Abstract
The IncP-1ε subgroup is a recently identified phylogenetic clade within IncP-1 plasmids, which plays an important role in the spread of antibiotic resistance and degradation of xenobiotic pollutants. Here, four IncP-1ε plasmids were exogenously captured from a petroleum-contaminated habitat in China and compared phylogenetically and genomically with previously reported IncP-1ε and other IncP-1 plasmids. The IncP-1ε plasmids can be clearly subdivided into two subclades, designated as ε-I and ε-II, based on phylogenetic analysis of backbone proteins TraI and TrfA. This was further supported by comparison of concatenated backbone genes. Moreover, the two subclades differed in the transposon types, phenotypes and insertion locations of the accessory elements. The accessory genes on ε-I plasmids were inserted between parA and traC, and harbored ISPa17 and Tn402-like transposon modules, typically carrying antibiotic resistance genes. In contrast, the accessory elements on ε-II plasmids were typically located between trfA and oriV, and contained IS1071, which was commonly inserted within the Tn501-like transposon, typically harboring a cluster of genes encoding mercury resistance and/or catabolic pathways. Our study is one of the first to compare IncP-1 plasmid genomes from China, expands the available collection of IncP-1ε plasmids and enhances our understanding of their diversity, biogeography and evolutionary history.
Keywords: IncP-1ε plasmid, comparative genome analysis, antibiotic resistance, horizontal gene transfer, petroleum polluted soil, wastewater
The present work is the first to isolate and describe IncP-1ε plasmids in China, greatly expands their available collection, and proposes the striking two phylogenetic subclades within IncP-1ε group.
Graphical Abstract Figure.
The present work is the first to isolate and describe IncP-1ε plasmids in China, greatly expands their available collection, and proposes the striking two phylogenetic subclades within IncP-1ε group.
INTRODUCTION
Horizontal gene transfer (HGT) between distantly related organisms plays a crucial role in prokaryotic evolution and adaptation (Davison 1999). Conjugative gene transfer mediated by plasmids with broad host ranges (BHR) is generally considered as one of the most important contributors to HGT (Thomas 2000). BHR self-transferable plasmids refer to the plasmids that can be introduced and stably maintained in the bacterial species from at least two different classes of Proteobacteria (Szpirer et al. 1999). They can be typically composed of two regions (Thomas 2000; Schlüter et al. 2003). The ‘plasmid backbone’ encodes genes involved in the replication, maintenance, control and transfer of the plasmid (Thorsted et al. 1998). The second region is comprised of various ‘accessory’ genes that confer specific phenotypic characteristics to the host organism, such as resistance to antibiotics, metabolic functions (e.g. those required for the degradation of various man-made pollutants) and virulence determinants (Rhodes et al. 2004; Schlüter et al. 2007, 2008).
With the emergence and development of next-generation sequencing methods, more complete BHR plasmid sequences have been added to the publically available databases (Sen et al. 2011; Brown et al. 2013; Fernández-López et al. 2014), which has greatly increased our understanding of their phylogenetic diversity. BHR plasmids have been shown to belong to the incompatibility groups IncP (Thorsted et al. 1998), IncW (Fernández-López et al. 2006), IncU (Rhodes et al. 2004), IncQ (Dehio and Meyer 1997) and the recently defined PromA group (Van der Auwera et al. 2009). Among these, the IncP plasmids are considered as one of the most promiscuous and best studied plasmid groups (Heuer and Smalla 2012). Since IncP is equivalent to IncP-1 in the Pseudomonas classification system, we will refer to these plasmids as IncP-1 from here on (Jacoby 1977). Early characterization of IncP-1 plasmids demonstrated two subgroups, designated as IncP-1α and IncP-1β, with RK2 and R751 as the representative archetypes, respectively (Macartney et al. 1997; Thomas 2000). With the increasing number of completely sequenced IncP-1 plasmids, seven evolutionary clades have been described for IncP-1 plasmids to date, based on backbone gene sequence phylogenies: the IncP-1-α, -β, -γ, -δ, -ε, -ζ and -η subgroups (Thorsted et al. 1998; Tennstedt et al. 2005; Norberg et al. 2011; Sen et al. 2011; Brown et al. 2013). This shows a much higher heterogeneity in the plasmid backbones within the IncP-1 plasmids than previously expected.
IncP-1ε plasmids have been firstly proposed by Bahl et al. (2007), as they differed clearly in their backbone from typical IncP-1α and IncP-1β plasmids. This subgroup was represented by plasmid pKJK5 (Bahl et al. 2007) and only contained two members when they were first recommended. To date, at least 13 IncP-1ε plasmids have been completely sequenced (Kim et al. 2013; Oliveira et al. 2013), most of which are regarded as important vectors for antibiotic resistance (Heuer et al. 2012). For example, plasmids pKJK5, pMLUA1, pMLUA3, pMLUA4, pKS77, pHH3414 and pHH128 all carry the class 1 integron and encode resistance towards tetracycline (Bahl et al. 2007; Sen et al. 2011; Oliveira et al. 2013), and the first four plasmids also code for multiple antibiotic resistances. Moreover, plasmids pAKD16, pAKD25 and pAKD34 encoded mercury resistance (Sen et al. 2011). Four IncP-1ε plasmids carried catabolic pathway genes responsible for herbicide degradation, with plasmids pAKD25, p712 and pEMT3 being associated with 2,4-dichlorophenoxyacetic acid degradation, and pAKD34 carrying putative 2,4-dichlorophenoxypropionic acid-degrading genes (Top, Holben and Forney 1995; Sen et al. 2011; Kim et al. 2013). Antibiotic resistance gene clusters, heavy metal resistance determinants and catabolic gene clusters on plasmids IncP-1ε have frequently been found to be associated with mobile genetic elements, e.g. insertion sequences (IS elements) and integrons (Heuer et al. 2012; Oliveira et al. 2013). In view of the aspects described above, the IncP-1ε plasmids play important roles in dissemination of antibiotic resistance and catabolic pathways. Nevertheless, the evolutionary information of plasmid backbones and accessory regions (such as transposons, integrons and IS elements) within this subgroup is still limited. The phylogenetic information of backbone genes provides fundamental information on the ‘long-term’ evolutionary history of BHR plasmids. Analysis of the backbone of the IncP-1ε plasmids may reveal important information on how these plasmids evolve and adapt to their hosts (Norberg et al. 2011), and it will help us to better understand their genetic diversity. Further analysis on accessory elements will improve our understanding of how the BHR plasmids allow bacteria to adapt to environmental selection pressures and thus generate functional diversity among bacterial strains (Norman, Hansen and Sørensen 2009).
In this study, four novel IncP-1ε plasmids were isolated by the triparental mating method from the Shen-fu irrigation zone, the largest petroleum wastewater irrigation zone in northeast China. The partial genome sequences of these plasmids were determined by Illumina sequencing, and the backbone regions were completely assembled. The TraI and TrfA proteins, functional modules, and organization of the backbone regions and accessory regions were compared between the newly isolated plasmids and previously reported IncP-1ε plasmids. The present study aims to deepen our knowledge of the evolutionary history of IncP-1ε plasmids, and their roles in dissemination of antibiotic resistance and pollutant degradation in the environment, especially in contaminated habitats.
MATERIALS AND METHODS
Site description and sampling
The sampling area was located in the Shen-fu wastewater irrigation zone (41°44′∼41°52′N; 123°35′∼123°45′E), Shenyang, northeast China. After long-term exposure to petroleum-containing wastewater that mainly came from an oil refinery, soils in this area are contaminated to varying degrees (Li et al. 2005; Zhou et al. 2012). Fourteen sampling sites were selected to collect soil samples from upstream to downstream of the irrigation channel, and were sequentially numbered S1 to S14 (Fig. S1, Supporting Information). Five soil cores were selected from each site and mixed to form an independent sample, and each soil core was collected with a 5-cm diameter PVC core to a depth of 20 cm.
Five wastewater samples and five sediment samples, numbered W1 to W5 and A1 to A5, respectively, were collected near the corresponding sites where soil sample S1, S4, S8, S11 and S14 were collected. The water samples were collected 25–40 cm below the surface, using the organic glass hydrophore as described by Song et al. (2007), and were stored at 4°C before being transported to laboratory. Water samples were filtered by sterilized medium-speed filter paper before use to remove suspended solid particles from water. The sediment samples (0–10 cm) were collected with a shovel as described by Cook et al. (2001).
