Abstract
Prokaryotic mRNA turnover can be initiated by the removal of pyrophosphate from the 5′ end of a transcript using the RNA pyrophosphohydrolase enzyme RppH. Following the initial dephosphorylation step, RNaseE then degrades the message into small oligonucleotide segments. This study assessed the small RNA transcriptome of Neisseria gonorrhoeae strain MS11 in two genetic backgrounds; using wild type cells as well as cells carrying a rppH insertional mutation. It was found that the presence of the RppH enzyme affected both the quantity and length of small RNAs (sRNAs) in various chromosomal locations and involved sense transcripts (seRNAs), transcripts originating from the opposite strand (asRNAs) as well as inter-genic-derived RNAs (IGRs). In comparing the two transcriptomes, we found that not all small RNAs were expressed in both genetic backgrounds, suggesting that RppH apparently targets only a subset of transcripts. Overall, this study shows that small RNAs can be detected from the majority of genes within the chromosome, as well as from inter-genic regions, and that more sRNA transcripts are detected in the absence of the RppH enzyme.
Keywords: mRNA turnover, small RNAs, gonococci
The RNA processing enzyme RNA pyrophosphohydrolase influences both the quality and quantity of small RNAs in the pathogen Neisseria gonorrhoeae.
The RNA processing enzyme RNA pyrophosphohydrolase influences both the quality and quantity of small RNAs in the pathogen Neisseria gonorrhoeae.
INTRODUCTION
Neisseria gonorrhoeae causes gonorrhea, a strictly human infectious disease. Considering the importance of the organism for human health, very few regulatory systems have been defined (Mellin and Hill 2010). Moreover, little is known regarding mRNA turnover in gonococci even though the organism possesses many of the RNA degrading enzymes that are found in Gram-negative bacteria. RNA turnover in prokaryotes is regulated through a variety of mechanisms with the combined action of the enzymes RppH (an RNA pyrophosphohydrolase) and RNaseE (an endo-ribonuclease) being of major importance (Richards et al., 2008). Prokaryotic transcripts carry a triphosphate group at the 5′ end of the message. mRNA turnover is initiated by the removal of pyrophosphate from the 5′ end through the enzymatic action of RppH which then allows RNaseE to digest the dephosphorylated message into small oligoribonucleotides (Richards et al., 2008). The tendency for RppH to target specific transcripts for degradation appears to reside within the nucleotide sequence of the transcript, where experimental data suggest that targeted transcripts must contain at least two unpaired nucleotides at their 5′ end, with higher catalytic activity being displayed if guanine is present at the second and third position followed by an additional guanine or adenine residue (Hsieh et al., 2013). Additional factors may also affect the rate of RNA turnover, such as the presence of secondary structures, the presence of translating ribosomes and/or other RNA degrading enzymes (Deutscher 2006).
An alternative strategy for RNA turnover is targeting transcripts using regulatory small RNAs (sRNAs). Regulatory sRNAs exert control over the stability of RNA transcripts through a variety of mechanisms (Storz, Vogel and Wassarman 2011), with four general categories having been defined; riboswitches, RNA thermometers as well as cis- or trans-encoded sRNAs (Han et al., 2013). Typically, sRNAs act post-transcriptionally to either decrease mRNA stability by providing double-stranded RNA substrates that can be digested by RNase III, or, alternatively, by blocking access of the translational machinery with their targeted transcripts (Livny and Waldor 2007). cis- and trans-encoded sRNAs are the largest group of regulatory sRNAs that have been identified, with both regulating mRNA expression through complementary binding to their target transcripts (Waters and Storz 2009). cis-encoded regulatory sRNAs are often ∼75 nucleotides or longer in length and are derived following transcription from the opposite strand of their target mRNA. In contrast, trans-encoded sRNAs display limited complementarity to their target transcripts and are typically transcribed from within inter-genic regions (IGRs) (Waters and Storz 2009). Generally, in Gram-negative bacteria, trans-encoded sRNAs require the RNA-binding protein, Hfq, for function and stability (Storz, Vogel and Wassarman 2011). Regulatory sRNAs have been shown to control many processes within cells, including stress responses, metabolic reactions, as well as modulating pathogenesis (Hill 2011; Raghavan, Groisman and Ochman 2011). However, very few of these regulatory sRNAs have been identified in Neisseria. A recent transcriptional profiling study identified 253 novel potential regulatory sRNAs located within IGRs which were anti-sense to coding genes. Interestingly, 9 of the 11 opacity proteins, a major outer membrane protein that is intimately involved in gonococcal pathogenesis, apparently are regulated by inter-genically-derived anti-sense sRNAs (Remmele et al., 2014).
