Abstract
We developed and evaluated a multiplex antibody detection-based immunoassay for the diagnosis of prosthetic joint infections (PJIs). Sixteen protein antigens from three Staphylococcus species (Staphylococcus aureus, Staphylococcus epidermidis, and Staphylococcus lugdunensis) (8 antigens), Streptococcus agalactiae (4 antigens), and Propionibacterium acnes (4 antigens) were selected by comparative immunoproteomics using serum samples from PJI cases versus controls. A bead-based multiplex immunoassay that measured serum IgG against purified, recombinant forms of each of the 16 antigens was developed. We conducted a prospective study to evaluate the performance of the assay. A PJI was defined by the presence of a sinus tract and/or positive intraoperative sample cultures (at least one sample yielding a virulent organism or at least two samples yielding the same organism). A total of 455 consecutive patients undergoing revision or resection arthroplasty (hip, 66.3%; knee, 29.7%; shoulder, 4%) at two French reference centers for the management of PJI were included: 176 patients (38.7%) were infected and 279 (61.3%) were not. About 60% of the infections involved at least one of the species targeted by the assay. The sensitivity/specificity values were 72.3%/80.7% for targeted staphylococci, 75%/92.6% for S. agalactiae, and 38.5%/84.8% for P. acnes. The assay was more sensitive for infections occurring >3 months after arthroplasty and for patients with an elevated C-reactive protein (CRP) or erythrocyte sedimentation rate (ESR). However, it detected 64.3% and 58.3% of targeted staphylococcal infections associated with normal CRP and ESR values, respectively. This new multiplex immunoassay approach is a novel noninvasive tool to evaluate patients suspected of having PJIs and provides information complementary to that from inflammatory marker values.
INTRODUCTION
Joint replacements are among the most successful surgical procedures with the number of procedures performed in 2010 exceeding 1 million (1) and expected to exceed 4 million yearly by 2030 (2). Prosthetic joint infection (PJI) is a major complication associated with increased morbidity, poor functional outcome, and increased use of health care resources (3, 4). It occurs in up to 2.4% of primary arthroplasties and in 10 to 25% of revision arthroplasties (5, 6). Staphylococcus aureus and coagulase-negative staphylococci are the most frequent organisms involved, followed by streptococci-enterococci, aerobic Gram-negative bacilli, Propionibacterium acnes, and other anaerobes (7–9).
The distinction between PJI and mechanical prosthetic dysfunction preoperatively is often difficult (8) but is essential because the two diagnoses require radically different surgical strategies. The current diagnostic algorithms involve serological screening with the erythrocyte sedimentation rate (ESR) and C-reactive protein (CRP), followed by joint aspiration if either is elevated (10). However, the ESR and CRP are both markers of inflammation and do not provide information on the infecting agent. Furthermore, their values in PJI may be normal (10, 11), delaying the management of patients with infections and triggering costly investigations.
Advances in multiplexing technology have paved the way for in vitro diagnostic tests that quantify antibodies to multiple antigens in a single reaction (12). However, this approach has rarely been applied to bacterial infectious diseases (13, 14) and never to PJIs. The serological diagnosis of systemic staphylococcal infection has been previously attempted with as many as seven antigenic preparations evaluated in combination (15) but with no subsequent use of the assay, and recent evaluations have confirmed the lack of clinical benefit of the commercially available antistaphylolysin and antinuclease assays (16).
We developed a multiplex antibody detection-based immunoassay using a panel of recombinant antigens from major PJI pathogens (Staphylococcus spp.,Streptococcus agalactiae, and Propionibacterium acnes) that provides independent results for each of the targeted pathogens. We performed a prospective study to evaluate its performance in two French orthopedic centers specializing in the management of complex bone and joint infections.
(This work was presented in part at the 115th General Meeting of the American Society for Microbiology, New Orleans, LA, 30 May to 2 June 2015, and at the 25th European Congress of Clinical Microbiology and Infectious Diseases, Copenhagen, Denmark, 25 to 28 April 2015.)
MATERIALS AND METHODS
Study design and oversight.
This was a prospective, multicenter, noninterventional study performed in patients scheduled for revision or resection arthroplasty for a knee, hip, or shoulder implant. The objective of the study was to assess the performance of the multiplex immunoassay, using as reference the microbiological cultures performed on periprosthetic tissue samples obtained during surgery.
The protocol and information sheet were approved by the institutional review board (IRB) CPP Île-de-France XI. The database was authorized by the Commission Nationale Informatique Libertés (French privacy watchdog), and all patients were informed before inclusion and given the opportunity to opt out. The study was performed in accordance with the principles of the Declaration of Helsinki and good clinical practice guidelines.
Study population and procedures.
We included all consecutive adult patients with total hip, knee, or shoulder prosthesis who underwent revision or resection arthroplasty between 25 June 2012 and 23 June 2014 at two French reference centers for the management of bone and joint infections. The main exclusion criteria were joint revision surgery for more than one implant, HIV infection, and cancer chemotherapy.
Blood samples for the multiplex immunoassay were taken at the inclusion visit. Serum aliquots were frozen and stored at −20°C. All samples were blinded with respect to all patient information before processing. The multiplex immunoassay and microbiological cultures using intraoperative tissue samples were performed for all patients, regardless of clinical suspicion of PJI. Antibiotics were withheld at least 2 weeks prior to collection of intraoperative tissue samples.
Selection of antigens included in the multiplex immunoassay.
Antigens were selected by comparative immunoproteomics followed by enzyme-linked immunosorbent assay (ELISA) and Luminex assessments (Fig. 1). Serum samples were from PJI cases involving a single species (S. aureus, 37; Staphylococcus epidermidis, 45; Staphylococcus lugdunensis, 4; Streptococcus agalactiae, 13; P. acnes, 31); controls included healthy blood donors (n = 98) and patients (n = 66) who underwent bone and joint replacement surgery at least 2 years previously without any abnormal signs or symptoms at follow-up (“orthopedic controls”). Strains of the targeted species were S. aureus Mu50, COL, and USA 300, S. epidermidis RP62A (ATCC 35894) and ATCC 12228, S. lugdunensis CIP 103642, S. agalactiae CIP 82.45, and P. acnes KPA171202 (type IB).
FIG 1.
