Abstract
Quantification of HIV-1 RNA has become the standard of care in the clinical management of HIV-1-infected individuals. The objective of this study was to evaluate performance characteristics and relative workflow of the Aptima HIV-1 Quant Dx assay in comparison with the Abbott RealTime HIV-1 assay using plasma and cervicovaginal lavage (CVL) specimens. Assay performance was evaluated by using an AcroMetrix HIV-1 panel, AcroMetrix positive controls, Qnostics and SeraCare HIV-1 evaluation panels, 208 clinical plasma samples, and 205 matched CVL specimens on the Panther and m2000 platforms. The Aptima assay demonstrated good linearity over the quantification range tested (2 to 5 log10 copies/ml), and there was strong linear correlation between the assays (R2 = 0.99), with a comparable coefficient of variance of <5.5%. For the plasma samples, Deming regression analyses and Bland-Altman plots showed excellent agreement between the assays, with an interassay concordance of 91.35% (kappa = 0.75; 95% confidence interval [CI], 0.65 to 0.85), and on average, the viral loads determined by the Aptima assay were 0.21 log10 copies/ml higher than those determined by the RealTime assay. The assays differed in their sensitivity for quantifying HIV-1 RNA loads in CVL samples, with the Aptima and RealTime assays detecting 30% and 20%, respectively. Aptima had fewer invalid results, and on average, the viral loads in CVL samples quantified by the Aptima assay were 0.072 log10 copies/ml higher than those of the RealTime assay. Our results demonstrate that the Aptima assay is sensitive and accurate in quantifying viral loads in both plasma and CVL specimens and that the fully automated Panther system has all the necessary features suitable for clinical laboratories demanding high-throughput sample processing.
INTRODUCTION
Quantitation of HIV-1 RNA has been an established surrogate marker for monitoring viral suppression and drug efficacy during antiretroviral therapy (ART) in HIV-infected patients. Early diagnosis during the acute phase of infection represents a tremendous opportunity for treatment and prevention interventions, as the HIV-1 concentration in both plasma and genital fluids is intensely high during this phase (1, 2). It is apparent from previous studies that plasma viral load has been an important determinant in detecting HIV-1 in the genital tract (3–5). RNA levels in cervicovaginal lavage (CVL) specimens are usually lower than plasma HIV-1 RNA loads; thus, HIV-1 RNA suppression in the genital tract may occur more rapidly than in plasma after initiation of therapy (6). Nevertheless, more recent studies demonstrate that genital HIV-1 RNA shedding occurs in women on highly active antiretroviral therapy (HAART) even in the absence of plasma viremia (7, 8). Therefore, an HIV-1 assay that accurately quantifies both genital and plasma RNA levels is a powerful research tool to better understand HIV-1 pathogenesis in the genital tract. Measurement of HIV-1 loads in CVL samples is challenging due to the presence of inhibitors of amplification and the viscous nature of the specimens. The Boom extraction method has been used successfully (5, 9), but it would be very helpful if CVL specimens could be tested on a fully automated real-time PCR system.
Multiple PCR-based assays designed to monitor HIV-1 loads are currently available to monitor the effectiveness of antiretroviral therapy in conjunction with clinical presentation and other laboratory markers. Automated or semiautomated instruments offer standardized processing technology for specimen extraction, specimen amplification, and detection of molecular targets. Although the real-time PCR methods introduced in recent years have improved the sensitivity and precision of HIV-1 RNA assays, there is little experience using these assays with CVL specimens. The RealTime test (Abbott Molecular) has been adopted for both dried blood spot (DBS) and CVL specimens (10, 11). These studies demonstrated that the assay provided good sensitivity and reliability, especially for DBSs, with a limit of detection (LOD) estimated to be 550 copies/ml. The Hologic Aptima HIV-1 Quant Dx assay (Conformité Européenne In Vitro Diagnostics [CE-IVD]) (not U.S. Food and Drug Administration [FDA] approved at this time) is a real-time assay that is able to quantify viral loads for HIV-1 groups M (and subtypes), O, and N in plasma with equal efficiencies, with a precision of ≤0.25 log copies/ml and a lower limit of quantification (LLOQ) of 30 copies/ml. The assay is based on transcription-mediated amplification (TMA) technology that uses a dual-target approach against highly conserved regions in the HIV genome (12). It is also the first HIV-1 load assay with a dual claim for both diagnosis and treatment monitoring (CE-IVD) using plasma and serum.
The objective of this study was to evaluate the performance characteristics and relative workflow of the Aptima HIV-1 Quant Dx assay in comparison with the Abbott RealTime HIV-1 assay using plasma and CVL specimens.
MATERIALS AND METHODS
Clinical samples.
