Abstract
Next-generation sequencing (NGS) technology has shown promise for the detection of human pathogens from clinical samples. However, one of the major obstacles to the use of NGS in diagnostic microbiology is the low ratio of pathogen DNA to human DNA in most clinical specimens. In this study, we aimed to develop a specimen-processing protocol to remove human DNA and enrich specimens for bacterial and viral DNA for shotgun metagenomic sequencing. Cerebrospinal fluid (CSF) and nasopharyngeal aspirate (NPA) specimens, spiked with control bacterial and viral pathogens, were processed using either a commercially available kit (MolYsis) or various detergents followed by DNase prior to the extraction of DNA. Relative quantities of human DNA and pathogen DNA were determined by real-time PCR. The MolYsis kit did not improve the pathogen-to-human DNA ratio, but significant reductions (>95%; P < 0.001) in human DNA with minimal effect on pathogen DNA were achieved in samples that were treated with 0.025% saponin, a nonionic surfactant. Specimen preprocessing significantly decreased NGS reads mapped to the human genome (P < 0.05) and improved the sensitivity of pathogen detection (P < 0.01), with a 20- to 650-fold increase in the ratio of microbial reads to human reads. Preprocessing also permitted the detection of pathogens that were undetectable in the unprocessed samples. Our results demonstrate a simple method for the reduction of background human DNA for metagenomic detection for a broad range of pathogens in clinical samples.
INTRODUCTION
Clinical microbiology is one of the most rapidly changing areas of laboratory medicine today due to the introduction of new technologies and automation (1). Molecular testing, such as PCR, is becoming the de facto gold standard for the detection of pathogens that are difficult to culture by offering high sensitivity and specificity and a rapid turnaround time (2). Syndrome-based multiplex molecular assays can detect up to 30 of the most common pathogens associated with respiratory infections, gastroenteritis, and central nervous system (CNS) infections (3–6). However, the complete list of infectious agents associated with these infections greatly exceeds the capabilities of even the best multiplex assays. These less common organisms, for which tests are not readily available, are likely responsible for many cases of undiagnosed illness, particularly in critically ill patients and those with compromised immunity (7, 8). Therefore, there is increasing interest in the application of novel technologies, such as next-generation sequencing (NGS), for unbiased detection of pathogens in clinical samples.
Among the various challenges with the implementation of NGS for routine pathogen detection using metagenomics, the presence of an overwhelming amount of host DNA is one of the most important problems to be addressed. A previous metagenomic study on nasopharyngeal aspirate samples from patients with acute lower respiratory tract infections revealed that up to ∼95% of raw NGS reads were of human DNA (9). The subtraction of human sequences from large NGS data sets can be a lengthy process, requiring significant computational power which may not be available to most clinical laboratories. The high background of human DNA also affects the sensitivity for pathogens that occur in low abundance in clinical specimens.
In this study, we aimed to preprocess clinical specimens to selectively deplete human DNA while minimizing the effect on pathogen DNA. We tested a commercial kit as well as a variety of detergents to permeabilize human cells in nasopharyngeal aspirate (NPA) and cerebrospinal fluid (CSF) specimens spiked with bacterial and viral control strains, followed by treatment with DNase prior to DNA extraction and analysis by real-time PCR and metagenomic sequencing on the Illumina MiSeq. Our results indicate that preprocessing of clinical samples with the nonionic surfactant saponin results in a significant reduction of background human DNA and improved the sensitivity of NGS to detect pathogens. The methods described in this study will facilitate the use of NGS in clinical microbiology laboratories.
MATERIALS AND METHODS
Bacterial and viral strains.
Escherichia coli (American Type Culture Collection [ATCC] 25922), Streptococcus pneumoniae (ATCC 49619), and Streptococcus agalactiae (ATCC 12386) strains were grown on blood agar plates (Oxoid) overnight at 37°C in a 5% CO2 atmosphere. Haemophilus influenzae (ATCC 10211) and Neisseria meningitidis (ATCC 13090) strains were grown on chocolate agar plates (Oxoid) under the same conditions. Bordetella pertussis (ATCC BAA-589) was grown in a charcoal agar medium at 37°C for 72 h in a humidified environment. Herpes simplex virus type 2 (HSV2) (ATCC VR-540) and human adenovirus type 7 (ATCC VR-7) strains, and a patient isolate of influenza A virus, were used from stocks maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% dimethyl sulfoxide (DMSO) at −80°C.
Specimens.
CSF specimens and NPA specimens submitted to the Microbiology and Virology Laboratory of BC Children's Hospital for PCR testing between August 2013 and July 2014 were used in this study. Specimens used for spiking experiments were all negative by PCR for all pathogens that were used for spiking. In addition, one NPA specimen was used, which was originally positive for human adenovirus by PCR. Following initial PCR testing, residual specimens were kept at 4°C and processed further in the same day. To maintain patient anonymity, all patient identifiers were removed by personnel, who were unaware of the current study results. Ethics approval for the study was obtained from the research ethics board of the University of British Columbia.
Spiking.
Representative species of Gram-positive and Gram-negative bacteria as well as enveloped and nonenveloped viruses were used for spiking. Freshly prepared bacterial suspensions were adjusted to a turbidity equivalent to a 0.5 McFarland standard and further diluted in phosphate-buffered saline (PBS) as necessary (S. pneumoniae, 10-fold; E. coli, 100-fold; and H. influenzae, 1,000-fold). Previously determined titers of HSV2 (2.8 × 105 TCID50/ml), adenovirus (2.8 × 106 TCID50/ml), and influenza A virus (PCR threshold cycle [CT] = 24) were directly added from cultured stocks (10, 11). Then, 5 μl of each of bacterial and viral preparation was spiked into 1 ml of CSF or NPA samples and vortexed for 10 s before further processing.
Specimen processing.
