The intra-axonal events governing formation of presynaptic terminals are still poorly understood. Pinto et al. reveal a mechanism by which a localized decrease in proteasome degradation and resultant accumulation of polyubiquitinated proteins at nascent sites signal assembly of presynaptic terminals.
Abstract
Differentiation of the presynaptic terminal is a complex and rapid event that normally occurs in spatially specific axonal regions distant from the soma; thus, it is believed to be dependent on intra-axonal mechanisms. However, the full nature of the local events governing presynaptic assembly remains unknown. Herein, we investigated the involvement of the ubiquitin–proteasome system (UPS), the major degradative pathway, in the local modulation of presynaptic differentiation. We found that proteasome inhibition has a synaptogenic effect on isolated axons. In addition, formation of a stable cluster of synaptic vesicles onto a postsynaptic partner occurs in parallel to an on-site decrease in proteasome degradation. Accumulation of ubiquitinated proteins at nascent sites is a local trigger for presynaptic clustering. Finally, proteasome-related ubiquitin chains (K11 and K48) function as signals for the assembly of presynaptic terminals. Collectively, we propose a new axon-intrinsic mechanism for presynaptic assembly through local UPS inhibition. Subsequent on-site accumulation of proteins in their polyubiquitinated state triggers formation of presynapses.
Introduction
Throughout development, establishment of functional synaptic contacts is pivotal for the correct wiring of neurons and ultimately proper brain function in adulthood. Therefore, it is of utmost importance to fully comprehend the cascade of events comprising synapse formation. One such event, presynaptic differentiation, corresponds to the organized clustering of presynaptic material in specific locations along the axon (Jin and Garner, 2008), which is induced by a cohort of presynaptic differentiating proteins, including adhesive or secreted factors (Chia et al., 2013). However, the intra-axonal on-site downstream events triggering assembly of presynaptic material at spots of axodendritic contact are still poorly understood. Control of protein turnover by the ubiquitin–proteasome system (UPS) has been shown to act locally at synapses (Segref and Hoppe, 2009), but its involvement in vertebrate presynapse formation is still unknown.
Steady-state levels of ubiquitin (Ub) are required for proper synapse formation. The ataxia mice Ups14axJ, with a loss-of-function mutation in the deubiquitinase Usp14, display severe structural and functional alterations at the neuromuscular junction (NMJ; Chen et al., 2009, 2011). These defects are completely rescued by restoration of neuronal Ub levels (Chen et al., 2009, 2011). A pioneering study in the Drosophila melanogaster NMJ concluded that a tight ubiquitination/deubiquitination balance is crucial for synapse development, thus revealing a role for synaptic ubiquitinated proteins (DiAntonio et al., 2001). In fact, ubiquitinated proteins are highly enriched at the vicinity of the active zone of Drosophila NMJs (Tian and Wu, 2013). Moreover, the presynaptic ubiquitinated proteome includes both structural and signaling proteins as well as proteins with known roles in synaptogenesis (Franco et al., 2011; Na et al., 2012). Despite the wealth of knowledge on UPS degradation at the synapse, the physiological significance of such a complex presynaptic ubiquitinated proteome is far from being understood. In the present study, we demonstrate that the UPS acts locally to control the assembly of new presynapses by regulating accumulation of an on-site pool of polyubiquitinated proteins that acts as a hub for presynaptic assembly.
Results and discussion
Inhibition of the proteasome in isolated axons has a synaptogenic effect
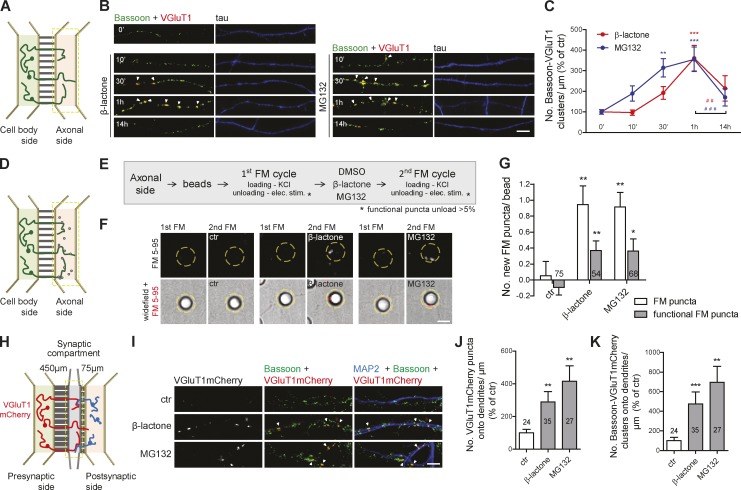
To understand the axonal intrinsic processes underlying formation of presynaptic clusters, we relied on microfluidic devices for the isolation of axons (Fig. S1 A and Fig. 1 A; Taylor et al., 2005, 2009; Cristovão et al., 2014; Neto et al., 2014). We used this platform to specifically inhibit the proteasome in axons with clasto-lactacystin β-lactone or MG132 (Fig. S1, B and C), here referred to as local or axonal proteasome inhibition. To evaluate the involvement of UPS in presynaptic differentiation, we first characterized the time course of presynaptic clustering upon local proteasome inhibition. Both MG132 and β-lactone caused a robust increase in the density of presynaptic clusters that peaked at 1 h with a decrease afterward (Fig. 1, B and C; and Fig. S1 D), which is likely caused by disassembly of the newly generated orphan presynapses (Yamada et al., 2013). The rapid assembly of presynaptic clusters (1 h) is in agreement with the proposed time line for synapse formation (Friedman et al., 2000). We therefore used this time point in subsequent experiments. We validated the clustering specificity of our presynaptic phenotype by excluding the possibility of a random increase in markers’ total levels caused by less degradation (Fig. S1, E–G).
