A fluorescence-based screen was used to analyze 70 LAT transporter mutants and identify variants with improved stability and monodispersity.
Abstract
The knowledge of three-dimensional structures at atomic resolution of membrane transport proteins has improved considerably our understanding of their physiological roles and pathological implications. However, most structural biology techniques require an optimal candidate within a protein family for structural determination with (a) reasonable production in heterologous hosts and (b) good stability in detergent micelles. SteT, the Bacillus subtilis l-serine/l-threonine exchanger is the best-known prokaryotic paradigm of the mammalian l–amino acid transporter (LAT) family. Unfortunately, SteT’s lousy stability after extracting from the membrane prevents its structural characterization. Here, we have used an approach based on random mutagenesis to engineer stability in SteT. Using a split GFP complementation assay as reporter of protein expression and membrane insertion, we created a library of 70 SteT mutants each containing random replacements of one or two residues situated in the transmembrane domains. Analysis of expression and monodispersity in detergent of this library permitted the identification of evolved versions of SteT with a significant increase in both expression yield and stability in detergent with respect to wild type. In addition, these experiments revealed a correlation between the yield of expression and the stability in detergent micelles. Finally, and based on protein delipidation and relipidation assays together with transport experiments, possible mechanisms of SteT stabilization are discussed. Besides optimizing a member of the LAT family for structural determination, our work proposes a new approach that can be used to optimize any membrane protein of interest.
INTRODUCTION
Membrane transport proteins are crucial for maintaining the homeostasis of amino acids in the different organs and tissues as they are responsible for the absorption and distribution of amino acids across the different cellular membranes and intracellular compartments (Christensen, 1990). Because of the large variety of biological functions of amino acids, dysfunctions of these transporters are associated with a large number of disorders such as neuronal excitability dysfunctions or metabolic diseases (Bröer and Palacín, 2011).
Amino acid transporters are found in 11 out of the 50 different SLC (solute carrier) families. The l–amino acid transporter (LAT) family of amino acid transporters, included in the SLC7 cluster, contains eight proteins found in humans (Fotiadis et al., 2013). These transporters are the catalytic subunit of the heteromeric amino acid transporters (HATs; Fotiadis et al., 2013), the only family of amino acid transporters composed by two subunits linked by a disulfide bridge: the LAT, also named light subunit, and an accessory protein from the SLC3 cluster, also known as heavy subunit because of the large glycosylated extracellular domain. Congenital mutations of HATs are responsible for two pathological disorders: cystinuria and lysinuric protein intolerance (Palacín et al., 2005). Moreover, LAT1 and xCT are often overexpressed in tumor cells, and their role on tumor growth is being studied (Fuchs and Bode, 2005; McCracken and Edinger, 2013). At the moment, there is no atomic structure of the HAT complex. The crystal structure of the ectodomain of the heavy subunit human 4f2 (Fort et al., 2007) reveals a bacterial α-glycosidase fold, although lacking the catalytic activity. Recently, a low-resolution structural model combined with functional and biochemical assays have provided important insights regarding the structural arrangement of the two heterodimers of HATs (Rosell et al., 2014). Nevertheless, getting insights into the functional mechanism of these transporters requires an atomic-resolution structure of the LAT subunit. LATs exhibit a 12–transmembrane domain (TMD) topology, where the N- and C-terminal ends reside in the cytoplasm (Fotiadis et al., 2013). The closest structural models of LATs are represented by the x-ray crystal structures of three prokaryotic transporters: AdiC, ApcT, and GadC (Fang et al., 2009; Gao et al., 2009, 2010; Shaffer et al., 2009; Kowalczyk et al., 2011; Ma et al., 2012), all of them belonging to the large amino acid, polyamines, and organocation (APC) superfamily (Jack et al., 2000), where the LAT family is included. These structures exhibit the 5 + 5 inverted repeat fold shared by different and nonrelated families of transporters (Shi, 2013). The structural conformations represented by these crystal structures have provided an overall view of the structural rearrangement of LATs during transport. However, the low amino acid identity (<20%) of LATs with AdiC, ApcT, or GadC makes necessary the search for better paradigms of LATs with closer amino acid sequence identity to identify the molecular determinants of substrate recognition and translocation.
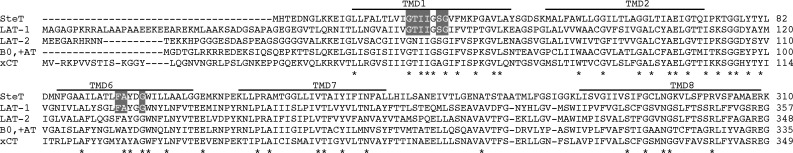
SteT, the l-serine/l-threonine antiporter of Bacillus subtilis is the first characterized prokaryotic member of LATs (Reig et al., 2007; Bartoccioni et al., 2010). Sequence alignments of SteT with LATs reveal amino acid identities ranging from 26 to 30% (Fig. 1; Reig et al., 2007). Also, a phylogenetic tree of the APC superfamily situates SteT within the LAT family (Reig et al., 2007). Most importantly, SteT shares the transport mechanism of the majority of LATs, that is, obligatory amino acid exchange activity together with a low substrate specificity (Reig et al., 2007; Bartoccioni et al., 2010). These characteristics situate SteT as an ideal bacterial paradigm of LATs. WT SteT can be heterologously expressed in Escherichia coli in sufficient amount for functional characterization using radiolabeled substrates (Reig et al., 2007; Bartoccioni et al., 2010). Unfortunately, SteT is highly unstable in detergent micelles, as judged by its low solubility and large tendency to aggregate at concentrations >2 mg/ml, impassable obstacles for the majority of structural biology techniques and, in particular, for protein crystallization.
Figure 1.
Multiple alignments of SteT and human LATs. The figure only shows the alignment of the regions where SteT and human LATs are more conserved (TMDs 1, 2, 6, 7, and 8). The full sequence of these transporters have ∼30% amino acid identity (Reig et al., 2007). Lines above the sequences define the SteT TMDs. Residues totally conserved among these transporters are indicated by an asterisk. Residues of LAT-1 predicted to interact with the substrate (Geier et al., 2013) and conserved in SteT are highlighted with a gray background.
Protein engineering is one of the most common and successful strategies for providing the desirable physical chemistry properties to a membrane protein for structural studies (Bill et al., 2011). In particular, point mutations within the TMDs are known to drastically increase protein stability in detergent (Smirnova and Kaback, 2003; Tate and Schertler, 2009). Also, single replacements are enough to stabilize a particular structural conformer of a membrane protein (Abramson et al., 2003; Kowalczyk et al., 2011), lowering the conformational heterogeneity in detergent micelles and increasing the probability of forming ordered crystals for x-ray diffraction and structure determination. Unfortunately, finding such mutations is often tedious and unsuccessful. Because of the still low amount of structural information at high resolution of membrane proteins, the prediction of stabilizing mutations using rational approaches very often fails. In this scenario, the screening of large libraries of mutants generated either by site-directed mutagenesis (Serrano-Vega et al., 2008; Penmatsa et al., 2013) or by random mutagenesis (Schlinkmann and Plückthun, 2013) has emerged as an excellent strategy to find engineered evolved versions of a particular membrane protein with optimal properties to initiate structural studies such as reasonable expression yield (typically >1 mg of pure and active protein per liter of culture) and stability after solubilizing in different detergents.
Here, aiming to advance in the structural and functional knowledge of LATs, we have defined a strategy to build evolved versions of SteT with improved expression yield and stability in detergent, as a mandatory step toward tridimensional structure determination of this transporter. First, we have generated a library of SteT random mutants. Second, optimized versions of SteT were identified using a screening method that combined the measurement of protein expression yield in E. coli cells and the analysis of its aggregation in detergent micelles as a measure of stability. Finally, the two best-optimized mutants were purified and their stability challenged in conditions typically used for membrane protein crystallization: high protein concentration and stability in detergents more suitable for crystallization. The possible mechanisms of SteT stabilization caused by these mutations are also discussed.
MATERIALS AND METHODS
Construction of a SteT random library
A codon-optimized cDNA encoding SteT for expressing in E. coli (GenScript) was cloned into the pTETGFP11 plasmid (provided by G.S. Waldo, Bioscience Division, Los Alamos National Laboratory, Los Alamos, NM; Cabantous and Waldo, 2006), generating pTET-SteT-GFP11, which includes the first 15 residues of the superfolder GFP (GFP11 fragment; Pédelacq et al., 2006) at the C-terminal end of SteT. The resulting plasmid pTET-SteT-GFP11 was subjected to error-prone PCR random mutagenesis using the GeneMorph II EZClone Domain Mutagenesis kit (Agilent Technologies), focusing only on the SteT coding region. We optimized the PCR setting conditions according the manufacturer’s instructions to obtain the desired mutational rate (1–2 amino acid substitution on each clone). The PCR product was then used as megaprimers in a second PCR reaction using pTET-SteT-GFP11 as a template to obtain pTET-SteTmutants-GFP11. This second PCR reaction containing pTET-SteTmutants-GFP11 was directly transformed in E. coli BL21(DE3) cells harboring the plasmid pETGFP1–10, which encodes residues 16–230 of the superfolder GFP (also donated by G.S. Waldo; Cabantous and Waldo, 2006). Transformed cells were plated overnight at 37°C in a nitrocellulose filter paper (Amersham Hybond-N; GE Healthcare) lying on top of a Luria broth (LB) agar plate containing the antibiotics spectinomycin and kanamycin. Once E. coli colonies appeared, we tested in the same colonies the expression of the corresponding SteTmutants-GFP11. Expression was initiated by transferring the filter paper into a new LB-agar plate containing 0.3 µg/ml anhydrotetracycline (ANTET) and incubating for 3 h at 30°C. After the incubation, the filter paper was moved to a new LB agar plate containing no inducing agent to remove the inducing agent and therefore to stop SteTmutants-GFP11 expression. Next, GFP1–10 was induced by transferring the filter paper into a new LB agar plate containing 0.4 mM isopropyl β-d-thiogalactoside (IPTG) followed by 3-h incubation at 30°C. Green E. coli colonies indicating the expression of a given SteTmutant-GFP11 after complementation with GFP1–10 were observed under UV or blue light using a stereo fluorescence microscope (Leica Biosystems). These green colonies were cultured and plasmids were isolated for DNA sequencing to identify and localize the position of each point mutation. Only single or double mutants with amino acid substitutions in TMDs were included in the SteT random library. SteT TMDs were defined using a previous 3-D model that used the APC bacterial amino acid transporter AdiC as template (Bartoccioni et al., 2010).
