Abstract
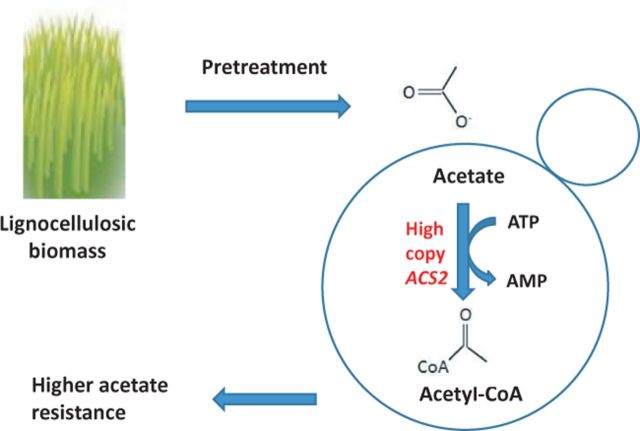
Acetic acid-mediated inhibition of the fermentation of lignocellulose-derived sugars impedes development of plant biomass as a source of renewable ethanol. In order to overcome this inhibition, the capacity of Saccharomyces cerevisiae to synthesize acetyl-CoA from acetic acid was increased by overexpressing ACS2 encoding acetyl-coenzyme A synthetase. Overexpression of ACS2 resulted in higher resistance to acetic acid as measured by an increased growth rate and shorter lag phase relative to a wild-type control strain, suggesting that Acs2-mediated consumption of acetic acid during fermentation contributes to acetic acid detoxification.
Keywords: Saccharomyces cerevisiae, acetyl-CoA synthetase, Acs2, acetic acid, lignocellulose
An approach to overcoming one of the fermentation-related problems that impedes use of plant biomass for generating renewable ethanol is described.
An approach to overcoming one of the fermentation-related problems that impedes use of plant biomass for generating renewable ethanol is described.
INTRODUCTION
Inefficient conversion of lignocellulose-derived sugars from plant biomass into ethanol and other biofuels has hindered large-scale biofuel production from this renewable energy source. One major bottleneck is the release of significant amounts of acetic acid from acetylated hemicellulose and lignin during pre-fermentation processing of lignocellulose (Klinke, Thomsen and Ahring 2004; Del Río et al., 2007). Although acetic acid is a normal yeast metabolite, it inhibits growth at the elevated concentrations expected in lignocellulosic hydrolysates. An estimate based on corn stover which comprises about 2.9% acetyl groups (Aden et al., 2002), 100% hydrolysis during pre-treatment and a 30% (w/w) solids loading for fermentation yields a hydrolysate containing in the range of 250 mM.
In Saccharomyces cerevisiae, acetic acid is taken up both by passive diffusion of the undissociated acid and via the Fps1 channel (Mollapour and Piper 2007). Once acetic acid enters the cell, it dissociates into an acetate anion and a proton because its pKa (4.78) is much lower than the near-neutral pH of the cytoplasm. Acetate is a substrate for acetyl-CoA synthetase (E.C.6.2.1.1), a homodimeric enzyme with a subunit of about 75 kDa (Frenkel and Kitchens 1977). In S. cerevisiae, the reaction catalyzed by acetyl-CoA synthetase (Berg 1956) is the major route for acetyl-CoA synthesis during fermentative growth (Van den Berg et al., 1996):
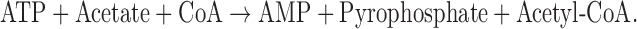 |
Although the reaction consumes a single ATP per acetyl-CoA formed, the net consumption is greater because AMP rather than ADP is generated. In contrast to organisms with a strictly aerobic metabolism and which synthesize acetyl-CoA via pyruvate dehydrogenase (PDH), S. cerevisiae prefers a fermentative metabolism during which PDH expression is largely repressed (Sierkstra,Verbakel and Verrips 1992). The S. cerevisiae genes ACS1 (De Virgilio et al., 1992) and ACS2 (Van den Berg and Steensma 1995) encode two immunologically distinct forms of acetyl-CoA synthetase: Acs1 and Acs2, respectively (Satyanarayana and Klein 1973; Satyanarayana, Mandel and Klein 1974; Frenkel and Kitchens 1977). Van den Berg and Steensma (1995) reported that loss of either gene alone did not interfere with growth, but that simultaneous loss of both was lethal, indicating that acetyl-CoA synthetase activity is essential. While ACS1 expression is subject to glucose repression, ACS2 is expressed constitutively during growth on glucose (van den Berg et al., 1996). A number of studies have focused on genetic manipulations to modulate acetyl-CoA content in yeast to increase ethanol yields or tolerance (Chen et al., 2010; Medina et al., 2010; Wei et al., 2013), to decrease acetic acid levels (De Jong-Gubbels et al., 1998; Remize, Andrieu and Dequin 2000; Medina et al., 2010; Wei et al., 2013), to increase yields of acetyl-CoA-derived metabolites at reduced energy cost (Kozak et al., 2014) or to increase lipid or sterol content (Shiba et al., 2007; Scalcinati et al., 2012; Chen et al., 2013; Runguphan and Keasling 2014). To our knowledge, none have focused directly on how such changes might alter tolerance to high levels of exogenous acetic acid.