In this study, we used transcriptional profiling to compare the sRNA transcriptomes of N. gonorrhoeae strain MS11 in two different genetic backgrounds; wild type and an rppH mutant. By comparing the two small transcriptomes, we have identified sRNAs from all regions of the chromosome which has allowed us to further evaluate sRNAs that are derived from IGRs as well as sRNAs that are sense as well as sRNAs that are anti-sense to sense transcripts within a cis-configuration. Moreover, we have also been able to assess the role of the RppH enzyme on both the abundance and quality of sRNAs within the cell.
METHODS
Bacterial strains and growth conditions
Neisseria gonorrhoeae strain MS11 strain 7/30:2 was the wild-type strain used in this study (Bergstrom et al., 1986). An N. gonorrhoeae ΔrppH::ermC insertional mutant was created by sequential ligation of two PCR fragments with the ermC gene. The ermC gene contains a transcriptional terminator thus eliminating any downstream polar effects. The PCR fragments were made using the following primer pairs; RppHfor1 5′-GGATCCGATGCTTGAACCATACCGTCCGATTT-3′ and RppHrev1 5′-ACTAGTCGTCGTAACGCAGCCAGTCGCGC-3′; RppHfor2 5′-CTCGAGTGCCGAACAACTGGGTGCGCCGCG-3′ and RppHrev2 5′-TCTAGATTACCGGTTGCCGGAAGGTTG-3′. Gonococci were passaged on gonococcal typing medium (Swanson 1982) at 37ºC in 5% CO2.
Rifampicin treatment of exponentially growing cells
Cells were grown in liquid medium until log phase was reached (OD600 = 0.15) at which point rifampicin was added to a final concentration of 200 μg mL−1. At various time points thereafter, rifampicin inhibition was terminated by immediately suspending the cells in an equal volume of methanol/phenol (50:1). The cells were then harvested by centrifugation. Cell pellets were frozen and total RNA was extracted.
RNA preparation, library construction and sequencing
Neisseria gonorrhoeae strain MS11 and the MS11 ΔrppH mutant were grown on solid medium for 12 hrs. Cells were harvested and sRNA was extracted utilizing the MirVana Mira Kit AM 1560 (Life Technologies, Inc) according to the manufacturer's specifications. Consequently, transcripts larger than 250–300 bp were excluded from further analysis by size fractionation. Two sRNA libraries (wild type and ΔrppH) were constructed using the ‘Small RNA v1.5 Sample Preparation Kit’ (Illumina). RNAs within both sets of samples were ligated to 5′ adaptors containing a 4 nt barcode and were subsequently sequenced on an Illumina HiSeq2000 with chemistry version 4 and analyzed with pipeline 1.8. For the wild-type sRNA library, the coverage was 24 592 832 reads; for the rppH sRNA library, the coverage was 11 021 353 reads. The error rate of the runs was <0.3%. For analysis, the adaptor sequences were subtracted from the reads. Only sequenced RNA transcripts that mapped to N. gonorrhoeae strain MS11 chromosome assembly GCA_000156855.2 were kept. Analysis focused solely on unambiguous reads (i.e. transcript reads needed to map to regions of the chromosome with no lapse in read depth). Both transcriptomes are available at the GEO accession number GSE62926.