Expression screening of SteT mutants using the GFP split system
20 ml E. coli BL21(DE3) harboring each pTET-SteTmutant-GFP11 and pETGFP1–10 were grown, and SteTmutant-GFP11 expression was induced overnight with 0.3 µg/ml ANTET at 30°C. Thereafter, GFP1–10 was induced after adding 0.4 mM IPTG and incubating for 1 h at 30°C. Cells were collected by centrifugation, washed twice with PBS, and resuspended in PBS at a final OD600 of 0.2. Finally, 200 µl of each sample was transferred into a 96-well plate to measure the GFP fluorescence generated after GFP complementation. Fluorescence background from noninduced cells was subtracted from each measurement. Values of expression were annotated as relative values of SteT WT expression.
Analysis of stability of mutants by fluorescence size exclusion chromatography (SEC [FSEC]) in n-dodecyl-β-d-maltopyranoside (DDM)
200 ml of a bacterial culture harboring a given pTET-SteTmutant-GFP11 and pETGFP1–10 were sequentially induced with ANTET and IPTG as described in the previous section, and plasma membranes were isolated as described earlier (Bartoccioni et al., 2010). In brief, cells were pelleted after centrifuging at 5,000 g for 15 min at 4°C, washed once with ice-cold lysis buffer (20 mM Tris-Base, pH 8.0, and 350 mM NaCl), resuspended with the same buffer at 0.2 g of cells/ml of buffer and disrupted by passage through a cell disruptor (UK Constant Systems), three times at 20,000 psi. Cell debris and unbroken cells were removed after a first centrifugation (1 h, 15,000 g, and 4°C), and the supernatant was then ultracentrifuged (2 h, 200,000 g, and 4°C) to isolate the membranes. The resulting membrane pellet was resuspended and homogenized in resuspension buffer (20 mM Tris-Base, pH 8.0, 150 mM NaCl, and 10% [wt/vol] glycerol), adjusting the total protein concentration at 10 mg/ml. Membranes were flash frozen in liquid nitrogen and stored at −80°C until use. Membrane suspensions were solubilized in 1% (wt/vol) DDM (Affymetrix) for 1 h at 4°C. Insoluble material was removed after ultracentrifugation (120,000 g, 1 h, 4°C), and 500 µl of the DDM-solubilized membranes was injected into a Superose 6 10/300 GL column (GE Healthcare) equilibrated with 20 mM Tris-Base, pH 7.6, 150 mM NaCl, and 0.05% (wt/vol) DDM. 200-µl fractions from the column elution were collected in a 96-well plate, and GFP fluorescence was measured in a fluorescent plate reader and plotted versus the elution time to construct the FPLC chromatogram. To quantify the degree of monodispersity of each SteT mutant with respect to WT, a parameter called index of monodispersity (IM) was calculated from each FSEC chromatogram according the following expression:
| (1) |
where ∫ refers to the area under the curve of the normalized FSEC chromatogram (N − FSEC) between the 13 and the 17 ml of elution. This interval of elution volume includes the main elution peak of SteT WT monomer (around 15 ml).
Detergent screening
10 mg/ml of plasma membranes prepared as described in the previous section was solubilized in 1% (wt/vol) n-decyl-β-d-maltopyranoside (DM), n-octyl-β-d-glucopyranoside (OG), 6-cyclohexyl-1-hexyl-β-d-maltoside (Cymal-6), or n-dodecyl-N,N-dimethylamine-N-oxide (LDAO; Affymetrix) for 1 h at 4°C. After ultracentrifugation (120,000 g, 1 h, and 4°C), solubilization efficiency was calculated from the GFP fluorescence ratio of the solubilized membrane fraction versus the nonsolubilized pellet. FSEC analysis was performed as in the experiments with DDM. In all of the experiments, the column was equilibrated with 20 mM Tris-Base, pH 7.6, 150 mM NaCl, and 0.05% (wt/vol) DDM.
Cloning SteT in pTTQ18-GFP-His(x10) for large-scale expression and protein purification
The cDNAs encoding SteT WT, L210Q/M229V, and I134V/A377T were cloned between the EcoRI and PstI sites of the pTTQ18-GFP-His(x10) vector to generate the corresponding SteT variant containing the whole GFP at the C-terminal end followed by a tail of 10 His. A HRV 3C protease recognition site (Leu-Glu-Val-Leu-Phe-Gln-Gly-Pro) was also introduced between each SteT variant and the GFP. All constructs were verified by DNA sequencing. For protein expression, 4.8 liters of E. coli BL21(DE3) harboring the pTTQ18-SteTmutant-GFP-His(x10) plasmid were grown at a cell density of OD600 = 0.5; at this point, protein expression was induced after adding 0.5 mM IPTG and incubating overnight at 37°C. After protein induction, bacterial pellets were resuspended in 20 mM Tris-Base, pH 8.0, and 350 mM NaCl supplemented with cOmplete EDTA-free protease inhibitor cocktail and Pefabloc (Roche) at 0.2 g cell pellet/ml buffer. Cells were disrupted by passage through a cell disruptor (UK Constant Systems), three times at 20,000 psi. Cell debris and unbroken cells were removed by centrifugation (1 h, 15,000 g, and 4°C). The supernatant was then ultracentrifuged (2 h, 200,000 g, and 4°C), and the membrane pellet was resuspended and homogenized in resuspension buffer (20 mM Tris-Base, pH 8.0, 150 mM NaCl, and 10% [wt/vol] glycerol). Membrane suspensions were aliquoted, frozen in liquid nitrogen, and stored at −80°C until use. Expression of the different SteT variants was tested by Western blot analysis using the HisProbe-HRP (Thermo Fisher Scientific) against the His-tag epitope.
Membrane suspension containing 3 mg/ml of total protein concentration was solubilized in 1% (wt/vol) DDM under agitation for 1 h at 4°C. Insolubilized material was removed after ultracentrifugation at 120,000 g, 1 h, and 4°C. The resulting supernatant was then incubated for 2 h and 4°C with Ni-NTA beads (QIAGEN) previously equilibrated with resuspension buffer containing 0.01% (wt/vol) DDM and 10 mM imidazole. After incubation, resin was washed sequentially with 10 bead volumes of washing buffer (20 mM Tris-Base, pH 8.0, 150 mM NaCl, 0.01% [wt/vol] DDM, 10 mM imidazole, and 10% [wt/vol] glycerol), 10 bead volumes of washing buffer plus 20 mM imidazole, and finally, 10 bead volumes of washing buffer with 40 mM imidazole. Protein was eluted after incubating the beads with elution buffer (20 mM Tris-Base, pH 8.0, 150 mM NaCl, 10% [wt/vol] glycerol, 0.01% [wt/vol] DDM, and 350 mM imidazole) for 30 min. After elution, imidazole was removed using PD-10 desalting columns (GE Healthcare) and incubated with HVR 3C protease (1:20 mol/mol protease/protein) with agitation in protease digestion buffer (20 mM Tris-Base, pH 8.0, 150 mM NaCl, 0.01% [wt/vol] DDM, 10% [wt/vol] glycerol, 0.5 mM EDTA, and 1 mM DTT) for 22 h at 4°C. After protease digestion, His(x10)-tagged GFP as well as His(x10)-tagged nondigested protein were removed by incubating the sample with Ni-NTA beads for 2 h at 4°C. Nonbound purified digested protein was concentrated in Vivaspin 100 MWCO concentrators (Sartorius) and after an ultracentrifugation step (100,000 g, 30 min) to remove possible aggregates, injected in the SEC columns (Superdex 200 5/150 or Superdex 200 10/300 GL; GE Healthcare). Columns were equilibrated with 20 mM Tris-Base, pH 8.0, 150 mM NaCl, and 0.01% (wt/vol) DDM. Detergent exchange of purified proteins was performed during protein concentration by successive cycles of protein concentration and dilution in the new detergent and during SEC using a final concentration of 2× critical micelle concentration (CMC) of each detergent in the running buffer.
Reconstitution of SteT WT, L210Q/M299, and I134V/A377T into proteoliposomes
E. coli polar lipids (Avanti Polar Lipids, Inc.) solubilized in chloroform at 50 mg/ml concentration were dried in a glass tube under a stream of nitrogen to obtain a thin layer. The dried lipids were resuspended in dialysis buffer (120 mM KPi, pH 7.4, 0.5 mM EDTA, 1 mM MgSO4, 5 mM TrisSO4, and 1% [wt/vol] glycerol) and 4 mM l-Ser to yield a final lipid concentration of 40 mg/ml. After four cycles of sonication and vortexing, the liposomes were extruded in a LiposoFast-Pneumatic Actuator (Avestin) through a 400-nm polycarbonate filter to obtain unilamellar vesicles of homogeneous size. These vesicles were destabilized after adding 1.25% (wt/vol) OG, mixed with the purified SteT version at a 1:100 protein/lipid ratio (wt/wt), and incubated on ice with occasional agitation for 5 min. Detergents were removed by dialysis (40 h and 4°C) using 100 sample volumes of dialysis buffer. Finally, the dialyzed proteoliposomes were pelleted by ultracentrifugation (100,000 g, 1 h, and 4°C) and resuspended in one third of the initial volume of dialysis buffer.
Transport measurements in proteoliposomes
Transport measurements were performed after a previously described protocol (Reig et al., 2007) with minor changes. 10 µl of proteoliposomes containing no amino acids or 4 mM l-Ser were quickly mixed with 180 µl of transport buffer (150 mM choline chloride, 10 mM Tris-HEPES, pH 7.4, 1 mM MgCl2, 1 mM CaCl2, 0.5 µCi l-[3H]Ser, and 10 µM l-Ser) and incubated at room temperature for different periods of time. Reactions were stopped by the addition of 850 µl of ice-cold stop buffer (10 mM Tris-HEPES, pH 7.4, 150 mM choline chloride, and 5 mM l-Ser) and quickly filtrated through nitrocellulose membrane filters (Sartorius; 0.45-µm pore size). Filters were then washed three times with 2 ml of stop buffer and dried, and finally, the trapped radioactivity was quantified by scintillation counting. All experimental values were corrected by subtracting the zero time values obtained by adding the stop solution in the transport buffer. Protein concentration in the proteoliposomes was determined using the amide black protein assay (Schaffner and Weissmann, 1973), and transport yield was expressed as pmol l-Ser/µg of protein and reported as the mean ± SE.
