Abstract
Perirectal surveillance cultures and a stool culture grew Aeromonas species from three patients over a 6-week period and were without epidemiological links. Detection of the blaKPC-2 gene in one isolate prompted inclusion of non-Enterobacteriaceae in our surveillance culture workup. Whole-genome sequencing confirmed that the isolates were unrelated and provided data for Aeromonas reference genomes.
TEXT
Aeromonas species are Gram-negative environmental bacteria that can cause infections in both immunocompetent and immunocompromised persons (1, 2) and have been isolated from a wide range of aquatic environments, including marine ecosystems, freshwater, hospital effluent, and drinking water (1, 3–6). The global rise of multidrug-resistant bacteria raises concern that a ubiquitous organism, such as an Aeromonas sp., could acquire antibiotic resistance genes derived from human isolates (3, 4, 7, 8). A variety of antibiotic resistance phenotypes, including carbapenem resistance (3, 4, 9–15), have been found in both clinical and environmental Aeromonas isolates. Although rare, clinical Aeromonas isolates containing blaKPC genes have been reported (16, 17).
Active microbial surveillance for multidrug-resistant Gram-negative organisms is integral to inpatient care at the NIH Clinical Center. Perirectal swabs are collected upon hospital admission, twice weekly in the intensive care unit, and monthly from all inpatients (excluding those in behavioral health wards). Perirectal swabs were inoculated onto HardyCHROM CRE (Hardy Diagnostics, Santa Maria, CA) and incubated at 35°C ambient air for 18 to 24 h. Isolates were identified using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS), and isolates belonging to Enterobacteriaceae underwent PCR for detection of blaKPC and blaNDM-1. Stool specimens were plated onto sheep blood, MacConkey, and xylose lysine deoxycholate agars with incubation at 35°C and 5% CO2 for 48 h, and on Campy CVA agar with incubation at 42°C under microaerophilic conditions. Results from the surveillance cultures that suggest possible nosocomial acquisition of carbapenemase-producing bacteria trigger epidemiological investigations, including environmental sampling.
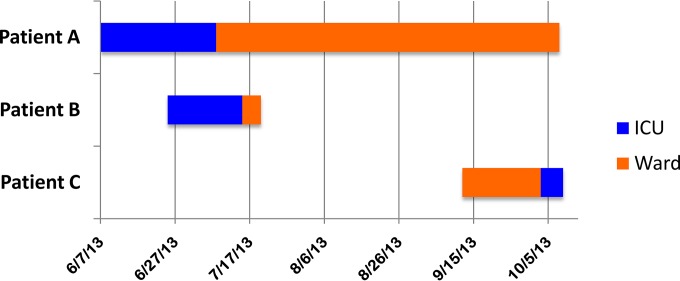
Perirectal surveillance cultures from two hospitalized adult patients and a stool culture from a third patient grew Aeromonas spp. within a 6-week period. The stool culture isolate (from patient A) was resistant to ertapenem and susceptible to imipenem and meropenem; the perirectal isolates (from patients B and C) were resistant to all three carbapenems. MALDI-TOF MS could not reliably identify the isolates to the species level. On average, clinical Aeromonas spp. had been isolated from clinical cultures in only four to five patients per year in our institution since 2010; this was the first time that Aeromonas had been isolated from perirectal surveillance cultures since they were introduced in 2011. This apparent cluster of Aeromonas colonization prompted an investigation. A patient trace (TheraDoc) revealed that patients A and B had each received care in the intensive care unit for 13 days but were never housed in the same room. Both were subsequently transferred to different rooms on another medical ward; patient B was discharged from ward room 1 five days later, and patient C was admitted to ward room 1 seven weeks after patient B had vacated the room (Fig. 1). An extensive chart review identified no other epidemiological links among the three patients, and extensive environmental samples, including sink water and swabs of faucet aerators for culture on multiple media, were negative for Aeromonas spp. and carbapenem-resistant organisms.
FIG 1.
Patient trace.
To investigate possible genetic relatedness, whole-genome sequencing was performed using Illumina MiSeq and assembled with MIRA for all three Aeromonas isolates (18). 16S rRNA sequences indicated that patient B's isolate was related to Aeromonas hydrophila, while isolates from patients A and C were most closely related to Aeromonas veronii, thus narrowing the epidemiological investigation.