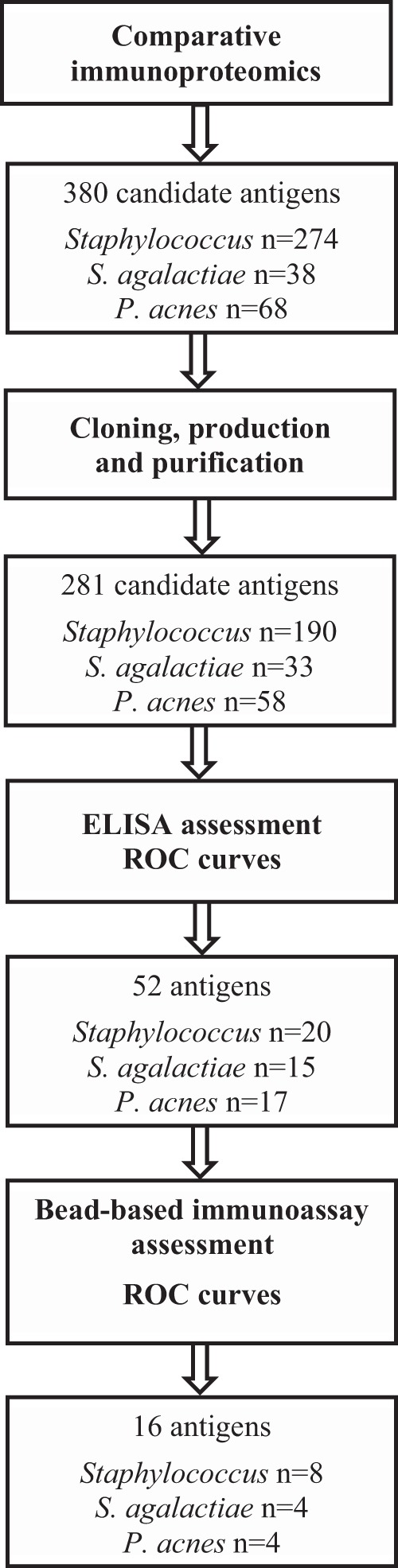
Selection of antigens included in the multiplex immunoassay.
A total of 380 candidate antigens (staphylococci, 274; S. agalactiae, 38; P. acnes, 68) were identified by comparative immunoproteomics using pooled serum samples from PJI cases and controls collected during the course of a separate multicentric prospective study to develop the assay (for further details, see the supplemental material). Among them, 281 antigens (staphylococci, 190; S. agalactiae, 33; P. acnes, 58) were successfully expressed in Escherichia coli. The antigens were produced as fusion proteins with a His6 tag at their N-terminal end; they were purified with the Äkta Xpress chromatography system (GE Healthcare, Velizy-Villacoublay, France) at a minimum purity of 95%.
A first screening of the purified recombinant antigens was performed using an ELISA. The purified antigens were coated onto the wells of Nunc MaxiSorp microtiter plates (Thermo Fisher Scientific, MA, USA). Serum IgG binding was detected with a goat anti-human IgG horseradish peroxidase-conjugated antibody. The optical density at 450 nm (OD450) values were analyzed with TANAGRA software, version 1.4.31 (http://eric.univ-lyon2.fr/∼ricco/tanagra/en/tanagra.html). The ability of each candidate antigen to discriminate PJI cases from controls was evaluated using receiver operator characteristic (ROC) curves, allowing the identification of 52 relevant antigens (staphylococci, 20; S. agalactiae, 15; P. acnes, 17).
The 52 antigens identified by the ELISA were further assessed using the Luminex technology. Purified recombinant proteins (50 μg/ml) were coupled to Magplex beads (Luminex, Austin, TX, USA). Serum IgG binding was detected with R-phycoerythrin-conjugated AffiniPure goat anti-human IgG (Moss, Inc., Pasadena, MD). Median fluorescence intensity (MFI) values were determined using a Magpix instrument and xPONENT software. ROC curves were used to select the 16 final proteins included in the immunoassay: 8 staphylococcal proteins (3 proteins involved in biofilm formation, 3 lipoproteins, 1 virulence factor, 1 putative adherence factor), 4 S. agalactiae proteins (1 virulence factor, 1 protein putatively involved in metabolism, 2 proteins with unknown function), and 4 P. acnes proteins (2 membrane proteins with transport functions, 1 protein involved in metabolism, 1 putative oxidoreductase).
Multiplex immunoassay.
The immunoassay (research use only version of BJI InoPlex; Diaxonhit, Paris, France) is a bead-based multiplex assay (Magplex beads) designed to target three Staphylococcus species (S. aureus, S. epidermidis, and S. lugdunensis), Streptococcus agalactiae, and P. acnes. It quantifies the patient's IgG binding to the purified, recombinant forms of 16 proteins (see above). The serum samples were diluted 1:70. IgG binding was detected with R-phycoerythrin-conjugated goat anti-human IgG. Positive-control, negative-control, and calibrating sera were included in each series. Measurements were performed on a Magpix instrument. MFI values were processed using proprietary software (Diaxonhit). Results are expressed as positive, negative, or undetermined for staphylococci, S. agalactiae, and P. acnes.
Bacteriological methods.
At least three intraoperative periprosthetic tissue samples or small hardware obtained using a sterile instrument set were placed in sterile doubly wrapped 30-ml containers and processed within 2 h as previously described (17). Briefly, samples were mechanically disrupted in sterile water with stainless steel beads using a Retsch MM 400 bead mill (Verder, Cergy-Pontoise, France). Bead-milled sample suspensions were then cultured on solid medium (Columbia sheep blood agar incubated under aerobic and anaerobic atmospheres or chocolate agar under 5% CO2) and on liquid aerobic (brain heart infusion) and anaerobic (Schaedler's) broths terminally subcultured after 15 days. Enumeration and differential counting were performed on intraoperative synovial aspirations, and an identical culture scheme was applied to these samples. All manipulations were performed using a safety cabinet. Isolates were identified by mass spectrometry using a Microflex LT instrument and the current CE-marked IVD Biotyper software (Bruker Daltonique, Wissenbourg, France).
Definition of prosthetic joint infection.
An infection was defined by (i) the presence of a sinus tract and/or (ii) at least one intraoperative sample positive in culture with a virulent organism or at least two intraoperative samples positive in culture with the same microorganism (same species and same susceptibility profile). The absence of infection was defined as no sinus tract and no positive culture for any sample or a single culture positive for a nonvirulent organism. An infection was considered polymicrobial when multiple organisms fulfilled the microbiological criteria for infection. This definition takes into account the major infection criteria of the Infectious Diseases Society of America (IDSA) (10) and the Musculoskeletal Infection Society (MSIS) (18, 19) guidelines. Of the supportive criteria, only single isolates of virulent microorganisms have been considered due to incomplete data sets.
