Abstract
The retroviral integrases are virally encoded, specialized recombinases that catalyze the insertion of viral DNA into the host cell’s DNA, a process that is essential for virus propagation. We have learned a great deal since the existence of an integrated form of retroviral DNA (the provirus) was first proposed by Howard Temin in 1964. Initial studies focused on the genetics and biochemistry of avian and murine virus DNA integration, but the pace of discovery increased substantially with advances in technology, and an influx of investigators focused on the human immunodeficiency virus (HIV). We begin with a brief account of the scientific landscape in which some of the earliest discoveries were made, and summarize research that led to our current understanding of the biochemistry of integration. A more detailed account of recent analyses of integrase structure follows, as they have provided valuable insights into enzyme function and raised important new questions.
Keywords: prophage, provirus, reaching dimer, core dimer, intasome, integrase
EARLY HISTORY
A New Type of Virus-Cell Interaction: Prophage and Provirus
The study of retroviruses dates back to the early 1900s, with the discovery of a filterable agent associated with equine anemia by Vallée and Carré in 1904 and the identification of similar agents that cause leukemia or solid tumors in chickens by Vilhelm Ellerman and Olaf Bang in 1908 and Peyton Rous in 1911, respectively (Table 1). Remarkably, the discovery of retroviruses as “filterable infectious agents” predated the discovery of the simpler bacterial viruses by Frederick Twort in 1915, which Félix d’Hérelle named bacteriophages in 1917. However, it was members of the Phage Group, a loose association of bacteriophage researchers coalesced around the physicist Max Delbrück at Caltech and the Cold Spring Harbor Laboratory in the 1940s and 1950s, who developed the quantitative approaches and tools of molecular biology that would eventually be applied to the study of all viruses. Initially, the Phage Group focused solely on the bacteriophages that kill their host cells, as the life cycles of these viruses were straightforward and easy to analyze. Although the phenomenon of lysogeny had been proposed by Eugène and Elisabeth Wollman at the Pasteur Institute in the 1930s (1), this more temperate manifestation of virus biology was at that time considered an irrelevant diversion by Delbrück and others. Nevertheless, André Lwoff began to study isolated clones of Wollman’s lysogenic Bacillus megaterium under the microscope. By 1949, he and his colleagues at the Pasteur Institute had discovered conditions in which the genetic material of the virus, which they presumed to have somehow incorporated itself into the host’s chromosome (he called this a prophage), could be induced to produce progeny virus and lyse the cell (reviewed in 2). François Jacob and the Wollmans’ son, Élie, extended these investigations with another bacteriophage, λ, which could be propagated lytically or establish lysogeny in Escherichia coli; this system was to become a workhorse for molecular biology. Although the λ DNA was initially thought to be somehow hooked on to the chromosomes of lysogenic bacteria (3), the position of attachment was mapped to a specific site. Subsequent mapping of λ genes by others revealed that the unintegrated and integrated genomes comprised circular permutations of one another, laying the groundwork for a unique model for λ DNA integration, put forth by Alan Campbell in 1961 (4). The Campbell model invoked a circular DNA intermediate, and the 1963 report by Alfred Hershey and colleagues that λ phage DNA possessed short complementary single-stranded ends provided the means for such circularization (5).
Table 1.
Integrase history: chronology of selected discoveries and milestones
| Year | Discovery | Virus/notes | Reference(s)a |
|---|---|---|---|
| Viruses as infectious agents; discovery of reverse transcriptase and integrase | |||
| 1904–1936 | Discovery of retroviruses as infectious agents |
EIAV Chicken leukemia (ALV) Sarcomas (ASV) Mouse mammary tumors (MMTV) |
Vallée & Carré (133b) Ellermann & Bang (134) Rous (135) Bittner (136) |
| 1951–1957 | Leukemia virus isolation | MLV | Gross (137, 138) |
| 1954 | Cell culture isolation | PFV | Enders & Peebles (139) |
| 1957–1967 | Many new virus discoveries | Infecting rodents, sheep, chickens, cats, etc. |
— |
| 1958 | Focus assay and cell transformation |
Rous ASV | Temin & Rubin (140) |
| 1959 | X-ray-induced virus | MLV | Lieberman & Kaplan (141) |
| 1963–1964 | Provirus hypothesis | ASV: transformation | Svoboda, Hilgert, and colleagues (8); Temin (7) |
| 1965 | Inherited, endogenous virus | MMTV | Muhlbock (142) |
| 1970 | Reverse transcriptase in virus particles |
MLV ASV |
Baltimore (9) Temin & Mizutani (10) |
| 1978–1980 | Molecular characterization of viral DNA and provirus in infected cells |
MLV MSV ASV |
Hughes, Varmus, and colleagues (143) Dhar, Vande Woude, and colleagues (144) Ju & Skalka (27) |
| 1978 | First integrase purified from virus particles |
ALV: pp32 endonuclease | Grandgenett et al. (145) |
| 1981 | Discovery of the first human retrovirus |
HTLV-1 | Poiesz, Gallo, and colleagues (127) |
| 1983 | Discovery of the AIDS virus | HIV-1 | Gallo et al. (129); Barré-Sinoussi, Montagnier, and colleagues (128) |
| Integrases and their biochemical activities | |||
| 1984 | Integrase is essential for virus propagation |
MLV: integrase mutation MLV: integrase deletion SNV: integrase deletion |
Donehower & Varmus (146) Schwartzberg & Goff (147) Panganiban & Temin (148) |
| 1987 | Integration demonstrated with preintegration complexes from cells |
MLV | Brown, Bishop, and colleagues (149) |
| 1989 | In vitro integrase assays show specificity for DNA ends |
ASLV (ASV and ALV): short oligonucleotides |
Katzman, Leis, and colleagues (35) |
| 1990 | Bacterially produced integrase catalyzes processing and joining in vitro |
MLV ASLV |
Craigie (33) Katz, Skalka, and colleagues (34) |
| Domain structure and integrase functions | |||
| 1991 | Domain organization | Sequence alignments Deletion studies |
Khan, Skalka, and colleagues (56) Bushman, Craigie, and colleagues (54) |
| 1992 | Identification of catalytic residues | Conserved D,D(35)E motif | Kulkosky, Skalka, and colleagues (59) |
| 1992–1993 | Functional form is a multimer | Biochemistry Complementation |
Jones, Skalka, and colleagues (71) van Gent, Plasterk, and colleagues (86); Engelman, Bushman & Craigie (72) |
| 1994–1995 | First crystal structures | HIV: CCD ASLV: CCD |
Dyda, Davies, and colleagues (60) Bujacz, Wlodawer, and colleagues (61) |
| 1995–1997 | NMR structures of the terminal domains |
HIV-1: CTD HIV-1: NTD HIV-2: NTD |
Lodi, Gronenborn, and colleagues (66) Cai, Gronenborn, and colleagues (57) Eijkelenboom, Plasterk, and colleagues (58) |
| 2000–2009 | Two-domain crystal structures | HIV-1: CCD+CTD SIV: CCD+CTD ASLV: CCD+CTD HIV: NTD+CCD MVV: NTD+CCD |
J.C. Chen, Stroud, and colleagues (76) Z. Chen, Kuo, and colleagues (77) Yang, Hyde, and colleagues (78) Wang, Craigie, and colleagues (75) Hare, Cherepanov, and colleagues (79) |
| 2002–2004 | Integration site selection | HIV-1: human cells ASLV: human cells |
Schroder, Bushman, and colleagues (150) Narezkina, Katz, and colleagues (151) |
| 2003–2006 | Integrase tetramer for concerted integration |
Biochemistry/biophysics | Bao, Skalka, Wong, and colleagues (70); Li, Craigie, and colleagues (74) |
| 2003–2005 | Integrase tethering protein | HIV-1: CCD+LEDGF | Cherepanov, Debyzer, and colleagues (152); Cherepanov, Engelman, and colleagues (153) |
| 2007 | First FDA-approved HIV-1 integrase antiviral drug |
Raltegravir (active site inhibitor) | Merck |
| Full-length integrase structures and inhibitor mechanisms | |||
| 2010 | First intasome structure | PFV | Hare, Cherepanov, and colleagues (83) |
| 2010 | Intasome structure with active site inhibitor |
PFV | Hare, Cherepanov, and colleagues (83) |
| 2010 | Intasome with target DNA | PFV | Maertens, Hare & Cherepanov (88) |
| 2011–2013 | First full-length architectures of apo-integrase |
ASLV: monomer and dimer HIV-1: dimers and tetramer |
Andrake, Skalka, and colleagues (94) Bojja, Skalka, and colleagues (93) |
| 2010–2014 | Allosteric inhibitor | HIV-1: CCD | Christ, Debyzer, and colleagues (118); Sharma, Kvaratskhelia, and colleagues (154) |
For space, only the first publication of a specific discovery is cited in this chronology. The many, equally important follow-up and confirmation studies could not be included, but the interested reader is urged to investigate these further.
Domain abbreviations: CCD, core catalytic domain; CTD, C-terminal domain; NTD, N-terminal domain.
Virus abbreviations (and genera): ALV, avian leukosis virus (Alpharetrovirus); ASLV, ALV and ASV family (Alpharetrovirus); ASV, avian sarcoma/leukosis virus (Alpharetrovirus); EIAV, equine infectious anemia virus (Lentivirus); HIV, human immunodeficiency virus (Lentivirus); HTLV-1, human lymphotropic virus type 1 (Deltaretrovirus); MLV, murine leukemia virus (Gammaretrovirus); MMTV, mouse mammary tumor virus (Betaretrovirus); MMV, maedi-visna virus (Lentivirus); MSV, Moloney sarcoma virus (Gammaretrovirus); PFV, prototype foamy virus (Spumavirus); SIV, simian immunodeficiency virus (Lentivirus); SNV, spleen necrosis virus (Gammaretrovirus).
A long hiatus followed the discovery of the avian retroviruses as infectious agents (Table 1), as chickens were not thought to be an important model for human disease and leukemias were not recognized as cancer. This changed in the 1950s with the identification of viruses that infect a variety of animals. Efforts to apply quantitative methods to the study of animal viruses began in the mid-1950s. One of the major centers of such research was the laboratory of Renato Dulbecco. It was in this Caltech laboratory that Harry Rubin and Howard Temin developed a focus assay for the Rous sarcoma virus (RSV) with cultures of chicken embryo fibroblasts. They noticed that foci of cells transformed with different strains of RSV had distinct morphologies, and that viruses produced by each type of transformed cell passed on that phenotype. From such observations Temin concluded that viral genes must control the morphology of the infected cells and that “transformation was a conversion analogous to lysogenic conversion” (6, p. 1075). He surmised that the RNA genome of RSV must somehow produce a DNA encoded form, which he called a provirus, in analogy with the prophages (7). A similar idea was put forth independently by Jan Svoboda in Prague, based on studies with RSV-transformed rat cells (8). But this concept was heretical, as it contradicted the accepted (unidirectional) dogma of molecular biology: DNA → RNA → protein. Although Temin acquired evidence consistent with his proposal, experimental tools available at the time were not quite up to the task, and the provirus hypothesis was widely ignored (and even derided) for the next six years. The situation changed only when, inspired by the discovery of polymerases in other animal virus particles, David Baltimore (9) as well as Temin and Satoshi Mizutani (10) independently discovered reverse transcriptase (RT) in retrovirus particles. That a retroviral provirus is actually embedded in host cell DNA was confirmed most convincingly in 1972 by Miroslav Hill and Jana Hillova, who showed that chicken cells treated with DNA purified from RSV-transformed rat cells produced progeny identical to the virus with which the rat cells were originally transformed (11). In the ensuing years it became apparent that the genomes of humans and other animals are littered with retroviral proviruses [as well as the genes of other viruses (12, 13)] that are relics of ancient infections of germline cells, and that such integrations are occurring even in contemporary time (14). Furthermore, the process is not unique to viruses; retrotransposition, DNA mobilization through an RNA intermediate, is a property of a number of noninfectious genetic elements, which, together with retroviruses, have had profound effects on evolution (summarized most recently in 15, 16).