Here, ACS2 was chosen as a target for overexpression to test the hypothesis that increasing acetyl-CoA biosynthetic capacity would increase yeast tolerance for exogenous acetic acid.
MATERIALS AND METHODS
Yeast strains, media, growth conditions, transformation
The yeast strains used in this study are listed in Table 1. Yeast transformations were performed as described (Gietz et al., 1995). Cells were grown in YNB + 2% glu (bacto yeast nitrogen base without amino acids to which 2% glucose were added) or in YNB-4.8 + 2% glu (bacto yeast nitrogen base without amino acids adjusted to pH 4.8 with HCl, to which 2% glucose were added). Liquid YNB and agar-based media were sterilized by autoclaving. Liquid YNB-4.8 + 2% glu was sterilized by filtration through a 0.45 micron filter. Where indicated, YNB-4.8 containing >2% glucose was also used. A 2N acetic acid stock was prepared using reagent grade glacial acetic acid and was adjusted to pH 4.8 using NaOH. The stock was replaced monthly.
Table 1.
Yeast strains.
| Strains | Genotype | Source |
|---|---|---|
| S288c | MATα SUC gal mal mel flo1 flo8-1 hap bio1 bio6 | ATCC 204508, Manassas, VA |
| S288c his3Δ | MATα SUC gal mal mel flo1 flo8-1 hap bio1 bio6 his3Δ::KanMX | Ding et al. (2013) |
| S288c his3ΔpXP420 | MATα SUC gal mal mel flo1 flo8-1 hap bio1 bio6 his3Δ::KanMX/pXP420 | This study |
| S288c his3ΔpXP420-ACS2 | MATα SUC gal mal mel flo1 flo8-1 hap bio1 bio6 his3Δ::KanMX/pXP420::ACS2 | This study |
Plasmid construction and yeast transformation
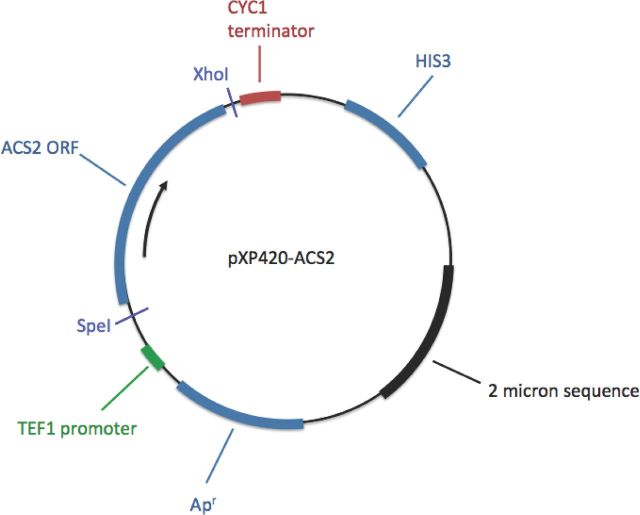
A high-copy, 2μ-based yeast expression vector with a strong promoter, TEF1, was used to ensure overexpression of the ACS2 open reading frame (ORF) (Fang et al., 2011). A 1846 bp DNA fragment consisting of the ACS2 ORF was amplified by PCR from S288c genomic DNA, using primers SpeIACS2Up and ACS2XhoILo (Table 2), which were designed with 5′-proximal SpeI and 3′-proximal XhoI sites. The ACS2 amplicon was initially cloned into a TOPO vector (Invitrogen, Carlsbad, CA) and subsequently digested with SpeI and XhoI to release the ORF. The released ORF was then subcloned into SpeI- and XhoI-digested pXP420 (Addgene, Cambridge, MA) to yield pXP420-ACS2 (Fig. 1). Saccharomyces cerevisiae S288c his3Δ was then transformed with pXP420 (control) or pXP420-ACS2, and transformants were selected on YNB + 2% glu plates.
Table 2.
Primers.
| Primer | Sequencea (5′ → 3′) | Use |
|---|---|---|
| SpeIACS2Up | CGCCACTAGTATGACAATCAAGGAACATAAA | ACS2 insertion |
| ACS2XhoILo | GGGGGGCTCGAGTTATTTCTTTTTTTGAGAG | ACS2 insertion |
| ACS2Up | TGCTAATCCCGACAAGCCAG | ACS2 overexpression verification |
| AmpUp | CCGGCGTCAATACGGGATAA | ACS2 overexpression verification |
| LacLo | CCAGCTGGCGTAATAGCGAA | ACS2 overexpression verification |
| Act1-F | TGGATTCCGGTGATGGTGTT | RT-PCR |
| Act1-R | CGGCCAAATCGATTCTCAA | RT-PCR |
| Acs2-F | CTGCTGTTGTCGGTATTCCA | RT-PCR |
| Acs2-R | TGTGTTCTGCATCACCTTCA | RT-PCR |
aThe underlined sequences are added SpeI and XhoI restriction sites.
Figure 1.
ACS2 overexpression construct pXP420-ACS2.
