Abstract
A single cycle of strain improvement was performed in Saccharopolyspora erythraea mutB and 15 genotypes influencing erythromycin production were found. Genotypes generated by transposon mutagenesis appeared in the screen at a frequency of ∼3%. Mutations affecting central metabolism and regulatory genes were found, as well as hydrolases, peptidases, glycosyl transferases and unknown genes. Only one mutant retained high erythromycin production when scaled-up from micro-agar plug fermentations to shake flasks. This mutant had a knockout of the cwh1 gene (SACE_1598), encoding a cell-wall-associated hydrolase. The cwh1 knockout produced visible growth and morphological defects on solid medium. This study demonstrated that random transposon mutagenesis uncovers strain improvement-related genes potentially useful for strain engineering.
Keywords: transposon, strain improvement, Actinomyces, directed evolution, erythromycin
A gene influencing the production level of the antibiotic erythromycin was uncovered using a random transposon mutagenesis procedure.
Graphical Abstract Figure.
A gene influencing the production level of the antibiotic erythromycin was uncovered using a random transposon mutagenesis procedure.
INTRODUCTION
Commercial production of antimicrobial products by large-scale submerged fermentation began with penicillin in the 1940s. Since then, hundreds of natural products have been produced for medicine and agriculture including antibiotics, anti-cancer agents, immunosuppressants and active pharmaceutical ingredients (Bérdy 2005). When microorganisms from the soil are transferred to the laboratory, they tend to produce only trace amounts of the active compound. Therefore, strain improvement is often a necessary step of the commercialization process (Vinci and Byng 1999; Demain and Adrio 2008).
Traditional strain improvement that leads to improved production levels is an empirical stepwise process performed through multiple labor-intensive cycles of random mutation and screening using higher antibiotic production as the selective criterion. It requires only basic microbiological methods (Demain and Adrio 2008).
Typically the strongest positive mutation steps are found during the first few cycles of the process followed by cycles that, despite a larger screening effort, produce smaller increases in yield (Barrick and Lenski 2013). The underlying genetics of strain improvement were never characterized during the ‘golden age’ when the first commercial antibiotic-producing strains were generated, but genomic technology today allows for the reverse engineering of the process, opening it up to scientific study. Thus, high-producing strains resulting from multiple cycles of mutation and selection have been compared to wild-type reference strains (Peano et al. 2012; Li et al. 2013). And randomly generated yield improvement mutations have been reverse engineered, advancing knowledge-based engineering of yield (Gehring et al. 2000; Santos and Stephanopoulos 2008).
For this project, in vitro transposition (Goryshin and Reznikoff 1998) was used to generate random mutations. Transposition creates single mutations that can be identified and mapped by plasmid rescue and DNA sequencing, generating a direct link between the higher yield (phenotype) and the transposon insertion (genotype). Once the genotype is known, it can be incorporated into different genetic backgrounds. Experiments were performed directly in a Saccharopolyspora erythraea mutB strain (FL2302), circumventing the need to use Aeromicrobium erythreum as a model host as was done in the past (Reeves et al. 2004). A single round of random transposon mutagenesis was used on a mutB strain of S. erythraea (FL2302) and a sample was knocked out comprising ∼7% of the genes in the chromosome. The mutants were screened by static micro-agar plug fermentations for changes in erythromycin yield and 15 mutants of interest were found. DNA sequence analysis revealed the site of insertions of the transposon. The function of the mutant genes was queried using the BLASTP algorithm. One of the mutant strains was scaled-up to flask fermentations making it a strong candidate for further study.
MATERIALS AND METHODS
Bacterial strains and growth conditions
General materials and methods are described in Kieser et al. (2000) and Sambrook, Fritsch and Maniatis (1989). Saccharopolyspora erythraea mutB FL2302 (Reeves et al. 2006) is a derivative of S. erythraea FL2267 which was obtained from a lyophilized vial of S. erythraea ATCC 11635. Saccharopolyspora erythraea FL2267 was the source of the genomic DNA used in the formation of Library 1. For more detail, see Supplementary Materials and Methods.