Specimen processing was performed as soon as the samples were available. For each set of data, specimens were processed and analyzed in at least three independent experiments. One milliliter each of unfrozen NPA and CSF specimens (n = 3, for each specimen type) was first spiked with S. pneumoniae and influenza A virus and divided into 0.2-ml aliquots. Control specimens were left untreated at room temperature (RT) until extraction. Remaining aliquots were processed by using the MolYsis basic kit (Molzym GmbH & Co. KG, Bremen, Germany) according to the manufacturer's instructions or with minor modifications. Briefly, with MolYsis method I, specimen aliquots were centrifuged at 13,000 rpm for 5 min at 4°C, and pellets were resuspended in 0.2 ml of PBS. Supernatants were saved and kept on ice. Then, 50 μl of MolYsis buffer CM was added to resuspended pellets, vortexed for 10 s, and incubated at RT for 5 min. Next, 50 μl of buffer DB1, 10 μl of MolDNase, and 2 μl of RNase cocktail enzyme mix (Thermo Fisher Scientific, Inc.) were added, vortexed for 10 s, and incubated at RT for 15 min. Specimens were then centrifuged again at 13,000 rpm for 10 min at 4°C, and the pellet was washed once with 1 ml of buffer RS. The final, washed pellet was resuspended with previously saved supernatant. In MolYsis method II, MolYsis buffer CM was directly added to each specimen aliquot instead of centrifugation and processed similarly. The volume of each sample was adjusted to 0.35 ml with TE8 buffer prior to extraction. With an aim to further optimize the MolYsis protocol, NPA specimens spiked with suspensions of S. pneumoniae, adenovirus, and HSV2 were processed according to MolYsis method II. However, in this case, RNase was excluded and DNase treatment was performed under the following conditions: (i) no treatment; (ii) 15 min at RT; (iii) 15 min at 37°C; (iv) 30 min at RT; or (v) 2 h at RT.
To test various detergents for selective lysis of human cells, NPA specimens spiked with S. pneumoniae, adenovirus, and HSV2 were divided into 0.2-ml aliquots and mixed with 1% saponin (Sigma), 1% Triton X-100 (Sigma), 5% Tween 20 (Sigma), and Chaps cell extract buffer (10×) (New England BioLabs) to final concentrations of 0.1%, 0.025%, 0.1%, and 1×, respectively. All working solutions were prepared or diluted in sterile, deionized water and filter sterilized before use. Samples were vortexed for 10 s and incubated for 5 min at RT, followed by the addition of 10× Turbo DNase buffer (Thermo Fisher Scientific, Inc.) to a final concentration of 1× and of 2 μl of Turbo DNase (Thermo Fisher Scientific, Inc.) to all tubes. Samples were gently mixed and incubated at 37°C for 30 min. For use as controls, specimens were also processed simultaneously according to MolYsis method II but with Turbo DNase instead of MolDNase, with only DNase treatment, or with no treatment.
To determine the optimum concentration of saponin for selective lysis of human cells, CSF specimens spiked with S. pneumoniae, E. coli, H. influenzae, adenovirus, and HSV2 were processed in the same way but with final saponin concentrations of 0.1%, 0.05%, and 0.025%. NPA specimens spiked with S. pneumoniae, H. influenzae, B. pertussis, and adenovirus were also tested with 0.1% and 0.025% saponin. For simultaneous analysis of specimens by PCR and NGS, 3 spiked CSF specimens, 2 spiked NPA specimens, and 1 positive original NPA specimen were left untreated or processed under optimum conditions, using saponin to final concentration of 0.025%.
DNA extraction and PCR.
DNA from 0.2 ml of processed or unprocessed specimens was extracted using the QIAsymphony virus/bacteria kit in an automated DNA extraction platform QIAsymphony SP (Qiagen). DNA concentration was measured in a Qubit 2.0 fluorometer using the Qubit dsDNA HS assay kit (Thermo Fisher Scientific, Inc.). For PCR analysis of spiked pathogens, TaqMan PCR assays used for routine diagnostic purposes were employed. To analyze human RNA, a commercially available TaqMan gene expression assay for β-2-microglobulin (β2M) (Thermo Fisher Scientific, Inc.) was used according to the manufacturer's instructions. For PCR analysis of human DNA, a new TaqMan assay was designed based on the third intron sequence of the actin gene. For all DNA targets, 5 μl of sample extract was mixed with 20 μl of a master mix containing 12.5 μl of TaqMan universal PCR master mix (Thermo Fisher Scientific, Inc.) as well as primers and and probes to final concentrations shown in Table 1. Thermal cycling was performed in an ABI7500 Fast instrument (Thermo Fisher Scientific, Inc.) with 1 cycle of 95°C for 10 min, followed by 40 cycles consisting of 95°C for 15 s and 60°C for 60 s. For the influenza A PCR, 5 μl of sample extract was mixed with 20 μl of a master mix containing 12.5 μl of 2× QuantiTect probe RT-PCR master mix and 0.25 μl of QuantiTect RT mix (Qiagen) as well as primers and probes to final concentrations shown in Table 1. Thermal cycling was performed in an ABI7500 Fast instrument (Thermo Fisher Scientific, Inc.) with 1 cycle of 50°C for 30 min, 1 cycle of 95°C for 10 min, followed by 40 cycles consisting of 95°C for 15 s and 60°C for 60 s. Fold changes (FC) in the relative quantity of human- and pathogen-specific amplification targets were calculated based on CT values using the equation FC = 2ΔCT, where ΔCT = (CTunprocessed − CTprocessed) (12). The percent DNA of different targets in the processed specimens was calculated from fold changes from the quantity of targets in the unprocessed specimens. Statistical significance was calculated by the paired Student's t test (two-tailed), and a P value of <0.05 was considered statistically significant.
TABLE 1.