Establishment of Landmarks
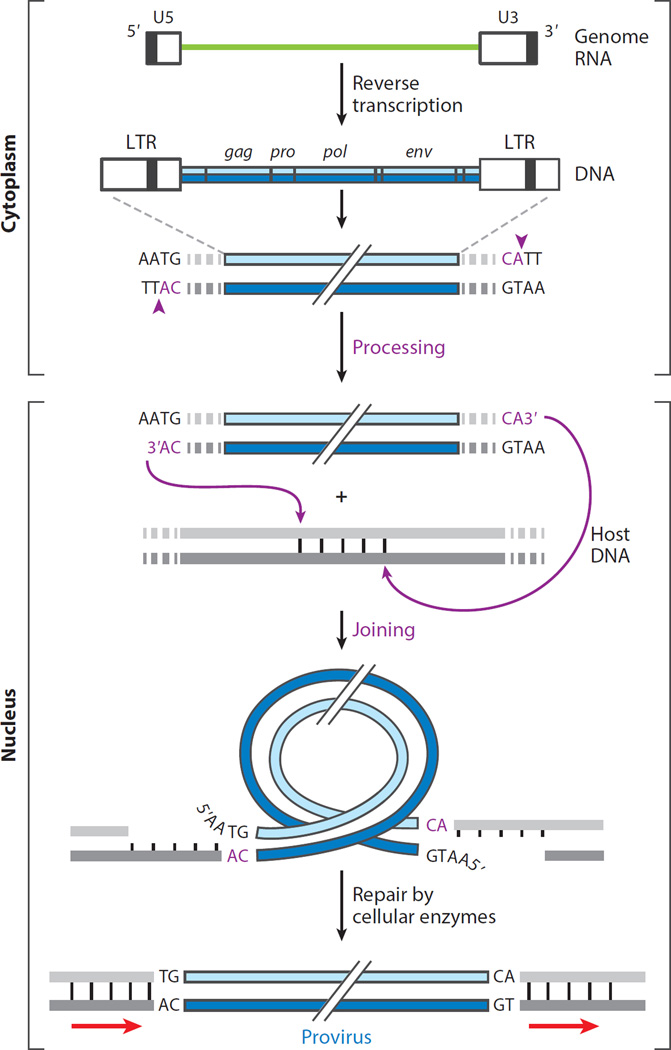
The discovery of RT, the concept of a provirus, and the implications of proviruses for genome evolution and cancer encouraged many researchers, including several phage biologists (the authors of this review included), to enter the retrovirus field. The application of new tools, such as molecular cloning and DNA sequencing, led to a burst of activity in the late 1970s and on into the 1980s, focused on elucidation of molecular aspects of retroviral DNA integration (Table 1). Initial analyses of retroviral DNA (vDNA) synthesized in cells infected with avian or murine retroviruses revealed several unique features but also created some interesting red herrings. The latter included the notion that, like bacteriophage λ, a closed circular form (or forms) of RSV DNA was an intermediate in the integration reaction (17, 18). This initially attractive idea proved to incorrect. It was subsequently established that a linear form of vDNA is the substrate for integration (19) and that the two circular forms normally present in very small amounts in the nucleus are nonfunctional, dead-end products (20, 21). Indeed, the more appropriate phage model for retroviral integration is bacteriophage Mu transposition (22). Subsequent analyses (summarized in 23) showed that vDNA and the provirus are colinear with the viral RNA genome and that upon integration the vDNA is cropped, usually by 2 bp from each end. The relationship with known transposable elements was established by the observation that, as with these elements, the ends of all proviruses include the same dinucleotide base pair, CA/GT. This dinucleotide is embedded, usually 2 bp from the termini of vDNA, in an imperfect inverted repeat that can be as long as 20 bp for some viruses (24–30). As with known transposable elements, small direct repeats of host DNA flank each proviral end (31). The fact that the sizes of the duplications are characteristic of the virus [4 bp for murine leukemia virus (MLV), 5 bp for human immunodeficiency virus type 1 (HIV-1), 6 bp for avian sarcoma/leukosis virus (ASLV)] implicated a viral rather than a host enzyme in their production. The genesis of these features (summarized in Figure 1) became clear with the isolation and characterization of retroviral integrases.
Figure 1.
Steps in the synthesis of retroviral DNA and its integration into host DNA. The viral RNA genome (green line) is reverse transcribed in the cytoplasm of the cell within a subviral nucleoprotein structure (called the reverse transcription complex) to form a duplex DNA containing long terminal repeats (LTRs) of sequences unique to the 5′ (U5) and 3′ (U3) ends of the viral RNA. The organization of the genes common to all retroviruses (gag, pro, pol, and env) is colinear with the RNA genome. Imperfect inverted repeats at the LTR duplex termini are recognized and nicked by cognate integrase (IN) proteins, following a conserved CA dinucleotide at each 3′ end (vertical arrows in inset), producing recessed 3′-OH ends. This first reaction catalyzed by IN is called processing and takes place within a nucleoprotein assembly called a preintegration complex. A tetramer of IN bound to the processed viral DNA (vDNA) ends enters the nucleus where a joining reaction catalyzed by IN connects the 3′-OH ends of the vDNA to staggered phosphates at a target site in the host DNA. The length of the stagger (4–6 bp) is characteristic of the viral IN protein. The conserved CA dinucleotide and steps catalyzed by IN are highlighted in magenta. Removal of the noncomplementary 5′ nucleotides of vDNA and repair of the gaps in host DNA by host enzymes generate a covalently integrated provirus, shorter by 2 bp on either end and flanked by duplications of the target site (arrows).
Discovery and Study of Integrase Activity In Vitro
If RT is present in virus particles, it made sense to young biochemist Duane Grandgenett that a viral protein that mediates integration might also be present in these particles. In pioneering work conducted from the 1970s to early 1980s (summarized in 32), Grandgenett purified and characterized the first retroviral integrase protein, initially called pp32, from avian myoblastosis virus (AMV) by virtue of its DNA-binding and endonuclease activities. AMV is an ASLV family member that was supplied to many in the scientific community via the Virus Cancer Program at the National Cancer Institute. Subsequent genetic studies with ASLV, MLV, and spleen necrosis virus (SNV) confirmed that a protein encoded at the end of the viral pol gene is indeed required for integration. This protein, subsequently named integrase (IN), is one of three enzymes encoded in all retroviral genomes [protease (PR), RT, and IN].
All three viral enzymes are contained in virus particles, along with RNA genomes, and are carried into the cell following virus entry. The vDNA is synthesized by RT within a subviral particle that retains capsid proteins and is called the reverse transcription complex. The integration reaction catalyzed by IN was first detected in vitro in 1987, by providing a target (e.g., DNA from a plasmid or bacteriophage λ) to subviral nucleoprotein structures derived from the reverse transcription complex, called preintegration complexes (PICs), which were isolated from the cytoplasm of MLV-infected cells (19, 21). However, the ability to molecularly clone IN DNA and purify catalytically active IN proteins (33, 34) made it possible to delineate many mechanistic details of integration. Specificity of the purified IN proteins for cognate vDNA ends was first demonstrated in 1989, by use of a simple assay in which short oligonucleotide duplexes represent cognate vDNA ends (35, 36). This assay, and its derivatives, proved invaluable in elucidating the biochemistry of retroviral integrases, analyzing their structure, and developing therapeutic drugs (37).
BIOCHEMICAL REACTIONS CATALYZED BY INTEGRASE
Action at the Ends of Retroviral DNA
Numerous studies with infected cells and purified retroviral IN proteins established that two biochemically and temporally distinct steps are catalyzed by these enzymes (Figure 1). In the first step, the 3′ ends of the vDNA are nicked, such that nucleotides (usually two) following the conserved CA are removed from each 3′ end. This processing step requires duplex DNA termini and therefore can occur only when synthesis of vDNA ends is completed by RT. In addition to the conserved CA, other nucleotides in the short terminal inverted repeats and some upstream in the vDNA also affect the efficiency of the reaction. Experiments with infected cells demonstrated that the processing step can take place in the cytoplasm (38) and that PICs include other viral and host proteins in addition to IN and vDNA (39). The second catalytic step is a concerted cleavage and ligation reaction in which the two newly processed 3′ vDNA ends are joined to staggered (by 4 to 6 bp) phosphates at a target site in host DNA. The product of the joining step is a gapped intermediate in which the 5′-PO4 ends of the provirus are not linked to host DNA.
Interruption of host cell chromatin by insertion of a large stretch of newly synthesized, naked vDNA comprises a major assault on the genomic integrity of the cell. Not surprisingly, such disruption induces a DNA damage response (40). Host enzymes are assumed to accomplish postintegration repair, which generates the short direct repeats of the target sequence that flank the provirus (40–43). In MLV-infected cells, covalent joining of the 5′ ends of the integrated vDNA to host DNA can be detected within an hour after 3′ end-joining by IN (44), although the details by which such repair occurs are not yet known.
Target Site Selection
Although retroviral DNA integration can occur at many loci in host cell genomes, site selection is not random and varies among retroviruses. The availability of whole-genome sequence data for human, mouse, and avian species and the application of high-throughput sequencing methods made it possible in the late 1990s to determine integration site preferences for representatives of most of the seven retroviral genera (Alpharetrovirus, Betaretrovirus, Gammaretrovirus, Deltaretrovirus, Epsilonretrovirus, Lentivirus, and Spumavirus) (reviewed in 45). For example, MLV, a gammaretrovirus, shows a marked preference for integration in, or near, transcription start sites and CpG islands, a feature common to the promoter regions of highly expressed genes. The lentivirus HIV-1 also shows a strong preference for integration within active transcription units (but not start sites or CpG islands), and ASLVs, which are alpharetroviruses, have only a weak preference for transcription units and CpG islands (but not transcription start sites). Finally, the betaretrovirus mouse mammary tumor virus (MMTV) exhibits no significant deviation from random in integration site preference. Dissimilar requirements for chromatin structure or accessibility may explain some of these differences (46–48). In at least two cases, it is now clear that attachment to specific chromatin protein tethers is a major determinant for integration site selection: A loop in a C-terminal domain of the chromatin-targeting transcription factor lens epithelium–derived growth factor (LEDGF/p75) binds tightly in a pocket formed at a dimer interface of HIV-1 IN, and the conserved extraterminal domain of Brd proteins, commonly found in transcriptional promoter regions, attaches to a short sequence in the C-terminal tail of the MLV and feline leukemia virus (FeLV) IN proteins (49–52). It is likely that chromatin tethering plays an important role in the integration of other retroviruses.
DNA sequence also influences integration site selection. Weak but characteristic palindromic consensus sequences have been identified in the integration target sites of members of the different retroviral families (for review, see 45). This preference is functionally distinct from that influenced by IN tethering. For example, although HIV-1 DNA integration frequency is greatly reduced and transcription units are no longer preferred in cells that lack LEDGF/p75, the palindromic consensus for these integrations is the same as in cells that possess the targeting protein (53). It appears therefore that IN binding to chromatin protein tethers brings the enzyme-vDNA complex into close proximity with distinct regions of host chromatin, and that target sites are then selected based on the ability of the host DNA to be accommodated in the enzyme’s active site.
