Stress granule condensation (SGC) of translationally arrested mRNAs requires G3BP, and G3BP-mediated SGC is inhibited by serine 149 phosphorylation, regulated by mutually exclusive binding of Caprin1 and USP10, and requires its RGG region for SGC and for interactions with 40S ribosomal subunits.
Abstract
Mammalian stress granules (SGs) contain stalled translation preinitiation complexes that are assembled into discrete granules by specific RNA-binding proteins such as G3BP. We now show that cells lacking both G3BP1 and G3BP2 cannot form SGs in response to eukaryotic initiation factor 2α phosphorylation or eIF4A inhibition, but are still SG-competent when challenged with severe heat or osmotic stress. Rescue experiments using G3BP1 mutants show that phosphomimetic G3BP1-S149E fails to rescue SG formation, whereas G3BP1-F33W, a mutant unable to bind G3BP partner proteins Caprin1 or USP10, rescues SG formation. Caprin1/USP10 binding to G3BP is mutually exclusive: Caprin binding promotes, but USP10 binding inhibits, SG formation. G3BP interacts with 40S ribosomal subunits through its RGG motif, which is also required for G3BP-mediated SG formation. We propose that G3BP mediates the condensation of SGs by shifting between two different states that are controlled by the phosphorylation of S149 and by binding to Caprin1 or USP10.
Introduction
Stress granules (SGs) are dynamic assemblies of stalled 48S preinitiation complexes, triggered by stresses causing polysome disassembly (Anderson and Kedersha, 2008, 2009; Buchan and Parker, 2009; Erickson and Lykke-Andersen, 2011). In mammalian cells, SG condensation is typically initiated when one of several stress-activated serine/threonine kinases phosphorylate eukaryotic initiation factor (eIF)2α to inhibit translation initiation (Kedersha et al., 1999), resulting in a sudden excess of mRNA released from polysomes. Alternatively, drugs targeting eIF4A, the DEAD-box helicase that allows the 48S preinitiation complex to reach the start codon, similarly trigger SG formation by inhibiting translation initiation (Bordeleau et al., 2005; Dang et al., 2006). Other proteins and posttranslational modifications of proteins acting downstream of translational arrest are required for SG assembly, including O-GlcNAc modification of ribosomal subunits (Ohn et al., 2008), and arginine methylation, polyribosylation, ubiquitin modification, acetylation, and phosphorylation of diverse SG proteins (Ohn and Anderson, 2010). In all cases, agents that stabilize polysomes prevent or reverse SG assembly, highlighting the dynamic relationship between SGs and polysomes (Kedersha et al., 2000).
SGs are dynamic entities that are transitional and temporary. Their formation is regulated by two related parameters: (a) SG-nucleating proteins, aggregating in response to overexpression (Gilks et al., 2004; Anderson and Kedersha, 2008), denaturing stresses (heat shock), or molecular crowding (osmotic stress), independent of phospho-eIF2α (p-eIF2α; Bevilacqua et al., 2010; Bounedjah et al., 2012), and (b) RNA, typically a sudden excess of nonpolysomal mRNA resulting from interrupted translation initiation (Bounedjah et al., 2014). Specific stresses and pharmacologic treatments also modulate signaling cascades (eIF2α kinases, eIF4F levels, and mTOR) that can affect either or both parameters to influence whether SGs form or dissolve.
Overexpression of some SG proteins nucleate SGs in the absence of stress or drugs (Kedersha and Anderson, 2007) and may provide mechanistic insights into various neurological pathologies associated with aggregation of SG proteins (Vanderweyde et al., 2013). Prion-like, low complexity (LC), and intrinsically unstructured/intrinsically disordered (ID) protein regions mediate protein aggregation (Tompa, 2002), and the SG protein TIA-1 contains a prion-like domain that forms insoluble aggregates when expressed alone (Gilks et al., 2004). TIA-1 requires both its prion-like domain and its RNA binding domains to assemble dynamic and reversible SGs (Gilks et al., 2004). Many properties associated with SGs and other RNA granules (concentration and temperature dependence, and fusion and fission) have led to an evolving “liquid–liquid phase separation” (LLPS) model (Brangwynne et al., 2009; Elbaum-Garfinkle et al., 2015) that posits that RNA granules are composed of immiscible liquid droplets, held apart from the cytosol by fleeting and multiple low-affinity interactions between LC/ID proteins and RNA (Weber and Brangwynne, 2012; Kroschwald et al., 2015; Lin et al., 2015; Molliex et al., 2015; Nott et al., 2015; Zhang et al., 2015). Recent in vitro studies with selected RNPs suggest that RNA granules adopt a spectrum of different structural states including more solid-like phases such as hydrogels (Han et al., 2012; Kato et al., 2012). The situation in cells is more complex; RNA granules, composed of hundreds of proteins and RNAs, show dynamic behavior (Kedersha et al., 2005; Bley et al., 2015) compatible with the LLPS model but also may exhibit aberrant transition to more static and solid-like states, as seen in neurodegeneration-associated protein inclusions (Patel et al., 2015). The relative contributions of protein–protein interactions and protein–RNA interactions in RNA granules and pathological aggregates are areas of active research.
G3BP1 or 2 (hereafter referred to jointly as G3BP) nucleate SG assembly, but the mechanism is unclear. Unlike many SG-nucleating proteins, G3BP lacks a prion-like domain. Although it contains predicted RNA-binding domains (RGG and RRM) and possesses RNA/DNA helicase activity in vitro (Costa et al., 1999), whether it directly binds specific mRNAs is not clear (Solomon et al., 2007). Importantly, phosphorylation of G3BP1 at S149 impairs both its ability to nucleate SGs and self-aggregate (Tourrière et al., 2003), suggesting that G3BP-mediated SG assembly results from phosphorylation-regulated homoaggregation. G3BP1 and G3BP2 are coexpressed in most cells as homodimers and heterodimers (Matsuki et al., 2013). G3BP interacts with many proteins, some of which are SGs nucleators (Caprin1; Solomon et al., 2007) and others that are not (OGFOD1; Wehner et al., 2010). Here we show that G3BP is essential for SG condensation initiated by p-eIF2α or eIF4A inhibition, but dispensable for SGs induced by hyperosmolarity or severe heat shock. G3BP-mediated SG assembly is inhibited by serine 149 phosphorylation, regulated by mutually exclusive binding of Caprin1 and USP10, and requires its RGG region that allows interactions with 40S subunits. Caprin1 binding to G3BP promotes SG formation, whereas USP10 binding to G3BP inhibits SG assembly. We propose a model in which G3BP shuttles between two states, possibly reflecting conformational changes, to mediate SG condensation.
Results
Role of G3BP and its partners in SG assembly
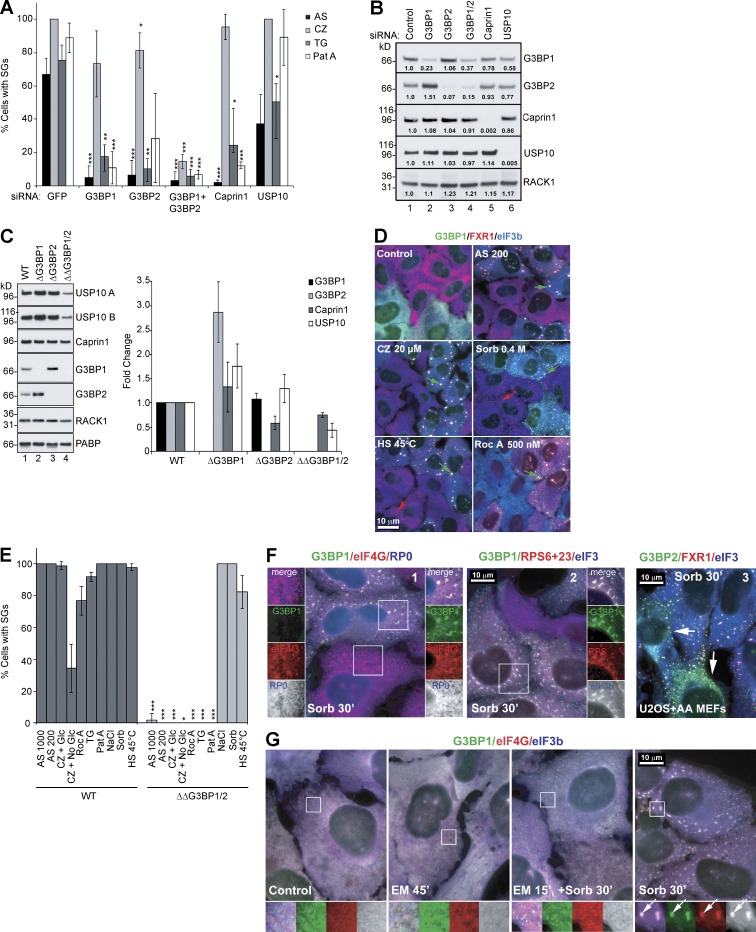
To understand the mechanistic role of G3BP and its partners Caprin1 and USP10 in SG formation, we used siRNA to knockdown (KD) expression of G3BP1, G3BP2, G3BP1 and G3BP2, Caprin1, or USP10 and then challenged cells with SG-inducing drugs and scored them for SGs using eIF4G and eIF3b as markers (Fig. 1 A). KD of either G3BP1 or G3BP2 alone significantly reduces sodium arsenite (AS)– and thapsigargin (TG)-induced SGs, but it is less effective at suppressing Pateamine A (Pat A)–induced SGs and largely ineffective against clotrimazole (CZ)-induced SGs. However, combined G3BP1/2 KD significantly reduces SGs in response to all treatments. Caprin1 KD inhibits SGs induced by AS, TG, and Pat A, but it is ineffective against CZ. USP10 KD does not significantly change SGs induced by AS, CZ, or Pat A, and it only modestly blocks TG-induced SGs. However, USP10 KD also reduces expression of G3BP1 and G3BP2 (to 58% and 77% of control values, respectively; Fig. 1 B), suggesting that the SG inhibition caused by USP10 KD may be caused by reduced levels of G3BP. In addition, KD of G3BP1 induces a compensatory increase in G3BP2, but not vice versa. G3BP, Caprin1, and USP10 all contain extensive predicted LC/ID regions (Fig. S1), which could contribute to SG assembly.
Figure 1.
Role of G3BP and partners in SG assembly. (A) U2OS-WT cells treated with indicated siRNAs were stressed for 1 h with 100 µM AS, 20 µM CZ, 1.0 µM TG, or 50 nM Pat A and then stained for SG markers eIF4G/eIF3b and scored. Data shown are mean ± SEM and are analyzed using the unpaired t test. *, P < 0.05; **, P < 0.01; ***, P < 0.005. siGFP control versus target siRNA treatments, n = 3. (B) Western blot analysis of siRNA-treated U2OS-WT cells. Proteins were quantified from blots using densitometry and normalized to RACK1, n = 3. Fold increase/decrease versus control is indicated. Mr (kD) are shown. (C) Western blot analysis of ΔG3BP1, ΔG3BP2, or ΔΔG3BP1/2 cell lysates, blotted for G3BP1, G3BP2, USP10 (A and B indicate different antibodies), Caprin1, RACK1, and PABP. Mr (kD) are shown. Proteins were quantified from blots using densitometry and normalized to RACK1. Fold change of WT versus KO was plotted. Data shown are mean ± SEM; no statistical analysis was performed, n = 4. (D) SGs in U2OS-WT and ΔΔG3BP1/2 cells, cocultured and treated as indicated, and then stained for G3BP1 (green), FXR1 (red), and eIF3b (blue). SGs in U2OS-WT cells (green arrows) and in ΔΔG3BP1/2 cells (red arrows) are indicated. Bar, 10 µm. (E) SG quantification in U2OS-WT cells (dark bars) and ΔΔG3BP1/2 (light bars), treated as indicated and scored for SGs using eIF4G and eIF3 as markers. Data shown are mean ± SEM and analyzed using the unpaired t test, WT versus ΔΔG3BP1/2. *, P < 0.05; ***, P < 0.005; n = 3. (F) Sorbitol-treated cocultured U2OS-WT (green) and ΔΔG3BP1/2 cells (1 and 2), stained as indicated. 3, sorbitol-treated cocultured mouse eIF2α-S51A MEFs (AA MEFs) and U2OS-WT, stained for G3BP2 (green), FXR1 (red), and eIF3b (blue). SGs in AA MEFs are indicated by white arrows. Insets zoomed 1.3× with separated colors. (G) U2OS-WT cells were untreated (a), EM treated for 45 s (b), EM alone for 15 s and then 0.4 M sorbitol for 30 s (c), or sorbitol alone for 30 s (d). Cells were stained for G3BP1 (green), eIF4G (red), and eIF3b (blue). Insets zoomed 2.3× with separated colors. Arrows indicate SGs. Bars, 10 µm.