Description of primers and probes used in this study
| Organism | Target gene | Primer/probe | Sequence (5′ to 3′) | Working concentration (μM) | Reference or source |
|---|---|---|---|---|---|
| Human | β-Actin | Forward | CGGCCTTGGAGTGTGTATTAAGTA | 0.3 | This study |
| Reverse | TGCAAAGAACACGGCTAAGTGT | 0.3 | |||
| Probe | FAM-TCTGAACAGACTCCCCATCCCAAGACC-BHQ | 0.2 | |||
| Streptococcus pneumoniae | lytA | Forward | ACGCAATCTAGCAGATGAAGCA | 0.2 | 17 |
| Reverse | TCGTGCGTTTTAATTCCAGCT | 0.2 | |||
| Probe | FAM-TGCCGAAAACGCTTGATACAGGGAG-BHQ1 | 0.2 | |||
| Haemophilus influenzae | Hpd | Forward | AGATTGGAAAGAAACACAAGAAAAAGA | 0.3 | 25 |
| Reverse | CACCATCGGCATATTTAACCACT | 0.3 | |||
| Probe | FAM-AAACATCCA/ZEN/ATCGTAATTATAGTTTACCCA ATAACCC-3IABkFQ | 0.2 | |||
| Escherichia coli | ompT | Forward | CAAGCCAATGTAGGGCATTTTAA | 0.3 | This study |
| Reverse | TTCAGAGATGATATCGGCTCCTT | 0.3 | |||
| Probe | FAM-ACGTTGTTTGTAGCCGATTGCTCTTTCTCC-BHQ1 | 0.2 | |||
| Neisseria meningitidis | ctrA | Forward | GCTGCGGTAGGTGGTTCAA | 0.33 | This study |
| Reverse | TTGTCGCGGATTTGCAACTA | 0.33 | |||
| Probe | FAM–TGTGCAGCTGACACGTGGCAATGT–BHQ1 | 0.2 | |||
| Streptococcus agalactiae | dltS | Forward | TTTAGGAATACCAGGCGATGAAC | 0.3 | This study |
| Reverse | GCTTTGAATCTTAACCATCTTTTGG | 0.3 | |||
| Probe | FAM-ATTGCTTTGGTGACTATAG-MGB | 0.2 | |||
| Bordetella pertussis | Porin gene | Forward | TGAACCATGCATACAACCTATTGA | 0.33 | 26 |
| Reverse | CCTGTCCCCTTAATCCGGAAT | 0.33 | |||
| Probe | FAM-TCTTCACAGTTAGCCCGCGCGC-BHQ1 | 0.2 | |||
| Herpes simplex virus 2 | Glycoprotein D gene | Forward | CCACATTCAGCCGAGCCT | 0.3 | 27 |
| Reverse | CTCGTCCGAAGCCCCG | 0.3 | |||
| Probe | 6FAM-TGTGTACTACGCAGTGCTGGAACGTGC-IABkFQ | 0.1 | |||
| Adenovirus | Hexon gene | Forward | GCCACGGTGGGGTTTCTAAACTT | 0.5 | 28 |
| Reverse | GCCCCAGTGGTCTTACATGCACATC | 0.5 | |||
| Probe | FAM-TGCACCAGACCCGGGCTCAGGTACTCCGA-3IABkFQ | 0.4 | |||
| Influenza A virus | Segment 7 matrix protein 2 (M2) and matrix protein 1 (M1) genes | Forward | GACCRATCCTGTCACCTCTGAC | 0.9 | 11 |
| Reverse | AGGGCATTTTGGACAAAKCGTCTA | 0.9 | |||
| Probe | FAM-TGCAGTCCT/ZEN/CGCTCACTGGGCACG-3IABkFQ | 0.25 |
Metagenomics library construction and sequencing.
The NexteraXT DNA sample preparation kit (Illumina) was used to prepare indexed, paired-end libraries from 1 ng of DNA extracted from clinical samples, according to the manufacturer's instructions. PCR amplification to add Illumina indices was performed in PCR strip tubes in a GeneAmp 9700 thermal cycler (Thermo Fisher Scientific, Inc.). Library DNA was purified for size selection and removal of very small library fragments by using Agencourt AMPure XP beads (Beckman Coulter, Inc.) according to the instructions provided in Nextera XT sample preparation guide, with the exception that the procedure was performed in 1.5-ml microcentrifuge tubes using a magnetic 6-tube stand, the Agencourt SPRIStand (Beckman Coulter, Inc.), instead of in 96-well plates. A total of 90 μl of AMPure XP beads was used for each of the 50-μl PCR products, and the purified libraries were eluted using 52.5 μl of resuspension buffer provided with the Nextera XT kit. DNA concentration of purified libraries was measured in a Qubit 2.0 fluorometer using the Qubit dsDNA HS assay kit (Thermo Fisher Scientific, Inc.). All sequencing libraries were sent to McGill University and the Génome Québec Innovation Centre, Montréal (Québec), Canada for quality control and paired-end 250-bp sequencing on an Illumina MiSeq sequencer.
Bioinformatics.
Quality control metrics for raw whole-genome sequencing reads were calculated with FastQC (13). Based on the quality report, filtration (adapter trimming, PHRED quality score >20, minimum length >100 bases) was performed with Cutadapt (14). MetageniE, a metagenomic data analysis software, was utilized to first remove filtered reads aligned against human reference genome (Hg19) with Burrows-Wheeler Aligner (15, 16). The pathogen detection module of MetaGeniE-generated statistics of the mapped reads aligned against bacterial (ftp://ftp.ncbi.nlm.nih.gov/genomes/bacteria) and viral (ftp://ftp.ncbi.nih.gov/refseq/release/viral) databases.
RESULTS
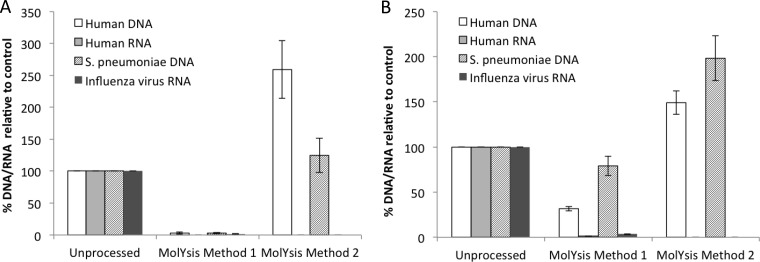
For human DNA subtraction, we first used the MolYsis kit to preprocess the NPA and CSF specimens spiked with S. pneumoniae and influenza A virus to represent bacterial DNA and viral RNA, respectively. We designed a new real-time PCR assay for human DNA by targeting an intron sequence in the β-actin gene to avoid the amplification of the corresponding RNA. For human RNA, a commercially available real-time PCR assay specific for β2M mRNA was used, because this assay is validated to not detect DNA. Specific PCR assays to detect S. pneumoniae DNA and influenza A RNA were described elsewhere (11, 17). MolYsis method I involves a centrifugation step intended to separate the viral supernatant prior to DNase and RNase treatment of the pellet. Based on PCR analysis, both human- and pathogen-associated DNA and RNA were eliminated by >95% in CSF specimens. In NPA samples, the depletion of human DNA compared to S. pneumoniae DNA was not statistically significant (Fig. 1A and B), and both human RNA and influenza A RNA were removed by >95%, similar to CSF samples.