Construction of plasmid pFL2073
A previously described cloning vector, pFL8 (Reeves et al. 2002; NCBI sequence accession: BankIt1812822 KR061299), was doubly digested with endonucleases BglII and BssHII; the short single-stranded ends were blunted and ligated to delete the neo (neomycin/kanamycin resistance) gene thus creating pFL2073 (Fig. 1). This plasmid has both Streptomyces and Escherichia coli origins of replication (pIJ101 and pBR322, respectively), the thiostrepton (tsr) and ampicillin (amp) resistance genes and the lacZ gene with a multicloning site for blue/white screening on X-gal-containing media. Plasmid pFL2073 was used as the parent plasmid for the generation of Library 1. Despite the fact that pFL2073 is a high copy autonomously replicating plasmid in Streptomyces lividans, it functions as an integrative plasmid in S. erythraea. When pFL2073 contains S. erythraea DNA clones, it integrates into the S. erythraea chromosome by homologous recombination (if the fragment is >0.4 kb) and can be readily used for single-crossover integration to knock out genes, or it can be used for double-crossover gene replacements (Weber et al. 1990) which is how it was used in this study to deliver transposon mutations into the chromosome.
Figure 1.
Map of plasmid pFL2073. Enzymes with unique restriction endonuclease sites are shown (except for PstI whose three sites are shown). Abbreviations: amp, ampicillin resistance gene; ori, origin of replication; lacZ, beta-galactosidase gene; f1, f1 phage; tsr, thiostrepton resistance gene; pIJ101, Streptomyces plasmid; rep, replicon.
Library 1: DNA cloning
Saccharopolyspora erythraea chromosomal DNA was partially digested into ∼10–15 Kb fragments and ligated into pFL2073 (Fig. 1). Ligation mixtures were transformed into E. coli DH5α by electroporation. A high-purity DNA preparation was generated from transformants. Plasmids carrying fragments larger than 9 kb and with similar copy numbers were combined in pools of approximately 20 plasmids each. Eleven different plasmid pools were used in the in vitro transposition reactions to create Library 2. For more detail, see Supplementary Materials and Methods.
Library 2: transposon mutagenesis
Eleven plasmid pools from Library 1 were mutagenized in vitro using the EZ-Tn5 <R6Kγori/Kan-2> kit from Epicentre Biotechnologies (Madison, WI). The mutagenized plasmid reaction mixture was used to transform competent E. coli cells, the cells were plated on LB and selected for ampicillin and kanamycin-resistant transformants. Primary transformants were harvested and high-purity DNA was prepared from them to create Library 3. For more detail, see Supplementary Materials and Methods.
Library 3: S. erythraea transformation and DNA replacement
Library 3 was generated by transforming plasmid DNA from Library 2 into protoplasts of S. erythraea mutB FL2302 using a modification of the method of Weber and Losick (1988). Eight highly transformable protoplast preparations were transformed with DNA from different pools of Library 2. Selection was for thiostepton resistance. Spores were harvested from confluent lawns of S. erythraea, diluted and plated for single colonies then replica plated onto agar media to determine their kanamycin- and thiostrepton-resistance phenotypes. Kanamycin-resistant and thiostrepton-sensitive colonies were chosen. Mutants were analyzed by plasmid rescue, taking advantage of the R6Kγ origin of replication present in the transposon, followed by DNA sequence analysis to confirm the randomness of the library. In S. erythraea, pIJ101-derived plasmids such as pFL2073 integrate into the chromosome via homologous recombination when they carry an S. erythraea genomic fragment >0.4 kb. The transposon with neighboring S. erythraea DNA is twice delivered into the genome by double-crossover gene replacement; no plasmid DNA is permanently incorporated into the genome by this procedure (Weber et al. 1990).
Transposon insertions made by double crossovers in the chromosome (kanr and thios) were found in ∼5–10% of the spores from these confluent lawns. This created a library of S. erythraea mutant strains (Library 3) which was then screened for increased or decreased erythromycin production. The remaining spores were made up of single-crossover plasmid insertions (∼50%, thior and kanr) or spores with no plasmid or transposon DNA (∼45%, thios and kans). These were not used further.
