Abstract
The Picornaviridae family comprises a large group of non-enveloped viruses that have a major impact on human and veterinary health. The viral genome contains one open reading frame encoding a single polyprotein that can be processed by viral proteinases. The crucial 3C proteinases (3Cpros) of picornaviruses share similar spatial structures and it is becoming apparent that 3Cpro plays a significant role in the viral life cycle and virus host interaction. Importantly, the proteinase and RNA-binding activity of 3Cpro are involved in viral polyprotein processing and the initiation of viral RNA synthesis. In addition, 3Cpro can induce the cleavage of certain cellular factors required for transcription, translation and nucleocytoplasmic trafficking to modulate cell physiology for viral replication. Due to interactions between 3Cpro and these essential factors, 3Cpro is also involved in viral pathogenesis to support efficient infection. Furthermore, based on the structural conservation, the development of irreversible inhibitors and discovery of non-covalent inhibitors for 3Cpro are ongoing and a better understanding of the roles played by 3Cpro may provide insights into the development of potential antiviral treatments. In this review, the current knowledge regarding the structural features, multiple functions in the viral life cycle, pathogen host interaction, and development of antiviral compounds for 3Cpro is summarized.
Keywords: 3Cpro, structure, viral replication, translation initiation, pathogenesis, protease inhibitor
1. Introduction
Members of the Picornaviridae family are positive-strand viruses that have a major impact on the health of humans and animals. This family is composed of 29 genera, including Aphthovirus, Cardiovirus, Kunsagivirus, Enterovirus, Hepatovirus and Sicinivirus [1,2,3]. The Picornaviridae family consists of at least 285 different picornaviruses that can infect various hosts, and outbreaks of several viruses have caused serious diseases and substantial economic burden [4,5,6]. This review mainly focuses on the 3C proteinases (3Cpros) of the Aphthovirus, Enterovirus and Hepatovirus genera.
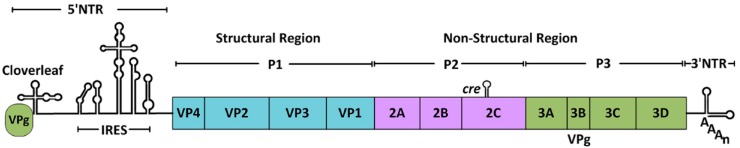
Despite the diversity of picornaviruses, their genome structures and translation processes are highly conserved. Picornaviruses are small, non-enveloped viruses containing a single-stranded RNA genome with a length of 7.0–8.5 kb. The viral genome contains one open reading frame that encodes a single polyprotein comprising a structural protein P1 region and non-structural protein P2 and P3 regions. Different from cap-dependent initiation of translation, the 5′ end of picornavirus genomic RNA is linked to a small viral-encoded protein (VPg), instead of 7-methylguanosine, which is necessary for initiating viral RNA replication (Figure 1). The release of mature and functional proteins from the polyprotein is primarily mediated by viral proteinases including 3Cpro, 2Apro and leader proteinase. Most processing is performed by 3Cpro and the 3CD precursor.
Figure 1.
The entire genome structure of poliovirus (PV) [9]. This RNA genome contains a 5′-nontranslated region (NTR), a large open reading frame, a 3′NTR and a poly (A) tail. A small viral-encoded protein, 3B (VPg), is linked to the 5′ terminus of the RNA. The 5′NTR consists of a cloverleaf structure and a type II internal ribosome entry site (IRES). The open reading frame encodes a single polyprotein comprising the structural protein P1 region and the non-structural protein P2 and P3 regions.
In this review, we summarize how 3Cpro is involved in polyprotein processing, protein-primed RNA synthesis initiation and the switch from viral translation to replication. We discuss the multiple roles that 3Cpro plays in the host cells, including shutting off transcription, inhibiting protein synthesis, blocking nucleocytoplasmic transport and inducing cell death. In addition, we also compare the functions of 3Cpro in the pathogenesis process of different picornaviruses. Discoveries have recently been made concerning effective and broad-spectrum inhibitors of picornaviruses. As a protease inhibitor for rhinovirus (RV) 3Cpro, rupintrivir (AG7088) has been the subject of clinical trials [7], however, AG7088 failed in a natural infection study [8]. Efforts to develop effective antiviral compounds are still ongoing. The synthesis of AG7088 analogues, the exploration of non-covalent inhibitors and research on natural medicine are the main strategies currently being used to develop 3Cpro inhibitors (3CPIs).
2. The Mechanism of Proteolysis
Although it is an unusual chymotrypsin-like cysteine protease, 3Cpro adopts a fold similar to that of the serine protease chymotrypsin; indeed, 3Cpro combines features of both serine and cysteine enzymes. Structural studies on picornaviral 3Cpro may identify unique features, providing useful information on protease inhibitors.
Since the 1990s, crystal structures have been determined for the 3Cpros of human rhinovirus (HRV), poliovirus (PV), hepatitis A virus (HAV), foot-and-mouth disease virus (FMDV) and enterovirus 71 (EV71) [10,11,12,13]. These studies have revealed two equivalent β-barrel domains in 3Cpro located approximately 90° from each other and composed of six antiparallel strands. Moreover, an extended shallow groove for substrate binding is located between the two domains [14]. A flexible surface loop, called the β-ribbon, has been observed in several picornaviral 3Cpros, including those of HRV (12 residues), PV (12 residues), and HAV (21 residues) [14,15]. The 13-residue β-ribbon of FMDV was considered in previous studies to be disordered but was subsequently demonstrated to be flexible [16]. The β-ribbon folds over the peptide-binding groove of the enzyme and accommodates a portion of the N-terminal peptide substrate. Thus, the β-ribbon plays a crucial role in substrate recognition. According to studies of EV71 3Cpro, the β-ribbon is thought to alternate between an open conformation and a closed conformation with the aid of hinge residues (Gly123 and His133) [17]. Before binding a substrate, the β-ribbon adopts an open conformation to increase substrate accessibility by exposing more substrate binding clefts; once bound, the interaction between the β-ribbon and N-terminal end of the substrate stabilizes the closed conformation to form an enzyme-substrate complex.
Picornavirus 3Cpros possesses a conserved Cys-His-Asp/Glu catalytic triad within the active site. This structure is similar to the Ser-His-Asp catalytic triad of serine proteases, except that third residue of the triad plays a less important catalytic role than in a serine protease. The catalytic residues in HAV include Cys-172 as a nucleophile and His-44 as a general base to form a Cys-His dyad [18]. Proteolytic mechanism studies in EV71 have shown that an uncharacterized residue Arg39 is involved in the proteolysis reaction by altering the distance to Glu71 as a charge neutralizer. Arg39 residue is conserved among CVB3, simian enterovirus (SEV) and FMDV 3Cpro whereas a conserved substitution of R39K is present in the 3Cpro of porcine enterovirus (PEV). In addition, Arg39 is replaced by an uncharged threonine or serine in the 3Cpro of PV, HRV and HAV, thereby indicating that Arg39 cannot be a neutralizer [19]. His40, a catalytic residue of triad, adopts two conformations as follows: a productive conformation that has been observed in most picornaviral 3Cpros and accounts for 39%; and a nonproductive conformation in which an imidazole side chain is positioned outside the hydrogen bonding distance to Cys-147, accounting for 61%. Thus, the negatively charged carboxylate side chain of Glu71 plays a critical role in maintaining the entire architecture of the active site and in stabilizing the productive conformation of His40 [19].
The cleavage specificity of 3Cpro has largely been investigated via extensive sequence analysis of cleavage junctions in polyprotein and cleavage assays using protein or peptide substrates in vitro. Investigations have shown that the 3Cpro of PV preferentially cleaves polypeptides with P1-Gln/P1’-Gly junctions. With the exception of FMDV 3Cpro, other picornaviruses have less strict cleavage, with a small residue at P1’, such as Gly, Ser, and Ala or a hydrophobic residue at P4. The 3Cpro of FMDV exhibits some variability at P1’, however, the probability of P1-Gln or P1-Glu is similar [20]. Specific studies on the 3Cpro of FMDV have shown that when P1-Gln is present, 3Cpro prefers large hydrophobic residues at P1 (Leu, Ile, and Thr) and P2-Lys. Nonetheless, it is possible for a small amino acid (Gly and Ser) to be strongly associated with P1-Glu. The 3Cpro of HAV can tolerate a large residue at P1′ and exhibits a preference for a small hydrophilic residue at P2 [21].
3. The Role of 3Cpro in the Viral Life Cycle
3.1. Processing of the Viral Polyprotein
Multiple viral proteins are generated through primary polyprotein processing and secondary polyprotein processing, and the analysis of polyprotein cleavage has shown that 3Cpro is responsible for most of the cleavages among the 29 genera of the picornavirus family (Figure 2). However, several exceptions have been found at the following four cleavage sites: L-VP4, VP4-VP2, P1-2A, and 2A-2B. In aphthovirus and erbovirus, the leader protein frees itself from VP0, whereas cardiovirus, kobuvirus, teschovirus, sapelovirus and senecavirus 3Cpros function in the primary cleavage [22,23]. Moreover, 2Apro exhibits proteinase activity or translational recoding activity during primary cleavage. The junction between P1 and 2A is cleaved by proteolytic 2Apro in enteroviruses and potentially in sapelovirus type 2 (SV2) as well as PEV8. The separation of capsid proteins from replication proteins is mediated by ribosome skipping at the Asn-Pro-Gly-Pro site, inducing cleavage of 2A–2B in aphthoviruses, cardioviruses, teschoviruses, cosaviruses, sencaviruses and erboviruses [24]. Under other conditions, this cleavage is achieved by 3Cpro [24,25]. 3Cpro presents in all picornaviruses, and is responsible for almost all secondary cleavages. For Aichi virus, in which L and 2Apro are not proteolytic, all cleavages are completed by only 3Cpro and the 3CD precursor. A series of studies on enteroviruses and apthoviruses have revealed that cleavage at VP2-VP3 and VP3-VP1 can be accomplished more efficiently by 3CDpro than 3Cpro [26,27,28].
Figure 2.
Polyprotein processing in different genera of the picornavirus family [23,28]. The release of mature and functional proteins from the polyprotein is dependent on primary and secondary cleavages and polyprotein processing differs slightly among diverse picornaviruses. In primary cleavage, Lpro functions in apthoviruses by freeing itself (blue arrow), whereas the cleavage of P1-P2 is mediated by ribosome skipping in apthoviruses and cardioviruses (red triangle) [29]. The proteolytic 2Apro of enteroviruses is involved in primary cleavage and 3Dpol cleavage to produce 3D’ intermediate (red arrows). 3Cpro and 3CD are responsible for a proportion of primary cleavages and almost all secondary cleavages (black arrow). The hepatovirus VP1-2A cleavage can be achieved by a host cell proteinase (green arrow). See the text for details.
