Abstract
The Epistem Genedrive assay rapidly detects the Mycobacterium tuberculosis complex from sputum and is currently available for clinical use. However, the analytical and clinical performance of this test has not been fully evaluated. The analytical limit of detection (LOD) of the Genedrive PCR amplification was tested with genomic DNA; the performance of the complete (sample processing plus amplification) system was tested by spiking M. tuberculosis mc26030 cells into distilled water and M. tuberculosis-negative sputum. Specificity was tested using common respiratory pathogens and nontuberculosis mycobacteria. A clinical evaluation enrolled adults with suspected pulmonary tuberculosis, obtained three sputum samples from each participant, and compared the accuracy of the Genedrive to that of the Xpert MTB/RIF assay using M. tuberculosis cultures as the reference standard. The Genedrive assay had an LOD of 1 pg/μl (100 genomic DNA copies/reaction). The LODs of the system were 2.5 × 104 CFU/ml and 2.5 × 105 CFU/ml for cells spiked into water and sputum, respectively. False-positive rpoB probe signals were observed in 3/32 (9.4%) of the negative controls and also in few samples containing Mycobacterium abscessus, Mycobacterium gordonae, or Mycobacterium thermoresistibile. In the clinical study, among 336 analyzed participants, the overall sensitivities for the tuberculosis case detection of Genedrive, Xpert, and smear microscopy were 45.4% (95% confidence interval [CI], 35.2% to 55.8%), 91.8% (95% CI, 84.4% to 96.4%), and 77.3% (95% CI, 67.7% to 85.2%), respectively. The sensitivities of Genedrive and Xpert for the detection of smear-microscopy-negative tuberculosis were 0% (95% CI, 0% to 15.4%) and 68.2% (95% CI, 45.1% to 86.1%), respectively. The Genedrive assay did not meet performance standards recommended by the World Health Organization for a smear microscopy replacement tuberculosis test. Epistem is working on modifications to improve the assay.
INTRODUCTION
Pulmonary tuberculosis (TB) is commonly diagnosed on the basis of patient history, clinical presentation, radiological findings, and sputum smear microscopy in high-TB-burden countries, but these approaches have limited sensitivity and specificity (1, 2). Culture-based diagnosis is more reliable but is labor-intensive, costly, and slow, which results in diagnostic delay even when available (3).
Epistem (Manchester, United Kingdom) developed and recently self-certified for CE-IVD a rapid molecular TB detection test that has the potential to speed up and simplify the diagnosis of pulmonary TB. This test uses a simple paper-based DNA extraction method coupled with PCR amplification and detection on Epistem's Genedrive instrument, a lightweight, portable, bench-top PCR platform with real-time PCR and melting temperature analysis capabilities. Epistem's Genedrive MTB/RIF (Genedrive) assay detects the Mycobacterium tuberculosis complex by targeting two different regions of the M. tuberculosis complex genome, a short repetitive region, rep13E12, and a segment of the rpoB gene (4–6). This test is CE marked for sale in European Economic Area (EEA) member states and is also licensed for distribution in India (7–9). However, this test's performance has not been extensively evaluated or reported in peer-reviewed journals apart from a single study published by the manufacturer and collaborators (6).
We assessed the analytical performance, biosafety, and diagnostic accuracy of the Genedrive assay for detection of M. tuberculosis. In the analytical study, we used DNA and M. tuberculosis cells spiked into buffer or M. tuberculosis-negative human sputum samples. In a multicenter prospective diagnostic accuracy clinical study, adults with suspected pulmonary TB were recruited, and the accuracy of Genedrive was compared to that of Xpert MTB/RIF (Cepheid, Sunnyvale, CA) using a reference standard of mycobacterial culture.
MATERIALS AND METHODS
Ethics statement.
The clinical study was approved by institutional review boards at Johns Hopkins Medicine, Boston University Medical Center, and each local study site. Participants provided written informed consent.
Analytic studies.
The complete protocol of the analytic studies is described in the supplemental material.
Analytic sensitivity.
Initial limit of detection (LOD) studies were performed with genomic M. tuberculosis DNA (ATCC 25618) to test the Genedrive PCR assay and the instrument without using the assay's sample processing cassette. For these studies, 19 μl of nuclease-free water and 1 μl diluted DNA were added to each PCR vial of the Genedrive PCR cartridge as described in the supplemental Materials and Methods. Studies designed to test the sample processing and PCR detection capacity of the Genedrive system used quantified M. tuberculosis mc26030 cells. For studies using cells without spiking into sputum, 20 μl of each dilution was applied at the center of the sample loading cassette. Twenty microliters of cell dilution was also spiked into 480 μl of distilled water and was tested for the presence of M. tuberculosis using the Xpert assay. For spiking studies into sputum, deidentified discarded sputum samples were collected from patients who were not suspected of having TB (10). Thirty-two microliters of homogenized sputum was spiked with 8 μl of the mc26030 strain, where the last dilution was adjusted as needed to obtain the final cell concentration desired. Twenty microliters of this mixture was then applied to the center of the Genedrive sample loading cassette. Eight microliters of each cell dilution was also spiked into 492 μl of sputum and was tested for the presence of M. tuberculosis using the Xpert assay as described in the supplemental Materials and Methods. At least one positive and one negative control were included in each set of experiments.
