Abstract
Rapid antigen detection tests (RADTs) for group A streptococci (GAS) are widely used for diagnosing acute pharyngitis, which has led to a considerable reduction in antibiotic prescriptions over the past decade. Beyond this intended use, their reassessment on invasive samples may be relevant in the management of life-threatening GAS infections. To this end, we evaluated the performances of three RADTs, culture, GAS PCR, and 16S rRNA gene PCR assays, and compared them with a composite gold standard (GAS-PCR assay and/or culture) for the diagnosis of severe GAS infection. A total of 192 specimens from deep-tissue (mostly normally sterile) sites enriched for 75 GAS-positive samples were enrolled in the study. The three evaluated RADTs showed sensitivities ranging from 88.0% to 94.7% versus 98.7% for GAS PCR, 84% for 16S rRNA gene PCR, and 77.3% for culture. The sensitivities of the ImmunoCard STAT! Strep A test (Meridian Bioscience) and the NADAL Strep A strip (Nal Von Minden) were similar to that of GAS PCR (P = 0.25 and 0.03, respectively) and higher than that of culture (P = 0.001 and 0.006, respectively), whereas the SD Bioline Strep A test strip (Standard Diagnostics) showed a performance similar to that of culture (P = 0.02). The three RADTs detected 10 distinct emm types, including a predominance of emm 1 (33.3%), emm 89 (10.6%), and emm 12 (7.6%). No false-positive results were observed, leading to a specificity of 100% for all the evaluated RADTs. The GAS RADTs turned out to be sensitive, specific, and easy-to-use tools that may aid in the management of invasive GAS infections in 24/7 point-of-care laboratories by enabling early diagnosis and focused therapy.
INTRODUCTION
Group A streptococci (GAS) are common bacterial pathogens responsible for mild diseases, such as pharyngitis, and invasive and life-threatening infections resulting from their dissemination into deep tissues. The worrying increase in the prevalence of these severe infections and their high fatality rates emphasize the need for rapid diagnostic tests to aid in patient management (1, 2).
Over the past 2 decades, manufacturers have developed immunochromatographic rapid antigen detection tests (RADTs) that detect the polysaccharide C cell-wall antigen of bacteria. These tests are now widely used for diagnosing pharyngitis in pediatric outpatient clinics and private practices, which has considerably reduced the number of antibiotic prescriptions and resulted in fewer throat cultures performed by laboratories (3–5).
Extending their indication beyond this intended use, namely, on deep-seated samples, may reduce delays in the diagnosis of invasive infections. To this end, we evaluated the performances of three commercially available RADTs approved for pharyngitis in the diagnosis of invasive GAS infections and compared them with those of conventional culture and molecular techniques.
MATERIALS AND METHODS
Patients and clinical specimens.
This survey was conducted by three teaching hospitals (in Lyon, Grenoble, and Saint-Etienne, France) between May 2013 and April 2014 on a population enriched for GAS-positive samples. With one exception, the GAS-positive specimens enrolled in this study were selected retrospectively (within 1 week after plating) on the basis of their positivity for GAS by culture or 16S rRNA gene PCR. All GAS-negative specimens and a single GAS-positive specimen were selected over the same period according to the site of infection on the basis of a GAS-negative result by culture or 16S rRNA gene PCR.
In this study, invasive GAS infection was defined by the isolation of Streptococcus pyogenes and/or the detection of GAS DNA by 16S rRNA gene or GAS-specific PCR from a sterile site or a deep pulmonary sample.
Repeated samples from the same patient were excluded. Data regarding any antibiotic therapy before and at the time of the sampling were collected. Oral informed consent was obtained from each patient or his or her legal representative.
Conventional culture.
After Gram staining, samples were inoculated after receipt in each laboratory on at least horse blood agar (bioMérieux, Marcy l'Etoile, France) incubated under aerobic conditions, chocolate agar (bioMérieux) incubated in a 5% CO2 atmosphere, and Schaedler broth (bioMérieux) at 35°C. A supplementary Gram-positive selective agar plate containing colistin and nalidixic acid (bioMérieux) was added for respiratory samples. Culture media were incubated for at least 48 h. All isolates were identified by colony morphology, hemolysis, Gram staining, oxidase and catalase tests, and matrix-assisted laser desorption/ionization–time of flight (MALDI-TOF) mass spectrometry (Vitek MS mass spectrometer [bioMérieux] or Biotyper [Bruker, Germany]) or latex agglutination (diagnostic reagents; Oxoid, Dardilly, France).
Specimens were stored at 4°C for 1 week after culturing and then frozen at −20°C. A 500- to 1,000-μl volume of DNA-free water was added to swabs, low-volume liquid samples, and soft samples (i.e., tissular biopsy specimens) to reach a 1,000-μl final volume, which was required to perform the RADTs and PCR assays.
Rapid antigen detection tests.