Two hundred eight EDTA plasma samples from 48 patients were selected from samples originally collected for two research projects between September 2003 and September 2008. Plasma specimens were aliquoted and stored at −80°C. Patients with 2 to 10 longitudinal samples were chosen to represent a wide spectrum of HIV concentrations (28 samples with peak plasma viral load levels of <80 copies/ml, 78 samples with 50 to 2,000 copies/ml, 36 samples with 2,000 to 10,000 copies/ml, 52 samples with 10,000 to 100,000 copies/ml, and 14 samples with >100,000 copies/ml) based on viral load data from previously reported studies (13, 14). Two hundred five matched CVL samples from the 48 patients were selected for the study as well. CVL specimens were collected by gently washing the cervicovaginal area with 10 ml of sterile normal saline (pH 7.2) and aspirating the saline pooling in the posterior fornix. Following CVL specimen collection, samples were aliquoted and stored at −80°C. On the day of the assay, samples were thawed to room temperature, vortexed, and briefly spun down at 5,000 rpm for 2 min, after which the samples were assayed. Both plasma and CVL specimens were stored at −70°C to −80°C in a controlled environment with a backup generator and an alarm notification system for out-of-range temperatures. The study was approved by the Institutional Review Board of The Miriam Hospital.
HIV-1 RNA load assays.
The Aptima HIV-1 Quant Dx assay (Hologic, San Diego, CA) was performed according to the manufacturer's recommendations. The Aptima assay is a TMA-based assay performed on the fully automated Panther system; the assay targets the HIV-1 Pol and long terminal repeat (LTR) regions and is able to quantify with equal efficiency HIV-1 groups M (and subtypes), O, and N, with an LLOQ of 30 copies/ml and a linear range from 30 copies/ml to 10,000,000 copies/ml, using a 0.5-ml reaction mixture volume (12). While plasma was tested according to the manufacturer's instructions, testing on CVL specimens was carried out based on a protocol developed in our laboratory for research purposes. The Abbott RealTime HIV-1 assay (Abbott Molecular Inc., Des Plaines, IL), a reverse transcription-PCR (RT-PCR) assay, was performed on the automated Abbott m2000 platform, which consist of the m2000sp instrument for automated extraction of RNA and the m2000rt instrument for real-time PCR analysis, according to the manufacturer's recommendations. The target sequence for the RealTime assay is the highly conserved integrase gene. The linear range of the assay is 40 to 10,000,000 copies/ml when using the protocol for the 0.6-ml version. The assay was performed by using the m2000 0.6 ml HIV-1 RNA 96 version 5 software application (15).
Quality assessment panels/analytical standards.
The Qnostics HIV-1 evaluation panel (catalog number QNCM14-038-HIV-1), which consists of 8 members, including subtypes B, C, and AG, was analyzed by both assays. The SeraCare HIV RNA genotype performance panel (SeraCare Life Sciences, Milford, MA) was used to determine the ability of the assays to detect HIV-1 subtypes. The panel represents HIV-1 genotypes A, B, C, D, AE, F, AG, G, and H. The AcroMetrix HIV-1 RNA quantification panel (Thermo Fischer Scientific, Waltham, MA), consisting of HIV-1 standards (subtype B) at concentrations of 100, 500, 5,000, 50,000, 500,000, and 5,000,000 copies/ml, was used for analytical evaluation of both assays. In addition, AcroMetrix HIV-1 low- and high-positive-control panels (Thermo Fischer Scientific, Waltham, MA) were tested in quadruplicate on both platforms on the same day. The HIV-1 high-positive control has a viral load reported to be ∼6.24 log10 IU/ml (catalog number 96-4003) (1.7 × 106 IU/ml; 600,000 copies/ml using a conversion factor of 0.35 for the Aptima assay), and the HIV-1 low-positive control has a viral load of ∼2.19 log10 IU/ml (catalog number 96-4001) (155 IU/ml; 54 copies/ml using a conversion factor of 0.35 for the Aptima assay).
Linearity and precision.
Linearity and interassay reproducibility were assessed by preparing serial dilutions of the AcroMetrix positive control with EDTA plasma. Five aliquots of the AcroMetrix HIV-1 RNA high positive control were pooled to obtain a homogenous sample for testing (600,000 copies/ml). This material was diluted to concentrations of 100,000, 10,000, 1,000, and 100 copies/ml of viral RNA in Basematrix human plasma diluent (SeraCare, Milford, MA). An additional 2-fold dilution was created to 50 copies/ml to assess precision near the limit of quantification. Each member was tested in triplicate on each of 3 days with the Aptima and RealTime tests.
Assay limit of detection.
To determine the limit of detection, four virus concentrations from an AcroMetrix HIV-1 panel member were prepared in Basematrix human plasma diluent. Nine replicates at each concentration (100, 50, 25, and 12.5 copies/ml) were tested on three consecutive days. All the dilutions were stored frozen at −20°C prior to testing.
During this study, workflow and maintenance characteristics of both instruments were determined. Daily, weekly, and monthly maintenance required for each platform was assessed.
Statistical analysis.