INTEGRASE STRUCTURE
Three Common Domains
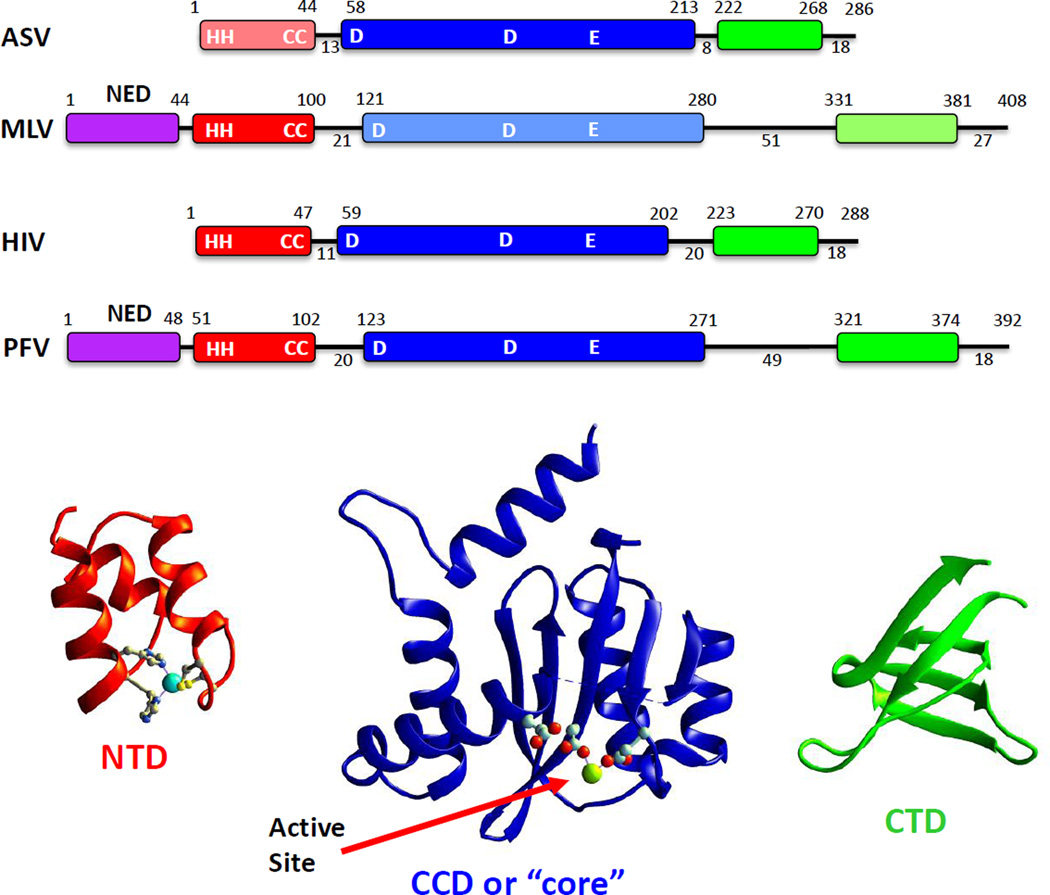
The domain structure of IN proteins was first delineated by protein sequence analyses, limited proteolysis, and deletion studies in the early 1990s (54–56) (Table 1). Retroviral IN proteins comprise approximately 300 to 400 amino acids and include three common domains, connected by linkers of varying length (Figure 2). The N-terminal domain (NTD) includes two pairs of invariant, Zn2+-chelating histidine and cysteine residues (HHCC motif). The bound Zn2+ ion stabilizes a helix-turn-helix structural motif in this domain (57, 58).
Figure 2.
Domain organization of integrase (IN) proteins from different retroviruses. (a) Maps for the organization of IN proteins from the alpharetrovirus avian sarcoma/leukosis virus (ASLV), the gammaretrovirus murine leukemia virus (MLV), the lentivirus human immunodeficiency virus type 1 (HIV-1), and the spumavirus prototype foamy virus (PFV). Amino acid numbers delineate the start and end of each domain: the N-terminal extension domain (NED; purple); the N-terminal domain (NTD; red), including the HHCC motif; the catalytic core domain (CCD; blue), including the D,D(35)E motif; and the C-terminal domain (CTD; green). The lengths of linkers that connect the domains are indicated below the lines between domains. Domains for which there is no experimentally determined structure from crystallography are in muted colors. (b) Domain models from crystal structures of the HIV-1 NTD (PDB 1K6Y), CCD (PDB 1BIU), and CTD (PDB 1EX4). The Zn2+ ion in the NTD is shown as an aqua sphere; one of the two Mg2+ ions bound in the active site of the CCD is shown as a green sphere. The conserved Glu residue of the D,D(35)E motif is presumed to chelate the second metal ion together with the first conserved Asp residue.
The catalytic core domain (CCD) is characterized by a constellation of three invariant acidic amino acids, the last two separated by 35 residues: the D,D(35)E motif (59). These acidic amino acids chelate the two metal ions that are required for both processing and joining. Solution of the crystal structures of the isolated CCDs of HIV-1 IN and ASLV IN (60, 61) revealed that these proteins are members of a large superfamily of nucleases and recombinases, including the RNase H domain of RT (reviewed in 62) and the cellular enzymes that catalyze V(D)J recombination required for antibody production (63, 64). During catalysis by these two-metal enzymes, one cation helps to position the incoming nucleophile and the other activates the leaving hydroxyl group (65). In the case of IN proteins, the two cations bound in the active site switch roles in the processing and joining reactions, in a ping-pong effect. The retroviral proteins are unusual, however, in that they have very low turnover rates (~0.1 s−1 in vitro). Given that only one concerted joining reaction is required to attach viral to host DNA in an infected cell, this is not likely to be a limitation in vivo.
The amino acid sequence of the C-terminal domain (CTD) is not well conserved among IN proteins from different retroviral genera. Nevertheless, similar tertiary structures are seen in the CTDs of the retroviral INs analyzed to date. All comprise β-strand barrels that resemble known SH3 domains (66, 67) and include DNA-binding and multimerization determinants (68, 69). Some retroviral IN proteins [e.g., those of MLV and prototype foamy virus (PFV) (Figure 2)] have an additional domain at their N termini, called the N-terminal extension domain (NED).
Integrase Multimers Are Required for Catalysis
Biochemical, genetic, and complementation studies with purified bacterially produced proteins established that ASLV IN and HIV-1 IN function as multimers (70–74). Whereas a dimer appears to be sufficient to perform the processing reaction in vitro, a tetramer is required for the concerted integration of two vDNA ends into a target DNA (70, 74), and the HIV-1 IN tetramer was shown to be stabilized by interaction with a pair of vDNA ends (74). As illustrated in the following section, our understanding of the organization of a functional IN tetramer, and the mechanics of the integration steps, took a giant leap forward in 2010 with the crystallization of PFV IN together with bound DNA substrates and active site inhibitors. These structures afford a framework for consolidation of much of the knowledge gained from earlier biochemical studies, but they also expose new questions.
Structure of the PFV Intasome
Numerous attempts by many laboratories to obtain crystals suitable for structural analysis of full-length IN proteins of the most intensely studies viruses—ASLV, MLV, and HIV-1— have, to date, met with failure. The HIV-1 and ASLV IN proteins tend to aggregate at concentrations required for crystallization, a phenomenon that is generally exacerbated in the presence of DNA. The presence of flexible linkers between the domains is also likely to contribute to these difficulties. The best that had been accomplished were structures of two-domain fragments of IN proteins from HIV-1 (75, 76), the related simian immunodeficiency virus (SIV) (77), ASLV (78), the lentivirus maedi-visna virus (MVV) (79), and, more recently, a full-length ASLV IN in which only two domains are resolved (80). These structures were nonetheless extremely valuable, as they confirmed essential features of the protein domains and suggested possible domain interactions. Undaunted by the formidable technical difficulties, Cherepanov and colleagues (81) systematically tested IN proteins from samples of all of the known genera to find one that might be amenable to crystallographic analysis. The winner was the somewhat unusual PFV IN, a protein that is monomeric at high concentrations but is highly active for concerted joining of viral oligonucleotides as short as 16 bp to a target DNA in vitro. Nevertheless, the observation that PFV IN is sensitive to active site inhibitors of HIV-1 indicated that their active sites were likely to be quite similar (81). Because only the right end of PFV DNA needs to be processed for integration (82), oligonucleotide duplexes representing that end of the unintegrated vDNA were included in the crystallization trials. Even so, “over 30 DNA constructs were tested in initial trials with full-length wild type and several mutant-derived PFV IN proteins in ~40,000 initial sparse matrix conditions” before the crystal structure of a tetramer of PFV IN bound to two viral oligonucleotides was obtained (83, p. 236).
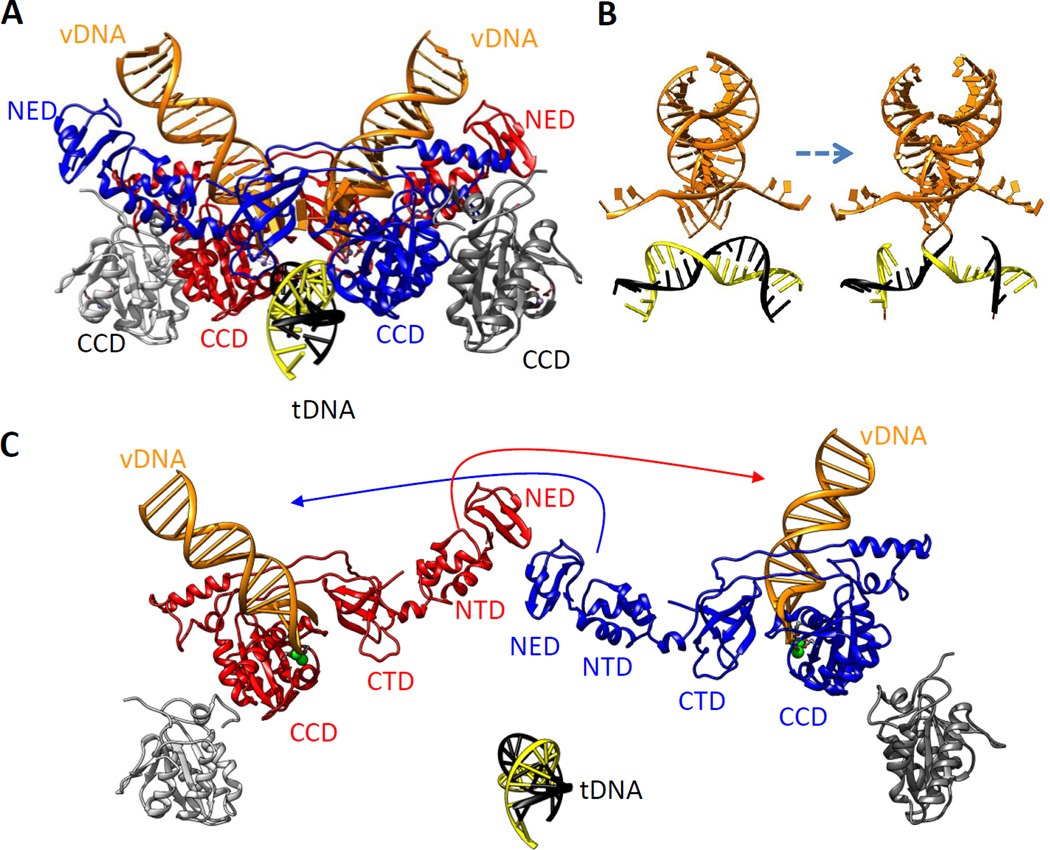
The PFV structure, called an intasome, showed somewhat surprisingly that the two inner IN protomers provide both DNA-binding and catalytic functions (Figure 3a). There are no clues to the function of the outer protomers, of which only the CCDs were resolved. As predicted from earlier studies (84, 85), the last three base pairs of the vDNA ends do not remain double stranded in the active sites of the complex; rather, each is distorted and partially unwound. The frayed 5′-PO4 end of the noncleaved vDNA strand is threaded between the CCD and CTD, and the strand with the 3′-OH end is positioned in the active site for cleavage. Consistent with the reciprocity suggested by earlier complementation experiments (72, 86), the NED and NTD of one inner monomer bind the vDNA that is acted upon in the catalytic center of the other. Each CTD of these two inner subunits makes contacts with both vDNAs, thus contributing both cis and trans DNA-binding functions. Results from subsequent small-angle X-ray and neutron scattering (SAXS/SANS) of PFV IN-vDNA complexes in solution were consistent with the intasome crystal structure and, in addition, showed that the disordered domains of the outer protomers are extended outward, making no contact with the substrates or inner dimers (87).