Library 3 (composed of 1048 mutant strains made by double-crossover recombination of the mutagenized plasmids from Library 2) was screened by micro-agar fermentation twice. Since half of the mutants in Library 3 were estimated to be mutants that shared a common parent, ∼524 genes (or 7% of the total in the genome) were knocked out in this study.
Identification of S. erythraea sequences flanking transposon insertions
This method has been described previously (Reeves and Weber 2012). Briefly, chromosomal DNA was prepared from the high-producing strain and digested with a frequent cutting endonuclease enzyme that did not cut within the transposon. The digested DNA was ligated to create circular DNA from individual fragments and electroporated with E. coli and plated on LB agar with kanamycin (40 μg ml−1) and the oriV inducer. The resulting Kanr colonies were grown for isolation of plasmid DNA which was subjected to DNA sequence analysis using the primers at the ends of the transposon.
Micro-agar fermentations and erythromycin titer determinations
The wells of a flat-bottomed 96-well microtiter plate were partially filled with 250 μl of E20A agar. Spores from mutant and control strains were inoculated into the center of the micro-agar plugs with sterile toothpicks. Each mutant was screened in duplicate. The plates were incubated for 5 days at 32°C under controlled humidity. After 5 days, the micro-agar plugs were transferred to the top of 20 cm agar plates seeded with the sensitive indicator organism, Bacillus subtilis. After overnight incubation at 32°C, the erythromycin titer determination plates showed zones of growth inhibition around the base of the micro-agar plugs in proportion to the amount of erythromycin being made. The inhibition zone diameters were measured and the amount of erythromycin produced was calculated compared to an erythromycin A reference standard. Methods for the determination of erythromycin yields in liquid culture broths have been described previously (Reeves et al. 2006).
Shake flask fermentation screen
Mutant cwh1 was tested in 250-ml shake flask cultures containing 25 ml of OFM1 medium (Reeves et al. 2006). Mutant number S6.07–03 and control FL2302 were compared in triplicate cultures; mutant S6.07–06 and control FL2302 were compared in duplicate fermentations (Fig. 2C).
Figure 2.

(A) Micro-agar plug fermentation screen. Erythromycin production levels of 15 transposon-insertion mutants are compared to the parent strain FL2302 (P). The dotted lines indicate the base-line level of erythromycin production by the parental strain; brackets represent standard deviation, n = 2. (B) The repeated microgel fermentation screen; brackets represent standard deviation, n = 2. (C) Scale-up from micro-agar plug to shake flask fermentation results for cwh1 (SACE_1598) mutants. Cultures were grown in 25-ml of OFM1 (oil-based medium) in 250-ml shake flasks. Bar numbers 1 and 3 represent parental strain FL2302; bar numbers 2 and 4 represent cwh1 mutant strains, S6.07–03 and S6.07–06, respectively.
RESULTS
A collection of mutants from a single cycle of strain improvement
Thirty-five mutants influencing erythromycin production were obtained from the screening of 1048 transposon-generated mutants of S. erythraea representing ∼7% of the genes in the genome. DNA sequence analysis of the transposon insertion sites revealed 15 unique genotypes; siblings and multiple mutations in the same gene accounted for the duplicate genotypes. Of the 15 knockout mutant strains found, 13 showed a >25% improved yield and 1 genotype had reduced yield and 1 genotype was neutral but showed reduced yield upon later scale-up analysis (Fig. 2). The mutants from the first screen showed mean increases in erythromycin yield of 34%–109%.
DNA sequence analysis revealed 15 gene targets affecting erythromycin production with transposons falling into coding regions in 13 cases, and promoter regions in 2 cases; nucleotide numbers of the transposon insertion sites are given (Table 1). Nine mutants showed sporulation or pigmentation defects in addition to influencing erythromycin production. The 15 targeted genes fell into six general functional categories determined by BLASTP analysis: transcriptional regulators (acrR and rho1), cell wall biogenesis (cps2I, cwh1 and possibly gtf1), hydrolases (tsp1, dppII and cwh1), metabolism, (citA4, hpcH, fabG, cysH, arsC), antibiotic biosynthesis (eryAII and possibly gtf1) and unknown (unk1597) (Table 1). Tn5 insertion into the eryAII gene produced the expected complete blockage in erythromycin production. A neutral phenotype was produced by insertion into the cccA gene coding for a cytochrome-c related protein involved in energy production; however, in shake flasks this mutant showed reduced erythromycin yield (data not shown).