Study endpoints.
The sensitivity for each microorganism targeted by the immunoassay was defined as the proportion of patients found seropositive for this microorganism among those infected with the same microorganism. The specificity for each targeted microorganism was defined as the proportion of patients seronegative for this microorganism among noninfected patients.
Statistical analysis.
The sensitivity and specificity of the immunoassay were estimated along with their two-sided 95% Clopper-Pearson exact confidence intervals for binomial proportions. The categorical and continuous variables were compared using Fisher's exact test, Student's t test, and the Wilcoxon rank sum test as appropriate. Two-sided P values of <0.05 were considered statistically significant. All calculations were performed using R version 2.13 (20).
RESULTS
Study population.
A total of 481 consecutive patients were enrolled in the study, and 455 were included in the analysis (Fig. 2); 279 (61.3%) were defined as noninfected and 176 (38.7%) were defined as infected. The characteristics of the infected and noninfected patients were significantly different except for age, excess weight, and the proportion undergoing a first prosthesis replacement (Table 1). In particular, the ratio of men to women was higher among infected than noninfected patients, and infected patients were more likely to have had a prosthesis inserted in the past 3 months (Table 1).
FIG 2.

Flowchart of patients' inclusion.
TABLE 1.
Baseline characteristics of the population studied
| Characteristica | Total (n = 455) | Noninfected (n = 279) | Infected (n = 176) | Pb |
|---|---|---|---|---|
| Age (mean ±SD) (yr) | 69.6 ± 12.1 | 70.5 ± 11 | 68.3 ± 13.6 | 0.083 |
| Female (no./total no. [%]) | 237/453 (52.3) | 162/277 (58.5) | 75/176 (42.6) | 0.001 |
| Wt (median [IQR]) (kg) | 75 (64 to 87) | 74 (62 to 85) | 79.5 (65.8 to 92.2) | 0.003 |
| Excess wt (median [IQR]) (kg) | 5.7 −3.6 to 15.8 | 5.2 −3.7 to 14.3 | 6.4 −3.2 to 19.1 | 0.077 |
| First implant revision (no./total no. [%]) | 264/409 (64.5) | 172/257 (66.9) | 92/152 (60.5) | 0.23 |
| Site of prosthesis (no./total no. [%]) | ||||
| Hip | 301/454 (66.3) | 202/278 (72.7) | 99/176 (56.2) | <0.001 |
| Knee | 135/454 (29.7) | 71/278 (25.5) | 64/176 (36.4) | |
| Shoulder | 18/454 (4) | 5/278 (1.8) | 13/176 (7.4) | |
| Time since prosthesis insertion (no./total no. [%]) | ||||
| ≤3 mo | 37/441 (8.4) | 7/272 (2.6) | 30/169 (17.8) | <0.001 |
| >3 mo | 404/441 (91.6) | 265/272 (97.4) | 139/169 (82.2) | |
| Sinus tract (no./total no. [%]) | 46/455 (10.1) | - | 46/176 (26.1) | |
| CRP of ≥10 mg/literc | 217/442 (49.1) | 71/268 (26.5) | 146/174 (83.9) | <0.001 |
| ESR of ≥30 mm/hc | 169/389 (41.6) | 50/250 (20) | 112/139 (80.6) | <0.001 |
IQR, interquartile range; CRP, serum C-reactive protein; ESR, erythrocyte sedimentation rate.
Infected versus non-infected cases.
The cutoffs are taken from reference 8.
Organisms recovered from intraoperative tissue cultures.
The species most frequently cultured from infected cases (Table 2) were S. aureus (33% of infections), S. epidermidis (22.2%), P. acnes, and Enterococcus faecalis (7.4% each); S. agalactiae was involved in 4.5% of infections and was the most frequent streptococcal species. S. aureus (54.2%) and S. epidermidis (37.5%) were also the most frequent species recovered from polymicrobial infections, which accounted for 14.4% (24/167) of the culture-positive cases (see Table S1 in the supplemental material). Overall, 59.2% (100/169) of the culture-positive infections involved at least one of the organisms targeted by the immunoassay.
TABLE 2.
Microbial species involved in the infected casesa
| Microbial species | All sites (n = 176)b | Hip (n = 99) | Knee (n = 64) | Shoulder (n = 13) |
|---|---|---|---|---|
| Staphylococcus species | ||||
| S. aureus | 58 (33) | 33 (33.3) | 22 (34.4) | 3 (23.1) |
| S. epidermidis | 39 (22.2) | 22 (22.2) | 14 (21.9) | 3 (23.1) |
| S. lugdunensis | 9 (5.1) | 3 (3) | 6 (9.4) | 0 |
| S. capitis | 7 (4) | 5 (5) | 2 (3.1) | 0 |
| S. caprae | 1 (0.6) | 1 (1) | 0 | 0 |
| S. haemolyticus | 1 (0.6) | 0 | 1 (1.6) | 0 |
| S. hominis | 1 (0.6) | 1 (1) | 0 | 0 |
| S. xylosus | 1 (0.6) | 0 | 1 (1.6) | 0 |
| Staphylococcus sp. | 0 | 0 | 0 | 0 |
| Streptococcus species | ||||
| S. agalactiae | 8 (4.5) | 3 (3) | 4 (6.2) | 1 (8) |
| S. bovis | 1 (0.6) | 0 | 1 | 0 |
| S. dysgalactiae | 1 (0.6) | 1 (1) | 0 | 0 |
| S. oralis | 2 (1.1) | 0 | 2 (3.1) | 0 |
| S. pneumoniae | 2 (1.1) | 1 | 0 | 1 (8) |
| S. sanguinis | 1 (0.6) | 1 (1) | 0 | 0 |
| Propionibacterium species | ||||
| P. acnes | 13 (7.4) | 6 (6.1) | 2 (3.1) | 5 (38.5)c |
| P. avidum | 3 (1.7) | 2 (2) | 0 | 1 (8) |
| Other bacterial species | ||||
| Acinetobacter ursingii | 1 (0.6) | 0 | 1 (1.6) | 0 |
| Anaerococcus hydrogenalis | 1 (0.6) | 0 | 1 (1.6) | 0 |
| Corynebacterium striatum | 3 (1.7) | 2 (2) | 1 (1.6) | 0 |
| Enterobacter cloacae | 8 (4.5) | 6 (6.1) | 2 (3.1) | 0 |
| Enterococcus faecalis | 13 (7.4) | 10 (10.1) | 2 (3.1) | 1 (8) |
| Enterococcus faecium | 2 (1.1) | 1 (1) | 1 (1.6) | 0 |
| Escherichia coli | 6 (3.4) | 6 (6.1) | 0 | 0 |
| Finegoldia magna | 4 (2.3) | 2 (2) | 1 (1.6) | 1 (8) |
| Granulicatella adjacens | 1 (0.6) | 1 (1) | 0 | 0 |
| Klebsiella pneumoniae | 2 (1.1) | 2 (2) | 0 | 0 |
| Morganella morganii | 1 (0.6) | 1 (1) | 0 | 0 |
| Peptoniphilus harei | 2 (1.1) | 1 (1) | 1 (1.6) | 0 |
| Pseudomonas aeruginosa | 5 (2.8) | 2 (2) | 3 (4.7) | 0 |
| Salmonella sp. | 2 (1.1) | 2 (1) | 0 | 0 |
| Fungal species | ||||
| Candida albicans | 1 (0.6) | 0 | 1 (1.6) | 0 |
Microbial species recovered from at least one intraoperative sample (virulent organisms) or from at least two intraoperative samples (nonvirulent organisms). Data are number (%) of cases; the percentage represents the proportion of all infected cases, culture-proven or not.
