Abstract
Early diagnosis of urinary tract infections (UTIs) is essential to avoid inadequate or unnecessary empirical antibiotic therapy. Microbiological confirmation takes 24 to 48 h. The use of screening methods, such as cytometry and automated microscopic analysis of urine sediment, allows the rapid prediction of negative samples. In addition, matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) is a widely established technique in clinical microbiology laboratories used to identify microorganisms. We evaluated the ability of MALDI-TOF MS to identify microorganisms from direct urine samples and the predictive value of automated analyzers for the identification of microorganisms in urine by MALDI-TOF MS. A total of 451 urine samples from patients with suspected UTIs were first analyzed using the Sysmex UF-1000i flow cytometer, an automatic sediment analyzer with microscopy (SediMax), culture, and then processed by MALDI-TOF MS with a simple triple-centrifuged procedure to obtain a pellet that was washed and centrifuged and finally applied directly to the MALDI-TOF MS plate. The organisms in 336 samples were correctly identified, mainly those with Gram-negative bacteria (86.10%). No microorganisms were misidentified, and no Candida spp. were correctly identified. Regarding the data from autoanalyzers, the best bacteriuria cutoffs were 1,000 and 200 U/μl for UF-1000i and SediMax, respectively. It was concluded that the combination of a urine screening method and MALDI-TOF MS provided a reliable identification from urine samples, especially in those containing Gram-negative bacteria.
INTRODUCTION
Urinary tract infections (UTIs) are among the most common nosocomial and community-acquired bacterial infections (1). The etiology is varied, but in approximately 90% of cases, enteric bacteria are implicated, especially Escherichia coli, which produces >70% of these infections. Other urinary tract pathogens are Klebsiella spp., Enterobacter spp., Proteus spp., Pseudomonas spp., Enterococcus spp., and Staphylococcus saprophyticus (2).
A rapid diagnosis of UTIs has a significant beneficial impact on patient health, since it reduces unnecessary or inadequate empirical antimicrobial therapy (3). Urine culture is still the gold standard for the microbiological confirmation of UTIs; however, it takes 24 to 48 h to provide results. The use of screening methods, such as traditional Gram staining or other methods for counting urine particles, like flow cytometry or automated microscopic urine sediment analysis, allow the prompt prediction of negative samples and a preliminary identification of microorganisms in positive samples (4–7). Nevertheless, this information obtained from these methods is insufficient and requires culture and other biochemical tests.
In recent years, proteomic techniques have achieved a relevant role in the identification of microorganisms in the field of clinical microbiology. Matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) has been suggested as a fast and reliable method for bacterial identification (8). This methodology has successfully been used for the rapid identification of microorganisms, both from culture plates and positive blood cultures, and also on other clinical samples, such as those from urine (9–14). A biomarker capable of predicting the ability of MALDI-TOF MS to identify microorganisms directly from clinical samples would be useful to improve the diagnostic procedure, reducing costs and time.
The objectives of our study were to (i) assess the ability of MALDI-TOF MS to identify microorganisms from direct urine samples, and (ii) evaluate the predictive value of cytometry and automated microscopic urine sediment analysis for the identification of microorganisms in urine by MALDI-TOF MS.
MATERIALS AND METHODS
Ethics statement.
Ethical approval from the research ethics committee of Germans Trias i Pujol University Hospital was obtained, and the need for informed consent was waived.
Settings.
Our institution is a university tertiary-care hospital that covers a population of >200,000 inhabitants and is the reference hospital of >900,000 inhabitants in the regions of North and Maresme Barcelonés (Barcelona, Spain). The clinical microbiology department at our hospital receives samples from different hospitals, primary care centers, and prisons, covering about 2,300,000 inhabitants.
Data collection.
The characteristics of patients whose samples were included in the study were analyzed by sex, age, department of origin, and urine collection technique.
Urine processing and conventional identification.