FIG 1.
Processing of clinical specimens using MolYsis reagents. CSF (A) or NPA (B) specimens were spiked with S. pneumoniae and influenza A virus and processed by MolYsis method I and MolYsis method II, as described in the Materials and Methods. Nucleic acids extracted from processed or unprocessed samples were analyzed by real-time PCR assays for β-actin DNA and β-2-microglobulin RNA for human DNA and RNA, respectively, and pathogen-specific targets, as described in Table 1. The percentage of DNA/RNA was obtained from fold changes calculated from CT values, with respect to CT values for unprocessed samples. The data are the average of data obtained from 3 independent experiments using a total of 3 CSF specimens and 3 NPA specimens. Error bars are the standard error of the mean; statistical significance was calculated by the pairwise Student's t test (two-tailed).
The MolYsis method II, which involved direct treatment of specimens, was not effective at all in removing either human DNA or bacterial DNA, but it completely eliminated human and viral RNAs from the NPA and the CSF specimens (Fig. 1A and B). MolDNase was not effective in depleting human DNA, even after increasing the incubation time to up to 2 h and increasing the incubation temperature to 37°C. No significant enrichment was observed for spiked S. pneumoniae and HSV2 DNA (data not shown).
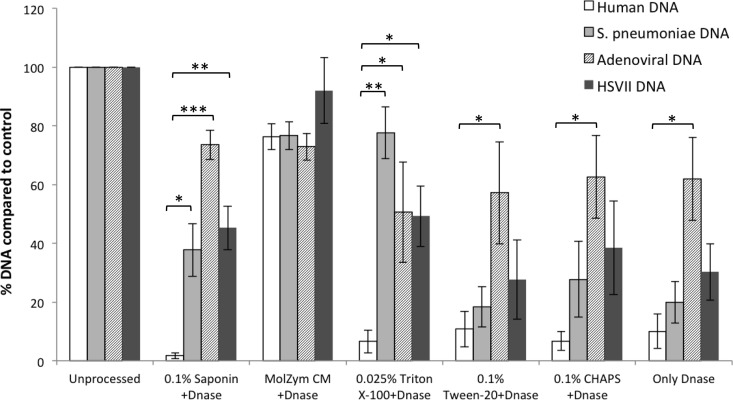
Various nonionic detergents, including saponin, Tween 20, and Triton X-100, or zwitterionic detergent, such as 3-([3-cholamidopropyl] dimethylammonio)-1-propanesulfonate hydrate [CHAPS], were assessed for their ability to selectively lyse human cells in NPA specimens. For postlysis DNase treatment, we used Turbo DNase, which is an engineered version of wild-type DNase I with much higher catalytic efficiency. Notable reduction of human DNA was observed in all samples compared to untreated controls except in the samples that were treated with MolZym CM plus Turbo DNase. The most impressive results were obtained with saponin and Triton X-100 (Fig. 2). With 0.1% saponin and DNase treatment, the average relative enrichment of S. pneumoniae, adenovirus, and HSV2 DNA compared to human DNA was >20-fold. When specimens were treated with only DNase, without any detergents, approximately 90% reduction of human DNA was observed. However, the relative enrichment of pathogen DNA was less significant compared to that of specimens treated with both saponin and DNase (Fig. 2).
FIG 2.
Processing of clinical specimens using various detergents for selective lysis of human cells. NPA specimens were spiked with S. pneumoniae, HSV2, and adenovirus and treated with saponin, Triton X-100, Tween 20, CHAPS buffer, or MolZym CM buffer to final concentrations indicated in the figure, followed by DNase treatment (see Materials and Methods). Nucleic acids extracted from processed or unprocessed samples were analyzed by real-time PCR assays for β-actin DNA for human DNA and pathogen-specific targets, as described in Table 1. The percentage of DNA was obtained from fold changes calculated from CT values, with respect to CT values for unprocessed samples. The data are the average of data obtained from 3 independent experiments using a total of 4 NPA specimens. Error bars are the standard error of the mean; statistical significance was calculated by the pairwise Student's t test (two-tailed). *, P < 0.05; **, P < 0.01; ***, P < 0.001.
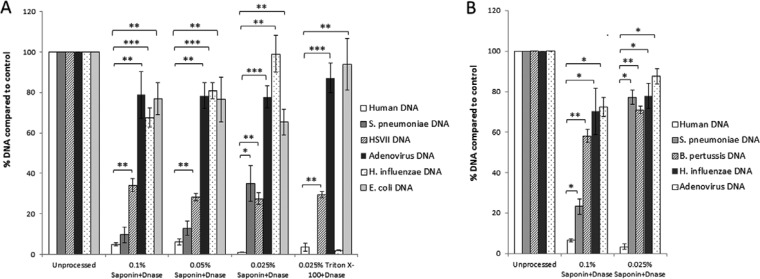
Spiked CSF specimens with different concentrations of saponin, along with an untreated control and Triton X-100, were analyzed. Although a significant (P < 0.001) decrease in human DNA content was observed in all treated samples, pathogen DNA-to-human DNA ratios were highest in the samples that were treated with saponin at a final concentration of 0.025%. At this concentration, approximately 30- to 100-fold enrichment of pathogen DNA was noted compared to human DNA (Fig. 3A). We also observed similar results in spiked NPA specimens, with 0.025% saponin at a final concentration being the most effective condition for selective depletion of human DNA and enrichment of pathogen DNA (Fig. 3B). The results obtained with Triton X-100 were inconsistent, with occasional, significant loss of the pathogen DNA (Fig. 3A).
FIG 3.