All viral load data were analyzed as log10-transformed values. Concordance on qualitative results between the Aptima assay and the RealTime test was established by Cohen's kappa statistic, and differences in detection rates were determined by Fisher's exact test. In the correlation analysis, only the viral load data in which both the Aptima HIV and RealTime assays had quantitative values were considered. The correlation between quantitative results was evaluated by using linear regression analysis and Bland-Altman plots (16). Assay results were plotted against each other, regression analysis was performed, and the correlation coefficient was calculated by using Pearson's correlation. All analyses were performed by using GraphPad Prism 6 (GraphPad Software Inc., San Diego, CA).
RESULTS
Analytical performance.
The limit of detection (LOD) was determined by using 36 replicates for the four nominal concentrations (12.5, 25, 50, and 100 copies/ml), as shown in Table 1. The Aptima assay detected all replicates at 12.5 copies/ml or higher, while the RealTime assay detected all replicates at 25 copies/ml and higher. The lower limit of quantification (LLOQ) was determined to be <50 copies/ml for both assays. The LLOQ was determined as the concentration at which both the total error (TE) and total analytical error (TAE) are ≤1. At 50 copies/ml, the Aptima assay had a TE of 0.46 and a TAE of 0.45, while the RealTime test had a TE of 0.61 and a TAE of 0.71.
TABLE 1.
Detection of HIV RNA in replicate samples by the Aptima and RealTime assays
| HIV RNA concn (copies/ml) | Aptima |
RealTime |
||||
|---|---|---|---|---|---|---|
| Total no. of samples | No. of samples in which RNA was detected | % of samples in which RNA was detected | Total no. of samples | No. of samples in which RNA was detected | % of samples in which RNA was detected | |
| 12.5 | 9 | 9 | 100.0 | 9 | 8 | 88.9 |
| 25 | 9 | 9 | 100.0 | 9 | 9 | 100.0 |
| 50 | 9 | 9 | 100.0 | 9 | 9 | 100.0 |
| 100 | 9 | 9 | 100.0 | 9 | 9 | 100.0 |
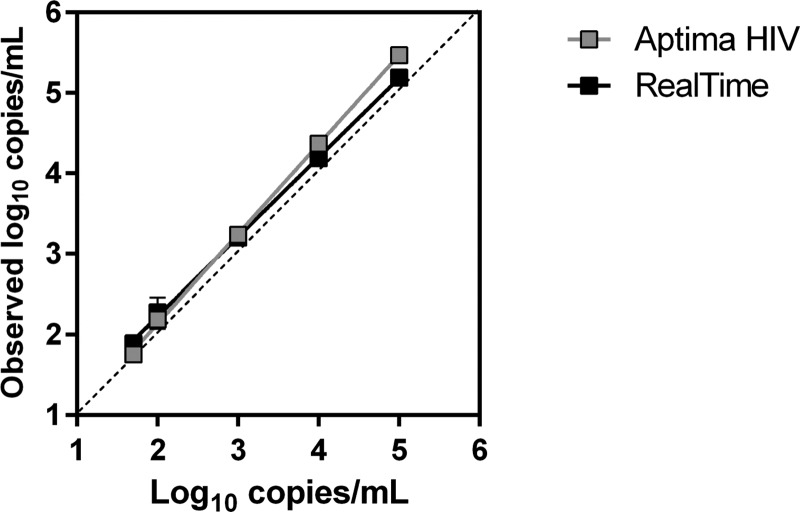
The linear dynamic range analysis showed coefficients of variation ranging from 1.3 to 5.4% for the Aptima assay and from 1.7 to 8.1% for the RealTime assay for the replicates in the nominal ranges of 2 to 5 log10 copies/ml. The interassay concordance for HIV-1 RNA at 50 copies/ml was 88.9% for both assays, as HIV RNA was detected in 8/9 replicates by both assays (Table 2). Both assays were linear across the dynamic range, and the results were highly correlated (Fig. 1).
TABLE 2.
Detection and quantification of HIV RNA in replicates by the Acrometrix HIV high positive control diluted to 1.7 to 5.0 log10 copies/mla
| HIV RNA load (log10 copies/ml) in replicates | Aptima HIV |
RealTime |
||||||
|---|---|---|---|---|---|---|---|---|
| Mean HIV RNA load (log10 copies/ml) detected | SD of HIV RNA load (log10 copies/ml) detected | % CV | No. of samples in which RNA was detected | Mean HIV RNA load (log10 copies/ml) detected | SD of HIV RNA load (log10 copies/ml) detected | % CV | No. of samples in which RNA was detected | |
| 1.7 | 1.76 | 0.10 | 5.64 | 8 | 1.89 | 0.10 | 5.51 | 8 |
| 2 | 2.19 | 0.12 | 5.36 | 9 | 2.28 | 0.18 | 8.11 | 9 |
| 3 | 3.23 | 0.08 | 2.33 | 9 | 3.20 | 0.09 | 2.93 | 9 |
| 4 | 4.37 | 0.07 | 1.66 | 9 | 4.19 | 0.06 | 1.53 | 9 |
| 5 | 5.47 | 0.07 | 1.29 | 9 | 5.19 | 0.09 | 1.67 | 9 |
CV, coefficient of variation.
FIG 1.