Real-time quantitative PCR
One mL cultures of YNB-4.8 + 2% glu were inoculated with cells taken from single colonies of S288c his3Δ/pXP420 and S288c his3Δ/pXP420-ACS2 on YNB + 2% glu plates and were incubated at 30°C and 200 rpm for 24 h. Cells were collected, rinsed twice with distilled water and suspended in 100 μL of distilled water. The washed cells were then used to inoculate fresh 1 mL YNB-4.8 + 2% glu cultures at a starting concentration of 6 × 106 cells mL−1 (estimated using a hemocytometer). Cells were incubated at 30°C and 200 rpm and were harvested in exponential phase at an A600 value of about 0.8 (2 × 107 cells mL−1). Total RNA isolation, reverse transcription and the real-time quantitative PCR reaction were performed as described (Ding et al., 2013). The PCR primers used are listed in Table 2. Gene expression levels were determined by the 2−ΔΔCT method (Pfaffl 2001) based on the ratio of fluorescence signals of ACS2 normalized to ACT1 expression in both wild-type and ACS2-overexpressing cells:
ACT1/ACS2 = 2CT(ACT1) − CT(ACS2) (2006 Real-Time PCR Applications Guide, Bio-Rad Laboratories, Inc.).
Protein extraction
Cells from single colonies of S288c his3Δ/pXP420 and S288c his3Δ/pXP420-ACS2 grown on YNB + 2% glu plates were used to inoculate 100 mL cultures of YNB-4.8 + 2% glu which were incubated at 30°C and 200 rpm for 24 h. Stationary-phase cells were harvested by centrifugation at 1500 g for 5 min at 4°C, after which cell wet weight was determined (approximately 900 mg wet weight per 100 mL culture). Cells were washed once with ice-cold water and resuspended in two volumes of glass bead disruption buffer [20 mM Tris-HCl, pH 7.9, 10 mM MgCl2, 1 mM EDTA, 5% glycerol, 1 mM DTT, 0.3 M ammonium sulfate, 1X protease inhibitor mix (Sigma-Aldrich, St. Louis, MO)] at 4°C. Cells were then disrupted after addition of four volumes of acid-washed 0.45 mm diameter glass beads (Sigma-Aldrich, St. Louis, MO) by vortexing for 30 s followed by holding cells for 30 s on ice. The vortexing/holding steps were repeated five times or until 70–80% of the cells appeared disrupted by microscopic observation. Cell debris was removed by centrifugation at 12 000 g for 60 min at 4°C. The clear supernatant was collected and stored at −70°C until analysis. Protein concentration was determined by the Bradford assay (Bradford 1976) using a commercial kit and BSA as a standard (Bio-Rad Protein Assay, Bio-Rad Laboratories, Hercules, CA).
Acetyl-CoA synthetase assay
Acetyl-CoA synthetase activity was determined in stationary-phase cells grown in YNB-4.8 + 2% glu using a commercially available protocol (Sigma-Aldrich, St. Louis, MO) based on a coupled assay (Berg 1956) in which activity was monitored spectrophotometrically at 546 nm. Briefly, the standard reaction mixture (1.1 mL) contained 136 mM potassium phosphate (pH 7.5), 4 mM magnesium chloride, 9.1 mM ATP, 45 mM potassium fluoride, 9.1 mM potassium acetate, 9.1 mM reduced glutathione, 0.35 mM coenzyme A, 182 mM hydroxylamine and 0.1 mg protein extract or the equivalent volume of distilled water (control). The reaction was started upon addition of 9.1 mM potassium acetate. All chemicals were reagent grade (Sigma-Aldrich, St. Louis, MO). One unit of specific enzyme activity was defined as that which formed 1 μmol of acetyl coenzyme A from acetate, ATP and coenzyme A per mg protein, per minute at pH 7.5 at 37°C. Experiments were conducted in triplicate.
Determination of growth parameters
Growth rates and duration of lag phase were determined by measuring A600 values in aerobic shake flask cultures grown in YNB-4.8 + 2% glu (30 mL/250 mL flask) incubated at 30°C and 200 rpm with or without added acetic acid (n = 3). Lag phase was defined as the time that elapsed before exponential growth was detectable by A600 measurement as described (Xu et al., 1994). Cell yield (A600) was also determined in the presence vs. absence of acetic acid after 48 h under non-strict anaerobic conditions (Ding et al., 2013). Briefly, cells from 24 h aerobic cultures grown at 30°C in YNB-4.8 + 2% glu were harvested and washed twice. The washed cells were suspended in an equal volume of distilled water and used to inoculate YNB-4.8 + 2% glu containing acetic acid at a range of concentrations (0 to 200 mM) at a starting concentration of about 2 × 105 cells mL−1 in a final volume of 1 mL in 1.7 mL screw-capped polypropylene tubes. Cultures were incubated at 30°C and 200 rpm (n = 3). Cell growth was measured as A600 values after 48 h. Glucose supplementation experiments were performed by inoculating YNB-4.8 containing 4, 6 or 8% glucose with and without 140 mM acetic acid and culturing cells as described above.