Mapping and visualization of reads
Initially, the sequencing data was mapped to the reference genome N. gonorrhoeae strain MS11 GenBank ID CP003909.1 (assembly GCA_00156855.2) submitted 02 September 2010. Subsequently, following the submission of the fully sequenced MS11 chromosome, the data were aligned to the MS11 sequence assembly GCA_000156855.2 version CP003909.1 GI:537637238 submitted 14 February 2014. The data were mapped to the reference genome using the Nesoni data analysis toolset available through the Victorian Bioinformatics Consortium (http://www.vicbioinformatics.com/software.nesoni.shtml). This toolset trims the reads and filters alignments through the SHRiMP read aligner, which is able to align short-read sequencing data to the reference genome regardless of small insertions or deletions (Rumble et al., 2009; David et al., 2011). Nesoni also creates files that may be supplied to Artemis in order to graphically visualize the coverage and read depth of transcripts (Carver et al., 2012). Figures including genomic locations of transcripts and histograms of transcript read depth were generated using the Circos software package version 0.66 (Krywinski et al., 2009).
Transcript quantification
Coordinates of all IGRs, protein-coding genes, tRNAs, rRNAs and ncRNAs were obtained from NCBI GenBank CP003909.1. The relative amount of transcripts were determined as the number of transcripts × 106 values (Wagner, Kin and Lynch 2012).
RESULTS
More sRNAs are observed in the absence of the RppH enzyme
Neisseria gonorrhoeae strain MS11 has 944 genes that are transcribed from the Watson strand and 1134 genes that are transcribed from the Crick strand. Analysis of sRNAs from both the N. gonorrhoeae MS11 wild-type and ΔrppH small transcriptomes revealed multiple sRNA transcripts that are distributed throughout the genome (Fig. 1; Fig. S1 and S2, Supporting Information). While sRNAs were observed in a similar number of chromosomal locations in both genetic backgrounds, more individual transcripts were detected in wild-type cells, yet greater quantities of sRNAs are observed in the ΔrppH mutant (Table 1). In comparing the two strains, (i) transcripts are observed at more chromosomal locations in wild-type cells, (ii) wild-type cells contained more chromosomal locations harboring longer transcripts and (iii) the ΔrppH mutation influences the quantity of transcripts.
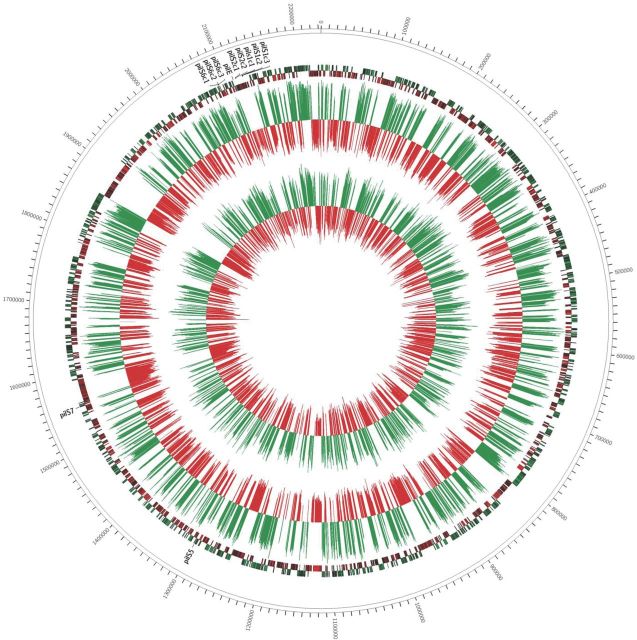
Figure 1.
The small seRNA transcriptome of N. gonorrhoeae strain MS11. The locations of genes originating from the Watson strand are shown as green and from the Crick strand as red. The quantity of sRNAs are presented as histograms inset to their corresponding locations on the chromosome of N. gonorrhoeae strain MS11. The wild-type transcriptome is displayed as the larger, outer histogram, while the ΔrppH transcriptome is presented as the smaller, inner histogram.