RESULTS
Construction and assessment of expression and folding of the SteT mutant library
A library of SteT random mutants was built using an error-prone PCR protocol previously optimized to produce a maximum of two amino acid substitutions per mutant. The low mutagenesis rate was intended to minimize any possible effect into the functional and structural integrity of SteT. Single or double mutants of SteT that expressed and folded properly in the host cytoplasmic membrane were selected using an assay based on the molecular complementation of the GFP (Rodríguez-Banqueri et al., 2012). In this method, a 15–amino acid fragment of an engineered superfolder GFP (GFP11; Pédelacq et al., 2006) is fused in the C-terminal end of SteT (Cabantous et al., 2005). GFP fluorescence is detected if the GFP11 fragment complements with the remaining nonfluorescent 215–amino acid fragment of the GFP (GFP1–10), sequentially coexpressed in the same cell (Rodríguez-Banqueri et al., 2012). Importantly, GFP complementation will only take place if the mutant is properly folded in the membrane (Rodríguez-Banqueri et al., 2012) because misfolded proteins are removed from the membrane and accumulated into inclusion bodies (Luirink et al., 2012). Consequently, if the random mutation drastically affects expression or membrane insertion and stability, no fluorescence will be detected. Sequential expression of SteT–random mutant–GFP11 and GFP1–10 was performed directly in agar LB plates containing E. coli colonies transformed with the cDNA product resulting from the random mutagenesis and cloning reaction; therefore, each colony contained the cDNA that encoded an individual SteT random mutant. This strategy quickly discards misfolded or nonexpressing mutants before DNA sequencing. In addition, attaching only a small portion of the GFP to SteT minimizes any effect related with the presence of the whole GFP during protein translation and membrane insertion. After colony selection and DNA sequencing, only mutants with substitutions localized in the TMDs of SteT were considered for the final library. The TMDs of SteT were defined using hydrophobicity plots and a 3-D structural model of SteT (Fig. 1; Bartoccioni et al., 2010). A final library of 70 random mutants of SteT (Table 1) with mutations distributed along the 12 TMDs of SteT was further analyzed for membrane expression and homogeneity in detergent micelles. In this library, 41 mutants (59%) contained single amino acid substitutions and 29 (41%) double amino acid substitutions.
Table 1.
SteT random mutants studied in this paper
| Mutant | TMD |
| L247M | 7 |
| L63R | 2 |
| L52Q | 2 |
| A305T/T410S | 8–12 |
| F49Y/L297V | 2–8 |
| I243P/A383S | 7–11 |
| P226Q | 7 |
| M392V | 11 |
| I235V | 7 |
| F89S/A105T | 3–3 |
| I235F | 7 |
| G61D/L78V | 2–3 |
| G62C/F304S | 2–8 |
| I285V | 8 |
| L199P | 6 |
| A197V/G232D | 6–7 |
| G152D/V370M | 1-9 |
| G69D | 2 |
| G87D | 3 |
| F49Y | 2 |
| F16L/I99V | 1–3 |
| R374C | 10 |
| T159I/S298T | 5–8 |
| P34Q | 1 |
| I132F | 4 |
| F371I/C415Y | 10–12 |
| V154E/G161S | 5–5 |
| L247V | 7 |
| G215D | 6 |
| G23R | 1 |
| A136E | 4 |
| G103S/L279P | 3–8 |
| A369G | 9 |
| G35R/G55D | 1–2 |
| L199M/L417M | 6–12 |
| A196T | 6 |
| C141W | 4 |
| E67K/E308K | 2–8 |
| F203S/R376P | 6–10 |
| F139L | 4 |
| G123D/I164T | 3–5 |
| I107F/H249Y | 3–7 |
| G54S | 2 |
| F49Y/N347Y | 2–9 |
| I336N/M413S | 9–12 |
| F402S | 11 |
| A60E/C168R | 2–5 |
| A267V | 8 |
| M32V/M342L | 1–9 |
| A424T | 12 |
| A109P | 3 |
| A398P | 11 |
| G283V | 8 |
| F31I | 1 |
| G27A/T156S | 1–5 |
| R374H | 10 |
| L53P | 2 |
| F391Y | 11 |
| W51R/L338Q | 2–9 |
| L14Q/T230A | 1–7 |
| F31L | 1 |
| A39F | 1 |
| A339D | 11 |
| L210Q/M229V | 6–7 |
| N193D | 6 |
| G161N | 5 |
| C291S | 8 |
| S303T | 8 |
| I134V/A377T | 4–10 |
| C168Y/L233M | 5–7 |
| Mutants were generated by error-prone PCR according the protocol described in Materials and methods. The TMD column indicates the number of TMDs where the substitutions are located. Mutants are ordered as in Fig. 3, on the basis of their relative expression yield with respect to WT. | |
Expression and detergent stability of SteT random mutants
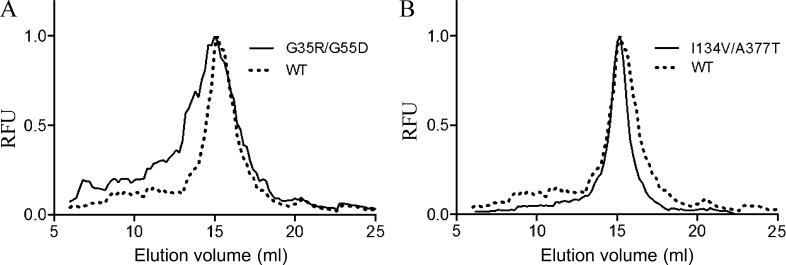
The 70 SteT random mutants (Table 1) were solubilized in DDM from E. coli membranes and subjected to FSEC analysis (Kawate and Gouaux, 2006). FSEC permits an accurate evaluation of the degree of monodispersity in detergent of nonpurified GFP-tagged membrane proteins by simply analyzing the shape and retention time of the chromatograms using the fluorescence of the GFP as readout. The aggregation state and therefore the stability of each SteT mutant in the conditions tested can be easily monitored from the chromatogram. DDM was the detergent of choice because it has shown before its ability to efficiently solubilize SteT in a functional state (Reig et al., 2007). As illustrated in Fig. 2, most of the SteT random mutants eluted as monomers with similar retention as WT; however, changes in the shape of the elution peak of the different mutants revealed different degrees of aggregation as a consequence of instability (examples in Fig. 2). These changes were evaluated and compared with WT’s chromatogram after assigning a numeric value to each chromatogram named IM (Fig. 3). This index is calculated by dividing the area under the curve of the elution peak of each mutant by the corresponding area of WT’s elution peak (Eq. 1; see Materials and methods for full description). Thus, mutants with IM >1 are more monodisperse and therefore more stable than WT in DDM, whereas mutants with IM <1 present less monodispersity in DDM than WT. In parallel, the expression yield of each mutant was quantified using the fluorescence of GFP after in vivo complementation. Measuring expression yields using the split GFP complementation assay ensured that the protein fraction present in the membrane and later solubilized in DDM micelles is the only fraction quantified because poorly stable mutants are removed from the membrane and stored in inclusion bodies that do not emit fluorescence (Rodríguez-Banqueri et al., 2012). Normalized values of expression yield versus WT of each mutant are plotted together with their respective IM in Fig. 3.
Figure 2.
Examples of normalized FSEC profiles of SteT-GFP variants. To compare the monodispersity in DDM of each SteT mutant, FSEC chromatograms were normalized and overlapped. (A) A typical result of a double mutation (G35R/G55R) that causes a decrease of monodispersity in DDM. (B) The improving effect of the double mutation I134V/A377T on SteT monodispersity in DDM. RFU, relative fluorescent units.
Figure 3.
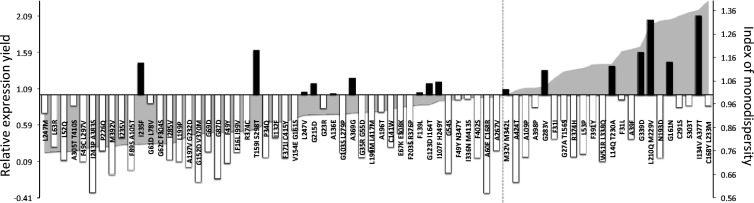
Analysis of expression and monodispersity in DDM of SteT random mutants. The values of relative expression yield in E. coli of each mutant with respect to WT (left axis) are represented as shadow areas. These values were calculated using the split GFP assay and normalized with respect to the corresponding value of SteT WT. Mutants situated on the left of the dashed line show lower expression yield than WT, whereas mutants that show better expression yield are situated on the right side of this line. Vertical bars in the graph represent the values of the IM of a giving mutant (see Materials and methods for description). White bars correspond to mutants with worse monodispersity in DDM than WT (IM <1) and black bars correspond to mutants with better monodispersity in DDM than WT (IM >1).
Overall effect of introducing random mutations in SteT TMDs
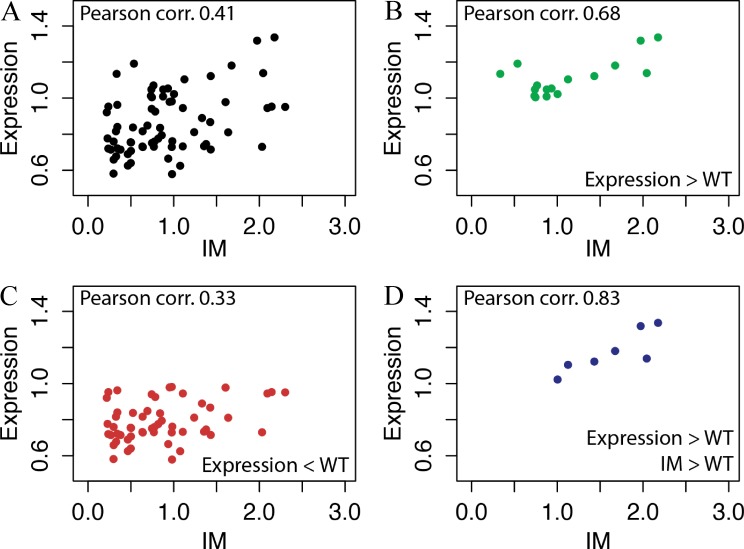
The results in Fig. 3 indicate that mutating residues located in TMDs more frequently resulted in a decrease of protein expression yield, in an IM <1, or both (Fig. 3). In particular, 69% of mutants expressed less than WT and 60% had an IM <1. Among the mutants expressing more than WT, 27% (6 out of 22) presented an IM >1. These better performing mutants were the following: G283V, L14Q/T230A, G339D, L210Q/M229V, G161N, and I134V/A377T. Among them, the double mutants L210Q/M229V and I134V/A377T were the ones that presented the best-combined improvement of expression and monodispersity in DDM (Fig. 3). In contrast, only 4% of mutants expressing less than WT (2 out of 48) presented an IM >1 (Fig. 3), suggesting that the stability in DDM micelles of a particular SteT mutant and its expression and stability in the host cytoplasmic membrane could present some correlation. We used Pearson’s coefficients to analyze the correlation between expression and IM (Fig. 4). A slight linear relationship of both variables was confirmed by a Pearson’s coefficient of 0.41 (Fig. 4 A), where 1 is total positive correlation and 0 is no correlation. We have analyzed separately the mutants regarding three subsets: expression levels higher than WT (Fig. 4 B), expression levels lower than WT (Fig. 4 C), and expression levels higher than WT and IM >1 (Fig. 4 D). The results showed a moderate (Pearson’s coefficient of 0.68; Fig. 4 B) and strong direct relationship (Pearson’s coefficient of 0.83; Fig. 4 D) between both variables for mutants expressing higher than WT and mutants with higher expression than WT and IM >1, respectively.