Data analysis
The statistical significance of differences was determined using Student's two-tailed, paired t-test (Microsoft Excel, Redmond, WA).
RESULTS AND DISCUSSION
ACS2 overexpression correlates with increased Acs2 activity
In order to confirm that ACS2 overexpression increased Acs2 activity in the S288c his3Δ/pXP420-ACS2 strain, ACS2 expression level was measured and acetyl-CoA synthetase activity assayed in cells grown in YNB-4.8 + 2% glu. By RT-qPCR analysis, ACS2 expression was 4-fold higher in the constructed strain than in the empty vector control, 4.45 ± 0.57 vs. 1.00 ± 0.07, P < 0.001, n = 3. Consistent with the increase in gene expression, acetyl-CoA synthetase activity was also found to be 4-fold higher in the constructed strain, 0.14 ± 0.01 vs. 0.03 ± 0.01 specific units, P < 0.001, n = 3.
Two relevant studies have also quantified acetyl-CoA synthetase activity in yeast. Both assayed activity using a coupled reaction involving malate dehydrogenase and citrate synthase for which specific activity was defined in terms of acetate-dependent NADH formed stoichiometrically equivalent to the specific activity unit used here (Van den Berg et al., 1996; Remize, Andrieu and Dequin 2000). Van den Berg et al. (1996) quantified protein using the Lowry assay with an unstated protein standard and detected 0.08 units of activity in a homothallic diploid strain grown under glucose-limited anaerobic conditions. This was about three times more activity than detected here in stationary-phase cells of the haploid control strain. Remize, Andrieu and Dequin (2000) measured activity in a haploid wine strain derivative and in an otherwise isogenic strain overexpressing ACS2 (PGK1 promoter, high-copy [2μ] plasmid) grown at high glucose concentration (20% to mimic grape juice) and also used the Bradford protein assay and BSA standard as used here. Activity in the overexpression strain was about 5–7-fold higher than in the control strain during log phase, but as observed in the present study, was about 4-fold higher during stationary phase.
ACS2 overexpression increases acetic acid tolerance
To test if overexpression of ACS2 increased acetic acid resistance, growth rate and lag-phase duration were determined in the ACS2 overexpression and empty vector control strains grown in aerobic shake flask cultures as a function of added acetic acid (Table 3). In the absence of added acetic acid, overexpression of ACS2 increased lag phase by 30% (P < 10−5), but increased growth rate by 10% (P < 10−3). In the presence of 140 mM acetic acid, the overexpression strain had an approximately 2-fold decrease in lag phase, 4.6 vs. 10.4 h (P < 10−4) and a 25% increase in growth rate, 0.25 vs. 0.20 h−1 (P < 10−2). Because both strains grew poorly or not at all at ≥160 mM acetic acid, no attempt was made to determine either growth rate or lag-phase duration at these higher concentrations. As has been pointed out by others (Swinnen et al., 2014), an increased lag observed in cells subjected to a potentially lethal treatment, e.g. acetic acid shock reflects a combination of at least two responses: death of a subset of treated cells and a delay in growth of survivors. Measurement of lag phase by monitoring A600 values as done here cannot distinguish these two effects. Nonetheless, on the basis of a 2-fold shorter lag phase and a 25% increase in growth rate in the presence of 140 mM acetic acid, the ACS2 overexpression strain exhibited greater acetic acid resistance than the empty vector control strain.
Table 3.
Growth rate (μ) and lag-phase duration in aerobic shake flask culturea.
| S288c his3Δ/pXP420 | S288c his3Δ/pXP420-ACS2 | |||
|---|---|---|---|---|
| No acid | 140 mM acetic acid | No acid | 140 mM acetic acid | |
| lag (h) | 1.23 ± 0.12 | 10.37 ± 0.49 | 1.63 ± 0.06 | 4.63 ± 0.15 |
| μ (h−1) | 0.34 ± 0.01 | 0.20 ± 0.00 | 0.38 ± 0.00 | 0.25 ± 0.01 |
aCultures were grown in YNB-4.8 + 2% glucose (30 mL/250 mL flask, n = 3) at 30°C and 200 rpm in the presence or absence of added acetic acid as described in the section ‘Materials and Methods’.