Table 1.
Transposon mutations from this study influencing erythromycin biosynthesis in S. erythraea.
| Gene | Mutant # | Transposon nucleotide (nt) insertion site | Ery1 | Spo2 | Pig3 | Predicted Function (reference where known) |
|---|---|---|---|---|---|---|
| FL2302 | Control | + | + | Parental strain control | ||
| acrR | S6.18–36 | In SACE_0303; 339,794 | + | + | + | acrR, regulator of multidrug efflux pump (Wu et al. 2014) |
| citA4 | S6.18–32 | In SACE_0632; 696,384 | + | + | + | citA4 citrate synthase (Viollier et al. 2001) |
| hpcH | S7.11–58 | In SACE_0699; 769,528 | + | – | – | hpcH 2,4-dihydroxyhept-2-ene-1,7-dioic acid aldolase |
| fabG | S6.07–125 | In SACE_0700; 770,333 | + | – | – | fabG, short-chain dehydrogenase/reductase family |
| eryAII | S5.17–06 | In SACE_0723; 802,912 | – | – | – | eryAII erythromycin polyketide synthase |
| cysH | S6.18–04 | In SACE_1474; 1,626,645 | + | + | + | cysH, sulfate adenylyltransferase subunit 2 |
| unk1597 | S6.07–72 | In SACE_1597; 1,752,052 | + | – | +/– | unk1597, hypotheical protein, polar effects expected on SACE_1598 |
| cwh1 | S6.07–03 | In SACE_1598; 1,752,509 | + | – | +/– | cwh1, cell-wall-associated hydrolase, NlpC/P60 superfamily (Anantharaman and Aravind 2003) |
| cccA | S5.17–12 | In SACE_1685; 1,852,856 | – | – | – | cccA - cytochrome c mono-and diheme variants |
| gtf1 | S7.05–53 | Upstream of SACE_2010; 2,197,132 | + | – | – | gtf1, glycosyltransferase- GT1_Gtf_like family (Liang and Qiao 2007) |
| cps2I | S6.15–149 | In SACE_3177; 3,507,747 | + | bld | + | cps2I, nucleotide-sugar-dependent glycosyltransferase, group 1 (Lakkitjaroen et al. 2014) |
| arsC | S6.18–12 | In SACE_5143; 5,750,836 | + | – | – | arsC, arsenical resistance protein, arsenate reductase, arsenic transporter (EC 1.20.4.1) |
| tsp1 | S6.18–17 | In SACE_5967; 6,700,519 | + | + | + | tsp1, secreted trypsin-like serine protease |
| rho1 | S7.12–136 | In SACE_6295; 7,045,657 | + | + | + | rho1, rho1-like transcription terminator (Cardinale et al. 2008) |
| dppII | S6.15–143 | Upstream of SACE_6505; 7,295,927 | + | + | + | dppII, X-Pro dipeptidyl-peptidase (Maes, Scharpé and De Meester 2007) |
Erythromycin production phenotype; increased production +, reduced production –, compared to parent strain FL2302.
Sporulation phenotype. +, wild type sporulation; –, makes aerial mycelium but no spores; bld, makes no aerial mycelium or spores.
Pigmentation. +, normal red pigmentation; –, no red pigmentation; +/–, reduced red pigmentation.
The transposon insertions were mapped broadly around the S. erythraea chromosome (Fig. 3A). Insertions influencing erythromycin production were most frequently found in the upper-half ‘core-metabolism’ region of the genome (0–2.5 and 5.5–8.0 Mb) and less frequently in the lower half ‘non-core’ region (2.5–5.5 Mb) (Oliynyk et al. 2007). Sibling mutants were found for six of the genes (acrR, fabG, eryAII, cwh1, gtf1 and rho1) and three insertion events occurred into the cell wall hydolase gene cwh1 (Fig. 3B). For 11 of the 15 genes in the collection, the closest NCBI GenBank homologs were found in S. spinosa.