Figure 4.
Correlation between expression and stability in detergent of the SteT random mutants. (A) Values of expression and IM from Fig. 3 were plotted and analyzed using Pearson’s coefficients. (B–D) Mutants were also analyzed separately according the following criteria: expression levels higher than WT (B), expression levels lower than WT (C), and expression levels higher than WT and IM >1 (D). The numerical values of Pearson’s coefficients of each dataset are indicated in each panel.
L210Q/M229V and I134V/A377T retain monodispersity in detergents used for membrane protein crystallization
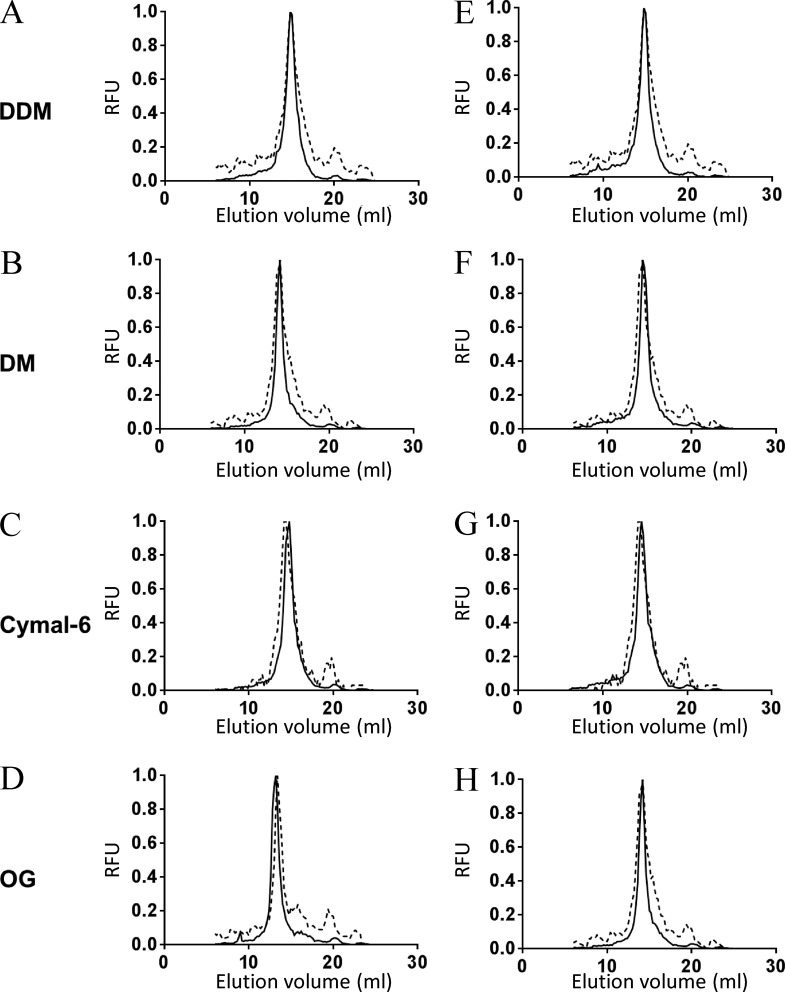
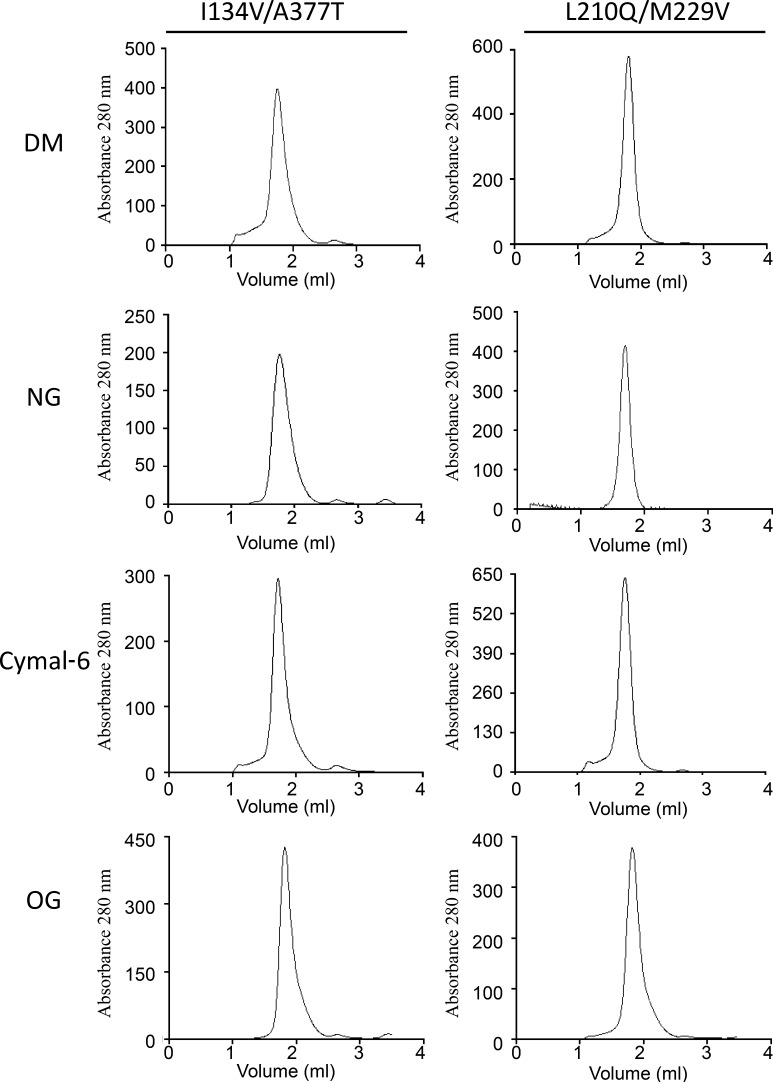
Protein–protein crystal contacts in membrane proteins are more likely to occur after reducing the size of protein–detergent micelles, normally achieved by using detergents with shorter hydrophobic tails and/or smaller head groups (Sonoda et al., 2011). However, these detergents can compromise protein stability in solution. Therefore, stability in these detergents is highly desirable when attempting to crystallize a membrane protein. In this line, we analyzed membrane solubilization and monodispersity in solution of SteT WT, L210Q/M229V, and I134V/A377T in five different detergents (including DDM as control) with different hydrophobic tails and head groups: DM, OG, Cymal-6, and LDAO. The solubilization efficiency of all SteT variants after 1 h of incubation in 1% (wt/vol) of a given detergent was fairly high (≥80%; not depicted), being slightly more efficient when solubilizing I134V/A377T (not depicted). Next, the monodispersity of nonpurified detergent-solubilized SteT WT, I134V/A377T, and L210Q/M229V in all detergents was monitored by FSEC. I134V/A377T and L210Q/M229V showed an improvement of SteT monodispersity in all detergents (Fig. 5) with the exception of LDAO, which induced a highly polydisperse behavior in all three SteT variants (not depicted).
Figure 5.
Normalized FSEC profiles of SteT WT, I134V/A377T, and L210Q/M229V solubilized in DDM, DM, Cymal-6, and OG. (A–H) Normalized FSECs of SteT-I134V/A377T (A–D) and SteT-L210Q/M229V (E–H) were overlapped with the corresponding normalized FSEC of SteT WT (dashed lines). SteT variants containing the GFP in the C-terminal end were solubilized from the membrane with DDM (A and E), DM (B and F), Cymal-6 (C and G), or OG (D and H) and injected into the gel filtration column equilibrated with a buffer containing 2× CMC of DDM. RFU, relative fluorescent units.