For DNA and sputum spiking studies, at least 10 samples were tested at each dilution. The highest dilution with 100% positive test results was then identified, after which the test was repeated with the same dilution for a total of at least 10 additional times. If any negative test results were obtained, then the next lowest dilution was tested in the same manner to find the percentage of positivity at that concentration. The lowest dilution with at least 20 test replicates that had 100% positive tests was used to define the limit of detection (LOD).
Analytic specificity.
Sixteen clinically occurring nontuberculosis mycobacterium (NTM) species (Table 1) and 11 Gram-positive and Gram-negative bacterial and fungal species (see Table S1 in the supplemental material) that are among those commonly reported as causes of respiratory infections or that have been identified as commonly isolated respiratory tract colonizers were analyzed at a concentration of 107 CFU/ml. All strains were obtained from either the ATCC or the microbiology laboratory at the New Jersey Medical School, Rutgers Biomedical and Health Sciences, Rutgers University.
TABLE 1.
Analytical specificity for nontuberculous mycobacteria panel
| NTM species | ATCC no. | Genedrive assay |
||
|---|---|---|---|---|
| rep13E12 target | rpoB target | Final result | ||
| M. abscessus | 19977 | Negative | 1/7 Positive | MTB detected Low |
| M. asiaticum | 25274 | Negative | Negative | Undetected |
| M. avium | 15769 | Negative | Negative | Undetected |
| M. celatum | 51131 | Negative | Negative | Undetected |
| M. chelonae | 35749 | Negative | Negative | Undetected |
| M. fortuitum | 35754 | Negative | Negative | Undetected |
| M. gordonae | 14470 | Negative | 2/6 Positive | MTB detected Low |
| M. intracellulare | 35771 | Negative | Negative | Undetected |
| M. kansasii | 12478 | Negative | Negative | Undetected |
| M. malmoense | 29571 | Negative | Negative | Undetected |
| M. scrofulaceum | 19073 | Negative | Negative | Undetected |
| M. smegmatis | Mc2155 | Negative | Negative | Undetected |
| M. simiae | 25275 | Negative | Negative | Undetected |
| M. szulgai | 23069 | Negative | Negative | Undetected |
| M. thermoresistibile | 19527 | Negative | 1/3 Positive | MTB detected Low |
| M. xenopi | 19250 | Negative | Negative | Undetected |
Biosafety studies.
We also studied the extent to which M. tuberculosis present in sputum samples was killed using the Genedrive DNA extraction cassette. Different concentrations (107 to 105) of M. tuberculosis mc26030 cells were applied onto the sample processing cassette directly or were first spiked into TB-negative sputum specimens and then applied to the cassette. The cassettes were allowed to stand at room temperature for 20 min. A 1 cm by 1 cm area of the membrane where the sample had been spotted in the center was cut out using a sterile scalpel and was inoculated into Bactec MGIT vials. Three different punches (1 mm by 1 mm in size) were also cut from the center of the membrane using a sterile disposable biopsy punch and were inoculated into MGIT vials. Similarly, 500 μl of 7H9 medium was added to the center of the sample loading cassette, the membrane was washed twice, and the membrane washings were inoculated in MGIT vials. All inoculated vials were incubated in a Bactec 320 MGIT instrument until they were flagged as positive by the instrument or remained negative for 42 days. Positive controls (M. tuberculosis mc26030 cultures of various concentrations with and without plain membrane) and negative controls (MGIT medium plus supplements plus panD) were incorporated in the study. Confirmation of M. tuberculosis growth in selected positive MGIT vials was performed by testing 1 ml of the culture using the Xpert assay.
Diagnostic accuracy clinical study. (i) Design, setting, and participants.
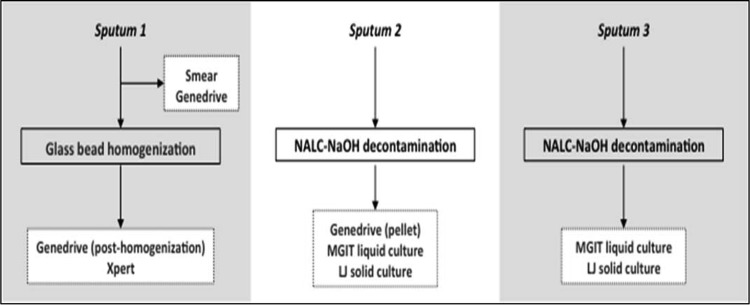
A multicenter, cross-sectional, blinded diagnostic accuracy study was conducted. Participants were enrolled at outpatient clinics in Brazil, South Africa, and Uganda; hospitalized individuals were also enrolled in Uganda. Key inclusion criteria included an age of ≥18 years, a cough for ≥2 weeks, and one or more of the following: fever, night sweats, or weight loss. Individuals were excluded if they had received ≥2 days of anti-TB treatment within the prior 60 days. Each study participant was asked to provide a total of two sputum samples over days 1 and 2; a third sputum sample was collected within 7 days after enrollment (Fig. 1).