Three colorimetric dipstick RADTs were evaluated at the same time in each laboratory on refrigerated or frozen samples, the ImmunoCard STAT! Strep A (Meridian Bioscience, Paris, France), the NADAL Strep A strip (Nal Von Minden, Moers, Germany), and the SD Bioline Strep A test strip (Standard Diagnostics, Jouy-en-Josas, France). Each specimen was absorbed on a swab provided by each manufacturer. In the case of a soft sample, the swab was also rubbed on the sample to discharge as many bacterial antigens as possible into the DNA-free water. Then, the RADTs were performed according to manufacturer instructions.
Double-blind readings of the result windows were performed to warrant an objective RADT evaluation. Any discrepant result led to a second test.
Genomic DNA extraction.
DNA was extracted from 200 μl of clinical samples within 1 week after plating by proteinase K enzymatic digestion and DNA isolation using the MagNA Pure compact nucleic acid isolation kit on a MagNA Pure compact system (Roche Diagnostics, Meylan, France) in Lyon or a QIAsymphony instrument (Qiagen, Courtaboeuf, France) in Saint-Etienne. The High Pure PCR template preparation kit (Roche Diagnostics, Meylan, France) was used for manual processing in the Grenoble laboratory. DNA extracts were collected in the Lyon laboratory to perform the PCR assays. They were all shipped and stored at −20°C until the PCR assays were performed.
16S rRNA gene PCR assay.
16S rRNA gene amplification was performed from 5 μl of DNA extract using the Taq'Ozyme HS system (Ozyme, St. Quentin-en-Yvelines, France), primers 91E (TCAAAKGAATTGACGGGGGC) and 13Bs (GCCCGGGAACGTATTCAC), both at 0.5 μM, and propidium monoazide to remove unspecific DNA background (6, 7). Amplified products were examined in 1.25% agar gel electrophoresis supplemented with SYBR Safe DNA (Invitrogen, Illkirch, France) and, if positive, were submitted to single-strand DNA sequencing using primer 13Bs (Biofidal, Vaulx-en-Velin, France). Bacterial identification was made by comparing the obtained sequences with Le BIBI (Quick BioInformatic Phylogeny of Prokaryotes) gene data bank (see https://umr5558-bibiserv.univ-lyon1.fr/lebibi/lebibi.cgi).
GAS-specific real-time PCR assay.
Amplification of the ntpB gene was performed from 2.5 μl of DNA extract on a SmartCycler (Orgentec, Trappes, France) using TaKaRa SYBR Premix Ex Taq (Ozyme) and primers StrA7-1 (GTCGATTTTGCCACGTACCG) and StrA7-2 (TGCATGGTCAACTCAATCATTTGC), both at 0.25 μM, for 45 cycles (8). A positive result was defined by the detection of a 178-bp-specific amplicon with a melting temperature of 80 ± 0.5°C.
emm type characterization.
All GAS isolates and positive DNA extracts were referred to the French National Reference Centre for Streptococci for emm sequence typing as previously described (9).
Statistical analysis.
The performance of RADTs, culture, and PCR assays was assessed, and the results were compared with those of a composite gold standard (GAS-PCR assay and/or culture) using the exact McNemar test. A P value of <0.01 indicated that the categories were statistically significantly different.
RESULTS
A total of 192 deep-seated specimens were enrolled in this study on the basis of their positivity or negativity for GAS. These samples were collected from patients suspected of having invasive infection and who were aged between 6 months and 92 years in Lyon (n = 136), Saint-Etienne (n = 39), or Grenoble (n = 17) hospitals. The samples consisted of deep respiratory (pleural fluid, pulmonary biopsy specimens, and bronchoalveolar lavage fluid; n = 44), osteoarticular (synovial fluid, hygroma, abscesses, discal biopsy specimens, and swabbed diabetic foot ulcers; n = 43), subcutaneous (abscesses and tissular biopsy specimens; n = 37), otolaryngologic (abscesses, tissue, and paracenteses fluid; n = 35), digestive (bile, peritoneal fluid, ascites, and abscesses; n = 16), cardiological (heart valves and pericardic fluid; n = 6), and neurological (cerebrospinal fluid and cerebral abscesses; n = 5) samples, gynecological abscesses (n = 3), a renal abscess (n = 1), a transplantectomy cavity (n = 1), and lymph fluid (n = 1). Among these samples, 75 (39.1%) tested positive for GAS by the gold-standard method (GAS PCR and/or culture) (Table 1), 9 of which (12%) were collected on a single swab that was plated before adding DNA-free water for performing RADTs.
TABLE 1.
