Abstract
Staphylococcus aureus is a major bacterial pathogen causing a variety of diseases ranging from wound infections to severe bacteremia or intoxications. Besides host factors, the course and severity of disease is also widely dependent on the genotype of the bacterium. Whole-genome sequencing (WGS), followed by bioinformatic sequence analysis, is currently the most extensive genotyping method available. To identify clinically relevant staphylococcal virulence and resistance genes in WGS data, we developed an in silico typing scheme for the software SeqSphere+ (Ridom GmbH, Münster, Germany). The implemented target genes (n = 182) correspond to those queried by the Identibac S. aureus Genotyping DNA microarray (Alere Technologies, Jena, Germany). The in silico scheme was evaluated by comparing the typing results of microarray and of WGS for 154 human S. aureus isolates. A total of 96.8% (n = 27,119) of all typing results were equally identified with microarray and WGS (40.6% present and 56.2% absent). Discrepancies (3.2% in total) were caused by WGS errors (1.7%), microarray hybridization failures (1.3%), wrong prediction of ambiguous microarray results (0.1%), or unknown causes (0.1%). Superior to the microarray, WGS enabled the distinction of allelic variants, which may be essential for the prediction of bacterial virulence and resistance phenotypes. Multilocus sequence typing clonal complexes and staphylococcal cassette chromosome mec element types inferred from microarray hybridization patterns were equally determined by WGS. In conclusion, WGS may substitute array-based methods due to its universal methodology, open and expandable nature, and rapid parallel analysis capacity for different characteristics in once-generated sequences.
INTRODUCTION
Staphylococcus aureus is a Gram-positive facultative pathogenic bacterium that is responsible for a high percentage of hospital- and community-acquired infections worldwide. An infection with S. aureus may manifest itself in a broad variety of diseases, ranging from rather harmless local skin infections to severe bacteremia or intoxications (1). This extensive spectrum of virulence is owed, in part, to the bacterium's individual equipment with virulence factors. Analyzing these virulence factors is difficult because purified staphylococcal toxins do not essentially cause distinctive symptoms when administered in the absence of the bacterium, and the specific knockout of single virulence factors does not necessarily reduce the bacterial virulence (2). Thus, it seems that the combination of different virulence factors, their regulation and transcription, and their allelic variants play a crucial role in determining the eventually expressed virulence phenotype. Therefore, it is important to determine not only the presence or absence of single key factors, such as, e.g., Panton-Valentine leucocidin (PVL) or certain enterotoxins, but to obtain a comprehensive picture of the exact allelic variants of as many virulence-associated genes and their regulatory systems as possible. With regard to treatment, it is also essential to know whether the bacterium is resistant to one or multiple antimicrobial agents. For S. aureus, especially the methicillin resistance status (methicillin-resistant S. aureus [MRSA] phenotype) and the identification of the responsible resistance-conferring mobile genetic element (staphylococcal cassette chromosome mec element [SCCmec]) are of interest. In addition to this essential information for patient treatment, it is also important to determine the bacterium's clonal lineage in order to trace its spread over time and space. One of the most frequently used molecular methods to determine clonal lineage is multilocus sequence typing (MLST), which classifies the isolates into sequence types (STs) and clonal complexes (CCs) (3, 4).
Many different single PCRs or phenotypic tests are available to obtain information about the different genetic features of S. aureus isolates. More extensive information about the bacterial genotype can be obtained by DNA microarrays, which allow the parallel identification of a variety of genes. One of them is the commercial Identibac S. aureus Genotyping DNA microarray (Alere Technologies, Jena, Germany), which queries 191 unique staphylococcal genes and automatically deduces the CC of the isolate and, if present, the SCCmec type (5–9). Its targets were selected either to encode clinically relevant information or to be of use for typing purposes (5). Since the costs of whole-genome sequencing (WGS) of prokaryotes have been dramatically dropping during the last few years, WGS is on its way to replace DNA microarrays in clinical and biological laboratories (10). Hence, the amount of complete bacterial genomes available in public databases like NCBI GenBank (http://www.ncbi.nlm.nih.gov/GenBank/) is growing rapidly, providing a massive amount of genomic information. However, sequence analysis, i.e., the extraction of relevant target gene sequences in WGS raw data, is not trivial and usually requires extensive knowledge in bioinformatics. In order to bridge this gap between available sequence raw data and precise genomic information with regard to applied (clinical) questions, we developed an easy-to-use WGS in silico typing scheme that, for the moment, queries the same target genes as the Alere Identibac DNA microarray. The commercial software SeqSphere+ (Ridom GmbH, Münster, Germany) was used for sequence analysis. In the future, this in silico typing scheme may be extended to other genes not present in the microarray; however, for a first accuracy validation of the new typing scheme, we assessed the concordance of the two methods.
MATERIALS AND METHODS
Bacterial isolates.
The analyzed study set contained 154 epidemiologically unrelated S. aureus isolates (145 methicillin-susceptible S. aureus [MSSA] isolates and 9 MRSA isolates) collected from healthy volunteers and various clinical specimens from six different hospitals in Germany (Münster [n = 19], Freiburg [n = 25], and Homburg [n = 22]) and sub-Saharan Africa (Gabon [n = 35], Mozambique [n = 17], Tanzania [n = 36]) between 2010 and 2012 in the framework of the African-German Network on Staphylococci (11) (www.African-German-Staph.net). The isolates originated from diverse human samples, including asymptomatic nasal colonization (n = 78), wound infections (n = 64), and bacteremia (n = 12). For clinical isolates, the inclusion criterion was community onset of disease, i.e., the samples were taken ≤48 h after admission. For nasal isolates, the inclusion criteria were (i) no hospitalization in the past 4 weeks, (ii) no antibacterial treatment in the past 4 weeks, and (iii) no antituberculous treatment in the past 4 weeks. No further exclusion criteria were applied. Until used for WGS, the bacteria were stored at −70°C.
Microarray genotyping and data processing.