All the soils and sediments were placed in plastic bags, and then were transported to the laboratory on ice. The soil samples were sieved (2 mm) and kept at 4°C for triparental exogenous plasmid isolation and the analysis of physicochemical properties. The fresh sediment samples were slightly air-dried and subsequently treated like soil. The principle chemical and physical characteristics of the environmental samples are determined by regular method (Table 1, only the samples from which IncP-1 plasmids were successfully isolated are shown). In brief, the total petroleum hydrocarbon (TPH) was determined by gravimetric method (Li et al. 2005). Soil pH was measured on soil slurry at 1:5 soil: water (w/v) ratio using a glass electrode. Fresh soil (10 g) was dried for 24 h at 110°C for determining soil moisture. SOC was determined with a TOC analyzer (Analytikjena HT1300, Germany) after removing soil carbonates with 1 M HCl. TN was determined using elemental analyzer (2400II CHN elemental analyzer; Perkin-Elmer, USA). Nitrate-N (NO3−-N) and ammonium-N (NH4+-N) were extracted with 2 M KCl, and then were analyzed on a continuous-flow ion auto-analyzer (Scalar SANplus segmented flow analyzer, the Netherlands).
Table 1.
Basic physical and chemical properties of the collected samples.
| Sample | TPHa | Moisture content | Organic matter | Total nitrogen | Available nitrogen | |||
|---|---|---|---|---|---|---|---|---|
| No. | Origin | Plasmids | (mg kg−1) | pHb | (%) | (g kg−1) | (g kg−1) | (mg kg−1) |
| S1 | Soil | pSFS12 | 713.3 | 6.7 | 20.7 | 39.0 | 1.1 | 202.5 |
| S2 | Soil | pSFS26 | 700.0 | 6.4 | 19.0 | 44.7 | 2.3 | 543.2 |
| S5 | Soil | pSFS52 | 1053.3 | 6.8 | 19.2 | 60.1 | 1.5 | 94.7 |
| W5 | Wastewater | pSFW53 | 18.32 | 6.3 | – | 13.8 | 9.2 | 30.6 |
TPH, total petroleum hydrocarbons.
pH (soil: water = 1:5).
Exogenous plasmid isolation
Plasmids were exogenously captured from soil, sediment and water samples by using the triparental mating method (Hill, Weightman and Fry 1992). In this method, the bacterial community of the samples are mixed with a plasmid-free recipient and a donor strain carrying a nonconjugative, mobilizable plasmid and incubated to allow mobilization to take place. Subsequently recipients that acquired the trait encoded on the mobilizable plasmids are selected for by plating. Thus, plasmids are captured based on their ability to mobilizable this nonconjugative plasmid and transfer themselves, and not on any phenotypic marker.
For the soil and air-dried sediment samples, plasmids were captured essentially as described previously (Li et al. 2015). Escherichia coli MG1685 (K12Rif) (Fox et al. 2008) and E. coli JM109 (pBBR1MCS-5) (Yanischperron, Vieira and Messing 1985) were used as recipient and donor, respectively. Each water sample was filtered to remove solid particles and phytoplankton, and 500 mL of the filtrate was centrifuged at 6000 × g at 4°C for 20 min. After centrifugation, the supernatant was discarded, and 3 mL of Luria Bertani (LB) liquid medium was added to the collected cell pellets. For the positive control, 500 mL of filtered water was mixed with 200 μL of a 10−1 dilution (in saline) of a fully grown E. coli DH5α (pB10) culture (about 107 cfu g−1 soil). To ensure operational consistency, 200 μL of saline was added to other samples. Individually, 500 μL of donor, recipient and water sample supernatant were dispensed into 1.5 mL Eppendorf tubes as the control for mating.
The presence of captured plasmids was confirmed by plasmid extraction using the alkaline lysis method, followed by gel electrophoresis. Restriction fragment length polymorphisms of captured plasmids were determined by restriction digestion with one or more restriction enzymes, and restriction fragments were analyzed on a 0.8% agarose gel essentially as described previously (Sambrook, Fritsch and Maniatis 1989). Comparison of the REP-PCR patterns (van Elsas et al. 1998) of the different transconjugants to that of recipient E. coli MG1685 was used to confirm that they were authentic E. coli MG1685 transconjugants.
Detection of antibiotic resistance of plasmids
The ability of the captured plasmids to confer antibiotic resistance on their host was tested by measuring their inhibition zones, as previously described (Li et al. 2015). The types and content of antibiotics per disc were the following: kanamycin (30 μg per disc), chloramphenicol (30), sulfamethoxazole (5), ceftazidime (30), polymyxin B (300), miramycin (100), tetracycline (30), Ciprofloxacin (5), erythromycin (15), amoxicillin (30), rifampicin (5), macrodantin (300), nalidixic acid (30), imipenem (10), gentamicin (10) and carbenicillin (100).
Tagging plasmids with the mini-Tn5 transposon
To facilitate selection of the plasmid in further analysis, the plasmids showing no resistance to any tested antibiotics were tagged with a miniTn5::Km1 transposon (De Lorenzo et al. 1990) using the biparental mating/mobilization strategy (Li et al. 2015). Plasmids carrying specific antibiotic resistance were directly transferred to E. coli EC100 (SmR), skipping the tagging step.
Host range test
To determine the host range of captured plasmids, the plasmids were transferred from E. coli EC100 (γ-Proteobacteria, SmR) to Agrobacterium tumefaciens C58 (α-Proteobacteria, RifR) (Wood et al. 2001) and Cupriavidus necator JMP228 (β-Proteobacteria, RifR) (Top, Holben and Forney 1995) by biparental matings as described previously (Binh et al. 2008).
Sequencing and annotation
Plasmid DNA was extracted from E. coli EC100 using the QIAGEN Plasmid Midi Kit (QIAGEN GmbH, Germany). The obtained plasmid DNA was sequenced by the Illumina Hiseq 2000 sequencing platform, using protocols described by Li et al. (2015). Primary sequence assembly and gene prediction were performed using the SOAPdenovo and GapCloser software as previously described (Li et al. 2015). Small gaps on the backbone region of plasmid pSFS26 were closed by general PCR method. The backbone regions of the other three newly isolated plasmids were fully assembled. Nevertheless, we were not able to close the entire plasmids due to HiSeq giving too short reads. The total genome size of the four plasmids was estimated by using restriction digestion with enzymes EcoRI, BamHI and PstI. The size of the gap was assessed by subtracting the confirmed bases from the estimated total genome size. The backbone genes of four plasmids (pSFS12, pSFS26, pSFS52 and pSFW53) were deposited in GenBank under accession numbers KM655308 to KM655311.
Bioinformatic analysis and software
Comparative genomic analysis was conducted between the newly isolated plasmids and 38 selected IncP-1 plasmids (Table S1, Supporting Information), including 13 previously reported IncP-1ε members. Amino acid sequences translated from the traI and trfA genes were aligned using ClustalX (Larkin et al. 2007), then the phylogenetic network was inferred using the SplitsTree program (Huson and Bryant 2006). The entire backbone regions and individual functional modules were also compared. The phylogenetic tree was inferred using the neighbor joining algorithm with aligned DNA sequences, and the best fit model for nucleotide substitution was selected using the ‘Find best DNA/Protein Models’ function in MEGA 6 (Tamura et al. 2013). A bootstrap analysis was performed to assess the reliability of the branching pattern. Schematic diagrams of multiple alignments of plasmids were performed by manually realigning the linear plasmid maps drawn using the software SnapGene Viewer (http://www.snapgene.com/products/snapgene_viewer/). Identity scores of pairwise comparisons (amino acid sequences and DNA sequences) were calculated by the BLAST algorithm bl2seq (Tatusova and Madden 1999). The insertion sequences from the DNA sequences of scaffolds including the accessory regions were searched using the ISfinder database (https://www-is.biotoul.fr) (Siguier et al. 2006), supplemented with the DNA sequences alignment by the BLAST algorithm bl2seq (Tatusova and Madden 1999). Integrons in the genomes of the four plasmids were searched using the INTEGRALL database (http://integrall.bio.ua.pt) (Moura et al. 2009).
RESULTS
Isolation and characterization of BHR plasmids from petroleum-contaminated habitats
A diverse set of plasmids with unique restriction profiles were captured from the Shen-fu petroleum wastewater irrigation zone. The draft genomes of seven representative plasmids were obtained by Illumina Hiseq 2000 high-throughput sequencing platform. Based on the plasmid backbone information, four of the seven sequenced plasmids were phylogenetically associated with IncP-1 plasmids, one plasmid was characterized as a PromA plasmid (Li et al. 2015), and two plasmids could not yet be grouped into any known incompatibility group (unpublished data). In the present study, we describe and discuss only the four IncP-1 plasmids. These plasmids were isolated from soil samples S1, S2, S5 and water sample W5, and were designated pSFS12, pSFS26, pSFS52 and pSFW53, respectively (Table 1). A plasmid with identical restriction fragment lengths as pSFS52 was also isolated from sediment sample A3, suggesting that this plasmid has a widespread distribution within the irrigation zone.