A total of 2,017 urine samples were processed during March to May 2015 from outpatients and inpatients with symptoms suggestive of UTIs. All samples were analyzed by the Sysmex UF-1000i flow cytometer (TOA Medical Electronics, Kobe, Japan), and a subgroup of 1,938 samples was also analyzed by the SediMax automatic sediment analyzer with microscopy (E77 Elektronika, Budapest, Hungary). On one hand, the Sysmex UF-1000i flow cytometer stains urine particles with a fluorescent dye, allowing their classification in leukocytes, red blood cells, epithelial cells, bacteria, and Candida spp. by impedance, scattering, and fluorescence. On the other hand, the SediMax automatic sediment analyzer homogenizes and transfers urine samples into special disposable cuvettes, which are centrifuged for a few seconds. Afterwards, whole-field high-definition images are obtained, and the inner software performs a morphological analysis of the particles, allowing their quantification and classification. All images are stored, which allows their review and further recounting if necessary. After that, 10 μl of urine was cultured onto chromogenic chromID CPS Elite agar plates (bioMérieux, Marcy-l'Étoile, France), and incubated under aerobic conditions at 37°C for 18 to 24 h; additionally, Gram staining was performed on those samples in which >40 leukocytes/μl were detected by cytometry, and these then were cultured in selective plates, according to microorganisms observed in the Gram stain (MacConkey agar [bioMérieux] for Gram-negative rods, colistin-nalidixic acid [CNA] agar [bioMérieux] for Gram-positive bacteria, Candida ID2 [CAN2] agar [bioMérieux] for yeast, and chocolate agar [bioMérieux] when no microorganism was observed). Identification from the colonies was done by MALDI-TOF MS, according to the manufacturer's procedures, and susceptibility testing was performed by disk diffusion method.
MALDI-TOF MS.
All samples with positive cultures, except those considered to be contaminated, were processed 24 h later, as follows: urine samples were vortexed, and 2 ml was centrifuged at 2,000 × g for 1 min to remove cellular debris, leukocytes, and mucus. The supernatant was centrifuged at 15,500 × g for 5 min and then discarded. Pellets were resuspended in 1 ml of sterile water and centrifuged again at 18,500 × g for 5 min. After discarding the supernatant, the pellet was spotted onto a polished steel MALDI-TOF MS target plate using a wooden toothpick and allowed to dry. Next, 1 μl of matrix solution (α-cyano-4-hydroxycinnamic acid solution in 50% acetonitrile and 2.5% trifluoroacetic acid) was added prior to the acquisition of spectra in a mass spectrometer. The samples tested the day after were stored at 4°C until the procedure was performed. Those urine samples for which the microorganism's identification using the direct transfer method was not possible were further tested according to the same procedure but samples were first covered with 1 μl of formic acid (70% [vol/vol]) and dried at room temperature before adding the matrix solution.
Protein analysis was carried out using the MALDI microflex LT mass spectrometer (Bruker Daltonik GmbH, Bremen, Germany) and FlexControl version 3.0 software (Bruker Daltonik GmbH), using the default settings.
When the results obtained by conventional microbiological methods and MALDI-TOF MS were inconsistent, both were repeated. According to the manufacturer, a score of ≥2.0 indicates species-level identification, a score between 1.7 and 1.99 indicates genus-level identification, and a score of <1.7 indicates no identification. However, we registered the first 10 identification options provided by the MALDI-TOF MS software, considering species-level correct identification to be those options that agreed with the urine culture.
The MALDI-TOF MS result was correlated with the culture result, and the results were classified into four groups: (i) insufficient pellet, when not enough pellet for MALDI-TOF MS analysis was obtained after urine processing; (ii) unidentified, when the software did not provide any microbiological identification; (iii) correctly identified, when identification by MALDI-TOF MS from colonies and direct samples were consistent (for mixed cultures, correct identification was considered if at least one of the two microorganisms isolated was correctly identified); and (iv) misidentified (MI), when identifications by both MALDI-TOF MS procedures were not consistent. MI results were verified by both methodologies.
Statistics.
Categorical variables are provided with their frequencies. Continuous variables were summarized as median and interquartile range (IQR) with the 95% confidence interval (95% CI). Chi-square test and the Kruskal-Wallis or Mann-Whitney test, as appropriate, were performed for qualitative and quantitative variables, respectively. The optimal cutoffs of bacteriuria provided by UF-1000i and SediMax for correct identification by MALDI-TOF MS were calculated using receiver operating characteristic (ROC) curves. Sensitivity (Se), specificity (Sp), and positive and negative predictive values (PPV and NPV, respectively) were calculated for these cutoffs. The data analysis was carried out using the Stata release 10.1 statistical package (StataCorp LP, TX, USA) and SPSS version 13.0 (SPSS, Inc., Chicago, IL) softwares.
RESULTS
In total, 451 out of 2,017 processed urine samples yielded positive cultures (22.34%) and were further tested by MALDI-TOF MS. The vast majority of these samples were clean-catch specimens (79.38%), followed by specimens collected by catheterization (19.96%), pediatric bags (0.44%), and suprapubic aspiration (0.22%). The median bacteriuria level estimated by UF-1000i was 4,242 U/μl (IQR, 1,326 to 10,957 U/μl). A subgroup of 379 urine samples was also analyzed by SediMax, with a median bacteriuria level of 614 U/μl (IQR, 142 to 1,401 U/μl).