L210Q/M229V and I134V/A377T catalyze l-Ser/l-Ser exchange in proteoliposomes
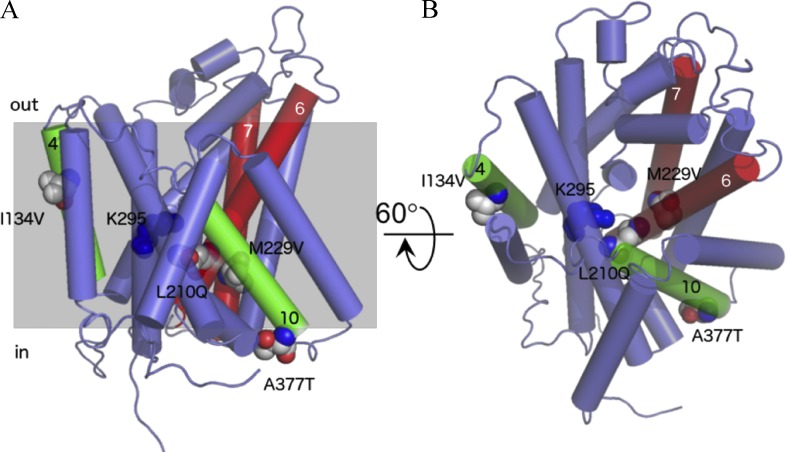
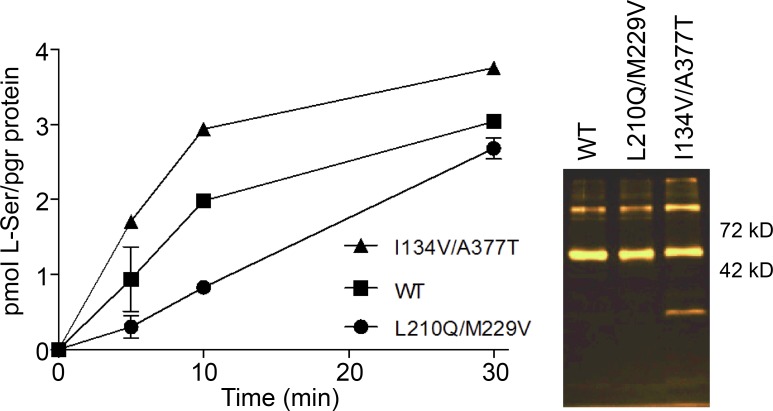
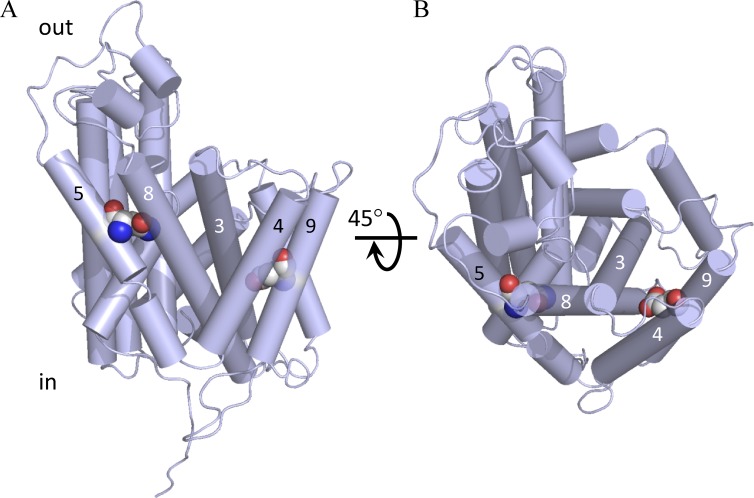
The residues replaced in L210Q/229V and I134V/A377T are sitting away from the proposed substrate-binding site of SteT, situated near residue K295 who plays a key role in substrate recognition (Fig. 6; Bartoccioni et al., 2010). Indeed, the crystal structures of AdiC show that K295 homologous residue (Trp293) interacts with the substrate (Gao et al., 2010). Transport experiments were performed to investigate any possible alteration of the structural integrity of SteT as a consequence of these mutations, thus affecting substrate recognition and/or translocation. SteT WT, L210Q/229V, and I134V/A377T with the GFP fused at the C-terminal end were purified by affinity chromatography, and their functional activity was tested after reconstitution into liposomes composed by E. coli lipids. The efficiency of reconstitution was similar in all three SteT variants, as judged by the GFP fluorescence in SDS-PAGE gels of reconstituted proteins (Fig. 7). Because SteT is an obligatory amino acid exchanger (as mammalian LATs), transport activity was measured monitoring the uptake of radioactive l-Ser into proteoliposomes previously loaded with a saturated concentration of cold l-Ser (Fig. 7). The net l-Ser uptake by SteT at different time intervals was calculated after subtracting counts obtained in the l-Ser–loaded proteoliposomes from the ones obtained in empty proteoliposomes. l-Ser/l-Ser exchange activity of SteT WT fused to the GFP was similar to that previously reported in the absence of the GFP (Fig. 7; Reig et al., 2007; Bartoccioni et al., 2010). The double mutants L210Q/M229V and I134V/A377T also displayed exchange activity as WT (Fig. 7), indicating that both sets of mutations do not substantially alter the overall fold of SteT, at least with regard to substrate binding. The transport activity of L210Q/M229V was somehow slower than that observed in WT and I134V/A377T (Fig. 7). These experiments were repeated three times with similar results.
Figure 6.
Position of the amino acid replaced in SteT mutants L210Q/M229V and I134V/A377T. Amino acid substitutions were modeled in a SteT 3-D structural model (Bartoccioni et al., 2010) and represented as spheres. Transmembrane segments are represented as cylinders, and only the TMDs containing mutations are numbered. TMDs 6 and 7 containing, respectively, the double substitution L210Q and M229V are colored in red, and TMDs 4 and 10 containing, respectively, the substitutions I134V and A377T are drawn in green. Residue K295 (TMD8), implicated in substrate recognition in SteT (Bartoccioni et al., 2010), is also labeled and represented as blue spheres. (A) A lateral perspective of the molecule. (B) A periplasmic view after 60° rotation of A through the x axis. The gray shadow square in A represents the putative position of the surrounding membrane.
Figure 7.
Transport activity of SteT WT and the double mutants I134V/A377T and L210Q/M229V. Transport activity of each SteT version was analyzed from the time-dependent uptake curves of 10 µM radiolabeled l-Ser into proteoliposomes formed after reconstituting each detergent-purified version of SteT, loaded with 4 mM l-Ser. The data points correspond to the net transport activity of each SteT version and were calculated after subtracting the data of nonloaded proteoliposomes from the data of proteoliposomes loaded with l-Ser. Each data dataset is the mean of three independent experiments, and the SD is also represented. The figure also shows the GFP fluorescence in SDS-PAGE gels of the three proteins after reconstituting in proteoliposomes. SteT monomer fused to the GFP is the main band between 42 and 72 kD.
Stability of purified I134V/A377T and L210Q/M229V
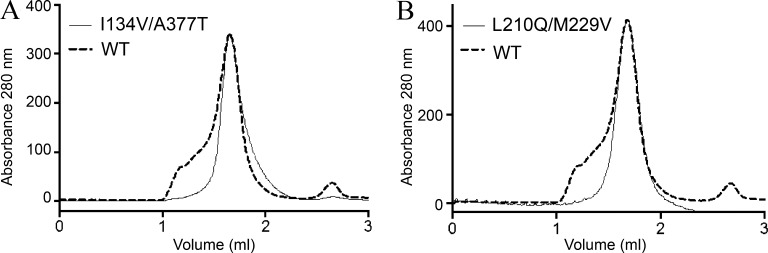
We next performed stability tests of purified SteT WT, L210Q/M229V, and I134V/A377T at protein concentrations and purity standards commonly used for structural studies. After DDM solubilization, affinity purification, and GFP-His tag removal by enzymatic digestion, SteT variants were concentrated up to 2 mg/ml (close to the limit of solubility of SteT WT [Reig et al., 2007]) and subjected to gel filtration chromatography (SEC) to evaluate their aggregation state. SEC profiles of L210Q/M229V and I134V/A377T showed a clear monodisperse behavior, as judged by their single and symmetric SEC elution peak consistent with monomeric SteT (Fig. 8). In contrast, SteT WT presented polydispersity in the same conditions displaying a broader elution peak as well as high–molecular weight peaks (Fig. 8). Differences between WT and mutants are better appreciated in the overlapped chromatograms (Fig. 8). As with the experiments with nonpurified GFP-tagged proteins, we also analyzed the stability of L210Q/M229V and I134V/A377T at 4 mg/ml in other detergents such as DM, NG, OG, and Cymal-6 (Fig. 9). As in the FSEC experiments, the SEC profiles of the two purified SteT mutants showed a monodisperse elution profile in all four detergents studied.
Figure 8.
SEC profiles of DDM-purified SteT WT, I134V/A377T, and L210Q/M229V. Purified proteins from His-tag affinity chromatography were concentrated up to 2 mg/ml and injected in a Superdex 200 50/1 50G column. (A and B) Monodispersity in these experimental conditions was analyzed by overlapping the normalized chromatograms of SteT WT with I134V/A377T (A) and L210Q/M229V (B).
Figure 9.
SEC profiles of purified SteT I134V/A377T and L210Q/M229V in DM, NG, Cymal-6, and OG. Affinity purified proteins in DDM were concentrated up to 4 mg/ml and injected into a Superdex 200 50/1 50G column equilibrated with 2× CMC of the indicated detergent.
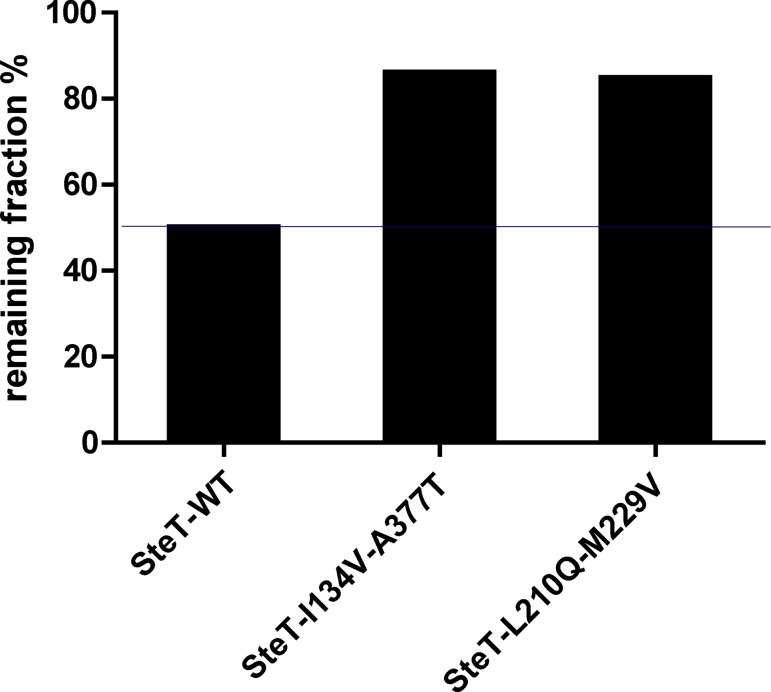
Stability in solution was also challenged after heating purified SteT WT, L210Q/M229V, and I134V/A377T in DDM. After incubating at 50°C for 30 min the purified proteins concentrated at 2 mg/ml, samples were ultracentrifuged to remove protein aggregates, and the remaining protein in the supernatant was quantified. Fig. 10 shows that after this treatment, only 50% of SteT WT remained in solution, whereas 80% of the two SteT mutants did not precipitate after this treatment.
Figure 10.
Stability of purified SteT WT, I134V/A377T, and L210Q/M229V after thermal denaturation. 2 mg/ml of purified proteins in DDM were incubated at 50°C for 30 min. The bars indicate the remaining percentage of each indicated SteT version present in the supernatant and measured after ultracentrifugation.