Table 1.
The total number of sRNA transcripts mapping to unique locations on the chromosome.
| Transcripts mapping Watson strand | Transcripts mapping to the Crick | |||||||
|---|---|---|---|---|---|---|---|---|
| of the chromosome | strand of the chromosome | |||||||
| Number of distinct | Average length | Relative | t-value | Number of distinct | Average length | Relative | t-value | |
| regions | (bp) | amount# | regions | (bp) | amount# | |||
| MS11 | 21 252 | 52 | 47 | 5.74* | 21 639 | 53 | 46 | 4.34* |
| MS11 ΔrppH | 13 855 | 41 | 72 | 14 796 | 42 | 67 | ||
| MS11 ≥ 100 bp | 2159 | 135 | 156 | 2.89* | 2288 | 135 | 153 | 3.09* |
| MS11 ΔrppH ≥ 100 bp | 537 | 132 | 340 | 636 | 132 | 365 | ||
#Relative amount reflects the number of transcripts × 106
*Significantly different (P < 0.05) between the two genetic backgrounds
Identification of different classes of sRNAs
The analysis was further refined by determining the orientation of the sRNAs with respect to the annotated genes which allowed the sRNAs to be classified as intra-genic sense transcripts (seRNAs), cis-intra-genic anti-sense (i.e. transcripts arising from the opposite strand) (asRNAs) and inter-genic transcripts (IGRs) (Table 2). This refined analysis revealed that a greater number of transcripts mapped as IGRs, as intra-genic asRNAs or as intra-genic seRNAs in a wild-type background, when compared to sRNAs detected in the mutant background (for IGRs, 2442 more locations; for intra-genic asRNAs, 7934 more locations; for intra-genic seRNAs, 3849 more locations Table 2). However, for ΔrppH cells, the overall abundance of sRNA transcripts is still much greater when compared to wild-type cells (for ΔrppH cells, there is ∼68% more IGR transcripts; ∼33% more asRNAs; ∼46% more seRNAs). Therefore, all classes of sRNAs are found in more chromosomal locations in wild-type cells yet in lower quantities than in the ΔrppH mutant. When statistical analysis was applied to the relative abundance of transcripts presented in Table 2, significantly greater amounts of IGR-derived sRNAs are detected in both genetic backgrounds when compared to either intra-genic seRNAs or intra-genic asRNAs (P < 0.05) (Table 3). Also, there were significantly more intra-genic seRNAs than intra-genic asRNAs (P < 0.05).
Table 2.
sRNAs mapped to the chromosome of N. gonorrhoeae MS11 in two genetic backgrounds.
| Transcripts mapped to | Transcripts mapped as | Transcripts mapped as | |||||||
|---|---|---|---|---|---|---|---|---|---|
| IGRs | intra-genic asRNAs | intra-genic seRNAs | |||||||
| Number mapped | Avg. | RA# | Number mapped | Avg. | RA# | Number mapped | Avg. | RA# | |
| to distinct | length | to distinct | length | to distinct | length | ||||
| regions | (bp) | distinct | (bp) | distinct | (bp) | ||||
| MS11 | 6422 | 45 | 70* | 16 242 | 48 | 33* | 20 175 | 59 | 48* |
| MS11 ΔrppH | 3980 | 37 | 118* | 8308 | 36 | 44* | 16 326 | 45 | 70* |
| MS11 ≥ 100 bp | 326 | 132 | 568* | 1205 | 133 | 100* | 2915 | 136 | 131* |
| MS11 ΔrppH ≥ 100 bp | 85 | 126 | 2488* | 169 | 131 | 177* | 919 | 133 | 189* |
#RA indicates the relative amount of RNA (the number of transcripts × 106).
