Abstract
We compared the newly approved BacT/Alert Virtuo blood culture system to the BacT/Alert 3D system using 115 clinical bacterial and fungal isolates in 784 simulated blood culture bottles. The time to detection was reduced by roughly 20% in the Virtuo system (P < 0.0001) while the detection rate did not differ.
TEXT
Bloodstream infections remain a leading cause of death and are associated with high mortality and morbidity (1, 2). Early intervention and initiation of appropriate treatment have been shown to improve patient outcome (3–5). Rapid detection and identification of the causing pathogen are therefore essential in the diagnosis of these invasive diseases.
Blood culture (BC) is the gold standard for detection of bacteria and fungi from blood or other normally sterile body fluids. It has previously been shown that the type of BC bottle used has a substantial impact on the microbiological diagnosis of bloodstream infections (6–10), as these media are complex formulations that provide nutrients and neutralizing antimicrobials in clinical blood samples (11). In addition, the BC system affects workflow and microbiological performance (11, 12). To date, the most widely distributed BC systems are the BacT/Alert 3D (bioMérieux; referred to as 3D hereafter), BD Bactec (Becton Dickinson), and VersaTREK (Thermo Scientific). bioMérieux has recently introduced a new BC system with automatic loading and unloading of BC bottles, the BacT/Alert Virtuo (Virtuo), for bacterial and fungal detection in BC. Enhanced colorimetric technology to detect microbial growth and improved temperature stability suggest improved culture conditions in the Virtuo compared to the 3D system. However, the microbiological performance of Virtuo has not been evaluated.
In this study, the Virtuo system was tested in direct comparison with the 3D system by parallel incubation of a total of 115 clinical bacterial and fungal isolates (Table 1). All isolates were originally collected from clinical BC samples sent for microbiological diagnosis to Karolinska University Laboratory (Stockholm, Sweden) from three tertiary care hospitals in the greater Stockholm area. Isolates were recovered from frozen stocks and were cultured overnight on appropriate agar medium. Approximately 15 CFU in phosphate-buffered saline was inoculated into each BC bottle with 8 ml defibrinated horse blood, and all BC bottles were incubated until positivity was reached or for a maximum of 5 days. We and others have previously demonstrated that the use of horse blood does not significantly influence the performance of BC systems (13–17). Growth was assessed in aerobic (FA and FA Plus) and anaerobic (FN and FN Plus) BacT/Alert BC bottles, except for Acinetobacter spp. and Candida spp., which were cultured under aerobic conditions only. While the CE-approved version of the Virtuo system is intended to operate with resin-based BC bottles (FA Plus and FN Plus) only, charcoal-based BC bottles (FA and FN) were evaluated in the 3D system and in the investigational-use-only version of Virtuo employed in this study. At the end of the incubation period, the BC medium was subcultured on blood (for bacteria) or Sabouraud dextrose (for yeast) agar plates, respectively, to exclude contamination and to confirm true-positive and true-negative detection results.
TABLE 1.
Clinical isolates tested in the BacT/Alert 3D and the BacT/Alert Virtuo blood culture systems
| Species/group | No. of isolatesa | No. with positive blood cultures within 5 days |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| FA |
FA Plus |
FN |
FN Plus |
||||||
| 3D | Virtuo | 3D | Virtuo | 3D | Virtuo | 3D | Virtuo | ||
| Gram-negative species | 39/32 | 38 | 38 | 39 | 38 | 32 | 32 | 32 | 32 |
| Escherichia coli | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
| Klebsiella oxytoca | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| Klebsiella pneumoniae | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 | 7 |
| Serratia marcescens | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 | 12 |
| Acinetobacter spp. | 7 | 6 | 6 | 7 | 6 | ||||
| Gram-positive species | 49 | 48 | 48 | 47 | 48 | 44 | 44 | 46 | 46 |
| Staphylococcus aureus | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 | 10 |
| CoNS | 5 | 5 | 5 | 5 | 5 | 2 | 2 | 5 | 5 |
| Enterococcus faecalis | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| Enterococcus faecium | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 | 5 |
| Group A streptococci | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 | 3 |
| Group G streptococci | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 | 2 |
| Streptococcus pneumoniae | 13 | 13 | 13 | 13 | 13 | 13 | 13 | 13 | 13 |
| Alpha-hemolytic streptococci | 6 | 5 | 5 | 4 | 5 | 4 | 4 | 3 | 3 |
| Yeast | 27/0 | 27 | 27 | 27 | 27 | ||||
| Candida albicans | 12 | 12 | 12 | 12 | 12 | ||||
| Candida glabrata | 15 | 15 | 15 | 15 | 15 | ||||
| Total (aerobic/anaerobic) | 115/81 | 113 | 113 | 113 | 113 | 76 | 76 | 78 | 78 |
Number of isolated tested under aerobic/anaerobic conditions.