I134V/A377T is stabilized with E. coli lipids
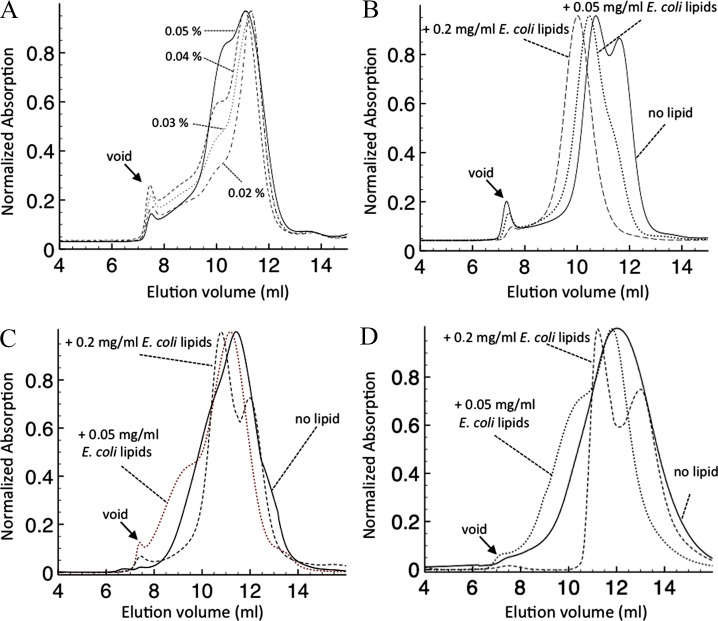
The 3-D model of SteT situates residues I134 and A377 in the periplasmic and cytoplasmic sides of the membrane, respectively, and likely oriented to the lipid phase (Fig. 6). We reasoned that changes in lipid–protein interactions caused by these mutations could explain (at least in part) the increase of expression and stability observed in I134V/A377T. To test this, we proceeded to analyze I134V/A377T monodispersity after sequential delipidation achieved by purifying I134V/A377T at different concentrations of DDM. Although DDM is considered as a mild detergent, increasing the number of detergent micelles in the washing buffer that passes through the protein bound to the Ni-NTA beads results in a progressive elimination of lipids from the mixed protein–detergent–lipid micelle (Harvey and Wysocki, 2015). Therefore, SEC chromatograms of I134V/A377T purified at 0.02, 0.03, 0.04, or 0.05% (wt/vol) DDM, respectively, were analyzed (Fig. 11 A). The results clearly revealed a progressive displacement from monodisperse to polydisperse behavior upon increasing the concentration of DDM (Fig. 11 A), indicating the appearance of high–molecular weight aggregates resulting from delipidation. Indeed, the elution profile of I134V/A377T in 0.05% (wt/vol) DDM resembled the SteT WT SEC chromatogram in nondelipidating conditions (0.02% [wt/vol] DDM). Moreover, SEC profiles of purified I134V/A377T in 0.05% (wt/vol) DDM but in the presence of 0.05, 0.1, or 0.2 mg/ml of E. coli lipids in both the purification and SEC buffers showed a progressive reversion of the polydisperse behavior, shifting from two to one protein elution peak upon increasing the concentration of E. coli lipids (Fig. 11 B). In contrast, when the same experiment was performed in SteT WT, we did not observe a complete shift from polydisperse to monodisperse elution profile upon adding the same amount of E. coli lipids in the buffers (Fig. 11 C). Similarly, relipidation of L210Q/M229V, a stabilized SteT version whose mutated residues are hidden from the lipid phase (Fig. 6) gave a similar result as WT, where a monodisperse elution profile could not be achieved after E. coli lipid addition (Fig. 11 D). The displacement of the retention time of the eluted protein toward higher apparent molecular mass in these experiments (Fig. 11, A–D) obeyed an increase of the apparent molecular mass of the mixed protein–detergent–lipid micelle caused by the incorporation of either detergent or lipid molecules. Although increasing DDM concentrations did not significantly change the apparent size of the micelle (Fig. 11 A), the E. coli lipids added in the buffer became quickly incorporated into the mixed micelle, increasing the apparent molecular mass of the micelle (Fig. 11, B–D). These experiments support the hypothesis that the I134V/A377T replacements might favor the interaction between SteT and one of several polar lipid components of the E. coli membrane, and this could play some important role during protein expression and stabilization after detergent solubilization.
Figure 11.
Effect of E. coli membrane lipids in the monodispersity of purified SteT I134V/A377T, L210Q/M229V, and WT. (A) SteT I134V/A377T was solubilized from the membrane with 1% (wt/vol) DDM and immobilized in Ni-NTA beads for affinity purification. Beads were divided and poured into four columns and washed with 5 column volumes of 0.02, 0.03, 0.04, and 0.05% (wt/vol) DDM, respectively, in each column. Each protein sample was eluted from the affinity column and subjected to gel filtration chromatography in a Superdex 200 10/30 column equilibrated with the same concentration of DDM used in the affinity purification. By gradually increasing the concentration of DDM during purification, SteT I134V/A377T became less monodisperse as a result of the delipidation effect of DDM. (B) Solubilized SteT I134V/A377T in 1% (wt/vol) DDM was immobilized in Ni-NTA beads for affinity purification. Beads were divided into three columns and washed with buffer containing no lipid or 0.05 and 0.2 mg/ml E. coli lipids, respectively, in each column, always in the presence of 0.05% (wt/vol) DDM (delipidating conditions). Each protein sample was eluted from the affinity column and subjected to gel filtration chromatography in a Superdex 200 10/30 column equilibrated with 0.05% (wt/vol) DDM and the same amount of E. coli lipids used in the affinity purification. As seen in the figure, even in the presence of delipidating conditions (washing the Ni-NTA beads with 0.05% [wt/vol] DDM), the presence of E. coli lipids kept SteT I134V/A377T monodisperse in a concentration-dependent manner. (C and D) Solubilized SteT WT (C) and L210Q/M229V (D) in 1% (wt/vol) DDM were immobilized in Ni-NTA beads for affinity purification. As in B, beads were divided into three columns and washed with buffer containing no lipid or 0.05 and 0.2 mg/ml E. coli lipids, respectively, in the presence of 0.02% and 0.05% (wt/vol) DDM for WT and L210Q/M229V, respectively. Each protein sample was eluted from the affinity column and subjected to gel filtration chromatography in a Superdex 200 10/30 column equilibrated with 0.05% (wt/vol) DDM (0.02% in the case of WT) and the same amount of E. coli lipids used in the affinity purification. Even in the presence of 0.2 mg/ml E. coli lipids (and lower concentration of DDM in the case of WT: 0.02% [wt/vol] instead of 0.05%), both SteT variants eluted in a polydisperse manner.
DISCUSSION
Finding general recipes to produce enough quantity of polytopic membrane proteins with enough purity and stability for structural studies is a remarkable challenge. This is probably more difficult for membrane transporters because in addition to their metastable nature, the large conformational heterogeneity inherent to their function prevents the formation of ordered crystals needed for structure determination using x-ray diffraction. Membrane transporters (either primary or secondary transporters) comprise ∼20% of the solved structures of membrane proteins, although in humans they represent ∼30% of membrane proteins (Hediger et al., 2013). Protein engineering and, in particular, individual side chain substitutions in suitable positions of the protein have overcome some of these limitations (Abramson et al., 2003; Gao et al., 2010; Kowalczyk et al., 2011), being fundamental in the crystallization of several transporters. Unfortunately, the molecular mechanism behind the stabilization effect of a point mutation is often difficult to interpret (Vaidehi et al., 2016; even after solving the 3-D structure), hampering the formulation of general rules for future predictions. One of the most efficient approaches to optimize selected targets for structural studies consists of generating large libraries of mutants, very often by random amino acid replacements, with the goal of finding an evolved version of the target using optimal screening methods. Indeed, the GPCR field has been largely benefited using this approach (Tate and Schertler, 2009; Schlinkmann and Plückthun, 2013).
In this work, we took the challenge of engineering SteT, the best-known prokaryotic paradigm of human LATs (Reig et al., 2007), to provide optimal expression and stability for crystallogenesis. SteT shares 30% of amino acid identity and transport mechanism with many LATs, particularly with LAT-1, LAT-2, or b0,+AT (Fig. 1; Reig et al., 2007). In addition, residues predicted to interact with the substrate are highly conserved between SteT and LAT-1 (Fig. 1; Geier et al., 2013). To find an optimized and stable version of SteT, we introduced random replacements within the TMDs, building a library composed of single and double mutants. Interactions between TMDs are the major determinant in the assembly and stability of integral membrane proteins; consequently, side chain substitutions within these domains are expected to produce a bigger impact on protein stability (Thévenin and Lazarova, 2008; Dalbey et al., 2011).
As described previously for soluble proteins (Cabantous and Waldo, 2006), using the split GFP assay to measure protein expression and solubility allows discarding nonexpressing or poorly stable membrane proteins (or mutants) confined in inclusion bodies by the host (Rodríguez-Banqueri et al., 2012). The GFP can be fluorescent in inclusion bodies (García-Fruitós et al., 2005); however, GFP11 and GFP1–10 do not complement inside them (Cabantous et al., 2005). Therefore, green colonies obtained after sequential coexpression of SteTmutants-GFP11 and GFP1–10 contained SteT versions with the ability to express and fold in the host cytoplasmic membrane.
Although resistance to thermal denaturation (or thermostability) is commonly used to analyze and compare protein stability upon the introduction of point mutations (Serrano-Vega et al., 2008; Miller and Tate, 2011), other methods to measure stability like the analysis of the SEC chromatogram after detergent solubilization are well accepted. A monodisperse elution profile of a membrane protein solubilized in a particular detergent is a direct proof of its stability in this particular detergent (Sonoda et al., 2011). Proteins nonoptimally solubilized tend rapidly to aggregate, leading to the appearance of wider and/or multiple peaks (polydispersity) in the gel filtration chromatogram. In fact, there is a direct relationship between degree of monodispersity and probability of protein crystallization (Sonoda et al., 2011). DDM is a mild detergent able to solubilize in a functional state many membrane proteins, including SteT (Reig et al., 2007; Bartoccioni et al., 2010). Therefore, this detergent was used to solubilize and compare stability of the SteT mutant library by FSEC.
The screening of a pool of 70 SteT mutants with substitutions exclusively localized in the TMDs (Table 1) revealed that most of these replacements lead to either an impairment of expression or an increase of polydispersity after DDM solubilization (IM <1; Fig. 3). This low tolerance of substituting residues situated in TMDs has been observed previously with other membrane proteins (Martinez Molina et al., 2008). The interactions between TMDs are the main determinants of protein folding and membrane insertion (Walters and DeGrado, 2006); consequently, amino acid substitutions in these domains are likely to negatively perturb the expression yield and stability of the protein. In addition, TMD interfaces tend to be highly packed as the result of the presence of small residues (Walters and DeGrado, 2006). This is consistent with our observations because mutations that resulted in the introduction of bulkier side chains normally resulted in the decrease of SteT expression and monodispersity in DDM (IM <1). Indeed, 69% of mutants expressing less than WT (31 out of 48) present this type of substitutions in at least one of the two mutations. In addition, the majority of these mutants (including those expressing more than WT) also presented an IM <1 (Fig. 3). This negative effect was even more pronounced when charged side chains were introduced (e.g., L63R, G67D/L78V, A197V/G232D, G152D/V370M, G69D, G87D, G23R, or A60E/C168R; Fig. 3). SteT contains a few glycines highly conserved in LATs and located in the TMDs (Fig. 1). Replacing some of these glycines (e.g., G23R, G23D/A197V, G35R/G55D, G61D/L78V, or G215D) normally resulted in an IM <1 (Fig. 3). In this regard, a notable decay of both expression and DDM monodispersity was found in the G87D mutant (Fig. 3). G87 is the homologous residue of G105 in b0,+AT. The congenital mutation G105R in b0,+AT induces protein misfolding and degradation during biogenesis, ultimately leading to a pathology called cystinuria (Bröer and Palacín, 2011). This position (G87 in SteT and G105 in b0,+AT) is located in the C-terminal end of the predicted reentrant loop between TMDs 2 and 3 (Gasol et al., 2004), suggesting a role of this reentrant loop for the stabilization of the overall fold of LATs. Expression and monodispersity were also reduced after introducing or replacing prolines (e.g., L53P, A109P, G103S/L279P, L279P, P34Q, or P226Q; Fig. 3), arguing for folding instability derived from the critical role of this side chain in transmembrane proteins (Perálvarez-Marín et al., 2008).
