Abstract
Objective
As genetic variation accounts for two-thirds of the variation in external apical root resorption (EARR) concurrent with orthodontic treatment, we analyzed the association of selected genetic and treatment-related factors with EARR concurrent with orthodontic treatment.
Setting and sample population
This case–control study of 134 unrelated, orthodontically treated Caucasian individuals was conducted in part at an Indiana Private Practice, Indiana University and the University of Kentucky.
Methods
Utilizing a research data bank containing information from ~1450 orthodontically treated patients, pre- and post-treatment radiographs from 460 individuals were evaluated for EARR of the four permanent maxillary incisors. Sixty-seven unrelated Caucasians with moderate to severe EARR were identified and were age-/sex-matched with orthodontically treated Caucasian controls yielding 38 females and 29 males per group. Factors tested for an association with EARR included the following: 1) treatment duration, 2) extraction of maxillary premolars, 3) numerous cephalometric measurements, and 4) DNA polymorphisms within/near candidate genes in a pathway previously implicated in EARR such as the purinergic-receptor-P2X, ligand-gated ion channel 7 (P2RX7; rs208294, rs1718119, and rs2230912), caspase-1 (CASP1; rs530537, rs580253, and rs554344), interleukin-1 beta (IL1B; rs1143634), interleukin-1 alpha (IL1A; rs1800587), and interleukin-1 receptor antagonist (IL1RA; rs419598) genes. Stepwise logistic regression was utilized to identify the factors significantly associated (significance taken at or less than the layered Bonferroni correction alpha) with the occurrence of EARR.
Results
A long length of treatment and the presence of specific genotypes for P2RX7 SNP rs208294 were significantly associated with EARR.
Conclusion
EARR occurrence was associated with both genetic and treatment-related variables, which together explained 25% of the total variation associated with EARR in the sample tested.
Keywords: caspase-1, external apical root resorption, interleukin-1, interleukin-1 receptor antagonist, purinergic-receptor-P2X ligand-gated ion channel 7
Introduction
External apical root resorption (EARR) is a reduction of root structure involving the apices. EARR occurrence has been reported with and without orthodontic treatment (1) and can be diagnosed by orthodontists during routine diagnostic, progress and post-treatment radiographs. It has been reported that 5 mm or more of apical root resorption may occur in 5% of orthodontic patients (1). Any unusual length of the roots should be noted on initial diagnostic radiographs and described to the patient, along with an explanation of how EARR may occur as an inflammatory process resulting in the loss of apical root structure and what factors are contributory. Although root resorption may introduce mobility of involved teeth in severe cases, it usually does not have a significant impact on the longevity of teeth (2). Orthodontic force magnitude, duration of the applied force, force direction, dilacerated, slender or pointed roots, allergy, history of trauma, age at start of treatment, excessive overjet, maxillary premolar extractions, sex, and length of treatment have been reported as risk factors for EARR during the course of orthodontic treatment (3–9).
Although orthodontic tooth movement, or ‘biomechanics’, has been found to account for approximately one-tenth to one-third of the total variation in EARR (10–12), Owman-Moll et al. showed that individual variation overshadowed the force magnitude and the force type in defining the susceptibility to histologic root resorption associated with orthodontic force (13). Individual variations were considerable regarding both extension and depth of histologic root resorption within individuals, and these were not correlated to the magnitude of tooth movement achieved (14). There was considerable individual variation in the EARR associated with orthodontic treatment, indicating an individual predisposition and multifactorial etiology (15–21). While the severity of EARR could not be fully explained by treatment-related factors, studies were initiated to investigate the association of genetic factors with the variation in EARR seen among individuals. Harris et al. (15) initially pointed out the involvement of genetic variation in EARR concurrent with orthodontia variation through a heritability study. Later, Hartsfield et al. (16) confirmed that genetic variation was associated with as much as 50–66% of the variation observed with EARR concurrent with orthodontia.
As compression of periodontal ligament (PDL) occurs when teeth are being moved, ATP is released from adjacent cells. During the process of binding this released extracellular ATP, the purinergic-receptor-P2X, ligand-gated ion channel 7 (P2RX7) on the cell surface of macrophage and/or monocytes change confirmation creating channels within the cell membrane that facilitate the exchange or changes of potassium (K), sodium (Na), and/or calcium (Ca) ions from within/outside of the cell. During this ion exchange process, K ions are lost from the cell while intracellular Ca ions increase. This leads to the activation of the inflammasome complex within the cell which contains the caspase-1 enzyme (also known as interleukin-converting enzyme or ICE, which is coded for by the CASP1 gene). Activated caspase-1 mediates IL1β maturation and the release of active IL1β (22). Based upon literature reports which suggested an association between adult onset chronic periodontitis and the +3953 genetic variant in the IL1B gene (23) (also termed +3954 or rs1143634), this variation was investigated and reported to be connected to EARR concurrent with orthodontia by trio genetic association and sib-pair linkage studies (24).
The IL1B +3953 genetic variant consists of individuals having either two arginine (AA) nucleic acid bases, two guanine (GG) nucleic acid bases, or a combination of the two (GA) at that particular genetic location. The other polymorphisms investigated for association with orthodontic patient EARR will also have the same possible combination of nucleic acids, or could have a combination of thymine (T), or cytosine (C) as illustrated in Table 2C. Some of the polymorphisms tested were functional, meaning the change in nucleic acids results in a change in protein function. These functional polymorphisms, as well as the nonfunctional ones, can also be markers for other genetic variations that may affect orthodontic patient EARR. In addition to the IL1B gene which codes for IL1β protein, the IL1 gene locus also contains the IL1A and IL1RN genes, which code for the IL1α and IL1 receptor antagonist proteins, respectively (25).
Table 2.