Clinical specimens collected for this study
| Clinical specimens (n = 192) | No. of GAS-positive specimens (total n = 75) | No. of GAS-negative specimens (total n = 117) |
|---|---|---|
| Respiratory | 14 | 30 |
| Osteoarticular | 15 | 28 |
| Subcutaneous | 21 | 16 |
| Otolaryngologic | 18 | 17 |
| Digestive | 2 | 14 |
| Cardiological | 0 | 6 |
| Neurological | 2 | 3 |
| Gynecological | 2 | 1 |
| Others | 1 | 2 |
The GAS-PCR assay was the most sensitive technique (sensitivity, 98.7%) with cycle thresholds ranging from 14.2 to 31.1, followed by RADTs (sensitivity, 88% to 94.7% according to the test), 16S rRNA gene PCR (sensitivity, 84%), and culture (sensitivity, 77.3%) (Table 2). Gram staining revealed typical Gram-positive cocci in pairs and/or chains for only 21 (28%) of the GAS-positive samples.
TABLE 2.
Comparison of methods for the detection of group A streptococci in patients suspected of having an invasive infectiona
| Diagnostic method | No. true positive | No. false positive | No. false negative | No. true negative | Sensitivityb |
Specificityb |
||
|---|---|---|---|---|---|---|---|---|
| % | 95% CIc | % | 95% CI | |||||
| RADTs | ||||||||
| ImmunoCard STAT! Strep A (Meridian Bioscience) | 71 | 0 | 4 | 117 | 94.7 | 86.9–98.5 | 100 | 96.9–100 |
| NADAL Strep A strip (Nal Von Minden) | 68 | 0 | 7 | 117 | 90.7 | 81.7–96.2 | 100 | 96.9–100 |
| SD Bioline Strep A test strip (Standard Diagnostics) | 66 | 0 | 9 | 117 | 88.0 | 78.4–94.4 | 100 | 96.9–100 |
| Molecular assays | ||||||||
| 16S rRNA gene PCR positive for GAS | 63d | 0 | 12 | 117 | 84.0 | 73.7–91.5 | 100 | 96.9–100 |
| GAS-PCR | 74 | 0 | 1e | 117 | 98.7 | 92.8–99.9 | 100 | 96.9–100 |
| Culture | 58 | 0 | 17 | 117 | 77.3 | 66.2–86.2 | 100 | 96.9–100 |
One hundred ninety-two samples were analyzed using all techniques.
With the limitation of an enriched population.
95% CI, 95% confidence interval.
The 63 samples were all positive with GAS PCR.
The sole sample that tested negative with GAS PCR was positive by culture only and grew few colonies.
Regarding the sensitivity of GAS PCR, statistically significant differences with culture (P = 0.0001) and 16S rRNA gene PCR (P = 0.001) were observed. Among the 75 GAS-positive samples, 11 had discrepant results between GAS and 16S rRNA gene PCR, although all of them were positive with 16S rRNA gene PCR. Seven of them yielded an undetermined identification due to plurimicrobial DNA sequences (contamination of samples or plurimicrobial infection). Three of them were limited to identification at the Streptococcus genus level, and the last DNA sequence led to the identification of Streptococcus intermedius which was associated with GAS by culture.
No significant differences were observed between the sensitivities of GAS PCR and those of the ImmunoCard STAT! Strep A test (P = 0.25) and NADAL Strep A strip (P = 0.03), whereas the SD Bioline Strep A test strip performed less well (P = 0.008). Only the ImmunoCard STAT! Strep A test and NADAL Strep A strip also yielded a sensitivity higher than that of culture (P = 0.001 and 0.006, respectively).
Within the 17 GAS-positive samples associated with a negative culture (Table 2), the data regarding any antibiotic therapy were collected for 14 of 17 patients. All of them were receiving antibiotic therapy before or at the time of sampling, which may explain these discrepant results. Among them, 3, 5, and 6 samples also tested negative with the ImmunoCard STAT! Strep A test, NADAL Strep A strip, and SD Bioline Strep A test strip, respectively, which decreased the respective RADT sensitivities from 94.7% to 82.4%, 90.7% to 70.6%, and 88% to 64.7% on culture-negative samples, suggesting that the performance of RADTs may be reduced if they are used as a second-line diagnostic test. For these culture-negative samples, the ImmunoCard STAT! Strep A test still demonstrated the highest sensitivity (Table 3).
TABLE 3.
Performance of culture and 16S rRNA gene and GAS-PCR assays on the nine samples showing at least one negative RADT
| Diagnostic method | Cerebrospinal fluid | Tissue biopsy specimen | Articular fluid 1 | Articular fluid 2 | Pleural fluid 1 | Pleural fluid 2 | Articular fluid 3 | Articular fluid 4 | Tonsil abscess |
|---|---|---|---|---|---|---|---|---|---|
| RADTs | |||||||||
| ImmunoCard STAT! Strep A (Meridian Bioscience) | + | + | − | + | + | + | − | − | − |
| NADAL Strep A strip (Nal Von Minden) | + | − | − | + | − | − | − | − | − |
| SD Bioline Strep A test strip (Standard Diagnostics) | − | − | − | − | − | − | − | − | − |
| Culture | + | + | +a | − | − | − | − | − | − |
| Molecular assays | |||||||||
| 16S rRNA gene PCR positive for GAS | + | + | − | + | + | + | + | + | −b |
| GAS PCR | + | + | − | + | + | + | + | + | + |
Less than five colonies.