FIG 1.
Specimen processing and testing schema for the prospective multicenter diagnostic accuracy clinical study. Each participant provided 3 sputum samples. Sputum 1 was used to perform direct smear microscopy and the Genedrive assay before being homogenized with glass beads. After homogenization, it was split into 2 portions for testing with the Genedrive assay and the Xpert MTB/RIF assay. Sputum 2 was decontaminated by the standard NALC-NaOH method, and then the resuspended pellet was used to perform liquid MGIT and solid Lowenstein-Jensen (LJ) cultures and the Genedrive assay. Sputum 3 was decontaminated by the standard NALC-NaOH method, and then the resuspended pellet was used to perform MGIT and LJ cultures.
(ii) Mycobacteriology testing.
Tests were performed at designated on-site labs, each of which had a quality assurance program in place. Lab staff was trained in Genedrive procedures by the manufacturer, and all laboratory tests were performed according to the manufacturer's protocol that is described in the supplemental material. Briefly, 20 μl of direct or processed sputum sample was placed on the Genedrive sample processing cassette and was allowed to dry at room temperature to extract nucleic acids from each sample. A 1-mm sterile disposable biopsy punch (Miltex, NY, USA) was used to cut 3 discs from the center of the cassette, and each disc was added individually to each of the 3 PCR vials of the Genedrive PCR cartridge. The sealed PCR cartridge was inserted in the Genedrive instrument, and the assay was run as per the manufacturer's instruction. Laboratory technologists performing Genedrive tests were blinded to the results of other tests for that specimen or participant. For each participant, the first sputum sample (sputum 1) was tested as a direct sample by fluorescence smear microscopy and Genedrive; the remaining portion of the sputum sample was then homogenized with 3-mm glass beads (Fisher Scientific, USA) and was tested using Xpert and Genedrive (Fig. 1). The second sputum sample (sputum 2) was digested and decontaminated using N-acetyl-l-cysteine (NALC)-NaOH (11). The resulting pellet was resuspended in 1.5 ml phosphate-buffered saline, mixed, and used for Genedrive (∼50 μl), Lowenstein-Jensen culture (LJ; 200 μl inoculum), and MGIT culture (500 μl inoculum). Sputum sample 3 was digested and decontaminated using NALC-NaOH; the pellet was resuspended as above and was used to inoculate an LJ culture and an MGIT culture. Genedrive and Xpert testing were performed per the manufacturer's instructions. A participant was considered to be culture positive if any study culture was positive for growth of M. tuberculosis and culture negative if no culture grew M. tuberculosis. All cultures growing NTM or other bacterial contaminants were excluded from the data analysis. Similarly, specimens with missing results for any of the smear microscopy, culture, Genedrive, or Xpert assays were excluded from the analysis.
(iii) Statistical analysis.
The LOD for the assay was analyzed by determining the percentage of positive tests for each concentration of M. tuberculosis isolates tested. The sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the Genedrive assay with 95% confidence intervals (CIs) were estimated for prehomogenized and posthomogenized sputum specimens using a reference standard of 4 valid culture results using online statistical software MedCalc, SigmaPlot (7.0), and Stata (Stata Corp., TX). Similarly, the sensitivities, specificities, PPVs, and NPVs of the Xpert assay and the acid-fast bacilli (AFB) smear were also calculated against culture standard. Additional secondary outcomes were sensitivity and specificity of Genedrive performed on raw sputum using Xpert as the reference standard.
RESULTS
Performance of Genedrive PCR for detection of M. tuberculosis genomic DNA.
The Genedrive PCR assay reproducibly detected as little as 1 pg of DNA per reaction, which is equivalent to approximately 100 genomic copies per reaction (Fig. 2A). One hundred femtograms of DNA (approximately 10 genomic copies) was detected with 45% positivity, and 10 fg of DNA (approximately 1 genomic copy) was detected with 8% positivity.
FIG 2.