(A) Environmental and treatment variables, (B) cephalometric variables, (C) genetic variables
| Subjects with moderate to severe EARR (affected) | Subjects with little to no EARR (unaffected) | |
|---|---|---|
| (A) | ||
| Sex | 38 females 29 males | 38 females 29 males |
| Age at start of treatment in years (average + std error) | 15.78 ± 1.13 | 15.79 ± 1.14 |
| Number of years in treatment (average + std error) | 2.49 + 0.10 | 1.97 + 0.08 |
| Number of subjects with upper 1st Premolars extracted or missing (U4's) | 20 of 67 subjects | 7 of 67 subjects |
| Number of subjects with upper 2nd Premolars extracted or missing (U5's) | 0 of 67 subjects | 3 of 67 subjects |
| Number of subjects who had 1 or more maxillary premolars extracted or missing (U4 or U5) | 20 of 67 subjects | 10 of 67 subjects |
| (B) | ||
| Average pre-treatment measures + std error | ||
| OJ (mm) | 4.21 + 0.31 | 4.56 + 0.25 |
| OB (mm) | 3.07 + 0.31 | 3.27 + 0.28 |
| ANB (°) | 3.31 + 0.33 | 3.37 + 0.25 |
| FMA (°) | 25.15 + 0.72 | 23.50 + 0.65 |
| U1-FH (°) | 112.17 + 0.94 | 112.40 + 0.94 |
| U1-NA (°) | 21.09 + 0.94 | 20.85 + 0.97 |
| U1-NA (mm) | 3.95 + 0.33 | 3.66 + 0.35 |
| U1-ptv (mm) | 59.68 + 0.74 | 60.23 + 0.68 |
| U1r-ptv (mm) | 51.31 + 0.60 | 51.38 + 0.70 |
| Average change (Δ) in variable at the end of treatment + std error | ||
| ΔOJ (mm) | 1.71 + 0.27 | 1.87 + 0.27 |
| ΔOB (mm) | 1.76 + 0.30 | 1.92 + 0.25 |
| ΔANB (°) | 0.66 + 0.19 | 0.42 + 0.15 |
| ΔFMA (°) | 0.65 + 0.33 | −0.30 + 0.37 |
| ΔU1-FH (°) | −3.58 + 1.01 | −3.38 + 0.94 |
| ΔU1-NA (°) | −3.67 + 1.01 | −3.34 + 1.01 |
| ΔU1-NA (mm) | −0.59 + 0.40 | −0.94 + 0.39 |
| ΔU1-ptv (mm) | −1.34 + 0.48 | −1.34 + 0.45 |
| ΔU1r-ptv (mm) | 0.06 + 0.25 | −0.42 + 0.49 |
| (C) | ||||||
| P2RX7, rs208294 (Functional polymorphism) | CC 28 (41.8%) | CT 33 (49.3%) | TT 6 (9.0%) | CC 15 (22.4%) | CT 34 (50.7%) | TT 18 (26.9%) |
| P2RX7, rs1718119 (Functional polymorphism) | GG 32 (47.8%) | GA 25 (37.3%) | AA 10 (14.9%) | GG 28 (41.8%) | GA 32 (47.8%) | AA 7 (10.4%) |
| P2RX7, rs2230912 (Functional polymorphism) | AA 53 (79.1%) | AG 13 (19.4%) | GG 1 (1.5%) | AA 43 (64.2%) | AG 21 (31.3%) | GG 3 (4.5%) |
| CASP1/ICE, rs530537 | TT 24 (35.8%) | TC 30 (44.8%) | CC 13 (19.4%) | TT 18 (26.9%) | TC 36 (53.7%) | CC 13 (19.4%) |
| CASP1/ICE, rs580253 (Functional polymorphism) | GG 52 (77.6%) | GA 14 (20.9%) | AA 1 (1.5%) | GG 46 (68.7%) | GA 17 (25.4%) | AA 4 (6.0%) |
| CASP1/ICE, rs554344 (Functional polymorphism) | GG 52 (77.6%) | GC 14 (20.9%) | CC 1 (1.5%) | GG 45 (67.2%) | GC 18 (26.9%) | CC 4 (6.0%) |
| IL1β, rs1143634 (Functional polymorphism) | GG 37 (55.2%) | GA 26 (38.8%) | AA 4 (6.0%) | GG 44 (65.7%) | GA 21 (31.3%) | AA 2 (3.0%) |
| IL1RA, rs419598 | TT 41 (61.2%) | TC 23 (34.3%) | CC 3 (4.5%) | TT 38 (56.7%) | TC 29 (43.3%) | CC 0 (–) |
| IL1α rs1800587 | GG 28 (41.8%) | GA 28 (41.8%) | AA 11 (16.4%) | GG 32 (47.8%) | GA 29 (43.3%) | AA 6 (9.0%) |
Functional polymorphism refers to a polymorphism which either has been shown to alter the expression of the gene, or leads to an amino acid coding change which can influence the function or quantity of protein produced from the gene.
For this study, we hypothesized that by combining the analysis of numerous polymorphisms within and/or near the P2RX7, CASP1, IL1B, IL1A, and IL1RA genes, together with distinct treatment parameters, specific risk factors could be defined which contribute to in EARR in a population of orthodontic patients. A combined analysis of both multiple genetic and clinical factors potentially associated with EARR with orthodontia was performed in a case–control design format.
Materials and methods
Subject population
Approval for the study was obtained from the Institutional Review Boards of both Indiana University-Purdue University Indianapolis (IUPUI, Indianapolis, IN, USA) and the University of Kentucky (Lexington, KY, USA). A research data bank consisting of patient records and DNA was generated within a northern Indiana private orthodontic practice and contained information from 1458 individuals who received standard phase II orthodontic treatment. All patients were treated using conventional fixed edgewise appliances with a general wire sequence of 0.016-inch round nickel–titanium (Ni-Ti) to 0.016-inch round stainless steel to 0.016 × 0.022 inch rectangular Ni-Ti, and were finished with 0.016 × 0.022 inch rectangular wire in 0.018-inch slot brackets. Pre- and post-treatment lateral cephalometric, panoramic, and occlusal radiographs were taken as a part of each patient's standard of care and were available for research purposes. Four hundred and sixty Caucasian individuals were randomly selected from the research data bank and evaluated for EARR the four permanent maxillary incisors after the completion of orthodontic treatment (methodology described below). Individuals diagnosed with a syndrome, cleft lip/palate, showing open apices, and/or with a previous history of trauma were excluded. Individuals with missing treatment records and/or having non-diagnostic quality radiographs were also excluded. Sixty-seven subjects (38 females and 29 males) were identified with moderate to severe EARR (affected group). Among the patients with no EARR or only minimal EARR, 67 age- and gender-matched individuals were selected to serve as the control (unaffected) group.