Out of 451 urine cultures, 44 (9.76%) resulted in polymicrobial cultures. Gram-negative microorganisms were isolated in 81.36%, Gram-positive in 16.43%, and yeast in 2.00% of the cultures. The most commonly isolated microorganisms were E. coli (56.51%), Enterococcus faecalis (9.42%), Klebsiella pneumoniae (7.01%), and Pseudomonas aeruginosa (6.01%). Three hundred thirty-nine urine samples (75.17%) yielded colony counts of >105 CFU/ml, 74 (16.41%) samples had between 5 × 104 and 105 CFU/ml, and 38 (8.42%) samples had ≤104 CFU/ml.
After processing urine samples for MALDI-TOF MS, 29 (6.43%) samples had insufficient pellet and so were not studied further, 86 (19.07%) samples were unidentified, and 336 (74.50%) samples were correctly identified. No microorganisms were misidentified.
Among the insufficient-pellet samples, 69.70% and 30.30% of the samples were Gram-negative and Gram-positive bacteria, respectively (Table 1). Among the unidentified samples, 64.52% were Gram-negative bacteria, 24.73% were Gram-positive bacteria, and 10.75% were yeast, which corresponded to all yeasts included in the study (n = 10) (Table 1). Therefore, samples with a positive culture for yeast could not be identified by MALDI-TOF MS. The majority of the correctly identified samples (86.10%) were Gram-negative bacteria, while 13.90% were Gram-positive bacteria (Tables 2 and 3). The correctly identified urine samples (91.07%) showed a score of >1.7, with an average score of 2.112 (95% CI, 2.092 to 2.132). The remaining 8.93% presented a score of <1.7, with an average score of 1.519 (95% CI, 1.468 to 1.570).
TABLE 1.
Conventional identification and bacteriuria/candiduria levels of insufficient pellet and unidentified urine samples processed for MALDI-TOF MS analysis
| Conventional identification result by sample category (no. of isolates) | Bacteriuria (median) (bacteria/μl) |
Colony count (median) (CFU/ml) | |
|---|---|---|---|
| UF-1000i | SediMax | ||
| Insufficient pellet (n = 29) | |||
| Monomicrobial cultures | |||
| E. coli (15) | 283 | 88 | 1 × 104–5 × 104 |
| E. faecalis (6) | 295 | 87 | 1 × 104–5 × 104 |
| S. saprophyticus (2) | 606 | 102 | 1 × 104–5 × 104 |
| E. faecium (1) | 159 | NAa | 1 × 104–5 × 104 |
| P. aeruginosa (1) | 2,212 | 413 | ≥1 × 105 |
| Polymicrobial cultures | |||
| Enterobacter aerogenes + P. aeruginosa (1) | 932 | 108 | ≥1 × 105 |
| Enterobacter cloacae + E. coli (1) | 515 | 65 | 1 × 104–5 × 104 |
| E. coli + Proteus spp. (1) | 250 | 48 | 1 × 104–5 × 104 |
| P. aeruginosa + E. faecalis (1) | 828 | 123 | ≥1 × 105 |
| Unidentified (n = 86) | |||
| Monomicrobial cultures | |||
| E. coli (32) | 1,507 | 243 | 1 × 104–5 × 104 |
| Candida albicans (9) | 1,294b | 449.46b | ≥1 × 105 |
| S. saprophyticus (7) | 1,728 | 341 | ≥1 × 105 |
| E. faecalis (7) | 1,059 | 111 | ≥1 × 105 |
| P. aeruginosa (7) | 460 | 104 | 1 × 104–5 × 104 |
| Proteus mirabilis (6) | 2,057 | 109 | 1 × 104–5 × 104 |
| K. pneumoniae (3) | 11,259 | 3,925 | ≥1 × 105 |
| Candida spp. (1) | 542b | 249.04b | 1 × 104–5 × 104 |
| Citrobacter freundii (1) | 926 | 1,342 | ≥1 × 105 |
| E. cloacae (1) | 726 | NA | 1 × 103 |
| E. faecium (1) | 20,703 | 4,752 | ≥1 × 105 |
| Haemophilus influenzae (1) | 304 | 51 | ≥1 × 105 |
| Proteus vulgaris (1) | 96 | 17 | 1 × 103–1 × 104 |
| Staphylococcus aureus (1) | 536 | 614 | 1 × 104–5 × 104 |
| Staphylococcus spp. (1) | 2,212 | 413 | 1 × 104–5 × 104 |
| Polymicrobial cultures | |||
| Citrobacter koseri + Streptococcus agalactiae (1) | 7,831 | 1,456 | ≥1 × 105 |
| E. faecalis + E. coli (1) | 271 | 164 | 1 × 102–1 × 103 |
| E. faecalis + P. mirabilis (1) | 2,215 | 118 | ≥1 × 105 |
| E. faecalis + P. aeruginosa (1) | 297 | 180 | 1 × 103–1 × 104 |
| E. faecalis + S. aureus (1) | 1,069 | 162 | ≥1 × 105 |
| K. pneumoniae + Morganella morganii (1) | 738 | 32 | ≥1 × 105 |
| Pseudomonas mendocina + Stenotrophomonas maltophilia (1) | 603 | 87 | ≥1 × 105 |
NA, not available.