Log10-transformed mean HIV load measurements (and standard deviations) in linearity samples determined by both the Aptima and RealTime assays. For linear regression analysis, the slope was 1.112 and the intercept was −0.087 for the Aptima assay (R2 = 0.996), and the slope was 0.986 and the intercept was +0.254 for the RealTime assay (R2 = 0.992). There were 9 replicates tested for each concentration.
Side-by-side testing of the Qnostics and SeraCare HIV-1 evaluation panels showed high concordance between the assays, with the Aptima assay quantifying all subtypes similarly to the RealTime assay with the exception of the SeraCare subtype F sample, where the viral load was 0.88 log10 copies/ml higher with the Aptima than with the RealTime assay. The value obtained with the Aptima assay was close to the nominal value provided in the SeraCare product insert. All other differences were 0.42 log10 copies/ml or lower (Table 3).
TABLE 3.
Detection and quantification of viral loads in Qnostics and SeraCare HIV-1 subtype panels by the Aptima and RealTime assays
| Panel | Subtype of panel member | Nominal HIV-1 RNA concn (log10 copies/ml) | HIV-1 RNA concn (log10 copies/ml) by Aptima | Difference between Aptima and nominal HIV-1 RNA concn (log10 copies/ml) | HIV-1 RNA concn by RealTime (log10 copies/ml) | Difference between RealTime and nominal HIV-1 RNA concn (log10 copies/ml) | Difference between Aptima and RealTime HIV-1 RNA concn (log10 copies/ml) |
|---|---|---|---|---|---|---|---|
| SeraCare | A | 4.74 | 4.88 | 0.14 | 4.67 | −0.07 | 0.21 |
| B | 4.81 | 4.91 | 0.10 | 4.69 | −0.12 | 0.22 | |
| C | 4.22 | 4.45 | 0.23 | 3.95 | −0.27 | 0.50 | |
| D | 4.81 | 5.05 | 0.24 | 4.87 | −0.06 | 0.18 | |
| AE | 4.60 | 4.70 | 0.10 | 4.62 | −0.02 | 0.08 | |
| F | 4.60 | 4.99 | 0.39 | 4.11 | −0.49 | 0.88 | |
| AG | 4.19 | 4.38 | 0.19 | 4.02 | −0.17 | 0.36 | |
| G | 4.48 | 4.79 | 0.31 | 4.52 | −0.04 | 0.27 | |
| H | 4.68 | 5.04 | 0.36 | 4.66 | −0.02 | 0.38 | |
| Qnostics | B | 3.10 | 2.98 | −0.12 | 2.94 | −0.16 | 0.04 |
| B | 3.50 | 3.36 | −0.14 | 3.27 | −0.23 | 0.09 | |
| B | 2.40 | 2.12 | −0.28 | 2.29 | −0.11 | −0.17 | |
| B | 3.50 | 3.46 | −0.04 | 3.3 | −0.20 | 0.16 | |
| C | 4.00 | 4.03 | 0.03 | 3.61 | −0.39 | 0.42 | |
| C | 3.00 | 2.77 | −0.23 | 2.64 | −0.36 | 0.13 | |
| AG | 4.50 | 4.73 | 0.23 | 4.61 | 0.11 | 0.12 |
Assay comparison using clinical samples.
A comparative evaluation of the Aptima and RealTime assays was performed on 208 plasma samples collected from 48 patients. The proportions of samples quantified by the assays were similar (79.0% for the Aptima assay and 80.3% for the RealTime test), while the proportion was dissimilar for the samples not detected: Aptima detected 4.3% of samples that were not detected by RealTime, whereas RealTime detected 0.5% of samples not detected by the Aptima assay (Table 4).
TABLE 4.
Comparison of the Aptima and RealTime assays for detection of HIV-1 RNA in plasma samplesa
| RealTime result | No. of plasma samples with Aptima result |
|||
|---|---|---|---|---|
| ND | Detection at <1.47 log10 copies/ml | Quantified | Total | |
| ND | 11 | 8 | 1 | 20 |
| Detection at <1.6 log10 copies/ml | 1 | 18 | 2 | 21 |
| Quantified | 0 | 6 | 161 | 167 |
| Total | 12 | 32 | 164 | 208 |
ND, not detected.
Of the 208 plasma specimens tested, 161 (77.4%) yielded quantifiable results by both the Aptima and RealTime assays. The mean viral load values (±standard deviations [SD]) determined by the Aptima assay were 3.4 ± 1.4 log10 copies/ml, while those obtained with the RealTime assay were 3.4 ± 1.2 log10 copies/ml. Deming regression analysis and a Bland-Altman plot are shown in Fig. 2; on average, the Aptima results were 0.21 log10 copies/ml higher than the RealTime results. The interassay concordance was 91.4% (kappa = 0.75; 95% confidence interval [CI], 0.65 to 0.85).
FIG 2.
Comparison of the Aptima HIV and RealTime assays by Bland-Altman analysis for the quantified plasma samples. For Deming regression analysis, the slope was 1.072 and the intercept was −0.06.