Figure 3.
(A) Genome map of S. erythraea showing transposon insertion sites of Library 3 mutants. Map is based on data generated by Oliynyk et al. (2007). Knockouts of genes highlighted in green gave increases in yield, and red shading is for decreases in yield. Map positions are shown in megabase pairs. (B) Map of the cwh1 (SACE_1598) region of the S. erythraea genome (Oliynyk et al. 2007). Transposon insertion and orientation are indicated by directional flags. Numbers above the flags refer to mutant numbers; all mutant numbers shown are from the S6.07 pool, for example, ‘03’ indicates mutant number S6.07–03. Map positions are shown in megabase pairs.
Interaction between genotype and environment
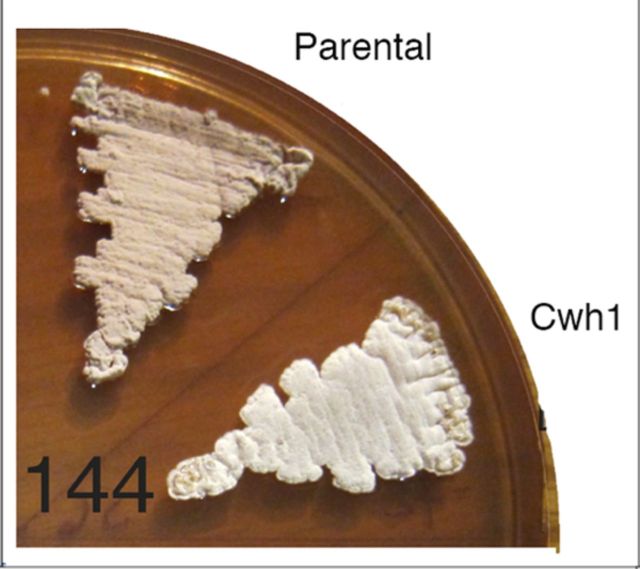
The fermentation growth environment determines which phenotypes can be scaled-up and which cannot. Genotypes influencing increased erythromycin production are potentially commercially viable. In this study, only one of the genotypes, mutant cwh1, showed increased erythromycin production after scale-up in shake flasks (Fig. 2C). Two independent cwh1 insertion mutants were tested: mutant S6.07–03 whose insertion was near the middle of the gene and mutant S6.07–06 whose insertion was near the 5′ end of the gene (Fig. 3B). Both mutant strains showed statistically significant increases in erythromycin production (Fig. 2C). Upstream of the cwh1 gene one insertion (‘72’, Fig. 3B) showed increased erythromycin production similar to that with insertions in cwh1; downstream of cwh1 one insertion (‘14’) was found that had a neutral phenotype—evidence that the Cwh1 phenotype is not due to an effect on the downstream gene. Based on DNA sequence data, Cwh1 is predicted to be cell-wall associated and to hydrolyze cell walls (see Supplementary Results and Fig. S1, Supporting Information). Therefore, it was not surprising that the Cwh1 phenotype included a growth defect (Fig. 4A) and non-sporulation (Fig. 4B) on solid medium.
Figure 4.
The parental and Cwh1 phenotypes are shown at 48 and 144 h (32°C, E20A agar). The parental phenotype of normal growth and sporulation was displayed by FL2302 as well as by mutants with insertions downstream of cwh1 (mutant S6.07–14 is shown). The Cwh1 phenotype of slower growth and no sporulation was displayed by mutants with insertions in cwh1 (S6.07–03 and −06), as well as by mutants with insertions upstream of cwh1 (S6.07–72 shown).
DISCUSSION
Improvement of antibiotic production, driven by multiple rounds of random mutagenesis and screening, is an empirical process that has been an integral part of fermentation manufacturing since the beginning of the penicillin era and is widely practiced to this day. This study used targeted mutagens to explore the genetics behind this process.