Median candiduria level (yeasts/μl).
TABLE 2.
MALDI-TOF MS identification and bacteriuria levels of 303 correctly identified urine samples with monomicrobial infections
| Identification by conventional methods and MALDI-TOF MS | No. of isolates | Bacteriuria (median) (bacteria/μl) |
|
|---|---|---|---|
| UF-1000i | SediMax | ||
| E. coli | 217 | 7,328 | 1,031 |
| K. pneumoniae | 23 | 7,098 | 1,168 |
| E. faecalis | 17 | 3,059 | 387 |
| P. mirabilis | 12 | 9,228 | 406 |
| P. aeruginosa | 8 | 2,858 | 248 |
| S. saprophyticus | 5 | 3,291 | 121 |
| E. cloacae | 4 | 4,913 | 722 |
| C. koseri | 3 | 1,319 | 1,118 |
| Providencia stuartii | 2 | 753 | 438 |
| Actinobaculum schaalii | 1 | 2,333 | 1,496 |
| C. freundii | 1 | 14,494 | NAa |
| E. aerogenes | 1 | 13,694 | NA |
| Enterobacter kobei | 1 | 6,268 | 192 |
| E. faecium | 1 | 3,890 | 1,037 |
| Klebsiella oxytoca | 1 | 10,723 | 1,651 |
| M. morganii | 1 | 1,752 | 2,190 |
| S. aureus | 1 | 13,708 | 5,474 |
| Staphylococcus spp. | 1 | 1,283 | 2,666 |
| S. agalactiae | 1 | 4,490 | 268 |
| Streptococcus gallolyticus subsp. pasteurianus | 1 | 11,983 | 1,651 |
| Streptococcus viridians | 1 | 664 | 59 |
NA, not available.
TABLE 3.
MALDI-TOF MS versus conventional identification and bacteriuria levels of 33 correctly identified urine samples with polymicrobial infections
| Identification by conventional methods | Identification by MALDI-TOF MS | No. of isolates | Bacteriuria (median) (bacteria/μl) |
|
|---|---|---|---|---|
| UF-1000i | SediMax | |||
| E. faecalis + P. aeruginosa | P. aeruginosa | 6 | 3,233 | 259 |
| E. faecalis + E. coli | E. faecalis | 2 | 2,261 | 81 |
| E. faecalis + E. coli | E. coli | 2 | 12,485 | 600 |
| Aerococcus urinae + P. stuartii | P. stuartii | 1 | 14,278 | NAa |
| C. freundii + K. pneumoniae | K. pneumoniae | 1 | 5,596 | 554 |
| Enterobacter asburiae + E. coli | E. coli | 1 | 1,870 | 152 |
| Enterobacter spp. + E. coli | E. coli | 1 | 29,763 | 2,640 |
| E. faecalis + E. coli + K. pneumoniae | K. pneumoniae | 1 | 4,629 | 4,906 |
| E. faecalis + E. coli + P. aeruginosa | E. coli | 1 | 6,871 | 27.72 |
| E. faecalis + K. pneumoniae + P. mirabilis | K. pneumoniae | 1 | 6,087 | NA |
| E. faecalis + P. mirabilis | P. mirabilis | 1 | 11,889 | 668 |
| E. faecalis + S. aureus | E. faecalis | 1 | 3,573 | 656 |
| E. faecium + M. morganii + P. stuartii | P. stuartii | 1 | 4,779 | 465 |
| Enterococcus spp. + E. coli + K. pneumoniae | K. pneumoniae | 1 | 7,579 | 257 |
| Enterococcus spp. + Staphylococcus spp. | E. faecalis | 1 | 4,242 | 333 |
| E. coli + K. oxytoca | E. coli | 1 | 21,257 | 1,912 |
| E. coli + K. pneumoniae | K. pneumoniae | 1 | 6,636 | 374 |
| E. coli + K. pneumoniae + P. aeruginosa | E. coli | 1 | 7,761 | NA |
| E. coli + K. pneumoniae + P. aeruginosa | K. pneumoniae | 1 | 17,754 | 98 |
| E. coli + K. pneumoniae + S. aureus | E. coli | 1 | 18,019 | 9,592 |
| E. coli + P. mirabilis | E. coli | 1 | 14,973 | 1,789 |
| E. coli + P. mirabilis | P. mirabilis | 1 | 9,802 | 497 |
| E. coli + S. agalactiae | E. coli | 1 | 1,791 | 216 |
| K. pneumoniae + P. mirabilis | P. mirabilis | 1 | 8,917 | NA |
| K. pneumoniae + P. aeruginosa | K. pneumoniae | 1 | 9,520 | 1,815 |
| P. stuartii + P. aeruginosa | P. aeruginosa | 1 | 6,098 | 2,107 |
NA, not available.