Nine infections with sinus tracts were associated with negative (n = 7) or nonsignificant (n = 2) microbiological cultures.
P. acnes was found significantly more frequently in the shoulder than in the hip (P = 0.0001) or knee (P = 0.0002).
The global distribution of species was similar for hip and knee infections (Table 2). However, the most frequent species after S. aureus and S. epidermidis were E. faecalis, P. acnes, Enterobacter cloacae, and E. coli in the hip and S. lugdunensis and S. agalactiae in the knee. P. acnes was significantly more frequent in shoulder infections than in hip (P = 0.0001) or knee (P = 0.0002) infections (Table 2).
P. acnes (20/279, 7.2%) and S. epidermidis (14/279, 5%) were the most frequent species recovered in single positive cultures from noninfected cases.
Performance of the immunoassay.
The sensitivity/specificity values for the immunoassay were calculated by excluding undetermined results and were 72.3%/80.7% (95% confidence intervals, 62.7 to 80.7/75.6 to 85.1) for staphylococci, 75%/92.6% (38.8 to 95.6/89 to 95.3) for S. agalactiae, and 38.5%/84.8% (15.7 to 65.9/80.2 to 88.7) for P. acnes (Table 3). The values calculated by classifying undetermined results as either positive or negative are presented in Table S2 in the supplemental material.
TABLE 3.
Performance of the multiplex immunoassaya
| Organism(s) | All cases |
Site of prosthesis |
Time since implantation |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hip |
Knee |
≤3 mo |
>3 mo |
|||||||
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | |
| Staphylococci targeted | 68/94 (72.3) [62.7–80.7] | 213/264 (80.7) [75.6–85.1] | 38/52 (73.1) [59.9–83.8] | 159/190 (83.7) [77.9–88.4] | 29/39 (74.4) [59–86.2] | 50/68 (73.5) [62.1–83] | 9/15 (60) [34.5–81.9] | 3/7 (42.9) [12.3–78.4] | 57/75 (76) [65.4–84.6] | 205/250 (82) [76.9–86.4] |
| S. aureus | 36/54 (66.7) [53.4–78.2] | 21/30 (70) [52–84.3] | 14/21 (66.7) [44.9–84.1] | 7/11 (63.6) [33.6–87.2] | 27/39 (69.2) [53.6–82.1] | |||||
| S. epidermidis | 26/35 (74.3) [58–86.7] | 16/21 (76.2) [54.9–90.7] | 10/13 (76.9) [49.1–93.8] | 2/4 (50) [9.4–90.6] | 24/31 (77.4) [60.4–89.6] | |||||
| S. lugdunensis | 9/9 (100) [71.7–100] | 3/3 (100) [36.8–100] | 6/6 (100) [60.7–100] | 9/9 (100) [71.7–100] | ||||||
| S. agalactiae | 6/8 (75) [38.8–95.6] | 250/270 (92.6) [89–95.3] | 3/3 (100) [36.8–100] | 181/195 (92.8) [88.5–95.9] | 2/4 (50) [9.4–90.6] | 63/69 (91.3) [82.8–96.4] | 0/1 (0) [0–95] | 7/7 (100) [65.2–100] | 6/7 (85.7) [47–99.3] | 237/256 (92.6) [88.9–95.3] |
| P. acnes | 5/13 (38.5) [15.7–65.9] | 235/277 (84.8) [80.2–88.7] | 1/6 (16.7) [0.8–59.1] | 172/200 (86) [80.7–89.5] | 1/2 (50) [2.5–97.5] | 60/71 (84.5) [74.7–91.6] | 7/7 (100) [65.2–100] | 4/12 (33.3) [11.6–62.3] | 222/263 (84.4) [79.6–88.4] | |
Data are no. (%) [95% confidence interval].
The sensitivity values were not significantly lower for S. aureus than for S. epidermidis infections (66.7% [95% confidence interval, 53.4 to 78.2] versus 74.3% [58 to 86.7]; P = 0.49); the immunoassay, however, detected 73.8% (31/42) and 85.7% (12/14) of S. aureus infections associated with two or more positive samples or presenting with a sinus tract, respectively (Table 3). All S. lugdunensis infections (9/9) were detected. The immunoassay tended to be more sensitive for staphylococcal infections occurring >3 months after arthroplasty (76% [65.4 to 84.6] versus 60% [34.5 to 81.9]; P = 0.214) (Table 3). The performance of the immunoassay was similar for the hip and knee (Table 3). Although the total number of shoulder infections was small (n = 13), the immunoassay was able to detect the single S. agalactiae case and three of the five P. acnes cases.
The immunoassay results were found to be “false” positive in 54.5% (6/11) of infections involving staphylococcal species other than S. aureus, S. epidermidis, or S. lugdunensis, 22.7% (5/22) of infections involving streptococci other than S. agalactiae and two of the three infections involving Propionibacterium avidum. The results were positive in 5 of the 13 cases with a single positive S. epidermidis culture (38.5%) and in only 2 of the 21 cases with a single positive P. acnes culture (9.5%).
Relationship with ESR and CRP.