*Indicates that the transcript quantities are significantly different (P < 0.05) between the wild-type and ΔrppH background.
Table 3.
Evaluating the relative amount of each class of sRNA.
| Wild-type | ΔrppH | |
|---|---|---|
| t-value | t-value | |
| Intergenic–intragenic seRNAs | 3.32 | 4.52 |
| Intergenic–intragenic asRNAs | 5.68 | 5.07 |
| Intragenic seRNAs – intragenic asRNAs | 7.09 | 14.14 |
All t-values indicate that the transcript comparisons are significantly different (P < 0.05).
Promoter analysis of seRNAs
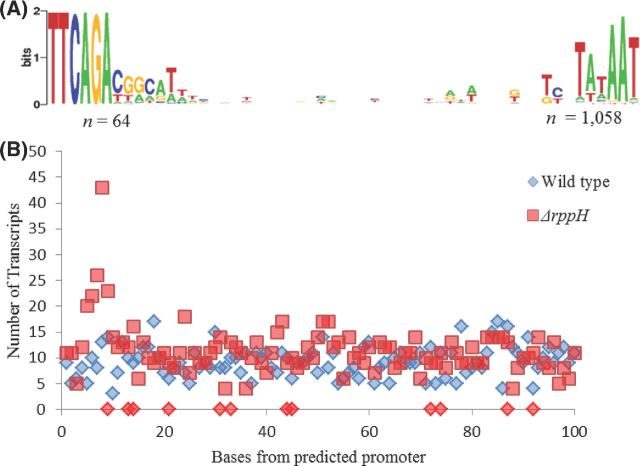
In order to determine promoter usage of the seRNAs mapped in Fig. 1, regions spanning 100 nucleotides upstream from all annotated CDS sequences were analyzed. Analysis of 1058 sRNA genes indicated that only 64 contained regions sharing homology to the previously identified extended −35 region (Remmele et al., 2014), whereas 994 only contained regions homologous to the canonical Pribnow box (Fig. 2A). This analysis also revealed that seRNAs from both cell types occurred with a similar frequency along the length of the CDS, yet there is a greater number of seRNAs in the ΔrppH background that are predicted to arise within 10 base pairs of the predicted promoter (Fig. 2B).
Figure 2.
Predicted promoter sequences of annotated CDS regions. (A) Regions 100 base pairs upstream of CDS start codons were analyzed for sequences containing homology to the Pribnow box and −35 promoter elements. Homologous regions were obtained, aligned and a sequence logo was constructed (Crooks et al., 2004). (B) All seRNAs were examined and graphed based on their location with respect to the putative promoter sequences; wild-type transcripts are presented as blue diamonds and ΔrppH transcripts as red squares. While, the majority of seRNAs do not appear to be affected in terms of location by the presence of the RppH enzyme, a greater number of seRNAs are located within 10 base pairs of the predicted promoter within a ΔrppH background. In addition, based on their location from the predicted promoter, a significant difference (P, 0.05) was detected in the quantity of seRNAs arising from a ΔrppH mutant background (red diamonds on x-axis) than from a wild-type background.
