8045469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim KH, Kim JG, Nozawa S, Sato T, Oang KY, Kim TW, Ki H, Jo J, Park S, Song C, Ogawa K, Togashi T, Tono K, Yabashi M, Ishikawa T, Kim J, Ryoo R, Ihee H, Adachi S. Nature. 2015;518:385–389. doi: 10.1038/nature14163. [DOI] [PubMed] [Google Scholar]
- 13.Oang KY, Kim JG, Yang C, Kim TW, Kim Y, Kim KH, Kim J, Ihee H. J. Phys. Chem. Lett. 2014;5:804–808. doi: 10.1021/jz4027425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim TW, Lee JH, Choi J, Kim KH, van Wilderen LJ, Guerin L, Kim Y, Jung YO, Yang C, Kim J, Wulff M, van Thor JJ, Ihee H. J. Am. Chem. Soc. 2012;134:3145–3153. doi: 10.1021/ja210435n. [DOI] [PubMed] [Google Scholar]
- 15.Kim TK, Lee JH, Wulff M, Kong QY, Ihee H. ChemPhysChem. 2009;10:1958–1980. doi: 10.1002/cphc.200900154. [DOI] [PubMed] [Google Scholar]
- 16.Ihee H. Acc. Chem. Res. 2009;42:356–366. doi: 10.1021/ar800168v. [DOI] [PubMed] [Google Scholar]
- 17.Cammarata M, Levantino M, Schotte F, Anfinrud PA, Ewald F, Choi J, Cupane A, Wulff M, Ihee H. Nat. Methods. 2008;5:881–886. doi: 10.1038/nmeth.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JH, Wulff M, Bratos S, Petersen J, Guerin L, Leicknam JC, Carnmarata M, Kong Q, Kim J, Moller KB, Ihee H. J. Am. Chem. Soc. 2013;135:3255–3261. doi: 10.1021/ja312513w. [DOI] [PubMed] [Google Scholar]
- 19.Henry JT, Crosson S. Annu. Rev. Microbiol. 2011;65:261–286. doi: 10.1146/annurev-micro-121809-151631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hellingwerf KJ, Hendriks J, Gensch T. J. Phys. Chem. A. 2003;107:1082–1094. [Google Scholar]
- 21.Cusanovich MA, Meyer TE. Biochemistry. 2003;42:4759–4770. doi: 10.1021/bi020690e. [DOI] [PubMed] [Google Scholar]
- 22.Sprenger WW, Hoff WD, Armitage JP, Hellingwerf KJ. J. Bacteriol. 1993;175:3096–3104. doi: 10.1128/jb.175.10.3096-3104.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jung YO, Lee JH, Kim J, Schmidt M, Moffat K, Srajer V, Ihee H. Nat. Chem. 2013;5:212–220. doi: 10.1038/nchem.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schotte F, Cho HS, Kaila VRI, Kamikubo H, Dashdorj N, Henry ER, Graber TJ, Henning R, Wulff M, Hummer G, Kataoka M, Anfinrud PA. Proc. Natl. Acad. Sci. U.S.A. 2012;109:19256–19261. doi: 10.1073/pnas.1210938109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vreede J, Juraszek J, Bolhuis PG. Proc. Natl. Acad. Sci. U.S.A. 2010;107:2397–2402. doi: 10.1073/pnas.0908754107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ihee H, Rajagopal S, Srajer V, Pahl R, Anderson S, Schmidt M, Schotte F, Anfinrud PA, Wulff M, Moffat K. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7145–7150. doi: 10.1073/pnas.0409035102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xie AH, Kelemen L, Hendriks J, White BJ, Hellingwerf KJ, Hoff WD. Biochemistry. 2001;40:1510–1517. doi: 10.1021/bi002449a. [DOI] [PubMed] [Google Scholar]
- 28.Brudler R, Rammelsberg R, Woo TT, Getzoff ED, Gerwert K. Nat. Struct. Biol. 2001;8:265–270. doi: 10.1038/85021. [DOI] [PubMed] [Google Scholar]