Positive 16S rRNA gene PCR assay showing a plurimicrobial sequence.
Three culture-positive samples tested negative in at least one RADT (Table 3). Among them, the sole articular fluid that tested negative in all the RADTs grew only a few colonies and tested negative in the 16S rRNA gene and GAS PCR.
Within GAS-negative specimens (n = 117), no etiological agent was identified in 26 samples. The other 91 samples proved to be positive by culture (n = 30), 16S rRNA gene PCR (n = 33), or both (n = 28) with at least one of the following pathogens: Streptococcus milleri group (n = 16), Streptococcus agalactiae (n = 8), Streptococcus pneumoniae (n = 7), Streptococcus dysgalactiae (n = 3), Streptococcus mitis/oralis (n = 2), Streptococcus parasanguinis (n = 2), Streptococcus gallolyticus (n = 1), Enterococcus faecalis (n = 2), Enterococcus faecium (n = 2), Staphylococcus aureus (n = 11), Staphylococcus epidermidis (n = 5), Peptostreptococcus micros (n = 1), Paracoccus spp. (n = 1), Escherichia coli (n = 7), Salmonella enteritidis (n = 1), Proteus mirabilis (n = 1), Enterobacter asburiae (n = 1), Hafnia alvei (n = 2), Pseudomonas aeruginosa (n = 2), Acinetobacter baumannii (n = 1), Porphyromonas spp. (n = 1), Fusobacterium spp. (n = 6), Bacteroides spp. (n = 1), Prevotella spp. (n = 5), Pasteurella multocida (n = 1), Mycobacterium tuberculosis (n = 1), Bartonella quintana (n = 1), Nocardia spp. (n = 1), Kingella kingae (n = 1), Candida spp. (n = 1), and Neisseria meningitidis (n = 1).
These samples were all negative for GAS by all of the methods used. It was notable that no false-positive results reflecting cross-reactivities with other streptococcal antigens were observed for the three evaluated RADTs that demonstrated specificities of 100%.
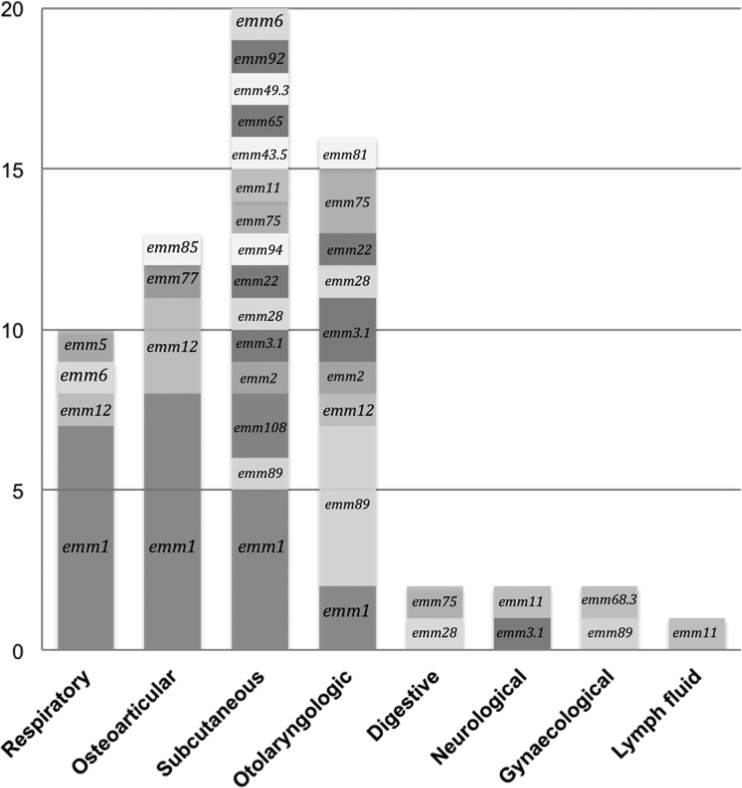
A total of 66 GAS were genotyped from the isolated strains (n = 4), directly from the DNA extracted from biological samples (n = 40), or both (n = 22). Twenty-one emm types were characterized, with the major one being emm 1 (33.3%; n = 22), followed by emm 89 (10.6%; n = 7) and emm 12 (7.6%; n = 5) (Fig. 1). Most of the GAS typed from respiratory (70%; n = 7) and osteoarticular (61.5%; n = 8) samples belonged to the emm 1 type. All emm 12 and most of emm 1 types (68%; n = 15) were found in children younger than 10 years. Nine GAS could not be typed due to both negative culture and an insufficient bacterial DNA amount in the clinical specimens.