An independent assessment of increased resistance was also undertaken based on growth (A600 values) after 48 h under non-strict anaerobic conditions (Table 4). In the absence of added acetic acid, the cell yield of the ACS2 overexpression and empty vector strains was the same in YNB-4.8 + 2% glu (A600 = 4.2 ± 0.5 vs. 4.1 ± 0.3, respectively). However, in the presence of 140 mM acetic acid, the cell yield of the overexpression strain was about 3-fold higher (A600 = 2.2 ± 0.1 vs. 0.8 ± 0.0, respectively, P < 10−5). At ≥160 mM acetic acid, growth (A600) of both strains after 48 h was insignificant, <2.5% of cell yields in the absence of acetic acid. Given the fact that acetic acid exposure results in ATP depletion in S. cerevisiae (Pampulha and Loureiro-Dias 2000; Ding et al., 2013) and that acetyl-CoA synthetase requires ATP to convert acetic acid to acetyl-CoA, we speculated that provision of more glucose during acetic acid treatment would help compensate for limiting ATP. Similarly, supplying the control strain with more glucose was also expected to increase its tolerance for acetic acid. Contrary to expectation, glucose supplementation beyond the 2% standard concentration in YNB-4.8 medium did not increase acetic acid resistance in either strain. In the absence of acetic acid, cell yields increased in response to the glucose supplementation for both strains, although the differences in yield per strain at 4, 6 and 8% glucose were statistically equivalent, suggesting that a nutrient other than glucose became growth limiting at ≥4% glucose. For the ACS2 overexpression strain, cell yields in the presence of 140 mM acetic acid were the same at 2, 4 and 8% glucose, but decreased about 10% (A600 = 2.2 to 2.0, P < 0.02) at 6% glucose relative to 2% glucose. For the empty vector control strain grown in the presence of 140 mM acetic acid, cell yields were the same at 2 and 4% glucose, but decreased significantly with increasing glucose addition: A600 = 0.8 at 2 and 4% glucose vs. 0.3 at 6% glucose (P < 10−5), vs. 0.1 at 8% glucose (P < 10−5). In sum, the additional glucose had either no effect or a minor negative impact on acetic acid response in the ACS2 overexpression strain, but increased the sensitivity of the empty vector control strain.
Table 4.
Growth as a function of glucose and acetic acid supplementation under non-strict anaerobic conditionsa.
| S288c his3Δ/pXP420 | S288c his3Δ/pXP420-ACS2 | |||
|---|---|---|---|---|
| A600 | ||||
| Glucose (%) | No acid | 140 mM acetic acid | No acid | 140 mM acetic acid |
| 2 | 4.13 ± 0.26 | 0.77 ± 0.02 | 4.20 ± 0.48 | 2.21 ± 0.07 |
| 4 | 6.12 ± 0.51 | 0.77 ± 0.08 | 5.69 ± 0.53 | 2.26 ± 0.22 |
| 6 | 5.54 ± 0.34 | 0.25 ± 0.01 | 6.29 ± 0.45 | 2.03 ± 0.03 |
| 8 | 5.95 ± 0.79 | 0.07 ± 0.02 | 6.60 ± 0.58 | 2.40 ± 0.14 |
aCultures were grown in YNB-4.8 + the indicated amount of glucose for 48 h at 30°C and 200 rpm in the presence or absence of added acetic acid in screw-capped 1.7 mL tubes (n = 3) as described in the section ‘Materials and Methods’.
Reducing the negative effect of lignocellulose-derived fermentation inhibitors such as acetic acid on the ethanol productivity of S. cerevisiae is expected to contribute to the development of a plant biomass-based source of renewable transportation fuels. Compared to physical and chemical methods of detoxification (e.g. absorbents, application of heat under vacuum to distill undissociated acetic acid), biological methods focusing on the in situ metabolism of acetic acid are relatively inexpensive to implement at the process level. The major investment is made prior to deployment—in the isolation or construction and development of such mutants. Here, we found that overexpression of an ATP-dependent, acetic acid-utilizing enzyme, Acs2, that is active under fermentation conditions in yeast, resulted in a shortened lag phase, an increased growth rate and an increase in growth yield in the presence of 140 mM acetic acid, but not at ≥160 mM. The net effect of glucose supplementation as a source of additional ATP was not found to compensate for the presumed increase in ATP demand. Nonetheless, ACS2 overexpression provided a moderate increase in resistance that might be coupled with other genetic modifications to provide production strains of yeast with greater tolerance.
What other genetic modifications that increase acetic acid tolerance are potential candidates for coupling with overexpression of ACS2? Table 5 lists mutations identified through directed studies, some of which have also been identified in genome-wide screens (Kawahata et al., 2006; Ding et al., 2013). Because acetic acid has been shown to inhibit nutrient uptake, auxotrophy itself has been found to increase yeast sensitivity to acetic acid-mediated stress (Ding et al., 2013). Hence, the genetic background of the various strains is provided as well. This is important because many genetic modifications that increase acetic acid tolerance in auxotrophic strains are involved in the response to nutritional starvation including downregulation of nutrient transporter turnover. Notably, mutations in this category that have been tested have not been found to increase tolerance in a prototrophic, but otherwise isogenic strain (Ding et al., 2013). Half of the genetic modifications listed in Table 5 (13 of 24) reflect this bias and are linked to overcoming acetic acid-mediated nutritional starvation (BAP2, GCN2, GCN4, GLC7, HIS3, LEU2, LYS2, PPH21, PPH22, SIT4, TOR1, TRP1, URA3). Other genetic modifications that increase acetic acid tolerance include three that interfere with apoptosis (ATP10, CTT1, CYC3), one that enhances V-ATPase activity (PEP3), two that modify histone proteins (HHF2, HHT2) and two that reduce acetic acid uptake (HAA1, FPS1). Medina et al. (2010) and Wei et al. (2013) have also described promising genetic modifications that increase ethanol yields via reactions that reduce acetyl-CoA and simultaneously consume acetic acid, and presumably increase tolerance for acetic acid.