To confirm the KD findings, we used an independent genetic approach based on CRISPR/Cas9 technology to create cells genetically ablated for G3BP1, G3BP2, or both G3BP1 and G3BP2 (Fig. 1 C and Fig. S2). Western blot analysis confirms the findings seen in the siRNA-treated cells (Fig. 1 B): ΔG3BP1 cells display 2.7-fold increase in G3BP2, whereas double-null ΔΔG3BP1/2 cells express reduced USP10 (∼50% of parental U2OS; Fig. 1 C). The ΔΔG3BP1/2 cells were tested for SG competence by coplating with parental (wild-type [WT]) U2OS (identified by staining for G3BP1), treating with the indicated stresses, and immunostaining for eIF4G or FXR1 and eIF3b as SG markers. As shown in Fig. 1 (D and E), ΔΔG3BP1/2 cells do not assemble SGs induced by agents causing p-eIF2α (AS, CZ, and TG) or inactivating eIF4A (Pat A and rocaglamide A; Sadlish et al., 2013). However, the ΔΔG3BP1/2 cells display small SG-like foci in response to hyperosmotic stress (NaCl, sorbitol) or robust (45°C for 45 min) heat shock (Fig. 1 D, green arrows show SGs in U2OS and red arrows indicate SGs in ΔΔG3BP1/2). Because the hyperosmotically induced foci appear smaller than other SGs, we examined them further. They contain eIF4G and small but not large ribosomal subunits (Fig. 1 F, 1 and 2), as do canonical SGs. They are independent of p-eIF2α because they form in mouse embryonic fibroblasts (MEFs) with knock-in eIF2α S51A mutation (Fig. 1 F, 3), consistent with previous studies (Bevilacqua et al., 2010; Bounedjah et al., 2012). Finally, they are prevented by emetine (EM)-induced polysome stabilization (Fig. 1 G). We conclude that hyperosmotically induced SGs are bona fide SGs that form independently of p-eIF2α and G3BP.
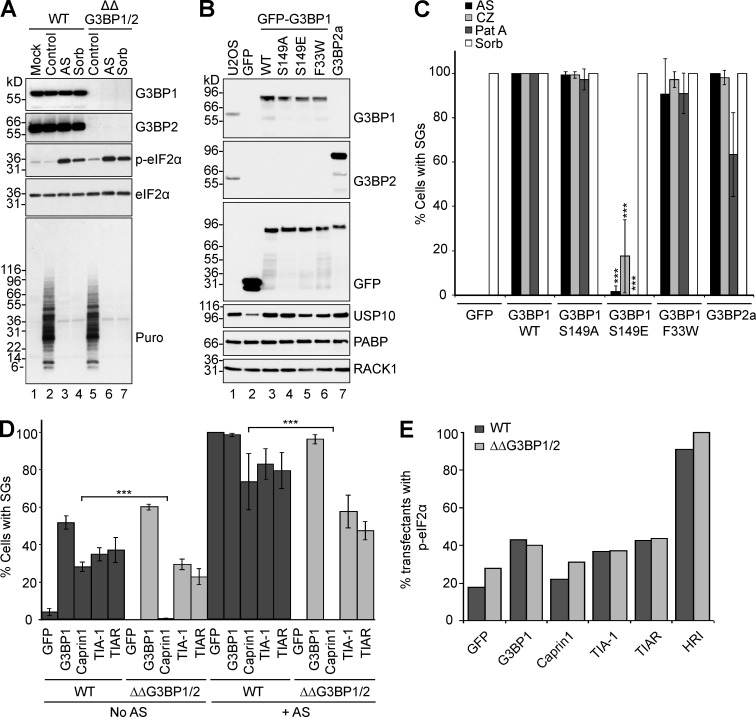
We next asked whether G3BP is required for stress-induced translational arrest. U2OS or ΔΔG3BP1/2 cells (Fig. 2 A) were stressed with AS (G3BP-dependent SGs) or sorbitol (G3BP-independent SGs), pulse-labeled with puromycin, and processed for Western blotting. Both stresses inhibit translation equally well in U2OS and ΔΔG3BP1/2 cells, as shown by reduced ribopuromycin incorporation (Fig. 2 A). Similarly, stress-induced phosphorylation of eIF2α is identical in U2OS and ΔΔG3BP1/2 cells, indicating that G3BP mediates SG formation but not translational arrest. We then attempted to rescue the SG response in ΔΔG3BP1/2 cells by genetic reconstitution with GFP, GFP-tagged WT, or mutant G3BP, using cloned cell lines expressing comparable levels of each protein (Fig. 2 B). Cells were stressed using AS, CZ, Pat A, or sorbitol, and SG formation was quantified. As shown in Fig. 2 C, SG competence is completely rescued by G3BP2, G3BP1-WT, and two mutants, (nonphospho)G3BP1-S149A and G3BP1-F33W (a Caprin1/USP10 nonbinding mutant; Reineke and Lloyd, 2015; Panas et al., 2015b). In contrast, the phosphomimetic G3BP1-S149E does not rescue SGs triggered by AS or Pat A and only partially restores CZ-induced SGs (Fig. 2 C). This agrees with an earlier study showing that G3BP1-S149E fails to nucleate SGs upon overexpression (Tourrière et al., 2003) and suggests that S149 comprises a regulatory switch. The sorbitol-induced SG response was unaffected. Collectively, the data indicate that G3BP is critical for SG assembly initiated via p-eIF2α and eIF4A, is regulated by S149 phosphorylation, but is dispensable for SG assembly induced by osmotic stress.
Figure 2.
G3BP is dispensable for translational arrest, required for SG formation, and regulated by S149. (A) U2OS-WT or ΔΔG3BP1/2 cells were untreated or exposed to 500 µM AS (1 h) or 0.4 M sorbitol (30 min) and lysed in SDS and resolved using SDS-PAGE/Western blotting. Blots were probed as indicated. Mr (kD) are shown. (B) ΔΔG3BP1/2 cells stably expressing the indicated constructs, analyzed by Western blot and probed as indicated. Mr (kD) are shown. (C) ΔΔG3BP1/2 cells stably expressing the indicated proteins, treated as indicated, stained, and scored for SGs and quantified. Data shown are mean ± SEM and analyzed using the unpaired t test, WT versus G3BP1-S149E. ***, P < 0.005; n = 3. (D) U2OS-WT (dark bars) or ΔΔG3BP1/2 cells (light bars) were transiently transfected with indicated plasmids, untreated or exposed to 500 µM AS for 1 h, and then stained and scored for SGs. Data shown are mean ± SEM and analyzed using the unpaired t test. ***, P < 0.005; n = 3. (E) U2OS-WT (dark bars) or ΔΔG3BP1/2 cells (light bars) were transiently transfected with indicated plasmids and then stained and scored for p-eIF2α–positive transfectants. Data represent mean. n = 2.
Transient overexpression of G3BP or Caprin1 nucleates SG assembly (Tourrière et al., 2003; Kedersha et al., 2005; Shiina et al., 2005; Solomon et al., 2007; Panas et al., 2012). We then asked whether transiently transfected GFP-tagged G3BP1 or Caprin1 nucleate SGs in ΔΔG3BP1/2 cells, compared with TIA-1 and TIAR, proteins that nucleate SGs but do not interact with G3BP (Fig. 2 D). Enforced expression of all of these proteins induces SGs in U2OS-WT cells (dark bars). In ΔΔG3BP1/2 cells (light bars), G3BP1 nucleates SGs in 60% of transfectants and fully rescues SGs in 100% of transfectants with AS treatment. Despite inducing similar levels of p-eIF2α in ΔΔG3BP1/2 cells as in U2OS (Fig. 2 E), Caprin1 overexpression does not rescue SG formation in ΔΔG3BP1/2 cells, without or with AS treatment (Fig. 2 D, light bars). In contrast, TIA-1 or TIAR overexpression nucleates SGs in the ΔΔG3BP1/2 cells, but does not rescue SG competence to 100%. We conclude that Caprin1 requires G3BP to nucleate SGs, but that TIA-1/TIAR overexpression assembles SGs via a G3BP-independent mechanism.
USP10 and Caprin1 antagonistically bind G3BP to regulate SG formation
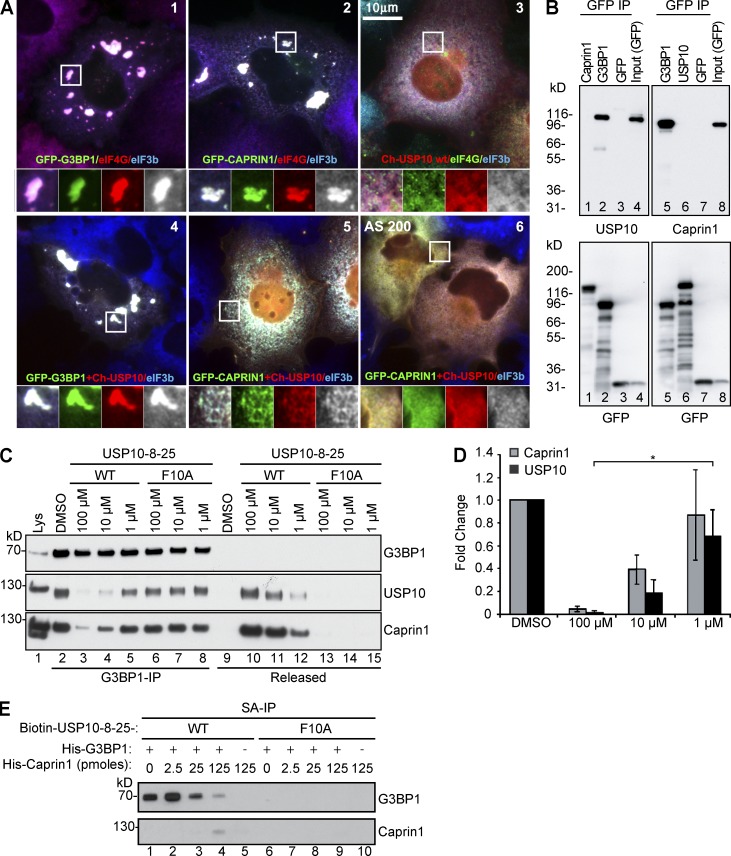
Our data suggest that G3BP is necessary to mediate SG assembly via p-eIF2α and eIF4A. To examine the roles of its interacting partners Caprin1 and USP10, we assessed their SG nucleating abilities alone or in combination. As expected, overexpressed GFP-G3BP1 (Fig. 3 A, 1) or GFP-Caprin1 (Fig. 3 A, 2) nucleates SGs, whereas overexpressed mCherry-USP10 (Fig. 3 A, 3) fails to nucleate SGs, and instead prevents AS-induced SG assembly (Fig. 3 A, 6). Therefore, although Caprin1 and USP10 both bind G3BP (Soncini et al., 2001; Solomon et al., 2007), they have antagonistic effects on SGs. When mCherry-tagged USP10 is cotransfected with GFP-G3BP1, both proteins colocalize in SGs (Fig. 3 A, 4), but mCherry-USP10 coexpressed with GFP-Caprin1 inhibits SGs even upon AS treatment (Fig. 3 A, 5 and 6), suggesting that USP10 and Caprin1 compete for limiting G3BP. To address this possibility, GFP-G3BP1, GFP-Caprin1, and GFP-USP10 were individually expressed, and each protein was immunoprecipitated from RNase-treated lysates and immunoblotted (Fig. 3 B). Whereas GFP-G3BP1 co-immunoprecipitates (IPs) both USP10 and Caprin1 (Fig. 3 B, lanes 2 and 5), GFP-Caprin1 does not co-IP USP10 and GFP-USP10 does not co-IP Caprin1 (lanes 1 and 6), suggesting that USP10 and Caprin1 are in separate G3BP-containing complexes, consistent with G3BP structural studies showing that USP10 and Caprin1 binding sites on G3BP are adjacent or overlapping (Vognsen et al., 2013; Panas et al., 2015b). USP10 contains a Phe-Gly-Asp-Phe (FGDF) motif that mediates G3BP binding and SG inhibition (Panas et al., 2015b), but Caprin1 has no FGDF motif. To assess whether Caprin1/USP10 binding sites into G3BP are adjacent or overlapping, IPs of endogenous G3BP1 were incubated with increasing amounts of USP108–25WT peptide containing the FGDF motif, or with USP108–25F10A, in which the G3BP-binding motif is inactivated by mutation (Fig. S1). Released and bound material was immunoblotted for Caprin1, G3BP1, and USP10. The WT FGDF peptide displaces both Caprin1 and USP10 from immobilized G3BP1, but mutant F10A peptide does not, suggesting that Caprin1 and USP10 bind within the same region of G3BP (Fig. 3, C and D). To obtain evidence of direct protein–protein interactions, recombinant His-tagged G3BP1 and His-Caprin1 were expressed in bacteria and purified to homogeneity (Fig. S5 C). His-G3BP1 was incubated with biotin-USP108–25WT or mutant peptide, bound to streptavidin beads, washed, incubated with increasing amounts of His-Caprin1, and then washed and subjected to SDS-PAGE (Fig. 3 E). This shows that Caprin1 prevents G3BP1 from binding to immobilized USP108–25WT in a dose-dependent manner, confirming that G3BP1–USP10 and G3BP1–Caprin1 complexes result from direct protein–protein binding and that competition between Caprin1 and USP10 for G3BP1 occurs in solution as well as in cells.