Traditional strain improvement mutations are created by chance and chosen on phenotypic performance alone, with no knowledge of genotype needed; however, no benefit is passed to future strain improvement programs. The transposon-based strategy used in this study allows the characterization of the high-performing genotypes so that these mutations might be rationally incorporated into other desired genetic backgrounds.
Another limitation of traditional mutagenic screening is that no information is generated concerning the frequency of unique strain improvement mutations. However, by using transposon mutagenesis it was possible to measure this frequency at ∼3%; mutations being both yield related and capable of scale-up occurred at ∼0.2%. In a typical actinomycete such as S. erythraea with over 7000 genes, this might mean that a saturating random mutagenesis could yield as many as 210 unique strain improvement mutations, 14 of which might have potential for scale-up under a certain set of conditions.
The large number of mutations yielding increases in erythromycin production may reflect the fact that the strain used had not previously been subjected to any rounds of mutations for improvement (except for the mutB knockout). As more cycles of the transposon-based procedure are performed, the diversity and number of additional genes that might be mutated to cause improved yield should gradually diminish (Barrick and Lenski 2013).
The transposition approach has additional value in that all mutations influencing erythromycin production are useful, even yield-reducing mutations because they can also be exploited for gain. Only one locus, cwh1, was found that could be scaled-up to a 100-fold increase in fermentation volume (0.25–25 ml). Nevertheless, mutations that could not be scaled-up under our desired conditions might scale-up under different conditions.
Mutations in cwh1 produced visible changes in growth on solid media, which is consistent with the predicted function of Cwh1 in cell wall biogenesis. If the cwh1 mutation affects the early stages of cell wall biosynthesis, then the yield improvement phenotype could result from the diversion of cell wall precursors such as NDP-rhamnose, from cell wall biosynthesis into erythromycin biosynthesis (Mikusová et al. 1996; Crick, Mahapatra and Brennan 2001). This could explain the increase in erythromycin production particularly when NDP-rhamnose is the limiting metabolite (Summers et al. 1997). This diversion of precursors would be aided in S. erythraea by the fact that cell wall biogenesis and erythromycin biosynthesis occur simultaneously (McDermott, Lethbridge and Bushell 1993; Wardell et al. 2002) unlike most other antibiotic producers that produce antibiotic only after rapid growth is completed (Chater 2006). Consistent with this hypothesis, the rhamnose biosynthetic pathway has been previously shown to supply precursors to both primary and secondary metabolism in the closely related organism S. spinosa (Madduri, Waldron and Merlo 2001).
Interestingly, neither cwh1 nor any of the other homologous hydrolase genes mentioned above were found to be mutated in either of the two high-producing S. erythraea genomes reported so far (Peano et al. 2012; Li et al. 2013). However, transcriptome analysis in these higher producing strains did show an under representation of cell wall/membrane/envelope biogenesis transcripts (Peano et al. 2012). One other result in common between our mutations and the mutations found in high-producing strains was for mutations in citrate synthase. In our study, a citA (SACE_0632) knockout led to increased erythromycin production; in the two high-producing strains reported thus far, citA knockouts were also found (in SACE_0633; Peano et al. 2012; Li et al. 2013). The fact that most of the genotypes from this study were not found in the genome sequences of high-producing strains, which in turn do not closely match one another (Peano et al. 2012; Li et al. 2013), may be due to the nature of strain improvement as a convergent evolutionary process (Wagner 2009; Kryazhimskiy et al. 2014).
Supplementary Material
Acknowledgments
We acknowledge the Small Business Innovation Research program and the National Institute of General Medical Sciences for grant R43-GM079893; also, we thank Andrew Reeves and Lien Chu for helpful discussions.
SUPPLEMENTARY DATA
Conflict of interest. None declared.