When testing the 205 CVL specimens, the Aptima assay gave one invalid result, and this sample was quantified by the RealTime assay, whereas the RealTime assay resulted in four invalid results, and these were tested successfully by the Aptima assay (3 quantified and 1 not detected). The five invalid samples were not included in the statistical analysis. The assays differed in quantifying the HIV-1 RNA levels in CVL samples, with Aptima detecting 30.0% and RealTime detecting 20.0% of the samples tested; however, the difference was not statistically significant (P = 0.07). Aptima quantified 15.0% of the samples that were not detected by the RealTime assay with an RNA viral load of >1.47 log10 copies/ml (mean viral load of 2.25 log10 copies/ml) and 9.5% of samples with a viral of <1.47 log10 copies/ml that were not detected by the RealTime assay. However, the RealTime assay quantified 2.0% of samples with viral loads of >1.60 log10 copies/ml that were not detected by the Aptima assay (mean viral load of 2.16 log10 copies/ml) and 1.0% of samples with viral loads of <1.60 log10 copies/ml that were not detected by the Aptima assay (Table 5).
TABLE 5.
Comparison of the Aptima and RealTime assays for detection of HIV-1 RNA in CVL samplesa
| RealTime result | No. of CVL samples with Aptima result |
|||
|---|---|---|---|---|
| ND | Detection at <1.47 log10 copies/ml | Quantified | Total | |
| ND | 107 | 19 | 30 | 156 |
| Detection at <1.6 log10 copies/ml | 2 | 3 | 0 | 5 |
| Quantified | 4 | 5 | 30 | 39 |
| Total | 113 | 27 | 60 | 200 |
ND, not detected.
A total of 30 CVL samples (15.0%) produced valid quantitative results by both assays (slope, 0.991; intercept, −0.099 [as determined by Deming regression analysis]) (Fig. 3). The mean viral load values (±SD) of the Aptima assay were 2.3 ± 1.0 log10 copies/ml, while those obtained with the RealTime assay were 2.8 ± 0.9 log10 copies/ml. On average, the viral load results determined by the Aptima assay were 0.072 log10 copies/ml higher than those of the RealTime assay. A 70% agreement between the assays was observed based on the samples detected or not detected by the assays (kappa = 0.40; 95% CI, 0.29 to 0.50).
FIG 3.
Comparison of the Aptima HIV and RealTime assays by Bland-Altman analysis for the quantified CVL samples. For Deming regression analysis, the slope was 0.991 and the intercept was −0.099.
Workflow and maintenance.
The time and labor required for each phase of assay processing for the Aptima and RealTime platforms are shown in Table 6. For convenience of calculating the hands-on time required for both assays, a 48-sample run was considered. Panther is a nonbatch, random-access system that can continuously accept samples and process them as they are loaded and requires 3 controls and a calibrator per run. The RealTime HIV-1 assay is a batch-based system that has a maximum capacity of 96 samples, including 3 controls (batch sizes of 24/48/96 samples, including 3 controls) per run. The sample volumes, including the dead volume required for Aptima and RealTime, were 0.70 ml and 1.1 ml, respectively.
TABLE 6.
Comparison of workflow and maintenance characteristics of Aptima and Abbott platforms
| Characteristic | Value for assay |
|
|---|---|---|
| Aptima (45 samples + 4 controls) | RealTime (45 samples + 3 controls) | |
| Hands-on time (min) | ||
| Instrument setup and run prepn | ∼5 | ∼15 |
| Reagent prepn and loading | ∼10 | ∼5 |
| Sample prepn and loading | ∼30 | ∼45 |
| Ease of use | Fully automated, walk away | 2 transfer steps (mandatory and time sensitive) |
| Time to result (h) | ∼2.5 | ∼6 |
| Postrun prepn (min) | ∼3 | ∼10 |
| Daily maintenance (min) | ∼3 | ∼15 |
| Weekly maintenance | ∼5 min | ∼1 h |
| Monthly maintenance | ∼1 h | <30 min |
For both assays, hands-on time included the manual transfer of specimen aliquots from their primary specimen collection tubes to instrument-compatible sample tubes prior to centrifugation and sample loading. There were substantial differences in the hands-on time required for daily maintenance of the systems, with Panther requiring less daily maintenance. Overall, Panther had the least hands-on-time from machine setup to postrun processing except for reagent preparation. There was more hands-on time required for reagent preparation for Aptima, as reconstitution of lyophilized reagents is required for each new reagent kit, which can process up to 100 samples. This does not include the time needed to bring reagents to room temperature before loading them onto the instrument. The RealTime m2000 system needs two mandatory transfer steps that are time sensitive: one to load master mix reagents following RNA extraction and another to transfer a 96-well optical reaction plate from the m2000sp instrument to the m2000rt instrument for amplification and detection. The Aptima assay generates the first result within 2.5 h and subsequent results every 5 min, whereas the time to result with the RealTime assay is ∼6 h for a 48-sample run.