cis-encoded sRNAs from opa genes
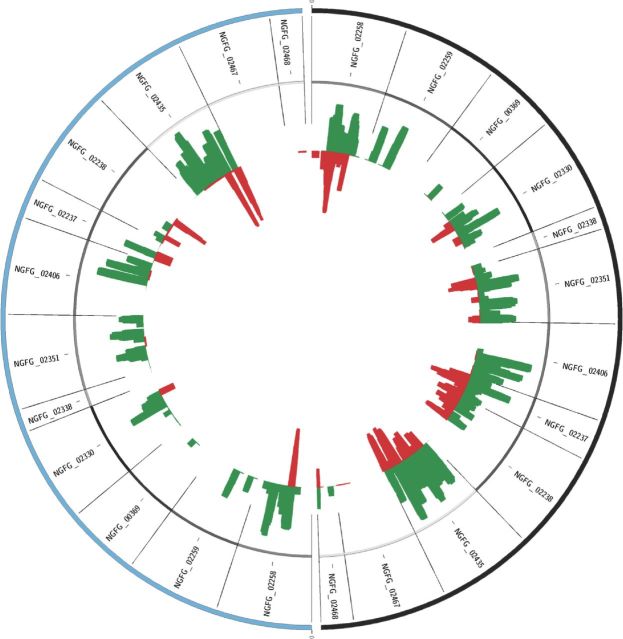
The amount of cis-encoded intra-genic asRNAs of opa were evaluated (Fig. 3), as a previous analysis had revealed anti-sense RNAs arising from 9 out of the 11 encoded opa genes of MS11 (Remmele et al., 2014). The current study corroborated these observations as cis-encoded intra-genic asRNAs were detected from ten opa genes of MS11 in a wild-type background and seven in a ΔrppH background (NGFG_02258, NGFG_02259, NGFG_00369, NGFG_02330, NGFG_02337, NGFG_02338, NGFG_02435 in both genetic backgrounds and NGFG_02406, NGFG_02467, NGFG_02468 in wild-type cells). For the majority of opa genes, it appears that the RppH enzyme affects the presence and quantity of asRNAs as there is an overall greater abundance of anti-sense transcripts in the mutant than compared to a wild-type background.
Figure 3.
sRNAs mapping to opa. The quantity of sRNAs mapping to the annotated opa genes of N. gonorrhoeae strain MS11 are shown as histograms inset to the genes. The wild-type genes are shown in black to the right while the genes from the rppH mutant are shown in blue to the left. sRNAs that map sense to the opa genes are shown in green, while sRNAs that map anti-sense to the opa genes are shown in red.
DISCUSSION
Transcriptome analysis allows for genome-wide detection and quantification of RNA transcripts. Furthermore, it also allows for the effect of specific genetic manipulations to be determined at the level of chromosome-wide transcription (Raghavan, Groisman and Ochman 2011). In this study, we assessed the sRNA transcriptome in two genetic backgrounds of N. gonorrhoeae MS11 (in wild-type cells and in a ΔrppH mutant background) with the analysis detecting major differences in sRNA composition, with the results indicating that sRNAs were found in more chromosomal locations in wild-type cells yet in lower abundance when compared to the ΔrppH mutant. Given the role that RppH plays in mRNA turnover, the latter observation was expected. The difference in abundance may reflect a difference in RNA half-lives in the two genetic backgrounds as wild-type mRNA has a shorter half-life than the half-life observed for ΔrppH mRNA (<5 min vs. <7.5 min, respectively, data not shown). When the sRNAs were further differentiated into IGRs, intra-genic asRNAs and seRNAs transcripts, it was found that the RppH enzyme also had an influence on all classes of sRNAs. However, a consistent feature in both genetic backgrounds was that the sRNAs overwhelmingly arose from genes where transcription utilized only a −10 promoter element (Fig. 2).
Whether in the presence or absence of the RppH enzyme many intra-genic asRNAs as well as intra-genic seRNAs were detected, with many of these sRNAs exhibiting complementarity (e.g. the opa gene family; Fig. 3). If the intra-genic sense and anti-sense RNAs were to pair, this would create double-stranded RNA molecules which would then substrates for RNase III digestion thus facilitating turnover (Callen, Shearwin and Egan 2004; Han et al., 2013; Wade and Grainger 2014). Alternatively, gene expression can also be regulated through convergent transcription with transcriptional interference occurring between the convergent sense and anti-sense promoters leading to the silencing of the weaker promoter (Shearwin, Callen and Egan 2005). Consequently, depending on the individual promoter strength, transcription of either sense or anti-sense RNA may be silenced in this manner. Thus, the abundance of asRNAs as well as seRNAs detected throughout the chromosome may have a major impact on global transcriptional regulation.