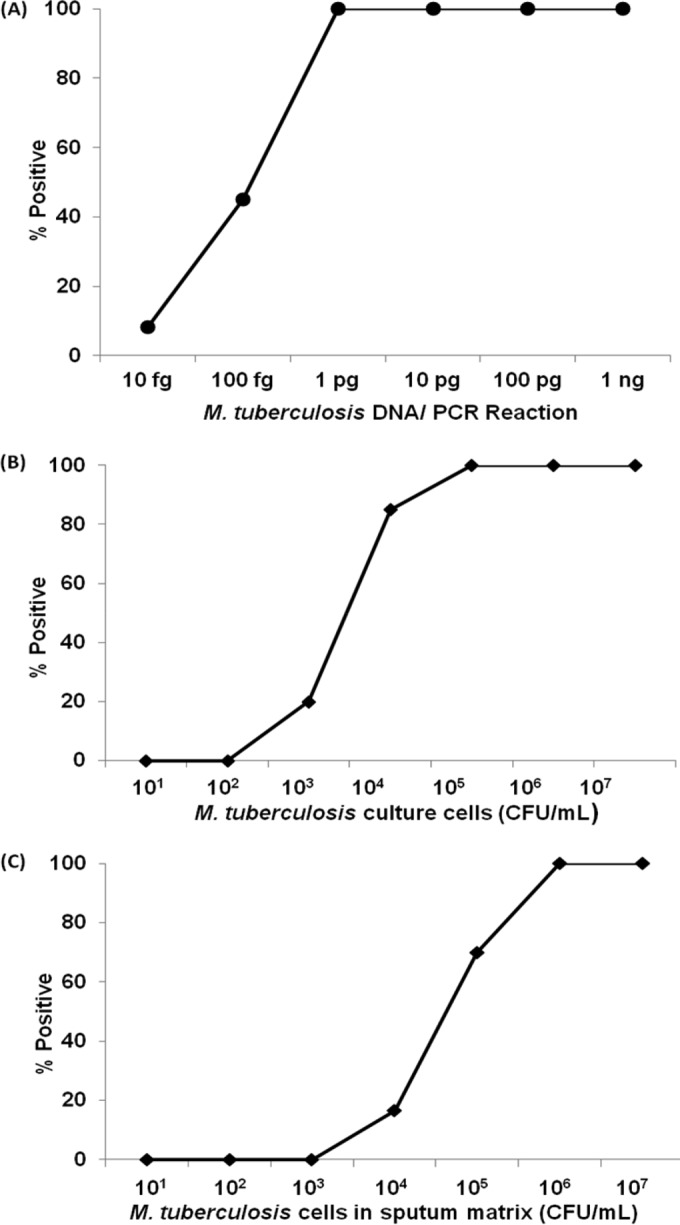
Limit of detection (LOD) of the Genedrive assay for detection of M. tuberculosis. LOD of Genedrive assay using (A) M. tuberculosis DNA. M. tuberculosis DNA at final concentration of 10 fg, 1pg, 10 pg, 100 pg, and 1 ng per PCR vial was loaded into Genedrive PCR cartridges and was processed according to the Genedrive protocol. (B) M. tuberculosis H37Rv mc26030 quantified culture cells in water. M. tuberculosis mc26030 quantified cells diluted in sterile water were tested by the Genedrive assay at final concentrations ranging from 101 to 107 CFU/ml (n = 15); the percentage of M. tuberculosis positive results was plotted for each dilution. (C) M. tuberculosis H37Rv mc26030 quantified culture cells in Sputum. M. tuberculosis mc26030 cells were added to TB negative sputum to final concentrations of 101 to 107 CFU/ml (n = 20) and were then processed according to the Genedrive assay protocol. The percentage of assays where M. tuberculosis was detected was then plotted for each concentration of cells.
Dynamic range and LOD of M. tuberculosis cell dilutions spiked into water.
M. tuberculosis cells were detected 100% of the time tested down to dilutions of 2.5 × 104 CFU/ml. This defined the Genedrive assay LOD for M. tuberculosis cells in a water matrix. The assay also detected 1 × 104 CFU/ml in 85% of the tests and 1 × 103 CFU/ml in 20% of the tests (Fig. 2B). As a comparator, tests performed with the Xpert assay on the same 20-μl sample volume of each concentration (brought up to a larger test volume with buffer as described in the Materials and Methods) detected 1 × 104 CFU/ml in 100% of tests and detected 1 × 103 CFU/ml in 60% of tests. We also tested the LOD of the Xpert assay using a 1-ml sample of each dilution (instead of the 20-μl sample of each dilution tested above) since this is the normal test volume for the Xpert assay. Using this normal test approach, the Xpert assay detected 250 CFU/ml in 100% of tests and detected 100 CFU/ml in 83.3% of tests, which is consistent with the previously reported Xpert LOD (10).
LOD of M. tuberculosis cell dilutions spiked into sputum.
We next tested the LOD of the Genedrive using known M. tuberculosis-negative clinical sputum specimens spiked with known numbers of M. tuberculosis cells. The assay correctly identified M. tuberculosis in sputum samples containing 107 and 106 CFU/ml and 5 × 105 CFU/ml in 100% of all tests, defining the LOD in sputum as 5 × 105 CFU/ml (Fig. 2C). The assay also detected 2.5 × 105 CFU/ml in 90% of tests, 1 × 105 CFU/ml in 70% of tests, and 1 × 104 CFU/ml in 16.7% of tests. The Xpert assay tested with the same volume (8 μl) of 5 × 104 CFU/ml cell dilution spiked into 492 μl of sputum (i.e., approximately 400 CFU per Xpert cartridge) detected TB in 100% of tests and the 5 × 103 CFU/ml dilution (i.e., approximately 40 CFU per Xpert cartridge) in 91.7% of tests. Using 1-ml samples of each dilution, the Xpert assay showed an LOD of 250 CFU/ml in 100% of tests.
Cross-reactivity with NTM and other bacterial species.