DISCUSSION
Quantification of HIV-1 RNA has become the standard of care in the clinical management of HIV-1-infected individuals. The interpretation of HIV-1 load values can be complex, especially at low viremia levels. Therefore, an understanding of the performance characteristics of HIV-1 load assays is necessary for appropriate interpretation of test results. There are two FDA-approved HIV-1 real-time assays that are used by most clinical laboratories, the Cobas AmpliPrep/Cobas TaqMan HIV-1 test (Roche Molecular Systems Inc., Pleasanton, CA) and the RealTime HIV-1 assay (Abbott Molecular). Studies have compared the performance characteristics of these assays (17–19); more recent reports (20–22) have assessed the performance characteristics of the Aptima assay and shown excellent overall performance in detection and accurate quantification of all the major HIV-1 subtypes, and viral load values agreed well with those of other established assays. Our results confirmed these findings; the Aptima assay had good linearity over the quantification range of the assay tested, and plasma viral load results were highly correlated with the RealTime results (R2 = 0.99), with a comparable coefficient of variance of <5.5%. The assays demonstrated intra-assay precision of 88.9%, which was consistent with results of a United Kingdom-based study that compared performance characteristics of the Aptima assay to those of the Abbott RealTime assay and two other assays (20). Since poor precision can affect the linearity of the assay (23), precision, especially at low viremia levels, is an important assay characteristic; the Aptima assay showed a tendency of less variability at low viral loads (1.76 and 2.19 log10 copies/ml) and greater variability at high viral loads (4.37 and 5.47 log10 copies/ml) than the RealTime assay. The Aptima assay appears to be more sensitive than the RealTime assay for the detection of all replicates tested at 12.5 copies/ml, and the Aptima assay detected >4% of the samples that were unable to be detected by the RealTime assay. In a study by Manak et al., the LOD calculated by probit analysis using 30 replicates (1.5 to 100 copies/ml) was 15 copies/ml at the 95% level (24). The Aptima assay precisely quantified all the major HIV-1 subtypes in group M, including subtypes A, B, C, D, F, G, H, AE, and AG. These findings are in agreement with those of previous studies that compared the performance of Aptima with those of the RealTime and Roche Cobas AmpliPrep/Cobas TaqMan assays in quantifying the major HIV-1 group M subtypes and group O subtypes (20–22).
When testing 208 plasma samples, there was good agreement between the assays, with an interassay concordance of 91.4%. Results were similar to those reported by two studies where 93.0% concordance was reported when the performance of the Aptima assay was compared to that of the RealTime assay or Roche Cobas TaqMan HIV-1 test v2.0 (20, 22).
A number of studies have attempted to compare HIV loads in plasma and genital secretions, especially CVL specimens, for patients on ART, many with conflicting results (6–8, 25). Hence, an assay that can quantify HIV-1 RNA without compromising the detection accuracy in both plasma and CVL specimens is important in HIV diagnostics as well as from a research perspective. Our comparison of the assays using CVL samples showed the efficiency of Aptima for detection of HIV-1 RNA levels in CVL specimens. Based on our results, Aptima may be more sensitive than RealTime in quantifying HIV-1 RNA in CVL specimens, with a lower rate of invalid results, and further testing with a larger sample size is needed to confirm these findings. This is an important feature when testing genital specimens, as these samples are quite viscous and can be inhibitory to molecular assays. In addition to having fewer invalid results, the Aptima assay detected 10% more samples than did the RealTime assay and nearly 25% of the samples not detected by the RealTime assay. Previous studies reported that RealTime can be used on CVL specimens (11, 26). The improved sensitivity seen with Aptima is particularly important when testing genital specimens, as viral load values tend to be quite low in these specimens. Our findings demonstrate that Aptima can accurately quantify RNA levels in both CVL and plasma samples, so it could serve as an important research tool to better study HIV-1 pathogenesis in the genital tract.
Automation of molecular testing has become routine in clinical laboratories, thus improving the overall efficiency of laboratory operations, with minimal operator time, improved workflow, and rapid turnaround. The cumulative hands-on time required for specimen processing and maintenance steps were consistently shorter with the Panther than with the m2000 system. The random-access feature of the Panther system allowed continuous loading of samples following the initial loading capacity of 120 samples, leading to a more rapid turnaround than that of the RealTime assay. In addition, it also has unique features such as controls that can be used over a 24-h period and stability of the reagents onboard for 72 h. Hence, the fully automated Panther system with random access has all the necessary features required for clinical laboratories demanding high-throughput sample processing with improved workflow and less turnaround time. Our findings were in concordance with those of other studies reporting workflow and maintenance characteristics (18, 27) of various HIV diagnostic platforms.
In conclusion, the Aptima HIV-1 Quant Dx assay has performance characteristics that are comparable with those of the RealTime assay for plasma samples and has fewer invalid results and a better sensitivity for testing of CVL specimens, making it a reliable test for both clinical and research purposes.
ACKNOWLEDGMENTS
This work was supported in part by Life Span/Tufts/Brown Center for AIDS Research (P30AI042853) and Hologic Inc., San Diego, CA.