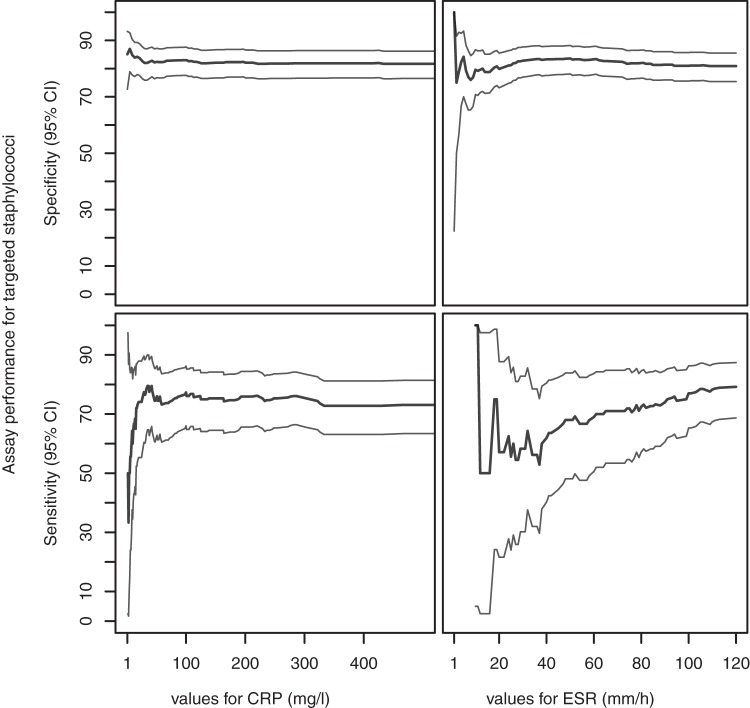
The immunoassay was more sensitive when the ESR or CRP was elevated, with unchanged specificity. The largest increases were observed with the ESR for staphylococcal species not included in the immunoassay (85.7% [95% confidence interval, 47 to 99.3] for an elevated ESR versus 0% [0 to 52.7] for a normal ESR; P = 0.0152) and S. epidermidis (90.9% [73.1 to 98.4] for an elevated ESR versus 50% [14.7 to 85.3] for a normal ESR; P = 0.05) (Table 4). Moreover, the ESR was directly correlated with the sensitivity of the immunoassay for staphylococci, but this was not the case for CRP (Fig. 3).
TABLE 4.
Performance of the multiplex immunoassay in relation to ESR and CRP resultsa
| Organism(s) | ESR elevatedb |
ESR normal |
CRP elevatedc |
CRP normal |
||||
|---|---|---|---|---|---|---|---|---|
| Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | Sensitivity | Specificity | |
| Staphylococci targeted | 50/60 (83.3) [72.3–91.2] | 33/45 (73.3) [59.1–84.7] | 7/12 (58.3) [30.2–82.8] | 157/190 (82.6) [76.7–87.5] | 59/79 (74.7) [64.2–83.3] | 54/68 (79.4) [68.6–87.8] | 9/14 (64.3) [37.6–85.6] | 152/185 (82.2) [76.1–87.2] |
| S. aureus | 26/34 (76.5) [60.2–88.4] | —d | 3/6 (50) [14.7–85.3] | — | 34/48 (70.8) [56.9–82.3] | — | 2/5 (40) [7.3–81.8] | — |
| S. epidermidis | 20/22 (90.9) [73.1–98.4] | — | 3/6 (50) [14.7–85.3] | — | 21/27 (77.8) [59.4–90.5] | — | 5/8 (62.5) [27.8–89.4] | — |
| S. lugdunensis | 6/6 (100) [60.7–100] | — | 1/1 (100) [5–100] | — | 7/7 (100) [65.2–100] | — | 2/2 (100) [22.4–100] | — |
| Other staphylococci | 6/7 (85.7) [47–99.3] | — | 0/4 (0) [0–52.7] | — | 5/9 (55.6) [24–84] | — | 1/2 (50) [2.5,97.5] | — |
| S. agalactiae | 6/8 (75) [38.8–95.6] | 43/49 (87.8) [76.3–94.9] | — | 180/192 (93.8) [89.6–96.6] | 4/5 (80) [33.4–99] | 64/70 (91.4) [83–96.5] | 1/2 (50) [2.5–97.5] | 176/189 (93.1) [88.8–96.1] |
| P. acnes | 3/6 (50) [14.7–85.3] | 45/50 (90) [79.2–96.2] | 0/4 (0) [0–52.7] | 166/198 (83.8) [78.2–88.5] | 4/8 (50) [18.4–81.6] | 59/71 (83.1) [73–90.5] | 1/5 (20) [1–66.6] | 167/195 (85.6) [80.2–90.1] |
Data are no. (%) [95% confidence interval].
Levels of ≥30 mm/h.
Levels of ≥10 mg/liter.
—, not applicable.
FIG 3.
Immunoassay sensitivity and specificity with respect to targeted staphylococci for all values of CRP and ESR. The plots show the sensitivities and specificities of the assay (versus targeted staphylococci) according to the values of CRP and ESR. The inner bold lines are estimated sensitivity and specificity values; the outer lines are pointwise 95% confidence bands (CI).
However, the immunoassay was also able to identify infections associated with normal levels of inflammatory markers (Table 4). It was positive in 64.3% and 58.3% of targeted staphylococcal infections associated with normal levels of CRP or ESR, respectively, and was also positive in one of the two S. agalactiae infections with a normal CRP.
DISCUSSION
This study provides evidence that the noninvasive detection of serum antibodies can indirectly diagnose PJI. Different carbohydrate antigen preparations of S. epidermidis have been considered in this indication, but the assays have not been commercialized or evaluated beyond preliminary studies (21, 22). The antigens included in the assay were all identified by comparative immunoproteomics, i.e., by comparing the antibody response profile of arthroplasty patients with PJIs to those without infection. The identification of bacterial targets of humoral immunity in PJI patients allowed us to select the antigens most relevant to PJI, in which pathogens commonly undergo phenotypic changes (23–25). All antigens included in our assay were synthesized as recombinant proteins, and some were further engineered to improve their solubility and stability or to enhance the specificity of the response through the deletion of nonrelevant epitopes.
Our main objective was to detect PJIs caused by staphylococci, which cause more than 50% of PJIs and are among the most deleterious and antibiotic-resistant agents (8). Antigens derived from S. aureus, S. epidermidis, and S. lugdunensis, the three most prevalent staphylococcal species (7, 8), were included in the assay. The sensitivity/specificity values of the assay for the three species were overall 72.3%/80.7%; the sensitivity was nonsignificantly lower for S. aureus, due to the weaker ability of the assay to detect cases with a single positive culture. The sensitivity/specificity values reached 76%/82% when the assay was performed ≥3 months after surgery (>80% of targeted staphylococcal infections). This performance is in the range of that reported with the ESR and CRP (26, 27), with the immunoassay showing a slightly lower sensitivity than either marker in our patients. However, the immunoassay detected around 60% of targeted staphylococcal infections associated with a normal ESR or CRP, demonstrating its ability to identify infection in the absence of systemic inflammation and to identifying patients otherwise overlooked by most diagnostic algorithms (10, 11, 18). The assay therefore improves the specificity of the currently available rule-out assays.