Processing of clinical specimens using saponin at different concentrations for selective lysis of human cells. CSF specimens were spiked with S. pneumoniae, H. influenzae, E. coli, HSV2, and adenovirus (A) and NPA specimens spiked with S. pneumoniae, H. influenzae, B. pertussis, and adenovirus (B) were treated with saponin and Triton X-100 to final concentrations indicated in the figure, followed by DNase treatment (see Materials and Methods). Nucleic acids extracted from processed or unprocessed samples were analyzed by real-time PCR assays for β-actin DNA for human DNA and pathogen-specific targets, as described in Table 1. The percentage of DNA was obtained from fold changes calculated from CT values, with respect to CT values for unprocessed samples. The data are the average of data obtained from 3 independent experiments using a total of 4 CSF specimens and 4 NPA specimens. Error bars are the standard error of the mean; statistical significance was calculated by the pairwise Student's t test (two-tailed); *, P < 0.05; **, P < 0.01; ***, P < 0.001.
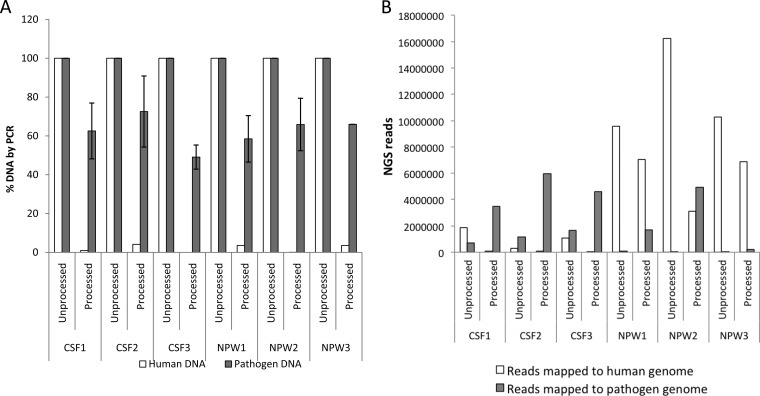
Spiked CSF and NPA were preprocessed with 0.025% saponin, alongside untreated controls, and analyzed simultaneously by both PCR and NGS. As determined by PCR, the relative quantity of human DNA (2.1% ± 1.9%) was significantly (P < 0.001) lower than the relative quantity of pathogen DNA (62.4% ± 8.0%) in the processed specimens (Fig. 4A). Similarly, in the processed specimens, there was a significant (P < 0.05) decrease in NGS reads that mapped to the human genome and a significant (P < 0.01) increase in NGS reads that mapped to different pathogen genomes, compared to the unprocessed specimens (Fig. 4B). On average, pathogen-associated NGS reads were enriched by ∼40-fold and ∼170-fold in CSF and NPA specimens, respectively, following processing. NGS results for specific pathogens were also highly correlated to PCR results (Table 2). The effectiveness of specimen preprocessing was more prominent for pathogens that were weakly positive by PCR. For example, S. pneumoniae and H. influenzae DNA with PCR CT values >35 had extremely low genome coverage (<1% genome coverage). However, following processing, genome coverage for these organisms improved to 9.8% and 26.6%, respectively, despite the fact that the PCR CT values were even weaker (>36).
FIG 4.
Analysis of clinical specimens processed with 0.025% saponin and DNase by PCR and NGS. CSF specimens (n = 3) spiked with S. pneumoniae, N. meningitidis, H. influenzae, E. coli, S. agalactiae, HSV2, and adenovirus and NPA specimens (n = 3) spiked with S. pneumoniae, H. influenzae, B. pertussis, HSV2, and adenovirus were treated with saponin to a final concentration of 0.025%, followed by DNase treatment (see Materials and Methods). (A) Nucleic acids extracted from processed or unprocessed samples were analyzed by real-time PCR assays for β-actin DNA for human DNA, and pathogen-specific targets, as described in Table 1. The percentage of DNA was obtained from fold changes calculated from CT values, with respect to CT values for unprocessed samples. Data for all spiked pathogens were averaged; error bars are the standard error of the mean. (B) Nucleic acids extracted from processed or unprocessed samples were analyzed by NGS on Illumina MiSeq. Sequencing libraries were constructed from 1 ng of DNA from specimens using Nextera XT. Raw data were filtered for quality and mapped to human genome and pathogen genomes, as described in the Materials and Methods. Specimens were processed and analyzed independently, as they were available.
TABLE 2.
Effect of specimen preprocessing on the analytical sensitivity of pathogen detection by NGS
| Organism | Samples | PCR result (CT) |
NGS result (No. of reads mapped to pathogen genome) |
NGS result (% genome coverage) |
||||
|---|---|---|---|---|---|---|---|---|
| Unprocessed | Processed | Unprocessed | Processeda | Enrichment factor (processed/unprocessed) | Unprocessed | Processedb | ||
| Spiked pathogens | ||||||||
| S. pneumoniae | NPW1 | 29.5 | 30.8 | 397 | 3,417 | 8.6 | 2.20 | 13.22 |
| CSF1 | 31.9 | 33.2 | 362 | 1,059 | 2.9 | 1.57 | 3.18 | |
| NPW2 | 35.9 | 36.4 | 6c | 2,086 | 347.7 | 0.08 | 9.76 | |
| E. coli | CSF1 | 29.2 | 29.9 | 57,204 | 275,178 | 4.8 | 65.14 | 93.19 |
| CSF2 | 34.6 | 34.3 | 7,024 | 46,010 | 6.6 | 6.89 | 37.37 | |
| CSF3 | 35.0 | 35.4 | 10,834 | 37,064 | 3.4 | 10.18 | 30.47 | |
| H. influenzae | CSF1 | 27.7 | 27.8 | 163,923 | 767,109 | 4.7 | 92.44 | 93.78 |
| NPW2 | 35 | 35 | 41 | 5,712 | 139.3 | 0.41 | 26.60 | |
| CSF2 | Undetermined | Undetermined | 8,963 | 41,250 | 4.6 | 2.55 | 5.26 | |
| CSF3 | Undetermined | Undetermined | 10,450 | 29,216 | 2.8 | 2.78 | 4.16 | |
| N. meningitidis | CSF2 | 23.7 | 24.2 | 840,062 | 4,895,057 | 5.8 | 95.63 | 97.60 |
| CSF3 | 24.3 | 25.1 | 1,283,102 | 3,692,081 | 2.9 | 96.54 | 97.36 | |
| S. agalactiae | CSF2 | 30.2 | 32.6 | 22,809 | 39,962 | 1.8 | 65.39 | 78.98 |
| CSF3 | 31.3 | 33.9 | 26,498 | 14,454 | 0.5 | 62.82 | 45.31 | |
| B. pertussis | NPW2 | 23.7 | 24.5 | 31,862 | 4,248,717 | 133.3 | 67.17 | 100.00 |
| Human adenovirus | NPW1 | 17.3 | 17.6 | 31,927 | 782,835 | 24.5 | 99.96 | 100.00 |
| CSF1 | 21.0 | 21 | 626,057 | 3,055,828 | 4.9 | 99.57 | 99.69 | |
| NPW2 | 22.2 | 23.7 | 8,243 | 729,057 | 88.4 | 98.83 | 99.52 | |
| Herpes simplex virus 2 | CSF2 | 22.3 | 23.3 | 260,048 | 991,123 | 3.8 | 98.55 | 99.18 |
| CSF3 | 23.2 | 24.3 | 343,310 | 798,024 | 2.3 | 98.76 | 99.18 | |
| NPW1 | 23.3 | 24.8 | 4,493 | 68,752 | 15.3 | 82.88 | 97.32 | |
| CSF1 | 24.6 | 26.6 | 51,468 | 118,020 | 2.3 | 95.26 | 97.60 | |