The 154 isolates were at first genotyped using the Identibac S. aureus Genotyping DNA microarray (hereafter referred to as “microarray”; Alere Technologies GmbH, Jena, Germany). The laboratory protocol was executed according to the manufacturer's instructions; subsequent data analysis was performed with the software Iconoclust (version 3.2.r1; Alere Technologies). This software automatically assigns the targets to the categories “positive” (present), “negative” (absent), or “ambiguous” based on the intensity of the representing spot in relation to the background using predefined thresholds. As recently described (7), targets that were determined as ambiguous (n = 2,788, 5.4%) were replaced by present or absent according to predictions made with latent factor models (LFM) (12, 13). Briefly, the LFM method reconstructs missing entries in a data matrix based on the entries in neighboring fields of the involved columns and rows. The accuracy of this approach was tested by a bootstrap approach (14) as follows: First, 5% of randomly selected entries that were known to be positive or negative were removed from the data set. This fraction corresponds to the typical number of targets typed as ambiguous in the microarray experiments (see above). Then, these missing entries were predicted using LFM and were compared to the original values. As a result, LFM yielded an accuracy of 97% against the original values. Thus, the error rate of predicted values can be estimated as about 3%.
The microarray includes 334 oligonucleotide probes in total, covering various genes for clinically relevant features and clonal lineage typing. The typing results of probes that covered different allelic variants of the same gene were summarized within one single result; if one of multiple probes for a single gene was positive, the gene was regarded as present. This summary resulted in 191 unique targets (101 virulence/persistence genes, 60 resistance genes, 15 regulatory genes, and 6 genes for species identification; see Table SA1 in the supplemental material). For comparison with WGS, the following nine unique targets were excluded from analysis. At first, the hypothetical proteins Q2FXC0, Q2YUB3, Q7A4X2, and Q9XB68 were excluded because it was impossible to find homologous sequences in NCBI GenBank using the applied criteria (Fig. 1) due to unspecified or ambiguous nomenclature. Moreover, targets hsdS1, hsdS2, hsdS3, and hsdSx were excluded due to insufficient annotation in GenBank and to high sequence diversity resulting in inconsistent groups (see Fig. SA1 in the supplemental material). Finally, the target spa was excluded because it was already implemented in SeqSphere+ as spa typing (15). In total, the typing results of 182 unique targets were used for comparison with the obtained WGS typing results.
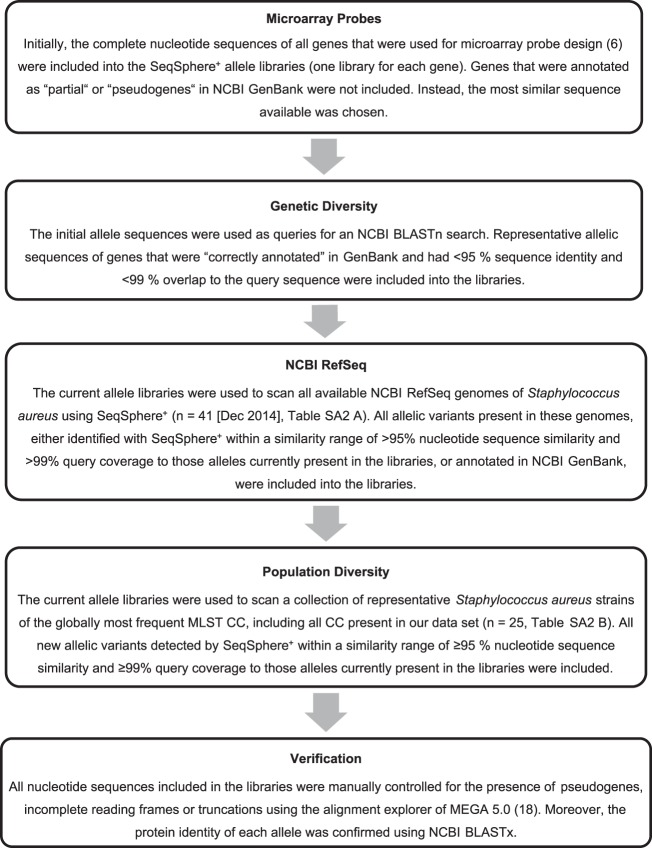
FIG 1.
Workflow of allele library creation. For each target gene, a FASTA allele library must be defined that includes a variety of nucleotide sequences that represent the allelic diversity of this gene. Only those sequences that are present in these libraries or differ within a defined similarity range can be found in the assembled genomes by gene-by-gene comparison.
Whole-genome sequencing, genome analysis, and data processing.
Staphylococcal chromosomal DNA was extracted using the MagAttract HMW DNA kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions for Gram-positive bacteria. Whole-genome shotgun sequencing of approximately 1 ng DNA and subsequent de novo assembly of the obtained reads were conducted as described previously (15, 16). The software SeqSphere+ version 2.3 beta (Ridom GmbH) was used for bioinformatic sequence analysis. The complete coding sequences of the 182 queried target genes were searched within the genomes using a similarity search based on the BLAST algorithm (17) implemented in SeqSphere+. These nucleotide sequences were stored within so-called FASTA allele libraries and represented each target's allelic diversity. They were grouped into four task templates, representing the functional categories of identification, regulation, resistance, and virulence. These task templates are implemented in SeqSphere+ and are accessible for all SeqSphere+ users; the FASTA raw files can be found in the supplemental material. The creation of allele libraries is essential for typing success; our study's approach is shown in Fig. 1. All allelic nucleotide sequences of each gene were aligned and were visually inspected for the presence of pseudogenes or incomplete open reading frames using MEGA 5.0 (18).
Targets were regarded as present if they were found in the genome within a range of ≥95% sequence identity and ≥99% query overlap to any of the nucleotide sequences stored in the allele library. In case of multiple matches for one target, the best match was chosen automatically. Targets that were only found partially in the genomes due to location on a cropped contig were not included; however, their cropped status was noted for further analyses. Internal stop codons, frame shifts, or nucleotide ambiguities were set to cause a “failed target” typing result. New alleles were assigned if the identified sequence was ≥95% identical and ≥99% overlapping to any of the allele sequences already present in one of the allele libraries. For the analysis of typing concordance of microarray and WGS, only the binary information gene present or gene absent was used. Cropped targets were regarded as absent, while failed targets were regarded as present. Further analysis of the functionality of different allelic variants, including failed targets (pseudogenes), was not conducted in this study. The allelic variants of the 13 genes coa, fnbB, fnbA, clfA, clfB, cna, ebh, vwb, sdrC, sdrD, sdrE, map (syn., eap), and hysA were highly variable between all examined isolates. Due to their low nucleotide sequence similarity to sequences already present in the allele libraries (<95% identity and <99% overlap), they were regarded as absent by SeqSphere+. In order to prevent such false-negative results, so-called “indicator targets” were designed for these genes that comprised partial nucleotide sequences (50 to 250 bp) of conserved regions of the genes. They enable the detection of the particular target gene but not the exact allelic determination.