Funding Statement
The work was supported in part by Lifespan/Tufts/Brown Center for AIDS Research (P30AI042853) and Hologic Inc., San Diego, CA. Hologic had no input in the study design, choice of instruments studied, data analysis, or preparation of the report.
REFERENCES
- 1.CDC. 2015. Sexually transmitted diseases treatment guidelines. MMWR Morb Mortal Wkly Rep 64:1–137.25590678 [Google Scholar]
- 2.Pilcher CD, Tien HC, Eron JJ Jr, Vernazza PL, Leu SY, Stewart PW, Goh LE, Cohen MS, Quest Study, Duke-UNC-Emory Acute HIV Consortium. 2004. Brief but efficient: acute HIV infection and the sexual transmission of HIV. J Infect Dis 189:1785–1792. doi: 10.1086/386333. [DOI] [PubMed] [Google Scholar]
- 3.Natividad-Villanueva GU, Santiago E, Manalastas RM Jr, Brown HW, Ingersoll J, Caliendo AM, Mayer KH, Cu-Uvin S. 2003. Human immunodeficiency virus in plasma and cervicovaginal secretions in Filipino women. Int J STD AIDS 14:826–829. doi: 10.1258/095646203322556165. [DOI] [PubMed] [Google Scholar]
- 4.Hart CE, Lennox JL, Pratt-Palmore M, Wright TC, Schinazi RF, Evans-Strickfaden T, Bush TJ, Schnell C, Conley LJ, Clancy KA, Ellerbrock TV. 1999. Correlation of human immunodeficiency virus type 1 RNA levels in blood and the female genital tract. J Infect Dis 179:871–882. doi: 10.1086/314656. [DOI] [PubMed] [Google Scholar]
- 5.Cu-Uvin SC, Caliendo AM. 1997. Cervicovaginal human immunodeficiency virus secretion and plasma viral load in human immunodeficiency virus-seropositive women. Obstet Gynecol 90:739–743. doi: 10.1016/S0029-7844(97)00411-0. [DOI] [PubMed] [Google Scholar]
- 6.Shepard RN, Schock J, Robertson K, Shugars DC, Dyer J, Vernazza P, Hall C, Cohen MS, Fiscus SA. 2000. Quantitation of human immunodeficiency virus type 1 RNA in different biological compartments. J Clin Microbiol 38:1414–1418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neely MN, Benning L, Xu J, Strickler HD, Greenblatt RM, Minkoff H, Young M, Bremer J, Levine AM, Kovacs A. 2007. Cervical shedding of HIV-1 RNA among women with low levels of viremia while receiving highly active antiretroviral therapy. J Acquir Immune Defic Syndr 44:38–42. doi: 10.1097/01.qai.0000248352.18007.1f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cu-Uvin S, DeLong AK, Venkatesh KK, Hogan JW, Ingersoll J, Kurpewski J, De Pasquale MP, D'Aquila R, Caliendo AM. 2010. Genital tract HIV-1 RNA shedding among women with below detectable plasma viral load. AIDS 24:2489–2497. doi: 10.1097/QAD.0b013e32833e5043. [DOI] [PubMed] [Google Scholar]
- 9.Boom R, Sol CJ, Salimans MM, Jansen CL, Wertheim-van Dillen PM, van der Noordaa J. 1990. Rapid and simple method for purification of nucleic acids. J Clin Microbiol 28:495–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arredondo M, Garrido C, Parkin N, Zahonero N, Bertagnolio S, Soriano V, de Mendoza C. 2012. Comparison of HIV-1 RNA measurements obtained by using plasma and dried blood spots in the automated Abbott real-time viral load assay. J Clin Microbiol 50:569–572. doi: 10.1128/JCM.00418-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Khopkar P, Sane S, Bichare S, Verma A, Kulkarni S. 2013. Association of plasma viremia with HIV-1 RNA levels in cervicovaginal lavage secretions in HIV-1 seropositive ART naïve women from Pune, India. J Clin Virol 58:730–732. doi: 10.1016/j.jcv.2013.10.018. [DOI] [PubMed] [Google Scholar]
- 12.Hologic Inc. 2014. Aptima HIV-1 Quant Dx assay package insert. Hologic Inc, San Diego, CA: http://www.hologic.com/sites/default/files/package%20inserts/AW-11853-001_002_01%20ENGLISH.pdf. [Google Scholar]
- 13.Moreira C, Venkatesh KK, DeLong A, Liu T, Kurpewski J, Ingersoll J, Caliendo AM, Cu-Uvin S. 2009. Effect of treatment of asymptomatic bacterial vaginosis on HIV-1 shedding in the genital tract among women on antiretroviral therapy: a pilot study. Clin Infect Dis 49:991–992. doi: 10.1086/605540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cu-Uvin S, Snyder B, Harwell JI, Hogan J, Chibwesha C, Hanley D, Ingersoll J, Kurpewski J, Mayer KH, Caliendo AM. 2006. Association between paired plasma and cervicovaginal lavage fluid HIV-1 RNA levels during 36 months. J Acquir Immune Defic Syndr 42:584–587. doi: 10.1097/01.qai.0000229997.52246.95. [DOI] [PubMed] [Google Scholar]