The four plasmids were tested for antibiotic resistance in their captured host using the paper disc method. Only pSFS26 conferred resistance to multiple antibiotic compounds, including chloramphenicol, spectinomycin and tetracycline. The other three newly isolated plasmids (pSFS12, pSFS52 and pSFW53) showed no resistance to any of the antibiotics tested (Table 3). To facilitate selection for the plasmids in further analyses, pSFS12, pSFS52 and pSFW53 were marked with a mini-Tn5 transposon encoding kanamycin resistance. All four plasmids were then separated from the mobilizable plasmid pBBR1MCS-5 by transferring them to E. coli EC100 (SmR).
Table 3.
Transposon/IS types and phenotypes of accessory elements within IncP-1ε plasmidsa.
| Plasmids | Origin | Antibiotic resistanceb | Mercury resistance | Catabolic pathwayc | Transposon types | References | |
|---|---|---|---|---|---|---|---|
| pSFS26 | Arable soil (irrigated using wastewater), China | CmR, SptR, TcR | − | − | ISPa17, IS26, Tn402-like | This study | |
| pSFS52 | Arable soil (irrigated using wastewater), China | –d | − | − | ISPa17, Tn402-like | This study | |
| pKJK5 | Manured soil in Denmark | TcR, TmR, SptR | − | − | ISPa17, IS1326, Tn402-like | Bahl et al. (2007); Oliveira et al. (2013) | |
| pHH128 | Manured soil, Germany | TcR | − | − | ISPa17, IS1326, Tn402-like | Oliveira et al. (2013) | |
| ε-I | pHH3408 | Manured soil, Germany | − | − | − | ISPa17, IS1326, Tn402-like | Oliveira et al. (2013) |
| pHH3414 | Manured soil, Germany | TcR | − | − | ISPa17, IS1326, Tn402-like | Oliveira et al. (2013) | |
| pKS77 | Manured soil, Germany | TcR | − | − | ISPa17, IS1326, Tn402-like | Oliveira et al. (2013) | |
| pMLUA1 | Estuarine water, Portugal | TcR, SmzR, SptR, SmR | − | − | ISPa17, IS1326, IS26, Tn402-like | Oliveira et al. (2013) | |
| pMLUA3 | Estuarine water, Portugal | TcR, EryR, SmzR, SptR, SmR | − | − | ISPa17, ISUnCu17, IS26, Tn402-like | Oliveira et al. (2013) | |
| pMLUA4 | Estuarine water, Portugal | TcR, EryR, SmzR | − | − | ISPa17, IS26, Tn402-like | Oliveira et al. (2013) | |
| pSFS12 | Arable soil (irrigated using wastewater), China | − | − | − | IS1071 | This study | |
| pSFW53 | Wastewater, China | − | − | − | IS1071 | This study | |
| pAKD25 | Arable soil (treated with HgCl2), Norway | − | + | 2, 4-D | IS1071, ISPps1, ISCsp2, Tn501-like | Sen et al. (2011) | |
| ε-II | pAKD34 | Arable soil (treated with HgCl2), Norway | − | + | 2, 4-DP | IS1071, ISPps1, ISCsp2, Tn501-like | Sen et al. (2011) |
| pEMT3 | Arable soil, USA | − | − | 2, 4-D | IS1071 | Top, Holben and Forney (1995) | |
| P712 | Arable soil, USA | − | − | 2, 4-D | IS1071 | Kim et al. (2013) | |
| pAKD16e | Arable soil (treated with HgCl2), Norway | − | + | − | IS1071, ISPps1, ISCsp2, Tn501-like, Tn6048-like | Sen et al. (2011) |
The insertion sites of accessory elements on ε-I plasmids are between parA and traC. The insertion sites of accessory elements on ε-II plasmids are between trfA and oriV.
Cm, chloramphenicol; Spt, spectinomycin; Tc, tetracycline; Gm, gentamicin; Sm, streptomycin; Tm, trimethoprim; Ery, erythromycin; Smz, sulfamethoxazole.
Putative and functional pathways; 2, 4- D: 2, 4- dichlorophenoxyacetic acid; 2, 4- DP: 2, 4- dichlorophenoxypropionic acid.
A kanamycin resistance gene was found on pSFS12, pSFS52 and pSFW53 but it was part of the minitransposon that was artificially inserted during marking of the plasmids. Therefore, it is not listed among the antibiotic resistances in column 3.
Plasmid pAKD16 is the only plasmid from clade II that has two accessory regions, but the natural mercury resistance operon was located in the trfA region as all the other plasmids. The transposon in the parA locus seemed identical to Tn6048 in the genome of the host that was used to capture this plasmid, and thus likely represents a transposition event that occurred in the laboratory.
To determine the host range of the four plasmids, we tested their ability to transfer from the γ-Proteobacterium E. coli and replicate in A. tumefaciens C58 and C. necator JMP228, members of the α- and β-Proteobacteria, respectively. These host range tests indicated that the four plasmids were self-transferrable and could replicate in the tested strains of α-, β- and γ-Proteobacteria.
Genetic organization of four novel plasmids revealed a typical IncP-1 plasmid backbone
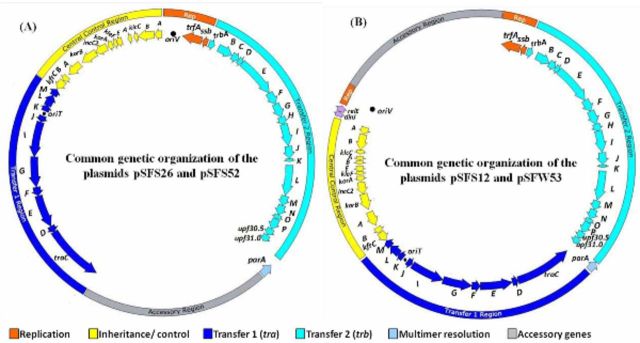
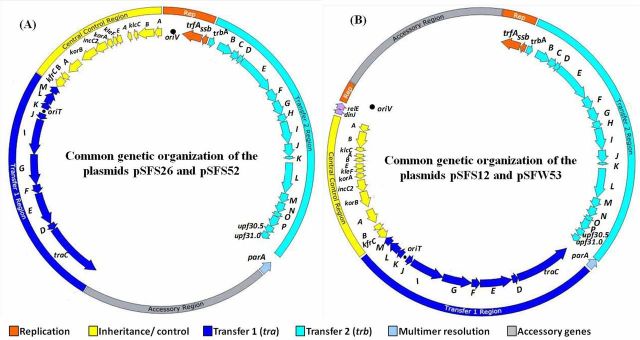
The total genome size of the four presumed IncP-1 plasmids were about 54, 51, 56 and 53 kb for pSFS12, pSFW53, pSFS26 and pSFS52, respectively, estimated by restriction enzyme digestion. The draft genome sequences of the four novel plasmids revealed the typical genetic organization that was unique to IncP-1 plasmids: regions for conjugative DNA transfer (here referred to as ‘Transfer 1’), mating-pair formation (‘Transfer 2’), the origin of conjugative transfer (oriT), the central control region (ctl), the replication genes trfA and ssb, and the origin of vegetative replication (oriV). The gene parA, encoding multimer resolution, was found in between the two transfer regions. The putative common backbone structure of these four novel IncP-1ε plasmids was illustrated in Fig. 1, which clearly showed that the genetic organization of plasmids pSFS26 and pSFS52 was different from that of plasmids pSFS12 and pSFW53. The fully assembled backbone regions of the four newly isolated plasmids were approximately 40 kb (Table 2). The Transfer 1 regions of the four plasmids all had 10 orfs. The Transfer 2 regions of plasmids pSFS12, pSFS26 and pSFS52 all contained 18 orfs, while that of the pSFW53 had 19 orfs. The ctl regions of pSFS12 and pSFW53 both had 13 orfs, while the corresponding regions of pSFS26 and pSFS52 (kleB absent) included 14 and 12 orfs, respectively (Table 2). Two genes (dinJ, relE/stbE) coding for a toxin–antitoxin system (Gotfredsen and Gerdes 1998) were found in between klcA and oriV of plasmids pSFS12 and pSFW53 (Fig. 1B). They could also be considered as backbone genes because of their function and their presence on some other IncP-1ε plasmids (e.g. pAKD16, pAKD25, pAKD34, pEMT3, p712).