Comparison of typing results and examination of discrepancies.
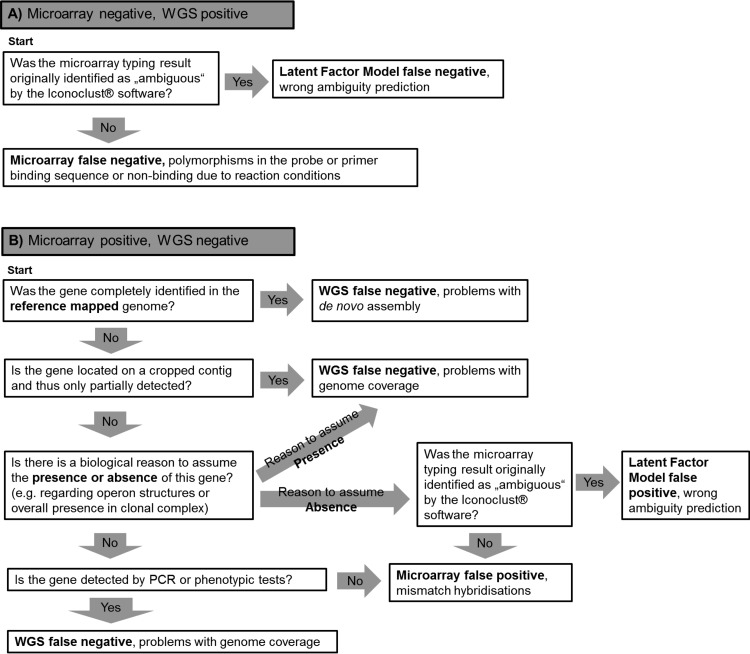
The concordance of typing results obtained by the microarray and WGS, respectively, was quantified by comparing the respective presence/absence determination of the analyzed targets. This resulted in four categories: (i) target present in WGS and microarray, (ii) target present in WGS and absent in microarray, (iii) target absent in WGS and present in microarray, and (iv) target absent in WGS and microarray. Targets that were only detected with one of the typing methods (categories ii and iii) were regarded as discrepant and investigated in further detail (Fig. 2).
FIG 2.
Decision tree for the explanation of discrepancies. Genes that were only detected with one of the applied typing methods (WGS or microarray) in one sample were regarded as discrepant. In order to find out which method was causative for the discrepancy, the depicted decision criteria were applied.
In order to detect false-negative results caused by de novo assembly errors, reference mapping was performed using CLC Genomics Workbench version 8 (Qiagen CLC bio, Aarhus, Denmark). The reads of all 154 samples were mapped against an artificial reference sequence comprising the representative nucleotide sequences of all investigated target genes, using default parameters with exception of length fraction (“0.8”) and nonspecific match handling (“Ignore”). Variable and diverse genes were represented by the nucleotide sequences of various alleles, and rather conserved genes were represented by only one allelic variant. Individual allelic sequences were separated by a sequence of 250 Ns (i.e., any base). The obtained bam files were analyzed with SeqSphere+ analogous to the de novo assembled ace files by performing a sequence similarity search for the 182 targets. Moreover, it was checked whether targets that were not identified in the de novo assembled or reference-mapped genomes had been partially identified on cropped contigs. Thus, the final WGS data set comprised four categories: (i) target present in de novo assembly, (ii) target absent in de novo assembly but detected by reference mapping, (iii) target absent in de novo assembled and reference-mapped genomes but partially identified on a cropped contig, and (iv) target not identified at all in whole-genome data. Similarly, information about initially obtained ambiguous microarray typing results (determined by Iconoclust software) was checked in order to identify discrepant typing results caused by mispredictions of the LFM. This resulted once more in four categories: (i) target detected as positive (present), (ii) target detected as ambiguous and predicted to be present by LFM, (iii) target detected as ambiguous and predicted to be absent by LFM, and (iv) target not detected (absent). In the following, these two data sets were compared in order to identify the causes of discrepant typing results (Fig. 2).
Discrepant typing results that were positively identified with WGS but that were negative in the microarray were always assigned to be typed false negatively by the microarray (or predicted false negatively by LFM). This was done because the extensive review and verification of implemented WGS allele libraries was assumed to prevent false-positive identifications in de novo assembled WGS data. Equally, targets that were positively identified in the reference-mapped genomes and with the microarray, but not in the de novo assembled genomes, were in principle regarded as typed false negatively by WGS due to de novo assembly errors. The remaining discrepancies were resolved manually based on their most probable biological explanation and the following assumptions: (i) All targets used for species identification should be positive; (ii) agr and cap genes should be present completely but only in one variant; (iii) targets hld, saeS, sarA, vraA, ebpS, icaC, isdA, clfA, hla, map, setB1, setB2, setB3, splA, splB, and splP should be present in all samples; (iv) operons like bla (blaI, blaZ, blaR), arginine catabolic mobile elements (ACMEs) (arcA, acrB, arcC, arcD), or the egc cluster (sei, seg, sem, sen, seo, seu) should be present completely; (v) single SCCmec-associated genes (like ugpQ, pls, or xylR) were only regarded as potentially positive if a mecA gene was present in the respective sample; (vi) leukotoxins (lukE and lukD, hlgB and hlgC, lukF-PV and lukS-PV, lukM and lukF-PV83, lukX and lukY) and merA and merB should only be present in pairs; and (vii) setC and fib were regarded as present or absent depending on their CC (for fib, only ST152 was negative in the used set of isolates; setC was not detected in isolates of CC30 and ST152 [as described in reference 5]). Discrepancies in ssl genes were resolved according to the expected CC patterns described by Monecke et al. in 2008 (5). In cases in which a target was detected in the reference-mapped genomes, but not in the de novo assembled genomes or with the microarray, their nucleotide sequences and assembly results were investigated in further detail in order to verify the correctness of the reference mapping result. If the reference mapping result was considered correct, a false-negative result was attested for WGS and microarray or LFM typing. The same was true for targets located on a cropped contig if there was a biological reason to assume the presence of this target. Remaining discrepancies were checked by target-specific PCRs or phenotypic tests. These included PCRs for qacC (19, 20), sdrC, sdrD, sdrE, (21) fnbB (22), sasG (23), etB, sea, seb, sed, seg, seh (24), and hlb (25) and phenotypic resistance tests for the presence of fosB (fosfomycin resistance) and cat (chloramphenicol resistance) in accordance with EUCAST clinical breakpoints (version 5.0). Discrepancies that still remained unsolved after this were categorized as unknown.