Figure 3.
PFV IN tetramer in complex with two vDNA oligonucleotides and a tDNA. (a) The assembled complex (PDB 4E7K) (89; see also supplemental movies in 88) is shown in ribbon representation with the inner subunits in red and blue. Only the CCDs of the outer subunits (gray) are resolved. vDNA oligonucleotides are in orange ribbon ladder representation and the tDNA in yellow and black. The locations of the NEDs and CCDs are indicated. (b) DNA components of the complex portrayed before and after joining, rotated 90° about the y axis from panel a. Processed vDNA ends are shown prior to joining (left) and after joining (right) to the target DNA. (c) The complex shown in panel a is pulled apart to show the positions of all domains in the inner subunits. Interactions between the distal NTD and NED of one inner subunit and the vDNA held in the CCD of the other inner subunit are indicated by arrows. Assembly of the complex is shown in Video 1. Abbreviations: CCD, catalytic core domain; CTD, C-terminal domain; IN, integrase; NED, N-terminal extension domain; NTD, N-terminal domain; PFV, prototype foamy virus; tDNA, target DNA; vDNA, viral DNA.
Crystallographic analysis of the PFV intasome together with bound active site inhibitors of HIV-1 IN showed that these drugs block the joining reaction by disorienting the 3′ A nucleotide and preventing target DNA access, affording new insight into the inhibitory mechanisms of current FDA-approved HIV antivirals (83). Furthermore, as the same structure is observed in PFV IN crystals with bound processed or unprocessed vDNA ends, no conformational changes are associated with the first step in the reaction. To determine the structure of a complex with target DNA, Cherepanov and colleagues (88) prepared crystals that included a 30-bp palindromic DNA duplex that contained the consensus PFV integration sequence. By omitting the metal cofactors or including vDNA oligonucleotides that lacked free 3′-OH ends, they solved the structure of a PFV integration intermediate with two viral ends and target DNAs bound but not joined (88). The overall topology of the IN-DNA complex did not change upon target DNA binding. However, as predicted from earlier biochemical studies, the target DNA was bent in the active site (Figure 3b). The physiological relevance of the structure was confirmed by observation of both processing and joining reactions in crystallo when the metal cofactor and hydrolysable vDNA ends were provided (89).
The PFV IN structures have afforded valuable templates for constructing models of other retroviral IN-DNA complexes, in particular for HIV-1 IN (90). However, the differences in properties, linkers, and domains of PFV IN and other retroviral IN proteins have presented some challenges to modeling efforts. Furthermore, whereas the PFV structures have illuminated critical details of enzyme architecture and mechanism, they have also brought several new questions into focus. The function of and requirement for the outer monomers and the pathway by which functional tetramers assemble are two at the forefront.
Properties of Integrase Proteins in the Absence of DNA
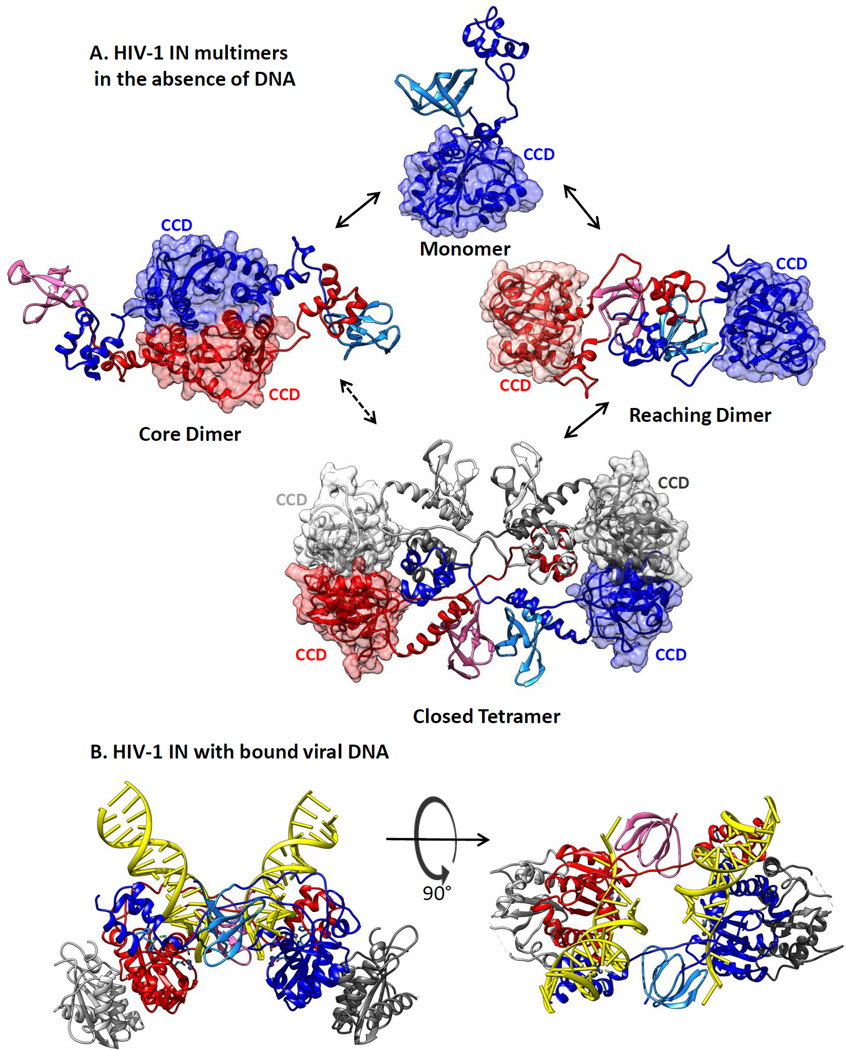
The topologies of the domains of different retroviral INs are similar, but aside from the conserved motifs, their surface charges and hydrophobicity vary, as do the lengths and compositions of the linkers by which the domains are joined. Such differences contribute to the features that distinguish their behaviors in solution. IN proteins from several retroviruses differ in both solubility and multimerization properties (91, 92). For example, in the absence of DNA and at similar protein concentration (~60 µM), ASLV IN is a dimer and HIV-1 IN is a tetramer, whereas PFV IN is a monomer even at twice this concentration (93). Interestingly, the architecture of the full-length ASLV dimer in solution, determined in our laboratory by SAXS and protein cross-linking analyses, resembles that of the inner dimer in the PFV intasome. The CCDs are located at the outer edges of this dimer, and the NTD of one protomer reaches out via its linker to contact the CCD and CTD of the second protomer, in a configuration called a reaching dimer (94). The CTDs of this ASLV dimer make strong hydrophobic contacts, a feature that is distinct from the inner dimer of the PFV intasome, in which the CTDs bind vDNA ends rather than each other. Similar SAXS and protein cross-linking analyses of HIV-1 IN revealed that this protein can form two apparently equally stable dimers in solution (Figure 4a). One dimer (called a core dimer) assembles via CCD-CCD interactions, as have been seen in crystals of isolated CCDs and two-domain fragments. The architecture of the other dimer resembles that of the ASLV IN reaching dimer. The SAXS-determined HIV-1 IN tetramer envelope can accommodate two stacked reaching dimers. (93). However, the apo-IN tetramer in this model has no room for the DNA substrates without undergoing substantial conformational change.
Figure 4.
Models for architectures of full-length human immunodeficiency virus type 1 (HIV-1) apo-integrase (IN) protein in solution. (a) Structures for HIV-1 IN protein in the absence of DNA substrates were derived by HADDOCK data-driven modeling of the HIV-1 IN monomer, dimer, and tetramer in solution, based on small-angle X-ray scattering and protein cross-linking data (93). Catalytic core domains (CCDs) are rendered in muted surface representation to emphasize their locations in the structures. It is not yet known which of these forms is competent for the viral DNA binding that leads to assembly of an HIV-1 intasome. (b) An HIV-1 intasome model (90) with DNA shown in yellow ladder representation. Structures, colored as in Figure 3, are in a ribbon rendering and were generated using Chimera software (133).
In one hypothetical intasome assembly pathway, IN monomers might first bind vDNA ends and partially separate the tips by threading the noncleaved strand between the CCD and CTD (Figure 3c, Video 1). Such an association would be consistent with the observation that end fraying by ASLV IN occurs in cis with the active site and, though detectable with isolated CCDs, is most efficient with fragments that contain both the CCD and CTD (85). As a dimer appears to be the minimal form able to process vDNA ends (73), full fraying and end processing may require formation of an inner dimer-like structure in which each NTD makes contact with the DNA held by the other inner protomer in trans. Addition of the outer monomers, before or after vDNA binding, may provide the increased stability required for target DNA capture, as CCD-CCD interactions are known to be required for concerted integration (94). Alternatively, formation of a reaching dimer might precede vDNA binding. In this case a conformational change in which CTDs are repositioned to bind and fray the vDNA ends would be required, as has been suggested for ASLV IN (93) (Figure 4b).
The PFV intasome assembles from IN monomers in vitro, with vDNA oligonucleotides imparting stability to the complex in the crystals. Some evidence suggests that HIV-1 intasomes are also assembled from monomers in vitro (95), but whether this is the case for other retroviral INs has not yet been determined. Moreover, nothing is known about how intasomes assemble in infected cells. It seems possible that additional molecules of IN and other viral or cellular proteins may promote such assembly. Consequently, until more is known about the internal organization of the reverse transcription complexes in which vDNA is synthesized, and the PICs in which IN processes the ends of that DNA, the significance of differences in solution properties of retroviral IN proteins is impossible to gauge. Indeed, there are many indications that much remains to be learned about how retroviral IN is organized and functions in vivo.
QUESTIONS REMAINING
Posttranslational Modification
Aside from partial phosphorylation of a serine near the C terminus of the ASLV IN (which has no known function) (96), the incidence and effect of modifications on IN proteins isolated from retrovirus particles have not been investigated in detail. It has been speculated, however, that biologically relevant modifications might be introduced following virus entry. HIV-1 IN purified from bacteria can be SUMOylated at sites in all three domains, and ubiquitination of residues in the CTD and preceding linker, acetylation in the CTD, and phosphorylation of a serine in the CCD have also been reported (reviewed in 97). The introduction of such changes could affect not only IN stability or catalytic activity but also interactions with other proteins at various stages of the retroviral life cycle. For example, HIV-1 IN is subject to degradation by the ubiquitin-proteasome pathway, and it has been suggested that such degradation may be required so that cellular components for postintegration repair and gene transcription can gain access to the provirus after its integration (98). Two histone acetyltransferases (p300 and GCN5) have been shown to bind to HIV-1 IN, both in vitro and in infected cells, and acetylation of lysines in the CTD is reported to enhance integration in vitro by increasing DNA binding affinity (99, 100). Finally, mutants of HIV-1 that express IN proteins with substitutions that prevent SUMOylation are defective in a step after reverse transcription but before integration, leading to the proposal that addition of SUMO may enhance IN binding to cellular proteins that are required for efficient virus reproduction (101). The cellular pathways for the placement and removal of posttranslational modifications are complex, but the interplay among them and the net downstream effects do regulate critical events in cell biology, including gene expression, DNA repair, and the cell cycle. Consequently, further study may reveal mechanisms by which such modifications can influence critical interactions of retroviral INs with other viral or cellular proteins.