Determination of EARR
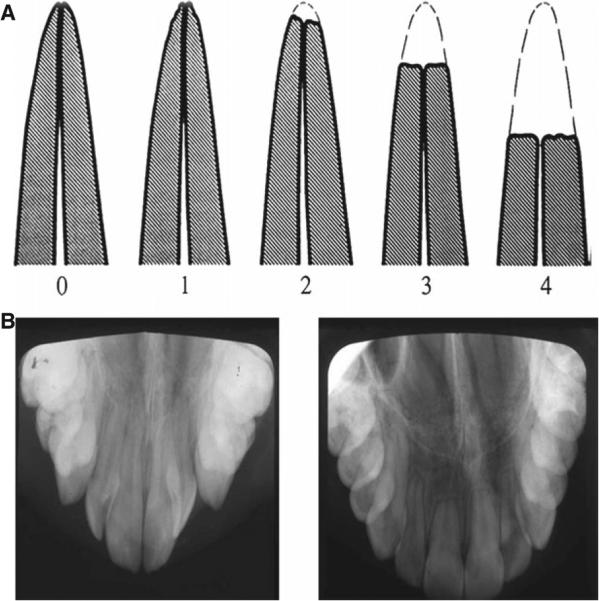
Digitized pre- and post-treatment panoramic and occlusal radiographs for each subject were compared to determine whether EARR had occurred to 1 or more of the four maxillary incisor roots during orthodontic treatment. The qualitative measures of Malmgren's grading system were used (26), where a rating of 0, 1, or 2 was considered control and a rating of 3 or 4 was graded as moderate or severe EARR (Fig. 1). Ratings of 3 or 4 represented EARR beyond blunting that involved the apical third or more of the tooth root. The evaluation of EARR was performed by three independent investigators in a blinded manner. When at least two of the three examiners agreed that the subject had EARR of 1 or more maxillary incisors, the patient was classified as being affected. All others were classified as being non-affected, even if slight blunting was present.
Fig. 1.
Evaluation method of EARR. (A). Classification scale of Malmgren (26) for EARR; where 0 = no resorption, 1 = slight irregularities, 2 = resorption of less than apical third, 3 = resorption of apical third, and 4 = resorption beyond apical third. (B). An example of the pre-orthodontic (left) and post-orthodontic treatment (right) occlusal radiographs for a subject diagnosed with EARR.
Measurement and/or assessment of clinical and treatment-related factors
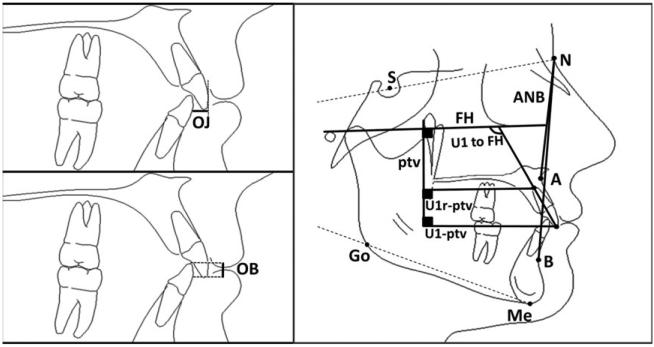
The length of treatment in months and type of treatment (extraction vs. non-extraction treatment) were recorded from patient records and confirmed on radiographs. Extraction of maxillary first premolars or maxillary second premolars was analyzed together and as unique variables in the statistical modeling. Digitalized cephalometric tracings were analyzed with Dolphin Imaging software (version 10.0; Dolphin Software Inc., Lake Oswego, OR, USA). Nine cephalometric variables were chosen as outlined in Table 1 and Fig. 2. The pre-treatment cephalometric values and the change (Δ) in cephalometric values at the end of treatment were considered unique variables in the statistical modeling. A decrease in the cephalometric value for incisor angulation with a negative (−) sign indicates an average retraction of those teeth.
Table 1.
Definitions of the cephalometric landmarks used
| Cephalometric variable | Description |
|---|---|
| Overjet (OJ) | Horizontal distance between the incisal edges of the maxillary and mandibular central incisors. |
| Overbite (OB) | Vertical distance between the incisal edges of the maxillary and mandibular central incisors |
| ANB | Angle formed by hard tissue A-point (A) to Nasion (N) to hard tissue B-point (B) |
| FMA | Angle formed between Frankfort horizontal (FH) plane and Gonion (Go) -Menton (Me) plane |
| U1- FH | Angle formed between the long axis of most procumbent maxillary central incisor (Upper1, U1) and FH. |
| U1-NA | Angle formed between the long axis of U1 and a line connecting hard tissue Nasion and A-point (NA). |
| U1-NA | Distance between the most anterior labial point of U1 and NA line |
| U1-ptv | Distance between U1 tip and the pterygoid vertical (ptv) perpendicular to FH |
| U1r-ptv | Distance between U1 root tip (U1r) and the pterygoid vertical (ptv) perpendicular to FH |
Fig. 2.
An illustration showing the selected cephalometric landmarks used in the study.
Genetic analyses
Buccal swab samples were obtained from each patient by having the subject brush the inside of each cheek 10 times with two sterile nylon bristle brushes. Genomic DNA was isolated with the Puregene method (Gentra Systems, Minneapolis, MN, USA), and DNA concentrations were measured on the NanoDrop-1000 spectrophotometer (Thermo Fisher Scientific, Wilmington, DE). Taq-Man®-based genotyping was utilized for the allelic discrimination of all polymorphisms tested on the LightCycler480® Instrument (Roche, Indianapolis, IN, USA) using standard genotyping reagents (Life Technologies/Applied Biosystems, Grand Island, NY, USA). Single nucleotide polymorphisms (SNPs) were examined within and/or near the purinergic-receptor-P2X, ligand-gated ion channel 7 (P2RX7; rs208294, rs1718119, and rs2230912), caspase-1/interleukin-converting enzyme (CASP1/ICE; rs580253, rs554344, and rs530537), interleukin-1 beta (IL1B; rs1143634), interleukin-1 alpha (IL1A; rs1800587), and interleukin-1 receptor antagonist (IL1RA, rs419598) genes.