Determination of MLST CCs and SCCmec types.
In addition to molecular characterization regarding the presence or absence of specific virulence and resistance genes, the microarray is also able to infer the isolates' MLST CCs from the obtained hybridization patterns (5). Moreover, if present, the SCCmec type is similarly deduced. From WGS data, we extracted the sequences of the seven genes comprising the allelic profile of the S. aureus MLST scheme and queried them against the S. aureus MLST database (http://saureus.mlst.net) in order to assign the classical ST in silico. The respective CC was calculated using the eBURST algorithm (http://saureus.mlst.net/eburst/). SCCmec elements were determined using presence/absence detection of nomenclature-relevant genes (based on http://www.sccmec.org/Pages/SCC_TypesEN.html and references 26 and 27).
Nucleotide sequence accession number.
All generated raw reads were submitted to the European Nucleotide Archive (http://www.ebi.ac.uk/ena/) under the study accession number PRJEB11627.
RESULTS
In this study, the whole-genome sequences of 154 diverse S. aureus isolates were determined and analyzed with regard to 182 targets relevant for clonal lineage typing and staphylococcal virulence and resistance, resulting in 28,028 individual typing results. The results of WGS typing were compared to those obtained with the microarray (Tables 1 and 2; see also Table SA3 in the supplemental material). A total of 96.8% of these 28,028 individual typing results (n = 27,119) were identically assigned to be either “present” (n = 11,374; 40.6%) or “absent” (n = 15,745; 56.2%) by microarray and WGS. Hence, 3.2% (n = 909) of all typing results were only positive in one typing approach and were thus regarded as discrepant. Discrepant results were related to errors caused by WGS (n = 489; 1.7%), to errors caused by the microarray (n = 359; 1.3%), or to errors caused by wrong ambiguity predictions by LFM (n = 33; 0.1%). In 28 cases (0.1%), the cause of error remained unknown. All WGS errors were caused by false-negative detection of targets in the de novo assembled genomes. They were subdivided into three distinct sources of error: (i) failures of de novo assembly (target was identified in reference-mapped genome), (ii) insufficient genome coverage during shotgun sequencing (target was identified partially on a cropped contig), and (iii) either insufficient genome coverage or missing reference allele/sequence similarity below the applied thresholds (not detected in WGS data at all although it should be present based on biological assumptions) (Tables 1 and 2). Microarray errors can be either false positive or false negative; false-positive results were presumably caused by mismatch hybridizations of similar probes with the same amplicon; false-negative results were presumably caused by polymorphisms in the gene sequence that prevented binding to the probe or primer. Errors of LFM were of a statistical nature and corresponded to the expected statistical error calculated from the total number of ambiguous microarray typing results (n = 2,788; 5.4%) and the experimentally determined LFM error rate of 3% (Tables 1 and 2). Very few discrepancies could not be resolved, even after traditional single-target PCRs or phenotyping (n = 28; 0.1%; targets fnbB [n = 3], fosB [n = 3], ugpQ [n = 1], ssl10 [n = 2], ssl01 [n = 2], ssl04 [n = 1], ssl06 [n = 1], and lukY [n = 15]). All of them were identified with the microarray but not with WGS. We found that fnbB (n = 3) has a high intrinsic allelic sequence diversity, which may have prevented the PCR primers of the applied diagnostic PCRs from binding (19, 20). Ssl10 could not be resolved because all other ssl genes were absent in the respective samples, and thus they did not correspond to the data provided by Monecke et al. (5). The remaining discrepancies in ssl genes could not be resolved because the respective samples belonged to clonal complexes that were not present in the data of Monecke et al. (5) (CC80 and CC9). No PCR was available for ugpQ. Target lukY was recognized in 15 of 16 CC152 isolates by the microarray but not by WGS. lukY is one of the targets that does thus not follow a universal nomenclature in NCBI, which hampers the identification of homologous sequences according to the applied criteria (Fig. 1). CC152 was generally found to possess differing alleles compared to those of other CCs; thus, it is possible that lukY was present in a new allelic variant in these samples but was not found by SeqSphere+ due to low sequence similarity. However, it is expected that lukY only occurs in combination with lukX, which was not detected with microarray or with WGS in CC152; this was due either to sequence diversity of CC152 or to real absence. All other examined isolates from other CCs possessed lukX and lukY. Thus, it could not be decided whether lukY was detected false positively by the microarray or false negatively by WGS. Phenotypic testing for fosfomycin resistance according to EUCAST breakpoints resulted in a susceptible phenotype for all three isolates. However, detailed analysis showed that all other isolates from Münster that harbored the fosB gene were fosfomycin susceptible according to EUCAST breakpoints. For the remaining 90 fosB-positive isolates present in our data set, no phenotypic antibiotic resistance testing results were available. Thus, it could not be decided based on the phenotypic data whether fosB should be present or not.
TABLE 1.
The percentage of concordantly typed (WGS and microarray identify a gene as present or absent, respectively) and discrepantly typed results (either only WGS or only microarray identifies a gene as present) for each functional target categorya
| Result category | Result caused by | Result by functional category of genes (no.) |
Total no. | Total, % | ||||
|---|---|---|---|---|---|---|---|---|
| Identification | Regulation | Resistance | Virulence | |||||
| Concordant (n = 27,119; 96.8 %) | ||||||||
| Positive | Microarray and WGS (de novo) | 829 | 990 | 1,060 | 8,495 | 11,374 | 40.6 | |
| Negative | Microarray and WGS (de novo) | 0 | 1,159 | 8,100 | 6,486 | 15,745 | 56.2 | |
| Discrepant (n = 909; 3.2 %) | ||||||||
| False positive | Microarray | Mishybridizations | 0 | 78 | 21 | 103 | 202 | 0.7 |
| LFM | Misprediction | 0 | 17 | 2 | 9 | 28 | 0.1 | |
| False negative | Microarray | Polymorphisms | 0 | 3 | 14 | 140 | 157 | 0.6 |
| LFM | Misprediction | 0 | 0 | 0 | 5 | 5 | <0.1 | |
| WGS | Assembly error | 88 | 42 | 16 | 164 | 310 | 1.1 | |
| Cropped contig | 1 | 12 | 15 | 28 | 56 | 0.2 | ||
| Not sequenced or aberrant allele | 6 | 9 | 8 | 100 | 123 | 0.4 | ||
| Unknown | 0 | 0 | 4 | 24 | 28 | 0.1 | ||
| Total no. of typing results | 924 | 2,310 | 9,235 | 15,554 | 28,028 | 100 | ||
Comparison of typing concordance of WGS typing and the traditional DNA microarray. In total, 182 targets in 154 samples were analyzed, resulting in 28,028 individual typing results. The discrepant results are subdivided by false-positive and false-negative typing results and different causes of error. Results that were assumed to be false negatively typed by the two methods are not considered in this table (n = 4).