Table 5.
Genetic modifications reported in non-comprehensive screens of S. cerevisiae mutants that increase acetic acid tolerance.
| Target gene | Function | Modification | Strain backgrounda | Reference |
|---|---|---|---|---|
| ACS2 | Acetyl-CoA synthetase | Overexpression | S288c | This study |
| ATP10 | Mitochondrial ATPase | Deletion | W303-1A | Ludovico et al. (2002) |
| BAP2 | Leucine transporter | Complementation of deletion allele | BY4741 | Hueso et al. (2012) |
| CTT1 | Cytosolic catalase T | Overexpression | W303-1B | Guaragnella et al. (2008) |
| CYC3 | Cytochrome c | Deletion | W303-1A | Ludovico et al. (2002) |
| FPS1 | Plasma membrane aquaglyceroporin | Deletion | BY4741 | Mollapour and Piper (2007) |
| GCN2 | Protein kinase phosphorylates eIF2 | Deletion | BY4742 | Almeida et al. (2009) |
| GCN4 | Transcriptional activator of amino acid biosynthetic genes | Deletion | BY4742 | Almeida et al. (2009) |
| GLC7′ | Type 1 ser/thr protein phosphatase | Overexpression of dominant negative allele | BWG1-7A | Hueso et al. (2012) |
| HAA1 | Transcriptional activator | Overexpression | S288c | Tanaka et al. (2012) |
| HHF2 | Histone 4 | Point mutation | H3 WT | Liu, Zhang and Zhang (2014) |
| HHT2 | Histone 3 | Point mutation | H3 WT | Liu, Zhang and Zhang (2014) |
| HIS3 | Histidine biosynthesis | Complementation of deletion allele | S288c | Ding et al. (2013) |
| LEU2 | Leucine biosynthesis | Complementation of deletion allele | BWG1-7A | Hueso et al. (2012) |
| LEU2 | Leucine biosynthesis | Complementation of deletion allele | S288c | Ding et al. (2013) |
| LYS2 | Lysine biosynthesis | Complementation of deletion allele | S288c | Ding et al. (2013) |
| PEP3 | CORVET and HOPS complexes | Overexpression | S288c | unpublished data, Ding J, Holzwarth G, Bakalinsky AT (2014) |
| PPH21 | Protein phosphatase 2A subunit | Deletion | BY4742 | Almeida et al. (2009) |
| PPH22 | Protein phosphatase 2A subunit | Deletion | BY4742 | Almeida et al. (2009) |
| SIT4 | Protein phosphatase 2A-related ser/thr phosphatase | Deletion | BY4742 | Almeida et al. (2009) |
| TOR1 | Kinase and rapamycin target | Deletion | BY4742 | Almeida et al. (2009) |
| TRP1 | Tryptophan biosynthesis | Complementation of deletion allele | S288c | Ding et al. (2013) |
| URA3 | Uracil biosynthesis | Complementation of deletion allele | S288c | Ding et al. (2013) |
aBY4741: MATa his3Δ0 leu2Δ0 met15Δ0 ura3Δ0; BY4742: MATa his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0; BWG1-7A: MATa ade1-100 his4-519 ura3-52 leu2-3,112; H3 WT: MATa his3Δ200 leu2Δ0 lys2Δ0 trp1Δ63 ura3Δ0 met15Δ0 can1::MFA1pr-HIS3 hht1-hhf1::NatMX4 hht2-hhf2::[HHTS-HHFS]*-URA3 (Baranwal et al., 2014); S288c: MATα SUC2 gal2 mal2 mel flo1 flo8-1 hap1 ho bio1 bio6; W303-1A: MATa ade2 leu2 his3 trp1 ura3; W303-1B: MATa ade2 leu2 his3 trp1 ura3.
Combining ACS2 overexpression with candidate genetic modifications that do not involve nutritional starvation in a prototrophic strain is a reasonable approach for further increasing acetic acid resistance. Clearly, the availability of libraries of deletion or overexpression mutants in appropriate industrial strain backgrounds would provide a much-needed resource to facilitate the development of acetic acid resistance and related traits of industrial importance. The recently described prototrophic version of the S288c-based collection of deletion mutants is a promising start (Mülledar et al., 2012).
FUNDING
This work was funded by grant no. 2010-5504-20345 from the United States Department of Agriculture National Institute of Food and Agriculture (USDA-NIFA) to ATB and MHP and by grant no. R15GM104876 from the United States Department of Health and Human Services National Institutes of Health (NIH) to JP-V.
Conflict of interest statement. None declared.