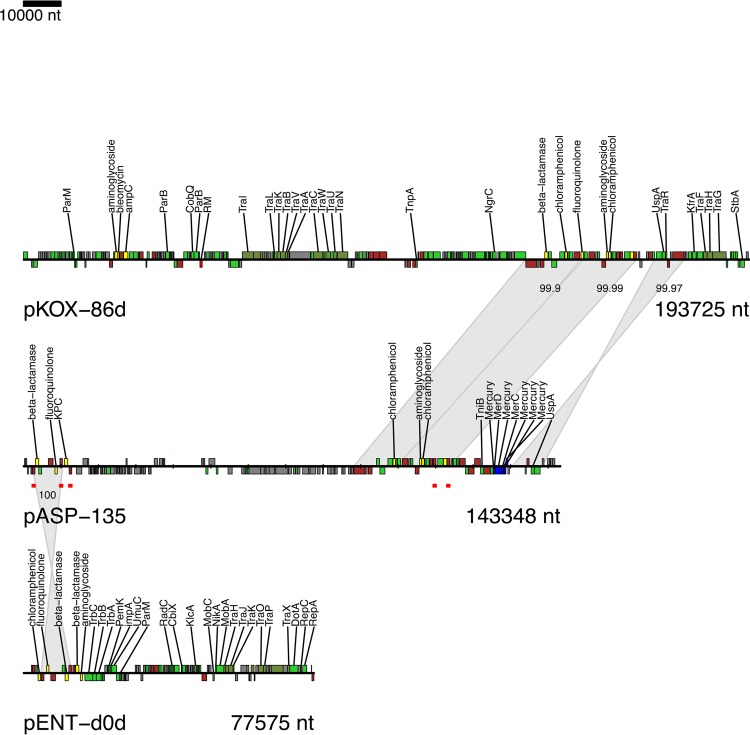
The A. hydrophila isolate from patient B (AHNIH1) was sequenced on the PacBio RSII instrument to generate a fully contiguous genome. AHNIH1 has a 4.91-Mb chromosome and carries a 143-kb plasmid (pASP-135) (Fig. 2). Unexpectedly, this plasmid carries a blaKPC-2 gene (retrospectively confirmed by blaKPC PCR) and TEM β-lactamase and genes encoding resistance to aminoglycosides, chloramphenicol, fluoroquinolones, macrolides, and mercury. We speculate that patient B acquired the isolate from an aqueous source within the hospital environment; however, the origin of the blaKPC-2 plasmid is unknown, as its sequence does not match either those of other blaKPC-2 bacteria identified in our institution or other publically available references. The blaKPC-2 gene is not carried on the commonly associated Tn4401 element and is instead flanked by IS26 elements, which are important factors in the mobility of resistance determinants (19). All three isolates contain chromosomally encoded homologs of CphA, a metallo-β-lactamase that is a frequent etiology of carbapenem resistance in Aeromonas species (20).
FIG 2.
The pASP-135 plasmid from the A. hydrophila isolate has short regions of identity to previously sequenced plasmids (pKOX-86d and pENT-d0d), largely demarcated by mobile elements (in brown). Genes are colored by function: conjugation, dark green; antibiotic resistance, yellow; metal resistance, blue; transposase/integrase/resolvase, brown; hypothetical, gray; other, light green. IS26 elements are highlighted on pASP-135 with red underlining. Aligned regions between plasmids are indicated by gray ribbons, with percent identities indicated.
To explore a possible epidemiological link between the A. veronii isolates from patients A and C, we undertook a genetic analysis. As no finished reference genome for A. veronii is available, we generated a fully contiguous reference genome on the PacBio RSII instrument. The A. veronii isolate from patient C (AVNIH1) has a 4.76-Mb chromosome and a 198-kb plasmid (pASP-a58) carrying genes encoding resistance to aminoglycosides, β-lactams, fluoroquinolones, macrolides, mercury, and rifampin, possibly explaining the recovery on selective surveillance media. The genome of the A. veronii-like isolate from patient A (AVNIH2) was assembled using AVNIH1 (from patient C) as the reference. The isolates from patients A and C are only 98.7% identical across aligned regions, excluding the possibility of transmission between these two patients. Neither isolate was found to contain a blaKPC gene.
Although epidemiological data alone were unable to establish or refute a link among the three Aeromonas isolates, despite the temporal clustering, whole-genome microbial sequencing established that these isolates were unrelated to each other and did not represent a true nosocomial cluster. The ability of sequencing data to resolve this epidemiological uncertainty enabled us to avoid further investigation, which would likely have been fruitless, labor-intensive, and expensive.
Analysis of sequencing data demonstrated that the Aeromonas isolates in this report belong to two different species, and we unexpectedly found that one isolate carried the blaKPC-2 gene on a plasmid. Few complete genome sequences exist for Aeromonas spp., making these a valuable addition to the reference database.