TABLE 2.
Samples grouped by their clonal complexes and the concordant and discrepant WGS and microarray typing results calculated for each CC (percentages per CC given in parentheses)a
| Clonal complex (no. of isolates) | No. concordant (n = 27,119; 96.8 %) |
No. discrepant (n = 909; 3.2 %) |
No. unknown | Total no. | |||||
|---|---|---|---|---|---|---|---|---|---|
| Positive | Negative | False positive |
False negative |
||||||
| Microarray | LFM | Microarray | LFM | WGS | |||||
| CC1 (7) | 541 (42.5) | 703 (55.2) | 0 | 0 | 4 (0.3) | 0 | 26 (2.0) | 0 | 1,274 |
| CC5 (17) | 1,375 (44.4) | 1,615 (52.2) | 2 (0.1) | 1 (<0.1) | 1 (<0.1) | 0 | 99 (3.2) | 1 (<0.1) | 3,094 |
| CC6 (6) | 448 (41.0) | 611 (56.0) | 5 (0.5) | 1 (0.1) | 0 | 0 | 27 (2.5) | 0 | 1,092 |
| CC7 (6) | 423 (38.7) | 633 (58.0) | 11 (1.0) | 1 (0.1) | 7 (0.6) | 0 | 15 (1.4) | 2 (0.2) | 1,092 |
| CC8 (16) | 1,258 (43.2) | 1,542 (53.0) | 38 (1.3) | 3 (0.1) | 0 | 1 (<0.1) | 70 (2.4) | 0 | 2,912 |
| CC9 (2) | 143 (39.3) | 210 (57.7) | 2 (0.5) | 0 | 1 (0.3) | 0 | 6 | 2 (0.5) | 364 |
| CC15 (28) | 2,152 (42.2) | 2,864 (56.2) | 33 (0.6) | 0 | 21 (0.4) | 0 | 26 (0.5) | 0 | 5,096 |
| CC22 (8) | 496 (34.1) | 831 (57.1) | 16 (1.1) | 5 (0.3) | 51 (3.5) | 2 (0.1) | 54 (3.7) | 1 (0.1) | 1,456 |
| CC30 (13) | 937 (39.6) | 1,352 (57.1) | 13 (0.5) | 6 (0.3) | 0 | 0 | 55 (2.3) | 3 (0.1) | 2,366 |
| CC45 (12) | 829 (38.0) | 1,271 (58.2) | 17 (0.8) | 1 (0.3) | 30 (1.4) | 2 (0.1) | 33 (1.5) | 1 (<0.1) | 2,184 |
| CC80 (1) | 75 (41.2) | 99 (54.4) | 2 (1.1) | 0 | 2 (1.1) | 0 | 1 (0.5) | 3 (1.6) | 182 |
| CC88 (4) | 312 (42.9) | 398 (54.7) | 0 | 0 | 1 (0.1) | 0 | 17 (1.6) | 0 | 728 |
| CC121 (18) | 1,469 (44.8) | 1,711 (52.2) | 33 (1.0) | 10 (0.3) | 3 (0.1) | 0 | 50 (1.5) | 0 | 3,276 |
| CC152 (16) | 916 (31.5) | 1,905 (65.4) | 30 (1.0) | 0 | 36 (1.2) | 0 | 10 (0.3) | 15 (0.5) | 2,912 |
| Total (154) | 11,374 | 15,745 | 202 | 28 | 157 | 5 | 489 | 28 | 28,028 |
Comparison of typing concordance of WGS typing and the traditional DNA microarray. In total, 182 targets in 154 samples were analyzed, resulting in 28,028 individual typing results. The discrepant results are subdivided by false-positive and false-negative typing results and different causes of error. Results that were assumed to be false negatively typed by the two methods are not considered in this table (n = 4). WGS results were summarized in one column irrespective of the cause of error.
The rate of discrepant results (n = 909 in total) differed between the four functional gene categories of (i) species identification (n = 95), (ii) regulation (n = 161), (iii) resistance (n = 80), and (iv) virulence (n = 573) (Table 1). Discrepancies in targets of (i) species identification were mainly caused by misassemblies of 23S rRNA sequences (identified in reference-mapped genomes but not in de novo assembled genomes in 83 of 154 samples). Discrepant typing results in (ii) regulatory targets were in large part either caused by mishybridizations between agr-I and agr-IV sequences in the microarray (n = 78) and related LFM mispredictions (n = 17) or by WGS nondetections (n = 63). The concordance of typing results for (iii) resistance targets was generally very high, with only few errors in the two methods. Microarray and WGS typing discrepancies in (iv) virulence genes were either caused by WGS nondetection of staphylococcal enterotoxins or superantigens (n = 169) or by microarray errors caused by polymorphisms in probe/primer binding sequences (n = 104), for example, in CC22 (setB1-B3, sdrM, ssl11, hlIII, ebh), CC45 (setB1, ssl03), CC152 (icaC, hl), and CC7 (ssl11). In four cases, the target was initially concordantly rated as negative by microarray and WGS but was detected in the reference-mapped genomes. After a manual investigation of the reference mapping result, it was decided that these targets had obviously been rated as false negative with microarray and WGS. This occurred in targets indicator-fnbB, ermC, icaC, and tetK. Table 2 shows the typing results per CC. CC22 contained the highest number of discrepancies (n = 129, 8.8% of all typing results within CC22); however, in general, the discrepancies were equally distributed across all CCs (Table 2).