ROC curves were calculated for the bacteriuria parameter. The area under the curve values were 0.85 (95% CI, 0.80 to 0.90) for UF-1000i and 0.76 (95% CI, 0.70 to 0.82) for SediMax (Fig. 1). According to these results, the cutoffs for the correctly identified samples by MALDI-TOF MS were 1,000 U/μl for UF-1000i (Se, 92.23%; Sp, 60.90%; NPV, 72.92%; PPV, 87.32%), and 200 U/μl for SediMax (Se, 82.56%; Sp, 60.20%; NPV, 54.63%; PPV, 85.61%).
FIG 1.
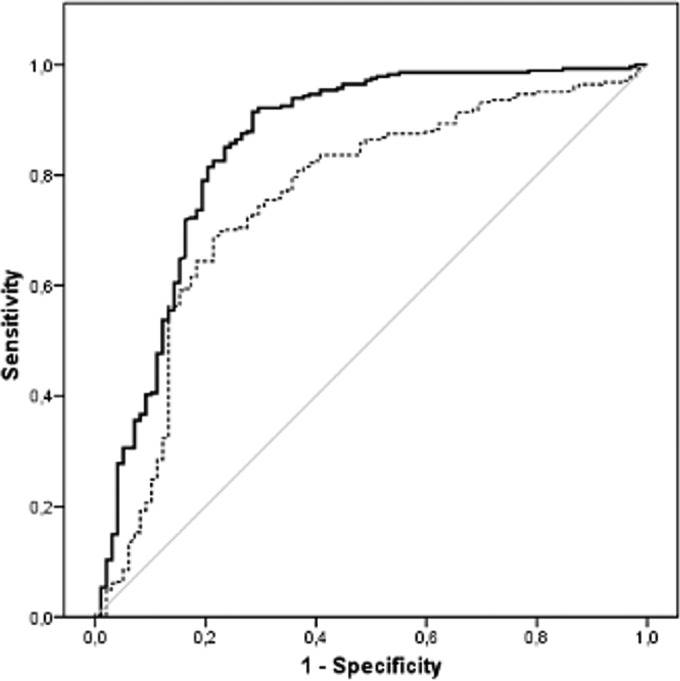
ROC curves of bacteriuria levels for UF-1000i and SediMax.
According to UF-1000i data, only 3 out of 29 insufficient-pellet samples (10.34%) presented a bacteriuria level of >1,000 U/μl (median bacteriuria level, 467 U/μl; IQR, 310 to 624 U/μl). Forty-two out of 86 unidentified samples presented a bacteriuria level of >1,000 U/μl, and 310 out of 336 correctly identified samples presented a bacteriuria level of >1,000 U/μl (Table 4). Out of these 336 correctly identified samples, 91.07% (n = 306) samples presented an identification score of >1.7 (average, 2.112; 95% CI, 2.092 to 2.132), while the remaining samples presented an identification score of <1.7 (average, 1.519; 95% CI, 1.468 to 1.570).
TABLE 4.
Bacteriuria levels and results of urine samples analyzed by MALDI-TOF MS
| Result of sample processing | No. (%) by bacteriuria level and instrument |
|||||
|---|---|---|---|---|---|---|
| UF-1000i |
SediMax |
|||||
| Total no. | >1,000 bacteria/μl | <1,000 bacteria/μl | Total no. | >200 bacteria/μl | <200 bacteria/μl | |
| Insufficient pellet | 29 | 3 (10.34) | 26 (89.66) | 25 | 2 (8) | 23 (92) |
| Unidentified | 86 | 42 (48.84) | 44 (51.16) | 73 | 37 (50.68) | 36 (49.32) |
| Correctly identified | 336 | 310 (92.26) | 26 (7.74) | 281 | 232 (82.56) | 49 (17.44) |
Urine samples processed also by SediMax presented percentages of insufficient-pellet, unidentified, and correctly identified samples similar to those of UF-1000i: 6.59%, 19.26%, and 74.14%, respectively. Out of these 25 insufficient-pellet samples, only 2 samples (8%) had a bacteriuria level of >200 U/μl. Thirty-seven out of 73 unidentified urine samples (50.68%) presented a bacteriuria level of >200 U/μl, and 232 out of 281 correctly identified urine samples (82.56%) presented a bacteriuria level of >200 U/μl (Table 4). The 90.39% of these urine samples that were correctly identified presented an identification score of >1.7 (average, 2.100; 95% CI, 2.088 to 2.132), and the remaining 9.61% of these samples presented an identification score of <1.7 (average, 1.522; 95% CI, 1.466 to 1.578).