REFERENCES
- Aden A, Ruth M, Ibsen K, et al. Technical report NREL/TP-510-32438. 2002. Lignocellulosic biomass to ethanol process design and economics utilizing co-current dilute acid prehydrolysis and enzymatic hydrolysis for corn stover. http://www.nrel.gov/docs/fy02osti/32438.pdf. [Google Scholar]
- Almeida B, Ohlmeier S, Almeida AJ, et al. Yeast protein expression profile during acetic acid-induced apoptosis indicates causal involvement of the TOR pathway. Proteomics. 2009;9:720–32. doi: 10.1002/pmic.200700816. [DOI] [PubMed] [Google Scholar]
- Baranwal S, Azad GK, Singh V, et al. Signaling of chloroquine-induced stress in the yeast Saccharomyces cerevisiae requires the Hog1 and Slt2 mitogen-activated protein kinase pathways. Antimicrob Agents Ch. 2014;58:5552–66. doi: 10.1128/AAC.02393-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg P. Acyl adenylates: an enzymatic mechanism of acetate activation. J Biol Chem. 1956;222:991–1013. [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chen Y, Daviet L, Schalk M, et al. Establishing a platform cell factory through engineering of yeast acetyl-CoA metabolism. Metab Eng. 2013;15:48–54. doi: 10.1016/j.ymben.2012.11.002. [DOI] [PubMed] [Google Scholar]
- Chen F, Zhou J, Shi Z, et al. Effect of acetyl-CoA synthase gene overexpression on physiological function of Saccharomyces cerevisiae. Acta Microbiol Sin. 2010;50:1172–9. [PubMed] [Google Scholar]
- De Jong-Gubbels P, van den Berg MA, Luttik MA, et al. Overproduction of acetyl-coenzyme A synthetase isoenzymes in respiring Saccharomyces cerevisiae cells does not reduce acetate production after exposure to glucose excess. FEMS Microbiol Lett. 1998;165:15–20. doi: 10.1111/j.1574-6968.1998.tb13121.x. [DOI] [PubMed] [Google Scholar]
- De Virgilio C, Bürckert N, Barth G, et al. Cloning and disruption of a gene required for growth on acetate but not on ethanol: the acetyl-coenzyme A synthetase gene of Saccharomyces cerevisiae. Yeast. 1992;8:1043–51. doi: 10.1002/yea.320081207. [DOI] [PubMed] [Google Scholar]
- Del Río JC, Marques G, Rencoret J, et al. Occurrence of naturally acetylated lignin units. J Agr Food Chem. 2007;55:5461–8. doi: 10.1021/jf0705264. [DOI] [PubMed] [Google Scholar]
- Ding J, Bierma J, Smith MR, et al. Acetic acid inhibits nutrient uptake in Saccharomyces cerevisiae: auxotrophy confounds the use of yeast deletion libraries for strain improvement. Appl Microbiol Biot. 2013;97:7405–16. doi: 10.1007/s00253-013-5071-y. [DOI] [PubMed] [Google Scholar]
- Fang F, Salmon K, Shen MWY, et al. A vector set for systematic metabolic engineering in Saccharomyces cerevisiae. Yeast. 2011;28:123–36. doi: 10.1002/yea.1824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frenkel EP, Kitchens RL. Purification and properties of acetyl coenzyme A synthetase from bakers’ yeast. J Biol Chem. 1977;252:504–7. [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, et al. Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast. 1995;11:355–60. doi: 10.1002/yea.320110408. [DOI] [PubMed] [Google Scholar]
- Guaragnella N, Antonacci L, Giannattasio S, et al. Catalase T and Cu, Zn-superoxide dismutase in the acetic acid-induced programmed cell death in Saccharomyces cerevisiae. FEBS Lett. 2008;582:210–4. doi: 10.1016/j.febslet.2007.12.007. [DOI] [PubMed] [Google Scholar]
- Hueso G, Aparicio-Sanchis R, Montesinos C, et al. A novel role for protein kinase Gcn2 in yeast tolerance to intracellular acid stress. Biochem J. 2012;441:255–64. doi: 10.1042/BJ20111264. [DOI] [PubMed] [Google Scholar]
- Kawahata M, Masaki K, Fujii T, et al. Yeast genes involved in response to lactic acid and acetic acid: acidic conditions caused by the organic acids in Saccharomyces cerevisiae cultures induce expression of intracellular metal metabolism genes regulated by Aft1p. FEMS Yeast Res. 2006;6:924–36. doi: 10.1111/j.1567-1364.2006.00089.x. [DOI] [PubMed] [Google Scholar]
- Klinke HB, Thomsen AB, Ahring BK. Inhibition of ethanol-producing yeast and bacteria by degradation products produced during pre-treatment of biomass. Appl Microbiol Biot. 2004;66:10–26. doi: 10.1007/s00253-004-1642-2. [DOI] [PubMed] [Google Scholar]
- Kozak BU, van Rossum HM, Luttik MAH, et al. Engineering acetyl coenzyme A supply: functional expression of a bacterial pyruvate dehydrogenase complex in the cytosol of Saccharomyces cerevisiae. mBio. 2014;5:e01696–14. doi: 10.1128/mBio.01696-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Zhang X, Zhang Z. Point mutation of H3/H4 histones affects acetic acid tolerance in Saccharomyces cerevisiae. J Biotechnol. 2014;187:116–23. doi: 10.1016/j.jbiotec.2014.07.445. [DOI] [PubMed] [Google Scholar]