The limited number of S. agalactiae PJI cases prevented us from definitively assessing the performance of the immunoassay for this pathogen, the most common streptococcal species causing PJIs (7, 28). However, the overall performance was similar to that found with targeted staphylococci and even better in cases occurring >3 months after arthroplasty (sensitivity/specificity, 85.7%/92.6%), which account for the vast majority of S. agalactiae PJIs (29). The sensitivity of the immunoassay for P. acnes was much lower at <40% when all PJIs were considered and 50% for PJIs in patients with elevated CRP or ESR. The antigens included in the assay may not be optimal. Bacteria were grown in liquid medium and not in biofilm, and the immunoproteomic evaluation might have not identified the relevant antigens solely expressed in sessile bacteria. The antigens were derived from an IB strain and might be less sensitive with other P. acnes types frequently isolated in PJIs such as type II (30). The low sensitivity of our assay may also reflect the low virulence of P. acnes. Indeed this organism is often isolated in cases of fortuitous intraoperative cultures (7, 31–34), and there are instances of clinically silent P. acnes PJIs defined bacteriologically and amenable to limited therapy (33). The correlation between serologically silent P. acnes and PJI diagnosed by fortuitous intraoperative culture should be explored. To better understand the relevance of the immunoassay, it would also be interesting to focus on shoulder arthroplasty, where the prevalence of P. acnes is highest (35, 36).
We cannot rule out a bias resulting from the recruitment of patients from infection reference centers, which guaranteed a large number of PJI cases and optimal bacteriological procedures. This may have resulted in an overrepresentation of the more challenging cases and may detract from the everyday use of the assay. Our data should therefore be validated by studies conducted in primary care settings, where the immunoassay would be used by practitioners less often confronted with PJIs, in conjunction with the ESR and CRP. These studies will allow determination of the positive and negative predictive values of the immunoassay, an analysis that would have been irrelevant in our study due to the abnormally high prevalence of PJIs.
Several culture methods have been proposed for the microbiological documentation of PJIs. The bead-milling of multiple intraoperative samples was routinely performed at both centers when the study was designed (17). This technique was subsequently evaluated independently and recognized in the literature (10). It mechanizes the process previously used to define the interpretation criteria of multiple intraoperative cultures (37). Implant sonication (38), which was not used routinely in either participating center, was not performed in the patients enrolled. The molecular detection of targeted organisms might have been an interesting comparator for discrepant results between culture and serology. However, 16S PCR and sequencing on bead-milled periprosthetic tissues has shown limited benefit in patients without previous antimicrobial treatment (39), and validated species-specific PCR protocols, including those for Staphylococcus spp., S. agalactiae, and P. acnes, were not available at the time of the study design. Molecular techniques would likely improve the etiologic documentation of cases not fulfilling the bacteriological diagnostic criteria, and future studies might benefit from their use. These techniques might also provide evidence of occult polymicrobial infection involving fastidious organisms such as P. acnes, likely to be overgrown by more hardy pathogens.
The immunoassay also has limitations. First, bacterial species not covered by the assay account for 40% of total PJIs. The assay sensitivity and specificity in diagnosing PJIs caused by any etiological agent, therefore, only reach 67.7% and 65.6%, respectively. However, the assay was developed to provide a noninvasive genus-level documentation of infection with selected microorganisms, in conjunction with the other tests used to establish infection; moreover, the multiplex nature of the assay allows for the subsequent broadening of the bacterial species included. Second, the sole detection of IgGs does not provide information on the time frame of the infection and may yield false-negative results when performed too soon after the onset of infection, as suggested by the lower performance of the assay within the first 3 months following the arthroplasty. Third, cross-reactivity between antigens from species belonging to the same genus (i.e., Staphylococcus) prevents species-level identification of the causative agent; however, it allows the diagnosis of PJIs caused by closely related species of the same genus. Further studies will allow us to determine the specificity of the antibody response observed in PJI cases involving nontargeted staphylococci, streptococci, and propionibacteria and might lead to broadening of the range of identified species. Finally, “real-life” patients with sera yielding undetermined results (6% of serum samples from PJI cases due to the targeted staphylococci in our series) would be retested after 4 to 6 weeks to provide a definitive result, a modality that was not possible within the framework of the blinded prospective study.
In summary, this novel noninvasive approach based on multiplex antibody detection provides information not captured by common serological inflammation markers, hinting at the identification of several major PJI pathogens at the genus level. It might provide help in showing infections in patients without elevated ESR or CRP, allowing earlier management while preventing invasive and costly investigations. It might facilitate the interpretation of inconclusive bacteriological results (i.e., negative or single positive cultures) or P. acnes cultures. This approach should be evaluated and validated by further studies performed in different patient populations and countries, and its medicoeconomic benefit should be determined. The usefulness of this assay both at presentation of patients and as a follow-up tool should be further evaluated by longitudinal studies.
Supplementary Material
ACKNOWLEDGMENTS
We thank the patients for their participation in the study and Layidé Méaude and Laurence Raizonville (Clinical Research Unit, Hospital Ambroise Paré) for their dedication to the study.
The study was funded by Diaxonhit. Diaxonhit personnel developed the immunoassay and participated in the redaction of the manuscript. J.-L.G. is a shareholder of Diaxonhit. J.-L.G. and M.R. served as consultants for Diaxonhit.
The study was designed by the authors in collaboration with the sponsor (Diaxonhit). The Clinical Research Unit Paris Île-de-France Ouest was responsible for clinical data management and statistical analysis. The laboratories of the participating centers performed the microbiological cultures and measured the ESR and CRP levels. Surgeon investigators recorded the clinical data in a case report form. The sponsor was responsible for monitoring the data and for performing the immunoassay. The authors verified the data, wrote all drafts of the manuscript, made the decision to submit the manuscript for publication, and vouch for the completeness of the data, the accuracy of the analyses, and the fidelity of the study to the protocol.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.02885-15.
REFERENCES
- 1.Anonymous. 2010. National hospital discharge survey: 2010 table, procedures by selected patient characteristics, number by procedure category and age. Centers for Disease Control and Prevention. http://www.cdc.gov/nchs/data/nhds/4procedures/2010pro4_numberprocedureage.pdf.