In order to understand which kind of samples were correctly identified with higher frequency, several variables from the patients (sex, age, department of origin, and urine collection technique) were analyzed. Statistically significant differences for qualitative variables were analyzed by two different groups, “correct identification” versus “no correct identification” (unidentified plus insufficient pellet). Statistical significance was detected (P < 0.001) in those samples from women, primary-care patients, and clean-catch midstream urine samples.
DISCUSSION
In this study, we evaluated the ability of MALDI-TOF MS to detect microorganisms from direct urine samples in order to provide early diagnosis of UTIs; performing this methodology on the day of sample collection reduces the identification of the etiological agent by 18 to 24 h and thereby inadequate or unnecessary empirical antimicrobial treatment. The incorporation of this methodology in clinical laboratories has been an important improvement in the etiologic diagnosis of all types of infections, as its speed and relatively low cost have displaced the classical biochemical identification. Previous studies have evaluated the ability of MALDI-TOF MS to identify microorganisms in positive blood cultures (10, 15–19). The next step was the identification of microorganisms from direct samples, without prior culture. One limitation is the large amount of protein necessary to obtain reliable profiles. Therefore, as suggested by Wang et al. (11), samples need to be preselected depending on the bacterial load. Another useful algorithm for the rapid diagnosis of UTIs is presented by Burillo et al. (20), who combined Gram stain with mass spectrometry.
In accordance with previous studies, the identification of Gram-negative bacteria has provided better results than Gram-positive and yeast (9, 21). We did not identify any yeast from the urine direct samples. In a study recently published by Galán et al. (22), the utility of this technology for the identification of clinically interesting yeast is highlighted. The 94.80% of the yeasts isolated in several clinical samples, mainly vaginal swabs, urine samples, respiratory secretions, and blood cultures, were correctly identified. However, in all cases, the identification was done directly from colonies. Therefore, the identification of yeasts from direct samples is still unresolved.
In our study, we found an association between the bacteriuria levels provided by the two analyzers, UF-1000i and SediMax, and the correct identification of microorganisms by MALDI-TOF MS. The data of the bacteriuria levels provided by UF-1000i and SediMax can be used for the direct identification on the same day of the sample collection, while conventional culture needs a minimum of 24 to 48 h. For our population, the established cutoffs were 1,000 U/μl for UF-1000i and 200 U/μl for SediMax. Additionally, the 89.66% and the 92% of insufficient-pellet urine samples showed bacteriuria levels of <1,000 U/μl and <200 U/μl, as estimated by UF-1000i and SediMax, respectively. Therefore, these data might be a good biomarker to provide better results in the identification of microorganisms with MALDI-TOF MS from direct urine. However, these cutoffs should be established for each population and for each urine particle screening system.
The analysis of mixed cultures did not provided successful results. Wang et al. (11) analyzed urine specimens containing two microorganisms in different ratios (E. coli and P. aeruginosa, and E. coli and Enterococcus faecium); two types of bacteria were simultaneously detected in a mixture at a ratio 1:1 or 1:2, but only the dominant bacteria were detected in a mixture at a ratio of 1:9. However, almost half of the polymicrobial clinical samples provided invalid results (11). On the other hand, in our study, the analysis of 75% of the polymicrobial urine samples provided the correct identification of one microorganism. The unreliable identification of the remaining 25% (15.91% unidentified and 9.10% insufficient-pellet urine samples) might be related to the low bacteriuria levels detected in these samples. Therefore, the identification of several microorganisms directly from urine samples is established as a still-unresolved diagnostic challenge.
In order to reduce costs, we analyzed only those samples considered positive for a urinary tract infection, so MALDI-TOF MS testing was performed when culture results were known the following day. Therefore, a limitation of our study is that screening of urine particles and urine cultures was performed the day before MALDI-TOF MS analysis was done. All urine samples were treated equally and correctly stored at 4°C in a sterile correctly closed container, so we consider that these samples had not undergone a significant change in such a short period of time. However, to confirm this, further studies would be necessary to evaluate the results provided by MALDI-TOF MS in urine samples on two consecutive days.
In general, the identification of microorganisms with MALDI-TOF MS from direct urine samples is a process for which standardization of the procedure is necessary. Several procedures have been described (11, 13, 23). Veron et al. (24) compared different methods, concluding that previous short-culture and dual-filtration methods provided the best results. In previous studies, we compared three urine processing methods (data not shown), establishing 2 ml as the optimum volume, as it is easily manipulated in Eppendorf tubes and centrifuges available in clinical microbiology laboratories, and it also requires limited hands-on time. The use of larger volumes also entails the use of larger tubes, and centrifugation at higher revolutions may require special equipment not present in routine clinical laboratories.