| Unspiked pathogens/organisms detected in NPA specimens | ||||||||
| Human adenovirus | NPW3 | 21.9 | 22.5 | 11,330 | 242,141 | 21.4 | 99.98 | 99.98 |
| M. catarrhalis | NPW1 | 599 | 904 | 1.5 | 3.47 | 4.32 | ||
| H. influenzae | NPW1 | 118 | 1025 | 8.7 | 0.91 | 5.59 | ||
| S. mitis | NPW1 | 76 | 220 | 2.9 | 0.51 | 1.37 | ||
| S. sanguinis | NPW1 | 10 | 271 | 27.1 | 0.08 | 1.21 | ||
| S. constellatus | NPW2 | 7 | 796 | 113.7 | 0.1 | 4.87 | ||
| A. xylosoxidans | NPW2 | 56,046 | 1,679,351 | 30.0 | 13.5 | 31.08 | ||
| Streptococcus intermedius | NPW2 | 18 | 3,134 | 174.1 | 0.18 | 16.77 | ||
| S. epidermidis | NPW2 | 43 | 1,278 | 29.7 | 0.49 | 5.82 | ||
| C. aurimucosum | NPW2 | 258 | 46,713 | 181.1 | 1.68 | 11.44 | ||
| C. diphtheriae | NPW2 | 117 | 37,642 | 321.7 | 0.02 | 6.16 | ||
| F. nucleatum | NPW2 | 4 | 1,030 | 257.5 | 1.48 | 8.84 | ||
Significantly different from unprocessed specimens (P < 0.01).
Significantly different from unprocessed specimens (P < 0.001).
Bold indicates spiked pathogen undetectable in unprocessed samples but detected in processed samples.
One of the NPA specimens (NPW3), which was originally positive for adenovirus by PCR, was also processed and analyzed by NGS simultaneously with its unprocessed counterpart. The number of NGS reads that mapped to the adenoviral genome was >20-fold higher in the processed specimen than in the unprocessed specimens (Table 2). However, because of the high abundance of NGS reads that mapped to human adenovirus in both specimens, the genome coverage was not different between these two specimens. Apart from the spiked pathogens, several other organisms were detected in the NPA specimens that were either normal flora, such as Moraxella catarrhalis, Haemophilus influenzae, Staphylococcus epidermidis and viridans streptococci, or potential opportunistic pathogens, such as Achromobacter xylosoxidans, which can be associated with pneumonia and other complications in immunosuppressed individuals (18). Consistent with data for spiked pathogens, we noted a significant improvement of sensitivity following preprocessing of specimens both in terms of number of NGS reads related to each taxa and in genome coverage.
DISCUSSION
NGS is increasingly being viewed as a technology that will be available for use in clinical microbiology laboratories within a few years. At present, NGS use in infectious disease diagnostics is very infrequent and limited to amplification-based, targeted sequencing for viral drug resistance and identification of isolated bacteria by 16S rRNA gene sequencing or by whole-genome sequencing. The potential application of NGS for the unbiased detection of pathogens through metagenomic sequencing of clinical specimens is an emerging concept. It is expected that the application of this approach will facilitate detection of uncommon, emerging, and unknown pathogens for which no routine testing currently is available (19–23). However, a number of challenges were identified, that include cost, turnaround time, infrastructure requirements, technical complexities, bioinformatics expertise, standardization, automation, and data quality. The predominance of host DNA in clinical specimens also necessitates additional processing of specimens for enrichment of pathogen-associated DNA or RNA and the use of computational tools for subtraction of human-derived sequences, which can be lengthy and cumbersome. So far, methods applied for pathogen enrichment for NGS are mostly for viruses and include costly and/or complex procedures, such as viral purification, viral genome amplification by rolling circle amplification, or target capture methods with specific oligonucleotide probes (19). While these approaches have been effective for specific purposes, they are not universally applicable to process specimens for the unbiased detection of a broad range of pathogens, including bacteria and viruses. Therefore, in this study, we aimed to develop a specimen-preprocessing protocol for metagenomic studies which is simple, inexpensive, and broadly applicable, so that samples can be enriched for nucleic acids derived from various pathogen types.
We aimed to exploit the differences in cell surface structures of human cells with bacteria and viruses for the selective lysis of human cells and subsequent nuclease treatment of the released DNA. We hypothesized that bacterial and viral DNA will be protected from nuclease digestion because of the presence of cell wall and viral capsids, respectively. Previously, Horz et al. described methods for the selective isolation of bacterial DNA from human oral samples based on two commercially available reagents, MolYsis (Molzym GmbH & Co. KG, Bremen, Germany) and Pureprove (SIRS-Lab GmbH, Jena, Germany) (24). Based on PCR quantitation of the β-2-microglobulin gene for human DNA, and 16S rRNA genes or the glycosyltransferase gene for bacteria, both methods were able to remove at least 90% of human DNA. The MolYsis kit works based on the principle that human cells are first selectively lysed by a chaotropic buffer and the released human DNA is degraded by MolDNase, which is active in the presence of chaotropes in the lysis buffer. Bacteria present in the specimen are then sedimented, washed, and subjected to DNA extraction. We initially attempted to remove both human DNA and RNA from NPA and CSF specimens using the MolYsis kit. However, adding RNase led to the depletion of both human RNA and influenza A RNA (Fig. 1A and B). This suggests that viral RNA was not adequately protected from RNase activity or that traces of RNase may have remained active during the extraction process.