While the amount of DNA comprising IGRs varies between bacteria, it is believed that bacterial genomes exhibit an evolutionary bias towards deleting IGR regions and maintaining only those regions that comprise open reading frames (Fukuda, Nakayama and Tomita 2003; Sridhar et al., 2011). Therefore, the maintenance of IGR-derived transcripts suggests that they are likely required to fulfill some biological function within the cell (Argaman et al., 2001). Many IGR transcripts act as trans-encoded regulatory sRNAs that either silence or enhance expression of their target mRNAs (Han et al., 2013). Consequently, IGR-derived sRNA transcripts that function in a crucial regulatory manner would be expected to be similar between wild-type and ΔrppH cells, especially as the RppH enzyme does not use regulatory sRNAs for RNA degradation (Storz, Vogel and Wassarman 2011). We found that the majority of transcripts that mapped to unique or analogous IGRs were found in similar quantities in both genetic backgrounds, but in fewer genomic locations in the absence of RppH. Interestingly, IGR-derived sRNAs were present in greater quantities when compared to both seRNAs and asRNAs (Table 3). The presence of RppH also influenced the quality of sRNAs within IGRs, as roughly two-thirds of the genomic locations giving rise to IGR-derived transcripts in one genetic background (either wild type or ΔrppH) lacked any sRNA transcripts in the other cell type (data not shown). Therefore, it appears as though RppH interacts with roughly two-thirds of IGR-derived transcripts and affects both their appearance and abundance.
We also examined sRNAs in two genetic systems that are intimately involved in gonococcal virulence; sRNAs within the opa gene family that encodes for a major outer membrane protein (Fig. 3) and within the pilE/pilS genes which are involved in PilE antigenic variation (data not shown). The analysis revealed the presence of seRNAs and asRNAs mapping to the majority of opa genes, as well as to most pil genes within the cell, indicating that these cis-encoded sRNAs may aid in the regulation of two important systems involved in the pathogenicity of the gonococcus.
CONCLUSION
The RppH enzyme was determined to regulate certain sRNAs present within the transcriptome of N. gonorrhoeae MS11. Not only did the absence of this enzyme increase the quantity of sRNA transcript detected within the chromosome, it also influenced the length and chromosomal locations giving rise to these transcripts. However, regardless of the presence of the RppH enzyme, the majority of sRNAs detected from MS11 arose from IGRs, implying that N. gonorrhoeae may use these IGR-derived sRNAs in a regulatory capacity.
SUPPLEMENTARY DATA
FUNDING
The work was supported by NIH grant 1R15 AI072720-01A1 to SAH and Northern Illinois University's graduate student grant program.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- Argaman L, Hershberg R, Vogel J, et al. Novel small RNA-encoding genes in the intergenic regions of Escherichia coli. Curr Biol. 2001;11:941–50. doi: 10.1016/s0960-9822(01)00270-6. [DOI] [PubMed] [Google Scholar]
- Bergstrom S, Robbins K, Koomey JM, et al. Piliation control mechanisms in Neisseria gonorrhoeae. P Natl Acad Sci USA. 1986;83:3890–4. doi: 10.1073/pnas.83.11.3890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callen BP, Shearwin KE, Egan JB. Transcriptional interference between convergent promoters caused by elongation over the promoter. Mol Cell. 2004;14:647–56. doi: 10.1016/j.molcel.2004.05.010. [DOI] [PubMed] [Google Scholar]
- Carver T, Harris SR, Berriman M, et al. Artemis: an integrated platform for visualization and analysis of high-throughput sequence-based experimental data. Bioinformatics. 2012;28:464–9. doi: 10.1093/bioinformatics/btr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, et al. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–90. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M, Dzamba M, Lister D, et al. SHRiMP2: sensitive yet practical short read mapping. Bioinformatics. 2011;27:1011–2. doi: 10.1093/bioinformatics/btr046. [DOI] [PubMed] [Google Scholar]