- 29.Genick UK, Borgstahl GEO, Ng K, Ren Z, Pradervand C, Burke PM, Srajer V, Teng TY, Schildkamp W, McRee DE, Moffat K, Getzoff ED. Science. 1997;275:1471–1475. doi: 10.1126/science.275.5305.1471. [DOI] [PubMed] [Google Scholar]
- 30.Wei LL, Wang HJ, Chen XB, Fang WH, Wang HB. Phys. Chem. Chem. Phys. 2014;16:25263–25272. doi: 10.1039/c4cp03495c. [DOI] [PubMed] [Google Scholar]
- 31.Hoff WD, van Stokkum IHM, van Ramesdonk HJ, van Brederode ME, Brouwer AM, Fitch JC, Meyer TE, van Grondelle R, Hellingwerf KJ. Biophys. J. 1994;67:1691–1705. doi: 10.1016/S0006-3495(94)80643-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hoshihara Y, Imamoto Y, Kataoka M, Tokunaga F, Terazima M. Biophys. J. 2008;94:2187–2193. doi: 10.1529/biophysj.107.115253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Y, Ujj L, Meyer TE, Cusanovich MA, Atkinson GH. J. Phys. Chem. A. 2001;105:5719–5726. [Google Scholar]
- 34.van der Horst MA, van Stokkum IH, Crielaard W, Hellingwerf KJ. FEBS Lett. 2001;497:26–30. doi: 10.1016/s0014-5793(01)02427-9. [DOI] [PubMed] [Google Scholar]
- 35.Hoff WD, Xie A, Van Stokkum IHM, Tang XJ, Gural J, Kroon AR, Hellingwerf KJ. Biochemistry. 1999;38:1009–1017. doi: 10.1021/bi980504y. [DOI] [PubMed] [Google Scholar]
- 36.Rubinstenn G, Vuister GW, Mulder FAA, Dux PE, Boelens R, Hellingwerf KJ, Kaptein R. Nat. Struct. Biol. 1998;5:568–570. doi: 10.1038/823. [DOI] [PubMed] [Google Scholar]
- 37.Changenet-Barret P, Plaza P, Martin MM, Chosrowjan H, Taniguchi S, Mataga N, Imamoto Y, Kataoka M. J. Phys. Chem. C. 2009;113:11605–11613. [Google Scholar]
- 38.Anderson S, Crosson S, Moffat K. Acta Crystallogr. Sect. D. 2004;60:1008–1016. doi: 10.1107/S090744490400616X. [DOI] [PubMed] [Google Scholar]
- 39.Borucki B, Devanathan S, Otto H, Cusanovich MA, Tollin G, Heyn MP. Biochemistry. 2002;41:10026–10037. doi: 10.1021/bi0256227. [DOI] [PubMed] [Google Scholar]
- 40.Devanathan S, Lin S, Cusanovich MA, Woodbury N, Tollin G. Biophys. J. 2000;79:2132–2137. doi: 10.1016/S0006-3495(00)76460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Genick UK, Devanathan S, Meyer TE, Canestrelli IL, Williams E, Cusanovich MA, Tollin G, Getzoff ED. Biochemistry. 1997;36:8–14. doi: 10.1021/bi9622884. [DOI] [PubMed] [Google Scholar]
- 42.Derix NM, Wechselberger RW, van der Horst MA, Hellingwerf KJ, Boelens R, Kaptein R, van Nuland NAJ. Biochemistry. 2003;42:14501–14506. doi: 10.1021/bi034877x. [DOI] [PubMed] [Google Scholar]
- 43.Malmerberg E, Bovee-Geurts PHM, Katona G, Deupi X, Arnlund D, Wickstrand C, Johansson LC, Westenhoff S, Nazarenko E, Schertler GFX, Menzel A, de Grip WJ, Neutze R. Sci. Signal. 2015:8. doi: 10.1126/scisignal.2005646. [DOI] [PubMed] [Google Scholar]