Figure 3.
Caprin1 and USP10 binding to G3BP is mutually exclusive and regulates SG formation. (A) COS7 cells were transiently transfected with the indicated GFP (green) or Cherry (red)-tagged constructs and stained for endogenous eIF4G (1 and 2, red; 3, green) and/or eIF3b (1–6, blue). In 6, the cells were treated with 200 µM AS and then fixed and stained. Bar, 10 µm. Insets zoomed 2.5× with separated colors. (B) COS7 were transiently transfected with the indicated constructs, lysed and immunoprecipitated. Lysates and IPs were resolved using SDS-PAGE and subjected to Western blotting for the indicated proteins. Lower panels, duplicate samples probed for GFP to confirm IP efficiency. Mr (kD) are shown. (C) Endogenous G3BP1 from U2OS-WT cell lysates was immunoprecipitated, and the bound complexes were incubated with the indicated amounts of USP108–25WT or mutant USP108–25F10A peptides. Bound and released material was subjected to Western blotting for endogenous G3BP1, USP10, or Caprin1. (D) Quantification of displaced Caprin1 and USP10. Western blots from samples in C were quantified using densitometry. The ratio of each protein relative to G3BP1 was determined. Data shown are mean ± SEM and are analyzed using the unpaired t test. *, P < 0.05; n = 3. (E) Purified recombinant His-G3BP was mixed with biotinylated USP108–25WT or mutant USP108–25F10A peptides and bound to streptavidin (SA) beads. Beads were then washed and incubated with the indicated amounts of purified recombinant His-Caprin1. Bound material was subjected to Western blotting for G3BP1 and Caprin1. n = 3. Mr (kD) are shown.
USP10 inhibits SG formation downstream of polysome disassembly
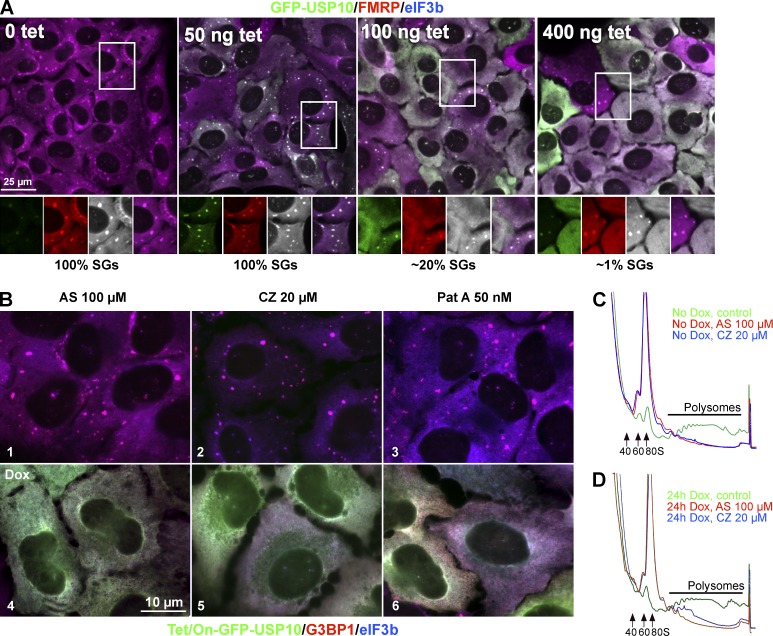
USP10 overexpression blocks SG assembly (Fig. 3 A), either by preventing polysome disassembly or by preventing mRNP condensation. To distinguish between these possibilities, we generated cells expressing tet-inducible GFP-USP10 and induced expression using increasing amounts of tetracycline. At low expression levels, GFP-USP10 is recruited into SGs, whereas higher levels of GFP-USP10 inhibit SG formation (Fig. 4 A), indicating that USP10 prevents or reverses SG formation in a dose-dependent manner. Uninduced tet-on GFP-USP10 cells assemble SGs normally in response to AS, CZ, or Pat A (Fig. 4 B, 1–3), whereas GFP-USP10 induction completely inhibits SG formation in response to these same treatments (Fig. 4 B, 4–6). To exclude the possibility that SGs form but disassemble before staining, we performed time-lapse microscopy using mixed cultures of cells expressing GFP-USP10 or mCherry-G3BP1 (Videos 1 and 2). As expected, cells expressing mCherry-G3BP1 exhibit SG formation, fusion, and enlargement when triggered by AS or Pat A. However, SGs do not form in GFP-USP10 overexpressing cells at any time.
Figure 4.
USP10 blocks SG assembly downstream of translational arrest. (A) Tet-on GFP-USP10 U2OS-WT cells were treated with increasing amounts of tetracycline for 22 h and then treated with 100 µM AS for 1 h and fixed and stained with SG markers (FMRP [FMR1], red; eIF3b, blue). Bar, 25 µm. Insets zoomed 1.3× with separated colors. The percentage of cells with SGs is indicated below each panel; n = 3. (B) Tet-on GFP-USP10 was uninduced (1–3) or induced with doxycycline (Dox) for 24 h (4–6) before indicated 1-h drug treatments. Cells were treated with 100 µM AS, (1 and 4), 20 µM CZ, (2 and 5), or 50 nM Pat A (3 and 6) and then stained for G3BP1 (red) and eIF3b (blue). Bar, 10 µm. (C) Polysome profiles obtained from uninduced tet-on GFP-USP10 cells as in B (1–3) and treated with control (green), 100 µM AS (red), or 20 µM CZ (blue); n = 3. (D) Polysome profiles of doxycycline-induced tet-on GFP-USP10 cells, treated as in B (4–6); n = 3.
We then asked whether USP10 overexpression prevents SG assembly by inhibiting polysome disassembly. In uninduced tet-on GFP-USP10 cells, AS and CZ induce SGs (Fig. 4 B), and cause polysome disassembly relative to unstressed control (Fig. 4 C). Tet-induced GFP-USP10 expression prevents AS and CZ induction of SGs (Fig. 4 B) but does not alter stress-induced polysome disassembly (Fig. 4 D), indicating that GFP-USP10 inhibits the cytoplasmic condensation of mRNPs into SGs. Because both USP10 overexpression and G3BP deletion similarly prevent SG formation, the data suggest that USP10 inhibits G3BP-mediated SG assembly downstream of translational arrest/polysome disassembly.
USP10 binding to G3BP1/2 via the FGDF motif inhibits SG formation
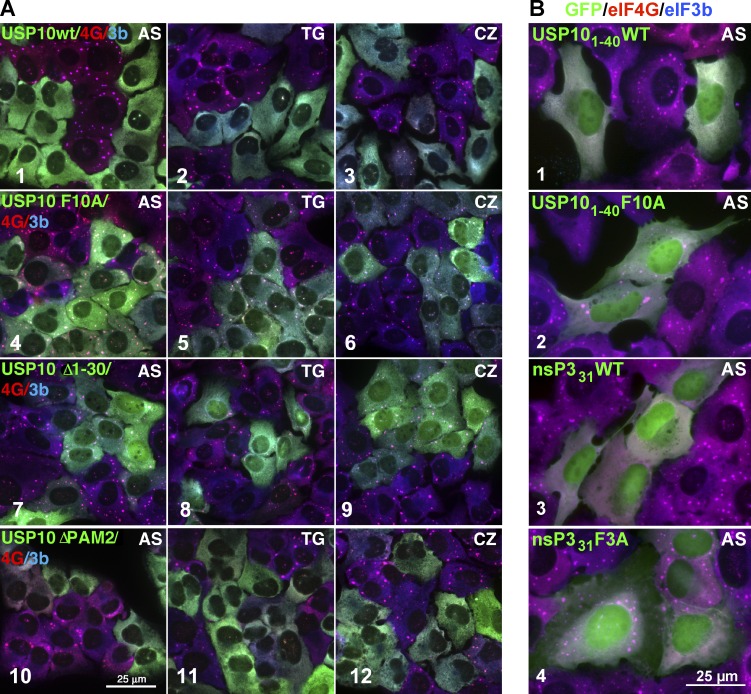
The FGDF motif in USP10 (aa 10–13) mediates G3BP binding and inhibits SG assembly (Panas et al., 2015b). The F10A mutation in full-length USP10 or a USP101–40 fragment completely prevents binding to G3BP (Fig. S4, A–C). USP10 also contains a PABP-binding site (PAM2 motif), adjacent to the FGDF motif. We then asked whether the PAM2 motif contributes to USP10-mediated SG inhibition. GFP-USP10 (WT) co-IPs endogenous G3BP1, G3BP2, PABP, and RPS6 (Fig. S4, A, 2), but not RPL4. Overexpressed USP10-WT inhibits AS-, TG-, or CZ-induced SGs (Fig. 5 A, 1–3), but mutant USP10-F10A does not (Fig. 5 A, 4–6). A truncation mutant lacking aa 1–30 (USP10-Δ1–30) does not bind G3BP, but co-IPs PABP and eIF4G (a PABP-binding protein; Otero et al., 1999; Fig. S4), yet does not inhibit SG assembly (Fig. 5 A, 7–9). USP10 lacking the PAM2 motif does not co-IP PABP or eIF4G, but still binds G3BP (Fig. S4, A–C) and blocks SGs (Fig. 5 A, 10–12). GFP-USP101–40 binds G3BP1 and G3BP2, but does not bind PABP (Fig. S4 A), confirming that the FGDF motif mediates USP10:G3BP binding and GFP-USP101–40 blocks SG formation (Fig. 5 B). Thus, the N-terminal 40 aa of USP10 containing the FGDF motif is sufficient to inhibit SG formation, and the functional PAM2 motif binds PABP but is not required for SG inhibition. Our data conflict with a previous study that USP10 knockout (KO) inhibits SG formation without affecting murine G3BP1 levels (Takahashi et al., 2013). However, this “USP10 KO” was an exon 3 deletion; the FGDF motif is encoded within exon 2 (Fig. S1). Cells with an exon 3 deletion may express an FGDF-containing inhibitory fragment of USP10, which would suppress SGs as does USP101–40 (Fig. 5 B) and explain the apparent contradiction.
Figure 5.
FGDF motif of USP10 or nsP331 is required to block SG formation. (A) U2OS-WT cells stably expressing GFP-tagged USP10-WT, F10A, Δ1–30, and ΔPAM2 (green) were cocultured with U2OS-WT cells (nongreen), treated with 200 µM AS (1, 4, 7, and 10), 1 µM TG (2, 5, 8, and 11), or 20 µM CZ (3, 6, 9, and 12) and stained for SG markers eIF4G (red) and eIF3b (blue). Bar, 25 µm. (B) U2OS-WT cells transiently transfected with GFP-USP101–40WT, GFP-USP101–40-F10A, GFP-nsP331WT, or GFP-nsP331F3A as indicated and treated with 500 µM AS for 1 h, fixed, and stained for eIF4G (red) and eIF3b (blue). Bar, 25 µm.
USP10 contains one FGDF motif, whereas Semliki Forest virus (SFV) nonstructural protein 3 (nsP3) contains two functional FGDF motifs (Fig. S1; Panas et al., 2015b). We tested F10A mutant versions of full-length and USP101–40 to confirm that this motif mediates binding to G3BP. Both full-length GFP-USP10-F10A and GFP-USP101–40F10A fail to bind G3BP, unlike their WT counterparts (Fig. S4 A, compare 3 and 7 with 2 and 6). Moreover, the USP10-F10A mutants fail to inhibit SG formation induced by AS, TG, or CZ (Fig. 5 A, 4–6). USP101–40WT blocks AS-induced SGs, whereas the USP101–40F10A does not (Fig. 5 B, 1 and 2). Similarly, WT nsP331 (containing two FGDF motifs) blocks SG formation, whereas the F3A mutant (in which both FGDF motifs are mutated) does not (Fig. 5 B, 3 and 4). Thus, G3BP binding to FGDF motifs in USP10 or nsP3 is critical for inhibiting SG assembly.