3.2. Initiation of Viral RNA Synthesis
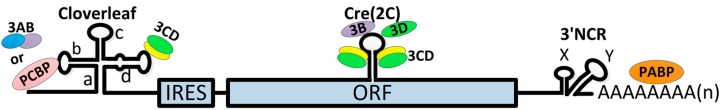
Members of the picornavirus family synthesize positive- and negative-stranded RNA by relying on viral RNA-dependent RNA polymerase. The 3Cpro alone and its precursors, namely 3ABC, 3BC, 3BCD and P3 (3ABCD), have been demonstrated to stimulate RNA synthesis at different levels in in vitro systems [30,31,32,33,34]. There are three RNA structures in the genome, which are important for RNA replication and which interact with 3Cpro and its precursor (Figure 3) [35]. These interactions are important for RNA replication in PV. First, a cloverleaf-like structure in the 5′-NCR of the viral RNA (5′CL) is important for RNA replication [36,37] and binding between 3CD and stem-loop d as well as binding between poly(rc)-binding protein 2 (PCBP2) and stem-loop b are required for viral replication in PV [38]. Mutation (K12N/R13N) in the 3C-binding region of PV results in complete inhibition of (−)strand synthesis. Complementation assays provide evidence that P3 is the preferred precursor that interacts with 5′CL. Upon binding to RNA, P3 can recruit another P3 to release 3D and VPg for efficient replication [30]. Second, the cis-acting replication RNA element (cre) is also important for RNA replication. During initiation of RNA synthesis, cre interacts with two molecules of 3CD, as shown by the “slide back” model for VPgpUpU synthesis in PV, and 3CD stimulates the cre-templated VPg uridylylation reaction. Although less effective, the function of 3CD can also be performed by 3Cpro, the 3BC precursor and the 3BCD precursor [31,32,33]. In FMDV, uridylylation of three VPgs can be enhanced by 3CD and in contrast to PV, the 3B33C and 3B1233C precursors of FMDV can serve as uridylylation substrates, even without added 3Cpro or 3CD [39,40]. Third, the 3′NTR-poly (A) tail is important for RNA replication. Interaction among 3CD, poly (A) binding-protein (PABP) and the poly (A) tail is required for negative-strand RNA synthesis to facilitate circularization of the RNA genome [41]. There are several viral and cellular proteins that have been found to bind to the 3′NCR of picornavirus genomic RNA. For PV, the 3AB and 3CD precursors have been reported to interact with the 3′NTR of genomic RNA [42]; for HAV, the 3AB and 3ABC precursors have been demonstrated to bind to the 3′NCR of genomic RNA [34].
Figure 3.
The RNA structures and proteins involved in PV RNA replication [46]. The a, b, c and d subelements form the cloverleaf structure. And the 3’NCR of RNA consists of two stem-loops (X, Y).There are three structures that can interact with 3Cpro and its precursor: namely the 5′ cloverleaf (5′ CL), cis-acting RNA elements (cre), and the 3′NTR poly (A). The ribonucleoprotein (RNP) complexes formed by the binding of 3CD to stem-loop d of 5′ CL and the binding of PCBP/3AB to stem-loop b are necessary for viral RNA replication. The cre hairpin of PV located in 2CATPase can interact with 3B, 3Dpol and 3CD to initiate RNA synthesis. The PABP bound to the poly (A) tail interacts with RNP for RNA replication.
Taken together, 3Cpro and the precursors exhibit essential functions in viral replication. Does proteinase activity or RNA-binding activity of the 3C domain stimulate RNA synthesis? There are three distinct regions within 3Cpro: the N-terminal region (K12 and R13), a central region (KFRDI86) and the C-terminal region (T153, G154 and K155) [43,44]. Mutation (R84S/I86A) within the 3C-binding region of PV prevents stimulation of virus synthesis [45] and mutation (K12N/R13N) within the 3C-binding region results in normal processing of viral translation but complete inhibition of (−)strand synthesis [34]. The mechanism by which 3Cpro is involved in RNA synthesis is closely related to its RNA-binding activity. Although both 3Cpro and 3CD can bind to viral RNA, there are differences. A wide range of compact conformations of 3CD induced by interaction between the 3C and 3D residues exhibit more activities than 3Cpro and 3Dpol and these interactions are conducive to the different RNA-binding activities of 3Cpro and 3CD [45].
3.3. Switch from Translation to RNA Replication
Numerous studies have focused on how picornaviruses switch from translation to replication, and the cleavage of PCBP2 by 3CDpro may provide mechanism [46]. For PV, a model has been proposed in which full-length PCBP2 functions in viral translation, but the cleavage product of PCBP2 functions in RNA replication [47]. Both 3Cpro and 3CDpro interact with phosphoinositide lipids to regulate the maturation of virus replication organelles and serve as the main protease to cleave virus and host proteins to regulate processes of both. However, the protease specificities and RNA-binding capabilities of 3Cpro and 3CDpro are different. The proteolytic processing of the C-terminal end of 3Cpro may cause the switch of activities from 3CDpro to 3Cpro to modulate functions during the virus cycle [48]. Above all, 3Cpro plays a major role in regulating translation and replication by binding to RNA sequences.
4. Host Cells Intervention by 3Cpro
The cleavage of host factors by 3Cpro in picornaviruses greatly alters several processes that are critical for cell viability, including transcription, initiation of protein synthesis, nucleocytoplasmic transport and growth, favoring the viral replication cycle.
4.1. Rapid Shut-off of Transcription in Host Cells
A rapid inhibition of RNA synthesis is observed in host cells infected by picornaviruses, which is mainly attributable to picornaviral 3Cpro cleavage of DNA-dependent RNA polymerases. It has been demonstrated that PV 3Cpro enters the nucleus via precursors and induces the cleavage of TATA-binding protein-associated factor 110 (TAF110), TATA box binding protein (TBP), cAMP response element-binding protein-1 (CREB-1), octamer binding protein-1 (Oct-1), p53 and transcription factor IIIC (TFIIIC) to shut off host cell transcription [49,50,51,52,53,54] (Table 1). In addition to these transcriptional factors associated with RNA polymerase, FMDV 3Cpro can also interrupt transcription by cleaving nuclear histone H3, and EV71 3Cpro can also cleave CstF64 of the polyadenylation machinery [55,56]. Replication of the host cell occurs in the nucleus, whereas replication of the virus occurs in the cytoplasm. Furthermore, 3Cpro interferes with the expression of host genes by inhibiting transcription and cap-dependent translation to provide cellular resources for viral replication.
Table 1.
Cleavage of cellular transcription factors by 3Cpro.
| Classification of RNA Polymerases | Host Cell Protein | Virus | Functions | Cleavage Site | Reference |
|---|---|---|---|---|---|
| I | TAF110 | PV | Regulates of transcriptional initiation | IQLQ265…ACAQ805G | [49] |
| II | TBP | PV | Involved in three RNA polymerase-mediated transcriptions and responsible for DNA binding | AAAVQQ104STSQQA | [50] |
| CREB-1 | PV | Binds to DNA elements for induction by cAMP | GQYIAITQ172GGAIQL | [51] | |
| Oct-1 | PV | Binds to the octamer sequence ATGCAAAT and activates the promoters of genes for some small nuclear RNAs | KLGFTQ329GDVGLA (Speculated) | [52] | |
| P53 | PV | Functions as a typical sequence specific transcription activator and a tumor suppressor | Not clear | [53] | |
| III | TFIIIC | PV | Binds to the B-box internal promoter element to process tRNA transcription | TSQPPVPQ732GEAEED | [54] |
| Other factors | CstF64 | EV 71 | Polyadenylation of host mRNA | MQASMQ251GGVPAPGQMP | [56] |
| Histone 3 | FMDV | DNA binding and involved in the structure of chromatin in eukaryotic cells | GKAPRKQL120ATKAAR | [55] |
4.2. Rapid Inhibition of Protein Synthesis Initiation in Host Cells
Protein synthesis in host cells depends on the 5′ terminal cap structure and several cap-binding proteins, but viral mRNA utilizes an IRES for translation (Figure 4). Firstly, the m7G(5′)ppp(5′)N structure is recognized by a translation initiation factor (eIF4F), which is composed of the eIF4G scaffolding protein, the eIF4E cap-binding protein and the eIF4A RNA helicase to initiate cellular mRNA translation. The 43S preinitiation complex (PIC) contains eIFs, including eIF1, eIF1A, eIF3, eIF5 and the ternary complex (eIF2-GTP-Met-tRNA), together with the 40S ribosomal subunit. The binding of eIF4F to the cap and the binding of PABP to the poly (A) tail circularizes the mRNA to activate the mRNA. The 43S ribosome then binds near the cap and scans the 5′-UTR for an AUG codon [57]. Identification of the AUG start codon constitutes the first stage of the translation of cellular mRNA (Figure 4B).
Figure 4.
Type II IRES-dependent and cap-dependent protein synthesis initiation. (A) The RNA of FMDV employs a type II internal ribosome entry site (IRES) to recruit the 40S subunit to start translation. Some eIFs and ITAFs are required for IRES-driven translational initiation [60]; (B) The host cell employs the cap-dependent protein system. The m7G(5′)ppp(5′)N structure is recognized by a translation initiation factor (eIF4F). The binding of eIF4F to the cap and the binding of PABP to the poly (A) tail circularize the mRNA to activate it [60]. Abbreviations include: eIF4G: eukaryotic translation initiation factor 4G; eIF4E: eukaryotic translation initiation factor 4E; PTB: polypyrimidine tract-binding protein; PCBP: poly (rC)-binding protein; PABP: poly(A)-binding protein; eIF3:translation initiation factor 3; m7G: 7-Methylguanosine.
In contrast to the eukaryotic translation mechanism, picornaviruses take advantage of the IRES element to recruit ribosomal subunits for initiation [58] (Figure 4A). Reconstitution assays have indicated that assembly of 48S initiation complexes on IRES elements of types I and II (FMDV and EMCV) requires eIF3, eIF4A and the C-terminal end of eIF4G [59]. In addition to eIFs, IRES-transacting factors (ITAFs) including PTB, PCBP2, and Germin 5 contribute to the modulation of IRES activity. Furthermore, 3Cpro plays a significant role by cleaving some eIFs and ITAFs to guarantee the internal initiation of translation (Table 1 and Table 2).
Table 2.
Cleavage of translation-related proteins by 3Cpro.
| Host Cell Protein | Virus | Functions | Cleavage Site | Reference |
|---|---|---|---|---|
| eIF4A I | FMDV | Binds capped mRNA to the 40S ribosomal subunit and unwinds double-stranded RNA | CIGGTNVRAE143VQKLQMEA | [63] |
| eIF4G I | FMDV | Brings mRNA to the 40S ribosome in translation initiation | RRSQQGPRKE712PRKIIATVL | [61] |
| eIF5B | PV | Positions the initiation methionine tRNA on the start codon of the mRNA | LCAAVEVMEQ478GVPEKEET | [64] |
| CV | ||||
| HRV | ||||
| G3BP1 | CV | RNA-binding protein that interacts with Ras-GAP | EAGEQ325GDIEP | [70] |
| PCBP2 | HAV | Translational activation and control of gene expression | IGRQ306GAKI (Speculated) | [66] |
| PV | AMQQ253SHFP…IGRQ306GAK | [65] | ||
| PABP | PV | Involved in poly (A) shortening and translation initiation | Not clear | [67] |
| HAV | Not clear | [68] | ||
| EMCV | VRPPAAIQ437GVQAGA | [69] | ||
| Sam68 | FMDV | Involved in cellular differentiation and proliferation | C-terminal portion | [71] |
Picornavirus-infected host cells experience a rapid shut-off of the cap-dependent protein and DNA repair systems. The cleavage of cellular factors as well as inhibition of their expression may explain this phenomenon (Table 2). The picornaviral proteinases, L, 2A and 3Cpro, have been reported to primarily function in the cleavage of host translation initiation factors (eIFs). Transient expression assays have shown that eIF4A and eIF4G can be cleaved when FMDV 3Cpro is expressed. Furthermore, cleavage by 3Cpro is different from that by the FMDV leader protein and 2Apro [61]. Subsequently, it has been demonstrated that Lpro and 3Cpro can function in the sequential cleavage of eIF4GI in BHK cells after infection with wild-type FMDV [62]. Two forms of eIF4A, namely, eIF4AI and eIF4AII, exist within mammalian cells. FMDV 3Cpro cleaves only eIF4AI, which is not related to eIF4AII even though they share 91% sequence identity [63]. In addition, during PV, CVB3 and HRV infection, eIF5B eukaryotic initiation factor is cleaved at a single site (VVEQ↓G), which separates the N-terminal domain from the conserved GTPase domain and C-terminal domain [64]. PCBP2, also known as hnRNP E2, has multiple functions in the post-transcriptional control of host and viral gene expression. The ability of PV to take advantage of PCBP2 in translation and replication has already been discussed [65], and it has also been observed that PCBP2 is cleaved by 3Cpro in HAV. Thus, HAV may regulate protein synthesis and RNA synthesis by 3Cpro [66]. In addition to PABP cleavage by 2Apro and PV, HAV and EMCV 3Cpros have also been reported to specifically cleave PABP [67,68,69]. A study of EMCV 3Cpro suggests that specific PABP cleavage is required for virus replication or that full-length PABP is not absolutely required for virus replication [69]. The cleavage of Ras GTPase-activating protein-binding proteins (G3BP1) may be induced by 3Cpro to facilitate viral replication after CVB3 infection [70]. Recently, it has been demonstrated that the Sam68 nuclear RNA-binding protein can be cleaved by 3Cpro of FMDV, thus allowing 3Cpro to redistribute Sam68 to the cytoplasm. Remarkably, Sam68 can interact with the IRES in FMDV during infection, which then can translate viral RNA. Thus, it is speculated that Sam68 plays a supportive role in the virus life cycle [71].