Interestingly, replacements of nonpolar by polar residues in a few positions resulted in an increase of protein expression yield and stability in DDM. This is the case of mutants G161N, A339D, and L210Q/M229V. The glycine in position 161 (TMD5) is fairly well conserved in LATs (Fig. 1). Modeling the G161N mutation situates the amide group of the asparagine in an H-bond distance of S287 (TMD8; Fig. 12), a residue highly conserved in LATs. We suggest that this potential hydrogen bond interaction might help stabilizing the protein, improving expression yield and monodispersity in DDM. Likewise, the alanine in position 339 (TMD9) is also highly conserved in LATs (Fig. 1) facing TMDs 3 and 4. It is conceivable that after introducing a negative charge in the middle of TMD9 (A339D), a new polar interaction might take place probably between TMD9 and TMDs 3 or 4 (Fig. 12), helping the observed increase of protein expression and stability in detergent. Similarly, in the double mutant L210Q/M229V, the L→Q substitution in position 210 (TMD6) might also favor the formation of new polar interactions between TMD6 and TMDs 3 and/or 10, explaining the observed increase of expression and stability of this mutant with respect to WT (Fig. 3). The apparent slow rate of this mutant to exchange l-Ser compared with WT (Fig. 7) also suggests that these mutations could stabilize the protein in a particular conformer, probably through new intra-TMD interactions (Smirnova and Kaback, 2003), resulting in a decrease of the translocation rate. In any case, more experimental evidence is needed to verify this hypothesis. As stated earlier (Gratkowski et al., 2001; Walther and Ulrich, 2014), our results also support the idea that the energy cost of introducing polar residues in specific positions within TMDs is compensated by the energy of stabilizing the overall fold or specific structural conformers of the protein through the formation of polar interactions between TMDs.
Figure 12.
Position of mutants G161N and A339D in a SteT 3-D model. Amino acid substitutions G161N and A339D were modeled in a SteT 3-D structural model (Bartoccioni et al., 2010) and represented as spheres. TMDs are represented as cylinders, and for clarity, only some of them are numbered. The G161N replacement (TMD5) introduces a new asparagine residue suggested to interact with TMD8. Similarly, the introduced aspartate in the A339D mutation (TMD9) is suggested to interact with residues in TMDs 3 and/or 4. (A) A lateral perspective of the molecule. (B) A periplasmic view after 45° rotation of A through the x axis.
Pearson coefficients revealed a correlation between the yield of stably expressed protein in the membrane (measured using the split GFP complementation assay) and stability after detergent solubilization (measured using FSEC; Figs. 3 and 4). This correlation was particularly strong in those mutants with expression yield higher than WT and IM >1 (Fig. 4 D). Optimal miscibility between the nascent protein and the host’s membrane lipids appears to be a prerequisite to gain stability in DDM micelles as well as in other less mild detergents commonly used in membrane protein crystallization as DM, NG, OG, or Cymal-6 (Figs. 5 and 9). In this regard, it is documented that well-folded membrane proteins are more efficiently solubilized using mild detergents like DDM (Thomas and Tate, 2014) than partially misfolded ones.
From our screening, the double mutant I134V/A377T is the most robust version of SteT, as judged by its production yield in E. coli and monodispersity behavior in various detergents (Figs. 3, 5, 8, and 9) or even stability against thermal denaturation (Fig. 10). In the SteT 3-D model, residues I334 and A377 are situated in opposite leaflets of the membrane where the side chains are likely oriented toward the phospholipid phase rather than toward other TMDs of the protein as in the case of L210Q/M229V (Fig. 6). In addition, I134V/A377T shows a higher l-Ser/l-Ser exchange activity than SteT WT (Fig. 7), probably as a consequence of its higher resistance to aggregate during the reconstitution process. We reasoned that I134V/A377T might have a different or an additional mechanism of protein stabilization than just stabilizing a particular conformer of SteT through interactions between TMDs, as suggested for L210Q/M229V or G161N. The fact that the monodispersity behavior of I334V/A337T after delipidating with DDM can be rescued after adding E. coli lipids in contrast to WT or L210Q/M229V (Fig. 11, B–D) provided arguments to suggest that the double substitution I334V/A337T enables SteT to interact with one or several lipid components of the E. coli membrane, conferring the observed improvement of expression and stability in several detergents. SteT is a protein of B. subtilis, expressed heterologously in E. coli, and although both prokaryotic organisms share similar mechanisms of membrane protein biogenesis, their phospholipid composition in the cytoplasmic membrane differs (Barák and Muchová, 2013), a possible handicap when trying to produce heterologous B. subtilis membrane proteins like SteT in E. coli. Certainly our experiments are unable to provide the identity of the lipid component (or lipids) of the E. coli membrane that presumably stabilizes I334V/A337T or the molecular mechanism of stabilization. Nevertheless, as suggested in an engineered thermostable GPCR (Lee et al., 2015), it seems clear that by modifying the interactions between SteT and the host’s lipids using mutagenesis, an improvement of both protein production and stability after detergent solubilization can be achieved.
Acknowledgments
We are very grateful to Geoffrey S. Waldo and members of his laboratory for sending us the split GFP expression plasmids.
This work was supported in part by the Spanish Ministry of Science and Innovation (grants BFU2008-04637 [to J.L. Vázquez-Ibar] and SAF2015-64869-R [to M. Palacín]), by the Generalitat de Catalunya (grant 2014 SGR 298 to M. Palacín), and by Fundació La Marató TV3 (grant 20132330 to M. Palacín). E. Errasti-Murugarrem is recipient of a Sara Borrell contract from the Instituto de Salut Carlos III. IRB Barcelona is the recipient of a Severo Ochoa Award of Excellence from MINECO (Government of Spain).
The authors declare no competing financial interests.
Merritt Maduke served as editor.
Footnotes
Abbreviations used in this paper:
- ANTET
- anhydrotetracycline
- CMC
- critical micelle concentration
- Cymal-6
- 6-cyclohexyl-1-hexyl-β-d-maltoside
- DDM
- n-dodecyl-β-d-maltopyranoside
- DM
- n-decyl-β-d-maltopyranoside
- FSEC
- fluorescence SEC
- HAT
- heteromeric amino acid transporter
- IM
- index of monodispersity
- IPTG
- isopropyl β-d-thiogalactoside
- LAT
- l–amino acid transporter
- LB
- Luria broth
- LDAO
- n-dodecyl-N,N-dimethylamine-N-oxide
- OG
- n-octyl-β-d-glucopyranoside
- SEC
- size exclusion chromatography
- TMD
- transmembrane domain
References
- Abramson J., Smirnova I., Kasho V., Verner G., Kaback H.R., and Iwata S.. 2003. Structure and mechanism of the lactose permease of Escherichia coli. Science. 301:610–615. 10.1126/science.1088196 [DOI] [PubMed] [Google Scholar]
- Barák I., and Muchová K.. 2013. The role of lipid domains in bacterial cell processes. Int. J. Mol. Sci. 14:4050–4065. 10.3390/ijms14024050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartoccioni P., Del Rio C., Ratera M., Kowalczyk L., Baldwin J.M., Zorzano A., Quick M., Baldwin S.A., Vázquez-Ibar J.L., and Palacín M.. 2010. Role of transmembrane domain 8 in substrate selectivity and translocation of SteT, a member of the L-amino acid transporter (LAT) family. J. Biol. Chem. 285:28764–28776. 10.1074/jbc.M110.116632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bill R.M., Henderson P.J.F., Iwata S., Kunji E.R.S., Michel H., Neutze R., Newstead S., Poolman B., Tate C.G., and Vogel H.. 2011. Overcoming barriers to membrane protein structure determination. Nat. Biotechnol. 29:335–340. 10.1038/nbt.1833 [DOI] [PubMed] [Google Scholar]
- Bröer S., and Palacín M.. 2011. The role of amino acid transporters in inherited and acquired diseases. Biochem. J. 436:193–211. 10.1042/BJ20101912 [DOI] [PubMed] [Google Scholar]
- Cabantous S., and Waldo G.S.. 2006. In vivo and in vitro protein solubility assays using split GFP. Nat. Methods. 3:845–854. 10.1038/nmeth932 [DOI] [PubMed] [Google Scholar]
- Cabantous S., Terwilliger T.C., and Waldo G.S.. 2005. Protein tagging and detection with engineered self-assembling fragments of green fluorescent protein. Nat. Biotechnol. 23:102–107. 10.1038/nbt1044 [DOI] [PubMed] [Google Scholar]
- Christensen H.N. 1990. Role of amino acid transport and countertransport in nutrition and metabolism. Physiol. Rev. 70:43–77. [DOI] [PubMed] [Google Scholar]
- Dalbey R.E., Wang P., and Kuhn A.. 2011. Assembly of bacterial inner membrane proteins. Annu. Rev. Biochem. 80:161–187. 10.1146/annurev-biochem-060409-092524 [DOI] [PubMed] [Google Scholar]
- Fang Y., Jayaram H., Shane T., Kolmakova-Partensky L., Wu F., Williams C., Xiong Y., and Miller C.. 2009. Structure of a prokaryotic virtual proton pump at 3.2 Å resolution. Nature. 460:1040–1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fort J., de la Ballina L.R., Burghardt H.E., Ferrer-Costa C., Turnay J., Ferrer-Orta C., Usón I., Zorzano A., Fernández-Recio J., Orozco M., et al. 2007. The structure of human 4F2hc ectodomain provides a model for homodimerization and electrostatic interaction with plasma membrane. J. Biol. Chem. 282:31444–31452. 10.1074/jbc.M704524200 [DOI] [PubMed] [Google Scholar]