Protein-Protein Interactions
As the product of RT is the substrate for IN, functional interactions between the two proteins at early stages in the retrovirus life cycle may be expected, and several studies with purified enzymes support this idea. Indeed, the larger subunit of the ASLV RT dimer has IN fused to its C-terminal end, an arrangement that is likely to contribute importantly to reverse transcription, as an RT dimer that lacks IN exhibits reduced processivity and RNase H activity in vitro (102). Physical associations between the RT and IN proteins of MLV and HIV-1 have been detected by a variety of assays (103–109), and investigations with purified HIV-1 RT indicate that IN can stimulate both the initiation and elongation activities of RT by enhancing its processivity (110). Site-directed mutagenesis studies have identified residues in the CTD of HIV-1 IN that are critical for binding to RT (111). However, interpretation of these results is complicated by the fact that RT-IN interactions may occur in the context of viral polyproteins during progeny particle assembly in infected cells as well as in their mature, proteolytically processed forms.
Numerous cellular proteins have been identified as candidate participants in the integration reactions of MLV or HIV-1, based on their association with PICs and/or their ability to bind to these IN proteins (112–114). The relevance of most such associations to retroviral DNA integration, or to other possible functions of IN, has yet to be investigated carefully. Notable exceptions include the chromatin tethering proteins LEDGF/p75 and members of the Brd family, mentioned above, and two cellular proteins: IN interactor 1 (INi-1) and barrier to autointegration factor (BAF). INi-1, a core component of the SWI/SNF chromatin-remodeling complex, was discovered in a search for cellular proteins that would bind to HIV-1 IN (115). Although the protein can stimulate IN catalysis in vitro, this did not prove to be a physiologically important activity. HIV-1 IN mutants that cannot bind to INi-1 exhibit defects in virus particle morphology and reverse transcription rather than in integration per se (116). The cellular BAF protein is a salt-extractable component of MLV and HIV-1 PICs isolated from infected cells. Purified BAF forms DNA-binding dimers and can produce intermolecular bridges that compact vDNA, a reaction that prevents an autointegration reaction (52). Purified virus particles do not contain BAF, which must therefore be acquired after entry. Experiments with MLV PICs support the notion that BAF facilitates normal integration by inhibiting autointegration, but it is still unclear whether this cellular protein provides the same function for HIV-1 or, indeed, any other retrovirus.
Virus Particle Morphogenesis
It has been known for some time that that HIV-1 IN mutants are pleiotropic (117): Some mutations (called Class I) block integration specifically—for example, by affecting residues in the active site—or are important for substrate binding. Others (Class II) can be mapped throughout the IN coding region and affect other reactions, including reverse transcription and virus particle assembly, as noted above. Viral RNA is excluded from the capsid in particles formed by some Class II mutants, as well as in particles that lack IN entirely (107). However, the importance of IN to particle morphogenesis was brought home most dramatically by analysis of certain allosteric inhibitors of HIV-1 IN that target the LEDGF/p75 binding pocket in the CCD-CCD dimer interface (118–121). These compounds, selected for their ability to block LEDGF/p75 binding or individual catalytic steps in vitro, were found to function by stabilizing nonfunctional multimers. Surprisingly, experiments with infected cells revealed that the most physiologically relevant activity of these inhibitors is not on the integration reaction but rather on virus particle assembly (122– 124). The compounds were found to elicit the same phenotype in HIV-producing cells as does the complete absence of IN protein: Viral RNA is excluded from capsids in progeny particles. Discovering how the lack of IN and its aberrant multimerization can produce the same phenotype may provide important clues to the mechanism of retroviral particle morphogenesis.
Whereas the in vitro studies indicate that only four molecules of IN are required to catalyze concerted integration, each retrovirus particle is estimated to contain some 50–100 monomer equivalents of this protein, embedded in the Gag-Pol precursor during particle assembly or as processed IN in mature virus particles. Footprinting analyses with MLV and HIV-1 PICs indicate that ~1 kb of each vDNA end is normally protected from enzymatic digestion, and such protection is absent in mutants that lack IN (125, 126). It seems possible therefore that numerous molecules of IN may be bound to vDNA ends, or that the four bound at the tips of vDNA recruit other proteins. Clearly, there is much more to be discovered concerning both the catalytic and noncatalytic roles of IN proteins as details of PIC architecture and viral morphogenesis are unraveled and a more holistic vision of viral reproduction emerges.
PERSPECTIVES
In the almost half century since the discovery of RT and the validation of the provirus hypothesis, we have learned a great deal about retroviral DNA integration and the enzyme that catalyzes this essential reaction (Table 1). Early insights were derived from studies of chicken or mouse retroviruses. Indeed, no human retroviruses had been identified until the discovery of human lymphotropic virus type 1 (HTLV-1) in 1981 (127) and HIV-1 in 1983 (128, 129), ending what now seems an incredible debate about whether human retroviruses existed at all. Today, given the devastation of AIDS and the need to develop methods to prevent and treat the disease, most retrovirologists are HIV researchers. The availability of three FDA-approved active site inhibitors of HIV IN was made possible by knowledge gained from intense studies of the biochemistry of this enzyme by the many researchers who entered this field (see 130). More recent understanding of the structure of IN proteins and their multimerization properties has motivated the search for novel allosteric IN inhibitors (reviewed in 131). In addition, the discovery that IN tethering cellular proteins govern host site preferences and are required for optimal efficiency of MLV and HIV-1 integration has suggested possible new targets for inhibition, as well as strategies for directing the integration of retroviral vectors to predetermined sites. Such knowledge may also inform the development of safer retroviral vectors for human gene therapy (132). Finally, as illustrated by the analyses of PFV, ASLV, and MLV IN proteins, comparisons of the properties of different IN proteins, and appreciation of the possible variations on the integration theme (15), can teach us much about essential features of the biology of retroviruses and the cells that they infect.
SUMMARY POINTS.
Although there are several examples among the bacterial viruses, retroviruses are the only animal viruses that encode an IN protein.
Retroviral IN is required for insertion of a DNA copy of the viral RNA genome into the chromosome of its host cell, a process that is essential for virus reproduction. Retroviral particles include 50–100 copies of IN.
The integration reaction comprises two temporally distinct steps, end processing (in the cytoplasm) and end joining (in the nucleus), which take place in a single active site and require two metal cofactors.
Integrated retroviral DNA, the provirus, is cropped at both ends (by 2 bp) and flanked by a short duplication of the host target sequence. The length (4–6 bp) and preferred target sequence are characteristic for different IN proteins.
Different retroviral genera exhibit distinct integration region preferences; for HIV-1 and MLV, preference is determined by IN tethering to specific host chromatin-binding proteins (LEDGF/p75 and Brd, respectively).
IN proteins comprise three common domains joined by linkers of varying length. The geometry of the active site in the central CCD establishes the relationship of IN to a large superfamily of polynucleotidyl transferases.
IN is a dynamic protein with multimerization properties that differ among the retroviruses, but the functional form for joining in vitro is a tetramer. Crystallographic and solution-derived structures of PFV IN indicate that only two (inner) protomers of this functional tetramer provide substrate binding and catalytic activity.
PFV IN crystals reveal that current active site inhibitors block target DNA binding to IN. Compounds that promote IN multimerization in the absence of DNA are allosteric inhibitors of IN catalysis and also prevent normal particle morphogenesis.
FUTURE ISSUES.
Pathways for intasome assembly are yet to be determined. The high concentration of IN within virus particles would predict that multimers are present before vDNA is made in an infected cell.
Detailed knowledge of the architectures, and viral and host components of PICs could provide clues to critical protein-protein associations, the significance of potential protein modifications, and the function, if any, of the many other IN molecules in an infecting virus particle.
Non-catalytic functions of IN, whether processed or embedded in a polyprotein precursor, remain to be elucidated.
Atomic-level structures of additional retroviral intasomes, as well as their interactions with other proteins, need to be solved. Such information will distinguish general features from those that are virus-specific
Acknowledgments
We thank our colleagues Drs. Richard A. Katz, Eileen Jaffe, and Katherine Buettner for helpful comments. Our research is supported by National Institutes of Health grants AI 040385, CA 71515, CA 06927, and AI 118589 and the W.W. Smith Charitable Trust.
KEY TERMS AND DEFINITIONS
- ASLV
avian sarcoma/leukosis virus
- HIV-1
human immunodeficiency virus type 1
- IN
integrase
- Intasome
a tetramer of integrase protein bound to two viral DNA ends and poised for joining to host (target) DNA
- MLV
murine leukemia virus
- PFV
prototype foamy virus
- Preintegration complex (PIC)
large nucleoprotein assembly derived from the RTC, capable of inserting the viral DNA into host/target DNA
- Provirus
retroviral DNA that is integrated into host DNA
- Reverse transcription complex (RTC)
cytoplasmic subviral particle, comprising a partially dissociated capsid, in which RNA is reverse transcribed into DNA
- RT
reverse transcriptase
Footnotes
Note Added in Proof
After our review was completed, a cryo-electron microscopy structure of the PFV intasome bound to a nucleosome was reported (Maskell et al, 2015). Interactions between both catalytic and non-catalytic IN subunits were described, providing new functions for at least some of the catalytic and “extra” IN molecules, noted in Future Issue point 2.
Ref. to add: Maskell DP, Renault L, Serrao E, Lesbats P, Matadeen R, Hare S, Lindemann D, Engelman AN, Costa A, Cherepanov P. Structural basis for retroviral integration into nucleosomes. Nature. 2015 Jul 16;523(7560):366-9. doi: 10.1038/nature14495. Epub 2015 Jun 10. PubMed PMID: 26061770; PubMed Central PMCID: PMC4530500.