The rep13E12-targeting portion of the Genedrive assay did not show any cross-reactivity with nonmycobacterial respiratory pathogens or bacteria (Table 1; see also Table S1 in the supplemental material). However, false-positive M. tuberculosis results were observed due to a melting temperature (Tm) shift in the rpoB probe of three NTM species, Mycobacterium abscessus, Mycobacterium thermoresistibile, and Mycobacterium gordonae (Table 1; see also Fig. S1a to d in the supplemental material). False positive reports due to Tm shifts of the rpoB probe were also observed in 3 of 32 (9.4%) no-target or negative-control tests, of which one was in sputum matrix and two were with nonsputum negative controls (see Fig. S2 in the supplemental material).
Biosafety.
M. tuberculosis growth occurred in all of the MGIT vials inoculated with an entire membrane, membrane washings, or three punches with cell concentrations ranging from 107 to 105 cells/ml (Table 2). Only the negative controls were culture negative. MGIT vials inoculated with the highest bacterial load, i.e., 107 cells/ml, took less time (6 to 8 days) to turn positive than the MGIT vials with the lowest load, i.e., 105 cells/ml of membrane (10 to 25 days).
TABLE 2.
Biosafety experiment: ability to detect viability of M. tuberculosis after paper-based DNA extraction
| Inoculum (CFU/ml) | Amount of material inoculated into MGIT viala | MGIT culture | Time to positivity | Bacterial load/MGIT vial | Confirmation (CT)b |
|---|---|---|---|---|---|
| 107 | 3 punchesc (replicate a) | Positive | 7 days, 1 h | 3 × 104 | Xpert MTB (17.8) |
| 107 | 3 punchesc (replicate b) | Positive | 8 days, 10 h | 3 × 104 | |
| 107 | Entire membrane (1 by 1 cm) | Positive | 6 days, 1 h | 1 × 105 | |
| 107 | Washings from SLCd | Positive | 8 days, 10 h | ||
| 107 | 20-μl culture + plain membrane | Positive | 4 days, 10 h | 2 × 105 | |
| 107 | 20-μl culture (positive control) | Positive | 5 days | 2 × 105 | |
| 106 | 3 punchesc (replicate a) | Positive | 9 days, 13 h | 3 × 103 | Xpert MTB (22.3) |
| 106 | 3 punchesc (replicate b) | Positive | 12 days, 7 h | 3 × 103 | |
| 106 | Entire membrane (1 by 1 cm) | Positive | 7 days, 17 h | 1 × 104 | Xpert MTB (18.2) |
| 106 | Washings from SLC | Positive | 9 days, 9 h | ||
| 106 | 20-μl culture + plain membrane | Positive | 7 days | 2 × 104 | |
| 105 | 3 punchesc (replicate a) | Positive | 25 days, 1 h | 3 × 102 | |
| 105 | 3 punchesc (replicate b) | Positive | 25 days, 13 h | 3 × 102 | |
| 105 | Entire membrane (1 cm by 1 cm) | Positive | 10 days, 11 h | 1 × 103 | Xpert MTB (20.1) |
| 105 | 20 μl Culture (positive control) | Positive | 7 days, 17 h | 2 × 103 | |
| Negative control (replicate a) | Negative | No growth at the end of 42 days | |||
| Negative control (replicate b) | Negative | ||||
Experiments conducted in duplicate are labeled as replicate a and b.
CT, Xpert cycle threshold defined as the number of cycles required for the fluorescent signal to cross the threshold (i.e., exceed background level).
Three punches were cut from the center of the Genedrive sample loading cassette and inoculated into the MGIT vial.
SLC, sample loading cassette.
Characteristics of participants in the clinical study.
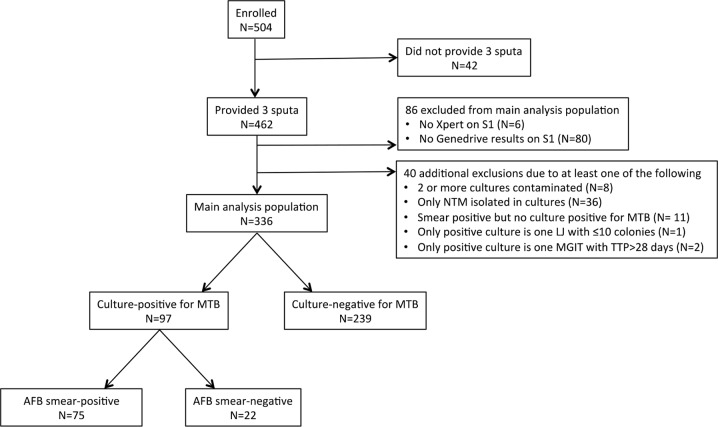
Between August 2014 and December 2014, 504 participants were enrolled. A total of 42 participants did not submit three sputum samples and were therefore excluded from the analysis (Fig. 3). An additional 80 participants did not have a Genedrive result for sputum 1, and 6 participants did not have an Xpert result for sputum 1. Forty participants were excluded from the analysis for other reasons per the protocol (Fig. 3). The resulting 336 participants comprised the main analysis population; 120 (36%) were enrolled in Brazil, 121 (36%) in Uganda, and 95 (28%) in South Africa. Of these patients, 202/336 (60%) were male, 60/336 (18%) were documented to be HIV-positive, and 120/336 (36%) had cavitation on chest radiograph (see Table S2 in the supplemental material). Among 336 analyzed participants, 97 (28.9%) had at least one culture that was positive for M. tuberculosis.