Statistical methods
The kappa statistic was utilized to measure the variation associated with three radiographic examiners independently determining a patient's EARR status. Genotyping departures in the unaffected group from Hardy–Weinberg equilibrium (HWE) were assessed using the chi-square test. Stepwise logistic regression analysis applying the p-value threshold stopping rule with JMP® PRO Statistical Analysis Software (version 10; SAS Institute Inc., Cary, NC, USA) was used to test for variables associated with EARR. In addition to the variables analyzed, the presence of an interaction between the following pairs of variables was determined: 1) between the length of treatment and P2RX7 SNP rs208294 genotype; and 2) between extraction of maxillary first premolars and P2RX7 SNP rs208294 genotype. Statistical significance was when p was equal to or <0.05 as the initial alpha value, corrected for multiple comparisons as indicated by the layered Bonferroni technique of the nine variables in the final linear regression analysis of key variables influencing EARR (27).
Results
The kappa value for the three EARR examiners involved in this study was 0.08, indicting a good agreement between the examiners regarding the EARR status of each subject. There were no age restrictions for study enrollment; subjects’ age ranged from 8.4 to 55.4 years. The average age at the start of treatment ± std error for the affected group was 15.78 ± 1.13 years (median age = 12.87 years, mode = 12.23 years) and for the unaffected group was 15.79 ± 1.14 years (median age = 12.89 years, mode = 12.15 years) (Table 2A). The average length of treatment ± std error for the case and control groups was 2.49 ± 0.10 years and 1.97 ± 1.14 years, respectively. In the final linear regression modeling, the length of treatment was the largest variable influencing the occurrence of EARR in the population studied where longer treatment times increased the probability of EARR (Table 3; p < 0.0001, significant at a layered Bonferroni correction of p = 0.006).
Table 3.
Final linear regression analysis of key variables influencing EARR
| Variables | p-value | Layered Bonferroni corrected significant alpha |
|---|---|---|
| Length of time in treatment (years) | <0.0001* | 0.006 |
| P2RX7, rs208294 {CC&CT vs. TT} | 0.0028* | 0.006 |
| Upper 1st Premolars were extracted or missing | 0.0469 | 0.007 |
| IL1β, rs1143634 {AA&GA vs. GG} | 0.0533 | 0.008 |
| Change in FMA | 0.0591 | 0.01 |
| P2RX7, rs1718119 {AA&GG vs. GA} | 0.0813 | 0.01 |
| Change in Ur-pvt | 0.0908 | 0.02 |
| CASP1/ICE, rs530537 {TT vs. CC&TC} | 0.1527 | 0.03 |
| P2RX7, rs208294 {CC vs. CT} | 0.1564 | 0.05 |
Statistically significant compared to alpha corrected by layered Bonferroni procedure; Extraction of second premolars was not included in the analysis. The factors summarized in the table explain 25.13% of the variation observed in this sample.
Of the 67 EARR-affected individuals, 20 had maxillary first premolar extractions compared to only seven controls with first premolar extractions and three controls with maxillary second premolar extractions. Although this type of extraction appeared to play a significant role in influencing the occurrence of EARR (Table 3; p = 0.0469), this was not significant when compared to the layered Bonferroni correction of p = 0.007 for that variable. Due to the low number of second premolar extractions in this study population, and the fact that second premolar extractions only occurred within the control population, second premolar extractions were not factored into the final modeling analysis.
The average overall change in FMA did not have a statistically significant influence in EARR occurrence during orthodontic treatment (Table 3; p = 0.0591, compared to being statistically significant if at or less than the layered Bonferroni correction of p = 0.01). Average pre-treatment ANB angles ± std error between the case and control groups were comparable at 3.31° ± 0.33° and 3.37° ± 0.25°, respectively (Table 2B), with the range of ANB measurements spanning from 12° to −4°. The average retraction distance for the central incisor(s) ± std error, as measured by the ΔU1-NA, was −0.59 ± 0.40 mm for the case group, compared to −0.94 ± 0.39 mm for the controls (Table 2B). Within the case group, however, individuals who received maxillary first premolar extractions (n = 20) had an average ΔU1-NA of 1.62 mm compared to an average ΔU1-NA of −1.54 mm for individuals in the same group who did not receive maxillary first premolar extractions (n = 47). Similarly, individuals in the control group who received maxillary first premolar extractions (n = 7) had an average ΔU1-NA of 3.31 mm, compared to −1.44 mm for those in the control group who did not have maxillary first premolars removed during orthodontic treatment (n = 60).
The average ΔU1-NA angle (°) for individuals in the case group ± std error was −3.67° ± 1.01°, compared to −3.34° ± 1.01° in the control group (Table 2B). Within the case group, individuals who received maxillary first premolar extractions (n = 20) had an average ΔU1-NA angle of 1.88° compared to an average ΔU1-NA of −6.08° for individuals in the same group who did not receive maxillary first premolar extractions (n = 47). Individuals in the control group who received maxillary first premolar extractions (n = 7) had an average ΔU1-NA angle of 8.46°, compared to −4.72° for those in the control group who did not have maxillary first premolars removed during orthodontic treatment (n = 60).