4.3. Inhibition of Nucleocytoplasmic Trafficking by the Nuclear Pore Complex
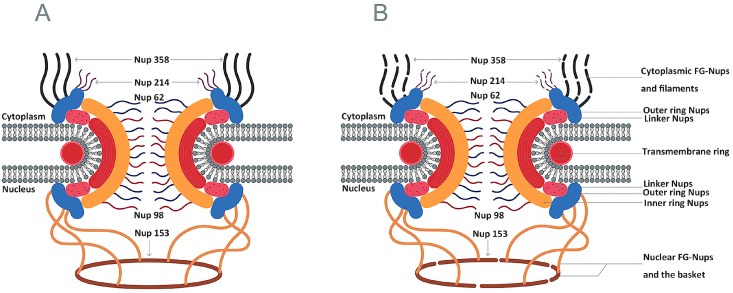
In eukaryotic cells, DNA replication and gene transcription occur in the nucleus, whereas protein production occurs in the cytoplasm. The spatial separation of these major processes guarantees cellular transcription and translation with the help of a transport network. The nucleocytoplasmic trafficking network is responsible for transporting nuclear-resident and cytoplasmic-resident proteins between these two compartments. Translation initiation and replication in the cytoplasm depend on some nuclear-resident proteins, and picornaviruses have recently been shown to shut down the nucleocytoplasmic trafficking of cells to promote viral replication. Five nucleoporins (Nups) have been reported to be targets for picornaviral proteins to disrupt macromolecule trafficking through the nuclear pore complex (NPC) (Figure 5A). The 2Apro and 3Cpro viral proteinases of enteroviruses are responsible for the specific proteolysis of Nup 62, Nup 98, and Nup 153, whereas the leader protein of cardioviruses induces hyper-phosphorylation of Nup 62, Nup 98, Nup 153 and Nup 214 (Table 3) [72,73,74,75]. Poliovirus 2Apro causes degradation of Nup 62, Nup 98, and Nup153 in cells, and even a low concentration of 2Apro can lead to Nup 98 degradation [76]. Furthermore, purified HRV 2Apro cleaves Nup 62 to release FG-rich regions [77]. Expression of 3Cpro or 3CD in HRV 16 causes degradation of Nup 153, Nup 214 and Nup 358 in cells [78] [Figure 5B]. Moreover, the cleavage of Nup 62 is partially induced in vitro by a high concentration of purified HRV 3C [78]. These studies suggest that picornaviruses may normally alter nucleocytoplasmic trafficking to meet the demands of nuclear-resident proteins for viral replication but that they block the export of cellular mRNA.
Figure 5.
Alteration of the nuclear pore complex induced by 3Cpro [72,79,80]. (A) The nuclear pore complex is composed by the outer ring Nups, the linker ring Nups, the inner ring Nups, cytoplasmic phenylalanine-glycine (FG)-Nups, nuclear FG-Nups and the nuclear basket; (B) The 3Cpro of RV can induce cleavage of Nup 153 and Nup 214 and Nup 358.
Table 3.
Picornavirus-induced alterations to the nuclear pore complex.
| Genus | Virus | Viral Protein | Host Protein | Reference |
|---|---|---|---|---|
| Enterovirus | Rhinovirus | 2Apro | Nup 62 | [77] |
| 3Cpro/3CD | Nup 153, Nup 214, Nup 358 | [78] | ||
| 3Cpro | Nup 62 | [78] | ||
| Poliovirus | 2Apro | Nup 62, Nup 98, Nup 153 | [76] | |
| Cardiovirus | Encephalomyocarditis virus | Leader protein | Nup 62, Nup 153, Nup 214 | [81] |
| Theiler’s murine encephalomyelitis virus | Leader protein | Nup 62, Nup 98 | [82] |
4.4. Induction of Cell Death
Picornaviral 3Cpro can induce apoptosis through cleavage of transcription and translation initiation factors that control cell survival and death pathways. Expression of poliovirus 3Cpro results in the degradation of cellular DNA and generation of apoptotic bodies [83]. Moreover, the transcriptional activator p53 is degraded by 3Cpro with cellular activity [53]. p53 can also function in the cytosol and mitochondria to promote apoptosis by transcription-independent mechanisms. VAD-fmk caspase 1 inhibitor interferes with 3Cpro-induced apoptosis [53]. For EV 71, 3Cpro-induced apoptosis is related to the proteolytic activity of 3Cpro, and apoptosis can be blocked by DEVD-fmk and VAD-fmk inhibitors [84]. These results suggest that caspase activation participates in the apoptotic process induced by 3Cpro. In addition, 2Apro and 3Cpro induce apoptosis through multiple pathways in CVB3. Both 2Apro and 3Cpro can induce the death receptor-mediated pathway through caspase 8-mediated activation of caspase 3 and cleavage of Bid [85]. Only 3Cpro can upregulate Bax and release cytochrome c from mitochondria in the intrinsic death pathway. Unlike these picornaviral 3Cpros, HAV 3Cpro can induce cell death independent of the caspase pathway, accompanied by cytoplasmic vacuolization [86]. Cell death is necessary for the release of virus, and 3Cpro influences this step through different mechanisms.
4.5. Other Functions
Except for their crucial role in the replication of picornaviruses and inhibition of protein synthesis in host cells, the 3Cpros of PV and FMDV have been previously reported to mediate cleavage of microtubule-associated protein 4(MAP-4) and cause disruption of the microtubule-organizing centre (MTOC) [87,88]. Subsequent studies have revealed that FMDV 3Cpro leads to fragmentation of the Golgi apparatus, which blocks the transport of proteins from the Golgi to the plasma membrane. Some scholars have studied the connection between Golgi fragmentation, microtubule depolymerization and secretion blockade. Through a series of experiments, it has been confirmed that FMDV 3Cpro blocks secretion in the following two ways: 3Cpro processes 2BC into 2B and 2C, which bind to the Golgi compartment to block intra-Golgi transport, and instead of physical binding, 3Cpro blocks secretion by degrading proteins necessary for intra-Golgi transports. Both of these routers depend on the proteolytic activity of 3Cpro [89]. Further research is needed to understand whether there is a connection between cleavage and the reduction of immune responses or between cleavage and the transmission of virus.
5. The Role of the Pathogenic Process
The host intrinsic immune defense against invading pathogens is closely related to the activation of parallel pattern recognition receptor (PRR) pathways that facilitate the production of type I interferon (IFN). PRRs detect conserved molecular signatures of pathogens, termed pathogen-associated molecular patterns (PAMPs), to distinguish host components from pathogens [90]. Vertebrate hosts encode several classes of PRRs, such as RIG-I-like receptors (RLRs), NOD-like receptors, C-type lectin receptors and Toll-like receptors (TLRs) [91,92]. Two well-known pattern recognition receptors are Toll-like receptors (PPRs) and RIG-I-like RNA helicases (RLHs). Notably, all TLRs are type I transmembrane proteins composed of a leucine-rich N-terminal of an ectodomain, a transmembrane domain, and a linker to a cytoplasmic C-terminal Toll-interleukin-I receptor (TIR) homology domain [93,94]. The RLR family, comprising RIG-I, MDA5 and LGP2, has a conserved DExD/H-box helicase domain and a C-terminal domain (CTD).
When toll-like receptor 3 (TLR3) recognizes double-stranded RNA, it recruits TIR domain-containing adapter inducing interferon-β (TRIF) [95], which together activate downstream IKK-related kinases. These kinases phosphorylate interferon regulatory factor 3 (IRF3), leading to the expression of IFN-α/β [96,97,98,99,100]. MDA5 and RIG-I, two activated DExD/H box RNA helicases, transmit signals to downstream adaptor IPS-I, leading to stimulation of NF-κB and IFN-β. The expression of IFN-α/β depends on the activation of IRF3, and this process is closely related to the interactions among IPS-I, TRF3 and TBK1/IKKi [101,102].
During EV71 infection, the expression of IFN-β, tumor necrosis factor (TNF-N), and IFN-stimulated gene 54(ISG54) is inhibited. Among EV71 proteins, 3Cpro blocks the virus-induced activation of IFN-β by targeting the cytosolic helicase I RIG-I. However, 3Cpro exerts no inhibitory activity against IFN-β promoter activation, which is mediated by MDA5 in EV71 infection [92]. Enteroviral 2Apro proteolytically targets MDA5 and MAVS to block the expression of IFN-I [103]. Notably, members of picornavirus family modulate MDA5 and RIG-I differently. RV and echovirus target RIG-I, whereas PV and EMCV mediate RIG-I and MDA5 [104,105,106]. Subsequent studies have revealed that EV71 3Cpro induces TRIF cleavage to inhibit NF-κB and IFN-β promoter activation in a caspase-independent manner (Figure 6) [107]. The use of 3Cpro mutants has shown that the Q312–S313 junction within TRIF might be a preferred cleavage site [104]. With regard to the constitutive activation domain of IRF7, EV71 3Cpro mediates cleavage of IRF7, rather than IRF3, at the Q189–S190 junction when expressed in mammalian cells (Table 4) [108]. EV71 3Cpro also inhibits NF-κB via cleavage of the transforming growth factor-β-activated kinase 1 (TAK1) and TAB2 complex [109]. The type I IFN antiviral response is closely related to the JAK-STAT signaling pathway, consisting of Janus kinase (JAK) family members and the signal transducers and activators of transcription (STAT) family. EV71 inhibits the type I IFN response through another mechanism of JAK1 downregulation. However, roles for the viral proteinases 2Apro and 3Cpro as antagonists have been ruled out [110]. Recently, it has been shown that the PARP9-DTX3L complex which is formed by poly (ADP-ribose) polymerase PARP9 and the DTX3L E3 ubiquitin ligase, can promote STAT1-mediated ISG expression and can induce viral 3Cpro degradation [111]. These observations provide guidance for studies of interferon signaling and viral infection. The mechanisms by which picornaviruses inhibit cellular type I IFN and the strategies by which host cells adopt a defense system are complicated.
Figure 6.
Mechanisms of 3Cpro from different picornaviruses suppress the immune response. It has been demonstrated that 3Cpro interferes with the host type I interferon (IFN) response and nuclear transcription factor kappa B (NF-κB) signaling pathway by cleaving several essential proteins. However, the mechanisms adopted by picornaviruses are slightly different. The immune response is depicted in black. Inhibition of immune response by viral proteinases is highlighted in red. Abbreviations include: MDA5: melanoma differentiation-associated gene 5; RIG-I: retinoic acid-inducible gene I; MAVS: mitochondrial antiviral signaling protein; TRIF: TIR domain-containing adapter inducing interferon-β; TLR3: toll-like receptor 3; TRAF: TNF receptor associated factor; NEMO: NF-κB essential modulator; IKK: IκB kinase; IRF: interferon regulatory factor; Tyk: tyrosine kinase; JAK1: janus kinase 1; STATs: signal transducers and activators of transcription; KPNA1: karyopherin α1.
Table 4.