- Fotiadis D., Kanai Y., and Palacín M.. 2013. The SLC3 and SLC7 families of amino acid transporters. Mol. Aspects Med. 34:139–158. 10.1016/j.mam.2012.10.007 [DOI] [PubMed] [Google Scholar]
- Fuchs B.C., and Bode B.P.. 2005. Amino acid transporters ASCT2 and LAT1 in cancer: partners in crime? Semin. Cancer Biol. 15:254–266. 10.1016/j.semcancer.2005.04.005 [DOI] [PubMed] [Google Scholar]
- Gao X., Lu F., Zhou L., Dang S., Sun L., Li X., Wang J., and Shi Y.. 2009. Structure and mechanism of an amino acid antiporter. Science. 324:1565–1568. 10.1126/science.1173654 [DOI] [PubMed] [Google Scholar]
- Gao X., Zhou L., Jiao X., Lu F., Yan C., Zeng X., Wang J., and Shi Y.. 2010. Mechanism of substrate recognition and transport by an amino acid antiporter. Nature. 463:828–832. 10.1038/nature08741 [DOI] [PubMed] [Google Scholar]
- García-Fruitós E., González-Montalbán N., Morell M., Vera A., Ferraz R.M., Arís A., Ventura S., and Villaverde A.. 2005. Aggregation as bacterial inclusion bodies does not imply inactivation of enzymes and fluorescent proteins. Microb. Cell Fact. 4:27 10.1186/1475-2859-4-27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasol E., Jiménez-Vidal M., Chillarón J., Zorzano A., and Palacín M.. 2004. Membrane topology of system xc− light subunit reveals a re-entrant loop with substrate-restricted accessibility. J. Biol. Chem. 279:31228–31236. 10.1074/jbc.M402428200 [DOI] [PubMed] [Google Scholar]
- Geier E.G., Schlessinger A., Fan H., Gable J.E., Irwin J.J., Sali A., and Giacomini K.M.. 2013. Structure-based ligand discovery for the Large-neutral Amino Acid Transporter 1, LAT-1. Proc. Natl. Acad. Sci. USA. 110:5480–5485. 10.1073/pnas.1218165110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratkowski H., Lear J.D., and DeGrado W.F.. 2001. Polar side chains drive the association of model transmembrane peptides. Proc. Natl. Acad. Sci. USA. 98:880–885. 10.1073/pnas.98.3.880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harvey S.R., and Wysocki V.H.. 2015. Mass spectrometry: Bound in flight. Nat. Chem. 7:189–190. 10.1038/nchem.2192 [DOI] [PubMed] [Google Scholar]
- Hediger M.A., Clémençon B., Burrier R.E., and Bruford E.A.. 2013. The ABCs of membrane transporters in health and disease (SLC series): introduction. Mol. Aspects Med. 34:95–107. 10.1016/j.mam.2012.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jack D.L., Paulsen I.T., and Saier M.H.. 2000. The amino acid/polyamine/organocation (APC) superfamily of transporters specific for amino acids, polyamines and organocations. Microbiology. 146:1797–1814. 10.1099/00221287-146-8-1797 [DOI] [PubMed] [Google Scholar]
- Kawate T., and Gouaux E.. 2006. Fluorescence-detection size-exclusion chromatography for precrystallization screening of integral membrane proteins. Structure. 14:673–681. 10.1016/j.str.2006.01.013 [DOI] [PubMed] [Google Scholar]
- Kowalczyk L., Ratera M., Paladino A., Bartoccioni P., Errasti-Murugarren E., Valencia E., Portella G., Bial S., Zorzano A., Fita I., et al. 2011. Molecular basis of substrate-induced permeation by an amino acid antiporter. Proc. Natl. Acad. Sci. USA. 108:3935–3940. 10.1073/pnas.1018081108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Bhattacharya S., Tate C.G., Grisshammer R., and Vaidehi N.. 2015. Structural dynamics and thermostabilization of neurotensin receptor 1. J. Phys. Chem. B. 119:4917–4928. 10.1021/jp510735f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luirink J., Yu Z., Wagner S., and de Gier J.-W.. 2012. Biogenesis of inner membrane proteins in Escherichia coli. Biochim. Biophys. Acta. 1817:965–976. 10.1016/j.bbabio.2011.12.006 [DOI] [PubMed] [Google Scholar]
- Ma D., Lu P., Yan C., Fan C., Yin P., Wang J., and Shi Y.. 2012. Structure and mechanism of a glutamate-GABA antiporter. Nature. 483:632–636. 10.1038/nature10917 [DOI] [PubMed] [Google Scholar]
- Martinez Molina D., Cornvik T., Eshaghi S., Haeggström J.Z., Nordlund P., and Sabet M.I.. 2008. Engineering membrane protein overproduction in Escherichia coli. Protein Sci. 17:673–680. 10.1110/ps.073242508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCracken A.N., and Edinger A.L.. 2013. Nutrient transporters: the Achilles’ heel of anabolism. Trends Endocrinol. Metab. 24:200–208. 10.1016/j.tem.2013.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.L., and Tate C.G.. 2011. Engineering an ultra-thermostable β1-adrenoceptor. J. Mol. Biol. 413:628–638. 10.1016/j.jmb.2011.08.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palacín M., Nunes V., Font-Llitjós M., Jiménez-Vidal M., Fort J., Gasol E., Pineda M., Feliubadaló L., Chillarón J., and Zorzano A.. 2005. The genetics of heteromeric amino acid transporters. Physiology (Bethesda). 20:112–124. 10.1152/physiol.00051.2004 [DOI] [PubMed] [Google Scholar]
- Pédelacq J.-D., Cabantous S., Tran T., Terwilliger T.C., and Waldo G.S.. 2006. Engineering and characterization of a superfolder green fluorescent protein. Nat. Biotechnol. 24:79–88. 10.1038/nbt1172 [DOI] [PubMed] [Google Scholar]
- Penmatsa A., Wang K.H., and Gouaux E.. 2013. X-ray structure of dopamine transporter elucidates antidepressant mechanism. Nature. 503:85–90. 10.1038/nature12533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perálvarez-Marín A., Lórenz-Fonfría V.A., Simón-Vázquez R., Gomariz M., Meseguer I., Querol E., and Padrós E.. 2008. Influence of proline on the thermostability of the active site and membrane arrangement of transmembrane proteins. Biophys. J. 95:4384–4395. 10.1529/biophysj.108.136747 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reig N., del Rio C., Casagrande F., Ratera M., Gelpí J.L., Torrents D., Henderson P.J.F., Xie H., Baldwin S.A., Zorzano A., et al. 2007. Functional and structural characterization of the first prokaryotic member of the L-amino acid transporter (LAT) family: a model for APC transporters. J. Biol. Chem. 282:13270–13281. 10.1074/jbc.M610695200 [DOI] [PubMed] [Google Scholar]
- Rodríguez-Banqueri A., Kowalczyk L., Palacín M., and Vázquez-Ibar J.L.. 2012. Assessment of membrane protein expression and stability using a split green fluorescent protein reporter. Anal. Biochem. 423:7–14. 10.1016/j.ab.2011.12.044 [DOI] [PubMed] [Google Scholar]
- Rosell A., Meury M., Álvarez-Marimon E., Costa M., Pérez-Cano L., Zorzano A., Fernández-Recio J., Palacín M., and Fotiadis D.. 2014. Structural bases for the interaction and stabilization of the human amino acid transporter LAT2 with its ancillary protein 4F2hc. Proc. Natl. Acad. Sci. USA. 111:2966–2971. 10.1073/pnas.1323779111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaffner W., and Weissmann C.. 1973. A rapid, sensitive, and specific method for the determination of protein in dilute solution. Anal. Biochem. 56:502–514. 10.1016/0003-2697(73)90217-0 [DOI] [PubMed] [Google Scholar]
- Schlinkmann K.M., and Plückthun A.. 2013. Directed evolution of G-protein-coupled receptors for high functional expression and detergent stability. Methods Enzymol. 520:67–97. 10.1016/B978-0-12-391861-1.00004-6 [DOI] [PubMed] [Google Scholar]
- Serrano-Vega M.J., Magnani F., Shibata Y., and Tate C.G.. 2008. Conformational thermostabilization of the β1-adrenergic receptor in a detergent-resistant form. Proc. Natl. Acad. Sci. USA. 105:877–882. 10.1073/pnas.0711253105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer P.L., Goehring A., Shankaranarayanan A., and Gouaux E.. 2009. Structure and mechanism of a Na+-independent amino acid transporter. Science. 325:1010–1014. 10.1126/science.1176088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi Y. 2013. Common folds and transport mechanisms of secondary active transporters. Annu. Rev. Biophys. 42:51–72. 10.1146/annurev-biophys-083012-130429 [DOI] [PubMed] [Google Scholar]
- Smirnova I.N., and Kaback H.R.. 2003. A mutation in the lactose permease of Escherichia coli that decreases conformational flexibility and increases protein stability. Biochemistry. 42:3025–3031. 10.1021/bi027329c [DOI] [PubMed] [Google Scholar]
- Sonoda Y., Newstead S., Hu N.-J., Alguel Y., Nji E., Beis K., Yashiro S., Lee C., Leung J., Cameron A.D., et al. 2011. Benchmarking membrane protein detergent stability for improving throughput of high-resolution X-ray structures. Structure. 19:17–25. 10.1016/j.str.2010.12.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate C.G., and Schertler G.F.X.. 2009. Engineering G protein-coupled receptors to facilitate their structure determination. Curr. Opin. Struct. Biol. 19:386–395. 10.1016/j.sbi.2009.07.004 [DOI] [PubMed] [Google Scholar]
- Thévenin D., and Lazarova T.. 2008. Stable interactions between the transmembrane domains of the adenosine A2A receptor. Protein Sci. 17:1188–1199. 10.1110/ps.034843.108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J., and Tate C.G.. 2014. Quality control in eukaryotic membrane protein overproduction. J. Mol. Biol. 426:4139–4154. 10.1016/j.jmb.2014.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaidehi N., Grisshammer R., and Tate C.G.. 2016. How can mutations thermostabilize G-protein-coupled receptors? Trends Pharmacol. Sci. 37:37–46. 10.1016/j.tips.2015.09.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters R.F.S., and DeGrado W.F.. 2006. Helix-packing motifs in membrane proteins. Proc. Natl. Acad. Sci. USA. 103:13658–13663. 10.1073/pnas.0605878103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther T.H., and Ulrich A.S.. 2014. Transmembrane helix assembly and the role of salt bridges. Curr. Opin. Struct. Biol. 27:63–68. 10.1016/j.sbi.2014.05.003 [DOI] [PubMed] [Google Scholar]