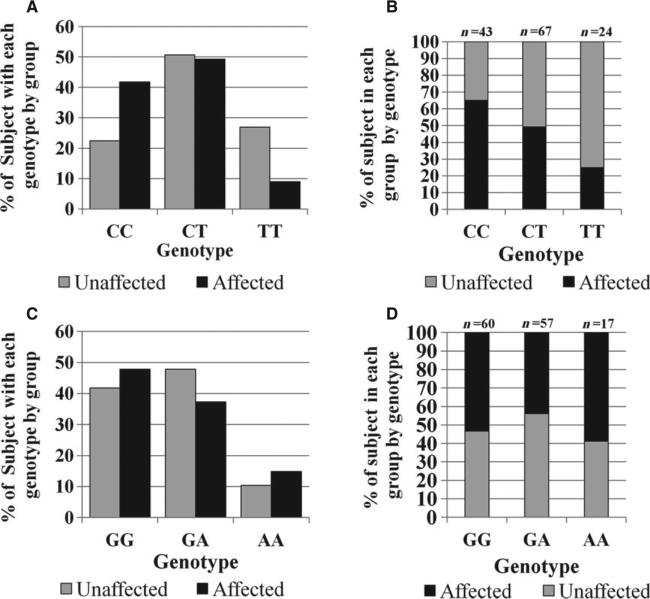
Within the genetic analyses included in this study, no departures from Hardy–Weinberg equilibrium (HWE) were detected within the control population for all of the tested SNPs (data not shown). The complete set of genetic data is summarized in Table 2C. Based on the final linear regression modeling, variations in the P2RX7 gene showed the greatest influence on EARR (Table 3). A comparison of the genotypic distributions of the P2RX7 polymorphisms rs208294 and rs1718119 for the case and control groups is illustrated in Fig. 3A,C, respectively. For the rs208294 polymorphism, patients carrying the CC or CT genotype were more likely to develop EARR than individuals carrying the TT genotype (Table 3; p = 0.0028), which is significant at the layered Bonferroni correction of p = 0.006. The percentage of individuals from each group with a specific P2RX7 rs208294 genotype is illustrated in Fig. 3B. The percentage of individuals from each group with a specific P2RX7 rs1718119 genotype is shown in Fig. 3D.
Fig. 3.
Genotyping results from P2RX7 polymorphisms rs208294 and rs1718119 for EARR-affected and unaffected individuals. (A). Genotypic distribution of P2RX7 SNP rs208294 by group. (B). Percentage of individuals from each group shown by P2RX7 rs208294 genotype. (C). Genotypic distribution of P2RX7 SNP rs1718119 by group. (D). Percentage of individuals from each group shown by P2RX7 rs1718119 genotype.
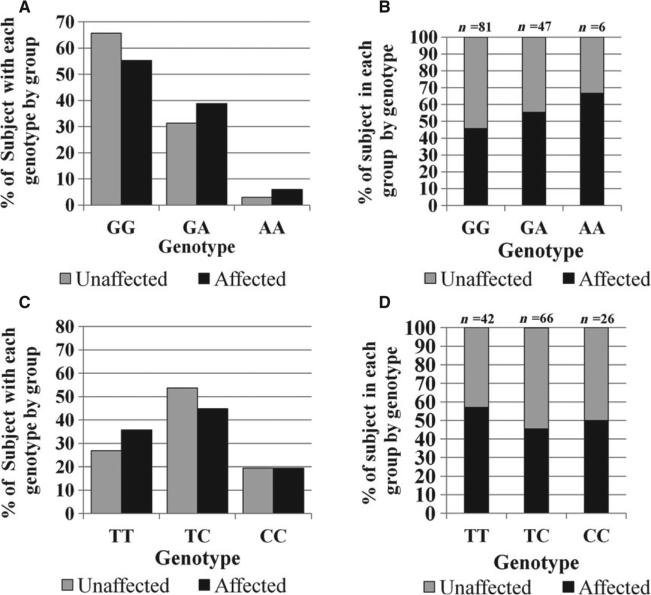
Although findings related to the IL1B polymorphism, rs1143634, were not statistically significant in this model (p = 0.0533 compared to the layered Bonferroni corrected significant alpha of p = 0.008), IL1β likely plays a downstream role in the etiology of EARR given the significance of the P2RX7 gene for the upstream P2RX7 receptor on EARR. A comparison of the genotypic distributions of the IL1B polymorphism rs1143634 for the case group vs. the control group is illustrated in Fig. 4A. The percentage of individuals from each group with a specific IL1B rs1143634 genotype is illustrated in Fig. 4B. Similarly, a comparison of the genotypic distributions of the CASP1 polymorphism rs580253 is shown by group in Fig. 4C for both the case and control group. The percentage of individuals from each group with a specific CASP1 rs580253 genotype is shown in Fig. 3D.
Fig. 4.
Genotyping results from IL1B SNP rs1143634 and CASP1 SNP rs580253 for EARR-affected and unaffected individuals. (A). Genotypic distribution of IL1B SNP rs1143634 by group. (B). Percentage of individuals from each group shown by IL1B rs1143634 genotype. (C). Genotypic distribution of CASP1 SNP rs530537 by group. (D). Percentage of individuals from each group shown by CASP1 rs530537 genotype.
In summary, the stepwise logistic regression analysis indicated that the nine modeled variables shown in Table 3 were able to explain ~25.13% of the variation of the EARR among individuals tested. The strongest factor associated with the occurrence of (EARR) was a longer length of treatment, explaining ~10% of the outcome by itself (Table 3). Of the tested genetic variables, the P2RX7 SNP rs1143634 CC&CT vs. TT genotypes showed a significant difference between affected and control groups. Extraction of the maxillary first premolars also explained a portion of the variation associated with EARR occurrence, although compared to the layered Bonferroni corrected alpha this variable was not significant by itself (Table 3).
The analysis showed no statistically significant interaction between the length of treatment and the P2RX7 SNP rs208294 (p = 0.8). Similarly, there was no interaction between the extraction of maxillary first premolars and the P2RX7 SNP rs208294.
Discussion
Research has aided in identifying many treatment-related factors that influence the occurrence of EARR during orthodontic treatment. It remains a major challenge, however, to resolve why certain subjects are more susceptible to EARR than others. Genetics plays a role in this patient-to-patient difference in susceptibility, with heritability studies indicating genetic variation accounting for approximately two-thirds of the EARR variation seen in orthodontic patients (15, 16).
The novelty of this particular study lays in the multivariate analysis of numerous genetic variables in the IL1B expression pathway, some of which were previously implicated as factors in root resorption, and treatment-related variables to generate a more comprehensive understanding of their collective influence on the occurrence of EARR concurrent with standard comprehensive 0.018-inch slot edgewise appliance orthodontic treatment. While the influence of age and gender on the etiology of EARR has not consistently been demonstrated in the literature (4, 10, 21, 28), age- and gender-matched case and control groups were utilized in this study to eliminate these potential cofactors from the equation.