Statistical analyses were performed using GraphPad Prism 6. The times to detection (TTD) between BC systems were compared using the Wilcoxon matched-pair signed-rank test or 2-way matched analysis of variance (ANOVA) and the TTD between groups of microorganisms by Mann-Whitney U test. The relation between the TTD values achieved by the two systems was assessed by Spearman correlation. Differences with P values of <0.05 were regarded as statistically significant.
A total of 784 BC bottles were included in this study. The vast majority (n = 760; 97%) signaled positive within the 5-day incubation period (Table 1). There was no difference in the number of negative bottles per BC system (n = 12 each). Mostly, the paired BC bottles from one isolate were affected (alpha-hemolytic streptococci, n = 7 bottle pairs; coagulase-negative staphylococci [CoNS], n = 3 bottle pairs; Acinetobacter, n = 1 bottle pair), except for one alpha-hemolytic streptococcal isolate that grew in the Virtuo system and one Acinetobacter isolate that grew only in the 3D system. Subcultures of the BC medium confirmed the absence of bacterial growth in these bottles. Thus, false-negative BC results were not observed.
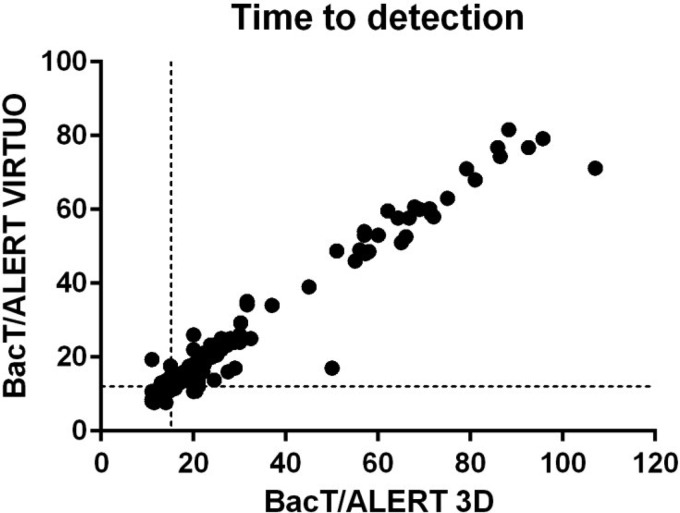
The TTD values were compared among the 379 bottle pairs that signaled positive in the two systems (Table 2; Fig. 1). Overall, the TTD was significantly shorter for bottles incubated in Virtuo than those incubated in 3D (median 12 h and 15 h, respectively; P < 0.0001) and was reduced by 18.5% (median) overall without striking differences between bacterial species or bottle types. Among bacterial cultures (n = 325 bottle pairs), 90% of the bottles reached positivity within 21 h in the 3D system, while this was already achieved after 16 h in the Virtuo system. The TTD was reduced by 2.8 h (median) in the Virtuo system (P < 0.0001). Among cultures with Candida (n = 54 bottle pairs), 90% reached positivity at the fourth day (81 h) in the 3D system and within 3 days (71 h) in the Virtuo system. The median TTD was reduced by 4.8 h (P < 0.0001). The two Candida species included in this investigation showed marked differences in TTD, with 24 h for Candida albicans and 66 h for Candida glabrata (P < 0.0001). C. glabrata is an increasingly common pathogen that is isolated from bloodstream infections (18, 19). Treatment of these infections is challenging due to reduced sensitivity of C. glabrata to antifungal agents (20). It has previously been observed that C. glabrata was more strongly affected by BC system and medium than C. albicans (6, 21), which in general grew faster and with less variable results (21–24). Similarly, TTD for C. glabrata was reduced by 14.7% (median, 9 h), while TTD for C. albicans was only 9.1% (2 h) shorter in Virtuo.