Our initial attempts to selectively remove human DNA with MolYsis reagents were also unsuccessful, but these reagents were found to be effective in a previous study (24). While the loss of both human and spiked S. pneumoniae DNA was observed with MolYsis method I, no loss of DNA was observed when the samples were processed by MolYsis method II. This suggests that the loss of DNA by MolYsis method I is more likely to be associated with centrifugation and washing instead of with MolDNase activity. Although it is not known what type of DNase is used as MolDNase, according to the manufacturer, the effectiveness of MolDNase was ascertained by a β-actin PCR with a target product size of >500 bp, which is much larger than the PCR assays used in this study (N. Murphy, personal communication). If the fragment size is not small enough, the effect of the DNase on the human and pathogen DNA concentrations will not be detectable either by TaqMan PCR or by short-read Illumina sequencing. It is also important to note that MolYsis basic kit is designed to be used for the pretreatment of whole blood, which may not be suitable for other specimen types, such as those used in this study. As a result, our specimen-processing and analytical approaches differ from those described in manufacturer's instructions and in previously published studies (24).
For the selective lysis of human cells, we sought methods based on mild detergents that are typically used to permeabilize mammalian cell lines for protein extraction or for the recovery of intracellular pathogens. Tween 20, Triton X-100, and CHAPS buffer are all very common reagents used in cell culture studies, while saponin is commonly used in hematology laboratories for hemolysis of human erythrocytes. Interestingly, we noted that Turbo DNase, which worked in the presence of all of these detergents, was ineffective in the presence of the chaotrophic buffer MolZym CM. On the other hand, the results of using mild detergents for cell lysis, followed by DNase treatment, were highly encouraging, with more human DNA than pathogen DNA being degraded. Saponin at a final concentration of 0.025% was most effective in its differential effect on human versus pathogen DNA in both the NPA and the CSF specimens. Overall, Gram-negative bacteria and nonenveloped viruses appeared more stable to sample preprocessing than Gram-positive bacteria and enveloped virus, respectively. These results were also reproducible in independent sets of spiked CSF and NPA specimens that were analyzed by both PCR and NGS for spiked pathogens. Based on data on NGS reads mapped to the genomes of specific pathogens, as well as on their genome coverage, most spiked pathogens were detectable in the unprocessed samples. However, preprocessing clearly enriched samples for pathogen DNA and improved the analytical sensitivity of pathogen detection by NGS. The benefit of specimen preprocessing was particularly seen in specimens that were weakly positive for a pathogen by PCR. For example, in one of the unprocessed NPA specimens, NGS reads mapped to S. pneumoniae and H. influenzae genomes were only 6 and 41, respectively, which would likely be considered insignificant without prior knowledge that these organisms were spiked in this specimen. However, following processing, NGS reads mapped to these genomes were increased 348- and 139-fold (Table 2), respectively.
While the results with spiked pathogens were encouraging, we also verified these results in an NPA sample that was originally positive for human adenovirus by PCR and noted that the number of NGS reads mapped to the adenovirus genome was significantly increased by sample processing (Table 2). Similarly, a number of organisms that were detected in NPA specimens in addition to the spiked pathogens were enriched by a factor of about 100-fold on average. Specimen processing also improved the genome coverage of these organisms. It is crucial to note that pathogens that naturally occur in clinical specimens may have damage to their cell walls or membranes due to the host immune system, antibiotics, or damage during sample transportation and handling, unlike spiked pathogens that were grown in the laboratory. These organisms would likely be susceptible to the pretreatment and DNase digestion discussed in this study. Additional studies using a larger number of clinical samples will be required to confirm and extend the observations made in this study.
Limitations of this study include the fact that the optimized specimen-processing protocol we discussed may not be effective for latent DNA viruses, bacteria that lack cell walls, and protozoal pathogens. In fact, in one of our spiking experiments, we noted the loss of Mycoplasma pneumoniae DNA along with human DNA (data not shown). Also, it has not been tested whether fungal DNA would be protected from DNase digestion under the experimental conditions discussed in this study. Thus, while we have demonstrated concentration and detection of some common bacterial and viral pathogens in this study, further work is required to assess the method for a wider range of respiratory pathogens.
In this study, we have used specimens that were not previously frozen because of the possibility that freezing and thawing may disrupt both human cells and potential pathogens and prevent the selective effect of saponin and DNase on human cells. However, clinical microbiology laboratories commonly freeze specimens to save them or transport them to other facilities for additional analysis. Whether the specimen-processing protocol we discussed would work for specimens that have undergone one or more freeze-thaw cycle requires experimental verification. Further optimization and validation is also necessary with specimen types other than NPA and CSF specimens, such as blood, sputum, and stool specimens. Another limitation of this study is that viruses with RNA genomes were not extensively tested. Future studies could employ an RNA-specific protocol with reduction of human RNA using one of the commercially available rRNA removal kits followed by sequencing of cDNA. Nevertheless, we demonstrate a simple and inexpensive procedure for selective depletion of human DNA from human metagenomic specimens with only ∼40 min of hands-on time and without requiring any specialized reagents or instruments. The results of this study can serve as the basis for further validation and standardization of an NGS protocol for routine detection of pathogens in a relatively unbiased manner.
ACKNOWLEDGMENTS
This study was supported by the British Columbia Clinical Genomics Network (BCCGN), Canada, grant BCCGN00032 and by a Telethon grant (KRZ28061) from BC Children's Hospital Foundation, Vancouver, BC, Canada. The funders had no role in study design, data collection, and interpretation or the decision to submit the work for publication.