- 44.Malmerberg E, Omran Z, Hub JS, Li XW, Katona G, Westenhoff S, Johansson LC, Andersson M, Cammarata M, Wulff M, van der Spoel D, Davidsson J, Specht A, Neutze R. Biophys. J. 2011;101:1345–1353. doi: 10.1016/j.bpj.2011.07.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kathuria SV, Guo L, Graceffa R, Barrea R, Nobrega RP, Matthews CR, Irving TC, Bilsel O. Biopolymers. 2011;95:550–558. doi: 10.1002/bip.21628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cho HS, Dashdorj N, Schotte F, Graber T, Henning R, Anfinrud P. Proc. Natl. Acad. Sci. U.S.A. 2010;107:7281–7286. doi: 10.1073/pnas.1002951107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Haldrup K, Christensen M, Cammarata M, Kong QY, Wulff M, Mariager SO, Bechgaard K, Feidenhans'l R, Harrit N, Nielsen MM. Angew. Chem. Int. Ed. 2009;48:4180–4184. doi: 10.1002/anie.200900741. [DOI] [PubMed] [Google Scholar]
- 48.Schmidt M, Pahl R, Srajer V, Anderson S, Ren Z, Ihee H, Rajagopal S, Moffat K. Proc. Natl. Acad. Sci. U.S.A. 2004;101:4799–4804. doi: 10.1073/pnas.0305983101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Svergun DI. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- 50.Chen EF, Gensch T, Gross AB, Hendriks J, Hellingwerf KJ, Kliger DS. Biochemistry. 2003;42:2062–2071. doi: 10.1021/bi020577o. [DOI] [PubMed] [Google Scholar]
- 51.Mitrea DM, Kriwacki RW. FEBS Lett. 2013;587:1081–1088. doi: 10.1016/j.febslet.2013.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ramachandran PL, Lovett JE, Carl PJ, Cammarata M, Lee JH, Jung YO, Ihee H, Timmel CR, van Thor JJ. J. Am. Chem. Soc. 2011;133:9395–9404. doi: 10.1021/ja200617t. [DOI] [PubMed] [Google Scholar]
- 53.Rajagopal S, Anderson S, Srajer V, Schmidt M, Pahl R, Moffat K. Structure. 2005;13:55–63. doi: 10.1016/j.str.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 54.Tenboer J, Basu S, Zatsepin N, Pande K, Milathianaki D, Frank M, Hunter M, Boutet S, Williams GJ, Koglin JE, Oberthuer D, Heymann M, Kupitz C, Conrad C, Coe J, Roy-Chowdhury S, Weierstall U, James D, Wang DJ, Grant T, Barty A, Yefanov O, Scales J, Gati C, Seuring C, Srajer V, Henning R, Schwander P, Fromme R, Ourmazd A, Moffat K, Van Thor JJ, Spence JCH, Fromme P, Chapman HN, Schmidt M. Science. 2014;346:1242–1246. doi: 10.1126/science.1259357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kumauchi M, Kaledhonkar S, Philip AF, Wycoff J, Hara M, Li YX, Xie AH, Hoff WD. J. Am. Chem. Soc. 2010;132:15820–15830. doi: 10.1021/ja107716r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kim Y, Ganesan P, Ihee H. Protein Sci. 2013;22:1109–1117. doi: 10.1002/pro.2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mertens HDT, Svergun DI. J. Struct. Biol. 2010;172:128–141. doi: 10.1016/j.jsb.2010.06.012. [DOI] [PubMed] [Google Scholar]
- 58.Hura GL, Menon AL, Hammel M, Rambo RP, Poole FL, Tsutakawa SE, Jenney FE, Classen S, Frankel KA, Hopkins RC, Yang SJ, Scott JW, Dillard BD, Adams MWW, Tainer JA. Nat. Methods. 2009;6:606–U683. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Henry ER, Hofrichter J. Methods Enzymol. 1992;210:129–192. [Google Scholar]
- 60.Svergun DI. Biophys. J. 1999;77:2896–2896. [Google Scholar]
- 61.Choi J, Yang C, Kim J, Ihee H. J. Phys. Chem. B. 2011;115:3127–3135. doi: 10.1021/jp106588d. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.