Figure 1.
Common genetic organization of four novel IncP-1ε plasmids. (A) Plasmids pSFS26 and pSFS52; (B) plasmids pSFS12 and pSFW53. The insertion sites of accessory elements in plasmids pSFS26 and pSFS52 are between parA and traC, while in pSFS12 and pSFW53 the insertion site was between trfA and klcA. Orfs are shown by arrows indicating direction of transcription. Different colors indicate replication (orange) and conjugative DNA transfer (dark blue), mating-pair formation (light blue) and central control (yellow). Hypothetical coding regions are shown in gray. The positions of the origins of vegetative (oriV) and transfer (oriT) are marked by black circles. The genes shared by the four IncP-1 plasmids are in the inner perimeter.
Table 2.
Structural organization of the four IncP-1ε plasmids isolated in this study.
| Total plasmid | Backbone region | No. of orfs | No. of orfs | No. of | Presumed accessory | Presumed accessory | |
|---|---|---|---|---|---|---|---|
| Plasmids | genome sizea | size | transfer 1 | transfer 2 | orfs ctl b | region sizec | gene locationd |
| pSFS12 | About 54 kb | 39 444 bp | 10 | 18 | 13 | About 15 kb | trfA |
| pSFS26 | About 56 kb | 40 306 bp | 10 | 18 | 14 | About 16 kb | parA |
| pSFS52 | About 53 kb | 40 011 bp | 10 | 18 | 12 | About 13 kb | parA |
| pSFW53 | About 51 kb | 39 973 bp | 10 | 19 | 13 | About 11 kb | trfA |
The total genome size of the four plasmids was preliminarily evaluated by using restriction digestion with enzymes EcoRI, BamHI and PstI.
ctl: central control and maintenance region.
The size of the accessory region was predicted by subtracting the backbone region from total genome size.
The gap locations are presumed to be insertion sites of accessory elements; parA: between parA and traC; trfA: between trfA and oriV.
Although the sequences of these four plasmids were not completely closed, the fully assembled backbone regions enabled us to infer the insertion sites of the partially sequenced accessory elements (Fig. 1 and Fig. S2, Supporting Information). For plasmids pSFS26 and pSFS52, accessory elements were located adjacent to parA and traC. By subtracting the backbone regions from total genome size, the length of the accessory regions of pSFS26 and pSFS52 was approximately 16 and 13 kb, respectively (Table 2). In contrast, accessory regions of plasmids pSFS12 and pSFW53 were inserted between the replication and control regions, and were estimated to be 15 and 11 kb in length, respectively (Table 2).
Phylogenetic analysis of selected IncP-1 plasmids based on TraI and TrfA
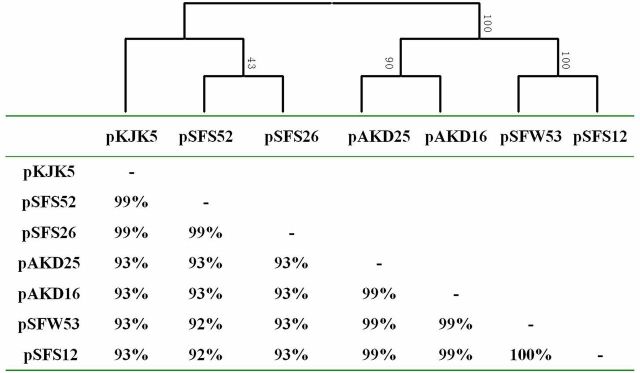
To visualize the phylogenetic position of the newly isolated plasmids within the IncP-1 plasmid family, we built phylogenetic networks together with 38 previously reported IncP-1 plasmids, including 13 IncP-1ε members, based on the conjugative relaxase protein TraI (Fig. 2A) and the replication initiation protein TrfA (Fig. 2B). The network topology of both TraI and TrfA indicated that the four newly captured plasmids fell within the IncP-1ε subgroup. Furthermore, as clearly visualized in Fig. 2, the ε-clade of IncP-1 plasmids could be divided into two separate subclades, designated as ε-I and ε-II. Within the ε-I subclade, plasmids pSFS26 and pSFS52 showed the highest similarity to pKJK5, the first published completely sequenced IncP-1ε plasmid isolate in Denmark (Bahl et al. 2007), estimated by either TrfA (100% amino acid identity) or TraI (99% identity). For the ε-II subclade, the TrfA protein encoded by pSFS12 showed 100% identity to that of pAKD16, and TrfA protein of pSFW53 was 100% identical to that of pAKD25, both from Norwegian soils. The TraI proteins of pSFS12 and pSFW53 both show 100% identity to that of pAKD16.
Figure 2.
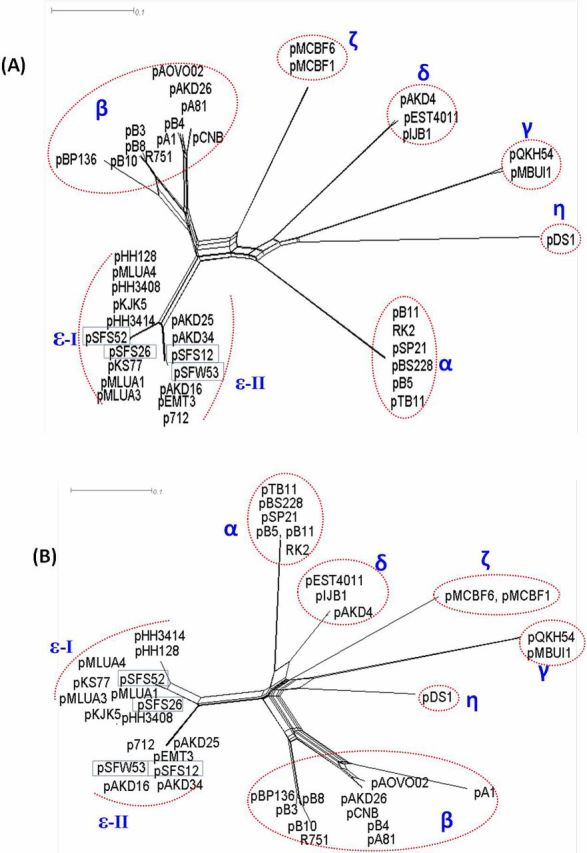
Phylogenetic analysis based on proteins (A) TraI and (B) TrfA of IncP-1 plasmids. The phylogenetic network was constructed using the neighbor joining algorithm on protein distances.
Comparison of the backbone regions between four novel IncP-1ε plasmids and their closest relatives
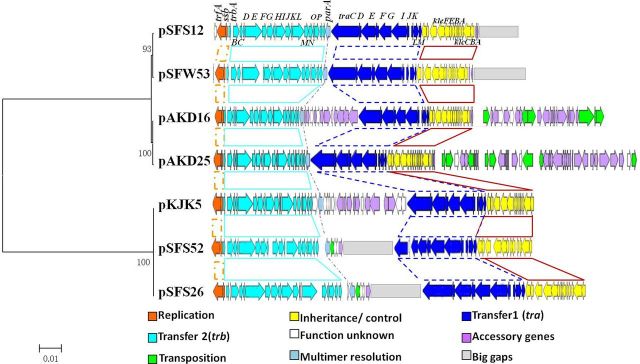
We further compared the entire backbone regions between the four novel IncP-1ε plasmids and their closest relatives, pKJK5, pAKD16 and pAKD25 (Fig. 3). Although the sequences of our four plasmids were not completely closed, the backbone regions were fully assembled (Fig. S2, Supporting Information). Using the Tamura-Nei + G sequence evolution model, a cluster analysis of the nucleotide sequences of 40 backbone genes common to all seven plasmids in Fig. 3 supported the phylogenetic analysis based on protein sequences for TraI and TrfA.
Figure 3.
Schematic diagram of backbone structures of our four and three most similar IncP-1ε plasmids. Orfs are represented by block arrows. Predicted functions are indicated in the color key below the figure. Key backbone genes and accessory genes are annotated at the top.