Regarding the studied population, correct identifications were made in those samples from women, samples from primary-care center and emergency departments, and in clean-catch midstream urine samples, and statistical significance was found (P < 0.001). Although the results were good, we cannot establish that the rest of the samples were not suitable for work-up, because the study was not designed for this purpose. This point should be analyzed in detail, leading to the design of a study that includes a similar number of patients from different departments and a similar number of urine samples collected by different methods.
In conclusion, the combination of urine screening methods, such as flow cytometry or automated microscopic urine sediment analysis, and MALDI-TOF MS provided a reliable bacterial identification from urine samples, mainly of Gram-negative microorganisms. In the future, it will be necessary to optimize the methodology for the study of polymicrobial infections and the UTIs caused by Gram-positive bacteria or yeasts.
ACKNOWLEDGMENTS
This research did not receive specific grants from any funding agency in the public, commercial, or not-for-profit sectors. This study was partially funded by Menarini, S.A., distributor of SediMax in Spain. The funder had no role in the study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We declare no conflicts of interest.
REFERENCES
- 1.Flores-Mireles AL, Walker JN, Caparon M, Hultgren SJ. 2015. Urinary tract infections: epidemiology, mechanisms of infection and treatment options. Nat Rev Microbiol 13:269–284. doi: 10.1038/nrmicro3432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andreu A, Planells I, Grupo Cooperativo Español para el Estudio de la Sensibilidad Antimicrobiana de los Patógenos Urinarios. 2008. Etiology of community-acquired lower urinary infections and antimicrobial resistance of Escherichia coli: a national surveillance study. Med Clin (Barc) 130:481–486. (In Spanish.) doi: 10.1157/13119488. [DOI] [PubMed] [Google Scholar]
- 3.McIsaac WJ, Hunchak CL. 2011. Overestimation error and unnecessary antibiotic prescriptions for acute cystitis in adult women. Med Decis Making 31:405–411. doi: 10.1177/0272989X10391671. [DOI] [PubMed] [Google Scholar]
- 4.Lewis JF, Alexander J. 1976. Microscopy of stained urine smears to determine the need for quantitative culture. J Clin Microbiol 4:372–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manoni F, Fornasiero L, Ercolin M, Tinello A, Ferrian M, Hoffer P, Valverde S, Gessoni G. 2009. Cutoff values for bacteria and leukocytes for urine flow cytometer Sysmex UF-1000i in urinary tract infections. Diagn Microbiol Infect Dis 65:103–107. doi: 10.1016/j.diagmicrobio.2009.06.003. [DOI] [PubMed] [Google Scholar]
- 6.Karakukcu C, Kayman T, Ozturk A, Torun YA. 2012. Analytic performance of bacteriuria and leukocyturia obtained by UriSed in culture positive urinary tract infections. Clin Lab 58:107–111. [PubMed] [Google Scholar]
- 7.Sterry-Blunt RE, Randall SK, Doughton JM, Aliyu HS, Enoch AD. 2015. Screening urine samples for the absence of urinary tract infection using the SediMax automated microscopy analyser. J Med Microbiol 64:605–609. doi: 10.1099/jmm.0.000064. [DOI] [PubMed] [Google Scholar]
- 8.Seng P, Drancourt M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]
- 9.Ferreira L, Sánchez-Juanes F, González-Avila M, Cembrero-Fuciños D, Herrero-Hernández A, González-Buitrago JM, Muñoz-Bellido JL. 2010. Direct identification of urinary tract pathogens from urine samples by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 48:2110–2115. doi: 10.1128/JCM.02215-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stevenson LG, Drake SK, Murray PR. 2010. Rapid identification of bacteria in positive blood culture broths by matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 48:444–447. doi: 10.1128/JCM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang XH, Zhang G, Fan YY, Yang X, Sui WJ, Lu XX. 2013. Direct identification of bacteria causing urinary tract infections by combining matrix-assisted laser desorption ionization-time of flight mass spectrometry with UF-1000i urine flow cytometry. J Microbiol Methods 92:231–235. doi: 10.1016/j.mimet.2012.12.016. [DOI] [PubMed] [Google Scholar]