- 2.Kurtz S, Ong K, Lau E, Mowat F, Halpern M. 2007. Projections of primary and revision hip and knee arthroplasty in the United States from 2005 to 2030. J Bone Joint Surg Am 89:780–785. doi: 10.2106/JBJS.F.00222. [DOI] [PubMed] [Google Scholar]
- 3.Kurtz SM, Lau E, Schmier J, Ong KL, Zhao K, Parvizi J. 2008. Infection burden for hip and knee arthroplasty in the United States. J Arthroplasty 23:984–991. doi: 10.1016/j.arth.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 4.Kurtz SM, Lau E, Watson H, Schmier JK, Parvizi J. 2012. Economic burden of periprosthetic joint infection in the United States. J Arthroplasty 27(8 Suppl):61–65.e1. [DOI] [PubMed] [Google Scholar]
- 5.Bozic KJ, Kamath AF, Ong K, Lau E, Kurtz S, Chan V, Vail TP, Rubash H, Berry DJ. 2015. Comparative epidemiology of revision arthroplasty: failed THA poses greater clinical and economic burdens than failed TKA. Clin Orthop Relat Res 473:2131–2138. doi: 10.1007/s11999-014-4078-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamath AF, Ong KL, Lau E, Chan V, Vail TP, Rubash HE, Berry DJ, Bozic KJ. 2015. Quantifying the burden of revision total joint arthroplasty for periprosthetic infection. J Arthroplasty 30:1492−1497. doi: 10.1016/j.arth.2015.03.035. [DOI] [PubMed] [Google Scholar]
- 7.Peel TN, Cole NC, Dylla BL, Patel R. 2015. Matrix-assisted laser desorption ionization time of flight mass spectrometry and diagnostic testing for prosthetic joint infection in the clinical microbiology laboratory. Diagn Microbiol Infect Dis 81:163–168. doi: 10.1016/j.diagmicrobio.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 8.Tande AJ, Patel R. 2014. Prosthetic joint infection. Clin Microbiol Rev 27:302–345. doi: 10.1128/CMR.00111-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zimmerli W, Trampuz A, Ochsner PE. 2004. Prosthetic-joint infections. N Engl J Med 351:1645–1654. doi: 10.1056/NEJMra040181. [DOI] [PubMed] [Google Scholar]
- 10.Osmon DR, Berbari EF, Berendt AR, Lew D, Zimmerli W, Steckelberg JM, Rao N, Hanssen A, Wilson WR. 2013. Diagnosis and management of prosthetic joint infection: clinical practice guidelines by the Infectious Diseases Society of America. Clin Infect Dis 56:e1–e25. doi: 10.1093/cid/cis803. [DOI] [PubMed] [Google Scholar]
- 11.Della Valle C, Parvizi J, Bauer TW, DiCesare PE, Evans RP, Segreti J, Spangehl M, Watters WC III, Keith M, Turkelson CM, Wies JL, Sluka P, Hitchcock K. 2011. American Academy of Orthopaedic Surgeons clinical practice guideline on: the diagnosis of periprosthetic joint infections of the hip and knee. J Bone Joint Surg Am 93:1355–1357. doi: 10.2106/JBJS.9314ebo. [DOI] [PubMed] [Google Scholar]
- 12.Pei R, Lee J, Chen T, Rojo S, Terasaki PI. 1999. Flow cytometric detection of HLA antibodies using a spectrum of microbeads. Hum Immunol 60:1293–1302. doi: 10.1016/S0198-8859(99)00121-4. [DOI] [PubMed] [Google Scholar]
- 13.Dessau RB, Moller JK, Kolmos B, Henningsson AJ. 2015. Multiplex assay (Mikrogen recomBead) for detection of serum IgG and IgM antibodies to 13 recombinant antigens of Borrelia burgdorferi sensu lato in patients with neuroborreliosis: the more the better? J Med Microbiol 64:224–231. doi: 10.1099/jmm.0.000009. [DOI] [PubMed] [Google Scholar]
- 14.Marangoni A, Nardini P, Foschi C, Moroni A, D'Antuono A, Bacchi Reggiani L, Cevenini R. 2013. Evaluation of the BioPlex 2200 syphilis system as a first-line method of reverse-sequence screening for syphilis diagnosis. Clin Vaccine Immunol 20:1084–1088. doi: 10.1128/CVI.00316-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryding U, Espersen F, Soderquist B, Christensson B. 2002. Evaluation of seven different enzyme-linked immunosorbent assays for serodiagnosis of Staphylococcus aureus bacteremia. Diagn Microbiol Infect Dis 42:9–15. doi: 10.1016/S0732-8893(01)00311-X. [DOI] [PubMed] [Google Scholar]
- 16.Elston J, Ling M, Jeffs B, Adams K, Thaker H, Moss P, Meigh R, Barlow G. 2010. An evaluation of the usefulness of Staphylococcus aureus serodiagnosis in clinical practice. Eur J Clin Microbiol Infect Dis 29:737–739. doi: 10.1007/s10096-010-0907-1. [DOI] [PubMed] [Google Scholar]
- 17.Roux AL, Sivadon-Tardy V, Bauer T, Lortat-Jacob A, Herrmann JL, Gaillard JL, Rottman M. 2011. Diagnosis of prosthetic joint infection by beadmill processing of a periprosthetic specimen. Clin Microbiol Infect 17:447–450. doi: 10.1111/j.1469-0691.2010.03359.x. [DOI] [PubMed] [Google Scholar]
- 18.Parvizi J, Zmistowski B, Berbari EF, Bauer TW, Springer BD, Della Valle CJ, Garvin KL, Mont MA, Wongworawat MD, Zalavras CG. 2011. New definition for periprosthetic joint infection: from the Workgroup of the Musculoskeletal Infection Society. Clin Orthop Relat Res 469:2992–2994. doi: 10.1007/s11999-011-2102-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parvizi J, Gehrke T. 2013. Proceedings of the international consensus meeting on periprosthetic joint infection. Musculoskeletal Infection Society, Rochester, MN: http://www.msis-na.org/international-consensus/ [Google Scholar]
- 20.R Core Team. 2008. R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 21.Lambert PA, Van Maurik A, Parvatham S, Akhtar Z, Fraise AP, Krikler SJ. 1996. Potential of exocellular carbohydrate antigens of Staphylococcus epidermidis in the serodiagnosis of orthopaedic prosthetic infection. J Med Microbiol 44:355–361. doi: 10.1099/00222615-44-5-355. [DOI] [PubMed] [Google Scholar]