FIG 3.
Overview of patient enrollment and study outcomes. A total 504 suspected pulmonary TB patients were recruited in this study, of which 42 patients were unable to produce sputum specimens. Of the remaining 462, 80 participants did not have a determinate Genedrive result on sputum 1 (12 with test result of “error” and 68 with Genedrive testing not performed due to instrument inoperability or no intact cartridges at the testing site), and 6 participants did not have a determinate Xpert result for sputum 1 (2 with test result of ‘error’ and 4 with Xpert testing not performed due to human error) and hence were excluded from the study. Of the 336 participants who completed this study, 97 were culture positive and 239 were culture negative. Among culture positives, 75 were AFB positive and 22 were AFB negative by smear microscopy. S1, sputum 1; NTM, nontuberculous mycobacteria; MTB, M. tuberculosis; LJ, Lowenstein-Jensen culture; MGIT, mycobacterial growth indicator tube culture; TTP, time to positivity; AFB, acid-fast bacilli.
Clinical performance of single rapid tests versus culture.
Using culture as a reference standard, the sensitivity for TB case detection of a single Genedrive assay prehomogenization (44/97 [45.4%]) was lower than that of Xpert (89/97 [91.8%]) (Table 3). The sensitivity of Genedrive performed prehomogenization was also lower than that of smear microscopy (75/97 [77.3%]) for a difference favoring smear microscopy of 32.0 percentage points (95% CI, 22.5 to 41.4). Genedrive failed to detect M. tuberculosis in any of 22 smear-negative/culture-positive specimens and detected M. tuberculosis in only 44/75 (58.7%) smear-positive/culture-positive specimens (Table 3). There was no difference in specificity between Genedrive performed prehomogenization (232/239 [97.5%]) and Xpert (234/239 [97.9%]).
TABLE 3.
Diagnostic accuracy clinical study: accuracy of single rapid tests using as a gold standard four cultures performed from two sputum samples
| Prospective diagnostic accuracy | Compared to 4 culture results (2 MGIT, 2 LJ) performed on 2 sputum specimens |
AFB Sm+/Cx+, Sensitivity (n/n [%] [95% CI])a | AFB Sm−/Cx+, Sensitivity (n/n [%] [95% CI]) | |||
|---|---|---|---|---|---|---|
| Sensitivity (n/n [%] [95% CI]) | Specificity (n/n [%] [95% CI]) | PPV (n/n [%] [95% CI]) | NPV (n/n [%] [95% CI]) | |||
| Sputum 1 result vs culture gold standard (n = 336 participants) | ||||||
| Genedrive, prehomogenization, sputum 1 | 44/97 (45.4) (35.2–55.8) | 233/239 (97.5) (94.6–99.1) | 44/50 (88.0) (75.7–95.5) | 233/286 (81.5) (75.7–85.8) | 44/75 (58.7) (46.7–70.0) | 0/22 (0) (0–15.4) |
| Genedrive, posthomogenization, sputum 1 | 57/97 (58.8) (48.3–68.7) | 233/239 (97.5) (94.6–99.1) | 57/63 (90.5) (80.4–96.4) | 233/273 (85.3) (80.6–89.3) | 56/75 (74.7) (63.3–84.0) | 1/22 (4.5) (0.0–22.8) |
| Xpert MTB/RIF, sputum 1 | 89/97 (91.8) (84.4–96.4) | 234/239 (97.9) (95.2–99.3) | 89/94 (94.7) (88.0–98.3) | 234/242 (96.7) (93.6–98.6) | 74/75 (98.7) (92.8–99.9) | 15/22 (68.2) (45.1–86.1) |
| AFB smear microscopy, sputum 1 | 75/97 (77.3) (67.7–85.2) | 239/239 (100) (98.5–100) | 75/75 (100) (95.1–100) | 239/261 (91.6) (87.5–94.6) | N/A | N/A |
| Sputum 2 result vs culture gold standard (n = 326 participants) | ||||||
| Genedrive, pellet, sputum 2 | 64/95 (67.4) (57.0–76.6) | 225/231 (97.4) (94.4–99.0) | 64/70 (97.0) (81.5–96.6) | 225/256 (87.9) (83.3–91.6) | 63/74 (85.14) (74.9–92.3) | 1/21 (4.7) (0–23.8) |
Cx, culture; Sm, smear; N/A, not available.
Compared with the sensitivity of Genedrive performed prehomogenization on sputum 1 (44/97 [45.4%]), the sensitivities were higher for Genedrive performed posthomogenization on sputum 1 (57/97 [58.8%]; P = 0.004) and for Genedrive performed on the sputum 2 pellet (64/95 [67.4%]; P < 0.001). However, the sensitivity for TB case detection of a single Genedrive assay performed on the sputum 2 pellet remained lower than that of Xpert (a difference favoring Xpert of 24.4 percentage points [95% CI, 15.8 to 34.6]) and also significantly different from that of smear microscopy (difference favoring smear microscopy of 9.9 percentage points [95% CI, 3.5 to 24.4]).