Given that the evolutionary history may differ between different regions of the plasmids, we carried out a comparative analysis based on the DNA sequences of each functional module. The Transfer 1 (tra gene cluster) and Transfer 2 (trb gene cluster) regions are both backbone segments responsible for conjugative transfer, and therefore these two regions were combined as a large functional module for phylogenetic analysis. As shown in Fig. 4, the phylogenetic tree (Tamura-Nei method) also clearly demonstrates two subclades, with plasmids pKJK5, pSFS26 and pSFS52 clustering together, and pAKD16, pAKD25, pSFW53 and pSFS12 grouping into the other clade. Pairwise comparison of DNA sequences of the gene clusters tra (traC–traM) and trb (trbA–upf 31.0) showed that this region is highly conserved, with at least 92% sequence identity score among all plasmids (Fig. 4). Within the ε-I subclade, the tra genes and trb gene clusters of pKJK5 are very closely related to the corresponding regions of plasmids pSFS26 (99%) and pSFS52 (99%). The four ε-II plasmids (pAKD16, pAKD25, pSFW53 and pSFS12) also showed high identities (Fig. 4). The tra and trb regions of plasmid pSFS12 even showed 100% identity to that of pSFW53 (Fig. 4).
Figure 4.
Identity scores from pairwise comparison of the genes of the Transfer 1 (traC–traM) and Transfer 2 (trbA–upf 31.0) regions.
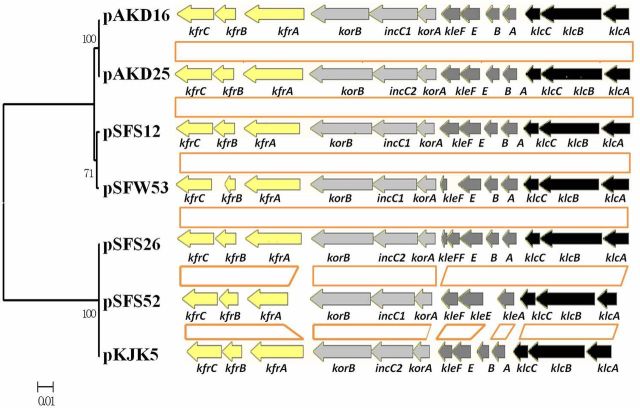
The central control (ctl) backbone genes were also compared between the four new IncP-1ε plasmids and their closest relatives (the kleB gene was excluded because it was unrecognizable on the plasmid pSFS52) (Fig. 5 and Fig. S3, Supporting Information). The ctl regions of the seven plasmids showed high similarity in composition, containing four putative transcriptional units, klc, kle, korA-incC-korB and kfr. Cluster analysis based on homologous genes of ctl region (Fig. 5) was consistent with the classification based on the entire backbone region. Further in-depth analysis showed phylogenetic divergences in specific loci of the central control regions, indicating complex evolutionary histories. For instance, kleF and kleF* present in pSFS26 showed evidence of duplication, while kleB on plasmid pSFS26 and pKJK5 was not found on pSFS52, though they were all classified into the ε-I subclade (Fig. 5). However, these small differences in backbone genes require further evidence due to unavoidable sequencing errors. The fragment flanking the trfA-oriV-klcA region still showed different organization. In specific, two putative backbone genes (dinJ, relE/stbE) coding for a toxin–antitoxin system were found in between klcA and oriV of ε-II plasmids, whereas they were absent in the corresponding site of ε-I plasmids.
Figure 5.
Genetic organization of the central control regions located on the IncP-1ε plasmids pSFS26, pSFS52, pKJK5, pAKD25, pAKD16, pSFS12 and pSFW53. Coding regions are marked by arrows indicating direction of transcription. Different putative transcriptional units (klc, kle, korA- incC- korB, kfr) of the stable inheritance and central control regions are presented in different colors. Homologous regions, labeled by orange boxes, were used for cluster analysis.
Comparison of accessory elements between ε-I and ε-II subclades
The observed genome divergence among the two IncP-1ε subclades was further supported by differences in insertion sites, phenotypes and IS/integron elements of the accessory genes. By comparing the four new plasmids from this study and the 13 previously sequenced IncP-1ε plasmids, we found that the parA locus (between parA and traC) could be considered as a hotspot for insertion of accessory elements in the ε-I subclade (Fig. 1A and Table 3). The insertion may have disrupted the N-terminus of the parA genes in ε-I plasmids (630 bp remaining for pSFS26 and pSFS52, and 606 bp remaining for other plasmids) compared to parA from ε-II ones (all 651 bp). The ε-II plasmids also have preferred spots for insertion of accessory elements. The accessory genes and transposons were located between trfA and oriV (Table 3). Plasmid pAKD16 contained two accessory regions at both parA and trfA sites (Table 3).
Besides the insertion sites, the two IncP-1ε subclades differed greatly in their phenotypes. We found that eight of ten ε-I plasmids harbored antibiotic resistance genes (Table 3), while none of them carried HgR or catabolic genes, which were frequently detected on plasmids of the ε-II subclade. In contrast, five of seven ε-II plasmids carried mercury resistance (HgR) genes and/or genes for degradation of herbicides (such as 2,4- dichlorophenoxyacetic acid and 2,4- dichlorophenoxypropionic acid) (Table 3). Our newly isolated ε-II plasmid pSFW53 carried a gene possibly encoding a putative glycosyl hydrolase, a gene coding for acriflavin resistance protein B and a gene encoding a putative Cupin2 conserved barrel protein with unknown function. The gene encoding a Cupin2 was also detected on plasmid pSFS12, together with a gene encoding an RND family efflux transporter (MFP subunit). Because the complete sequences were not closed, we could not determine the exact position of these accessory genes. In combination with our phenotypic assays, it was however clear that, unlike the ε-I plasmids, the ε-II subclade did not carry antibiotic resistance genes.
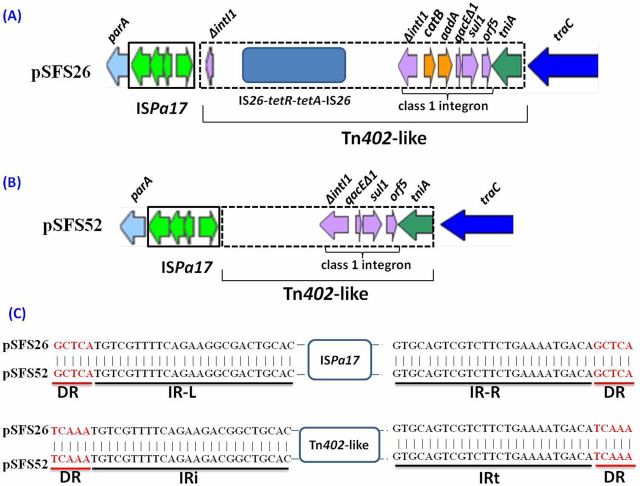
Further analysis of the IS/integron elements showed that the ε-I plasmids pSFS26 and pSFS52 carried typical ISPa17 insertion sequences and a Tn402-like transposon containing a class 1 integron (Fig. 6), which are typical of plasmids in the ε-I subclade (Table 3). The ISPa17 insertion sequence contained four orfs and was flanked by 25 bp inverted repeats (IRs, IR-L and IR-R) and 5 bp direct repeats (DRs, the sequences were ‘GCTCA’) (Fig. 6C). Tn402-like transposon was flanked by two 25 bp inverted repeats (IRi and IRt) and two 5 bp DRs (the sequences were ‘TCAAA’) (Fig. 6C). For plasmid pSFS26, the Tn402-like transposon included a composite transposon IS26-tetR-tetA-IS26 and a class 1 integron (ΔintI1-catB-aadA-qacEΔ1-sul1-orf5) (Fig. 6A). The composite transposon contained genes conferring resistance to tetracycline (tetR-tetA). The integrase gene (ΔintI1) was disrupted by the composite transposon flanked by IS26 copies. The variable integron regions of pSFS26 consisted of the gene cassettes catB-aadA, encoding chloramphenicol acetyltransferase (catB) and aminoglycoside adenylyltransferase (aadA). This was consistent with the observed resistance to chloramphenicol and spectinomycin encoded by this plasmid. The Tn402-like transposon of pSFS26 harbored the 3′ conserved segment (3′-CS: qacEΔ1-sul1-orf5) which is common in typical class 1 integrons, with qacEΔ1 conferring resistance to quaternary ammonium compounds, sul1 encoding a dihydropteroate synthetase resistant to sulfonamides, and orf5 encoding a putative acetyltransferase (Fig. 6A). In contrast, the class 1 integron of plasmid pSFS52 contained only a truncated ΔintI1 gene and the 3′ conserved segment (3′-CS), devoid of any gene cassettes (Fig. 6B), which was similar to the integron structure of IncP-1ε-I plasmid pHH3408 (Heuer et al. 2012). While the two plasmids pSFS26 and pSFS52 harbored similar IS/integron elements, carrying the typical ISPa17 insertion sequence and a Tn402-like transposon containing class 1 integron (Fig. 6), no antibiotic resistance genes were detected on pSFS52, which instead harbors a series of genes encoding hypothetical proteins.