FIG 1.
emm type distribution among group A streptococci responsible for invasive infections (n = 66). Genotyping was performed from the isolated strains (n = 4), the DNA extracted from the biological samples (n = 40), or both (n = 22).
DISCUSSION
In this study, we assessed the role of RADTs for the diagnosis of invasive GAS infections. Neither RADTs nor GAS PCR reached a sensitivity of 100%, but both of them turned out to be more sensitive than conventional culture and 16S rRNA gene PCR, which are routinely performed in microbiology laboratories for diagnosing invasive infection.
The samples that tested negative for RADTs but positive for GAS PCR all had a PCR cycle threshold greater than 30.5, reflecting a low bacterial load. These findings are consistent with the detection limits of the two techniques: 104 to 105 CFU for RADTs (according to the manufacturers) and 25 genome copies per assay for GAS PCR (determined in our laboratory).
We observed differences between the performances of the three RADTs, with sensitivities ranging from 88.0% to 94.7%, although this difference did not reach statistical significance. The ImmunoCard STAT! Strep A and the NADAL Strep A strip were significantly more sensitive than culture, which was not expected due to the respective detection limits announced by the manufacturers.
As with GAS PCR and culture, RADTs had demonstrated specificities of 100%, outperforming the expected results for pharyngitis (95.8% to 97.8% according to the manufacturers). No false-positive results, in particular with other streptococci were observed in our study, although RADT cross-reactivities with the Streptococcus milleri group have been described (10, 11). These cross-reactivities must be interpreted with caution, because RADTs were compared with culture only and GAS PCR was not performed. Cohen et al. (12) recently showed that most of the samples with false-positive RADTs were PCR positive, suggesting specificity close to 100% in pharyngitis. Moreover, Rubin et al. (11) assessed the lack of specificity of RADTs by directly testing colonies, which provides a high bacterial load compared with that of clinical samples and may generate cross-reactivity.
To our knowledge, we performed the largest study to date on the reassessment of GAS RADTs on invasive samples. Literature data on extrapharyngeal specimens are scarce and limited to superficial infections, such as perianal cellulitis, impetigo, whitlows, and rhinitis (13–16). All these data confirmed that RADTs designed for GAS pharyngitis can be used to diagnose extrapharyngeal GAS infection accurately. A single study on 120 pleural fluid samples but limited to 10 GAS-positive specimens from pediatric patients has already been published (17). Sensitivities of 100%, 90%, and 10% have been reported for GAS PCR, RADTs, and culture, respectively. With the exception of culture, the results of our work show similar trends, but we allowed the assessment of RADTs on a large number of positive samples and a broad range of patients and biological matrices in which the presence of GAS is considered abnormal. Our study also included other major sources of variability, such as site-to-site variations and the effect of different operators.
The determination of emm type distribution is a major tool for epidemiological investigations of GAS infection, because invasive strains markedly differ from strains responsible for mild infections in Europe. Because RADTs were developed for diagnosing pharyngitis, we aimed to characterize the emm types implicated in this study. emm 1, emm 89, and emm 12 have been the most frequently identified sequence types described in invasive infections in Europe, the United States, and Australia (18–20). Nonetheless, the increased prevalence of emm type 28 strains was not observed in our study. emm type 12 strains were isolated exclusively from children, which correlated with literature data that showed a predominance of emm types 1 and 12 in pediatric invasive infections (21, 22). Finally, GAS strains detected from respiratory and osteoarticular samples were predominantly emm type 1. Plainvert et al. described such an association for respiratory samples only (with the emm type predominant in septic arthritis being emm 28), but the limited number of osteoarticular samples in this study may restrict our interpretation regarding emm distribution (18).
Our study had several biases. The relatively low incidence of invasive GAS infection (75 cases in 3 teaching hospitals over a 1-year period) and the limited number of RADTs provided by the manufacturers mandated the use of repository samples. Although the combination of culture and PCR sounds like a reasonable and well-admitted reference standard, the enrichment for positive specimens may potentially bias the sensitivity and specificity calculations. In this setting, the prevalence of invasive GAS infection was also not evaluated, and thus the positive and negative predictive values of the three evaluated RADTs were not determined.
Many GAS-positive samples were also selected according to the culture results, which probably overestimated the performance of culture and minimized the contribution of RADTs as potential 24/7 point-of-care (POC) laboratory tests. It would have been relevant to assess the performance of RADTs on more specimens containing noncultivable GAS collected from patients who had received an antimicrobial treatment before sampling.
Furthermore, 12% of the positive samples were collected on a single swab that was plated before adding water for performing the RADTs. This also contributed to minimize the sensitivity of RADTs compared with that of culture, because we observed that their sensitivities had decreased by a value of 12.3% to 23.3% (depending on the test) among culture-negative samples compared with those of culture-positive samples.