Long treatment length and the presence of the CC&CT genotypes for P2RX7 SNP rs208294 were significantly associated with EARR in this study. Together, the nine cofactors tested in the final linear regression modeling explained ~25.13% of the variation of the EARR observed in the sample tested. Long lengths of treatment proved to be the strongest risk factor for EARR in this study, accounting for ~10% of the variation. This observation is supported by several previously published studies (7, 10, 29). Prolonged stress on the roots increases the chance of root resorption. Therefore, minimizing treatment time when possible or, in some cases, altering the treatment plan to perform a shorter treatment might be the best option.
Based on mouse gene knockout (KO) experiments, there has been supporting evidence in the literature to suggest that IL1B expression pathway has a significant role in the occurrence EARR. In the mouse Il1b gene KO model, the absence of Il1b was shown to be associated with increased orthodontically induced histologic root resorption of the mouse molars (30, 31). In a separate mouse KO model of the P2x7 receptor (P2rx7), which lies upstream of Il1b maturation and release, no difference was observed at baseline between the wild-type (‘normal’) and KO mice histologic root resorption levels. With the application of force to the teeth of both wild-type and P2rx7 gene KO mice, however, there was a significant increase in histologic root resorption on the KO mouse teeth over the levels of resorption observed in wild-type animals (32, 33).
Pertinent to this study in humans, the second most influential risk factor associated with EARR was the genetic influence of the CC&CT genotypes for the P2RX7 polymorphism, rs208294. In contrast, the IL1B polymorphism rs1143634 A-allele, which had been linked to increased IL1β secretion in vitro (34), and was thought to be somewhat protective against EARR based on the initial studies in humans (24, 35, 36), was not significant by itself. It is not clear why this study found that the rs1143634 AA&GA genotypes were associated with EARR, in contrast to previous reports showing that the IL1B SNP rs1143634 G-allele was more likely to be associated with orthodontic patients who developed EARR (24, 35, 36).
In a recent published meta-analysis, no association was apparent between EARR and IL1B SNP +3954 (37); however, the studies included in the meta-analysis only focused on genetic factors and did not take into account other treatment and environmental factors that could influence the occurrence of EARR in a multivariate analysis. In other studies, the IL1A polymorphism rs1800587 was associated with EARR in a German sample, while the rs1143634 IL1B SNP was not (38). IL1B rs1143634 was reported to be associated with velocity of tooth movement along with IL1RA, suggesting a possible connection between velocity of tooth movement, and EARR (39). Taking together the findings of both mouse and human studies, the role of IL1B in EARR may be more complicated than first anticipated. Clearly, the role of the IL1β expression pathway on influencing EARR warrants further study.
The finding in this study that the genetic factors tested only account for a small amount of the variation associated with EARR when compared to the influence of the length of treatment does not appear to be consistent with the earlier estimation of heritability suggesting that approximately two-thirds of the variation in EARR with orthodontia is due to genetic factors (15, 16). This may be due in part to the inherent limitations of determining heritability in the narrow sense and its application to clinical questions (40), and/or to the limited number of genetic factors that we analyzed for this complex trait. Future studies should look for additional genetic factors that may be involved in orthodontic patient EARR utilizing a non-biased whole-exome or whole-genome sequencing approach when possible instead of the current candidate gene models.
Conclusion
It can be concluded that increased length of treatment was a strong risk factor of post-orthodontic EARR. The functional SNP, rs208294, located in P2RX7 gene was also associated with EARR in this model, indicating that the downstream expression of IL1B also affects EARR during orthodontic treatment.
Clinical relevance.
A better understanding of the etiology of EARR is needed to improve the ability of predicting its occurrence. Although EARR may arise in the absence of orthodontic treatment, studies suggest that its incidence may be increased during orthodontic care. When obtaining a patient's consent for orthodontic treatment, clinicians should explain the potential risk factors that lead to EARR and the odds of it occurring. Studies have indicated that two-thirds of the clinical variation in EARR is associated with genetic variation in orthodontic patients. However, determination of these genetic factors is incomplete, requiring studies to include more genetic and clinical factors. By understanding how a combination of both genetic and treatment factors may place a patient at higher risk for EARR during orthodontics, clinicians should be better equipped to explain the risk and minimize treatment-related EARR occurrence.
Acknowledgements
The authors would like to thank Dr Pinar Emecen Huja and Dr Riyad Al-Qawasmi for their insightful reviews. The authors would also like to thank Dr McKenzie Woods, Dr Mutlaq Alotaibi, Dr Hemina Desai, and Kelly Ren for assisting with the digitization of radiograph, importing radiographs into Dolphin, and/or gathering tooth extraction information from patient records. This work was funded in part by The Indiana University Bixler Fund for Research in Genetics, the Southern Association of Orthodontists, and The University of Kentucky College of Dentistry E. Preston Hicks Endowed Chair. The authors have no conflict of interest associated with this study or any future research pertaining to this subject.
Footnotes
Sharab L. Y., Morford L. A., Dempsey J., Falcão-Alencar G., Mason A., Jacobson E., Kluemper G. T., Macri J. V., Hartsfield J. K. Jr. Genetic and treatment-related risk factors associated with external apical root resorption (EARR) concurrent with orthodontia
Contributor Information
L. Y. Sharab, Department of Oral Health Practice, University of Kentucky College of Dentistry, Lexington, KY, USA
L. A. Morford, Department of Oral Health Science, University of Kentucky College of Dentistry, Lexington, KY, USA Hereditary Genomics Laboratory, Center for Oral Health Research, University of Kentucky College of Dentistry, Lexington, KY, USA.