REFERENCES
- Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol. 2003;4:R11. doi: 10.1186/gb-2003-4-2-r11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrick JE, Lenski RE. Genome dynamics during experimental evolution. Nat Rev Genet. 2013;14:827–39. doi: 10.1038/nrg3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bérdy J. Bioactive microbial metabolites. J Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- Cardinale CJ, Washburn RS, Tadigotla VR, et al. Termination factor Rho and its cofactors NusA and NusG silence foreign DNA in E. coli. Science. 2008;320:935–8. doi: 10.1126/science.1152763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chater KF. Streptomyces inside-out: a new perspective on the bacteria that provide us with antibiotics. Philos T R Soc B. 2006;361:761–8. doi: 10.1098/rstb.2005.1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crick DC, Mahapatra S, Brennan PJ. Biosynthesis of the arabinogalactan-peptidoglycan complex of Mycobacterium tuberculosis. Glycobiology. 2001;11:107R–18R. doi: 10.1093/glycob/11.9.107r. [DOI] [PubMed] [Google Scholar]
- Demain AL, Adrio JL. Strain improvement for production of pharmaceuticals and other microbial metabolites by fermentation. In: Petersen F, Amstutz R, editors. Natural Products as Drugs (part of Progress in Drug Research vol 65) Basel: Birkhaeuser; 2008. pp. 251–90. [DOI] [PubMed] [Google Scholar]
- Gehring AM, Nodwell JR, Beverley SM, et al. Genome wide insertional mutagenesis in Streptomyces coelicolor reveals additional genes involved in morphological differentiation. P Natl Acad Sci USA. 2000;97:9642–7. doi: 10.1073/pnas.170059797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goryshin IY, Reznikoff WS. Tn5 in vitro transposition. J Biol Chem. 1998;273:7367–74. doi: 10.1074/jbc.273.13.7367. [DOI] [PubMed] [Google Scholar]
- Kieser T, Bibb MJ, Buttner MJ, et al. Practical Streptomyces Genetics. Norwich: John Innes Foundation; 2000. [Google Scholar]
- Kryazhimskiy SK, Rice DP, Jerison ER, et al. Global epistasis makes adaptation predictable despite sequence-level stochasticity. Science. 2014;344:1519–22. doi: 10.1126/science.1250939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakkitjaroen N, Takamatsu D, Okura M, et al. Capsule loss or death: the position of mutations among capsule genes sways the destiny of Streptococcus suis. FEMS Microbiol Lett. 2014;354:46–54. doi: 10.1111/1574-6968.12428. [DOI] [PubMed] [Google Scholar]
- Li YY, Chang X, Yu WB, et al. Systems perspectives on erythromycin biosynthesis by comparative genomic and transcriptomic analyses of S. erythraea E3 and NRRL23338 strains. BMC Genomics. 2013;14:523. doi: 10.1186/1471-2164-14-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang D, Qiao J. Phylogenetic analysis of antibiotic glycosyltransferases. J Mol Evol. 2007;64:342–53. doi: 10.1007/s00239-006-0110-2. [DOI] [PubMed] [Google Scholar]
- McDermott JF, Lethbridge G, Bushell ME. Estimation of the kinetic constants and elucidation of trends in growth and erythromycin production in batch and continuous cultures of Saccharopolyspora erythraea using curve-fitting techniques. Enzyme Microb Technol. 1993;15:657–63. doi: 10.1016/0141-0229(93)90065-a. [DOI] [PubMed] [Google Scholar]
- Madduri K, Waldron C, Merlo DJ. Rhamnose biosynthesis pathway supplies precursors for primary and secondary metabolism in Saccharopolyspora spinosa. J Bacteriol. 2001;183:5632–8. doi: 10.1128/JB.183.19.5632-5638.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maes MB, Scharpé S, De Meester I. Dipeptidyl peptidase II (DPPII), a review. Clin Chim Acta. 2007;380:31–49. doi: 10.1016/j.cca.2007.01.024. [DOI] [PubMed] [Google Scholar]
- Mikusová K, Mikus M, Besra GS, et al. Biosynthesis of the linkage region of the mycobacterial cell wall. J Biol Chem. 1996;271:7820–8. doi: 10.1074/jbc.271.13.7820. [DOI] [PubMed] [Google Scholar]