Figure 6.
Analysis of transposon types and integrons (between parA and traC) of plasmids pSFS26 and pSFS52. (A) Plasmid pSFS26 harbors the ISPa17 insertion sequence and a typical Tn402-like transposon. The Tn402-like transposon contains a composite transposon (IS26-tetR-tetA- IS26) and a class 1 integron. (B) Structure of transposon structure in plasmid pSFS52, composed of the ISPa17 and Tn402-like transposon. (C) The nucleotide sequences at the ends of ISPa17 and the Tn402-like transposon are shown. IRs are marked in black and DRs are marked in red. IR-L and IR-R represent the left inverted repeat and right inverted repeat of ISPa17, respectively. IRi and IRt represent the inverted repeats found at the integron end and the transposition module end of class 1 integrons/transposons, respectively.
Only one insertion sequence, IS1071, was found for plasmids pSFS12 and pSFW53 (Table 3). IS1071 is often found as part of Tn501-like transposons, which usually contain other insertion sequences. For example, Tn501-like transposons were detected on pAKD plasmids that contained the IS1071, ISPps1 and ISCsp2, harboring the mercury resistance (mer) operon and/or the tfd gene cluster region (responsible for the degradation of 2, 4-D) (Sen et al. 2011; Kim et al. 2013).
DISCUSSION
Despite the general agreement on the importance of BHR plasmids in the adaptive evolution of bacteria (Jechalke et al. 2013b; Shintani, Sanchez and Kimbara 2015), their evolutionary history and genetic diversity are still not very well understood, limited by an insufficient pool of BHR plasmid sequences. Thus, isolation and characterization of new BHR plasmids from environmental samples, especially from understudied locations like China, is still fundamental to understanding the nature and evolutionary history of these important mobile genetic elements and their role in HGT. The present work is among the first to identify BHR plasmids in China (Li et al. 2015). The isolation of very similar BHR plasmids from geographically distinct sites further demonstrates the wide distribution of these mobile genetic elements, and provides an opportunity for comparing genomics of plasmids collected worldwide.
Polluted habitats have been recognized as hotspots for self-transmissible plasmids due to environmental selection pressures imposed by the pollutants. A large number of plasmids were previously captured from contaminated soils (Gstalder et al. 2003), polluted sludges (Top et al. 1994), wastewaters (Schlüter et al. 2007), etc. In particular, wastewater treatment plants have been described as reservoirs for plasmids and other mobile genetic elements (Schlüter et al. 2003). With long-term exposure to petroleum pollutants, the Shen-fu petroleum-wastewater irrigation zone is a potential ‘dream site’ for BHR plasmid isolation, and we hypothesized that this habitat should be rich in such plasmids. The present study indeed captured a diverse set of BHR plasmids from this irrigation zone, and thereby increased the currently available BHR plasmid genome sequence pool. In our present work, the plasmids were isolated by the donor/recipient system E. coli JM109 (pBBR1MCS-5)/E. coli MG1685 (Yanischperron, Vieira and Messing 1985; Fox et al. 2008), both of which belong to the γ- Proteobacteria. While it has been previously suggested that using donor and recipient strains belonging to different Proteobacteria may increase the frequency of obtaining plasmids with a BHR (Top et al. 1994), we obtained BHR plasmids using E. coli strains only.
We performed phylogenetic analyses based on the amino acid sequences of TraI and TrfA for 38 selected IncP-1 plasmids (including the four newly isolated IncP-1 plasmids). The TrfA (Vedler, Vahter and Heinaru 2004; Sen et al. 2012) and TraI (Garcillán-Barcia, Francia and De La Cruz 2009) are frequently selected as the representative backbone gene product for phylogenetic analysis of the IncP-1 plasmids. Our results are quite consistent with previous classification of IncP-1 plasmids, which divided 25 IncP-1 plasmids into seven subgroups (Norberg et al. 2011), designated IncP-1α to IncP-1η. Moreover, two separate subclades within the IncP-1ε subgroup were clearly demonstrated by split network analysis. Preliminary signs of the presence of two clusters within the IncP-1ε subgroup were pointed out by previous studies (Sen et al. 2012; Dealtry et al. 2014). Here, by comparing more IncP-1ε members, we designated these two striking subclades as ε-I and ε-II. We still compared the trfA genes between ε-I and ε-II plasmids by constructing a phylogenetic tree (data not shown). As expected, the two subclades were clearly separated, implying that these two subclades could be distinguished from each other by hybridization using a possible probe of each trfA gene. From another point of view, since DNA amplification with trfA primers followed by sequencing is often used in environmental studies to assess the presence and diversity of IncP-1 plasmids, we tested the trfA primers developed by Bahl et al. (2009) in silico against the IncP-1ε plasmids. They should amplify all ε-I and ε-II plasmids described here, because there are no mismatches at the 3′end (data not shown). Similarly, primers developed for amplification of korB genes (Jechalke et al. 2013a) should amplify both of these two IncP-1ε plasmid subclades. We will put our efforts into quantifying the abundance of IncP-1ε plasmids in the contaminated soil or wastewater in the future.
Comparative genomic analysis based on the concatenation of all shared backbone genes further strengthened the suggestion for separating IncP-1ε plasmids into two subclades. This strategy may not be suitable for evolutionarily distinct plasmids since concatenation may ignore the multiple histories of the underlying data and thus represent an incorrect single history for all backbone genes (Sen et al. 2012). However, in our study concatenation of backbone genes was an effective method for inferring the evolutionary history of our IncP-1ε plasmids, since they have highly similar backbones (Norberg et al. 2011).
It was observed that 16 of 17 IncP-1ε members, including the four new plasmids captured in this study, harbored accessory elements in only one of two typical insertion sites. Interestingly, we found clear differences in the phenotypes, IS/integron elements, and insertion location of the accessory elements between the ε-I and ε-II subclades. The ε-I plasmids encoded only antibiotic resistance genes, and carried a typical ISPa17 and a typical Tn402-like transposon containing a class 1 integron. In contrast, several ε-II plasmids tended to carry mercury resistance genes and herbicide degradation genes, which are usually inserted in a Tn501-like transposon. Moreover, the IS1071 insertion sequence, which is commonly located within the Tn501-like transposon, was unique to the ε-II plasmids. In fact, IS1071 has often been associated with catabolic genes (Tan 1999). In addition, the accessory elements of ε-I plasmids were inserted between parA and traC, a typical insertion site for IncP-1 plasmids (Sota et al. 2007). In contrast, accessory regions of ε-II plasmids are inserted between trfA and oriV, a second typical insertion hotspot (Thorsted et al. 1998). Three plausible mechanisms have been proposed for site preference of parA and/or trfA loci. First, the 20-bp IRs with a consensus sequence of CATCGCCANNTCYGRCGATG in both trfA and parA locus was suggested to be responsible for the region-specific acquisition of transposons (Thorsted et al. 1998; Heuer et al. 2004). However, the reported IRs were not found on our newly isolated plasmid. Second, transposition occurs randomly, but plasmids with insertions in the two specific regions are most stable or transferable, or least costly to their host, and thereby persist longer over evolutionary time than the cognate plasmids with insertions in other sites (Simonsen 1991; De Gelder et al. 2007). Third, a combination of region-specific insertion and selection explains the common plasmid structure (Sota et al. 2007).
The difference in accessory gene content between the ε-I and ε-II plasmids is intriguing and may be explained by the differences in selective pressures the plasmid hosts experienced in their environments. Almost all of the ε-I plasmids carry antibiotic resistance genes and five of the ten encode multiple drug resistance, including our newly isolated plasmid pSFS26 (Table 3). These ε-I plasmids were isolated from manured soil (pKJK5, pHH128, pHH3408, pHH3414 and pKS77), polluted soils irrigated by wastewater (pSFS26 and pSFS52), aquatic environments (pMLUA1, pMLUA3 and pMLUA4), which were all susceptible to pollution with antibiotic residues (Binh et al. 2008; Rizzo et al. 2013). In contrast, most of the ε-II plasmids were isolated from arable soils treated with mercuric chloride or herbicides, and as a result mainly conferred mercury resistance genes and genes for degradation of herbicides. Good examples are the mercury resistance plasmids pAKD16 and pAKD25, closest relatives to two of our new IncP-1ε plasmids, which were both isolated from arable soils treated with mercury in the laboratory (Sen et al. 2011). Our two ε-II plasmids were from soil and wastewater in the Shen-fu petroleum wastewater irrigation zone, which likely contained a variety of organic and inorganic pollutants (Zhou et al. 2012).