Finally, all CCs and SCCmec types were identically determined by microarray and WGS typing; however, WGS typing was more detailed regarding clonal lineage typing by determining the ST in contrast to the CC (see Table SA3 in the supplemental material). Nine previously undescribed STs of five different CCs were identified in seven German and two Tanzanian isolates. The novel allelic profiles were submitted to the S. aureus MLST database (http://saureus.mlst.net/) and were assigned to STs 3196 to 3204 (see Table SA3).
DISCUSSION
We successfully established and evaluated a novel easy-to-use in silico typing method for the detection of 182 unique target genes in staphylococcal WGS data. Validation of the in silico typing was performed by comparing the obtained results to those of an established microarray that queries the same targets. Overall, there was a high concordance between the two methods; only 3.2% (n = 909) of the 28,028 total typing results were discrepant.
The microarray was assumed to have produced erroneous typing results in 1.3% of all cases. False-negative nondetections of particular targets were observed especially in samples of CC22, CC45, CC152, and CC7. They were presumably caused by nonbinding of the sample amplicon to the microarray's probe or primer oligonucleotide due to polymorphisms in the respective target gene (for the exemplary misdetection of icaC in CC152, see Fig. SA2 in the supplemental material). CC22 and CC45 are known to have diverse alleles compared to those of other pandemic lineages like CC1, CC8, and CC5 and are thus prone to yield “aberrant,” “irregular,” or “weak” signals in microarray typing (5, 6). CC152 is a clonal lineage frequently found in West and Central Africa and the Balkan region but is rarely isolated in the rest of the world (28). It was described as an “aberrant and isolated strain” (5), thus causing some problems in microarray as well as in WGS typing due to diverse alleles (6). CC7 isolates were only rarely detected in previous microarray evaluation studies (6), which indicates that maybe not the entire genetic diversity of this CC can be considered for microarray probe design. False-positive results occurred between highly similar probe and amplicon sequences, e.g., between agrI and agrIV. In these cases, WGS typing was much more precise. Typing errors of the microarray can be generally explained by the fact that the microarray is a closed system; only those targets can be detected whose PCR-amplified region is complementary to one of the available oligonucleotide probes. This is a limitation that the creators of the microarray are well aware of and that cannot be prevented in this kind of hybridization procedure. Although the microarray's probe set is constantly curated, its expansion is rather time-consuming and complicated because one always has to consider potential cross-reactions with other probes and targets. In contrast, WGS allele libraries may be more easily extended as soon as a novel genetic variant is found (with the sequence databases growing rapidly). However, the diversification of WGS allele libraries also involves the risk of blurring frontiers between different, though related, genes, e.g., between paralogs. Evolution is a gradual process, and new genes evolve from older ones; thus, there is always a gray zone between two distinct genes, depending on their divergence time. In some cases, it is impossible (or very difficult) to decide which sequence is still an allelic variant of a gene A and which should already be assigned to gene B. Every nucleotide sequence identified within a similarity range of ≥95% identity and ≥99% overlap to any allele within the library will automatically be detected as a new allele of the respective target, although it may actually belong to another, highly similar gene. This was, for example, observed for targets blaI and mecI. Thus, careful curation of the libraries is needed in order to prevent false-positive results in WGS typing.
Furthermore, 1.8% of all typing results were regarded to have been falsely rated as negative by WGS (n = 489). The majority of these errors were caused by identified assembly failures (n = 310), which were attested if a target was identified in the reference-mapped genome but not in the de novo assembled data. These failures predominantly occurred in genes or operons that exist in multiple copies per genome (e.g., 23S rRNA) or share too much nucleotide sequence similarity among each other (e.g., enterotoxins, ssl genes). The remaining errors were presumably caused by incomplete genome coverage of shotgun sequencing (>97% instead of 100%) (29), which implies the possibility to lose some sequence information. In some cases, at least parts of the target gene can be identified on a cropped contig (n = 56). The remaining 121 typing results that were supposed to be present were not detected with any of the WGS analyses (de novo assembly, reference mapping, cropped contigs). For these typing results, it is also possible that the respective target was not detected in the genomes because it is present in an allelic variant that has <95% identity to alleles implemented in the respective allele library. This may be the case especially for genes with nonuniversal nomenclature like, for example, the ssl genes; setB1, setB2, and setB3; or lukX and lukY. Four cases were identified by reference mapping in which the microarray and the de novo assembled WGS produced false-negative typing results. It cannot be excluded that there are more double-negative results, which are also negative in the reference mapping and thus may not be detected in our approach. These technical limitations (incomplete genome coverage and failures of de novo assembly) are likely to be overcome in the future with the development of third-generation single molecule sequencing techniques that provide longer sequence reads, which can then bridge these repetitive structures and lead to a correct assembly (30). Moreover, assembly errors may also be overcome by direct mapping of reads to reference sequences of genes of interest as was performed, for example, by Zhang et al. in a recent study (31). False-positive results did not occur in de novo WGS typing because all allelic sequences were checked manually with a BLASTn and BLASTx search if they corresponded to “correctly” annotated genes in the NCBI nucleotide database. However, the reference mapping was shown to provide false-positive typing results for target hlb-intact because both the 3′ and the 5′ end of the phage-disrupted gene mapped to the intact hlb reference sequence.
Compared to WGS with its irreproachable advantages, the microarray is still a valuable tool to examine the S. aureus gene repertoire. Nevertheless, it is very likely that the anticipated gap between technological profiles and development/curating of the two techniques will soon result in the replacement of array-based genotyping methods by WGS in the majority of laboratories due to its universal methodology and open system. Moreover, sequencing costs are constantly dropping and are already comparable to the costs for microarray analysis. In contrast to other molecular typing methods, WGS typing allows for the infinite use of once generated raw data for many different studies and for the easy combination of accessory typing results with epidemiologically relevant typing data like core genome MLST (cgMLST) (32). A major benefit of WGS typing is that genes can be discriminated into allelic variants, which is of particular interest for virulence and resistance genes. Minor changes in the DNA sequence may result in major changes in the expressed protein and thus in the respective phenotype. This also relates to the easy detection of truncated genes or pseudogenes using our in silico typing method, which may either result in nonfunctional proteins or may present new functional variants with new phenotypic properties. The importance of allele typing is exemplified by the phenotyping and genotyping of fosB in this study. Unfortunately, due to repetitive regions and high diversity among different alleles, not all targets can be distinguished into allelic variants, including the staphylococcal surface proteins Ebh, Map (syn., Eap), Coa, Cna, SdrC, SdrD, SdrE, ClfA, ClfB, FnbA, FnbB, and Vwb, which influence the bacterium's interaction with the host (33, 34). Different allelic variants likely represent specific adaptations to the human immune system, and ongoing coevolution probably caused the extensive number of detectable diverse allelic variants. It is important to note that it cannot be ruled out that certain targets were not found with the microarray or with the WGS typing scheme. This may be, for example, the case for hlgC in CC152 (n = 16) because this target was identified in samples of all other clonal lineages and does usually occur in combination with hlgB, whose presence was identically identified with microarray and WGS in CC152 samples.