DISCUSSION
We report the first comprehensive evaluation of the Epistem Genedrive assay for detection of M. tuberculosis. The sensitivity of Genedrive was low compared to other methods, and this finding was consistent across the analytic studies using sputum spiked with M. tuberculosis and a clinical study using fresh sputum from adults with signs/symptoms of pulmonary TB. Analytic study results indicated that the observed suboptimal sensitivity can be traced to the sample processing component of the assay. The amplification portion of the assay appeared to perform well, with an LOD of 100 genomes when tested with purified M. tuberculosis DNA that did not require any additional sample processing. Therefore, it is likely that the assay's limited performance is due to the relatively small amount of sample that is processed on the paper cassette and then placed into the assay amplification vial. Insufficient cell lysis or insufficient removal of PCR inhibitors from sputum by the cassette may also adversely affect sensitivity, although we did not specifically study these parameters. Our results differ markedly from the only other paper published on this assay to date (6).
The Genedrive assay did not show any positive results when tested with common Gram-positive and Gram-negative respiratory pathogens. However, there were false-positive rpoB peaks when tested with M. abscessus, M. gordonae, and M. thermoresistibile. False-positive results for M. tuberculosis detection were also observed in 9% of negative controls, although the reasons are unclear. A test design concern is that the assay internal control is run in a different assay tube than the two M. tuberculosis detection tubes. This control format may make it difficult to detect tube-specific reagent issues or uneven distribution of PCR inhibitors from the test sample.
The analytical component of our study demonstrated that the Genedrive sample-processing cassette did not result in a reduction in viable M. tuberculosis CFU sufficient to make the sample biosafe. At least a six-log reduction in viability is generally required to consider that a procedure has sterilizing ability (12, 13). We recommend that biosafety level 3 (BSL3) lab facilities or at least a BSL2 facility with a biosafety-2 cabinet be used to process samples for this assay until additional work can be done to confirm that assay procedures are biosafe. Sample processing cassettes should be treated with 20% bleach or another acceptable decontaminant for 20 to 30 min before being discarded.
Our analytic LOD study was critically dependent on proper mixing and dispersion of the spiked M. tuberculosis cells into the test matrix. Only a small (20 μl) volume of this matrix was applied to the processing cassette, and even smaller portions (two small punches) were placed into the assay amplification tubes. Therefore, poor mixing of the sample may have resulted in some 20-μl aliquots having unexpectedly low numbers of spiked M. tuberculosis cells. However, our procedures addressed this issue as much as possible by vortexing and sonicating samples for homogenization. Furthermore, the Xpert assay was able to detect M. tuberculosis from dilution aliquots containing as little as 103 CFU/ml when we placed the same volume of cell suspension (20 μl) in the Xpert and on the Genedrive sample processing cassette. This parallel study with the Xpert assay demonstrated that M. tuberculosis was detectable even from the 20-μl aliquots.
Findings from the diagnostic accuracy clinical study confirmed the low sensitivity observed in the analytic study. Genedrive sensitivity was inferior to that of Xpert overall and was also substantially lower than that of smear microscopy. Genedrive's sensitivity was well below minimum targets (60% for smear-negative/culture-positive TB and 99% for smear-positive/culture-positive TB) recommended by the World Health Organization for high-priority new TB diagnostics (14). It is worth noting, however, that the observed Genedrive performance generally met or exceeded the WHO targets with respect to specificity, number of assay steps, time to result, and instrument and power requirements.
In the clinical study, the sensitivity of Genedrive was lower in direct sputum specimens tested before homogenization versus those tested after homogenization and was highest when NALC-NaOH-processed sediment was used. This is likely a consequence of heterogeneous distribution of bacilli in nonmanipulated sputum, a challenge also encountered in the analytic study. While it is encouraging that sensitivity can be increased through sputum manipulation, a requirement for sputum manipulation would nevertheless be a disadvantage in practice and limit the test's use at point-of-care. A limitation of the current study is that we did not assess the ability of the Genedrive assay to detect genotypic changes associated with rifampin resistance.
In summary, results from analytical and clinical studies showed low Genedrive sensitivity for the detection of M. tuberculosis. There was cross-reactivity with three nontuberculosis mycobacterial species, and the sample-processing cassette did not result in a reduction in viable M. tuberculosis CFU sufficient to make the sample biosafe. The Genedrive assay is undergoing further development, and the test developers are aiming to address the different issues highlighted in this work.
Supplementary Material
ACKNOWLEDGMENTS
We thank Epistem Holdings Plc for donation of instruments and supplies for the Genedrive assay. We thank the clinical study participants, as well as the TB-CDRC Data Coordinating Center.
In addition to the authors, the contributions of the following individuals are gratefully acknowledged: Carolyn Namaganda, Eric Bugumirwa, and Carol Kayiza (Infectious Disease Institute and Makerere University, Kampala, Uganda); Luiz Guilherme Schmidt, Paolo Poloni, and Pedro Sousa (Nucleo de Doencas Infecciosas, Universidade Federal do Espirito Santo, Vitoria, Brazil); Slindile Mbele, Layla Hendricks, Michelle Mento, and Judi van Heerden (University of Cape Town, Cape Town, South Africa); and David Hom and Rachel Kubiak (Boston University Medical Center, Boston, Massachusetts, USA).