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Wollman E, Wollman E. Les «phases» des bactériophages (facteurs lysogènes) C.R. Soc. Biol. 1937;124:931–934. [Google Scholar]
- 2.Lwoff A. Lysogeny. Bacteriol. Rev. 1953;17:269–337. doi: 10.1128/br.17.4.269-337.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jacob F, Wollman E. Sexuality and the Genetics of Bacteria. New York: Academic; 1961. [Google Scholar]
- 4.Campbell A. Episomes. Adv. Genet. 1961;11:101–145. [Google Scholar]
- 5.Hershey AD, Burgi E, Ingraham L. Cohesion of DNA molecules isolated from phage lambda. PNAS. 1963;49:748–755. doi: 10.1073/pnas.49.5.748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Temin HM. The DNA provirus hypothesis. Science. 1976;192:1075–1080. doi: 10.1126/science.58444. [DOI] [PubMed] [Google Scholar]
- 7.Temin HM. Text of a presentation at the International Conference on Avian Tumor Viruses, Duke University, Durham, NC, entitled “Nature of the provirus of Rous sarcoma virus,” reproduced with introduction and comments by GM Cooper. In: Cooper GM, Greenberg R, Sugden W, editors. DNA Provirus: Howard Temin’s Scientific Legacy. Washington, DC: ASM Press; 1995. pp. 25–40. (1964) [Google Scholar]
- 8.Svoboda J, Chyle P, Simkovic D, Hilgert I. Demonstration of the absence of infectious Rous virus in rat tumour XC, whose structurally intact cells produce Rous sarcoma when transferred to chicks. Folia Biol. 1963;9:77–81. [PubMed] [Google Scholar]
- 9.Baltimore D. RNA-dependent DNA polymerase in virions of RNA tumour viruses. Nature. 1970;226:1209–1211. doi: 10.1038/2261209a0. [DOI] [PubMed] [Google Scholar]
- 10.Temin HM, Mizutani S. RNA-dependent DNA polymerase in virions of Rous sarcoma virus. Nature. 1970;226:1211–1213. doi: 10.1038/2261211a0. [DOI] [PubMed] [Google Scholar]
- 11.Hill M, Hillova J. Virus recovery in chicken cells tested with Rous sarcoma cell DNA. Nat. New Biol. 1972;237:35–39. doi: 10.1038/newbio237035a0. [DOI] [PubMed] [Google Scholar]
- 12.Belyi VA, Levine AJ, Skalka AM. Unexpected inheritance: multiple integrations of ancient Bornavirus and Ebolavirus/Marburgvirus sequences in vertebrate genomes. PLOS Pathog. 2010;6:e1001030. doi: 10.1371/journal.ppat.1001030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Belyi VA, Levine AJ, Skalka AM. Sequences from ancestral single-stranded DNA viruses in vertebrate genomes: The Parvoviridae and Circoviridae are more than 40 to 50 million years old. J. Virol. 2010;84:12458–12462. doi: 10.1128/JVI.01789-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tarlinton RE, Meers J, Young PR. Retroviral invasion of the koala genome. Nature. 2006;442:79–81. doi: 10.1038/nature04841. [DOI] [PubMed] [Google Scholar]
- 15.Skalka AM. Retroviral DNA transposition: themes and variations. In: Sandmeyer S, Craig N, editors. Mobile DNA III. Washington, DC: ASM Press; 2015. In press. [Google Scholar]
- 16.Flint SJ, Racaniello VR, Rall G, Skalka AM, editors. Principles of Virology. 4th. Washington, DC: ASM Press; 2015. [Google Scholar]
- 17.Guntaka RV, Mahy BW, Bishop JM, Varmus HE. Ethidium bromide inhibits appearance of closed circular viral DNA and integration of virus-specific DNA in duck cells infected by avian sarcoma virus. Nature. 1975;253:507–511. doi: 10.1038/253507a0. [DOI] [PubMed] [Google Scholar]
- 18.Panganiban AT, Temin HM. Circles with two tandem LTRs are precursors to integrated retrovirus DNA. Cell. 1984;36:673–679. doi: 10.1016/0092-8674(84)90347-7. [DOI] [PubMed] [Google Scholar]
- 19.Brown PO, Bowerman B, Varmus HE, Bishop JM. Retroviral integration: structure of the initial covalent product and its precursor, and a role for the viral IN protein. PNAS. 1989;86:2525–2529. doi: 10.1073/pnas.86.8.2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown PO, Bowerman B, Varmus HE, Bishop JM. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 21.Fujiwara T, Mizuuchi K. Retroviral DNA integration: structure of an integration intermediate. Cell. 1988;54:497–504. doi: 10.1016/0092-8674(88)90071-2. [DOI] [PubMed] [Google Scholar]
- 22.Mizuuchi K. Transpositional recombination: mechanistic insights from studies of Mu and other elements. Annu. Rev. Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- 23.Coffin JM, Hughes SH, Varmus HE. Retroviruses. Cold Spring Harbor, NY: Cold Spring Harbor Press; 1997. [PubMed] [Google Scholar]
- 24.Dhar R, McClements WL, Enquist LW, Vande Woude GF. Nucleotide sequences of integrated Moloney sarcoma provirus long terminal repeats and their host and viral junctions. PNAS. 1980;77:3937–3941. doi: 10.1073/pnas.77.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hughes SH, Mutschler A, Bishop JM, Varmus HE. A Rous sarcoma virus provirus is flanked by short direct repeats of a cellular DNA sequence present in only one copy prior to integration. PNAS. 1981;78:4299–4303. doi: 10.1073/pnas.78.7.4299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ju G, Hishinuma F, Skalka AM. Nucleotide sequence analysis of avian retroviruses: structural similarities with transposable elements. Fed. Proc. 1982;41:2659–2661. [PubMed] [Google Scholar]
- 27.Ju G, Skalka AM. Nucleotide sequence analysis of the long terminal repeat (LTR) of avian retroviruses: structural similarities with transposable elements. Cell. 1980;22:379–386. doi: 10.1016/0092-8674(80)90348-7. [DOI] [PubMed] [Google Scholar]
- 28.Majors JE, Varmus HE. Nucleotide sequences at host-proviral junctions for mouse mammary tumour virus. Nature. 1981;289:253–258. doi: 10.1038/289253a0. [DOI] [PubMed] [Google Scholar]
- 29.Shimotohno K, Mizutani S, Temin HM. Sequence of retrovirus provirus resembles that of bacterial transposable elements. Nature. 1980;285:550–554. doi: 10.1038/285550a0. [DOI] [PubMed] [Google Scholar]
- 30.Van Beveren C, Goddard JG, Berns A, Verma IM. Structure of Moloney murine leukemia viral DNA: nucleotide sequence of the 5′ long terminal repeat and adjacent cellular sequences. PNAS. 1980;77:3307–3311. doi: 10.1073/pnas.77.6.3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hishinuma F, DeBona PJ, Astrin S, Skalka AM. Nucleotide sequence of acceptor site and termini of integrated avian endogenous provirus ev1: integration creates a 6 bp repeat of host DNA. Cell. 1981;23:155–164. doi: 10.1016/0092-8674(81)90280-4. [DOI] [PubMed] [Google Scholar]
- 32.Grandgenett DP. pp32 is hot. See Reference 130. 2011:15–22. [Google Scholar]
- 33.Craigie R, Fujiwara T, Bushman F. The IN protein of Moloney murine leukemia virus processes the viral DNA ends and accomplishes their integration in vitro. Cell. 1990;62:829–837. doi: 10.1016/0092-8674(90)90126-y. [DOI] [PubMed] [Google Scholar]
- 34.Katz RA, Merkel G, Kulkosky J, Leis J, Skalka AM. The avian retroviral IN protein is both necessary and sufficient for integrative recombination in vitro. Cell. 1990;63:87–95. doi: 10.1016/0092-8674(90)90290-u. [DOI] [PubMed] [Google Scholar]
- 35.Katzman M, Katz RA, Skalka AM, Leis J. The avian retroviral integration protein cleaves the terminal sequences of linear viral DNA at the in vivo sites of integration. J. Virol. 1989;63:5319–5327. doi: 10.1128/jvi.63.12.5319-5327.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merkel G, Andrake MD, Ramcharan J, Skalka AM. Oligonucleotide-based assays for integrase activity. Methods. 2009;47:243–248. doi: 10.1016/j.ymeth.2008.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Grobler JA, Stillmock K, Hu B, Witmer M, Felock P, et al. Diketo acid inhibitor mechanism and HIV-1 integrase: implications for metal binding in the active site of phosphotransferase enzymes. PNAS. 2002;99:6661–6666. doi: 10.1073/pnas.092056199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roth MJ, Schwartzberg PL, Goff SP. Structure of the termini of DNA intermediates in the integration of retroviral DNA: dependence on IN function and terminal DNA sequence. Cell. 1989;58:47–54. doi: 10.1016/0092-8674(89)90401-7. [DOI] [PubMed] [Google Scholar]
- 39.Farnet CM, Haseltine WA. Determination of viral proteins present in the human immunodeficiency virus type 1 preintegration complex. J. Virol. 1991;65:1910–1915. doi: 10.1128/jvi.65.4.1910-1915.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Daniel R, Kao G, Taganov K, Greger JG, Favorova O, et al. Evidence that the retroviral DNA integration process triggers an ATR-dependent DNA damage response. PNAS. 2003;100:4778–4783. doi: 10.1073/pnas.0730887100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sakurai Y, Komatsu K, Agematsu K, Matsuoka M. DNA double strand break repair enzymes function at multiple steps in retroviral infection. Retrovirology. 2009;6:114. doi: 10.1186/1742-4690-6-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Skalka AM, Katz RA. Retroviral DNA integration and the DNA damage response. Cell Death Differ. 2005;12:971–978. doi: 10.1038/sj.cdd.4401573. [DOI] [PubMed] [Google Scholar]
- 43.Daniel R, Ramcharan J, Rogakou E, Taganov KD, Greger JG, et al. Histone H2AX is phosphorylated at sites of retroviral DNA integration but is dispensable for postintegration repair. J. Biol. Chem. 2004;279:45810–45814. doi: 10.1074/jbc.M407886200. [DOI] [PubMed] [Google Scholar]
- 44.Roe T, Chow SA, Brown PO. 3′-End processing and kinetics of 5′-end joining during retroviral integration in vivo. J. Virol. 1997;71:1334–1340. doi: 10.1128/jvi.71.2.1334-1340.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Desfarges S, Ciuffi A. Retroviral integration site selection. Viruses. 2010;2:111–130. doi: 10.3390/v2010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Benleulmi MS, Matysiak J, Henriquez DR, Vaillant C, Lesbats P, et al. Intasome architecture and chromatin density modulate retroviral integration into the nucleosome. Retrovirology. 2015;12:13. doi: 10.1186/s12977-015-0145-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lesbats P, Botbol Y, Chevereau G, Vaillant C, Calmels C, et al. Functional coupling between HIV-1 integrase and the SWI/SNF chromatin remodeling complex for efficient in vitro integration into stable nucleosomes. PLOS Pathog. 2011;7:e1001280. doi: 10.1371/journal.ppat.1001280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taganov KD, Cuesta I, Daniel R, Cirillo LA, Katz RA, et al. Integrase-specific enhancement and suppression of retroviral DNA integration by compacted chromatin structure in vitro. J. Virol. 2004;78:5848–5855. doi: 10.1128/JVI.78.11.5848-5855.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Engelman A, Cherepanov P. The lentiviral integrase binding protein LEDGF/p75 and HIV-1 replication. PLOS Pathog. 2008;4:e1000046. doi: 10.1371/journal.ppat.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gupta SS, Maetzig T, Maertens GN, Sharif A, Rothe M, et al. Bromo- and extraterminal domain chromatin regulators serve as cofactors for murine leukemia virus integration. J. Virol. 2013;87:12721–12736. doi: 10.1128/JVI.01942-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sharma A, Larue RC, Plumb MR, Malani N, Male F, et al. BET proteins promote efficient murine leukemia virus integration at transcription start sites. PNAS. 2013;110:12036–12041. doi: 10.1073/pnas.1307157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Craigie R, Bushman FD. Host factors in retroviral integration and the selection of integration target sites. Microbiol. Spectr. 2014;2 doi: 10.1128/microbiolspec.MDNA3-0026-2014. MDNA3-0026-2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shun MC, Raghavendra NK, Vandegraaff N, Daigle JE, Hughes S, et al. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bushman FD, Engelman A, Palmer I, Wingfield P, Craigie R. Domains of the integrase protein of human immunodeficiency virus type 1 responsible for polynucleotidyl transfer and zinc binding. PNAS. 1993;90:3428–3432. doi: 10.1073/pnas.90.8.3428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Engelman A, Bushman FD, Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993;12:3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Khan E, Mack JP, Katz RA, Kulkosky J, Skalka AM. Retroviral integrase domains: DNA binding and the recognition of LTR sequences. Nucleic Acids Res. 1991;19:851–860. doi: 10.1093/nar/19.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cai M, Zheng R, Caffrey M, Craigie R, Clore GM, Gronenborn AM. Solution structure of the N-terminal zinc binding domain of HIV-1 integrase. Nat. Struct. Biol. 1997;4:567–577. doi: 10.1038/nsb0797-567. [DOI] [PubMed] [Google Scholar]