- 12.Barnini S, Ghelardi E, Brucculeri V, Morici P, Lupetti A. 2015. Rapid and reliable identification of Gram-negative bacteria and Gram-positive cocci by deposition of bacteria harvested from blood cultures onto the MALDI-TOF plate. BMC Microbiol 15:124. doi: 10.1186/s12866-015-0459-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.March Rosselló GA, Gutiérrez Rodríguez MP, Ortiz de Lejarazu Leonardo R, Orduña Domingo A, Bratos Pérez Ḿ A. 2014. New procedure for rapid identification of microorganisms causing urinary tract infection from urine samples by mass spectrometry (MALDI-TOF). Enferm Infecc Microbiol Clin 33:89–94. doi: 10.1016/j.eimc.2014.02.022. [DOI] [PubMed] [Google Scholar]
- 14.Demarco ML, Burnham CA. 2014. Diafiltration MALDI-TOF mass spectrometry method for culture-independent detection and identification of pathogens directly from urine specimens. 141:204–212. Am J Clin Pathol doi: 10.1309/AJCPQYW3B6JLKILC. [DOI] [PubMed] [Google Scholar]
- 15.Altun O, Botero-Kleiven S, Carlsson S, Ullberg M, Özenci V. 2015. Rapid identification of bacteria from positive blood culture bottles by MALDI-TOF MS following short-term incubation on solid media. J Med Microbiol 64:1346–1352. doi: 10.1099/jmm.0.000168. [DOI] [PubMed] [Google Scholar]
- 16.Ferreira L, Sánchez-Juanes F, Porras-Guerra I, García-García MI, García-Sánchez JE, González-Buitrago JM, Muñoz-Bellido JL. 2011. Microorganisms direct identification from blood culture by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Clin Microbiol Infect 17:546–551. doi: 10.1111/j.1469-0691.2010.03257.x. [DOI] [PubMed] [Google Scholar]
- 17.Schmidt V, Jarosch A, März P, Sander C, Vacata V, Kalka-Moll W. 2012. Rapid identification of bacteria in positive blood culture by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Eur J Clin Microbiol Infect Dis 31:311–317. doi: 10.1007/s10096-011-1312-0. [DOI] [PubMed] [Google Scholar]
- 18.Martinez RM, Bauerle ER, Fang FC, Butler-Wu SM. 2014. Evaluation of three rapid diagnostic methods for direct identification of microorganisms in positive blood cultures. J Clin Microbiol 52:2521–2529. doi: 10.1128/JCM.00529-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mestas J, Felsenstein S, Bard JD. 2014. Direct identification of bacteria from positive BacT/ALERT blood culture bottles using matrix-assisted laser desorption ionization-time-of-flight mass spectrometry. Diagn Microbiol Infect Dis 80:193–196. doi: 10.1016/j.diagmicrobio.2014.07.008. [DOI] [PubMed] [Google Scholar]
- 20.Burillo A, Rodríguez-Sánchez B, Ramiro A, Cercenado E, Rodríguez-Créixems M, Bouza E. 2014. Gram-stain plus MALDI-TOF MS (matrix-assisted laser desorption ionization-time of flight mass spectrometry) for a rapid diagnosis of urinary tract infection. PLoS One 9:e86915. doi: 10.1371/journal.pone.0086915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rodríguez-Sánchez B, Sánchez-Carrillo C, Ruiz A, Marín M, Cercenado E, Rodríguez-Créixems M, Bouza E. 2014. Direct identification of pathogens from positive blood cultures using matrix-assisted laser desorption-ionization time-of-flight mass spectrometry. Clin Microbiol Infect 20:O421–O427. doi: 10.1111/1469-0691.12455. [DOI] [PubMed] [Google Scholar]
- 22.Galán F, García-Agudo L, Guerrero I, Marín P, García-Tapia A, García-Martos P, Rodríguez-Iglesias M. 2015. Evaluation of mass spectrometry for the identification of clinically interesting yeasts. Enferm Infecc Microbiol Clin 33:372–378. (In Spanish.) doi: 10.1016/j.eimc.2014.10.003. [DOI] [PubMed] [Google Scholar]
- 23.Sánchez-Juanes F, Siller Ruiz M, Moreno Obregón F, Criado González M, Hernández Egido S, de Frutos Serna M, González-Buitrago JM, Muñoz-Bellido JL. 2014. Pretreatment of urine samples with SDS improves direct identification of urinary tract pathogens with matrix-assisted laser desorption ionization–time of flight mass spectrometry. J Clin Microbiol 52:335–338. doi: 10.1128/JCM.01881-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Veron L, Mailler S, Girard V, Muller BH, L'Hostis G, Ducruix C, Lesenne A, Richez A, Rostaing H, Lanet V, Ghirardi S, van Belkum A, Mallard F. 2015. Rapid urine preparation prior to identification of uropathogens by MALDI-TOF MS. Eur J Clin Microbiol Infect Dis 34:1787–1795. doi: 10.1007/s10096-015-2413-y. [DOI] [PubMed] [Google Scholar]