Cleavage of innate immune-related proteins by 3Cpro.
| Virus | Host Factors | Cleavage Site | Reference |
|---|---|---|---|
| FMDV | NEMO | LSSPLALPSQ383RRSPPEEPPD | [116] |
| EV71 | IRF7 | GDLLLQAVQQ189SCLADHLLTA | [108] |
| TRIF | TEGSAGPQ312SLPLPILEP | [104] | |
| TAK1 | KNQAKQQ360SESGRL | [109] | |
| TAB2 | SISDGQLQ113GGQSNSEL | [109] | |
| CVB3 | MAVS | SYPMPVQETQ148APESPGENSEQ | [120] |
| TRIF | Q190, Q653, Q659, Q671, Q702 (potential) | [120] | |
| HAV | NEMO | METVPVLKAQ304ADIYKADFQA | [115] |
| TRIF | SDWSQ190GCSLR…EQSQ554HLDGER | [114] | |
| MAVS | LASQ428VDSP…YKSE463GTFG (potential) | [113] |
The 3Cpro of another, HAV, has the capability to induce proteolysis of MAVS by the viral 3ABC precursor. The cleavage of this mitochondrially localized adaptor of RIG-I and MDA5 disrupts activation of IRF3 and abolishes type I IFN responses. The protease activities of 3Cpro and the transmembrane domain of 3A are both necessary for the proteolysis of MAVS [112,113]. For another precursor, HAV 3CD protease-polymerase disrupts TLR3 signaling by degrading TRIF (Figure 6) and this degradation is not dependent on only the protease activity of 3Cpro but also requires the 3D sequence to modulate the substrate specificity of 3Cpro [114]. NEMO, a bridging adaptor protein for the activation of NF-κB, is another substrate of HAV 3Cpro at the Q304 residue (Table 4) [115]. NEMO is also cleaved by FMDV 3Cpro at the Gln-383 residue but not by the 3Cpro of EV 71 [116].
Except for the cleavage of NEMO by 3Cpro [116], the Lpro of FMDV is also associated with the degradation of p65/RelA and decrease in IRF3/7 expression to suppress the immune response [117,118]. Subsequently, a study has shown that 3Cpro induces the degradation of karyopherin α1 (KPNA1) to antagonize interferon via the JAK-STAT signaling pathway by blocking the nuclear translocation of STAT1/STAT2 [119]. In CV, 3Cpro exhibits the capability of cleaving the proline-rich region of MAVS and two terminal domains of TRIF to escape host immunity (Figure 6) [120].
In addition to viral and host proteins, microRNAs (miRNAs) is also involved in posttranscriptional gene expression regulation and it has been confirmed that miRNAs play significant roles in the interaction between virus and host. The 3Cpro of EV71 downregulates miR-526a to suppress the RIG-I-dependent innate immune response [121].
A novel model shows that CVB3 manipulates transactive response DNA-binding protein-43 (TDP-43) to regulate viral pathogenesis. Normally, TDP-43 regulates RNA metabolism in the nucleus as a heterogeneous ribonucleoprotein (hnRNP), and a pathophysiological link between TDP-43 and amyotrophic lateral sclerosis (ALS) has been established [122]. When cells are infected by CVB3, 2Apro induces translocation of TDP-43 from the nucleus to the cytoplasm. TDP-43 is then cleaved by 3Cpro at Q327, generating two fragments. The C-terminal fragment of TDP-43 is rapidly degraded, whereas the N-terminal product negatively regulates the function of native TDP-43 [123]. These processes benefit viral replication. The cleavage of full-length TDP-43 is closely related to neurodegeneration. Studies of 3Cpro and TDP-43 will provide information regarding the pathogenesis of CV.
The development of effective immune evasion mechanisms is beneficial to viral infection. Evidence has shown that picornaviruses adopt multiple mechanisms to suppress host antiviral responses. EV encodes 2Apro and 3Cpro for polyprotein processing, whereas FMDV encodes Lpro and 3Cpro. These proteins with proteolytic activity target and cleave certain cellular proteins to down-regulate the type I IFN response, pro-inflammatory cytokines and ISG induction. HAV encodes 3Cpro for its polyprotein processing. The multipronged cleavage of innate immune proteins is a remarkable example of picornaviral evolution through 3Cpro and its 3ABC and 3CD precursors, and 3Cpro can bind to and cleave different cellular molecules to interfere with antiviral signaling pathways to evade host immune response.
6. Treatment
6.1. Inhibitors of Picornaviral 3Cpro
Picornaviral 3Cpro serves as an attractive target for antiviral drugs against picornaviruses because it plays an essential role in the virus replication cycle and it does not have cellular homologues. Some small peptides and a ketone-containing tripeptide, known as peptide aldehydes, can inhibit 3Cpro [124,125]. The peptidyl aldehyde NK-1.8 k targets 3Cpro to inhibit the replication of different EV71 strains and one EV68 strain [126].
Peptidyl Michael acceptors have been used to develop 3Cpro inactivators for inhibiting viral replication [127,128,129]. As a potent irreversible inhibitor of HRV 3Cpro, AG7088 contains an α,β-unsaturated ester at P1′ that serves as a Michael acceptor, a lactam ring at P1, a fluoro-phenylalanine at P2, a Val at P3 and a 5-methyl-3-isoxazole. AG7088 binds with the active site Cys residue to influence the functionality of 3Cpro [130]. Through a series of cell-based protease assays, the activity spectra of the 3Cpro inhibitors AG7088 and SG85 have been found to include inhibition of CVB3, EV71, PV, and HRV14 3Cpros. However, these inhibitors have no effect on the 3Cpro of EMCV [131,132]. The inhibition of HAV replication by AG7088 is strain-dependent [133]. As an analogue of AG7088, AG7404 has been demonstrated to inhibit HRV, PV and several EV types [134].
Although covalent binding results in strong inhibition, the electrophilic nature of irreversible 3Cpro inhibitors may increase off-target safety risks due to potential interactions with host cysteine proteases [135]. Thus, exploration of non-covalent inhibitors of picornaviral 3Cpros has already begun. Non-covalent inhibitors of RV 3Cpro and CVB3 3Cpro have been identified through fragment screening and FRET-based enzyme assay screening [136]. The chemical modification from water-insolubility to water-solubility shows a positive effect. Indeed, water-insoluble 3Cpro inhibitors of CV B3 dissolved in 100% dimethyl sulfoxide (DMSO) have been reported to inhibit viral proliferation and to decrease myocardial damage and mortality in a chronic myocarditis model [137,138]. With the exception of organic solvents, organic reagents exhibit potential as 3Cpro inhibitors. Interaction between the cyanohydrins moiety and 3Cpro catalytic site exhibits high selectivity and improved inhibitory activity [139].
6.2. Broad-Spectrum Inhibitors of 3Cpro and 3CLpro
3Cpro and 3C-like protease (3CLpro) encoded by the Picornaviridae, Caliciviridae, and Coronaviridae families, are necessary for virus replication. Sharing a typical chymotrypsin-like fold, an extended binding site, a Cys-His-Glu/Asp catalytic triad (EV, CV 3Cpro and NV 3Cpro) or Cys-His dyad (SARS-CoV 3CLpro), and a cleavage preference at the Gln-Gly (P1-P1′) junction, 3Cpro and 3CLpro have been investigated as broad-spectrum antiviral targets, though 3Cpro is a monomer and 3CLpro is a dimer [140]. The typical inhibitor of picornavirus 3Cpro AG7088 fails to inhibit SARS-CoV 3CLpro due to subtle differences in the active site structures [141]. However, a modified AG7088 shows good inhibition of 3CLpro and no inhibition of 3Cpro [142]. Using high-throughput screening, one compound that contains a central dihydropyrazole ring and four surrounding rings has been identified as equally inhibiting both the PV 3Cpro and CoV 3CLpro [143]. By designing tripeptidyl transition state inhibitors and mimics, highly potent inhibitors that function against picornaviruses, coronaviruses and noroviruses have been discovered with minimal or no ability to inhibit a panel of proteases such as trypsin, thrombin or α-chymotrypsin [144]. The first macrocyclic inhibitor of 3Cpro of CVB3, 3CLpro of SARS-CoV and 3CLpro of noroviruses has been reported, with inhibitory activities of 1.8, 15.5 and 5.1 μM, respectively [145]. Research on picornaviral 3Cpro can provide a reference for other viruses.
6.3. Natural Medicine
Some extracts of plants and microbes have also been found to inhibit the proteolytic activity of 3Cpro. As a member of the pyanonaphthoquinone family, the natural product (−)-thysanone has been reported to inhibit HRV 3Cpro with an IC50 value of 47 μM [146]. Through synthesis of analogues and the biological evaluation of (−)-thysanone, the 2-methylene analogue of (−)-thysanone has lost the capacity to inhibit HRV 3Cpro [147]. However, 9-deoxy analogues of (−)-thysanone display better inhibitory ability than the natural product [148]. Studies of herbs have demonstrated that flavonoids, such as fisetin and rutin, can inhibit the enzymatic activity of EV 71 3Cpro in a dose-dependent manner with IC50 values of 85 and 110 μM, respectively [149].
7. Conclusions
Due to their proteinase activities, 3Cpro and its precursor 3CD are involved in the processing of the polyprotein, but their catalytic efficiencies and specificities exhibit differences that are related to the different substrate-binding site residues in the 3C domain and the N-terminus of 3Cpro. The RNA-binding activity of 3Cpro is important for initiating viral RNA synthesis. 3Cpro and its 3ABC, 3BC, 3BCD and P3 precursors can stimulate RNA synthesis at different levels. Due to the limited encoding capacity of picornaviruses, essential functions are performed by modulation of 3Cpro and its precursors.
The translation and replication of the picornaviral genome utilize the host translation machinery and the alteration and cleavage of host proteins lead to inhibition of host translation and transcription and nucleus-cytoplasm transport. 3Cpro and the 3CD precursor are involved in many steps of gene expression to hijack cellular resources and to generate an optimal intracellular environment for the virus life cycle. Disrupting mRNA circularization by cleaving PABP has been observed in PV, CV and HAV. After cleavage by 3Cpro, some cellular factors and their fragments, such as eIF4G, PTB and PCBP2, function in distinct manners for the host cell and virus. These observations underscore the significant roles that 3Cpro plays in translation, replication, and the switch from translation to replication to guarantee viral replication. At the end of the replication cycle, the release and spread of virus are closely related to cell lysis and the formation of phosphatidylserine (PS) lipid-enriched vesicles [150]. 3Cpro not only induces cell death through caspase-dependent and caspase-independent pathways but also mediates fragmentation of MAP-4 and the Golgi apparatus. These findings clearly demonstrate the various mechanisms of 3Cpro that are required to subvert different processes in host cells to promote the viral life cycle.
Inhibition of host protein synthesis via cleavage of proteins required for transcription and cap-dependent translation can also block the production and transportation of antiviral proteins. In addition to innate immune responses, picornaviruses employ diverse mechanisms to suppress antiviral responses. There are some similarities and differences among various genera. With the help of precursors and the cooperation of other viral proteinases, 3Cpro has a crucial role in down-regulating type I IFN, pro-inflammatory cytokines and ISG induction. Picornavirus-infected cells engage multiple mechanisms. For example, the PARP9-DTX3L complex promotes ISG expression and induces degradation of viral 3Cpro to enhance the immune response. The cleavage of TDP-43 is a major molecular marker for some neurodegenerative diseases, and TDP-43 cleavage by the 3Cpro of CVB3 provides a novel model for viral pathogenesis research.