Hospital surveillance for blaKPC bacteria should not be limited to Enterobacteriaceae, as the blaKPC carbapenemase has been reported in numerous species belonging to a wide range of other bacteria, including non-Enterobacteriaceae (e.g., Pseudomonas, Aeromonas, and Acinetobacter) (21–23). As a result of this case, the standard workup of bacterial colonies growing on perirectal surveillance cultures at our institution now includes blaKPC and blaNDM screening for all Gram-negative bacilli, not limited to the Enterobacteriaceae.
In summary, the detection of blaKPC in a surveillance Aeromonas isolate led to a change in our hospital's multidrug-resistant Gram-negative surveillance protocol. Whole-genome microbial sequencing suggested that nosocomial transmission did not occur, prevented a more laborious evaluation, and provided reference genome sequences that might be useful for future epidemiological investigations.
Nucleotide sequence accession number.
The whole-genome sequencing data can be retrieved at NCBI BioProject no. PRJNA279868.
ACKNOWLEDGMENTS
This research was supported by the NIH Clinical Center and the intramural research programs of the National Human Genome Research Institute and the National Institute of Allergy and Infectious Diseases.
We thank Clay Deming for his underlying molecular diagnostic contribution, and Camille Hardiman and the staff of the NIH Clinical Center Microbiology Service for their contributions to this project. Sequencing was performed at NIH Intramural Sequencing Center and Leidos Biomedical Research, Frederick National Laboratory for Cancer Research. Isolates can be obtained from K.M.F.; a material transfer agreement is necessary.
REFERENCES
- 1.Janda JM, Abbott SL. 2010. The genus Aeromonas: taxonomy, pathogenicity, and infection. Clin Microbiol Rev 23:35–73. doi: 10.1128/CMR.00039-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parker JL, Shaw JG. 2011. Aeromonas spp. clinical microbiology and disease. J Infect 62:109–118. doi: 10.1016/j.jinf.2010.12.003. [DOI] [PubMed] [Google Scholar]
- 3.Walsh TR, Weeks J, Livermore DM, Toleman MA. 2011. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis 11:355–362. doi: 10.1016/S1473-3099(11)70059-7. [DOI] [PubMed] [Google Scholar]
- 4.Picão RC, Cardoso JP, Campana EH, Nicoletti AG, Petrolini FV, Assis DM, Juliano L, Gales AC. 2013. The route of antimicrobial resistance from the hospital effluent to the environment: focus on the occurrence of KPC-producing Aeromonas spp. and Enterobacteriaceae in sewage. Diagn Microbiol Infect Dis 76:80–85. doi: 10.1016/j.diagmicrobio.2013.02.001. [DOI] [PubMed] [Google Scholar]
- 5.Presley SM, Rainwater TR, Austin GP, Platt SG, Zak JC, Cobb GP, Marsland EJ, Tian K, Zhang B, Anderson TA, Cox SB, Abel MT, Leftwich BD, Huddleston JR, Jeter RM, Kendall RJ. 2006. Assessment of pathogens and toxicants in New Orleans, LA following Hurricane Katrina. Environ Sci Technol 40:468–474. doi: 10.1021/es052219p. [DOI] [PubMed] [Google Scholar]
- 6.Hiransuthikul N, Tantisiriwat W, Lertutsahakul K, Vibhagool A, Boonma P. 2005. Skin and soft-tissue infections among tsunami survivors in southern Thailand. Clin Infect Dis 41:e93–e96. doi: 10.1086/497372. [DOI] [PubMed] [Google Scholar]
- 7.Nguyen HN, Van TT, Nguyen HT, Smooker PM, Shimeta J, Coloe PJ. 2014. Molecular characterization of antibiotic resistance in Pseudomonas and Aeromonas isolates from catfish of the Mekong Delta, Vietnam. Vet Microbiol 171:397–405. doi: 10.1016/j.vetmic.2014.01.028. [DOI] [PubMed] [Google Scholar]
- 8.Dobiasova HKI, Piackova V, Vesely T, Cizek A, Dolejska M. 2014. Ornamental fish as a source of plasmid-mediated quinolone resistance genes and antibiotic resistance plasmids. Vet Microbiol 171:413–421. doi: 10.1016/j.vetmic.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 9.Neuwirth C, Siebor E, Robin F, Bonnet R. 2007. First occurrence of an IMP metallo-beta-lactamase in Aeromonas caviae: IMP-19 in an isolate from France. Antimicrob Agents Chemother 51:4486–4488. doi: 10.1128/AAC.01462-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fosse T, Giraud-Morin C, Madinier I, Mantoux F, Lacour JP, Ortonne JP. 2004. Aeromonas hydrophila with plasmid-borne class A extended-spectrum β-lactamase TEM-24 and three chromosomal class B, C, and D β-lactamases, isolated from a patient with necrotizing fasciitis. Antimicrob Agents Chemother 48:2342–2343. doi: 10.1128/AAC.48.6.2342-2343.