- 15.Abbott Molecular. 2016. RealTime HIV-1. Abbott Laboratories, Abbott Park, IL: https://www.abbottmolecular.com/products/realtime-hiv-1.html. [Google Scholar]
- 16.Bland JM, Altman DG. 1999. Measuring agreement in method comparison studies. Stat Methods Med Res 8:135–160. doi: 10.1191/096228099673819272. [DOI] [PubMed] [Google Scholar]
- 17.Braun P, Ehret R, Wiesmann F, Zabbai F, Knickmann M, Kühn R, Thamm S, Warnat G, Knechten H. 2007. Comparison of four commercial quantitative HIV-1 assays for viral load monitoring in clinical daily routine. Clin Chem Lab Med 45:93–99. [DOI] [PubMed] [Google Scholar]
- 18.Sloma CR, Germer JJ, Gerads TM, Mandrekar JN, Mitchell PS, Yao JD. 2009. Comparison of the Abbott RealTime human immunodeficiency virus type 1 (HIV-1) assay to the Cobas AmpliPrep/Cobas TaqMan HIV-1 test: workflow, reliability, and direct costs. J Clin Microbiol 47:889–895. doi: 10.1128/JCM.02231-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amendola A, Marsella P, Bloisi M, Forbici F, Angeletti C, Capobianchi MR. 2014. Ability of two commercially available assays (Abbott RealTime HIV-1 and Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 version 2.0) to quantify low HIV-1 RNA levels (<1,000 copies/milliliter): comparison with clinical samples and NIBSC working reagent for nucleic acid testing assays. J Clin Microbiol 52:2019–2026. doi: 10.1128/JCM.00288-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hopkins M, Hau S, Tiernan C, Papadimitropoulos A, Chawla A, Beloukas A, Geretti AM. 2015. Comparative performance of the new Aptima HIV-1 Quant Dx assay with three commercial PCR-based HIV-1 RNA quantitation assays. J Clin Virol 69:56–62. doi: 10.1016/j.jcv.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 21.Mor O, Gozlan Y, Wax M, Mileguir F, Rakovsky A, Noy B, Mendelson E, Levy I. 2015. Evaluation of the RealTime HIV-1, Xpert HIV-1, and Aptima HIV-1 Quant Dx assays in comparison to the NucliSens EasyQ HIV-1 v20 assay for quantification of HIV-1 viral load. J Clin Microbiol 53:3458–3465. doi: 10.1128/JCM.01806-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schalasta G, Börner A, Speicher A, Enders M. 18 August 2015. Comparative evaluation of the Aptima HIV-1 Quant Dx assay and COBAS TaqMan HIV-1 v2.0 assay using the Roche High Pure system for the quantification of HIV-1 RNA in plasma. Clin Chem Lab Med doi: 10.1515/cclm-2015-0522. [DOI] [PubMed] [Google Scholar]
- 23.Burd EM. 2010. Validation of laboratory-developed molecular assays for infectious diseases. Clin Microbiol Rev 23:550–576. doi: 10.1128/CMR.00074-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manak M, Hack H, Trsic T, Nair S, Worlock A, Peel S, Malla J, Jagodzinski L. 2014. Evaluation of the Hologic Aptima HIV-1 Quant Dx assay with HIV-1 subtypes, abstr 620, p 305 Abstr 2014 Conf Retroviruses Opportun Infect, Boston, MA. [Google Scholar]
- 25.Landay A, Golub ET, Desai S, Zhang J, Winkelman V, Anastos K, Durkin H, Young M, Villacres MC, Greenblatt RM, Norris PJ, Busch MP, Women's Interagency HIV Study. 2014. HIV RNA levels in plasma and cervical-vaginal lavage fluid in elite controllers and HAART recipients. AIDS 28:739–743. doi: 10.1097/QAD.0000000000000150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legoff J, Bouhlal H, Grésenguet G, Weiss H, Khonde N, Hocini H, Désiré N, Si-Mohamed A, de Dieu Longo J, Chemin C, Frost E, Pépin J, Malkin JE, Mayaud P, Bélec L. 2006. Real-time PCR quantification of genital shedding of herpes simplex virus (HSV) and human immunodeficiency virus (HIV) in women coinfected with HSV and HIV. J Clin Microbiol 44:423–432. doi: 10.1128/JCM.44.2.423-432.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ratnam S, Jang D, Gilchrist J, Smieja M, Poirier A, Hatchette T, Flandin JF, Chernesky M. 2014. Workflow and maintenance characteristics of five automated laboratory instruments for the diagnosis of sexually transmitted infections. J Clin Microbiol 52:2299–2304. doi: 10.1128/JCM.03549-13. [DOI] [PMC free article] [PubMed] [Google Scholar]