In conclusion, our WGS typing scheme reliably identified the presence of 182 clinically relevant genes in WGS data, including, for example, toxic shock toxin, Panton-Valentine leucocidin, or methicillin resistance. The number of investigated targets is easily and infinitely expandable and is not limited to the targets used in the Identibac microarray. Indeed, some targets of the microarray can be excluded for future WGS analyses, for example, 23S rRNA for species identification. On the other hand, other targets like dfrG, mecC, or speG should prospectively be included in our in silico typing scheme in order to expand the covered clinical characteristics. With few exceptions only, all targets can be discriminated into different allelic variants, enabling a detailed analysis of disease-associated factors in the future. Based on such analysis, we envision a risk assessment for every clinical S. aureus isolate based on the association of specific S. aureus genotypes with specific human disease progressions.
Supplementary Material
ACKNOWLEDGMENTS
We thank the members of the African-German StaphNet consortium, including Pedro Alonso (Barcelona, Spain), Alexander W. Friedrich (Groningen, The Netherlands), Jonas Hofmann-Eifler (Freiburg, Germany), Peter Kremsner (Tübingen, Germany), Sabine Schubert (Homburg, Germany), Marcel Tanner (Basel, Switzerland), Hagen von Briesen (St. Ingbert, Germany), Delfino Vubil (Manhiça, Mozambique), and Laura Wende (Homburg, Germany). We thank Ursula Keckevoet, Isabell Höfig, Thomas Böking, and Stefan Bletz (Institute of Hygiene, Münster, Germany) and Martina Schulte (Institute of Medical Microbiology, Münster, Germany) for performing the sequencing and molecular tests. Moreover, we thank Stefan Monecke (Dresden, Germany) for help with the interpretation of microarray data and probe design.
Support by the Münster Graduate School of Evolution (MGSE) to Lena Strauß is gratefully acknowledged.
We declare no conflicts of interest.
Funding Statement
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.03022-15.
REFERENCES
- 1.Lowy FD. 1998. Staphylococcus aureus infections. N Engl J Med 339:520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Bukowski M, Wladyka B, Dubin G. 2010. Exfoliative toxins of Staphylococcus aureus. Toxins 2:1148–1165. doi: 10.3390/toxins2051148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Maiden MC, Bygraves JA, Feil E, Morelli G, Russell JE, Urwin R, Zhang Q, Zhou J, Zurth K, Caugant DA, Feavers IM, Achtman M, Spratt BG. 1998. Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc Natl Acad Sci U S A 95:3140–3145. doi: 10.1073/pnas.95.6.3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol 38:1008–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Monecke S, Slickers P, Ehricht R. 2008. Assignment of Staphylococcus aureus isolates to clonal complexes based on microarray analysis and pattern recognition. FEMS Immunol Med Microbiol 53:237–251. doi: 10.1111/j.1574-695X.2008.00426.x. [DOI] [PubMed] [Google Scholar]
- 6.Monecke S, Coombs G, Shore AC, Coleman DC, Akpaka P, Borg M, Chow H, Ip M, Jatzwauk L, Jonas D, Kadlec K, Kearns A, Laurent F, O'Brien FG, Pearson J, Ruppelt A, Schwarz S, Scicluna E, Slickers P, Tan HL, Weber S, Ehricht R. 2011. A field guide to pandemic, epidemic and sporadic clones of methicillin-resistant Staphylococcus aureus. PLoS One 6:e17936. doi: 10.1371/journal.pone.0017936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruffing U, Akulenko R, Bischoff M, Helms V, Herrmann M, von Müller L. 2012. Matched-cohort DNA microarray diversity analysis of methicillin sensitive and methicillin resistant Staphylococcus aureus isolates from hospital admission patients. PLoS One 7:e52487. doi: 10.1371/journal.pone.0052487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Candès EJ, Recht B. 2009. Exact matrix completion via convex optimization. Found Comput Math 9:717–772. doi: 10.1007/s10208-009-9045-5. [DOI] [Google Scholar]
- 9.Koren Y, Bell R, Volinsky C. 2009. Matrix factorization techniques for recommender systems. Computer 8:30–37. [Google Scholar]
- 10.Efron B, Gong G. 1983. A leisurely look at the bootstrap, the jackknife, and cross-validation. Am Stat 37:36–48. doi: 10.1080/00031305.1983.10483087. [DOI] [Google Scholar]
- 11.Hamed M, Nitsche-Schmitz DP, Ruffing U, Steglich M, Dordel J, Nguyen D, Brink J-H, Chhatwal GS, Herrmann M, Nübel U, Helms V, von Müller L. 2015. Whole genome sequence typing and microarray profiling of nasal and blood stream methicillin-resistant Staphylococcus aureus isolates: clues to phylogeny and invasiveness. Infect Genet Evol 36:475–482. doi: 10.1016/j.meegid.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 12.Shittu AO, Oyedara O, Okon K, Raji A, Peters G, von Müller L, Schaumburg F, Herrmann M, Ruffing U. 2015. An assessment on DNA microarray and sequence-based methods for the characterization of methicillin-susceptible Staphylococcus aureus from Nigeria. Front Microbiol 6:1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Didelot X, Bowden R, Wilson DJ, Peto TE, Crook DW. 2012. Transforming clinical microbiology with bacterial genome sequencing. Nat Rev Genet 13:601–612. doi: 10.1038/nrg3226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Herrmann M, Abdullah S, Alabi A, Alonso P, Friedrich AW, Fuhr G, Germann A, Kern WV, Kremsner PG, Mandomando I, Mellmann AC, Pluschke G, Rieg S, Ruffing U, Schaumburg F, Tanner M, Peters G, von Briesen H, von Eiff C, von Müller L, Grobusch MP. 2013. Staphylococcal disease in Africa: another neglected ‘tropical’ disease. Future Microbiol 8:17–26. doi: 10.2217/fmb.12.126. [DOI] [PubMed] [Google Scholar]