- 58.Eijkelenboom AP, van den Ent FM, Vos A, Doreleijers JF, Hard K, et al. The solution structure of the amino-terminal HHCC domain of HIV-2 integrase: a three-helix bundle stabilized by zinc. Curr. Biol. 1997;7:739–746. doi: 10.1016/s0960-9822(06)00332-0. [DOI] [PubMed] [Google Scholar]
- 59.Kulkosky J, Jones KS, Katz RA, Mack JP, Skalka AM. Residues critical for retroviral integrative recombination in a region that is highly conserved among retroviral/retrotransposon integrases and bacterial insertion sequence transposases. Mol. Cell. Biol. 1992;12:2331–2338. doi: 10.1128/mcb.12.5.2331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dyda F, Hickman AB, Jenkins TM, Engelman A, Craigie R, Davies DR. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science. 1994;266:1981–1986. doi: 10.1126/science.7801124. [DOI] [PubMed] [Google Scholar]
- 61.Bujacz G, Jaskolski M, Alexandratos J, Wlodawer A, Merkel G, et al. High-resolution structure of the catalytic domain of avian sarcoma virus integrase. J. Mol. Biol. 1995;253:333–346. doi: 10.1006/jmbi.1995.0556. [DOI] [PubMed] [Google Scholar]
- 62.Nowotny M. Retroviral integrase superfamily: the structural perspective. EMBO Rep. 2009;10:144–151. doi: 10.1038/embor.2008.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kim DR, Dai Y, Mundy CL, Yang W, Oettinger MA. Mutations of acidic residues in RAG1 define the active site of the V(D)J recombinase. Genes Dev. 1999;13:3070–3080. doi: 10.1101/gad.13.23.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Landree MA, Wibbenmeyer JA, Roth DB. Mutational analysis of RAG1 and RAG2 identifies three catalytic amino acids in RAG1 critical for both cleavage steps of V(D)J recombination. Genes Dev. 1999;13:3059–3069. doi: 10.1101/gad.13.23.3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol. Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 66.Lodi PJ, Ernst JA, Kuszewski J, Hickman AB, Engelman A, et al. Solution structure of the DNA binding domain of HIV-1 integrase. Biochemistry. 1995;34:9826–9833. doi: 10.1021/bi00031a002. [DOI] [PubMed] [Google Scholar]
- 67.Eijkelenboom AP, Lutzke RA, Boelens R, Plasterk RH, Kaptein R, Hard K. The DNA-binding domain of HIV-1 integrase has an SH3-like fold. Nat. Struct. Biol. 1995;2:807–810. doi: 10.1038/nsb0995-807. [DOI] [PubMed] [Google Scholar]
- 68.Andrake MD, Skalka AM. Multimerization determinants reside in both the catalytic core and C terminus of avian sarcoma virus integrase. J. Biol. Chem. 1995;270:29299–29306. doi: 10.1074/jbc.270.49.29299. [DOI] [PubMed] [Google Scholar]
- 69.Engelman A, Hickman AB, Craigie R. The core and carboxyl-terminal domains of the integrase protein of human immunodeficiency virus type 1 each contribute to nonspecific DNA binding. J. Virol. 1994;68:5911–5917. doi: 10.1128/jvi.68.9.5911-5917.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bao KK, Wang H, Miller JK, Erie DA, Skalka AM, Wong I. Functional oligomeric state of avian sarcoma virus integrase. J. Biol. Chem. 2003;278:1323–1327. doi: 10.1074/jbc.C200550200. [DOI] [PubMed] [Google Scholar]
- 71.Jones KS, Coleman J, Merkel GW, Laue TM, Skalka AM. Retroviral integrase functions as a multimer and can turn over catalytically. J. Biol. Chem. 1992;267:16037–16040. [PubMed] [Google Scholar]
- 72.Engelman A, Bushman FD, Craigie R. Identification of discrete functional domains of HIV-1 integrase and their organization within an active multimeric complex. EMBO J. 1993;12:3269–3275. doi: 10.1002/j.1460-2075.1993.tb05996.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Faure A, Calmels C, Desjobert C, Castroviejo M, Caumont-Sarcos A, et al. HIV-1 integrase crosslinked oligomers are active in vitro. Nucleic Acids Res. 2005;33:977–986. doi: 10.1093/nar/gki241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Li M, Mizuuchi M, Burke TR, Jr, Craigie R. Retroviral DNA integration: reaction pathway and critical intermediates. EMBO J. 2006;25:1295–1304. doi: 10.1038/sj.emboj.7601005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang JY, Ling H, Yang W, Craigie R. Structure of a two-domain fragment of HIV-1 integrase: implications for domain organization in the intact protein. EMBO J. 2001;20:7333–7943. doi: 10.1093/emboj/20.24.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chen JC, Krucinski J, Miercke LJ, Finer-Moore JS, Tang AH, et al. Crystal structure of the HIV-1 integrase catalytic core and C-terminal domains: a model for viral DNA binding. PNAS. 2000;97:8233–8238. doi: 10.1073/pnas.150220297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chen Z, Yan Y, Munshi S, Li Y, Zugay-Murphy J, et al. X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50- 293)—an initial glance of the viral DNA binding platform. J. Mol. Biol. 2000;296:521–533. doi: 10.1006/jmbi.1999.3451. [DOI] [PubMed] [Google Scholar]
- 78.Yang ZN, Mueser TC, Bushman FD, Hyde CC. Crystal structure of an active two-domain derivative of Rous sarcoma virus integrase. J. Mol. Biol. 2000;296:535–548. doi: 10.1006/jmbi.1999.3463. [DOI] [PubMed] [Google Scholar]
- 79.Hare S, Di Nunzio F, Labeja A, Wang J, Engelman A, Cherepanov P. Structural basis for functional tetramerization of lentiviral integrase. PLOS Pathog. 2009;5:e1000515. doi: 10.1371/journal.ppat.1000515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Shi K, Pandey KK, Bera S, Vora AC, Grandgenett DP, Aihara H. A possible role for the asymmetric C-terminal domain dimer of Rous sarcoma virus integrase in viral DNA binding. PLOS ONE. 2013;8:e56892. doi: 10.1371/journal.pone.0056892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Valkov E, Gupta SS, Hare S, Helander A, Roversi P, et al. Functional and structural characterization of the integrase from the prototype foamy virus. Nucleic Acids Res. 2009;37:243–255. doi: 10.1093/nar/gkn938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Juretzek T, Holm T, Gartner K, Kanzler S, Lindemann D, et al. Foamy virus integration. J. Virol. 2004;78:2472–2477. doi: 10.1128/JVI.78.5.2472-2477.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hare S, Gupta SS, Valkov E, Engelman A, Cherepanov P. Retroviral intasome assembly and inhibition of DNA strand transfer. Nature. 2010;464:232–236. doi: 10.1038/nature08784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Katz RA, DiCandeloro P, Kukolj G, Skalka AM. Role of DNA end distortion in catalysis by avian sarcoma virus integrase. J. Biol. Chem. 2001;276:34213–34220. doi: 10.1074/jbc.M104632200. [DOI] [PubMed] [Google Scholar]
- 85.Katz RA, Merkel G, Andrake MD, Roder H, Skalka AM. Retroviral integrases promote fraying of viral DNA ends. J. Biol. Chem. 2011;286:25710–25718. doi: 10.1074/jbc.M111.229179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.van Gent DC, Vink C, Groeneger AA, Plasterk RH. Complementation between HIV integrase proteins mutated in different domains. EMBO J. 1993;12:3261–3267. doi: 10.1002/j.1460-2075.1993.tb05995.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Gupta K, Curtis JE, Krueger S, Hwang Y, Cherepanov P, et al. Solution conformations of prototype foamy virus integrase and its stable synaptic complex with U5 viral DNA. Structure. 2012;20:1918–1928. doi: 10.1016/j.str.2012.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Maertens GN, Hare S, Cherepanov P. The mechanism of retroviral integration from X-ray structures of its key intermediates. Nature. 2010;468:326–329. doi: 10.1038/nature09517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hare S, Maertens GN, Cherepanov P. 3′-Processing and strand transfer catalysed by retroviral integrase in crystallo. EMBO J. 2012;31:3020–3028. doi: 10.1038/emboj.2012.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Krishnan L, Li X, Naraharisetty HL, Hare S, Cherepanov P, Engelman A. Structure-based modeling of the functional HIV-1 intasome and its inhibition. PNAS. 2010;107:15910–15915. doi: 10.1073/pnas.1002346107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ballandras-Colas A, Naraharisetty H, Li X, Serrao E, Engelman A. Biochemical characterization of novel retroviral integrase proteins. PLOS ONE. 2013;8:e76638. doi: 10.1371/journal.pone.0076638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jenkins TM, Engelman A, Ghirlando R, Craigie R. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J. Biol. Chem. 1996;271:7712–7718. doi: 10.1074/jbc.271.13.7712. [DOI] [PubMed] [Google Scholar]
- 93.Bojja RS, Andrake MD, Merkel G, Weigand S, Dunbrack RL, Jr, Skalka AM. Architecture and assembly of HIV integrase multimers in the absence of DNA substrates. J. Biol. Chem. 2013;288:7373–7386. doi: 10.1074/jbc.M112.434431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bojja RS, Andrake MD, Weigand S, Merkel G, Yarychkivska O, et al. Architecture of a full-length retroviral integrase monomer and dimer, revealed by small angle X-ray scattering and chemical cross-linking. J. Biol. Chem. 2011;286:17047–17059. doi: 10.1074/jbc.M110.212571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pandey KK, Bera S, Grandgenett DP. The HIV-1 integrase monomer induces a specific interaction with LTR DNA for concerted integration. Biochemistry. 2011;50:9788–9796. doi: 10.1021/bi201247f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Horton R, Mumm S, Grandgenett DP. Avian retrovirus pp32 DNA endonuclease is phosphorylated on Ser in the carboxyl-terminal region. J. Virol. 1988;62:2067–2075. doi: 10.1128/jvi.62.6.2067-2075.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zheng Y, Yao X. Posttranslational modifications of HIV-1 integrase by various cellular proteins during viral replication. Viruses. 2013;5:1787–1801. doi: 10.3390/v5071787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mousnier A, Kubat N, Massias-Simon A, Segeral E, Rain JC, et al. Von Hippel-Lindau binding protein 1-mediated degradation of integrase affects HIV-1 gene expression at a postintegration step. PNAS. 2007;104:13615–13620. doi: 10.1073/pnas.0705162104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Cereseto A, Manganaro L, Gutierrez MI, Terreni M, Fittipaldi A, et al. Acetylation of HIV-1 integrase by p300 regulates viral integration. EMBO J. 2005;24:3070–3081. doi: 10.1038/sj.emboj.7600770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Terreni M, Valentini P, Liverani V, Gutierrez MI, Di Primio C, et al. GCN5-dependent acetylation of HIV-1 integrase enhances viral integration. Retrovirology. 2010;7:18. doi: 10.1186/1742-4690-7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Zamborlini A, Coiffic A, Beauclair G, Delelis O, Paris J, et al. Impairment of human immunodeficiency virus type-1 integrase SUMOylation correlates with an early replication defect. J. Biol. Chem. 2011;286:21013–21022. doi: 10.1074/jbc.M110.189274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Hizi A, Leis JP, Joklik WK. RNA-dependent DNA polymerase of avian sarcoma virus B77. II. Comparison of the catalytic properties of the α, β2, and αβ enzyme forms. J. Biol. Chem. 1977;252:2290–2295. [PubMed] [Google Scholar]