J. Dempsey, Department of Oral Health Science, University of Kentucky College of Dentistry, Lexington, KY, USA
G. Falcão-Alencar, Hereditary Genomics Laboratory, Center for Oral Health Research, University of Kentucky College of Dentistry, Lexington, KY, USA
A. Mason, Department of Oral Health Science, University of Kentucky College of Dentistry, Lexington, KY, USA
E. Jacobson, Hereditary Genomics Laboratory, Center for Oral Health Research, University of Kentucky College of Dentistry, Lexington, KY, USA
G. T. Kluemper, Department of Oral Health Science, University of Kentucky College of Dentistry, Lexington, KY, USA
J. V. Macri, Department of Orthodontics and Oral Facial Genetics, Indiana University School of Dentistry, Indianapolis, IN, USA
J. K. Hartsfield, Jr, Department of Oral Health Science, University of Kentucky College of Dentistry, Lexington, KY, USA; Hereditary Genomics Laboratory, Center for Oral Health Research, University of Kentucky College of Dentistry, Lexington, KY, USA; Department of Orthodontics, College of Dentistry, University of Illinois at Chicago, Chicago, IL, USA; Department of Orthodontics and Oral Facial Genetics, Indiana University School of Dentistry, Indianapolis, IN, USA.
References
- 1.Killiany DM. Root resorption caused by orthodontic treatment: an evidence-based review of literature. Semin Orthod. 1999;5:128–33. doi: 10.1016/s1073-8746(99)80032-2. [DOI] [PubMed] [Google Scholar]
- 2.Remington DN, Joondeph DR, Artun J, Riedel RA, Chapko MK. Long-term evaluation of root resorption occurring during orthodontic treatment. Am J Orthod Dentofacial Orthop. 1989;96:43–6. doi: 10.1016/0889-5406(89)90227-8. [DOI] [PubMed] [Google Scholar]
- 3.Weltman B, Vig KW, Fields HW, Shanker S, Kaizar EE. Root resorption associated with orthodontic tooth movement: a systematic review. Am J Orthod Dentofacial Orthop. 2010;137:462–76. doi: 10.1016/j.ajodo.2009.06.021. discussion 12A. [DOI] [PubMed] [Google Scholar]
- 4.Segal GR, Schiffman PH, Tuncay OC. Meta analysis of the treatment-related factors of external apical root resorption. Orthod Craniofac Res. 2004;7:71–8. doi: 10.1111/j.1601-6343.2004.00286.x. [DOI] [PubMed] [Google Scholar]
- 5.Martins DR, Tibola D, Janson G, Maria FR. Effects of intrusion combined with anterior retraction on apical root resorption. Eur J Orthod. 2012;34:170–5. doi: 10.1093/ejo/cjq178. [DOI] [PubMed] [Google Scholar]
- 6.Parker RJ, Harris EF. Directions of orthodontic tooth movements associated with external apical root resorption of the maxillary central incisor. Am J Orthod Dentofacial Orthop. 1998;114:677–83. doi: 10.1016/s0889-5406(98)70200-8. [DOI] [PubMed] [Google Scholar]
- 7.Nanekrungsan K, Patanaporn V, Janhom A, Korwanich N. External apical root resorption in maxillary incisors in orthodontic patients: associated factors and radiographic evaluation. Imaging Sci Dent. 2012;42:147–54. doi: 10.5624/isd.2012.42.3.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Acar A, Canyurek U, Kocaaga M, Er-verdi N. Continuous vs. discontinuous force application and root resorption. Angle Orthod. 1999;69:159–63. doi: 10.1043/0003-3219(1999)069<0159:CVDFAA>2.3.CO;2. discussion 63–4. [DOI] [PubMed] [Google Scholar]
- 9.Picanco GV, de Freitas KM, Cancado RH, Valarelli FP, Picanco PR, Feijao CP. Predisposing factors to severe external root resorption associated to orthodontic treatment. Dental Press J Orthod. 2013;18:110–20. doi: 10.1590/s2176-94512013000100022. [DOI] [PubMed] [Google Scholar]
- 10.Baumrind S, Korn EL, Boyd RL. Apical root resorption in orthodontically treated adults. Am J Orthod Dentofacial Orthop. 1996;110:311–20. doi: 10.1016/s0889-5406(96)80016-3. [DOI] [PubMed] [Google Scholar]
- 11.Horiuchi A, Hotokezaka H, Kobayashi K. Correlation between cortical plate proximity and apical root resorption. Am J Orthod Dentofacial Orthop. 1998;114:311–8. doi: 10.1016/s0889-5406(98)70214-8. [DOI] [PubMed] [Google Scholar]
- 12.Linge L, Linge BO. Patient characteristics and treatment variables associated with apical root resorption during orthodontic treatment. Am J Orthod Dentofacial Orthop. 1991;99:35–43. doi: 10.1016/S0889-5406(05)81678-6. [DOI] [PubMed] [Google Scholar]
- 13.Owman-Moll P, Kurol J, Lundgren D. Continuous vs. interrupted continuous orthodontic force related to early tooth movement and root resorption. Angle Orthod. 1995;65:395–401. doi: 10.1043/0003-3219(1995)065<0395:CVICOF>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 14.Kurol J, Owman-Moll P, Lundgren D. Time-related root resorption after application of acontrolled continuous orthodontic force. Am J Orthod Dentofacial Orthop. 1996;110:303–10. doi: 10.1016/s0889-5406(96)80015-1. [DOI] [PubMed] [Google Scholar]
- 15.Harris EF, Kineret SE, Tolley EA. A heritable component for external apical root resorption in patients treated orthodontically. Am J Orthod Dentofacial Orthop. 1997;111:301–9. doi: 10.1016/s0889-5406(97)70189-6. [DOI] [PubMed] [Google Scholar]
- 16.Hartsfield JK, Everett ET, Al-Qawasmi RA. Genetic factors in external apical root resorption and orthodontic treatment. Crit Rev Oral Biol Med. 2004;15:115–22. doi: 10.1177/154411130401500205. [DOI] [PubMed] [Google Scholar]