These observations suggest to us that the different plasmid subclades are evolving in different types of bacteria, which are under different selective pressures. For example, the hosts of ε-I plasmids might be enteric bacteria that thrive in environments such as manure, and those of the ε-II clade plasmids more typical soil bacteria, phylogenetically distinct from enterics. Thus, we hypothesize that these two subclades of plasmids have adapted to different ranges of hosts, which is reflected in the divergence of their backbone genes (Yano et al. 2013). Although using exogenous methods for isolating plasmids does not provide us information on the actual hosts of these plasmids, this hypothesis could be tested directly by more extensive comparisons of the host ranges of plasmids from the two subclades, as done by Yano et al. (2013) for other IncP-1 plasmids. Alternatively, hosts carrying ε-I and ε-II plasmids could be isolated directly from environments with antibiotic residues versus chemical pollutants, using a hybridization approach. In conclusion, the striking correlation of antibiotic resistance genes with ε-I plasmids suggests coevolution with specific hosts that benefit from these resistance traits.
The present work enhances our understanding of the phylogenetic diversity, biogeography and evolutionary history of the IncP-1 plasmids, and increases the available collection of IncP-1ε plasmid sequences. In this era of high-throughput sequencing, more BHR plasmid sequences will help us understand the reservoirs, evolutionary trajectories and host-beneficial traits of these important vectors of HGT, and will shed light on the alarmingly rapid worldwide spread of antibiotic resistance.
Supplementary Material
Acknowledgments
We thank for Dr Hao Sun from Institute of Applied Ecology, Chinese Academy of Sciences for helpful discussion of bioinformatics analysis.
SUPPLEMENTARY DATA
FUNDING
This work was supported by the Program of the National Science Foundation of China (31070102) for HL, the US National Science Foundation grant EF-0627988 and NIH grant no. R01 AI084918 from the National Institute of Allergy and Infectious Diseases (NIAID) to EMT, Strategic Priority Research Program of the Chinese Academy of Sciences (XDB15010103) to YJ, and also the Idaho INBRE Program, NIH grants P20RR016454 and P20GM103408 through support for CJB.
Conflict of interest. None declared.
REFERENCES
- Bahl MI, Burmolle M, Meisner A, et al. All IncP-1 plasmid subgroups, including the novel epsilon subgroup, are prevalent in the influent of a Danish wastewater treatment plant. Plasmid. 2009;62:134–9. doi: 10.1016/j.plasmid.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Bahl MI, Hansen LH, Goesmann A, et al. The multiple antibiotic resistance IncP-1 plasmid pKJK5 isolated from a soil environment is phylogenetically divergent from members of the previously established alpha, beta and delta sub-groups. Plasmid. 2007;58:31–43. doi: 10.1016/j.plasmid.2006.11.007. [DOI] [PubMed] [Google Scholar]
- Binh CT, Heuer H, Kaupenjohann M, et al. Piggery manure used for soil fertilization is a reservoir for transferable antibiotic resistance plasmids. FEMS Microbiol Ecol. 2008;66:25–37. doi: 10.1111/j.1574-6941.2008.00526.x. [DOI] [PubMed] [Google Scholar]
- Brown CJ, Sen D, Yano H, et al. Diverse broad-host-range plasmids from freshwater carry few accessory genes. Appl Environ Microb. 2013;79:7684–95. doi: 10.1128/AEM.02252-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook MA, Osborn AM, Bettandorff J, et al. Endogenous isolation of replicon probes for assessing plasmid ecology of marine sediment microbial communities. Microbiology. 2001;147:2089–101. doi: 10.1099/00221287-147-8-2089. [DOI] [PubMed] [Google Scholar]
- Davison J. Genetic exchange between bacteria in the environment. Plasmid. 1999;42:73–91. doi: 10.1006/plas.1999.1421. [DOI] [PubMed] [Google Scholar]
- De Gelder L, Ponciano JM, Joyce P, et al. Stability of a promiscuous plasmid in different hosts: no guarantee for a long-term relationship. Microbiology. 2007;153:452–63. doi: 10.1099/mic.0.2006/001784-0. [DOI] [PubMed] [Google Scholar]
- De Lorenzo V, Herrero M, Jakubzik U, et al. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–72. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dealtry S, Holmsgaard PN, Dunon V, et al. Shifts in abundance and diversity of mobile genetic elements after the introduction of diverse pesticides into an on-farm biopurification system over the course of a year. Appl Environ Microb. 2014;80:4012–20. doi: 10.1128/AEM.04016-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehio C, Meyer M. Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J Bacteriol. 1997;179:538–40. doi: 10.1128/jb.179.2.538-540.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-López R, del Campo I, Revilla C, et al. Negative feedback and transcriptional overshooting in a regulatory network for horizontal gene transfer. PLoS Genet. 2014;10:e1004171. doi: 10.1371/journal.pgen.1004171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-López R, Pilar Garcillán-Barcia M, Revilla C, et al. Dynamics of the IncW genetic backbone imply general trends in conjugative plasmid evolution. FEMS Microbiol Rev. 2006;30:942–66. doi: 10.1111/j.1574-6976.2006.00042.x. [DOI] [PubMed] [Google Scholar]
- Fox RE, Zhong X, Krone SM, et al. Spatial structure and nutrients promote invasion of IncP-1 plasmids in bacterial populations. ISME J. 2008;2:1024–39. doi: 10.1038/ismej.2008.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcillán-Barcia MP, Francia MV, De La Cruz F. The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol Rev. 2009;33:657–87. doi: 10.1111/j.1574-6976.2009.00168.x. [DOI] [PubMed] [Google Scholar]
- Gotfredsen M, Gerdes K. The Escherichia coli relBE genes belong to a new toxin–antitoxin gene family. Mol Microbiol. 1998;29:1065–76. doi: 10.1046/j.1365-2958.1998.00993.x. [DOI] [PubMed] [Google Scholar]
- Gstalder ME, Faelen M, Mine N, et al. Replication functions of new broad host range plasmids isolated from polluted soils. Res Microbiol. 2003;154:499–509. doi: 10.1016/S0923-2508(03)00143-8. [DOI] [PubMed] [Google Scholar]
- Heuer H, Binh CT, Jechalke S, et al. IncP-1ε plasmids are important vectors of antibiotic resistance genes in agricultural systems: diversification driven by class 1 integron gene cassettes. Front Microbiol. 2012;3:2. doi: 10.3389/fmicb.2012.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer H, Smalla K. Plasmids foster diversification and adaptation of bacterial populations in soil. FEMS Microbiol Rev. 2012;36:1083–104. doi: 10.1111/j.1574-6976.2012.00337.x. [DOI] [PubMed] [Google Scholar]
- Heuer H, Szczepanowski R, Schneiker S, et al. The complete sequences of plasmids pB2 and pB3 provide evidence for a recent ancestor of the IncP-1beta group without any accessory genes. Microbiology. 2004;150:3591–9. doi: 10.1099/mic.0.27304-0. [DOI] [PubMed] [Google Scholar]
- Hill K, Weightman A, Fry J. Isolation and screening of plasmids from the epilithon which mobilize recombinant plasmid pD10. Appl Environ Microb. 1992;58:1292–300. doi: 10.1128/aem.58.4.1292-1300.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH, Bryant D. Application of phylogenetic networks in evolutionary studies. Mol Biol Evol. 2006;23:254–67. doi: 10.1093/molbev/msj030. [DOI] [PubMed] [Google Scholar]
- Jacoby G. Classification of plasmids in Pseudomonas aeruginosa. In: Schlessinger D, editor. Microbiology 1977. Washington, DC: American Society for Microbiology; 1977. pp. 119–26. [Google Scholar]
- Jechalke S, Dealtry S, Smalla K, et al. Quantification of IncP-1 plasmid prevalence in environmental samples. Appl Environ Microb. 2013a;79:1410–3. doi: 10.1128/AEM.03728-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jechalke S, Schreiter S, Wolters B, et al. Widespread dissemination of class 1 integron components in soils and related ecosystems as revealed by cultivation-independent analysis. Front Microbiol. 2013b;4:420. doi: 10.3389/fmicb.2013.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DU, Kim MS, Lim JS, et al. Widespread occurrence of the tfd-II genes in soil bacteria revealed by nucleotide sequence analysis of 2, 4-dichlorophenoxyacetic acid degradative plasmids pDB1 and p712. Plasmid. 2013;69:243–8. doi: 10.1016/j.plasmid.2013.01.003. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown N, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–8. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li H, Zhang Y, Zhang C, et al. Effect of petroleum-containing wastewater irrigation on bacterial diversities and enzymatic activities in a paddy soil irrigation area. J Environ Qual. 2005;34:1073–80. doi: 10.2134/jeq2004.0438. [DOI] [PubMed] [Google Scholar]