USP10/nsP3 binding alters G3BP Biotin-isoxazole solubility
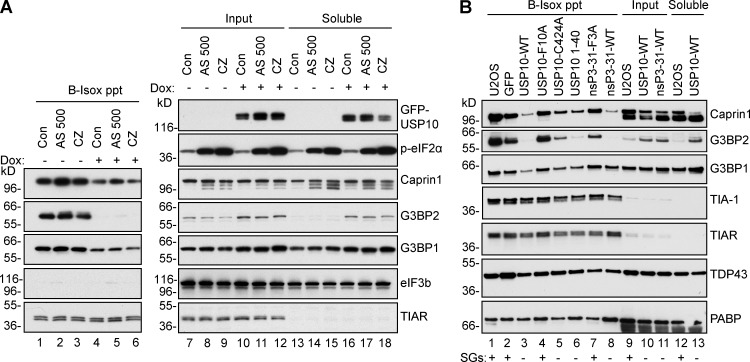
RNA granule formation appears linked to LC/ID sequences common to many RNA binding proteins. G3BP, Caprin1, and USP10 all contain extensive LC/ID regions (Fig. S1). Biotin-isoxazole (B-isox) is a reagent that coprecipitates many SG-associated proteins by interacting with LC/ID regions (Han et al., 2012; Kato et al., 2012), making it a useful tool to assess one property of SG-associated proteins. To determine the fraction of each protein that is B-isox precipitated, we lysed cells in ribosome-dissociating EDTA-EGTA (EE) buffer and compared input with the postprecipitation soluble material. B-isox precipitates 75–80% of Caprin1 and G3BP2 (Fig. S3, compare lanes 1–4 with 9–12), but only a trace fraction of USP10 and eIF3b. Other SG-associated proteins are differentially precipitated with B-isox, falling into three classes: largely or entirely soluble, which includes eIF3b, USP10, PABP, RPS6, RPS23, and RACK1; intermediately precipitated (50–80%), such as Caprin1, G3BP2, and TDP43; and largely/entirely precipitated (TIAR). Pretreatment of cells to induce SGs using CZ has no effect on the B-isox solubility of any protein (Fig. S3), suggesting that B-isox precipitation acts at the molecular level. To determine whether stress alters B-isox binding under ribosome-stabilizing conditions, we performed B-isox precipitation using 5 mM Mg-containing lysis buffer (Kato et al., 2012) and tet-inducible GFP-USP10 cells with or without induction (Fig. 6 A). These results show that the relative B-isox solubility of TIAR and TIA-1 (almost entirely precipitated by B-isox), G3BP, and Caprin1 (largely precipitated), or not precipitated (eIF3b), is unaltered by these buffer conditions, or by AS or CZ pretreatment (Fig. 6 A). Because G3BP binds many proteins, we note that its B-isox precipitation could result either from G3BP binding to other proteins that in turn bind directly to B-isox, or by direct G3BP–B-isox interactions.
Figure 6.
USP10/nsP3 binding to G3BP inhibits B-isox precipitation of G3BP and Caprin1. (A) Tet-on GFP-USP10 cells without (lanes 1–3, 7–9, and 13–15) or with induction (lanes 4–6, 10–12, and 16–18) were treated with AS (500 µM) or CZ (20 µM), or were untreated, before lysis in Kato buffer and B-isox fractionation. Precipitated material (lanes 1–6), input (lanes 7–12), and soluble material (lanes 13–18) were subjected to Western blotting for the indicated proteins. Mr (kD) are shown. (B) U2OS-WT cells stably expressing indicated GFP-tagged proteins were lysed in EE buffer, fractionated using B-isox, and Western blotted for the indicated proteins. SGs or lack thereof are indicated at the bottom. Lanes 1–8, precipitates; lanes 9–11, input; and lanes 12 and 13, soluble material. The C424A USP10 mutant is enzymatically inactive. Mr (kD) are shown.
Because USP10 overexpression blocks SG assembly in a manner that requires G3BP binding (Figs. 4 and 5), we asked whether USP10 overexpression alters the physical state of G3BP, using B-isox solubility as an assay. GFP-USP10 overexpression inhibits B-isox precipitation of G3BP2 (Fig. 6 A, lanes 4–6 vs. 1–3) and modestly decreases that of G3BP1 and Caprin1, whereas that of TIAR is unchanged. Expression of GFP-USP10 (lanes 4–6 and 16–18) increases G3BP2 expression (Fig. 6 A, lanes 10–12 vs. 7–9), but the increased G3BP2 is not precipitated by B-isox (lanes 4–6), suggesting that B-isox binds and/or precipitates a SG-competent form of G3BP that is rendered SG-incompetent and B-isox soluble upon USP10 binding. To test this hypothesis, we asked whether FGDF binding to G3BP alters B-isox solubility and whether this correlates with SG inhibition. Lysates from stable cell lines expressing GFP-tagged forms of USP10, its derivatives, or minimal G3BP-binding fragments of SFV nsP3 were precipitated with B-isox. G3BP-binding/SG-inhibiting variants of USP10 and SFV-nsP3 (Fig. 6 B, lanes 3, 5, 6, and 8) selectively decrease G3BP1/2 and Caprin1 B-isox precipitation without affecting that of TIA-1, TIAR, TDP43, or PABP. In contrast, G3BP-nonbinding USP10 or nsP3 mutants (Fig. 6 B, lanes 4 and 7) fail to alter G3BP1/2 or Caprin1 solubility. The data suggest that FGDF-mediated binding to G3BP alters the physical state of G3BP to prevent its precipitation/coprecipitation with B-isox, and this change in G3BP correlates with SG competence.
G3BP associates with 40S ribosomal subunits
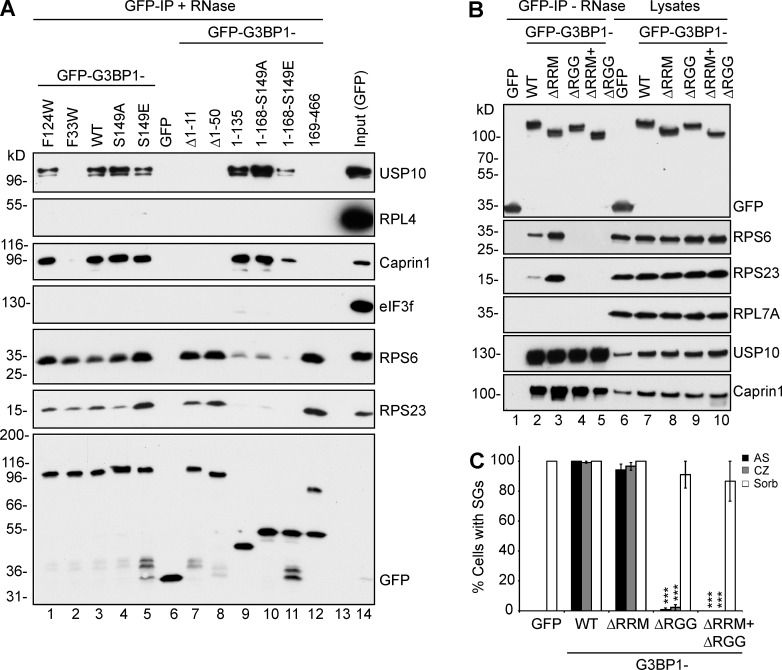
The finding that G3BP is required for SGs triggered by p-eIF2α or inhibition of eIF4A, events that directly alter translation initiation, led us to ask whether G3BP interacts with translation machinery components, such as eIF3 and ribosomes. GFP-tagged G3BP1-WT and mutant constructs (Fig. S1) were transiently expressed, immunoprecipitated from RNase-treated lysates, resolved using SDS-PAGE, and immunoblotted to identify coimmunoprecipitated proteins (Fig. 7 A). As expected, GFP-G3BP1-WT and a silent G3BP1 mutant F124W (Fig. 7 A, lanes 3 and 1; Vognsen et al., 2013; Panas et al., 2015b) co-IP endogenous USP10 and Caprin1. In contrast, G3BP1 mutants (G3BP1Δ1–11, G3BP1Δ1–50, and site-specific mutant G3BP1-F33W) unable to bind USP10 (Panas et al., 2015b) or Caprin1 (Reineke et al., 2015) fail to precipitate either protein (Fig. 7 A, lanes 2, 7, and 8). No G3BP1 variants co-IP eIF3 or the 60S ribosomal subunit protein RPL4. However, small ribosomal subunit proteins RPS6 and RPS23 coimmunoprecipitated with GFP-G3BP1-WT and GFP-G3BP1169–466 (Fig. 7 A, lanes 3 and 12). Coimmunoprecipitation of 40S proteins is not inhibited by G3BP1 mutations (F33W, Δ1–11, and Δ1–50) that abrogate USP10/Caprin1 binding (Fig. 7 A, lanes 2, 7, and 8) or by mutation of S149 (lanes 4 and 5). The finding that G3BP1 interacts with small ribosomal subunit proteins but not eIF3 is surprising because eIF3 is a SG-component that binds 40S subunits and is recruited to G3BP1-nucleated SGs. To avoid possible artifacts caused by overexpressed mutant G3BP forming dimers with endogenous G3BP-WT (Matsuki et al., 2013), we performed additional studies in the ΔΔG3BP1/2 cells. Because the 40S-interacting region of G3BP1 (residues 169–466) contains two putative RNA binding regions (one RRM domain and one RGG region; Fig. S1), we deleted the RRM domain, the RGG region, or both (Fig. S1) and then stably expressed the truncations in ΔΔG3BP1/2 cells. IP analysis reveals that the RGG region is required for association with the 40S subunits (Fig. 7 B, lane 4), whereas the RRM is not. Surprisingly, deletion of the RRM region enhances the G3BP:40S interaction (Fig. 7 B, lanes 3 and 2). The RGG domain is required to rescue AS- or CZ-induced SGs, whereas the RRM domain is dispensable (Fig. 7 C), suggesting that G3BP mediates SG assembly via RGG-mediated interaction with 40S ribosomal subunits.
Figure 7.
G3BP1 RGG motif is required for association with 40S ribosomal subunits and for SG competence. (A) GFP and GFP-tagged G3BP1 variants were expressed in COS7 cells, lysed in EE buffer, and immunoprecipitated with RNase treatment. Lysates and IPs were resolved using SDS-PAGE and subjected to Western blotting for the indicated proteins. Mr (kD) are shown. (B) GFP and GFP-tagged G3BP1 variants were stably expressed in ΔΔG3BP1/2 cells and immunoprecipitated. Lysates and IPs were resolved using SDS-PAGE and subjected to Western blotting for the indicated proteins. Mr (kD) are shown. (C) ΔΔG3BP1/2 cells expressing the indicated proteins were treated as indicated, stained, and scored for SGs using eIF4G and eIF3b. Data shown are mean ± SEM and are analyzed using the unpaired t test, WT versus G3BP1 variants. ***, P < 0.005; n = 3.
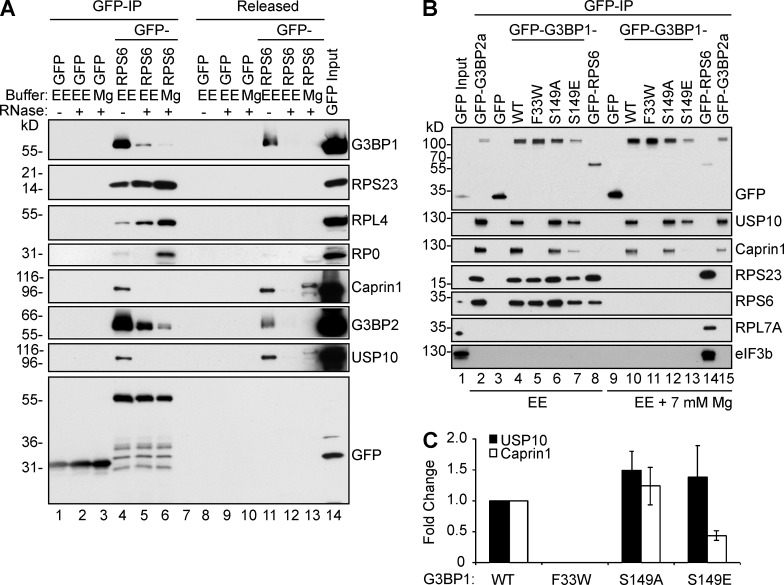
To validate the G3BP:40S interaction, we created U2OS cell lines stably expressing RPS6-GFP, which is functionally incorporated into ribosomes (Fig. S5 A) and recruited to SGs (Fig. S5 B). Cell lysates from the RPS6-GFP cells were made in either ribosome-dissociating EE buffer or ribosome stabilizing buffer (Mg2+), and RPS6-GFP complexes were immunoprecipitated. The bound complexes were treated with/without RNase A to distinguish between RNA-dependent and RNA-independent interactions. Fig. 8 A shows that (a) RPS23 remains associated with GFP-RPS6 under all conditions (lanes 4–6), indicating that the 40S subunit remains at least partially intact. (b) Co-IP of large ribosomal proteins RPL4 and RPLP0 requires Mg2+ (Fig. 8 A, compare lane 4 with 5) to keep 80S ribosomes intact. (c) The 40S interaction with G3BP1, G3BP2, Caprin1, and USP10 is restricted to EDTA-dissociated 40S subunits and does not occur when Mg2+ is present to stabilize 80S monosomes (Fig. 8 A, compare lanes 4 and 6). (d) The binding of G3BP to RPS6-GFP is partly RNase resistant (lane 5). Thus, G3BP associates preferentially to 40S ribosomal subunits rather than 80S ribosomes, and both Caprin1 and USP10 are bound to 40S-associated G3BP. A similar analysis was performed using the ΔΔG3BP1/2 cells engineered to express GFP or GFP-G3BP1 mutants (Fig. 8 B), lysed with or without Mg2+ in the absence of RNase. The results confirm that G3BP1 (lane 4) and G3BP2 (lane 2) interact only with dissociated 40S subunits (Fig. 8 A, compare lanes 2 and 4–7 with 10–13 and 15) and that the G3BP:40S association is independent of binding to Caprin1 or USP10 (G3BP1-F33W; Fig. 8 A, lane 5). The interaction between 40S subunits and G3BP is partially RNase labile (Fig. 8 A, compare lanes 4 and 5); hence, it is formally possible that G3BP interacts with mRNA bound to EDTA-dissociated 40S subunits but not to intact, Mg-stabilized 80S ribosomes (Fig. 8, A [lanes 4–6] and B). Another explanation is that G3BP binds directly to rRNA on the interface side of the 40S ribosome, which is not accessible in 80S ribosomes. Further studies using purified proteins and ribosomal subunits will be necessary to resolve this point. The data also show that S149E preferentially binds USP10 relative to Caprin1 (Fig. 8 C), possibly explaining the inability of G3BP-S149E to fully rescue SG competence in the ΔΔG3BP1/2 cells (Fig. 2 C).