Understanding the structure and functions of 3Cpro provides accurate information for the design of broad-spectrum anti-picornaviral inhibitors. The development of irreversible inhibitors and the discovery of non-covalent inhibitors of 3Cpro will accelerate the treatments for infection by picornaviruses and other viruses encoding 3CLpro. Due to the limitations involved with using killed whole virus, recombinant empty capsids, which are structural and immunogenic mimics of virus particles, have become a research hotspot for an alternate vaccine. The development and the production of empty capsids is dependent on 3Cpro levels [3,151]. 3Cpro is well known to be a crucial proteinase for processing polyprotein and RNA-binding protein in viral replication. However, there is little information to date regarding the molecular mechanism by which 3Cpro blocks various cellular pathways. Comparison of 3Cpro functions in the life cycles of the host cell and virus can provide more insight into the pathogenesis and regulatory mechanisms of picornavirus. Comparison of 3Cpro functions in different genera reveals that this proteinase still maintains distinct functions in these genera. In conclusion, a mechanistic understanding of 3Cpro will have a potential future impact on antiviral treatment.
Acknowledgments
The research was supported by grants from the National Natural Science Foundation of China (31472223), National Science and Technology Support Program (No. 2015BAD12B05), China Agricultural Research System (CARS-43-8) and the Innovative Research Team Program in Education Department of Sichuan Province (12TD005/2013TD0015).
Author Contributions
Di Sun wrote the paper; Shun Chen revised the paper; and Anchun Cheng and Mingshu Wang revised the paper.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Adams M.J., King A.M.Q., Carstens E.B. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2013) Arch. Virol. 2013;158:2023–2030. doi: 10.1007/s00705-013-1688-5. [DOI] [PubMed] [Google Scholar]
- 2.King A.M.Q., Adams M.J., Carstens E.B., Lefkowitz E.J. Virus Taxonomy: Classification and Nomenclature of Viruses: Ninth Report of the International Committee on Taxonomy of Viruses. Academic Press/Elsevier; London, UK: 2012. pp. 855–880. [Google Scholar]
- 3.Lewis S.A., Morgan D.O., Grubman M.J. Expression, processing, and assembly of foot-and-mouth disease virus capsid structures in heterologous systems: Induction of a neutralizing antibody response in guinea pigs. J. Virol. 1991;65:6572–6580. doi: 10.1128/jvi.65.12.6572-6580.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan A., Sharif S., Shaukat S., Khan S., Zaidi S. An outbreak of acute hemorrhagic conjunctivitis (AHC) caused by coxsackievirus A24 variant in pakistan. Virus Res. 2008;137:150–152. doi: 10.1016/j.virusres.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 5.Sapkal G.N., Bondre V.P., Fulmali P.V., Patil P., Gopalkrishna V., Dadhania V., Ayachit V.M., Gangale D., Kushwaha K.P., Rathi A.K., et al. Enteroviruses in patients with acute encephalitis, Uttar Pradesh, India. Emerg. Infect. Dis. 2009;15:295–298. doi: 10.3201/eid1502.080865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang Y., Tan X.J., Wang H.Y., Yan D.M., Zhu S.L., Wang D.Y., Ji F., Wang X.J., Gao Y.J., Chen L., et al. An outbreak of hand, foot, and mouth disease associated with subgenotype C4 of human enterovirus 71 in Shandong, China. J. Clin. Virol. 2009;44:262–267. doi: 10.1016/j.jcv.2009.02.002. [DOI] [PubMed] [Google Scholar]
- 7.Hayden F.G., Turner R.B., Gwaltney J.M., Chi-Burris K., Gersten M., Hsyu P., Patick A.K., Smith G.J., Zalman L.S. Phase II, randomized, double-blind, placebo-controlled studies of ruprintrivir nasal spray 2-percent suspension for prevention and treatment of experimentally induced rhinovirus colds in healthy volunteers. Antimicrob. Agents Chemother. 2003;47:3907–3916. doi: 10.1128/AAC.47.12.3907-3916.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Patick A.K. Rhinovirus chemotherapy. Antiviral Res. 2006;71:391–396. doi: 10.1016/j.antiviral.2006.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Paul A.V., Rieder E., Kim D.W., van Boom J.H., Wimmer E. Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J. Virol. 2000;74:10359–10370. doi: 10.1128/JVI.74.22.10359-10370.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Birtley J.R., Knox S.R., Jaulent A.M., Brick P., Leatherbarrow R.J., Curry S. Crystal structure of foot-and-mouth disease virus 3C protease. New insights into catalytic mechanism and cleavage specificity. J. Biol. Chem. 2005;280:11520–11527. doi: 10.1074/jbc.M413254200. [DOI] [PubMed] [Google Scholar]
- 11.Bergmann E.M., Cherney M.M., McKendrick J., Frormann S., Luo C., Malcolm B.A., Vederas J.C., James M.N. Crystal structure of an inhibitor complex of the 3C proteinase from hepatitis A virus (HAV) and implications for the polyprotein processing in HAV. Virology. 1999;265:153–163. doi: 10.1006/viro.1999.9968. [DOI] [PubMed] [Google Scholar]
- 12.Mosimann S.C., Cherney M.M., Sia S., Plotch S., James M.N. Refined X-ray crystallographic structure of the poliovirus 3C gene product. J. Mol. Biol. 1997;273:1032–1047. doi: 10.1006/jmbi.1997.1306. [DOI] [PubMed] [Google Scholar]
- 13.Matthews D.A., Smith W.W., Ferre R.A., Condon B., Budahazi G., Sisson W., Villafranca J.E., Janson C.A., McElroy H.E., Gribskov C.L., et al. Structure of human rhinovirus 3C protease reveals a trypsin-like polypeptide fold, RNA-binding site, and means for cleaving precursor polyprotein. Cell. 1994;77:761–771. doi: 10.1016/0092-8674(94)90059-0. [DOI] [PubMed] [Google Scholar]
- 14.Matthews D.A., Dragovich P.S., Webber S.E., Fuhrman S.A., Patick A.K., Zalman L.S., Hendrickson T.F., Love R.A., Prins T.J., Marakovits J.T., et al. Structure-assisted design of mechanism-based irreversible inhibitors of human rhinovirus 3C protease with potent antiviral activity against multiple rhinovirus serotypes. Proc. Nalt. Acad. Sci. USA. 1999;96:11000–11007. doi: 10.1073/pnas.96.20.11000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yin J., Bergmann E.M., Cherney M.M., Lall M.S., Jain R.P., Vederas J.C., James M.N. Dual modes of modification of hepatitis a virus 3C protease by a serine-derived β-lactone: Selective crystallization and formation of a functional catalytic triad in the active site. J. Mol. Biol. 2005;354:854–871. doi: 10.1016/j.jmb.2005.09.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sweeney T.R., Roqué-Rosell N., Birtley J.R., Leatherbarrow R.J., Curry S. Structural and mutagenic analysis of foot-and-mouth disease virus 3C protease reveals the role of the β-ribbon in proteolysis. J. Virol. 2007;81:115–124. doi: 10.1128/JVI.01587-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui S., Wang J., Fan T., Qin B., Guo L., Lei X., Wang J., Wang M., Jin Q. Crystal structure of human enterovirus 71 3C protease. J. Mol. Biol. 2011;408:449–461. doi: 10.1016/j.jmb.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bergmann E.M., Mosimann S.C., Chernaia M.M., Malcolm B.A., James M.N. The refined crystal structure of the 3C gene product from hepatitis a virus: Specific proteinase activity and RNA recognition. J. Virol. 1997;71:2436–2448. doi: 10.1128/jvi.71.3.2436-2448.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang J., Fan T., Yao X., Wu Z., Guo L., Lei X., Wang J., Wang M., Jin Q., Cui S. Crystal structures of enterovirus 71 3C protease complexed with rupintrivir reveal the roles of catalytically important residues. J. Virol. 2011;85:10021–10030. doi: 10.1128/JVI.05107-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curry S., Roqué-Rosell N., Zunszain P.A., Leatherbarrow R.J. Foot-and-mouth disease virus 3C protease: Recent structural and functional insights into an antiviral target. Int. J. Biochem. Cell Biol. 2007;39:1–6. doi: 10.1016/j.biocel.2006.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seipelt J., Guarné A., Bergmann E., James M., Sommergruber W., Fita I., Skern T. The structures of picornaviral proteinases. Virus Res. 1999;62:159–168. doi: 10.1016/S0168-1702(99)00043-X. [DOI] [PubMed] [Google Scholar]
- 22.Guarné A., Hampoelz B., Glaser W., Carpena X., Tormo J., Fita I., Skern T. Structural and biochemical features distinguish the foot-and-mouth disease virus leader proteinase from other papain-like enzymes. J. Mol. Biol. 2000;302:1227–1240. doi: 10.1006/jmbi.2000.4115. [DOI] [PubMed] [Google Scholar]
- 23.Salas E.M., Ryan M.D. Translation and protein process. In: Ehrenfeld E., Domingo E., Roos R., editors. The Picornaviruses. ASM Press; Washington, DC, USA: 2010. pp. 141–150. [Google Scholar]
- 24.Palmenberg A., Neubauer D., Skern T. Genome organization and encoded proteins. In: Ehrenfeld E., Domingo E., Roos R., editors. The Picornaviruses. ASM Press; Washington, DC, USA: 2010. pp. 3–17. [Google Scholar]
- 25.Castelló A., Alvarez E., Carrasco L. The multifaceted poliovirus 2A protease: Regulation of gene expression by picornavirus proteases. J. Biomed. Biotechnol. 2011 doi: 10.1155/2011/369648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amineva S.P., Aminev A.G., Palmenberg A.C., Gern J.E. Rhinovirus 3C protease precursors 3CD and 3CD′ localize to the nuclei of infected cells. J. Gen. Virol. 2004;85:2969–2979. doi: 10.1099/vir.0.80164-0. [DOI] [PubMed] [Google Scholar]
- 27.Ryan M.D., Belsham G.J., King A.M. Specificity of enzyme-substrate interactions in foot-and-mouth disease virus polyprotein processing. Virology. 1989;173:35–45. doi: 10.1016/0042-6822(89)90219-5. [DOI] [PubMed] [Google Scholar]
- 28.Ypma-Wong M.F., Dewalt P.G., Johnson V.H., Lamb J.G., Semler B.L. Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology. 1988;166:265–270. doi: 10.1016/0042-6822(88)90172-9. [DOI] [PubMed] [Google Scholar]
- 29.Guarné A., Tormo J., Kirchweger R., Pfistermueller D., Fita I., Skern T. Structure of the foot-and-mouth disease virus leader protease: A papain-like fold adapted for self-processing and eIF4G recognition. EMBO J. 1998;17:7469–7479. doi: 10.1093/emboj/17.24.7469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spear A., Ogram S.A., Morasco B.J., Smerage L.E., Flanegan J.B. Viral precursor protein P3 and its processed products perform discrete and essential functions in the poliovirus RNA replication complex. Virology. 2015;485:492–501. doi: 10.1016/j.virol.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yin J., Paul A.V., Wimmer E., Rieder E. Functional dissection of a poliovirus cis-acting replication element (PV-cre(2C)): Analysis of single- and dual-cre viral genomes and proteins that bind specifically to PV-cre RNA. J. Virol. 2003;77:5152–5166. doi: 10.1128/JVI.77.9.5152-5166.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pathak H.B., Arnold J.J., Wiegand P.N., Hargittai M.R., Cameron C.E. Picornavirus genome replication: Assembly and organization of the VPg uridylylation ribonucleoprotein (initiation) complex. J. Biol. Chem. 2007;282:16202–16213. doi: 10.1074/jbc.M610608200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pathak H.B., Oh H.S., Goodfellow I.G., Arnold J.J., Cameron C.E. Picornavirus genome replication: Roles of precursor proteins and rate-limiting steps in oriI-dependent VPg uridylylation. J. Biol. Chem. 2008;283:30677–30688. doi: 10.1074/jbc.M806101200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kusov Y.Y., Morace G., Probst C., Gauss-Müller V. Interaction of hepatitis A virus (HAV) precursor proteins 3AB and 3ABC with the 5′ and 3′ termini of the HAV RNA. Virus Res. 1997;51:151–157. doi: 10.1016/S0168-1702(97)00089-0. [DOI] [PubMed] [Google Scholar]