- Ludovico P, Rodrigues F, Almeida A, et al. Cytochrome c release and mitochondria involvement in programmed cell death induced by acetic acid in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:2598–606. doi: 10.1091/mbc.E01-12-0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina VG, Almering MJH, van Maris AJA, et al. Elimination of glycerol production in anaerobic cultures of a Saccharomyces cerevisiae strain engineered to use acetic acid as an electron acceptor. Appl Environ Microb. 2010;76:190–5. doi: 10.1128/AEM.01772-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollapour M, Piper PW. Hog1 mitogen-activated protein kinase phosphorylation targets the yeast Fps1 aquaglyceroporin for endocytosis, thereby rendering cells resistant to acetic acid. Mol Cell Biol. 2007;27:6446–56. doi: 10.1128/MCB.02205-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mülledar M, Capuoano F, Pir P, et al. A prototrophic deletion mutant collection for yeast metabolomics and systems biology. Nature Biotechnol. 2012;30:1176–8. doi: 10.1038/nbt.2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pampulha ME, Loureiro-Dias MC. Energetics of the effect of acetic acid on growth of Saccharomyces cerevisiae. FEMS Microbiol Lett. 2000;184:69–72. doi: 10.1111/j.1574-6968.2000.tb08992.x. [DOI] [PubMed] [Google Scholar]
- Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remize F, Andrieu E, Dequin S. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae: role of the cytosolic Mg2+ and mitochondrial K+ acetaldehyde dehydrogenases Ald6p and Ald4p in acetate formation during alcoholic fermentation. Appl Environ Microb. 2000;66:3151–9. doi: 10.1128/aem.66.8.3151-3159.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runguphan W, Keasling JD. Metabolic engineering of Saccharomyces cerevisiae for production of fatty acid-derived biofuels and chemicals. Metab Eng. 2014;21:103–13. doi: 10.1016/j.ymben.2013.07.003. [DOI] [PubMed] [Google Scholar]
- Satyanarayana T, Klein HP. Studies on acetyl-Coenzyme A synthetase of yeast: inhibition by long-chain acyl-Coenzyme A esters. J Bacteriol. 1973;115:600–6. doi: 10.1128/jb.115.2.600-606.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satyanarayana T, Mandel AD, Klein HP. Evidence for two immunologically distinct acetyl-co-enzyme A synthetase in yeast. Biochim Biophys Acta. 1974;341:396–401. doi: 10.1016/0005-2744(74)90232-0. [DOI] [PubMed] [Google Scholar]
- Scalcinati G, Partow S, Siewers V, et al. Combined metabolic engineering of precursor and co-factor supply to increase α-santalene production by Saccharomyces cerevisiae. Microb Cell Fact. 2012;11:117. doi: 10.1186/1475-2859-11-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiba Y, Paradise EM, Kirby J, et al. Engineering of the pyruvate dehydrogenase bypass in Saccharomyces cerevisiae for high-level production of isoprenoids. Metab Eng. 2007;9:160–8. doi: 10.1016/j.ymben.2006.10.005. [DOI] [PubMed] [Google Scholar]
- Sierkstra LN, Verbakel JM, Verrips CT. Analysis of transcription and translation of glycolytic enzymes in glucose-limited continuous cultures of Saccharomyces cerevisiae. J Gen Microbiol. 1992;138:2559–66. doi: 10.1099/00221287-138-12-2559. [DOI] [PubMed] [Google Scholar]
- Swinnen S, Fernández Niño M, González-Ramos D, et al. The fraction of cells that resume growth after acetic acid addition is a strain-dependent parameter of acetic acid tolerance in Saccharomyces cerevisiae. FEMS Yeast Res. 2014;14:642–53. doi: 10.1111/1567-1364.12151. [DOI] [PubMed] [Google Scholar]
- Tanaka K, Ishii Y, Ogawa J, et al. Enhancement of acetic acid tolerance in Saccharomyces cerevisiae by overexpression of the HAA1 gene, encoding a transcriptional activator. Appl Environ Microb. 2012;78:8161–3. doi: 10.1128/AEM.02356-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van den Berg MA, Jong-Gubbels Pde, Kortland CJ, et al. The two acetyl-coenzyme a synthetases of Saccharomyces cerevisiae differ with respect to kinetic properties and transcriptional regulation. J Biol Chem. 1996;271:28953–9. doi: 10.1074/jbc.271.46.28953. [DOI] [PubMed] [Google Scholar]
- Van den Berg MA, Steensma HY. ACS2, a Saccharomyces cerevisiae gene encoding acetyl-coenzyme A synthetase, essential for growth on glucose. Eur J Biochem. 1995;231:704–13. doi: 10.1111/j.1432-1033.1995.tb20751.x. [DOI] [PubMed] [Google Scholar]
- Wei N, Quarterman J, Kim SR, et al. Enhanced biofuel production through coupled acetic acid and xylose consumption by engineered yeast. Nat Commun. 2013;4:2580. doi: 10.1038/ncomms3580. [DOI] [PubMed] [Google Scholar]
- Xu X, Wightman JD, Geller BL, et al. Isolation and characterization of sulfite mutants of Saccharomyces cerevisiae. Curr Genet. 1994;25:488–96. doi: 10.1007/BF00351667. [DOI] [PubMed] [Google Scholar]