Figure 8.
G3BP1 and G3BP2 selectively associate with dissociated 40S ribosomal subunits. (A) Western blot of IPs from cells expressing either GFP (lanes 1–3) or RPS6-GFP (lanes 4–6), lysed in either ribosome-dissociating (EE) or ribosome-stabilizing (+5.0 mM MgCl2) buffer, washed, and incubated with RNase A or buffer. Released material was concentrated using acetone precipitation before SDS treatment; the bound material (GFP-IP) was eluted in SDS. IPs, lanes 1–6; released material, lanes 8–13. Mr (kD) are shown. (B) Stably expressed GFP-RPS6 in U2OS-WT cells, or GFP, GFP-tagged G3BP1 variants, or G3BP2a stably expressed in ΔΔG3BP1/2 cells were lysed in ribosome-dissociating EE or in ribosome stabilizing (EE + 7 mM MgCl2) buffer and immunoprecipitated and Western blotted as indicated. Mr (kD) are shown. (C) Caprin1, USP10, and GFP were quantified from EE/IP Western blots (C) using densitometry. The ratio of Caprin1 or USP10 relative to GFP-G3BP1 was determined, and the fold change was plotted. Data shown are mean ± SEM; no statistical analysis was performed; n = 3.
Discussion
G3BP1/2 are established regulators of SG assembly: (a) overexpression of either protein nucleates SGs (Tourrière et al., 2003; Matsuki et al., 2013), (b) KD of either inhibits SGs (White et al., 2007; Aulas et al., 2015), (c) specific viral proteins bind G3BP to inhibit SG formation and thus enhance SFV viral replication (Panas et al., 2012, 2015b), and (d) other viruses cleave G3BP1 to disrupt SG assembly (Piotrowska et al., 2010; Lloyd, 2013). We now show that (a) deletion of G3BP1/2 ablates SGs triggered by p-eIF2α or eIF4A inhibition, but not those induced by hyperosmolarity or severe heat shock, (b) the phosphomimetic G3BP1-S149E fails to rescue SG competence, (c) binding of G3BP to Caprin1 or USP10 is not required for SG competence, but may alter G3BP and USP10 protein levels, (d) Caprin1 and USP10 bind G3BP1/2 in a mutually exclusive manner with antagonistic effects on SGs, (e) G3BP1 interacts with 40S ribosomal subunits through its RGG motif, which is required for SG competence, (f) overexpression of USP10 inhibits SG formation downstream of polysome disassembly, and (g) USP10/FGDF binding to G3BP specifically alters the B-isox precipitation of G3BP1/2 and Caprin1, but not that of other SG-associated proteins.
Based on these data, we propose a model of G3BP–Caprin1–USP10 interactions in SG assembly (Fig. 9). G3BP is essential for SGs driven by “RNA excess” and requires its RGG motif for SG competence. Association of G3BP with 40S ribosomal subunits distinguishes G3BP from other SG-nucleating proteins, such as TIA-1/TIAR, proteins with sequence-specific mRNA binding properties enabling them to silence certain transcripts (Piecyk et al., 2000; Mazan-Mamczarz et al., 2006) and could enable G3BP to serve as a sentinel for stalled translation complexes. When G3BP is in excess because of transient transfection, or when stalled preinitiation complexes exceed a threshold, 40S:G3BP oligomers may synergize with other SG-nucleating mRNA binding proteins (Caprin1 or TIA-1 and TIAR) to select and coalesce specific transcripts. Coordinated aggregation of 40S subunits and associated mRNA may then effect SG condensation via LLPS. Hyperosmotic stress may bypass the requirement for G3BP by decreasing cell volume, thereby increasing the local concentration of both cellular proteins and mRNAs (Bounedjah et al., 2014), making G3BP dispensable. A role for G3BP as a condensase is consistent with its historical classification as “DNA/RNA helicase VIII” (Costa et al., 1999). It will be important to determine whether G3BP exhibits helicase activity in the context of SG formation, whether G3BP binds 40S subunits directly or other factors are involved, and whether G3BP transiently interacts with all 40S subunits or only with a specialized subset.
Figure 9.
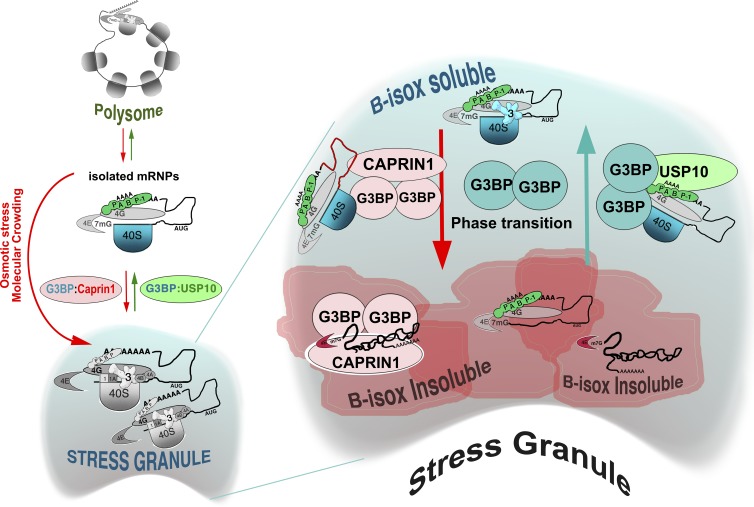
USP10 binding to G3BP regulates SG condensation by inhibiting a G3BP-mediated condensation event. Stress promotes polysome disassembly, thus exposing mRNA and converting polysomes into mRNPs. G3BP shuttles between two different phases, promoting a similar state change in 40S subunits, Caprin1, and their bound mRNAs. USP10 binding to G3BP stabilizes a soluble conformation of G3BP bound to 40S subunits (via G3BP C terminus) and to PABP (through USP10), causing SG disassembly. Some SG-associated factors, such as eIF3, remain in the cytoplasmic soluble state, but accumulate in the SG as individual mRNPs are mobilized back into active translation. For further details, see the Discussion section.
Overexpression of TIA-1 or TIAR partially rescues SGs in G3BP-null cells, but fails to rescue SG competence to 100% as does G3BP1. G3BP1-F33W is SG competent despite its inability to bind Caprin1 or USP10, suggesting that G3BP is only regulated by Caprin1/USP10. Caprin1 binds and silences specific mRNAs (Solomon et al., 2007), but unlike TIA-1/TIAR, its overexpression does not rescue SGs in the G3BP-null cells, suggesting that Caprin1 acts through G3BP to cause SG assembly. It was reported previously that overexpressed Caprin1 does not need G3BP to nucleate SGs because a GFP-Caprin1 fragment (GFP-Caprin1381–709) lacking the G3BP1-binding region formed granules when overexpressed (Solomon et al., 2007; Fig. 8 B). However, the GFP-Caprin1381–709 granules were not identified as SGs by counterstaining for SG markers nor by forced disassembly with elongation inhibitors, so whether these aggregates were in fact SGs is unclear.
USP10/FGDF overexpression acts downstream of polysome disassembly to block SG assembly while altering G3BP’s physical state, as assessed by coprecipitation with B-isox. B-isox-mediated precipitation is selective for LC regions and mediated through the formation of B-isox microcrystals that serve as templates to convert LC/ID regions into an insoluble form (Han et al., 2012; Kato et al., 2012). B-isox distinguishes between G3BP1/2 bound to FGDF proteins (SG incompetent), and G3BP1/2 that is not (SG competent), suggesting that FGDF binding alters the physical state of G3BP, in parallel with inhibiting SG formation in vivo. G3BP rapidly shuttles in and out of SGs (Kedersha et al., 2005; Bley et al., 2015) and is not part of a fixed SG scaffold. We speculate that the spatial shuttling in/out of SGs requires that G3BP shifts between conformations, one of which is B-isox soluble (USP10 bound) and one of which is B-isox insoluble (Caprin1-bound). Increasing USP10 to levels where it out-competes Caprin1 (and possibly other G3BP-binding proteins) may “lock” G3BP in one (B-isox soluble) conformation, unable to mediate the condensation of mRNPs into SGs. The SG-incompetent G3BP1-S149E (Fig. 2 C) may similarly assume a SG-incompetent conformation because it preferentially binds USP10 more than Caprin1 (Fig. 8 C). In the simplest scenario, G3BP–Caprin1 complexes could assemble mRNPs into SGs by escorting them through a demixing phase transition, and G3BP–USP10 complexes could reverse the process. This two-state model is appealing, but the in vivo situation is likely to be more complex. Several other proteins interact with G3BP (Hinton et al., 2010; Sahoo et al., 2012) or Caprin1 (Shiina and Nakayama, 2014) and regulate SG formation; it will be important to determine whether these proteins alter G3BP solubility or compete with USP10 and Caprin1 for G3BP.
We note that SGs include proteins that are not precipitated by B-isox, such as USP10, eIF3, and RACK1. Our model acknowledges this by envisioning SGs as mosaics of liquid–liquid insoluble aggregates embedded in a larger SG territory (Fig. 9, blue area), which houses both “condensing” proteins G3BP and Caprin1 and the “decondensing” protein USP10. This model is consistent with photobleaching data indicating that G3BP, PABP, TIA-1, and TIAR are only fleetingly present in SGs (Kedersha et al., 2002, 2005; Bley et al., 2015) and with morphologic EM data showing that SGs have heterogeneous substructures (Souquere et al., 2009). The diverse size and irregular morphology of SGs reflect the variable number of B-isox insoluble foci within a diffuse cloud of translation initiation factors. Hyperosmotically induced SG assembly via molecular crowding (Bounedjah et al., 2012) is consistent with this model. The extremely rapid dissolution of “cold shock SGs” upon warming, long before translation is resumed (Hofmann et al., 2012), also suggests a temperature-dependent phase transition from insoluble to soluble, in agreement with a two-stage model of SG formation in which polysome disassembly and SG condensation are linked but independent steps in the process. The condensation step in SG formation is temperature-dependent, whereas upstream events such as phosphorylation of eIF2α and polysome disassembly are not.
Whether the hypothetical G3BP SG-forming “condensase” activity requires proteins other than G3BP is unknown. G3BP is reported to possess an Mg-dependent DNA/RNA helicase activity (Costa et al., 1999) which could facilitate the SG condensation process. Helicase or RNA-chaperone activity mediating SG condensation is likely given the importance of other helicases in both SG and P-body assembly: inactivation of the helicase eIF4A by Hippuristanol or Pat A promotes SG assembly, suggesting that eIF4A antagonizes SG assembly (Dang et al., 2006; Mazroui et al., 2006), whereas DDX6/RCK helicase activity is required for P-body assembly (Ohn et al., 2008). By binding to both G3BP and PABP (a translational enhancer), USP10 could simultaneously retain mRNPs in a soluble state and prime them for translational activation and consequent removal from SGs. G3BP association with dissociated 40S subunits but not 80S monosomes is compatible with its potent ability to nucleate SGs, even before detectable translational inhibition or PKR activation (Reineke et al., 2012). A recent study (Berger et al., 2014) reported that the SRP heterodimer 9/14 regulates SG dynamics by binding 40S subunits in competition with Alu mRNA. As the G3BP1 binding protein OGFOD1 mediates the hydroxylation of RPS23 (Singleton et al., 2014), a role for G3BP in both ribosomal maturation and quality control seems likely. These studies establish the central role of the G3BP–Caprin1–USP10 axis in SG formation.
Materials and methods
Cell lines
COS7 and U2OS cells were maintained at 5.0–7.0% CO2 in DMEM containing 20 mM Hepes, 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin. Stable U2OS-derived cell lines constitutively expressing fluorescently tagged proteins (G3BP1-WT and mutants and RPS6-GFP) were made as described in detail elsewhere (Kedersha et al., 2008) by transfecting peGFPC1-G3BP1 or peGFPN1-RPS6 into U2OS cells, selecting with 0.5 µg/ml G418, cloning by limiting dilution, and screened using fluorescence microscopy and Western blotting. The Tet-on parent U2OS cell line used to make the tet-inducible cell lines was a gift from D. Schoenberg (Ohio State University, Columbus, OH). Tet-on cell lines were obtained using transfection of the tet-on pcDNA4-based plasmids containing GFP-tagged USP10 followed by zeocin selection at 250 µg/ml and cloning via limiting dilution.
siRNA transfection and transient transfection
U2OS-WT were transfected using Lipofectamine 2000 (Invitrogen) with the appropriate siRNA at a final concentration of 40 nM. Cells were treated twice with the siRNA complexes overnight, at time 0 and 48 h, reseeded at 72 h, and harvested at 96 h for immunofluorescence or protein determination to determine the efficiency of gene KD by Western blot. siRNAs (siGenome SMARTpool) directed against G3BP1, G3BP2, Caprin1, and USP10 were purchased from GE Healthcare; sequences are shown in Table S1. Control siRNA was against GFP (duplex sequence 5′-GGCTACGTCCAGGAGCG-3′). For transient transfections, COS7, U2OS-WT, and ΔΔG3BP1/2 cells at 90% confluency were transfected overnight using SuperFect (QIAGEN), replated, and processed at 36 h.