TABLE 2.
Time to detection in four different types of blood culture bottles incubated in two blood culture systems
| Species/group | No. of isolates | Time to positivity (median, h) |
P valuea | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| FA |
FA Plus |
FN |
FN Plus |
|||||||
| 3D | Virtuo | 3D | Virtuo | 3D | Virtuo | 3D | Virtuo | |||
| Gram-negative species | 38 | 14.5 | 11.4 | 14.0 | 11.4 | 13.2 | 10.7 | 14.5 | 11.7 | <0.0001 |
| Escherichia coli | 10 | 14.7 | 11.4 | 11.5 | 9.2 | 13.1 | 10.1 | 11.4 | 8.8 | <0.0001 |
| Klebsiella spp. | 10 | 12.6 | 9.8 | 14.0 | 11.9 | 12.1 | 9.7 | 15.0 | 12.1 | <0.0001 |
| Serratia marcescens | 12 | 15.3 | 11.9 | 15.5 | 12.0 | 14.5 | 11.5 | 15.5 | 12.5 | <0.0001 |
| Acinetobacter spp. | 6 | 16.5 | 14.0 | 14.0 | 12 | <0.0001 | ||||
| Gram-positive species | 43–48 | 16.3 | 13.3 | 13.0 | 11 | 16.8 | 13.7 | 14.0 | 11.0 | <0.0001 |
| Staphylococcus aureus | 10 | 16.6 | 13.8 | 13.1 | 11.3 | 18.7 | 14.3 | 13 | 10.9 | <0.0001 |
| CoNS | 2–5 | 21.8 | 16.0 | 19.6 | 15.5 | 33.5 | 30.0 | 19.6 | 17.7 | <0.0001 |
| Enterococcus spp. | 10 | 16.0 | 12.9 | 13.0 | 10.5 | 15.0 | 11.8 | 13.5 | 10.1 | <0.0001 |
| Beta-hemolytic streptococci | 5 | 21.0 | 14.0 | 12.0 | 10.0 | 17.0 | 13.8 | 14.0 | 12.1 | <0.0001 |
| Streptococcus pneumoniae | 13 | 15.0 | 11.0 | 13.0 | 10.3 | 16.0 | 13.0 | 14.0 | 11.0 | <0.0001 |
| Alpha-hemolytic streptococci | 3–5 | 21.0 | 15.0 | 17.9 | 14.6 | 18.9 | 14.1 | 19.4 | 13.6 | <0.0001 |
| Yeast | 27 | 50 | 39 | 56 | 48 | <0.0001 | ||||
| Candida albicans | 12 | 23.9 | 20.8 | 26.5 | 24.4 | <0.05 | ||||
| Candida glabrata | 15 | 79.2 | 68.0 | 65.0 | 54.0 | <0.0001 | ||||
| Total | 108–113 | 16.6 | 13.3 | 14.0 | 12.0 | 15.0 | 12.0 | 14.0 | 11.1 | <0.0001 |
Comparison of BacT/Alert 3D and BacT/Alert Virtuo using 2-way matched ANOVA.
FIG 1.
Relation between time to detection (TTD) in the BacT/Alert Virtuo and the BacT/Alert 3D blood culture systems. Time to detection was evaluated in 379 pairs of blood culture bottles incubated in parallel in the two BacT/Alert blood culture systems. TTD is expressed in hours, and median TTD are indicated by broken lines.
A limitation of this study is the usage of simulated BC. While our results indicate a systematic influence of the BC system across all microorganisms tested (Fig. 1) (r = 0.91; P < 0.001), it cannot be ruled out that the performance might differ in the presence of antibiotics and other inhibitory factors in clinical samples. Also, the effect on anaerobe bacteria and polymicrobial growth were not assessed. In addition, practical integration of the two different BacT/Alert systems into clinical routine and processing time prior to incubation were not considered.