- 103.Herschhorn A, Hizi A. Retroviral reverse transcriptases. Cell. Mol. Life Sci. 2010;67:2717–2747. doi: 10.1007/s00018-010-0346-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Herschhorn A, Oz-Gleenberg I, Hizi A. Quantitative analysis of the interactions between HIV-1 integrase and retroviral reverse transcriptases. Biochem. J. 2008;412:163–170. doi: 10.1042/BJ20071279. [DOI] [PubMed] [Google Scholar]
- 105.Lai L, Liu H, Wu X, Kappes JC. Moloney murine leukemia virus integrase protein augments viral DNA synthesis in infected cells. J. Virol. 2001;75:11365–11372. doi: 10.1128/JVI.75.23.11365-11372.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wu X, Liu H, Xiao H, Conway JA, Hehl E, et al. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J. Virol. 1999;73:2126–2135. doi: 10.1128/jvi.73.3.2126-2135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Engelman A. In vivo analysis of retroviral integrase structure and function. Adv. Virus Res. 1999;52:411–426. doi: 10.1016/s0065-3527(08)60309-7. [DOI] [PubMed] [Google Scholar]
- 108.Zhu K, Dobard C, Chow SA. Requirement for integrase during reverse transcription of human immunodeficiency virus type 1 and the effect of cysteine mutations of integrase on its interactions with reverse transcriptase. J. Virol. 2004;78:5045–5055. doi: 10.1128/JVI.78.10.5045-5055.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hehl EA, Joshi P, Kalpana GV, Prasad VR. Interaction between human immunodeficiency virus type 1 reverse transcriptase and integrase proteins. J. Virol. 2004;78:5056–5067. doi: 10.1128/JVI.78.10.5056-5067.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Dobard CW, Briones MS, Chow SA. Molecular mechanisms by which human immunodeficiency virus type 1 integrase stimulates the early steps of reverse transcription. J. Virol. 2007;81:10037–10046. doi: 10.1128/JVI.00519-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Wilkinson TA, Januszyk K, Phillips ML, Tekeste SS, Zhang M, et al. Identifying and characterizing a functional HIV-1 reverse transcriptase-binding site on integrase. J. Biol. Chem. 2009;284:7931–7939. doi: 10.1074/jbc.M806241200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Studamire B, Goff SP. Interactions of host proteins with the murine leukemia virus integrase. Viruses. 2010;2:1110–1145. doi: 10.3390/v2051110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Raghavendra NK, Shkriabai N, Graham R, Hess S, Kvaratskhelia M, Wu L. Identification of host proteins associated with HIV-1 preintegration complexes isolated from infected CD4+ cells. Retrovirology. 2010;7:66. doi: 10.1186/1742-4690-7-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jager S, Cimermancic P, Gulbahce N, Johnson JR, McGovern KE, et al. Global landscape of HIV–human protein complexes. Nature. 2012;481:365–370. doi: 10.1038/nature10719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kalpana GV, Marmon S, Wang W, Crabtree GR, Goff SP. Binding and stimulation of HIV-1 integrase by a human homolog of yeast transcription factor SNF5. Science. 1994;266:2002–2006. doi: 10.1126/science.7801128. [DOI] [PubMed] [Google Scholar]
- 116.Mathew S, Nguyen M, Wu X, Pal A, Shah VB, et al. INI1/hSNF5-interaction defective HIV-1 IN mutants exhibit impaired particle morphology, reverse transcription and integration in vivo. Retrovirology. 2013;10:66. doi: 10.1186/1742-4690-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Engelman A. Pleiotropic nature of HIV-1 integrase mutations. See Reference 130. 2011:67–81. [Google Scholar]
- 118.Christ F, Voet A, Marchand A, Nicolet S, Desimmie BA, et al. Rational design of small-molecule inhibitors of the LEDGF/p75-integrase interaction and HIV replication. Nat. Chem. Biol. 2010;6:442–448. doi: 10.1038/nchembio.370. [DOI] [PubMed] [Google Scholar]
- 119.Fenwick CW, Tremblay S, Wardrop E, Bethell R, Coulomb R, et al. Resistance studies with HIV-1 non-catalytic site integrase inhibitors. Antivir. Ther. 2011;16:A9. [Google Scholar]
- 120.Kessl JJ, Jena N, Koh Y, Taskent-Sezgin H, Slaughter A, et al. Multimode, cooperative mechanism of action of allosteric HIV-1 integrase inhibitors. J. Biol. Chem. 2012;287:16801–16811. doi: 10.1074/jbc.M112.354373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Tsiang M, Jones GS, Niedziela-Majka A, Kan E, Lansdon EB, et al. New class of HIV-1 integrase (IN) inhibitors with a dual mode of action. J. Biol. Chem. 2012;287:21189–21203. doi: 10.1074/jbc.M112.347534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Jurado KA, Wang H, Slaughter A, Feng L, Kessl JJ, et al. Allosteric integrase inhibitor potency is determined through the inhibition of HIV-1 particle maturation. PNAS. 2013;110:8690–8695. doi: 10.1073/pnas.1300703110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Desimmie BA, Schrijvers R, Demeulemeester J, Borrenberghs D, Weydert C, et al. LEDGINs inhibit late stage HIV-1 replication by modulating integrase multimerization in the virions. Retrovirology. 2013;10:57. doi: 10.1186/1742-4690-10-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Balakrishnan M, Yant SR, Tsai L, O’Sullivan C, Bam RA, et al. Non-catalytic site HIV-1 integrase inhibitors disrupt core maturation and induce a reverse transcription block in target cells. PLOS ONE. 2013;8:e74163. doi: 10.1371/journal.pone.0074163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wei SQ, Mizuuchi K, Craigie R. A large nucleoprotein assembly at the ends of the viral DNA mediates retroviral DNA integration. EMBO J. 1997;16:7511–7520. doi: 10.1093/emboj/16.24.7511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Chen H, Wei SQ, Engelman A. Multiple integrase functions are required to form the native structure of the human immunodeficiency virus type I intasome. J. Biol. Chem. 1999;274:17358–17364. doi: 10.1074/jbc.274.24.17358. [DOI] [PubMed] [Google Scholar]
- 127.Poiesz BJ, Ruscetti FW, Reitz MS, Kalyanaraman VS, Gallo RC. Isolation of a new type C retrovirus (HTLV) in primary uncultured cells of a patient with Sezary T-cell leukaemia. Nature. 1981;294:268–271. doi: 10.1038/294268a0. [DOI] [PubMed] [Google Scholar]
- 128.Barré-Sinoussi F, Chermann JC, Rey F, Nugeyre MT, Chamaret S, et al. Isolation of a T-lymphotropic retrovirus from a patient at risk for acquired immune deficiency syndrome (AIDS) Science. 1983;220:868–871. doi: 10.1126/science.6189183. [DOI] [PubMed] [Google Scholar]
- 129.Gallo RC, Sarin PS, Gelmann EP, Robert-Guroff M, Richardson E, et al. Isolation of human T-cell leukemia virus in acquired immune deficiency syndrome (AIDS) Science. 1983;220:865–867. doi: 10.1126/science.6601823. [DOI] [PubMed] [Google Scholar]
- 130.Neamati N, editor. HIV-1 Integrase: Mechanism and Inhibitor Design. Hoboken, NJ: Wiley; 2011. [Google Scholar]
- 131.Jurado KA, Engelman A. Multimodal mechanism of action of allosteric HIV-1 integrase inhibitors. Expert Rev. Mol. Med. 2013;15:e14. doi: 10.1017/erm.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kvaratskhelia M, Sharma A, Larue RC, Serrao E, Engelman A. Molecular mechanisms of retroviral integration site selection. Nucleic Acids Res. 2014;42:10209–10225. doi: 10.1093/nar/gku769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 133b.Vallée H, Carré H. Sur la nature infectieuse de l’anémie ducheval. C.R. Acad. Sci. 1904;139:331–333. [Google Scholar]
- 134.Ellermann V, Bang O. Experimentelle Leukämie bei Hühnern. Zent. Bakt. Parasitenkd. Infekt. 1908;46:595–609. [Google Scholar]
- 135.Rous P. A sarcoma of the fowl transmissible by an agent separable from the tumor cells. J. Exp. Med. 1911;13:397–411. doi: 10.1084/jem.13.4.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Bittner JJ. Some possible effects of nursing on the mammary gland tumor incidence in mice. Science. 1936;84:162. doi: 10.1126/science.84.2172.162. [DOI] [PubMed] [Google Scholar]
- 137.Gross L. “Spontaneous” leukemia developing in C3H mice following inoculation in infancy, with AK-leukemic extracts, or AK-embryos. Proc. Soc. Exp. Biol. Med. 1951;76:27–32. [PubMed] [Google Scholar]
- 138.Gross L. Development and serial cell-free passage of a highly potent strain of mouse leukemia virus. Proc. Soc. Exp. Biol. Med. 1957;94:767–771. doi: 10.3181/00379727-94-23080. [DOI] [PubMed] [Google Scholar]
- 139.Enders JF, Peebles TC. Propagation in tissue cultures of cytopathogenic agents from patients with measles. Proc. Soc. Exp. Biol. Med. 1954;86:277–286. doi: 10.3181/00379727-86-21073. [DOI] [PubMed] [Google Scholar]
- 140.Temin HM, Rubin H. Characteristics of an assay for Rous sarcoma virus and Rous sarcoma cells in tissue culture. Virology. 1958;6:669–688. doi: 10.1016/0042-6822(58)90114-4. [DOI] [PubMed] [Google Scholar]
- 141.Lieberman M, Kaplan HS. Leukemogenic activity of filtrates from radiation-induced lymphoid tumors of mice. Science. 1959;130:387–388. doi: 10.1126/science.130.3372.387. [DOI] [PubMed] [Google Scholar]
- 142.Muhlbock O. Note on a new inbred mouse-strain GR-A. Eur. J. Cancer. 1965;1:123–124. doi: 10.1016/0014-2964(65)90003-4. [DOI] [PubMed] [Google Scholar]
- 143.Hughes SH, Shank PR, Spector DH, Kung HJ, Bishop JM, et al. Proviruses of avian sarcoma virus are terminally redundant, co-extensive with unintegrated linear DNA and integrated at many sites. Cell. 1978;15:1397–1410. doi: 10.1016/0092-8674(78)90064-8. [DOI] [PubMed] [Google Scholar]
- 144.Dhar R, McClements WL, Enquist LW, Vande Woude GF. Nucleotide sequences of integrated Moloney sarcoma provirus long terminal repeats and their host and viral junctions. PNAS. 1980;77:3937–3941. doi: 10.1073/pnas.77.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Grandgenett DP, Vora AC, Schiff RD. A 32,000-Dalton nucleic acid-binding protein from avian retravirus cores possesses DNA endonuclease activity. Virology. 1978;89:119–132. doi: 10.1016/0042-6822(78)90046-6. [DOI] [PubMed] [Google Scholar]
- 146.Donehower LA, Varmus HE. A mutant murine leukemia virus with a single missense codon in pol is defective in a function affecting integration. PNAS. 1984;81:6461–6465. doi: 10.1073/pnas.81.20.6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Schwartzberg P, Colicelli J, Goff SP. Construction and analysis of deletion mutations in the pol gene of Moloney murine leukemia virus: a new viral function required for productive infection. Cell. 1984;37:1043–1052. doi: 10.1016/0092-8674(84)90439-2. [DOI] [PubMed] [Google Scholar]
- 148.Panganiban AT, Temin HM. The retrovirus pol gene encodes a product required for DNA integration: identification of a retrovirus int locus. PNAS. 1984;81:7885–7889. doi: 10.1073/pnas.81.24.7885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Brown PO, Bowerman B, Varmus HE, Bishop JM. Correct integration of retroviral DNA in vitro. Cell. 1987;49:347–356. doi: 10.1016/0092-8674(87)90287-x. [DOI] [PubMed] [Google Scholar]
- 150.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 151.Narezkina A, Taganov KD, Litwin S, Stoyanova R, Hayashi J, et al. Genome-wide analyses of avian sarcoma virus integration sites. J. Virol. 2004;78:11656–11663. doi: 10.1128/JVI.78.21.11656-11663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Cherepanov P, Maertens G, Proost P, Devreese B, Van Beeumen J, et al. HIV-1 integrase forms stable tetramers and associates with LEDGF/p75 protein in human cells. J. Biol. Chem. 2003;278:372–381. doi: 10.1074/jbc.M209278200. [DOI] [PubMed] [Google Scholar]
- 153.Cherepanov P, Ambrosio AL, Rahman S, Ellenberger T, Engelman A. Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75. PNAS. 2005;102:17308–17313. doi: 10.1073/pnas.0506924102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Sharma A, Slaughter A, Jena N, Feng L, Kessl JJ, et al. A new class of multimerization selective inhibitors of HIV-1 integrase. PLOS Pathog. 2014;10:e1004171. doi: 10.1371/journal.ppat.1004171. [DOI] [PMC free article] [PubMed] [Google Scholar]