- Oliynyk M, Samborskyy M, Lester JB, et al. Complete genome sequence of the erythromycin-producing bacterium Saccharopolyspora erythraea NRRL23338. Nat Biotechnol. 2007;25:447–53. doi: 10.1038/nbt1297. [DOI] [PubMed] [Google Scholar]
- Peano C, Talà A, Corti G, et al. Comparative genomics and transcriptional profiles of Saccharopolyspora erythraea NRRL2338 and a classically improved erythromycin over-producing strain. Microb Cell Fact. 2012;11:32. doi: 10.1186/1475-2859-11-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves AR, Brikun IA, Cernota WH, et al. Effects of methylmalonyl-CoA mutase gene knockouts on erythromycin production in carbohydrate-based and oil-based fermentations of Saccharopolyspora erythraea. J Ind Microbiol Biot. 2006;33:600–9. doi: 10.1007/s10295-006-0094-3. [DOI] [PubMed] [Google Scholar]
- Reeves AR, Cernota WH, Brikun IA, et al. Engineering precursor flow for increased erythromycin production in Aeromicrobium erythreum. Metab Eng. 2004;6:300–12. doi: 10.1016/j.ymben.2004.03.003. [DOI] [PubMed] [Google Scholar]
- Reeves AR, Weber G, Cernota WH, et al. Analysis of an 8.1-kb DNA fragment contiguous with the erythromycin gene cluster of Saccharopolyspora erythraea in the eryCI-flanking region. Antimicrob Agents Ch. 2002;46:3892–9. doi: 10.1128/AAC.46.12.3892-3899.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves AR, Weber JM. Methods in Molecular Biology. Vol. 834. Clifton, NJ: Springer; 2012. Metabolic engineering of antibiotic-producing actinomycetes using in vitro transposon mutagenesis; pp. 153–75. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Vol. 3. NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Santos CNS, Stephanopoulos G. Combinatorial engineering of microbes for optimizing cellular phenotype. Curr Opin Chem Biol. 2008;12:168–76. doi: 10.1016/j.cbpa.2008.01.017. [DOI] [PubMed] [Google Scholar]
- Summers RG, Donadio S, Staver MJ, et al. Sequencing and mutagenesis of genes from the erythromycin biosynthetic gene cluster of Saccharopolyspora erythraea that are involved in L-mycarose and D-desosamine production. Microbiology. 1997;143:3251–62. doi: 10.1099/00221287-143-10-3251. [DOI] [PubMed] [Google Scholar]
- Vinci VA, Byng G. Strain improvement by nonrecombinant methods. In: Demain AL, Davies JE, editors. Manual of Industrial Microbiology and Biotechnology. 2nd edn. Washington, DC: ASM Press; 1999. pp. 103–13. [Google Scholar]
- Viollier PH, Minas W, Dale GE, et al. Role of acid metabolism in Streptomyces coelicolor morphological differentiation and antibiotic biosynthesis. J Bacteriol. 2001;183:3184–92. doi: 10.1128/JB.183.10.3184-3192.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner A. Evolutionary constraints permeate large metabolic networks. BMC Evol Biol. 2009;9:231. doi: 10.1186/1471-2148-9-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wardell JN, Stocks SM, Thomas CR, et al. Decreasing the hyphal branching rate of Saccharopolyspora erythraea NRRL 2338 leads to increased resistance to breakage and increased antibiotic production. Biotechnol Bioeng. 2002;78:141–6. doi: 10.1002/bit.10210. [DOI] [PubMed] [Google Scholar]
- Weber JM, Leung JO, Maine GT, et al. Organization of a cluster of erythromycin genes in Saccharopolyspora erythraea. J Bacteriol. 1990;172:2372–83. doi: 10.1128/jb.172.5.2372-2383.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JM, Losick R. The use of a chromosome integration vector to map erythromycin resistance and production genes in Saccharopolyspora erythraea (Streptomyces erythreus) Gene. 1988;68:173–80. doi: 10.1016/0378-1119(88)90019-4. [DOI] [PubMed] [Google Scholar]
- Wu P, Pan H, Zhang C, et al. SACE_3986, a TetR family transcriptional regulator, negatively controls erythromycin biosynthesis in Saccharopolyspora erythraea. J Ind Microbiol Biot. 2014;41:1159–67. doi: 10.1007/s10295-014-1449-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.