- 22.Artini M, Romano C, Manzoli L, Scoarughi GL, Papa R, Meani E, Drago L, Selan L. 2011. Staphylococcal IgM enzyme-linked immunosorbent assay for diagnosis of periprosthetic joint infections. J Clin Microbiol 49:423–425. doi: 10.1128/JCM.01836-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jacqueline C, Caillon J. 2014. Impact of bacterial biofilm on the treatment of prosthetic joint infections. J Antimicrob Chemother 69(Suppl 1):i37−i40. doi: 10.1093/jac/dku254. [DOI] [PubMed] [Google Scholar]
- 24.Tande AJ, Osmon DR, Greenwood-Quaintance KE, Mabry TM, Hanssen AD, Patel R. 2014. Clinical characteristics and outcomes of prosthetic joint infection caused by small colony variant staphylococci. mBio 5:e01910–e01914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vandecasteele SJ, Peetermans WE, Merckx R, Van Eldere J. 2003. Expression of biofilm-associated genes in Staphylococcus epidermidis during in vitro and in vivo foreign body infections. J Infect Dis 188:730–737. doi: 10.1086/377452. [DOI] [PubMed] [Google Scholar]
- 26.Berbari E, Mabry T, Tsaras G, Spangehl M, Erwin PJ, Murad MH, Steckelberg J, Osmon D. 2010. Inflammatory blood laboratory levels as markers of prosthetic joint infection: a systematic review and meta-analysis. J Bone Joint Surg Am 92:2102–2109. doi: 10.2106/JBJS.I.01199. [DOI] [PubMed] [Google Scholar]
- 27.Piper KE, Fernandez-Sampedro M, Steckelberg KE, Mandrekar JN, Karau MJ, Steckelberg JM, Berbari EF, Osmon DR, Hanssen AD, Lewallen DG, Cofield RH, Sperling JW, Sanchez-Sotelo J, Huddleston PM, Dekutoski MB, Yaszemski M, Currier B, Patel R. 2010. C-reactive protein, erythrocyte sedimentation rate and orthopedic implant infection. PLoS One 5:e9358. doi: 10.1371/journal.pone.0009358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeller V, Lavigne M, Biau D, Leclerc P, Ziza JM, Mamoudy P, Desplaces N. 2009. Outcome of group B streptococcal prosthetic hip infections compared to that of other bacterial infections. Joint Bone Spine 76:491–496. doi: 10.1016/j.jbspin.2008.11.010. [DOI] [PubMed] [Google Scholar]
- 29.Sendi P, Christensson B, Uckay I, Trampuz A, Achermann Y, Boggian K, Svensson D, Widerstrom M, Zimmerli W. 2011. Group B streptococcus in prosthetic hip and knee joint-associated infections. J Hosp Infect 79:64–69. doi: 10.1016/j.jhin.2011.04.022. [DOI] [PubMed] [Google Scholar]
- 30.Sampedro MF, Piper KE, McDowell A, Patrick S, Mandrekar JN, Rouse MS, Steckelberg JM, Patel R. 2009. Species of Propionibacterium and Propionibacterium acnes phylotypes associated with orthopedic implants. Diagn Microbiol Infect Dis 64:138–145. doi: 10.1016/j.diagmicrobio.2009.01.024. [DOI] [PubMed] [Google Scholar]
- 31.Achermann Y, Goldstein EJ, Coenye T, Shirtliff ME. 2014. Propionibacterium acnes: from commensal to opportunistic biofilm-associated implant pathogen. Clin Microbiol Rev 27:419–440. doi: 10.1128/CMR.00092-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dramis A, Aldlyami E, Grimer RJ, Dunlop DJ, O'Connell N, Elliott T. 2009. What is the significance of a positive Propionibacterium acnes culture around a joint replacement? Int Orthop 33:829–833. doi: 10.1007/s00264-008-0534-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tsukayama DT, Estrada R, Gustilo RB. 1996. Infection after total hip arthroplasty. A study of the treatment of one hundred and six infections. J Bone Joint Surg Am 78:512–523. [DOI] [PubMed] [Google Scholar]
- 34.Updegrove GF, Armstrong AD, Kim HM. 2015. Preoperative and intraoperative infection workup in apparently aseptic revision shoulder arthroplasty. J Shoulder Elbow Surg 24:491–500. doi: 10.1016/j.jse.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 35.Dodson CC, Craig EV, Cordasco FA, Dines DM, Dines JS, Dicarlo E, Brause BD, Warren RF. 2010. Propionibacterium acnes infection after shoulder arthroplasty: a diagnostic challenge. J Shoulder Elbow Surg 19:303–307. doi: 10.1016/j.jse.2009.07.065. [DOI] [PubMed] [Google Scholar]
- 36.Sulkowski MS, Abolnik IZ, Morris EI, Granger DL. 1994. Infectious arthritis due to Propionibacterium acnes in a prosthetic joint. Clin Infect Dis 19:224–225. doi: 10.1093/clinids/19.1.224. [DOI] [PubMed] [Google Scholar]
- 37.Atkins BL, Athanasou N, Deeks JJ, Crook DW, Simpson H, Peto TE, McLardy-Smith P, Berendt AR. 1998. Prospective evaluation of criteria for microbiological diagnosis of prosthetic-joint infection at revision arthroplasty. The OSIRIS Collaborative Study Group. J Clin Microbiol 36:2932–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trampuz A, Piper KE, Jacobson MJ, Hanssen AD, Unni KK, Osmon DR, Mandrekar JN, Cockerill FR, Steckelberg JM, Greenleaf JF, Patel R. 2007. Sonication of removed hip and knee prostheses for diagnosis of infection. N Engl J Med 357:654–663. doi: 10.1056/NEJMoa061588. [DOI] [PubMed] [Google Scholar]
- 39.Bémer P, Plouzeau C, Tande D, Leger J, Giraudeau B, Valentin AS, Jolivet-Gougeon A, Vincent P, Corvec S, Gibaud S, Juvin ME, Hery-Arnaud G, Lemarie C, Kempf M, Bret L, Quentin R, Coffre C, de Pinieux G, Bernard L, Burucoa C, Centre de Reference des Infections Osteo-articulaires du Grand Ouest Study Team. 2014. Evaluation of 16S rRNA gene PCR sensitivity and specificity for diagnosis of prosthetic joint infection: a prospective multicenter cross-sectional study. J Clin Microbiol 52:3583–3589. doi: 10.1128/JCM.01459-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.