Nonetheless, this study provides further insight into ways of improving the management of invasive GAS infection. RADTs can be performed in <1 h, whereas culture or PCR requires at least 1 day for achieving results. As demonstrated for pharyngitis, their use may limit antibiotic overprescription and focus the treatment, limiting the side effects and the emergence of antimicrobial resistance (5, 23). It may also be helpful for optimizing therapy by adding antitoxin antibiotics, such as clindamycin, and intravenous immunoglobulin, which reduce the risk of fatal outcome (24, 25).
A suitable environment is required for performing RADTs and detecting weak positive samples. For pharyngitis, Park et al. highlighted that the main reason for many clinicians not using RADTs is a lack of time (26). Among several factors that affect RADT sensitivity, the training of the person performing the test is a key determinant. Cohen et al. showed that RADT sensitivity is strongly influenced by the physician performing the test and is higher for physicians with hospital-based clinical activity in addition to office-based practice (27). Toepfner et al. showed significant differences in their performance when performed by lab technicians versus physicians and demonstrated that the performance no longer differed between the two groups after implementing additional hands-on training (28).
Altogether, our results validate the use of RADTs for GAS on invasive samples. Because Gram staining demonstrated a limited contribution to the diagnosis and because the sensitivity of RADTs was decreased with culture-negative samples, we recommend their use as first-line diagnostic tests, at the time of plating, when clinical symptoms give rise to suspicion of invasive GAS infection. They can be used at any time and are less time-consuming, less expensive, and more rapid and suitable than PCR assays, which are usually performed in cases of negative culture. As tests that are used for diagnosing pneumococcal pleural empyema or meningitis, we showed that GAS RADTs are sensitive and specific tools that may be particularly useful when patients have received previous antibiotic therapy that usually leads to negative cultures (29). Nonetheless, data in the literature lead to the implementation of these RADTs in POC laboratories rather than hospitalization wards.
Although they demonstrated strong performance, any RADTs can have a lower sensitivity than GAS PCR for the diagnosis of invasive infections. It may be relevant to evaluate the performance of new fully automated tests on the basis of loop-mediated isothermal amplification technology (illumigene; Meridian Bioscience) or fluorometry (mariPOC; ArcDia, Turku, Finland), which may be more sensitive than colorimetric RADTs and comply with quality requirements (30, 31).
ACKNOWLEDGMENTS
We thank Mieke Vermander (Meridian Bioscience), Orianne Goudou, Antoine Schell (Nal Von Minden), and Laurent Ville (Alere) for providing us the RADTs free of charge and Sébastien Bailly for statistical analysis. We also thank Richard Hughes (Meridian Bioscience) and Frederica Swaine (Nal Von Minden) for English editing.
REFERENCES
- 1.Lamagni TL, Darenberg J, Luca-Harari B, Siljander T, Efstratiou A, Henriques-Normark B, Vuopio-Varkila J, Bouvet A, Creti R, Ekelund K, Koliou M, Reinert RR, Stathi A, Strakova L, Ungureanu V, Schalen C, Strep ESG, Jasir A. 2008. Epidemiology of severe Streptococcus pyogenes disease in Europe. J Clin Microbiol 46:2359–2367. doi: 10.1128/JCM.00422-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamagni TL, Efstratiou A, Vuopio-Varkila J, Jasir A, Schalen C, Strep E. 2005. The epidemiology of severe Streptococcus pyogenes associated disease in Europe. Euro Surveill 10:179–184. [PubMed] [Google Scholar]
- 3.Maltezou HC, Tsagris V, Antoniadou A, Galani L, Douros C, Katsarolis I, Maragos A, Raftopoulos V, Biskini P, Kanellakopoulou K, Fretzayas A, Papadimitriou T, Nicolaidou P, Giamarellou H. 2008. Evaluation of a rapid antigen detection test in the diagnosis of streptococcal pharyngitis in children and its impact on antibiotic prescription. J Antimicrob Chemother 62:1407–1412. doi: 10.1093/jac/dkn376. [DOI] [PubMed] [Google Scholar]