The present results strongly indicate that the Virtuo BC system allows faster detection of pathogens from bloodstream infections, which is of high relevance in clinical microbiological diagnostics. Further controlled clinical studies are warranted to evaluate the performance of the Virtuo system on clinical specimens.
ACKNOWLEDGMENT
We thank bioMérieux for providing the BacT/Alert Virtuo BC system during the study period.
REFERENCES
- 1.Goto M, Al-Hasan MN. 2013. Overall burden of bloodstream infection and nosocomial bloodstream infection in North America and Europe. Clin Microbiol Infect 19:501–509. doi: 10.1111/1469-0691.12195. [DOI] [PubMed] [Google Scholar]
- 2.Angus DC, Linde-Zwirble WT, Lidicker J, Clermont G, Carcillo J, Pinsky MR. 2001. Epidemiology of severe sepsis in the United States: analysis of incidence, outcome, and associated costs of care. Crit Care Med 29:1303–1310. doi: 10.1097/00003246-200107000-00002. [DOI] [PubMed] [Google Scholar]
- 3.Kumar A, Roberts D, Wood KE, Light B, Parrillo JE, Sharma S, Suppes R, Feinstein D, Zanotti S, Taiberg L, Gurka D, Kumar A, Cheang M. 2006. Duration of hypotension before initiation of effective antimicrobial therapy is the critical determinant of survival in human septic shock. Crit Care Med 34:1589–1596. doi: 10.1097/01.CCM.0000217961.75225.E9. [DOI] [PubMed] [Google Scholar]
- 4.Palmer HR, Palavecino EL, Johnson JW, Ohl CA, Williamson JC. 2013. Clinical and microbiological implications of time-to-positivity of blood cultures in patients with Gram-negative bacilli bacteremia. Eur J Clin Microbiol Infect Dis 32:955–959. doi: 10.1007/s10096-013-1833-9. [DOI] [PubMed] [Google Scholar]
- 5.Ferrer R, Martin-Loeches I, Phillips G, Osborn TM, Townsend S, Dellinger RP, Artigas A, Schorr C, Levy MM. 2014. Empiric antibiotic treatment reduces mortality in severe sepsis and septic shock from the first hour: results from a guideline-based performance improvement program. Crit Care Med 42:1749–1755. doi: 10.1097/CCM.0000000000000330. [DOI] [PubMed] [Google Scholar]
- 6.Ericson EL, Klingspor L, Ullberg M, Ozenci V. 2012. Clinical comparison of the Bactec Mycosis IC/F, BacT/Alert FA, and BacT/Alert FN blood culture vials for the detection of candidemia. Diagn Microbiol Infect Dis 73:153–156. doi: 10.1016/j.diagmicrobio.2012.02.020. [DOI] [PubMed] [Google Scholar]
- 7.Jekarl DW, Lee SY, Lee S, Park YJ, Lee J, Baek SM, An YJ, Ock SM, Lee MK. 2012. Comparison of the Bactec Fx Plus, Mycosis IC/F, Mycosis/F Lytic blood culture media and the BacT/Alert 3D FA media for detection of Candida species in seeded blood culture specimens containing therapeutic peak levels of fluconazole. J Clin Lab Anal 26:412–419. doi: 10.1002/jcla.21535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nylén T, Saeedi B, Borg C, Ullberg M, Özenci V. 2013. The performance of 4 different supplements and 5 blood culture bottles types in detection of bacteria and Candida spp. in simulated sterile body fluid cultures. Diagn Microbiol Infect Dis 77:1–4. doi: 10.1016/j.diagmicrobio.2013.05.017. [DOI] [PubMed] [Google Scholar]
- 9.Kirn TJ, Mirrett S, Reller LB, Weinstein MP. 2014. Controlled clinical comparison of BacT/Alert FA Plus and FN Plus blood culture media with BacT/Alert FA and FN blood culture media. J Clin Microbiol 52:839–843. doi: 10.1128/JCM.03063-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mirrett S, Petti CA, Woods CW, Magadia R, Weinstein MP, Reller LB. 2004. Controlled clinical comparison of the BacT/Alert FN and the standard anaerobic SN blood culture medium. J Clin Microbiol 42:4581–4585. doi: 10.1128/JCM.42.10.4581-4585.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weinstein MP. 1996. Current blood culture methods and systems: clinical concepts, technology, and interpretation of results. Clin Infect Dis 23:40–46. doi: 10.1093/clinids/23.1.40. [DOI] [PubMed] [Google Scholar]