- Li X, Top EM, Wang Y, et al. The broad-host-range plasmid pSFA231 isolated from petroleum-contaminated sediment represents a new member of the PromA plasmid family. Front Microbiol. 2015;5:777. doi: 10.3389/fmicb.2014.00777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macartney DP, Williams DR, Stafford T, et al. Divergence and conservation of the partitioning and global regulation functions in the central control region of the IncP plasmids RK2 and R751. Microbiology. 1997;143:2167–77. doi: 10.1099/00221287-143-7-2167. [DOI] [PubMed] [Google Scholar]
- Moura A, Soares M, Pereira C, et al. INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics. 2009;25:1096–8. doi: 10.1093/bioinformatics/btp105. [DOI] [PubMed] [Google Scholar]
- Norberg P, Bergstrom M, Jethava V, et al. The IncP-1 plasmid backbone adapts to different host bacterial species and evolves through homologous recombination. Nat Commun. 2011;2:268. doi: 10.1038/ncomms1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman A, Hansen LH, Sørensen SJ. Conjugative plasmids: vessels of the communal gene pool. Philos Trans R Soc Lond B Biol Sci. 2009;364:2275–89. doi: 10.1098/rstb.2009.0037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveira CS, Moura A, Henriques I, et al. Comparative genomics of IncP-1ε plasmids from water environments reveals diverse and unique accessory genetic elements. Plasmid. 2013;70:412–9. doi: 10.1016/j.plasmid.2013.06.002. [DOI] [PubMed] [Google Scholar]
- Rhodes G, Parkhill J, Bird C, et al. Complete nucleotide sequence of the conjugative tetracycline resistance plasmid pFBAOT6, a member of a group of IncU plasmids with global ubiquity. Appl Environ Microb. 2004;70:7497–510. doi: 10.1128/AEM.70.12.7497-7510.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizzo L, Manaia C, Merlin C, et al. Urban wastewater treatment plants as hotspots for antibiotic resistant bacteria and genes spread into the environment: a review. Sci Total Environ. 2013;447:345–60. doi: 10.1016/j.scitotenv.2013.01.032. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning. Vol. 2. New York: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schlüter A, Heuer H, Szczepanowski R, et al. The 64 508 bp IncP-1β antibiotic multiresistance plasmid pB10 isolated from a waste-water treatment plant provides evidence for recombination between members of different branches of the IncP-1β group. Microbiology. 2003;149:3139–53. doi: 10.1099/mic.0.26570-0. [DOI] [PubMed] [Google Scholar]
- Schlüter A, Krause L, Szczepanowski R, et al. Genetic diversity and composition of a plasmid metagenome from a wastewater treatment plant. J Biotechnol. 2008;136:65–76. doi: 10.1016/j.jbiotec.2008.03.017. [DOI] [PubMed] [Google Scholar]
- Schlüter A, Szczepanowski R, Pühler A, et al. Genomics of IncP-1 antibiotic resistance plasmids isolated from wastewater treatment plants provides evidence for a widely accessible drug resistance gene pool. FEMS Microbiol Rev. 2007;31:449–77. doi: 10.1111/j.1574-6976.2007.00074.x. [DOI] [PubMed] [Google Scholar]
- Sen D, Brown CJ, Top EM, et al. Inferring the evolutionary history of IncP-1 plasmids despite incongruence among backbone gene trees. Mol Biol Evol. 2012;30:154–66. doi: 10.1093/molbev/mss210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sen D, Van der Auwera GA, Rogers LM, et al. Broad-host-range plasmids from agricultural soils have IncP-1 backbones with diverse accessory genes. Appl Environ Microb. 2011;77:7975–83. doi: 10.1128/AEM.05439-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shintani M, Sanchez ZK, Kimbara K. Genomics of microbial plasmids: classification and identification based on replication and transfer systems and host taxonomy. Front Microbiol. 2015;6:242. doi: 10.3389/fmicb.2015.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Pérochon J, Lestrade L, et al. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:32–6. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonsen L. The existence conditions for bacterial plasmids: theory and reality. Microb Ecol. 1991;22:187–205. doi: 10.1007/BF02540223. [DOI] [PubMed] [Google Scholar]
- Song L, Chen W, Peng L, et al. Distribution and bioaccumulation of microcystins in water columns: a systematic investigation into the environmental fate and the risks associated with microcystins in Meiliang Bay, Lake Taihu. Water Res. 2007;41:2853–64. doi: 10.1016/j.watres.2007.02.013. [DOI] [PubMed] [Google Scholar]
- Sota M, Tsuda M, Yano H, et al. Region-specific insertion of transposons in combination with selection for high plasmid transferability and stability accounts for the structural similarity of IncP-1 plasmids. J Bacteriol. 2007;189:3091–8. doi: 10.1128/JB.01906-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szpirer C, Top E, Couturier M, et al. Retrotransfer or gene capture: a feature of conjugative plasmids, with ecological and evolutionary significance. Microbiology. 1999;145:3321–9. doi: 10.1099/00221287-145-12-3321. [DOI] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, et al. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan H. Bacterial catabolic transposons. Appl Microbiol Biot. 1999;51:1–12. doi: 10.1007/s002530051356. [DOI] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL. BLAST 2 Sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett. 1999;174:247–50. doi: 10.1111/j.1574-6968.1999.tb13575.x. [DOI] [PubMed] [Google Scholar]
- Tennstedt T, Szczepanowski R, Krahn I, et al. Sequence of the 68 869 bp IncP-1α plasmid pTB11 from a waste-water treatment plant reveals a highly conserved backbone, a Tn402-like integron and other transposable elements. Plasmid. 2005;53:218–38. doi: 10.1016/j.plasmid.2004.09.004. [DOI] [PubMed] [Google Scholar]
- Thomas CM. Paradigms of plasmid organization. Mol Microbiol. 2000;37:485–91. doi: 10.1046/j.1365-2958.2000.02006.x. [DOI] [PubMed] [Google Scholar]
- Thorsted PB, Macartney DP, Akhtar P, et al. Complete sequence of the IncPβ plasmid R751: implications for evolution and organisation of the IncP backbone. J Mol Biol. 1998;282:969–90. doi: 10.1006/jmbi.1998.2060. [DOI] [PubMed] [Google Scholar]
- Top E, De Smet I, Verstraete W, et al. Exogenous isolation of mobilizing plasmids from polluted soils and sludges. Appl Environ Microb. 1994;60:831–9. doi: 10.1128/aem.60.3.831-839.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Top EM, Holben WE, Forney LJ. Characterization of diverse 2, 4-dichlorophenoxyacetic acid-degradative plasmids isolated from soil by complementation. Appl Environ Microb. 1995;61:1691–8. doi: 10.1128/aem.61.5.1691-1698.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Krol JE, Suzuki H, et al. Plasmids captured in C. metallidurans CH34: defining the PromA family of broad-host-range plasmids. Anton Leeuw. 2009;96:193–204. doi: 10.1007/s10482-009-9316-9. [DOI] [PubMed] [Google Scholar]
- van Elsas JD, Gardener BBM, Wolters AC, et al. Isolation, characterization, and transfer of cryptic gene-mobilizing plasmids in the wheat rhizosphere. Appl Environ Microb. 1998;64:880–9. doi: 10.1128/aem.64.3.880-889.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vedler E, Vahter M, Heinaru A. The completely sequenced plasmid pEST4011 contains a novel IncP1 backbone and a catabolic transposon harboring tfd genes for 2, 4-dichlorophenoxyacetic acid degradation. J Bacteriol. 2004;186:7161–74. doi: 10.1128/JB.186.21.7161-7174.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood DW, Setubal JC, Kaul R, et al. The genome of the natural genetic engineer Agrobacterium tumefaciens C58. Science. 2001;294:2317–23. doi: 10.1126/science.1066804. [DOI] [PubMed] [Google Scholar]
- Yanischperron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mpl8 and pUC19 vectors. Gene. 1985;33:103–19. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yano H, Rogers LM, Knox MG, et al. Host range diversification within the IncP-1 plasmid group. Microbiology. 2013;159:2303–15. doi: 10.1099/mic.0.068387-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Li H, Zhang Y, et al. Abundance and diversity of Sphingomonas in Shenfu petroleum-wastewater irrigation zone, China. Environ Sci Pollut R. 2012;19:282–94. doi: 10.1007/s11356-011-0552-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.