- 4.Ayanruoh S, Waseem M, Quee F, Humphrey A, Reynolds T. 2009. Impact of rapid streptococcal test on antibiotic use in a pediatric emergency department. Pediatr Emerg Care 25:748–750. doi: 10.1097/PEC.0b013e3181bec88c. [DOI] [PubMed] [Google Scholar]
- 5.Llor C, Madurell J, Balague-Corbella M, Gomez M, Cots JM. 2011. Impact on antibiotic prescription of rapid antigen detection testing in acute pharyngitis in adults: a randomised clinical trial. Br J Gen Pract 61:e244–e251. doi: 10.3399/bjgp11X572436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hein I, Schneeweiss W, Stanek C, Wagner M. 2007. Ethidium monoazide and propidium monoazide for elimination of unspecific DNA background in quantitative universal real-time PCR. J Microbiol Methods 71:336–339. doi: 10.1016/j.mimet.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 7.Gauduchon V, Chalabreysse L, Etienne J, Celard M, Benito Y, Lepidi H, Thivolet-Bejui F, Vandenesch F. 2003. Molecular diagnosis of infective endocarditis by PCR amplification and direct sequencing of DNA from valve tissue. J Clin Microbiol 41:763–766. doi: 10.1128/JCM.41.2.763-766.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hung GC, Nagamine K, Li B, Lo SC. 2012. Identification of DNA signatures suitable for use in development of real-time PCR assays by whole-genome sequence approaches: use of Streptococcus pyogenes in a pilot study. J Clin Microbiol 50:2770–2773. doi: 10.1128/JCM.01155-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bidet P, Liguori S, Plainvert C, Bonacorsi S, Courroux C, d'Humieres C, Poyart C, Efstratiou A, Bingen E. 2012. Identification of group A streptococcal emm types commonly associated with invasive infections and antimicrobial resistance by the use of multiplex PCR and high-resolution melting analysis. Eur J Clin Microbiol Infect Dis 31:2817–2826. doi: 10.1007/s10096-012-1635-5. [DOI] [PubMed] [Google Scholar]
- 10.Johnson DR, Kaplan EL. 2001. False-positive rapid antigen detection test results: reduced specificity in the absence of group A streptococci in the upper respiratory tract. J Infect Dis 183:1135–1137. doi: 10.1086/319286. [DOI] [PubMed] [Google Scholar]
- 11.Rubin LG, Kahn RA, Vellozzi EM, Isenberg HD. 1996. False positive detection of group A Streptococcus antigen resulting from cross-reacting Streptococcus intermedius (Streptococcus milleri group). Pediatr Infect Dis J 15:715–717. doi: 10.1097/00006454-199608000-00021. [DOI] [PubMed] [Google Scholar]
- 12.Cohen JF, Cohen R, Bidet P, Levy C, Deberdt P, d'Humieres C, Liguori S, Corrard F, Thollot F, Mariani-Kurkdjian P, Chalumeau M, Bingen E. 2013. Rapid-antigen detection tests for group a streptococcal pharyngitis: revisiting false-positive results using polymerase chain reaction testing. J Pediatr 162:1282–1284, 1284.e1. doi: 10.1016/j.jpeds.2013.01.050. [DOI] [PubMed] [Google Scholar]
- 13.Cohen R, Levy C, Bonacorsi S, Wollner A, Koskas M, Jung C, Bechet S, Chalumeau M, Cohen J, Bidet P. 2015. Diagnostic accuracy of clinical symptoms and rapid diagnostic test in group a streptococcal perianal infections in children. Clin Infect Dis 60:267–270. doi: 10.1093/cid/ciu794. [DOI] [PubMed] [Google Scholar]
- 14.Wollner A, Levy C, Benani M, Thollot F, Bechet S, Cohen J, Bonacorsi S, Bidet P, Cohen R. 2014. Use of group A streptococcal rapid diagnostic test in extra-pharyngeal infections. Arch Pediatr 21(Suppl 2):S84–S86. doi: 10.1016/S0929-693X(14)72266-3. [DOI] [PubMed] [Google Scholar]
- 15.Clegg HW, Dallas SD, Roddey OF, Martin ES, Swetenburg RL, Koonce EW, Felkner MB, Ryan AG, Presbyterian Pediatric Research G . 2003. Extrapharyngeal group A Streptococcus infection: diagnostic accuracy and utility of rapid antigen testing. Pediatr Infect Dis J 22:726–731. doi: 10.1097/01.inf.0000078835.72497.ab. [DOI] [PubMed] [Google Scholar]
- 16.Kokx NP, Comstock JA, Facklam RR. 1987. Streptococcal perianal disease in children. Pediatrics 80:659–663. [PubMed] [Google Scholar]
- 17.Zheng X, O'Leary A, Uhl JR, Patel R, Shulman ST. 2012. Rapid detection of Streptococcus pyogenes in pleural fluid samples from pediatric patients with empyema. J Clin Microbiol 50:2786–2787. doi: 10.1128/JCM.00603-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plainvert C, Doloy A, Loubinoux J, Lepoutre A, Collobert G, Touak G, Trieu-Cuot P, Bouvet A, Poyart C, CNR-Strep Network . 2012. Invasive group A streptococcal infections in adults, France (2006-2010). Clin Microbiol Infect 18:702–710. doi: 10.1111/j.1469-0691.2011.03624.x. [DOI] [PubMed] [Google Scholar]