REFERENCES
- 1.Fournier PE, Drancourt M, Colson P, Rolain JM, La Scola B, Raoult D. 2013. Modern clinical microbiology: new challenges and solutions. Nat Rev Microbiol 11:574–585. doi: 10.1038/nrmicro3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nolte FS, Caliendo AM. 2010. Molecular microbiology. In Versalovic J, Carroll KC, Jorgensen JH, Funke G, Landry ML, Warnock DW (ed), Manual of clinical microbiology, vol 1 ASM Press, Washington, DC. [Google Scholar]
- 3.Salez N, Vabret A, Leruez-Ville M, Andreoletti L, Carrat F, Renois F, de Lamballerie X. 2015. Evaluation of four commercial multiplex molecular tests for the diagnosis of acute respiratory infections. PLoS One 10:e0130378. doi: 10.1371/journal.pone.0130378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhang H, Morrison S, Tang YW. 2015. Multiplex polymerase chain reaction tests for detection of pathogens associated with gastroenteritis. Clin Lab Med 35:461–486. doi: 10.1016/j.cll.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butt SA, Maceira VP, McCallen ME, Stellrecht KA. 2014. Comparison of three commercial RT-PCR systems for the detection of respiratory viruses. J Clin Virol 61:406–410. doi: 10.1016/j.jcv.2014.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pardo J, Klinker KP, Borgert SJ, Butler BM, Rand KH, Iovine NM. 2014. Detection of Neisseria meningitidis from negative blood cultures and cerebrospinal fluid with the FilmArray blood culture identification panel. J Clin Microbiol 52:2262–2264. doi: 10.1128/JCM.00352-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tregoning JS, Schwarze J. 2010. Respiratory viral infections in infants: causes, clinical symptoms, virology, and immunology. Clin Microbiol Rev 23:74–98. doi: 10.1128/CMR.00032-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayden RT, Carroll KC, Tang Y, Wolk DM. 2009. Diagnostic microbiology of the immunocompromized host. ASM Press, Washington, DC. [Google Scholar]
- 9.Yang J, Yang F, Ren L, Xiong Z, Wu Z, Dong J, Sun L, Zhang T, Hu Y, Du J, Wang J, Jin Q. 2011. Unbiased parallel detection of viral pathogens in clinical samples by use of a metagenomic approach. J Clin Microbiol 49:3463–3469. doi: 10.1128/JCM.00273-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reed LJ, Muench H. 1938. A simple method of estimating fifty percent endpoints. Am J Hygiene 27:493–497. [Google Scholar]
- 11.Selvaraju SB, Selvarangan R. 2010. Evaluation of three influenza A and B real-time reverse transcription-PCR assays and a new 2009 H1N1 assay for detection of influenza viruses. J Clin Microbiol 48:3870–3875. doi: 10.1128/JCM.02464-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-(delta delta CT) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 13.Andrews S. 2010. FastQC: a quality control tool for high-throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
- 14.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 15.Rawat A, Engelthaler DM, Driebe EM, Keim P, Foster JT. 2014. MetaGeniE: characterizing human clinical samples using deep metagenomic sequencing. PLoS One 9:e110915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H. 2013. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 [q-bio.GN] 00:1–3. [Google Scholar]
- 17.Carvalho Mda G, Tondella ML, McCaustland K, Weidlich L, McGee L, Mayer LW, Steigerwalt A, Whaley M, Facklam RR, Fields B, Carlone G, Ades EW, Dagan R, Sampson JS. 2007. Evaluation and improvement of real-time PCR assays targeting lytA, ply, and psaA genes for detection of pneumococcal DNA. J Clin Microbiol 45:2460–2466. doi: 10.1128/JCM.02498-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Swenson CE, Sadikot RT. 2015. Achromobacter respiratory infections. Ann Am Thorac Soc 12:252–258. doi: 10.1513/AnnalsATS.201406-288FR. [DOI] [PubMed] [Google Scholar]
- 19.Lefterova MI, Suarez CJ, Banaei N, Pinsky BA. 2015. Next-generation sequencing for infectious disease diagnosis and management: a report of the Association for Molecular Pathology. J Mol Diagn 17:623–634. doi: 10.1016/j.jmoldx.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 20.Kwong JC, McCallum N, Sintchenko V, Howden BP. 2015. Whole-genome sequencing in clinical and public health microbiology. Pathology 47:199–210. doi: 10.1097/PAT.0000000000000235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lecuit M, Eloit M. 2014. The diagnosis of infectious diseases by whole-genome next-generation sequencing: a new era is opening. Front Cell Infect Microbiol 4:25. doi: 10.3389/fcimb.2014.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Padmanabhan R, Mishra AK, Raoult D, Fournier PE. 2013. Genomics and metagenomics in medical microbiology. J Microbiol Methods 95:415–424. doi: 10.1016/j.mimet.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 23.Barzon L, Lavezzo E, Costanzi G, Franchin E, Toppo S, Palu G. 2013. Next-generation sequencing technologies in diagnostic virology. J Clin Virol 58:346–350. doi: 10.1016/j.jcv.2013.03.003. [DOI] [PubMed] [Google Scholar]
- 24.Horz HP, Scheer S, Vianna ME, Conrads G. 2010. New methods for selective isolation of bacterial DNA from human clinical specimens. Anaerobe 16:47–53. doi: 10.1016/j.anaerobe.2009.04.009. [DOI] [PubMed] [Google Scholar]
- 25.Wang X, Theodore MJ, Mair R, Trujillo-Lopez E, du Plessis M, Wolter N, Baughman AL, Hatcher C, Vuong J, Lott L, von Gottberg A, Sacchi C, McDonald JM, Messonnier NE, Mayer LW. 2012. Clinical validation of multiplex real-time PCR assays for detection of bacterial meningitis pathogens. J Clin Microbiol 50:702–708. doi: 10.1128/JCM.06087-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hasan MR, Tan R, Al-Rawahi GN, Thomas E, Tilley P. 2014. Evaluation of amplification targets for the specific detection of Bordetella pertussis using real-time polymerase chain reaction. Can J Infect Dis Med Microbiol 25:217–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Corey L, Huang ML, Selke S, Wald A. 2005. Differentiation of herpes simplex virus types 1 and 2 in clinical samples by a real-time Taqman PCR assay. J Med Virol 76:350–355. doi: 10.1002/jmv.20365. [DOI] [PubMed] [Google Scholar]
- 28.Heim A, Ebnet C, Harste G, Pring-Akerblom P. 2003. Rapid and quantitative detection of human adenovirus DNA by real-time PCR. J Med Virol 70:228–239. doi: 10.1002/jmv.10382. [DOI] [PubMed] [Google Scholar]