This work was supported by the Foundation for Innovative New Diagnostics and also by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services under contract HHSN2722000900050C, “TB Clinical Diagnostics Research Consortium.” Additional support was provided by NIH to S.E.D. under contract K24AI104830.
D.A. is one of a group of coinvestigators who invented molecular beacon technology and who receive income from licensees, including a license to Cepheid for M. tuberculosis detection. To manage this conflict of interest, the income attributable to the Xpert MTB/RIF assay, which he may receive, has been irrevocably capped at $4,999 per year. D.A. also reports receiving two research contracts from Cepheid. E.V., C.M.D., D.L.D., and C.B. are employed by FIND, a nonprofit organization that collaborates with industry partners, including BD, Cepheid, and Hain Lifesciences among others for the development, evaluation, and demonstration of new diagnostic tests for poverty-related diseases. M.P.N. and the University of Cape Town have received funding from Epistem (subsequent to the conduct of the study described here) to test a modified version of the assay. The remaining authors declare no conflict of interest.
Funding Statement
This work was supported by the Foundation for Innovative New Diagnostics and also by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, Department of Health and Human Services, under contract no. HHSN2722000900050C, “TB Clinical Diagnostics Research Consortium.” Additional support was provided by NIH K24AI104830 to S.E.D.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02847-15.
REFERENCES
- 1.Hobby GL, Holman AP, Iseman MD, Jones JM. 1973. Enumeration of tubercle bacilli in sputum of patients with pulmonary tuberculosis. Antimicrob Agents Chemother 4:94–104. doi: 10.1128/AAC.4.2.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Harries AD, Dye C. 2006. Tuberculosis. Ann Trop Med Parasitol 100:415–431. [DOI] [PubMed] [Google Scholar]
- 3.Urbanczik R, Rieder HL. 2009. Scaling up tuberculosis culture services: a precautionary note. Int J Tuberc Lung Dis 13:799–800. [PubMed] [Google Scholar]
- 4.Lee TY, Lee TJ, Belisle JT, Brennan PJ, Kim SK. 1997. A novel repeat sequence specific to Mycobacterium tuberculosis complex and its implications. Tuber Lung Dis 78:13–19. doi: 10.1016/S0962-8479(97)90011-3. [DOI] [PubMed] [Google Scholar]
- 5.Gordon SV, Heym B, Parkhill J, Barrell B, Cole ST. 1999. New insertion sequences and a novel repeated sequence in the genome of Mycobacterium tuberculosis H37Rv. Microbiology 145:881–892. [DOI] [PubMed] [Google Scholar]
- 6.Castan P, de Pablo A, Fernandez-Romero N, Rubio JM, Cobb BD, Mingorance J, Toro C. 2014. Point-of-care system for detection of Mycobacterium tuberculosis and rifampin resistance in sputum samples. J Clin Microbiol 52:502–507. doi: 10.1128/JCM.02209-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Epistem. 2012. Epistem press release. Epistem signs tuberculosis channel partner agreement. Epistem Plc., Manchester, UK: http://www.epistem.co.uk/press-releases/EpistemPressReleaseTBCollaborationAgreement05Mar12.pdf. [Google Scholar]
- 8.DDNews. August 2011. Epistem and Xcelris Labs partner on TB diagnostics. DDNews, Rocky River, OH: http://www.ddn-news.com/index.php?newsarticle=5251. [Google Scholar]
- 9.Labmate. 15 May 2015. India approves licence for TB diagnosis. Labmate, Hertfordshire, UK: http://www.labmate-online.com/news/news-and-views/5/epistem_ltd/india_approves_licence_for_tb_diagnosis/34643/. [Google Scholar]
- 10.Helb D, Jones M, Story E, Boehme C, Wallace E, Ho K, Kop J, Owens MR, Rodgers R, Banada P, Safi H, Blakemore R, Lan NT, Jones-Lopez EC, Levi M, Burday M, Ayakaka I, Mugerwa RD, McMillan B, Winn-Deen E, Christel L, Dailey P, Perkins MD, Persing DH, Alland D. 2010. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol 48:229–237. doi: 10.1128/JCM.01463-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kent PT, Kubica GP. 1985. Public health mycobacteriology, a guide for level III laboratory. Centers for Disease Control, Atlanta, GA. [Google Scholar]
- 12.Rutala WA, Weber DJ. 2001. New disinfection and sterilization methods. Emerg Infect Dis 7:348–353. doi: 10.3201/eid0702.010241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Association for the Advancement of Medical Instrumentation. 2003. Sterilization of health care products: requirements for products labled “sterile.” Standard ANSI/AAMI ST67. Association for the Advancement of Medical Instrumentation, Arlington, VA. [Google Scholar]
- 14.World Health Organization. 2014. High-priority target product profiles for new tuberculosis diagnostics: report of a consensus meeting. In Proceedings of the WHO/HTM/TB/2014.18. WHO, Geneva, Switzerland. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.