- 19.O'Loughlin RE, Roberson A, Cieslak PR, Lynfield R, Gershman K, Craig A, Albanese BA, Farley MM, Barrett NL, Spina NL, Beall B, Harrison LH, Reingold A, Van Beneden C, Active Bacterial Core Surveillance T. 2007. The epidemiology of invasive group A streptococcal infection and potential vaccine implications: United States, 2000-2004. Clin Infect Dis 45:853–862. doi: 10.1086/521264. [DOI] [PubMed] [Google Scholar]
- 20.O'Grady KA, Kelpie L, Andrews RM, Curtis N, Nolan TM, Selvaraj G, Passmore JW, Oppedisano F, Carnie JA, Carapetis JR. 2007. The epidemiology of invasive group A streptococcal disease in Victoria, Australia. Med J Aust 186:565–569. [DOI] [PubMed] [Google Scholar]
- 21.Plainvert C, Loubinoux J, Bidet P, Doloy A, Touak G, Dmytruk N, Collobert G, Bingen E, Bouvet A, Fouet A, Poyart C. 2014. Epidemiology of Streptococcus pyogenes invasive diseases in France (2007-2011). Arch Pediatr 21(Suppl 2):S62–S68. doi: 10.1016/S0929-693X(14)72262-6. [DOI] [PubMed] [Google Scholar]
- 22.Koutouzi F, Tsakris A, Chatzichristou P, Koutouzis E, Daikos GL, Kirikou E, Petropoulou N, Syriopoulou V, Michos A. 2015. Streptococcus pyogenes emm types and clusters during a 7-year period (2007 to 2013) in pharyngeal and nonpharyngeal pediatric isolates. J Clin Microbiol 53:2015–2021. doi: 10.1128/JCM.00301-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McIsaac WJ, Kellner JD, Aufricht P, Vanjaka A, Low DE. 2004. Empirical validation of guidelines for the management of pharyngitis in children and adults. JAMA 291:1587–1595. doi: 10.1001/jama.291.13.1587. [DOI] [PubMed] [Google Scholar]
- 24.Linner A, Darenberg J, Sjolin J, Henriques-Normark B, Norrby-Teglund A. 2014. Clinical efficacy of polyspecific intravenous immunoglobulin therapy in patients with streptococcal toxic shock syndrome: a comparative observational study. Clin Infect Dis 59:851–857. doi: 10.1093/cid/ciu449. [DOI] [PubMed] [Google Scholar]
- 25.Carapetis JR, Jacoby P, Carville K, Ang SJ, Curtis N, Andrews R. 2014. Effectiveness of clindamycin and intravenous immunoglobulin, and risk of disease in contacts, in invasive group a streptococcal infections. Clin Infect Dis 59:358–365. doi: 10.1093/cid/ciu304. [DOI] [PubMed] [Google Scholar]
- 26.Park M, Hue V, Dubos F, Lagree M, Pruvost I, Martinot A. 2013. Reasons for low usage of strep A rapid antigen detection tests for pharyngitis in private medical practice. Arch Pediatr 20:1083–1088. doi: 10.1016/j.arcped.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 27.Cohen JF, Chalumeau M, Levy C, Bidet P, Benani M, Koskas M, Bingen E, Cohen R. 2013. Effect of clinical spectrum, inoculum size and physician characteristics on sensitivity of a rapid antigen detection test for group A streptococcal pharyngitis. Eur J Clin Microbiol Infect Dis 32:787–793. doi: 10.1007/s10096-012-1809-1. [DOI] [PubMed] [Google Scholar]
- 28.Toepfner N, Henneke P, Berner R, Hufnagel M. 2013. Impact of technical training on rapid antigen detection tests (RADT) in group A streptococcal tonsillopharyngitis. Eur J Clin Microbiol Infect Dis 32:609–611. doi: 10.1007/s10096-012-1783-7. [DOI] [PubMed] [Google Scholar]
- 29.Picazo JJ, Contreras JR, Rios E, Culebras E, Rodriguez-Avial I, Mendez C, Betriu C, Heracles Study G. 2013. Rapid diagnosis of invasive pneumococcal disease in pediatric population. J Microbiol Methods 93:116–120. doi: 10.1016/j.mimet.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 30.Anderson NW, Buchan BW, Mayne D, Mortensen JE, Mackey TL, Ledeboer NA. 2013. Multicenter clinical evaluation of the illumigene group A Streptococcus DNA amplification assay for detection of group A Streptococcus from pharyngeal swabs. J Clin Microbiol 51:1474–1477. doi: 10.1128/JCM.00176-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vakkila J, Koskinen JO, Brandt A, Muotiala A, Liukko V, Soittu S, Meriluoto S, Vesalainen M, Huovinen P, Irjala K. 2015. Detection of Group A Streptococcus from pharyngeal swab samples by bacterial culture is challenged by a novel mariPOC point-of-care test. J Clin Microbiol 53:2079–2083. doi: 10.1128/JCM.00018-15. [DOI] [PMC free article] [PubMed] [Google Scholar]