- 35.Paul A.V., Wimmer E. Initiation of protein-primed picornavirus RNA synthesis. Virus Res. 2015;206:12–26. doi: 10.1016/j.virusres.2014.12.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Andino R., Rieckhof G.E., Trono D., Baltimore D. Substitutions in the protease (3Cpro) gene of poliovirus can suppress a mutation in the 5’noncoding region. J. Virol. 1990;64:607–612. doi: 10.1128/jvi.64.2.607-612.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andino R., Rieckhof G.E., Baltimore D. A functional ribonucleoprotein complex forms around the 5′ end of poliovirus RNA. Cell. 1990;63:369–380. doi: 10.1016/0092-8674(90)90170-J. [DOI] [PubMed] [Google Scholar]
- 38.Andino R., Rieckhof G.E., Achacoso P.L., Baltimore D. Poliovirus RNA synthesis utilizes an RNP complex formed around the 5’-end of viral RNA. EMBO J. 1993;12:3587–3598. doi: 10.1002/j.1460-2075.1993.tb06032.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nayak A., Goodfellow I.G., Belsham G.J. Factors required for the uridylylation of the foot-and-mouth disease virus 3B1, 3B2, and 3B3 peptides by the RNA-dependent RNA polymerase (3Dpol) in vitro. J. Virol. 2005;79:7698–7706. doi: 10.1128/JVI.79.12.7698-7706.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nayak A., Goodfellow I.G., Woolaway K.E., Birtley J., Curry S., Belsham G.J. Role of RNA structure and RNA binding activity of foot-and-mouth disease virus 3C protein in VPg uridylylation and virus replication. J. Virol. 2006;80:9865–9875. doi: 10.1128/JVI.00561-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Herold J., Andino R. Poliovirus RNA replication requires genome circularization through a protein-protein bridge. Mol. Cell. 2001;7:581–591. doi: 10.1016/S1097-2765(01)00205-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Harris K.S., Xiang W., Alexander L., Lane W.S., Paul A.V., Wimmer E. Interaction of poliovirus polypeptide 3CDpro with the 5’and 3’termini of the poliovirus genome. Identification of viral and cellular cofactors needed for efficient binding. J. Biol. Chem. 1994;269:27004–27014. [PubMed] [Google Scholar]
- 43.Franco D., Pathak H.B., Cameron C.E., Rombaut B., Wimmer E., Paul A.V. Stimulation of poliovirus synthesis in a HeLa cell-free in vitro translation-RNA replication system by viral protein 3CDpro. J. Virol. 2005;79:6358–6367. doi: 10.1128/JVI.79.10.6358-6367.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blair W.S., Parsley T.B., Bogerd H.P., Towner J.S., Semler B.L., Cullen B.R. Utilization of a mammalian cell-based RNA binding assay to characterize the RNA binding properties of picornavirus 3C proteinases. RNA. 1998;4:215–225. [PMC free article] [PubMed] [Google Scholar]
- 45.Moustafa I.M., Gohara D.W., Uchida A., Yennawar N., Cameron C.E. Conformational ensemble of the poliovirus 3CD precursor observed by md simulations and confirmed by saxs: A strategy to expand the viral proteome? Viruses. 2015;7:5962–5986. doi: 10.3390/v7112919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gamarnik A.V., Andino R. Switch from translation to RNA replication in a positive-stranded RNA virus. Genes Dev. 1998;12:2293–2304. doi: 10.1101/gad.12.15.2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chase A.J., Daijogo S., Semler B.L. Inhibition of poliovirus-induced cleavage of cellular protein PCBP2 reduces the levels of viral RNA replication. J. Virol. 2014;88:3192–3201. doi: 10.1128/JVI.02503-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chan Y.M., Boehr D.D. Allosteric functional switch in poliovirus 3C protease. FASEB J. 2015;29 doi: 10.1016/j.bpj.2014.11.2893. [DOI] [Google Scholar]
- 49.Banerjee R., Weidman M.K., Navarro S., Comai L., Dasgupta A. Modifications of both selectivity factor and upstream binding factor contribute to poliovirus-mediated inhibition of RNA polymerase I transcription. J. Gen. Virol. 2005;86:2315–2322. doi: 10.1099/vir.0.80817-0. [DOI] [PubMed] [Google Scholar]
- 50.Kundu P., Raychaudhuri S., Tsai W., Dasgupta A. Shutoff of RNA polymerase II transcription by poliovirus involves 3C protease-mediated cleavage of the TATA-binding protein at an alternative site: Incomplete shutoff of transcription interferes with efficient viral replication. J. Virol. 2005;79:9702–9713. doi: 10.1128/JVI.79.15.9702-9713.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yalamanchili P., Datta U., Dasgupta A. Inhibition of host cell transcription by poliovirus: Cleavage of transcription factor CREB by poliovirus-encoded protease 3Cpro. J. Virol. 1997;71:1220–1226. doi: 10.1128/jvi.71.2.1220-1226.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yalamanchili P., Weidman K., Dasgupta A. Cleavage of transcriptional activator Oct-1 by poliovirus encoded protease 3Cpro. Virology. 1997;239:176–185. doi: 10.1006/viro.1997.8862. [DOI] [PubMed] [Google Scholar]
- 53.Weidman M.K., Yalamanchili P., Ng B., Tsai W., Dasgupta A. Poliovirus 3C protease-mediated degradation of transcriptional activator p53 requires a cellular activity. Virology. 2001;291:260–271. doi: 10.1006/viro.2001.1215. [DOI] [PubMed] [Google Scholar]
- 54.Shen Y., Igo M., Yalamanchili P., Berk A.J., Dasgupta A. DNA binding domain and subunit interactions of transcription factor IIIC revealed by dissection with poliovirus 3C protease. Mol. Cell. Biol. 1996;16:4163–4171. doi: 10.1128/MCB.16.8.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Falk M.M., Grigera P.R., Bergmann I.E., Zibert A., Multhaup G., Beck E. Foot-and-mouth disease virus protease 3C induces specific proteolytic cleavage of host cell histone H3. J. Virol. 1990;64:748–756. doi: 10.1128/jvi.64.2.748-756.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Weng K.F., Li M.L., Hung C.T., Shih S.R. Enterovirus 71 3C protease cleaves a novel target CstF-64 and inhibits cellular polyadenylation. PLoS Pathog. 2009;5:82. doi: 10.1371/journal.ppat.1000593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sonenberg N., Hinnebusch A.G. Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lozano G., Martínez-Salas E. Structural insights into viral IRES-dependent translation mechanisms. Curr. Opin. Virol. 2015;12:113–120. doi: 10.1016/j.coviro.2015.04.008. [DOI] [PubMed] [Google Scholar]
- 59.Martínez-Salas E., Pacheco A., Serrano P., Fernandez N. New insights into internal ribosome entry site elements relevant for viral gene expression. J. Gen. Virol. 2008;89:611–626. doi: 10.1099/vir.0.83426-0. [DOI] [PubMed] [Google Scholar]
- 60.Martínez-Salas E., Francisco-Velilla R., Fernandez-Chamorro J., Lozano G., Diaz-Toledano R. Picornavirus IRES elements: RNA structure and host protein interactions. Virus Res. 2015;206:62–73. doi: 10.1016/j.virusres.2015.01.012. [DOI] [PubMed] [Google Scholar]
- 61.Belsham G.J., McInerney G.M., Ross-Smith N. Foot-and-mouth disease virus 3C protease induces cleavage of translation initiation factors eIF4A and eIF4G within infected cells. J. Virol. 2000;74:272–280. doi: 10.1128/JVI.74.1.272-280.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Gradi A., Foeger N., Strong R., Svitkin Y.V., Sonenberg N., Skern T., Belsham G.J. Cleavage of eukaryotic translation initiation factor 4GII within foot-and-mouth disease virus-infected cells: Identification of the L-protease cleavage site in vitro. J. Virol. 2004;78:3271–3278. doi: 10.1128/JVI.78.7.3271-3278.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li W., Ross-Smith N., Proud C.G., Belsham G.J. Cleavage of translation initiation factor 4AI (eIF4AI) but not eIF4AII by foot-and-mouth disease virus 3C protease: Identification of the eIF4AI cleavage site. FEBS Lett. 2001;507:1–5. doi: 10.1016/S0014-5793(01)02885-X. [DOI] [PubMed] [Google Scholar]
- 64.De Breyne S., Bonderoff J.M., Chumakov K.M., Lloyd R.E., Hellen C.U. Cleavage of eukaryotic initiation factor eIF5B by enterovirus 3C proteases. Virology. 2008;378:118–122. doi: 10.1016/j.virol.2008.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Perera R., Daijogo S., Walter B.L., Nguyen J.H., Semler B.L. Cellular protein modification by poliovirus: The two faces of poly (rC)-binding protein. J. Virol. 2007;81:8919–8932. doi: 10.1128/JVI.01013-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang B., Seitz S., Kusov Y., Zell R., Gauss-Müller V. RNA interaction and cleavage of poly (C)-binding protein 2 by hepatitis A virus protease. Biochem. Biophys. Res. Commun. 2007;364:725–730. doi: 10.1016/j.bbrc.2007.09.133. [DOI] [PubMed] [Google Scholar]
- 67.Kuyumcu-Martinez N.M., Van Eden M.E., Younan P., Lloyd R.E. Cleavage of poly (A)-binding protein by poliovirus 3C protease inhibits host cell translation: A novel mechanism for host translation shutoff. Mol. Cell. Biol. 2004;24:1779–1790. doi: 10.1128/MCB.24.4.1779-1790.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhang B., Morace G., Gauss-Müller V., Kusov Y. Poly(A) binding protein, C-terminally truncated by the hepatitis A virus proteinase 3C, inhibits viral translation. Nucleic Acids Res. 2007;35:5975–5984. doi: 10.1093/nar/gkm645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kobayashi M., Arias C., Garabedian A., Palmenberg A.C., Mohr I. Site-specific cleavage of the host poly (A) binding protein by the encephalomyocarditis virus 3C proteinase stimulates viral replication. J. Virol. 2012;86:10686–10694. doi: 10.1128/JVI.00896-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Fung G., Ng C.S., Zhang J., Shi J., Wong J., Piesik P., Han L., Chu F., Jagdeo J., Jan E., et al. Production of a dominant-negative fragment due to G3BP1 cleavage contributes to the disruption of mitochondria-associated protective stress granules during CVB3 infection. PLoS ONE. 2013;8:82. doi: 10.1371/journal.pone.0079546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lawrence P., Schafer E.A., Rieder E. The nuclear protein Sam68 is cleaved by the FMDV 3C protease redistributing Sam68 to the cytoplasm during FMDV infection of host cells. Virology. 2012;425:40–52. doi: 10.1016/j.virol.2011.12.019. [DOI] [PubMed] [Google Scholar]