- 17.Massler M, Malone AJ. Root resorption in human permanent teeth: a roentgenographic study. Am J Orthod. 1954;40:619–33. [Google Scholar]
- 18.Massler M, Perreault J. Root resorption in the permanent teeth of young adults. J Dent Child. 1954;21:158–64. [Google Scholar]
- 19.Newman WG. Possible etiologic factors in external root resorption. Am J Orthod. 1975;67:522–39. doi: 10.1016/0002-9416(75)90298-5. [DOI] [PubMed] [Google Scholar]
- 20.Reitan K. Some factors determining the evaluation of forces in orthodontics. Am J Orthod. 1957;43:32–45. [Google Scholar]
- 21.Sameshima GT, Sinclair PM. Predicting and preventing root resorption: Part I. Diagnostic factors. Am J Orthod Dentofacial Orthop. 2001;119:505–10. doi: 10.1067/mod.2001.113409. [DOI] [PubMed] [Google Scholar]
- 22.Ferrari D, Pizzirani C, Adinolfi E, Lemoli RM, Curti A, Idzko M, et al. The P2X7 receptor: a key player in IL-1 processing and release. J Immunol. 2006;176:3877–83. doi: 10.4049/jimmunol.176.7.3877. [DOI] [PubMed] [Google Scholar]
- 23.Kornman KS, Crane A, Wang HY, Giovlne FSD, Newman MG, Pirk FW, et al. The interleukin-1 geno-type as a severity factor in adult periodontal disease. J Clin Period. 1997;24:72–7. doi: 10.1111/j.1600-051x.1997.tb01187.x. [DOI] [PubMed] [Google Scholar]
- 24.Al-Qawasmi RA, Hartsfield JK, Jr, Everett ET, Flury L, Liu L, Foroud TM, et al. Genetic predisposition to external apical root resorption. Am J Orthod Dentofacial Orthop. 2003;123:242–52. doi: 10.1067/mod.2003.42. [DOI] [PubMed] [Google Scholar]
- 25.Dinarello CA. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood. 2011;117:3720–32. doi: 10.1182/blood-2010-07-273417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Malmgren O, Goldson L, Hill C, Orwin A, Petrini L, Lundberg M. Root resorption after orthodontic treatment of traumatized teeth. Am J Orthod. 1982;82:487–91. doi: 10.1016/0002-9416(82)90317-7. [DOI] [PubMed] [Google Scholar]
- 27.Darlington RB. Regression and Linear Models. McGraw-Hill; New York: 1990. [Google Scholar]
- 28.Kjaer I. Morphological characteristics of dentitions developing excessive root resorption during orthodontic treatment. Eur J Orthod. 1995;17:25–34. doi: 10.1093/ejo/17.1.25. [DOI] [PubMed] [Google Scholar]
- 29.DeShields RW. A study of root resorption in treated Class II. Division I malocclusions. Angle Orthod. 1969;39:231–45. doi: 10.1043/0003-3219(1969)039<0231:ASORRI>2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 30.Al-Qawasmi RA, Hartsfield JK, Hartsfield JK, Jr, Everett ET, Weaver MR, Foroud TM, et al. Root resorption associated with orthodontic force in IL-1Beta knockout mouse. J Musculoskelet Neuronal Interact. 2004;4:383–5. [PubMed] [Google Scholar]
- 31.Hartsfield JK., Jr Pathways in external apical root resorption associated with orthodontia. Orthod Craniofac Res. 2009;12:236–42. doi: 10.1111/j.1601-6343.2009.01458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Viecilli RF, Katona TR, Chen J, Hartsfield JK, Jr, Roberts WE. Orthodontic mechanotransduction and the role of the P2X7 receptor. Am J Orthod Dentofacial Orthop. 2009;135:694, e1–16. doi: 10.1016/j.ajodo.2008.10.018. discussion 94–5. [DOI] [PubMed] [Google Scholar]
- 33.Viecilli R, Katona T, Chen J, Roberts E, Hartsfield J., Jr Comparison of dentoalveolar morphology in WT and P2X7R KO mice for the development of biomechanical orthodontic models. Anat Rec (Hoboken) 2009;292:292–8. doi: 10.1002/ar.20822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pociot F, Molvig J, Wogensen L, Worsaae H, Nerup J. A TaqI polymorphism in the human interleukin-1β (IL-1β) gene correlates with IL-1β secretion in vitro. Eur J Clin Invest. 1992;22:396–402. doi: 10.1111/j.1365-2362.1992.tb01480.x. [DOI] [PubMed] [Google Scholar]
- 35.Bastos Lages EM, Drummond AF, Pretti H, Costa FO, Lages EJ, Gontijo AI, et al. Association of functional gene polymorphism IL-1beta in patients with external apical root resorption. Am J Orthod Dentofacial Orthop. 2009;136:542–6. doi: 10.1016/j.ajodo.2007.10.051. [DOI] [PubMed] [Google Scholar]
- 36.Iglesias-Linares A. Postorthodontic external root resorption is associated with IL1 receptor antagonist gene variations. Oral Dis. 2011;18:198–205. doi: 10.1111/j.1601-0825.2011.01865.x. [DOI] [PubMed] [Google Scholar]
- 37.Wu FL, Wang LY, Huang YQ, Guo WB, Liu CD, Li SG. Interleukin-1beta +3954 polymorphisms and risk of external apical root resorption in orthodontic treatment: a meta-analysis. Genet Mol Res. 2013;12:4678–86. doi: 10.4238/2013.October.18.6. [DOI] [PubMed] [Google Scholar]
- 38.Gülden N, Eggermann T, Zerres K, Beer M, Meinelt A, Diedrich P. Inter-leukin-1 polymorphisms in relation to External Apical Root Resorption (EARR). J Orofacial Orthop. 2009;70:20–38. doi: 10.1007/s00056-009-8808-6. [DOI] [PubMed] [Google Scholar]
- 39.Iwasaki LR, Gibson CS, Crouch LD, Marx DB, Pandey JP, Nickels JC. Speed of tooth movement is related to stress and IL-1 gene polymorphisms. Am J Orthod Dentofacial Orthop. 2006;130:e1–698. e9. doi: 10.1016/j.ajodo.2006.04.022. [DOI] [PubMed] [Google Scholar]
- 40.Harris EF. Interpreting heritability estimates in the orthodontic literature. Semin Orthod. 2008;14:125–34. [Google Scholar]