Cas9 deletion cell lines
U2OS-WT cells were plated and transfected with the pCas9-Guide (Origene GE100002) constructs (see Plasmids section) using Lipofectamine 2000 overnight, allowed to recover for ≥2 d, and then were reseeded and immunostained for the proteins of interest. Cultures with <5% KO cells were first “pool cloned” to enrich for KOs by plating at 5–10 cells per well in 24 well plates, allowing the cells to grow to >50% confluency before reseeding in duplicate (on coverslips in a 24-well plate for screening and in a 12-well plate for growth). When the cells on coverslips reached 80% confluency, they were treated with AS, fixed, and stained. Samples showing desired KO >5% were subcloned by limiting dilution. To create the double-null ΔΔG3BP1/2 KO cell line, G3BP1 was first knocked out and cloned, and then the ΔG3BP1 cells were transfected with the pCas9-Guide containing the guides for G3BP2.
Genotyping of Cas9 mutants
To identify Cas9-induced mutations in the G3BP1 coding sequence, genomic amplification was performed using the primers 5′-AGCTAAATGATTCGGTCTTTTCC-3′ (forward) and 5′-ATAAGTACCACATACTAAAAGACAGC-3′ (reverse). The G3BP2 coding region was amplified using 5′-TGAGGAGACAGGAAATGCAA-3′ (forward) and 5′-TTCATGGTGGTTGATGACAAA-3′ (reverse). Genomic DNA PCR was done with Invitrogen AccuPrime GC-Rich DNA Polymerase (buffer A). DNA was initially denatured at 95°C for 3 min, followed by denaturation at 95°C for 30 s, annealing at 60°C for 30 s, and extension at 72°C for 1 min for 30 cycles. Final extension was done at 72°C for 10 min. PCR products were adenylated using Taq polymerase and cloned into Promega pGEM-T Easy vector; individual clones were obtained and sequenced.
SG induction and quantification
SGs were induced by treatment with AS (concentration indicated in the figure legends), CZ (20 µM in serum-free media for 1 h), Pat A (50 nM for 1 h), rocaglamide A (500 nM for 1 h), or TG (1.0 µM for 1 h), sorbitol (0.4 M for 30 min), NaCl (0.2 M for 30 min), heat shock (45°C for 40 min), or by transient transfection of SG-nucleating proteins. Cells were scored for SGs by manual counting using fluorescent microscopy using eIF4G and eIF3b as SG markers; only cells with granules costaining for these markers were considered SGs, and a minimum of three granules per cell was required to score as positive.
Immunofluorescence
Cells were fixed and processed for fluorescence microscopy as described previously (Kedersha and Anderson, 2007). In brief, cells were grown on glass coverslips, stressed as indicated, and fixed using 4% paraformaldehyde in PBS for 15 min, followed by 5 min postfixation/permeabilization in cold methanol. Cells were blocked in 5% horse serum/PBS, and primary (Table S2) and secondary incubations were performed in blocking buffer for 1 h with rocking. All secondary antibodies (tagged with Cy2, Cy3, Cy5, or HRPO) were ML (multiple labeling) grade and obtained from Jackson Immunoresearch. After washes with PBS, cells were mounted in polyvinyl mounting media and viewed at RT using a Nikon Eclipse E800 microscope with a 40× Plan fluor (NA 0.75) or 100× Plan Apo objective lens (NA 1.4) and illuminated with a mercury lamp and standard filters for DAPI (UV-2A 360/40; 420/LP), Cy2 (FITC HQ 480/40; 535/50), Cy3 (Cy 3 HQ 545/30; 610/75), and Cy5 (Cy 5 HQ 620/60; 700/75). Images were captured using either a SPOT RT digital camera (Diagnostics Instruments) or a SPOT Pursuit digital Camera (Diagnostics Instruments) with the manufacturer’s software, and raw TIF files were imported into Adobe Photoshop CS3. Identical adjustments in brightness and contrast were applied to all images in a given experiment.
Ribopuromycylation assay
Ribopuromycylation assay was modified from David et al. (2012), as described in Panas et al. (2015a). In brief, cells were unstressed or stressed as indicated. 5 min before fixation, puromycin and EM were added to a final concentration of 9 and 91 µM, respectively, and the incubation continued for 5 min. Cells were then lysed subjected to Western blotting using anti-puromycin antibody (1:1,000 dilution; Millipore). Cells without puromycin treatment were used as negative controls.
Immunoprecipitation
The 150-mm dishes of near-confluent cells were treated as indicated, washed with cold HBSS, and scrape-harvested at 4°C into EE buffer (containing 50 mM Hepes, pH 7.0, 150 mM NaCl, 0.1% NP-40, 10% glycerol, 2 mM EDTA, 2.5 mM EGTA, 1 mM DTT, HALT phosphatase inhibitors, and protease inhibitors; Thermo Fisher Scientific). EE buffer containing additional MgCl2 was used in Figs. 7 and 8. Cells were rotated for 20 min at 4°C, cleared by centrifugation (10,000 g for 15 min), and incubated with Chromotek-GFP-Trap Beads (Allele Biotech) for 2 h with continuous rotation at 4°C. Beads were washed five times and either eluted directly into SDS-lysis buffer without RNase treatment or incubated with 40 µg/ml RNase A for 1 h at 4°C with rotation. Material released by RNase was recovered and precipitated with 60% acetone. Proteins were resolved on 4–20% gradient gels (Invitrogen), transferred to nitrocellulose, and blotted using standard procedures. Silver stain was performed using the Pierce Silver Stain kit.
Protein purification
His-Caprin1 and His-G3BP1 proteins were expressed in BL-21 Escherichia coli cells for 4 h at 37°C with 0.5 mM IPTG. Cells were lysed by sonication for 2 min in lysis buffer (0.1% Triton X-100, 10 mM imidazol, 300 mM NaCl, and 50 mM Na2PO4, pH 8), and lysates were clarified by centrifugation and incubated with Ni-NTA agarose (QIAGEN) for 2 h at 4°C, tumbling. Subsequently, Ni-NTA immobilized His-Caprin1 or His-G3BP1 proteins were washed and eluted with lysis buffer supplemented with 150 mM imidazol. His-Caprin1 and His-G3BP1 proteins were analyzed by Coomassie staining and SDS-PAGE. Protein concentrations were quantified by comparison with BSA standards.
USP10 peptide and Caprin1 competition
U2OS-WT cells were lysed in EE buffer and clarified by centrifugation at 10,000 g for 10 min at 4°C. The supernatants were incubated with a mixture of two mouse anti-G3BP1 antibodies (sc-365338 [Santa Cruz Biotechnology, Inc.] and TL611126 [BD]) for 15 min at RT. Washed protein A/G UltraLink Resin (Thermo Fisher Scientific) was added, and the samples were incubated for 90 min at 4°C. The resin was washed 5× with EE buffer and resuspended in 50 µl EE buffer, and then biotinylated USP108–25WT or USP108-25F10A peptides (100, 10, or 1 µM) were added and the samples were incubated for 60 min at 4°C. After incubation, the supernatants were collected, and the resins were washed 5× with EE buffer, eluted in 50 µl 2× SDS sample buffer, and heated for 5 min at 95°C. The IPs and supernatants were resolved in a 4–20% Mini-PROTEAN TGX Precast Gel (Bio-Rad) and transferred to nitrocellulose membranes using the Transfer-Blot Turbo transfer system (Bio-Rad). Chemiluminescent was detected using SuperSignal West Pico substrate (Thermo Fisher Scientific).
Purified His-G3BP1 (2.5 pmol) was incubated with biotinylated USP108–25WT or F10A peptide (250 pmol) and rotated for 1 h at 4°C in EE buffer + 0.5% NP-40. Then streptavidin agarose resin (Thermo Fisher Scientific) was added, and the mixture was incubated for 1 h at 4°C. The resin was washed 5× with EE buffer + 0.5% NP-40 and resuspended in 50 µl of EE buffer + 0.5% NP-40, and then purified His-Caprin1 (2.5, 25, or 125 pmol) was added and rotated for 1 h at 4°C. After incubation, the beads were washed 5× with EE buffer + 0.5% NP-40, eluted in 40 µl 2× SDS sample buffer, and heated for 5 min at 95°C; they were then subjected to Western blotting.
Polysome profile analysis
Cells were washed with cold HBSS, scrape-harvested directly into lysis buffer (10 mM Hepes, pH 7.5, 125 mM KCl, 5 mM MgCl2, 1 mM DTT, 100 µg/ml cycloheximide, 100 µg/ml heparin, and 1% NP-40 made in DEPC-treated water), and supplemented with RNAsin Plus inhibitor (Promega) and HALT phosphatase and protease inhibitors (Thermo Fisher Scientific). Lysates were rotated at 4°C for 15 min, cleared by centrifugation for 10 min at 12,000 g, and supernatants were loaded on preformed 17.5–50% sucrose gradients made in gradient buffer (10 mM Hepes, pH 7.5, 125 mM KCl, 5 mM MgCl2, and 1 mM DTT). Samples were centrifuged in a Beckman SW140 Ti rotor for 2.5 h at 35,000 rpm and then eluted using a Brandel bottom-piercing apparatus connected to an ISCO UV monitor, which measured the eluate at OD 254.
B-isox precipitation
B-isox (6-(5-(Thiophen-2-yl)isoxazole-3-carboxamido)hexyl 5-((3aS,4S,6aR)-2-oxohexahydro-1H-thieno(3,4-d)imidazol-4-yl)pentanoate) was obtained from Sigma-Aldrich. Cells were lysed in EE buffer or modified Kato buffer (20 mM Tris-HCl, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT 0.5% NP-40, and 10% glycerol), precleared at 16,000 g for 15 min at 4°C, and then were incubated with 100 µM B-isox or vehicle (DMSO) control in the cold for 2 h with rotation. Precipitates were obtained using a 10-min centrifugation at 16,000 g at 4°C. Precipitates were washed twice in EE buffer before SDS solubilization.
Confocal, live, and video microscopy
For video microscopy, cells were plated onto 35-mM FluoroDishes (World Precision Instruments) and pretreated with doxycycline (1.0 µg/ml), where indicated. Cells were grown in phenol-red free DMEM supplemented with 25 mM Hepes, pH 7.2, and 10% FBS and viewed in a custom-built heated chamber warmed to 37°C. Live cell images were obtained on a Nikon Eclipse TE2000U Inverted Microscope using an Eclipse EZ-C1 system (v. 3.90; Nikon) and a Plan Apo 60x Pan Apo (NA 1.40) objective lens. EGFP fluorescence was excited with the 488-nm line from a Melles Griot 488 Ion Laser and detected with a 515/30 emission filter; mRFP/cherry fluorescence was obtained using Melles Griot 543 laser excitation and a 590/50 emission filter. Z-series were collected every 1 min (20 optical sections with a step size of 0.8 µm). Z-series were volume- and time-rendered using C1 software (Nikon) and displayed as maximum z-projection AVI files. Gamma, brightness, and contrast were adjusted on the AVI files using Adobe Photoshop CS3, labels were added, and the files were rendered as MOV files.
Drugs and chemical reagents
DMD (desmethyl, desamino)-modified Pat A was a gift from J. Lui (Johns Hopkins, Baltimore, MD). AS, CZ, doxycycline, rocaglamide A, puromycin, tetracycline, TG EM, sorbitol, glucose-free DMEM, glucose, and Biotin-isox were obtained from Sigma-Aldrich. Zeocin was obtained from Life Technologies. C-Terminal biotinylated USP10 peptides were obtained from GeneScript.
Plasmids
Cas9 constructs: pCas9-Guide plasmid (Origene) was used to clone guide oligos targeting G3BP1 or G3BP2 genes according to the manufacturer’s recommendations (GE100001 pCas9-Guide kit; Origene). Target 20-bp sequences were chosen using the http://crispr.mit.edu/ site, targeting 100 nucleotides around ATG start codon G3BP1 or G3BP2. Top candidates with minimally predicted off-target effects were chosen (Table S1). Two synthetic oligos (IDT Technology) were annealed to each other to create duplex DNA for cloning into pCas9-Guide plasmid using BamHI and BsmBI sites. All genetic constructs were verified by DNA sequencing.