- 72.Flather D., Semler B.L. Picornaviruses and nuclear functions: Targeting a cellular compartment distinct from the replication site of a positive-strand RNA virus. Front. Microbiol. 2015;6:594. doi: 10.3389/fmicb.2015.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Watters K., Palmenberg A.C. Differential processing of nuclear pore complex proteins by rhinovirus 2A proteases from different species and serotypes. J. Virol. 2011;85:10874–10883. doi: 10.1128/JVI.00718-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Walker E.J., Younessi P., Fulcher A.J., McCuaig R., Thomas B.J., Bardin P.G., Jans D.A., Ghildyal R. Rhinovirus 3C protease facilitates specific nucleoporin cleavage and mislocalisation of nuclear proteins in infected host cells. PLoS ONE. 2013;8:82. doi: 10.1371/journal.pone.0071316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Caly L., Ghildyal R., Jans D.A. Respiratory virus modulation of host nucleocytoplasmic transport; target for therapeutic intervention? Front. Microbiol. 2015;6:848. doi: 10.3389/fmicb.2015.00848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Park N., Katikaneni P., Skern T., Gustin K.E. Differential targeting of nuclear pore complex proteins in poliovirus-infected cells. J. Virol. 2008;82:1647–1655. doi: 10.1128/JVI.01670-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Park N., Skern T., Gustin K.E. Specific cleavage of the nuclear pore complex protein Nup62 by a viral protease. J. Biol. Chem. 2010;285:28796–28805. doi: 10.1074/jbc.M110.143404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ghildyal R., Jordan B., Li D., Dagher H., Bardin P.G., Gern J.E., Jans D.A. Rhinovirus 3C protease can localize in the nucleus and alter active and passive nucleocytoplasmic transport. J. Virol. 2009;83:7349–7352. doi: 10.1128/JVI.01748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Alber F., Dokudovskaya S., Veenhoff L.M., Zhang W., Kipper J., Devos D., Suprapto A., Karni-Schmidt O., Williams R., Chait B.T. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- 80.Grossman E., Medalia O., Zwerger M. Functional architecture of the nuclear pore complex. Annu. Rev. Biophys. 2012;41:557–584. doi: 10.1146/annurev-biophys-050511-102328. [DOI] [PubMed] [Google Scholar]
- 81.Porter F.W., Palmenberg A.C. Leader-induced phosphorylation of nucleoporins correlates with nuclear trafficking inhibition by cardioviruses. J. Virol. 2009;83:1941–1951. doi: 10.1128/JVI.01752-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ricour C., Delhaye S., Hato S.V., Olenyik T.D., Michel B., van Kuppeveld F.J., Gustin K.E., Michiels T. Inhibition of mrna export and dimerization of interferon regulatory factor 3 by theiler’s virus leader protein. J. Gen. Virol. 2009;90:177–186. doi: 10.1099/vir.0.005678-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Barco A., Feduchi E., Carrasco L. Poliovirus protease 3Cpro kills cells by apoptosis. Virology. 2000;266:352–360. doi: 10.1006/viro.1999.0043. [DOI] [PubMed] [Google Scholar]
- 84.Li M.L., Hsu T.A., Chen T.C., Chang S.C., Lee J.C., Chen C.C., Stollar V., Shih S.R. The 3C protease activity of enterovirus 71 induces human neural cell apoptosis. Virology. 2002;293:386–395. doi: 10.1006/viro.2001.1310. [DOI] [PubMed] [Google Scholar]
- 85.Chau D.H., Yuan J., Zhang H., Cheung P., Lim T., Liu Z., Sall A., Yang D. Coxsackievirus B3 proteases 2A and 3C induce apoptotic cell death through mitochondrial injury and cleavage of eIF4GI but not DAP5/p97/NAT1. Apoptosis. 2007;12:513–524. doi: 10.1007/s10495-006-0013-0. [DOI] [PubMed] [Google Scholar]
- 86.Shubin A.V., Demidyuk I.V., Lunina N.A., Komissarov A.A., Roschina M.P., Leonova O.G., Kostrov S.V. Protease 3C of hepatitis A virus induces vacuolization of lysosomal/endosomal organelles and caspase-independent cell death. BMC Cell Biol. 2015;16:4. doi: 10.1186/s12860-015-0050-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Amero C.D., Arnold J.J., Moustafa I.M., Cameron C.E., Foster M.P. Identification of the oriI-binding site of poliovirus 3C protein by nuclear magnetic resonance spectroscopy. J. Virol. 2008;82:4363–4370. doi: 10.1128/JVI.02087-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Joachims M., Harris K.S., Etchison D. Poliovirus protease 3C mediates cleavage of microtubule-associated protein 4. Virology. 1995;211:451–461. doi: 10.1006/viro.1995.1427. [DOI] [PubMed] [Google Scholar]
- 89.Zhou Z., Mogensen M.M., Powell P.P., Curry S., Wileman T. Foot-and-mouth disease virus 3C protease induces fragmentation of the Golgi compartment and blocks intra-Golgi transport. J. Virol. 2013;87:11721–11729. doi: 10.1128/JVI.01355-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Takeda K., Akira S. Toll-like receptors in innate immunity. Int. Immunol. 2005;17:1–14. doi: 10.1093/intimm/dxh186. [DOI] [PubMed] [Google Scholar]
- 91.Deretic V. Autophagy: An emerging immunological paradigm. J. Immunol. 2012;189:15–20. doi: 10.4049/jimmunol.1102108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pålsson-McDermott E.M., O’Neill L.A. Building an immune system from nine domains. Biochem. Soc. Trans. 2007;35:1437–1444. doi: 10.1042/BST0351437. [DOI] [PubMed] [Google Scholar]
- 93.Akira S., Takeda K. Toll-like receptor signalling. Nat. Rev. Immunol. 2004;4:499–511. doi: 10.1038/nri1391. [DOI] [PubMed] [Google Scholar]
- 94.Bowie A., O’neill L.A. The interleukin-1 receptor/toll-like receptor superfamily: Signal generators for pro-inflammatory interleukins and microbial products. J. Leukoc. Biol. 2000;67:508–514. doi: 10.1002/jlb.67.4.508. [DOI] [PubMed] [Google Scholar]
- 95.Kawai T., Akira S. Innate immune recognition of viral infection. Nat. Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 96.Oganesyan G., Saha S.K., Guo B., He J.Q., Shahangian A., Zarnegar B., Perry A., Cheng G. Critical role of TRAF3 in the Toll-like receptor-dependent and -independent antiviral response. Nature. 2006;439:208–211. doi: 10.1038/nature04374. [DOI] [PubMed] [Google Scholar]
- 97.Häcker H., Redecke V., Blagoev B., Kratchmarova I., Hsu L.C., Wang G.G., Kamps M.P., Raz E., Wagner H., Häcker G., et al. Specificity in toll-like receptor signalling through distinct effector functions of TRAF3 and TRAF6. Nature. 2006;439:204–207. doi: 10.1038/nature04369. [DOI] [PubMed] [Google Scholar]
- 98.Sharma S., tenOever B.R., Grandvaux N., Zhou G.P., Lin R., Hiscott J. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 99.Oshiumi H., Matsumoto M., Funami K., Akazawa T., Seya T. TICAM-1, an adaptor molecule that participates in Toll-like receptor 3-mediated interferon-beta induction. Nat. Immunol. 2003;4:161–167. doi: 10.1038/ni886. [DOI] [PubMed] [Google Scholar]
- 100.Fitzgerald K.A., McWhirter S.M., Faia K.L., Rowe D.C., Latz E., Golenbock D.T., Coyle A.J., Liao S.M., Maniatis T. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat. Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 101.Arpaia N., Barton G.M. Toll-like receptors: Key players in antiviral immunity. Curr. Opin. Virol. 2011;1:447–454. doi: 10.1016/j.coviro.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Lei X., Liu X., Ma Y., Sun Z., Yang Y., Jin Q., He B., Wang J. The 3C protein of enterovirus 71 inhibits retinoid acid-inducible gene I-mediated interferon regulatory factor 3 activation and type I interferon responses. J. Virol. 2010;84:8051–8061. doi: 10.1128/JVI.02491-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Wang B., Xi X., Lei X., Zhang X., Cui S., Wang J., Jin Q., Zhao Z. Enterovirus 71 protease 2A pro targets MAVS to inhibit anti-viral type I interferon responses. PLoS Pathol. 2013;9:82. doi: 10.1371/journal.ppat.1003231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lei X., Sun Z., Liu X., Jin Q., He B., Wang J. Cleavage of the adaptor protein TRIF by enterovirus 71 3C inhibits antiviral responses mediated by toll-like receptor 3. J. Virol. 2011;85:8811–8818. doi: 10.1128/JVI.00447-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Papon L., Oteiza A., Imaizumi T., Kato H., Brocchi E., Lawson T.G., Akira S., Mechti N. The viral RNA recognition sensor RIG-I is degraded during encephalomyocarditis virus (EMCV) infection. Virology. 2009;393:311–318. doi: 10.1016/j.virol.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 106.Barral P.M., Morrison J.M., Drahos J., Gupta P., Sarkar D., Fisher P.B., Racaniello V.R. MDA-5 is cleaved in poliovirus-infected cells. J. Virol. 2007;81:3677–3684. doi: 10.1128/JVI.01360-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Huang H.I., Weng K.F., Shih S.R. Viral and host factors that contribute to pathogenicity of enterovirus 71. Future Microbiol. 2012;7:467–479. doi: 10.2217/fmb.12.22. [DOI] [PubMed] [Google Scholar]
- 108.Lei X., Xiao X., Xue Q., Jin Q., He B., Wang J. Cleavage of interferon regulatory factor 7 by enterovirus 71 3C suppresses cellular responses. J. Virol. 2013;87:1690–1698. doi: 10.1128/JVI.01855-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Lei X., Han N., Xiao X., Jin Q., He B., Wang J. Enterovirus 71 3C inhibits cytokine expression through cleavage of the TAK1/TAB1/TAB2/TAB3 complex. J. Virol. 2014;88:9830–9841. doi: 10.1128/JVI.01425-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Liu Y., Zhang Z., Zhao X., Yu R., Zhang X., Wu S., Liu J., Chi X., Song X., Fu L., et al. Enterovirus 71 inhibits cellular type I interferon signaling by downregulating jak1 protein expression. Viral Immunol. 2014;27:267–276. doi: 10.1089/vim.2013.0127. [DOI] [PubMed] [Google Scholar]
- 111.Zhang Y., Mao D., Roswit W.T., Jin X., Patel A.C., Patel D.A., Agapov E., Wang Z., Tidwell R.M., Atkinson J.J., et al. PARP9-DTX3L ubiquitin ligase targets host histone H2BJ and viral 3C protease to enhance interferon signaling and control viral infection. Nat. Immunol. 2015;16:1215–1227. doi: 10.1038/ni.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Yang Y., Liang Y., Qu L., Chen Z., Yi M., Li K., Lemon S.M. Disruption of innate immunity due to mitochondrial targeting of a picornaviral protease precursor. Proc. Natl. Acad. Sci. USA. 2007;104:7253–7258. doi: 10.1073/pnas.0611506104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Qu L. Ph.D. Thesis. University of Texas Medical Branch; Galveston, TX, USA: 2010. Evasion of RIG-I/MDA5 and TLR3-Mediated Innate Immunity by Hepatitis A Virus. [Google Scholar]