- 12.Opota O, Croxatto A, Prod'hom G, Greub G. 2015. Blood culture-based diagnosis of bacteraemia: state of the art. Clin Microbiol Infect 21:313–322. doi: 10.1016/j.cmi.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 13.Almuhayawi M, Altun O, Abdulmajeed AD, Ullberg M, Ozenci V. 2015. The performance of the four anaerobic blood culture bottles BacT/Alert-FN, -FN Plus, Bactec-Plus and -Lytic in detection of anaerobic bacteria and identification by direct MALDI-TOF MS. PLoS One 10:e0142398. doi: 10.1371/journal.pone.0142398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rimmer K, Cabot M. 1988. Comparison of Bactec NR-660 and Signal systems. J Clin Pathol 41:676–678. doi: 10.1136/jcp.41.6.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rogers MS, Oppenheim BA. 1998. The use of continuous monitoring blood culture systems in the diagnosis of catheter related sepsis. J Clin Pathol 51:635–637. doi: 10.1136/jcp.51.8.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stevens CM, Swaine D, Butler C, Carr AH, Weightman A, Catchpole CR, Healing DE, Elliott TS. 1994. Development of o.a.s.i.s., a new automated blood culture system in which detection is based on measurement of bottle headspace pressure changes. J Clin Microbiol 32:1750–1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weinbren MJ, Borthwick MA. 2005. Rapid detection of extended-spectrum beta-lactamase (ESBL)-producing organisms in blood culture. J Antimicrob Chemother 55:131–132. [DOI] [PubMed] [Google Scholar]
- 18.Guinea J. 2014. Global trends in the distribution of Candida species causing candidemia. Clin Microbiol Infect 20(Suppl):S5–S10. [DOI] [PubMed] [Google Scholar]
- 19.Pfaller MA, Diekema DJ. 2007. Epidemiology of invasive candidiasis: a persistent public health problem. Clin Microbiol Rev 20:133–163. doi: 10.1128/CMR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pfaller MA, Diekema DJ, Jones RN, Sader HS, Fluit AC, Hollis RJ, Messer SA, SENTRY Participant Group. 2001. International surveillance of bloodstream infections due to Candida species: frequency of occurrence and in vitro susceptibilities to fluconazole, ravuconazole, and voriconazole of isolates collected from 1997 through 1999 in the SENTRY antimicrobial surveillance program. J Clin Microbiol 39:3254–3259. doi: 10.1128/JCM.39.9.3254-3259.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Horvath LL, George BJ, Murray CK, Harrison LS, Hospenthal DR. 2004. Direct comparison of the BACTEC 9240 and BacT/Alert 3D automated blood culture systems for candida growth detection. J Clin Microbiol 42:115–118. doi: 10.1128/JCM.42.1.115-118.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fernandez J, Erstad BL, Petty W, Nix DE. 2009. Time to positive culture and identification for Candida blood stream infections. Diagn Microbiol Infect Dis 64:402–407. doi: 10.1016/j.diagmicrobio.2009.04.002. [DOI] [PubMed] [Google Scholar]
- 23.Huang L, Zhang YY, Sun LY. 2013. Time to positivity of blood culture can predict different Candida species instead of pathogen concentration in candidemia. Eur J Clin Microbiol Infect Dis 32:917–922. doi: 10.1007/s10096-013-1826-8. [DOI] [PubMed] [Google Scholar]
- 24.Lai CC, Wang CY, Liu WL, Huang YT, Hsueh PR. 2012. Time to positivity of blood cultures of different Candida species causing fungaemia. J Med Microbiol 61:701–704. doi: 10.1099/jmm.0.038166-0. [DOI] [PubMed] [Google Scholar]