- 15.Bletz S, Mellmann A, Rothgänger J, Harmsen D. 2015. Ensuring backwards compatibility: traditional genotyping efforts in the era of whole genome sequencing. Clin Microbiol Infect 21:347.e1–347.e4. [DOI] [PubMed] [Google Scholar]
- 16.Ruppitsch W, Pietzka A, Prior K, Bletz S, Fernandez HL, Allerberger F, Harmsen D, Mellmann A. 2015. Defining and evaluating a core genome multilocus sequence typing scheme for whole-genome sequence-based typing of Listeria monocytogenes. J Clin Microbiol 53:2869–2876. doi: 10.1128/JCM.01193-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 18.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noguchi N, Hase M, Kitta M, Sasatsu M, Deguchi K, Kono M. 1999. Antiseptic susceptibility and distribution of antiseptic-resistance genes in methicillin-resistant Staphylococcus aureus. FEMS Microbiol Lett 172:247–253. doi: 10.1111/j.1574-6968.1999.tb13475.x. [DOI] [PubMed] [Google Scholar]
- 20.Mayer S, Boos M, Beyer A, Fluit AC, Schmitz FJ. 2001. Distribution of the antiseptic resistance genes qacA, qacB and qacC in 497 methicillin-resistant and -susceptible European isolates of Staphylococcus aureus. J Antimicrob Chemother 47:896–897. doi: 10.1093/jac/47.6.896. [DOI] [PubMed] [Google Scholar]
- 21.Xue H, Lu H, Zhao X. 2011. Sequence diversities of serine-aspartate repeat genes among Staphylococcus aureus isolates from different hosts presumably by horizontal gene transfer. PLoS One 6:e20332. doi: 10.1371/journal.pone.0020332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paniagua-Contreras G, Monroy-Pérez E, Gutiérrez-Lucas R, Sainz-Espuñes T, Bustos-Martínez J, Vaca S. 2014. Genotypic characterization of methicillin-resistant Staphylococcus aureus strains isolated from the anterior nares and catheter of ambulatory hemodialysis patients in Mexico. Folia Microbiol (Praha) 59:295–302. doi: 10.1007/s12223-013-0300-4. [DOI] [PubMed] [Google Scholar]
- 23.Roche FM, Massey R, Peacock SJ, Day NP, Visai L, Speziale P, Lam A, Pallen M, Foster TJ. 2003. Characterization of novel LPXTG-containing proteins of Staphylococcus aureus identified from genome sequences. Microbiology 149:643–654. doi: 10.1099/mic.0.25996-0. [DOI] [PubMed] [Google Scholar]
- 24.Becker K, Roth R, Peters G. 1998. Rapid and specific detection of toxigenic Staphylococcus aureus: use of two multiplex PCR enzyme immunoassays for amplification and hybridization of staphylococcal enterotoxin genes, exfoliative toxin genes, and toxic shock syndrome toxin 1 gene. J Clin Microbiol 36:2548–2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Wamel WJ, Rooijakkers SH, Ruyken M, van Kessel KP, van Strijp JA. 2006. The innate immune modulators staphylococcal complement inhibitor and chemotaxis inhibitory protein of Staphylococcus aureus are located on beta-hemolysin-converting bacteriophages. J Bacteriol 188:1310–1315. doi: 10.1128/JB.188.4.1310-1315.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.International Working Group on the Classification of Staphylococcal Cassette Chromosome Elements (IWG-SCC). 2009. Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother 53:4961–4967. doi: 10.1128/AAC.00579-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hiramatsu K, Ito T, Tsubakishita S, Sasaki T, Takeuchi F, Morimoto Y, Katayama Y, Matsuo M, Kuwahara-Arai K, Hishinuma T, Baba T. 2013. Genomic basis for methicillin resistance in Staphylococcus aureus. Infect Chemother 45:117–136. doi: 10.3947/ic.2013.45.2.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schaumburg F, Alabi AS, Peters G, Becker K. 2014. New epidemiology of Staphylococcus aureus infection in Africa. Clin Microbiol Infect 20:589–596. doi: 10.1111/1469-0691.12690. [DOI] [PubMed] [Google Scholar]
- 29.Jünemann S, Sedlazeck FJ, Prior K, Albersmeier A, John U, Kalinowski J, Mellmann A, Goesmann A, von Haeseler A, Stoye J, Harmsen D. 2013. Updating benchtop sequencing performance comparison. Nat Biotechnol 31:1148. [DOI] [PubMed] [Google Scholar]
- 30.Clarke J, Wu HC, Jayasinghe L, Patel A, Reid S, Bayley H. 2009. Continuous base identification for single-molecule nanopore DNA sequencing. Nat Nanotechnol 4:265–270. doi: 10.1038/nnano.2009.12. [DOI] [PubMed] [Google Scholar]
- 31.Zhang S, Yin Y, Jones MB, Zhang Z, Deatherage Kaiser BL, Dinsmore BA, Fitzgerald C, Fields PI, Deng X. 2015. Salmonella serotype determination utilizing high-throughput genome sequencing data. J Clin Microbiol 53:1685–1692. doi: 10.1128/JCM.00323-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leopold S, Goering RV, Witten A, Harmsen D, Mellmann A. 2014. Bacterial whole-genome sequencing revisited: portable, scalable, and standardized analysis for typing and detection of virulence and antibiotic resistance genes. J Clin Microbiol 52:2365–2370. doi: 10.1128/JCM.00262-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clarke SR, Foster SJ. 2006. Surface adhesins of Staphylococcus aureus. Adv Microb Physiol 51:187–224. doi: 10.1016/S0065-2911(06)51004-5. [DOI] [PubMed] [Google Scholar]
- 34.Lindsay JA, Moore CE, Day NP, Peacock SJ, Witney AA, Stabler RA, Husain SE, Butcher PD, Hinds J. 2006. Microarrays reveal that each of the ten dominant lineages of Staphylococcus aureus has a unique combination of surface-associated and regulatory genes. J Bacteriol 188:669–676. doi: 10.1128/JB.188.2.669-676.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.