- 114.Qu L., Feng Z., Yamane D., Liang Y., Lanford R.E., Li K., Lemon S.M. Disruption of TLR3 signaling due to cleavage of TRIF by the hepatitis A virus protease-polymerase processing intermediate, 3CD. PLoS Pathog. 2011;7:82. doi: 10.1371/journal.ppat.1002169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Wang D., Fang L., Wei D., Zhang H., Luo R., Chen H., Li K., Xiao S. Hepatitis A virus 3C protease cleaves NEMO to impair induction of β interferon. J. Virol. 2014;88:10252–10258. doi: 10.1128/JVI.00869-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang D., Fang L., Li K., Zhong H., Fan J., Ouyang C., Zhang H., Duan E., Luo R., Zhang Z., et al. Foot-and-mouth disease virus 3C protease cleaves NEMO to impair innate immune signaling. J. Virol. 2012;86:9311–9322. doi: 10.1128/JVI.00722-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.De Los Santos T., Diaz-San Segundo F., Grubman M.J. Degradation of nuclear factor κB during foot-and-mouth disease virus infection. J. Virol. 2007;81:12803–12815. doi: 10.1128/JVI.01467-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wang D., Fang L., Luo R., Ye R., Fang Y., Xie L., Chen H., Xiao S. Foot-and-mouth disease virus leader proteinase inhibits dsRNA-induced type I interferon transcription by decreasing interferon regulatory factor 3/7 in protein levels. Biochem. Biophys. Res. Commun. 2010;399:72–78. doi: 10.1016/j.bbrc.2010.07.044. [DOI] [PubMed] [Google Scholar]
- 119.Du Y., Bi J., Liu J., Liu X., Wu X., Jiang P., Yoo D., Zhang Y., Wu J., Wan R., et al. 3Cpro of foot-and-mouth disease virus antagonizes the interferon signaling pathway by blocking STAT1/STAT2 nuclear translocation. J. Virol. 2014;88:4908–4920. doi: 10.1128/JVI.03668-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Mukherjee A., Morosky S.A., Delorme-Axford E., Dybdahl-Sissoko N., Oberste M.S., Wang T., Coyne C.B. The Coxsackievirus B 3C protease cleaves MAVS and TRIF to attenuate host type I interferon and apoptotic signaling. PLoS Pathog. 2011;7:82. doi: 10.1371/journal.ppat.1001311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Xu C., He X., Zheng Z., Zhang Z., Wei C., Guan K., Hou L., Zhang B., Zhu L., Cao Y., et al. Downregulation of MicroRNA miR-526a by enterovirus inhibits RIG-I-Dependent innate immune response. J. Virol. 2014;88:11356–11368. doi: 10.1128/JVI.01400-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J.C., Williams K.L., Buratti E., et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Fung G., Shi J., Deng H., Hou J., Wang C., Hong A., Zhang J., Jia W., Luo H. Cytoplasmic translocation, aggregation, and cleavage of TDP-43 by enteroviral proteases modulate viral pathogenesis. Cell Death Differ. 2015;22:2087–2097. doi: 10.1038/cdd.2015.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Dragovich P.S., Zhou R., Webber S.E., Prins T.J., Kwok A.K., Okano K., Fuhrman S.A., Zalman L.S., Maldonado F.C., Brown E.L., et al. Structure-based design of ketone-containing, tripeptidyl human rhinovirus 3C protease inhibitors. Bioorg. Med. Chem. Lett. 2000;10:45–48. doi: 10.1016/S0960-894X(99)00587-9. [DOI] [PubMed] [Google Scholar]
- 125.Shepherd T.A., Cox G.A., McKinney E., Tang J., Wakulchik M., Zimmerman R.E., Villarreal E.C. Small peptidic aldehyde inhibitors of human rhinovirus 3C protease. Bioorg. Med. Chem. Lett. 1996;6:2893–2896. doi: 10.1016/S0960-894X(96)00537-9. [DOI] [Google Scholar]
- 126.Wang Y., Yang B., Zhai Y., Yin Z., Sun Y., Rao Z. Peptidyl aldehyde NK-1.8 k suppresses enterovirus 71 and enterovirus 68 infection by targeting protease 3C. Antimicrob. Agents Chemother. 2015;59:2636–2646. doi: 10.1128/AAC.00049-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Liu S., Hanzlik R.P. Structure-activity relationships for inhibition of papain by peptide Michael acceptors. J. Med. Chem. 1992;35:1067–1075. doi: 10.1021/jm00084a012. [DOI] [PubMed] [Google Scholar]
- 128.Kong J.S., Venkatraman S., Furness K., Nimkar S., Shepherd T.A., Wang Q.M., Aubé J., Hanzlik R.P. Synthesis and evaluation of peptidyl Michael acceptors that inactivate human rhinovirus 3C protease and inhibit virus replication. J. Med. Chem. 1998;41:2579–2587. doi: 10.1021/jm980114+. [DOI] [PubMed] [Google Scholar]
- 129.Dragovich P.S., Webber S.E., Babine R.E., Fuhrman S.A., Patick A.K., Matthews D.A., Lee C.A., Reich S.H., Prins T.J., Marakovits J.T., et al. Structure-based design, synthesis, and biological evaluation of irreversible human rhinovirus 3C protease inhibitors. 1. Michael acceptor structure-activity studies. J. Med. Chem. 1998;41:2806–2818. doi: 10.1021/jm980068d. [DOI] [PubMed] [Google Scholar]
- 130.Patick A.K., Binford S.L., Brothers M.A., Jackson R.L., Ford C.E., Diem M.D., Maldonado F., Dragovich P.S., Zhou R., Prins T.J., et al. In vitro antiviral activity of AG7088, a potent inhibitor of human rhinovirus 3C protease. Antimicrob. Agents Chemother. 1999;43:2444–2450. doi: 10.1128/aac.43.10.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Tan J., George S., Kusov Y., Perbandt M., Anemüller S., Mesters J.R., Norder H., Coutard B., Lacroix C., Leyssen P., et al. 3C protease of enterovirus 68: Structure-based design of Michael acceptor inhibitors and their broad-spectrum antiviral effects against picornaviruses. J. Virol. 2013;87:4339–4351. doi: 10.1128/JVI.01123-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Van der Linden L., Ulferts R., Nabuurs S.B., Kusov Y., Liu H., George S., Lacroix C., Goris N., Lefebvre D., Lanke K.H., et al. Application of a cell-based protease assay for testing inhibitors of picornavirus 3C proteases. Antivir. Res. 2014;103:17–24. doi: 10.1016/j.antiviral.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Debing Y., Kaplan G.G., Neyts J., Jochmans D. Rapid and convenient assays to assess potential inhibitory activity on in vitro hepatitis A replication. Antivir. Res. 2013;98:325–331. doi: 10.1016/j.antiviral.2013.03.016. [DOI] [PubMed] [Google Scholar]
- 134.De Palma A.M., Pürstinger G., Wimmer E., Patick A.K., Andries K., Rombaut B., de Clercq E., Neyts J. Potential use of antiviral agents in polio eradication. Emerg. Infect. Dis. 2008;14:545–551. doi: 10.3201/eid1404.070439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Baxter A., Chambers M., Edfeldt F., Edman K., Freeman A., Johansson C., King S., Morley A., Petersen J., Rawlins P., et al. Non-covalent inhibitors of rhinovirus 3C protease. Bioorg. Med. Chem. Lett. 2011;21:777–780. doi: 10.1016/j.bmcl.2010.11.110. [DOI] [PubMed] [Google Scholar]
- 136.Kim B.K., Cho J.H., Jeong P., Lee Y., Lim J.J., Park K.R., Eom S.H., Kim Y.C. Benserazide, the first allosteric inhibitor of Coxsackievirus B3 3C protease. FEBS Lett. 2015;589:1795–1801. doi: 10.1016/j.febslet.2015.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Lim B.K., Yun S.H., Ju E.S., Kim B.K., Lee Y.J., Yoo D.K., Kim Y.C., Jeon E.S. Soluble Coxsackievirus B3 3C protease inhibitor prevents cardiomyopathy in an experimental chronic myocarditis murine model. Virus Res. 2015;199:1–8. doi: 10.1016/j.virusres.2014.11.030. [DOI] [PubMed] [Google Scholar]
- 138.Kim B.K., Kim J.H., Kim N.R., Lee W.G., Lee S.D., Yun S.H., Jeon E.S., Kim Y.C. Development of anti-coxsackievirus agents targeting 3C protease. Bioorg. Med. Chem. Lett. 2012;22:6952–6956. doi: 10.1016/j.bmcl.2012.08.120. [DOI] [PubMed] [Google Scholar]
- 139.Zhai Y., Zhao X., Cui Z., Wang M., Wang Y., Li L., Sun Q., Yang X., Zeng D., Liu Y., et al. Cyanohydrin as an Anchoring Group for potent and selective inhibitors of enterovirus 71 3C protease. J. Med. Chem. 2015;58:9414–9420. doi: 10.1021/acs.jmedchem.5b01013. [DOI] [PubMed] [Google Scholar]
- 140.Ramajayam R., Tan K.P., Liang P.H. Recent development of 3C and 3CL protease inhibitors for anti-coronavirus and anti-picornavirus drug discovery. Biochem. Soc. Trans. 2011;39:1371–1375. doi: 10.1042/BST0391371. [DOI] [PubMed] [Google Scholar]
- 141.Shie J.J., Fang J.M., Kuo T.H., Kuo C.J., Liang P.H., Huang H.J., Wu Y.T., Jan J.T., Cheng Y.S., Wong C.H. Inhibition of the severe acute respiratory syndrome 3CL protease by peptidomimetic α, β-unsaturated esters. Bioorg. Med. Chem. 2005;13:5240–5252. doi: 10.1016/j.bmc.2005.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Yang S., Chen S.J., Hsu M.F., Wu J.D., Tseng C.T., Liu Y.F., Chen H.C., Kuo C.W., Wu C.S., Chang L.W., et al. Synthesis, crystal structure, structure-activity relationships, and antiviral activity of a potent SARS coronavirus 3CL protease inhibitor. J. Med. Chem. 2006;49:4971–4980. doi: 10.1021/jm0603926. [DOI] [PubMed] [Google Scholar]
- 143.Kuo C.J., Liu H.G., Lo Y.K., Seong C.M., Lee K.I., Jung Y.S., Liang P.H. Individual and common inhibitors of coronavirus and picornavirus main proteases. FEBS Lett. 2009;583:549–555. doi: 10.1016/j.febslet.2008.12.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Prior A.M., Kim Y., Weerasekara S., Moroze M., Alliston K.R., Uy R.A., Groutas W.C., Chang K.O., Hua D.H. Design, synthesis, and bioevaluation of viral 3C and 3C-like protease inhibitors. Bioorg. Med. Chem. Lett. 2013;23:6317–6320. doi: 10.1016/j.bmcl.2013.09.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mandadapu S.R., Weerawarna P.M., Prior A.M., Uy R.A., Aravapalli S., Alliston K.R., Lushington G.H., Kim Y., Hua D.H., Chang K.O., et al. Macrocyclic inhibitors of 3C and 3C-like proteases of picornavirus, norovirus, and coronavirus. Bioorg. Med. Chem. Lett. 2013;23:3709–3712. doi: 10.1016/j.bmcl.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Singh S.B., Cordingley M.G., Ball R.G., Smith J.L., Dombrowski A.W., Goetz M.A. Structure of stereochemistry of thysanone: A novel human rhinovirus 3C-protease inhibitor from Thysanophora penicilloides. Tetrahedron Lett. 1991;32:5279–5282. doi: 10.1016/S0040-4039(00)92364-5. [DOI] [Google Scholar]
- 147.Schünemann K., Furkert D.P., Choi E.C., Connelly S., Fraser J.D., Sperry J., Brimble M.A. Synthesis of the 2-methylene analogue of the HRV 3C protease inhibitor thysanone (2-carbathysanone) Org. Biomol. Chem. 2014;12:905–912. doi: 10.1039/C3OB41951G. [DOI] [PubMed] [Google Scholar]
- 148.Young Jeong J., Sperry J., Taylor J.A., Brimble M.A. Synthesis and evaluation of 9-deoxy analogues of (−)-thysanone, an inhibitor of HRV 3C protease. Eur. J. Med. Chem. 2014;87:220–227. doi: 10.1016/j.ejmech.2014.09.063. [DOI] [PubMed] [Google Scholar]
- 149.Lin Y.J., Chang Y.C., Hsiao N.W., Hsieh J.L., Wang C.Y., Kung S.H., Tsai F.J., Lan Y.C., Lin C.W. Fisetin and rutin as 3C protease inhibitors of enterovirus A71. J. Virol. Methods. 2012;182:93–98. doi: 10.1016/j.jviromet.2012.03.020. [DOI] [PubMed] [Google Scholar]
- 150.Chen Y.H., Du W., Hagemeijer M.C., Takvorian P.M., Pau C., Cali A., Brantner C.A., Stempinski E.S., Connelly P.S., Ma H.C., et al. Phosphatidylserine vesicles enable efficient en Bloc transmission of enteroviruses. Cell. 2015;160:619–630. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Roosien J., Belsham G.J., Ryan M.D., King A.M., Vlak J.M. Synthesis of foot-and-mouth disease virus capsid proteins in insect cells using baculovirus expression vectors. J. Gen. Virol. 1990;71:1703–1711. doi: 10.1099/0022-1317-71-8-1703. [DOI] [PubMed] [Google Scholar]