USP10
A cDNA clone was obtained from Open Biosystems (IMAGE ID 3501606, Clone ID 3501606) for full-length human USP10 (NCBI accession no. BC000263.1). The coding region of USP10 was cloned into the pmCherry-C1 vector in fragments, in-frame with a mCherry tag within the vector. To clone full-length USP10, two separate constructs were made corresponding to an N-terminal region (aa 1–280) and the C-terminal portion (aa 270–798), which were of unequal length and overlapped by 10 aa and included an internal KpnI site. For pmCherry-USP10-NT and CT, both fragments were amplified by PCR using primers adding XhoI and SacII sites to the 5′ and 3′ terminal ends of both DNA fragments (5′-ATATCTCGAGCTATGGCCCTCCACAG-3′ and 5′-TGTGCCGCGGTTATTCAGTAGTATCG-3′; 5′-ATATCTCGAGCTGGGGCTCAGCCCTG-3′ and 5′-TATACCGCGGTTACAGCAGGTCCAC-3′), respectively, and each was cloned separately into the XhoI and SacII sites of pmCherry-C1, in-frame with the mCherry tag (a gift from the laboratory of R. Tsien, University of California, San Diego, San Diego, CA). To generate a full-length mCherry-tagged USP10 construct, pE-mCherry-USP10 NT was cut with XhoI and KpnI, which excised a fragment that was then cloned into pE-mCherry-USP10 CT that was similarly cut with XhoI and KpnI. Tet-on USP10 was obtained by replacing the coding region of pcDNA4TM/T/O-GFP-AcGFP1 with the coding region of USP10.
GFP-tagged SFV-nsP331WT and F3A and GFP-tagged USP101-40WT and F10A
WT sequences and corresponding alanine mutant sequences were obtained from GeneArt, ligated between the BglII and EcoRI sites of pEGFP-C1, and characterized in more detail previously (Panas et al., 2015b).
Caprin1
A cDNA template for the coding region of human CAPRIN1 (NCBI accession no. BC001731) was purchased from Open Biosystems (IMAGE ID 3355481, Clone ID 3355481). The coding region was amplified by PCR using primers adding XhoI and BamHI sites to the 5′- and 3′-terminal ends (5′-ATATCTCGAGCTATGCCCTCGGCCACC-3′ and 5′-CGCGGGATCCTTAATTCACTTGCTGAGTG-3′), respectively. The amplified full-length DNA target was then cloned into the XhoI and BamHI sites of the pAcGFP1-C1 vector (ClonTech), in-frame with the GFP tag. The plasmid encoding RPS6 was made from templates obtained from Open Biosystems and cloned into Clontech pAcGFP-N1 vector using the XhoI and SacII sites and PCR (primers 5′-GCGCCTCGAGATGAAGCTGAACATCTCC-3′ and 5′-ACACCCGCGGTTTCTGACTGGATTCAG-3′); constructs were verified by sequencing. Bold indicates restriction sites.
G3BP and mutants
GFP-G3BP1 (human) was obtained from J. Tazi (Institut de Génétique Moléculaire de Montpellier, Montpellier, France). The original construct was found to lack its native stop site and encoded a protein with an additional 16 aa at the C terminus (5′-GEFCSRRYRGPGIHRI-3′); we restored the native stop codon using PCR to insert the missing A, confirmed it by sequencing, and used the repaired construct as a template for the creation of all other mutants via PCR. GFP-G3BP2a was obtained from D. Kennedy (Griffith University, Nathan, Australia). A plasmid encoding the coding sequence of human Heme responsive inhibitor kinase was obtained from Open Bisystems and cloned into the XhoI–SacII sites in the Clontech peYFP-C1 vector. Bacterial expression constructs for Caprin1 and G3BP1 purification were generated by subcloning Caprin1 or G3BP1 into pET28 by using the BamHI and NotI restriction sites.
PCR mutations and truncations
In brief, the indicated primer pairs (Table S1; final concentration 500 nM) were mixed with 100 ng of plasmid in 1× Phusion PCR mix at a final volume of 50 µl. The reaction was denatured at 98°C for 30 s, followed by 24 cycles of the following: 98°C for 10 s, 60°C for 20 s, and 72°C for 5 min. There was a final extension step of 72°C for 10 min. PCR products were cleaned up with a column kit (QIAGEN) and brought to 18.5 µl in 1× T4 DNA ligase buffer (NEB). The PCR products were treated with 0.5 µl T4 PNK (NEB) for 30 min at 37°C, and then 0.5 µl T4 DNA ligase was added, and the mixture was incubated for 60 min at 23°C after an 0.5 µl DpnI digest (NEB) for 30 min at 37°C. The ligation mix was used for chemical transformation into high efficiency E. coli. Multiple clones for each reaction were picked and verified by sequencing.
GFP-G3BP1-ΔRRM, ΔRGG, and ΔRRM+ΔRGG
The 5′ phosphorylated primers (see Table S2 for sequences) were mixed with 1 ng of pEGFP-G3BP1-WT in a 1× Phusion PCR mastermix (Thermo Fisher Scientific) at a final volume of 25 µl. The mixture was denatured at 98°C for 30 s, followed by 25 cycles of the following: 98°C for 10 s, 60°C for 15 s, 72°C for 2 min 30 s, with a final extension step of 72°C for 5 min. 25 ng of the PCR product was ligated with T4 DNA ligase in a final volume of 10 µl for 1 h at RT. 5 µl of the ligation mix was used for chemical transformation into high-efficiency E. coli. Multiple clones were picked and verified by sequencing.
In silico analysis
Regions of LC (blue) were obtained from using the NCBI “conserved domains” graphic, which calculates LC regions using the SEG program (Wootton and Federhen, 1996). ID regions were determined using the programs of Dosztányi et al. (2005) on the ANCHOR website (http://anchor.enzim.hu/), in which regions of disorder >50% on the intrinsically unordered histograms were graphically rendered (red) for simplicity.
Statistical analyses
Statistical analyses were performed using Microsoft Excel. Statistical differences between the two groups in immunofluorescence or Western blot experiments were evaluated using unpaired t test. P < 0.05 was considered significant. All data are expressed as mean ± SEM.
Online supplemental material
Fig. S1 schematically depicts the G3BP, Caprin1, and USP10 constructs used in this study. Fig. S2 shows the genotype analysis of G3BP mutations introduced by CRISPR/Cas9 and the predicted possible protein products. Fig. S3 shows that B-isox fractionation is not affected by pretreated cells with SG-inducing and noninducing stresses. Fig. S4 shows the effect of different USP10 deletions on coprecipitation of G3BP, PABP, eIF4G, RPS6, and RPL4 and on total proteins revealed by silver staining. Fig. S5 shows the characterization of the RPS6-GFP stable U2OS cells and the purity of the recombinant G3BP1 and Caprin1 used in Fig. 3 E. Table S1 lists the oligo sequences used in this study (for CRISPR/Cas9, siRNA, and PCR); Table S2 lists the antibodies used in this study. Video 1 shows that GFP-USP10–expressing cells resist AS-induced SG assembly; Video 2 shows that GFP-USP10–expressing cells resist SG formation in response to Pat A. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201508028/DC1.
Supplementary Material
Acknowledgments
We thank the Brigham and Women’s Hospital Confocal Microscopy Core for live imaging.
This work was supported by National Institutes of Health (AI0655858 and CA168872) to P. Anderson, National Institutes of Health (NS094918) and Amyotrophic Lateral Sclerosis Association (N7W220) to P. Ivanov, and Swedish Cancer Society (CAN 2012/789) and Vetenskapsrådet (621-2014-4718) to G.M. McInerney.
The authors declare no competing financial interests.
Footnotes
Abbreviations used in this paper:
- AS
- sodium arsenite
- B-isox
- Biotin-isoxazole
- CZ
- clotrimazole
- EE
- EDTA-EGTA
- eIF
- eukaryotic initiation factor
- EM
- emetine
- ID
- intrinsically disordered
- IP
- immunoprecipitate
- KD
- knockdown
- KO
- knockout
- LC
- low complexity
- LLPS
- liquid–liquid phase separation
- MEF
- mouse embryonic fibroblast
- Pat A
- pateamine A
- p-eIF2α
- phospho-eIF2α
- SFV
- Semliki Forest virus
- SG
- stress granule
- TG
- thapsigargin
- WT
- wild type
References
- Anderson P., and Kedersha N.. 2008. Stress granules: the Tao of RNA triage. Trends Biochem. Sci. 33:141–150. 10.1016/j.tibs.2007.12.003 [DOI] [PubMed] [Google Scholar]
- Anderson P., and Kedersha N.. 2009. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell Biol. 10:430–436. 10.1038/nrm2694 [DOI] [PubMed] [Google Scholar]
- Aulas A., Caron G., Gkogkas C.G., Mohamed N.V., Destroismaisons L., Sonenberg N., Leclerc N., Parker J.A., and Vande Velde C.. 2015. G3BP1 promotes stress-induced RNA granule interactions to preserve polyadenylated mRNA. J. Cell Biol. 209:73–84. 10.1083/jcb.201408092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger A., Ivanova E., Gareau C., Scherrer A., Mazroui R., and Strub K.. 2014. Direct binding of the Alu binding protein dimer SRP9/14 to 40S ribosomal subunits promotes stress granule formation and is regulated by Alu RNA. Nucleic Acids Res. 42:11203–11217. 10.1093/nar/gku822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevilacqua E., Wang X., Majumder M., Gaccioli F., Yuan C.L., Wang C., Zhu X., Jordan L.E., Scheuner D., Kaufman R.J., et al. 2010. eIF2alpha phosphorylation tips the balance to apoptosis during osmotic stress. J. Biol. Chem. 285:17098–17111. 10.1074/jbc.M110.109439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bley N., Lederer M., Pfalz B., Reinke C., Fuchs T., Glaß M., Möller B., and Hüttelmaier S.. 2015. Stress granules are dispensable for mRNA stabilization during cellular stress. Nucleic Acids Res. 43:e26 10.1093/nar/gku1275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordeleau M.E., Matthews J., Wojnar J.M., Lindqvist L., Novac O., Jankowsky E., Sonenberg N., Northcote P., Teesdale-Spittle P., and Pelletier J.. 2005. Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc. Natl. Acad. Sci. USA. 102:10460–10465. 10.1073/pnas.0504249102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounedjah O., Hamon L., Savarin P., Desforges B., Curmi P.A., and Pastré D.. 2012. Macromolecular crowding regulates assembly of mRNA stress granules after osmotic stress: new role for compatible osmolytes. J. Biol. Chem. 287:2446–2458. 10.1074/jbc.M111.292748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bounedjah O., Desforges B., Wu T.D., Pioche-Durieu C., Marco S., Hamon L., Curmi P.A., Guerquin-Kern J.L., Piétrement O., and Pastré D.. 2014. Free mRNA in excess upon polysome dissociation is a scaffold for protein multimerization to form stress granules. Nucleic Acids Res. 42:8678–8691. 10.1093/nar/gku582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brangwynne C.P., Eckmann C.R., Courson D.S., Rybarska A., Hoege C., Gharakhani J., Jülicher F., and Hyman A.A.. 2009. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 324:1729–1732. 10.1126/science.1172046 [DOI] [PubMed] [Google Scholar]
- Buchan J.R., and Parker R.. 2009. Eukaryotic stress granules: the ins and outs of translation. Mol. Cell. 36:932–941. 10.1016/j.molcel.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M., Ochem A., Staub A., and Falaschi A.. 1999. Human DNA helicase VIII: a DNA and RNA helicase corresponding to the G3BP protein, an element of the ras transduction pathway. Nucleic Acids Res. 27:817–821. 10.1093/nar/27.3.817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y., Kedersha N., Low W.K., Romo D., Gorospe M., Kaufman R., Anderson P., and Liu J.O.. 2006. Eukaryotic initiation factor 2alpha-independent pathway of stress granule induction by the natural product pateamine A. J. Biol. Chem. 281:32870–32878. 10.1074/jbc.M606149200 [DOI] [PubMed] [Google Scholar]
- David A., Dolan B.P., Hickman H.D., Knowlton J.J., Clavarino G., Pierre P., Bennink J.R., and Yewdell J.W.. 2012. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J. Cell Biol. 197:45–57. 10.1083/jcb.201112145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dosztányi Z., Csizmok V., Tompa P., and Simon I.. 2005. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 21:3433–3434. 10.1093/bioinformatics/bti541 [DOI] [PubMed] [Google Scholar]
- Elbaum-Garfinkle S., Kim Y., Szczepaniak K., Chen C.C., Eckmann C.R., Myong S., and Brangwynne C.P.. 2015. The disordered P granule protein LAF-1 drives phase separation into droplets with tunable viscosity and dynamics. Proc. Natl. Acad. Sci. USA. 112:7189–7194. 10.1073/pnas.1504822112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson S.L., and Lykke-Andersen J.. 2011. Cytoplasmic mRNP granules at a glance. J. Cell Sci. 124:293–297. 10.1242/jcs.072140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., and Anderson P.. 2004. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 15:5383–5398. 10.1091/mbc.E04-08-0715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han T.W., Kato M., Xie S., Wu L.C., Mirzaei H., Pei J., Chen M., Xie Y., Allen J., Xiao G., and McKnight S.L.. 2012. Cell-free formation of RNA granules: bound RNAs identify features and components of cellular assemblies. Cell. 149:768–779. 10.1016/j.cell.2012.04.016 [DOI] [PubMed] [Google Scholar]