- Deutscher MP. Degradation of RNA in bacteria: comparison of mRNA and stable RNA. Nucleic Acids Res. 2006;34:659–66. doi: 10.1093/nar/gkj472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y, Nakayama Y, Tomita M. On dynamics of overlapping genes in bacterial genomes. Gene. 2003;323:181–7. doi: 10.1016/j.gene.2003.09.021. [DOI] [PubMed] [Google Scholar]
- Han Y, Liu L, Fang N, et al. Regulation of pathogenicity by non-coding RNAs in bacteria. Future Microbiol. 2013;8:579–91. doi: 10.2217/fmb.13.20. [DOI] [PubMed] [Google Scholar]
- Hill SA, Davies JK. Pilin gene variation in Neisseria gonorrhoeae: reassessing the old paradigms. FEMS Microbiol Rev. 2009;33:521–30. doi: 10.1111/j.1574-6976.2009.00171.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill SA. Stress response in the pathogenic Neisseria: overlapping regulons and sRNA regulation. In: Kidd SP, editor. Stress Response in Pathogenic Bacteria. UK: CAB International; 2011. pp. 115–32. [Google Scholar]
- Hsieh P, Richards J, Liu L, et al. Specificity of RppH-dependent RNA degradation in Bacillus subtilis. P Natl Acad Sci USA. 2013;110:8864–9. doi: 10.1073/pnas.1222670110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krywinski M, Schein JE, Birol I, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–45. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livny J, Waldor MK. Identification of small RNAs in diverse bacterial species. Curr Opin Microbiol. 2007;10:96–101. doi: 10.1016/j.mib.2007.03.005. [DOI] [PubMed] [Google Scholar]
- Mellin JR, Hill SA. Gene expression strategies of the pathogenic Neisseria. In: Genco CA, Wetzler L, editors. Neisseria: Molecular Mechanisms of Pathogenesis. Norfolk, UK: Horizon Scientific Press; 2010. pp. 3–18. [Google Scholar]
- Raghavan R, Groisman EA, Ochman H. Genome-wide detection of novel regulatory RNAs in E. coli. Genome Res. 2011;21:1487–97. doi: 10.1101/gr.119370.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remmele CW, Xian Y, Albrecht M, et al. Transcriptional landscape and essential genes of Neisseria gonorrhoeae. Nucleic Acids Res. 2014;42:10579–95. doi: 10.1093/nar/gku762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards J, Sundermeier T, Svetlanov A, et al. Quality control of bacterial mRNA decoding and decay. Biochim Biophys Acta. 2008;1779:574–82. doi: 10.1016/j.bbagrm.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumble SM, Lacroute P, Dalca AV, et al. SHRiMP: accurate mapping of short color-space reads. PLoS Comput Biol. 2009;5:e1000386. doi: 10.1371/journal.pcbi.1000386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearwin KE, Callen BP, Egan JB. Transcriptional interference - a crash course. Trends Genet. 2005;21:339–45. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridhar J, Sabarinathan R, Balan SS, et al. Junker: an intergenic explorer for bacterial genomes. Genomics Proteom Bioinformatics. 2011;9:179–82. doi: 10.1016/S1672-0229(11)60021-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storz G, Vogel J, Wassarman KM. Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell. 2011;43:880–91. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J. Colony opacity and protein II compositions of gonococci. Infect Immun. 1982;37:359–68. doi: 10.1128/iai.37.1.359-368.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wade JT, Grainger DC. Pervasive transcription: illuminating the dark matter of bacterial transcriptomes. Nat Rev. 2014;12:647–53. doi: 10.1038/nrmicro3316. [DOI] [PubMed] [Google Scholar]
- Wagner GP, Kin K, Lynch VJ. Measurement of mRNA abundance using RNA-seq data: RPKM measure is inconsistent among samples. Theor Biosci. 2012;131:281–85. doi: 10.1007/s12064-012-0162-3. [DOI] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in Bacteria. Cell. 2009;136:615–28. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.