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Libisch B, Giske CG, Kovacs B, Toth TG, Fuzi M. 2008. Identification of the first VIM metallo-beta-lactamase-producing multiresistant Aeromonas hydrophila strain. J Clin Microbiol 46:1878–1880. doi: 10.1128/JCM.00047-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sánchez-Céspedes J, Figueras MJ, Aspiroz C, Aldea MJ, Toledo M, Alperí A, Marco F, Vila J. 2009. Development of imipenem resistance in an Aeromonas veronii biovar sobria clinical isolate recovered from a patient with cholangitis. J Med Microbiol 58:451–455. doi: 10.1099/jmm.0.47804-0. [DOI] [PubMed] [Google Scholar]
- 13.Chen PL, Ko WC, Wu CJ. 2012. Complexity of beta-lactamases among clinical Aeromonas isolates and its clinical implications. J Microbiol Immunol Infect 45:398–403. doi: 10.1016/j.jmii.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 14.Marchandin H, Godreuil S, Darbas H, Jean-Pierre H, Jumas-Bilak E, Chanal C, Bonnet R. 2003. Extended-spectrum beta-lactamase TEM-24 in an Aeromonas clinical strain; acquisition from the prevalent Enterobacter aerogenes clone in France. Antimicrob Agents Chemother 47:3994–3995. doi: 10.1128/AAC.47.12.3994-3995.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Esteve C, Alcaide E, Giménez MJ. 2014. Multidrug-resistant (MDR) Aeromonas recovered from the metropolitan area of Valencia (Spain): diseases spectrum and prevalence in the environment. Eur J Clin Microbiol Infect Dis 34:137–145. [DOI] [PubMed] [Google Scholar]
- 16.Mathers AJ, Yeh A, Cox HL, Carroll J, Hoffman PS, Poulter MD, Sifri CD. 2013. Nosocomially acquired Klebsiella pneumoniae carbapenemase (KPC)-producing Aeromonas hydrophila associated with KPC transmission to multiple species of Enterobacteriaceae, abstr C2-1095. 53rd Int Conf Antimicrob Agents Chemother (ICAAC), 10 to 13 September 2013, Denver, CO. [Google Scholar]
- 17.Visconde MF, Zoccoli CM, Cardoso JP, Araujo V, Affini R, Tobouti NR, Pignantari AC, Gales A. 2013. First report of KPC-2-producing Aeromonas hydrophila on a clinical isolate: a surveillance report, abstr C2-1104. 53rd Int Conf Antimicrob Agents Chemother (ICAAC), 10 to 13 September 2013, Denver, CO. [Google Scholar]
- 18.Chevreux B, Wetter T, Suhal S. 1999. Genome sequence assembly using trace signals and additional sequence information, p 45–56. In Computer science and biology: proceedings of the German Conference on Bioinformatics, GCB 99. [Google Scholar]
- 19.He S, Hickman AB, Varani AM, Siguier P, Chandler M, Dekker JP, Dyda F. 2015. Insertion sequence IS26 reorganizes plasmids in clinically isolated multidrug-resistant bacteria by replicative transposition. mBio 6(3):e00762-15. doi: 10.1128/mBio.00762-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segatore B, Massidda O, Satta G, Setacci D, Amicosante G. 1993. High specificity of cphA-encoded metallo-beta-lactamase from Aeromonas hydrophila AE036 for carbapenems and its contribution to beta-lactam resistance. Antimicrob Agents Chemother 37:1324–1328. doi: 10.1128/AAC.37.6.1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Villegas MV, Lolans K, Correa A, Kattan JN, Lopez JA, Quinn JP. 2007. First identification of Pseudomonas aeruginosa isolates producing a KPC-type carbapenem-hydrolyzing beta-lactamase. Antimicrob Agents Chemother 51:1553–1555. doi: 10.1128/AAC.01405-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poirel L, Nordmann P, Lagrutta E, Cleary T, Munoz-Price LS. 2010. Emergence of KPC-producing Pseudomonas aeruginosa in the United States. Antimicrob Agents Chemother 54:3072. doi: 10.1128/AAC.00513-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Robledo IE, Aquino EE, Sante MI, Santana JL, Otero DM, Leon CF, Vazquez GJ. 2010. Detection of KPC in Acinetobacter spp. in Puerto Rico. Antimicrob Agents Chemother 54:1354–1357. doi: 10.1128/AAC.00899-09. [DOI] [PMC free article] [PubMed] [Google Scholar]