- Hinton S.D., Myers M.P., Roggero V.R., Allison L.A., and Tonks N.K.. 2010. The pseudophosphatase MK-STYX interacts with G3BP and decreases stress granule formation. Biochem. J. 427:349–357. 10.1042/BJ20091383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann S., Cherkasova V., Bankhead P., Bukau B., and Stoecklin G.. 2012. Translation suppression promotes stress granule formation and cell survival in response to cold shock. Mol. Biol. Cell. 23:3786–3800. 10.1091/mbc.E12-04-0296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M., Han T.W., Xie S., Shi K., Du X., Wu L.C., Mirzaei H., Goldsmith E.J., Longgood J., Pei J., et al. 2012. Cell-free formation of RNA granules: low complexity sequence domains form dynamic fibers within hydrogels. Cell. 149:753–767. 10.1016/j.cell.2012.04.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., and Anderson P.. 2007. Mammalian stress granules and processing bodies. Methods Enzymol. 431:61–81. 10.1016/S0076-6879(07)31005-7 [DOI] [PubMed] [Google Scholar]
- Kedersha N., Cho M.R., Li W., Yacono P.W., Chen S., Gilks N., Golan D.E., and Anderson P.. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151:1257–1268. 10.1083/jcb.151.6.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Chen S., Gilks N., Li W., Miller I.J., Stahl J., and Anderson P.. 2002. Evidence that ternary complex (eIF2-GTP-tRNA(i)(Met))-deficient preinitiation complexes are core constituents of mammalian stress granules. Mol. Biol. Cell. 13:195–210. 10.1091/mbc.01-05-0221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Stoecklin G., Ayodele M., Yacono P., Lykke-Andersen J., Fritzler M.J., Scheuner D., Kaufman R.J., Golan D.E., and Anderson P.. 2005. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 169:871–884. 10.1083/jcb.200502088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., Tisdale S., Hickman T., and Anderson P.. 2008. Real-time and quantitative imaging of mammalian stress granules and processing bodies. Methods Enzymol. 448:521–552. 10.1016/S0076-6879(08)02626-8 [DOI] [PubMed] [Google Scholar]
- Kedersha N.L., Gupta M., Li W., Miller I., and Anderson P.. 1999. RNA-binding proteins TIA-1 and TIAR link the phosphorylation of eIF-2 α to the assembly of mammalian stress granules. J. Cell Biol. 147:1431–1442. 10.1083/jcb.147.7.1431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroschwald S., Maharana S., Mateju D., Malinovska L., Nüske E., Poser I., Richter D., and Alberti S.. 2015. Promiscuous interactions and protein disaggregases determine the material state of stress-inducible RNP granules. eLife. 4:e06807 10.7554/eLife.06807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Protter D.S., Rosen M.K., and Parker R.. 2015. Formation and Maturation of Phase-Separated Liquid Droplets by RNA-Binding Proteins. Mol. Cell. 60:208–219. 10.1016/j.molcel.2015.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lloyd R.E. 2013. Regulation of stress granules and P-bodies during RNA virus infection. Wiley Interdiscip. Rev. RNA. 4:317–331. 10.1002/wrna.1162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuki H., Takahashi M., Higuchi M., Makokha G.N., Oie M., and Fujii M.. 2013. Both G3BP1 and G3BP2 contribute to stress granule formation. Genes Cells. 18:135–146. 10.1111/gtc.12023 [DOI] [PubMed] [Google Scholar]
- Mazan-Mamczarz K., Lal A., Martindale J.L., Kawai T., and Gorospe M.. 2006. Translational repression by RNA-binding protein TIAR. Mol. Cell. Biol. 26:2716–2727. 10.1128/MCB.26.7.2716-2727.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazroui R., Sukarieh R., Bordeleau M.E., Kaufman R.J., Northcote P., Tanaka J., Gallouzi I., and Pelletier J.. 2006. Inhibition of ribosome recruitment induces stress granule formation independently of eukaryotic initiation factor 2alpha phosphorylation. Mol. Biol. Cell. 17:4212–4219. 10.1091/mbc.E06-04-0318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molliex A., Temirov J., Lee J., Coughlin M., Kanagaraj A.P., Kim H.J., Mittag T., and Taylor J.P.. 2015. Phase separation by low complexity domains promotes stress granule assembly and drives pathological fibrillization. Cell. 163:123–133. 10.1016/j.cell.2015.09.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nott T.J., Petsalaki E., Farber P., Jervis D., Fussner E., Plochowietz A., Craggs T.D., Bazett-Jones D.P., Pawson T., Forman-Kay J.D., and Baldwin A.J.. 2015. Phase transition of a disordered nuage protein generates environmentally responsive membraneless organelles. Mol. Cell. 57:936–947. 10.1016/j.molcel.2015.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T., and Anderson P.. 2010. The role of posttranslational modifications in the assembly of stress granules. Wiley Interdiscip. Rev. RNA. 1:486–493. 10.1002/wrna.23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T., Kedersha N., Hickman T., Tisdale S., and Anderson P.. 2008. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 10:1224–1231. 10.1038/ncb1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otero L.J., Ashe M.P., and Sachs A.B.. 1999. The yeast poly(A)-binding protein Pab1p stimulates in vitro poly(A)-dependent and cap-dependent translation by distinct mechanisms. EMBO J. 18:3153–3163. 10.1093/emboj/18.11.3153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas M.D., Varjak M., Lulla A., Eng K.E., Merits A., Karlsson Hedestam G.B., and McInerney G.M.. 2012. Sequestration of G3BP coupled with efficient translation inhibits stress granules in Semliki Forest virus infection. Mol. Biol. Cell. 23:4701–4712. 10.1091/mbc.E12-08-0619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas M.D., Kedersha N., and McInerney G.M.. 2015a Methods for the characterization of stress granules in virus infected cells. Methods. 90:57–64. 10.1016/j.ymeth.2015.04.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panas M.D., Schulte T., Thaa B., Sandalova T., Kedersha N., Achour A., and McInerney G.M.. 2015b Viral and cellular proteins containing FGDF motifs bind G3BP to block stress granule formation. PLoS Pathog. 11:e1004659 10.1371/journal.ppat.1004659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A., Lee H.O., Jawerth L., Maharana S., Jahnel M., Hein M.Y., Stoynov S., Mahamid J., Saha S., Franzmann T.M., et al. 2015. A Liquid-to-Solid Phase Transition of the ALS Protein FUS Accelerated by Disease Mutation. Cell. 162:1066–1077. 10.1016/j.cell.2015.07.047 [DOI] [PubMed] [Google Scholar]
- Piecyk M., Wax S., Beck A.R., Kedersha N., Gupta M., Maritim B., Chen S., Gueydan C., Kruys V., Streuli M., and Anderson P.. 2000. TIA-1 is a translational silencer that selectively regulates the expression of TNF-alpha. EMBO J. 19:4154–4163. 10.1093/emboj/19.15.4154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piotrowska J., Hansen S.J., Park N., Jamka K., Sarnow P., and Gustin K.E.. 2010. Stable formation of compositionally unique stress granules in virus-infected cells. J. Virol. 84:3654–3665. 10.1128/JVI.01320-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke L.C., and Lloyd R.E.. 2015. The stress granule protein G3BP1 recruits protein kinase R to promote multiple innate immune antiviral responses. J. Virol. 89:2575–2589. 10.1128/JVI.02791-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke L.C., Dougherty J.D., Pierre P., and Lloyd R.E.. 2012. Large G3BP-induced granules trigger eIF2α phosphorylation. Mol. Biol. Cell. 23:3499–3510. 10.1091/mbc.E12-05-0385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke L.C., Kedersha N., Langereis M.A., van Kuppeveld F.J., and Lloyd R.E.. 2015. Stress granules regulate double-stranded RNA-dependent protein kinase activation through a complex containing G3BP1 and Caprin1. MBio. 6:e02486 10.1128/mBio.02486-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadlish H., Galicia-Vazquez G., Paris C.G., Aust T., Bhullar B., Chang L., Helliwell S.B., Hoepfner D., Knapp B., Riedl R., et al. 2013. Evidence for a functionally relevant rocaglamide binding site on the eIF4A-RNA complex. ACS Chem. Biol. 8:1519–1527. 10.1021/cb400158t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo P.K., Murawala P., Sawale P.T., Sahoo M.R., Tripathi M.M., Gaikwad S.R., Seshadri V., and Joseph J.. 2012. Wnt signalling antagonizes stress granule assembly through a Dishevelled-dependent mechanism. Biol. Open. 1:109–119. 10.1242/bio.2011023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina N., and Nakayama K.. 2014. RNA granule assembly and disassembly modulated by nuclear factor associated with double-stranded RNA 2 and nuclear factor 45. J. Biol. Chem. 289:21163–21180. 10.1074/jbc.M114.556365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina N., Shinkura K., and Tokunaga M.. 2005. A novel RNA-binding protein in neuronal RNA granules: regulatory machinery for local translation. J. Neurosci. 25:4420–4434. 10.1523/JNEUROSCI.0382-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton R.S., Liu-Yi P., Formenti F., Ge W., Sekirnik R., Fischer R., Adam J., Pollard P.J., Wolf A., Thalhammer A., et al. 2014. OGFOD1 catalyzes prolyl hydroxylation of RPS23 and is involved in translation control and stress granule formation. Proc. Natl. Acad. Sci. USA. 111:4031–4036. 10.1073/pnas.1314482111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon S., Xu Y., Wang B., David M.D., Schubert P., Kennedy D., and Schrader J.W.. 2007. Distinct structural features of caprin-1 mediate its interaction with G3BP-1 and its induction of phosphorylation of eukaryotic translation initiation factor 2alpha, entry to cytoplasmic stress granules, and selective interaction with a subset of mRNAs. Mol. Cell. Biol. 27:2324–2342. 10.1128/MCB.02300-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soncini C., Berdo I., and Draetta G.. 2001. Ras-GAP SH3 domain binding protein (G3BP) is a modulator of USP10, a novel human ubiquitin specific protease. Oncogene. 20:3869–3879. 10.1038/sj.onc.1204553 [DOI] [PubMed] [Google Scholar]
- Souquere S., Mollet S., Kress M., Dautry F., Pierron G., and Weil D.. 2009. Unravelling the ultrastructure of stress granules and associated P-bodies in human cells. J. Cell Sci. 122:3619–3626. 10.1242/jcs.054437 [DOI] [PubMed] [Google Scholar]
- Takahashi M., Higuchi M., Matsuki H., Yoshita M., Ohsawa T., Oie M., and Fujii M.. 2013. Stress granules inhibit apoptosis by reducing reactive oxygen species production. Mol. Cell. Biol. 33:815–829. 10.1128/MCB.00763-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. 2002. Intrinsically unstructured proteins. Trends Biochem. Sci. 27:527–533. 10.1016/S0968-0004(02)02169-2 [DOI] [PubMed] [Google Scholar]
- Tourrière H., Chebli K., Zekri L., Courselaud B., Blanchard J.M., Bertrand E., and Tazi J.. 2003. The RasGAP-associated endoribonuclease G3BP assembles stress granules. J. Cell Biol. 160:823–831. 10.1083/jcb.200212128 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Vanderweyde T., Youmans K., Liu-Yesucevitz L., and Wolozin B.. 2013. Role of stress granules and RNA-binding proteins in neurodegeneration: a mini-review. Gerontology. 59:524–533. 10.1159/000354170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vognsen T., Møller I.R., and Kristensen O.. 2013. Crystal structures of the human G3BP1 NTF2-like domain visualize FxFG Nup repeat specificity. PLoS One. 8:e80947 10.1371/journal.pone.0080947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber S.C., and Brangwynne C.P.. 2012. Getting RNA and protein in phase. Cell. 149:1188–1191. 10.1016/j.cell.2012.05.022 [DOI] [PubMed] [Google Scholar]
- Wehner K.A., Schütz S., and Sarnow P.. 2010. OGFOD1, a novel modulator of eukaryotic translation initiation factor 2alpha phosphorylation and the cellular response to stress. Mol. Cell. Biol. 30:2006–2016. 10.1128/MCB.01350-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White J.P., Cardenas A.M., Marissen W.E., and Lloyd R.E.. 2007. Inhibition of cytoplasmic mRNA stress granule formation by a viral proteinase. Cell Host Microbe. 2:295–305. 10.1016/j.chom.2007.08.006 [DOI] [PubMed] [Google Scholar]
- Wootton J.C., and Federhen S.. 1996. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 266:554–571. 10.1016/S0076-6879(96)66035-2 [DOI] [PubMed] [Google Scholar]
- Zhang H., Elbaum-Garfinkle S., Langdon E.M., Taylor N., Occhipinti P., Bridges A.A., Brangwynne C.P., and Gladfelter A.S.. 2015. RNA Controls PolyQ Protein Phase Transitions. Mol. Cell. 60:220–230. 10.1016/j.molcel.2015.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.