Figure 1.
Axonal proteasome inhibition boosts formation of presynaptic sites. (A) Microfluidic devices. (B) Local proteasome inhibition by 10 µM β-lactone and 1 µM MG132 (box in A) induced an increase in presynaptic clusters (arrowheads), assessed by immunostaining for the SV marker VGluT1 (red) and the active zone marker Bassoon (green). (C) Number of presynaptic clusters (colocalized VGluT1 and Bassoon punctum >0.05 µm2) per axonal length (percentage of 0 min). Three to five independent experiments. (D and E) Formation of new functional presynaptic sites on beads by repeated cycles of FM dye staining. (F) New FM dye puncta (red) appear on beads (dashed circles) after local β-lactone or MG132 treatment (1 h; box in D). (G) Number of new total and active FM puncta per bead. n, beads. (H) Synapse formation chambers. (I) Proteasome inhibition in the synaptic compartment (box in H) triggered presynaptic assembly (arrowheads) onto dendrites, assessed by presynaptic expression of VGluT1mCherry and staining for MAP2 (blue) and Bassoon (green). (J and K) Number of VGluT1mCherry puncta (J) and presynaptic clusters (K) along dendrites per length of transduced axons (percentage of control). n, dendrites. (G, J, and K) Three independent experiments. ***, P < 0.001; **, P < 0.01; and *, P < 0.05 compared with 0 min or control and ###, P < 0.001 and ##, P < 0.01 (Kruskal-Wallis test followed by Dunn’s multiple comparison test). Results are presented as mean values ± SEM. Bars, 5 µm.
To detect the appearance of newly formed functional presynaptic terminals upon local proteasome inhibition, we used poly-d-lysine (PDL)–coated beads, a synapse-inducing system (Lucido et al., 2009) that spatially restricts sites of presynaptic differentiation. Indeed, when added to the axonal side, they cluster presynaptic material (Taylor et al., 2013), which was enhanced by local proteasome inhibition (Fig. S1, H and I). Live formation of new active sites on beads (Fig. 1 D) was evaluated by repeated cycles of styryl (FM) dye staining (Fig. 1 E), as performed elsewhere (Friedman et al., 2000; Shen et al., 2006). Proteasome inhibitors increased the number of new FM puncta on beads in comparison with vehicle-treated axons (Fig. 1, F and G). Approximately half of these new puncta were capable of releasing the dye upon stimulation (Fig. 1 G), thus showing that local proteasome inhibition leads to the formation of new functional presynaptic sites.
To evaluate the relevance of localized proteasome inhibition for presynaptic differentiation in an axodendritic synapse, we used microfluidic chambers specialized for the compartmentalization of synapses (Fig. 1 H; Taylor et al., 2010). The excitatory synaptic vesicle (SV) marker VGluT1mCherry was presynaptically expressed with axons of transduced neurons crossing to the synaptic compartment (Fig. S1 J) and forming synaptic contacts with dendrites (Fig. S1, J and K). Proteasome inhibition in the synaptic compartment increased the number of presynaptic clusters formed on dendrites (Fig. 1, I–K). Analysis of VGluT1mCherry-containing presynaptic clusters, whose somatodendritic elements were not exposed to treatment, demonstrates that inhibition of the proteasome in distal axons enhances their capability of establishing synapses with a postsynaptic partner. Collectively, these results demonstrate a role for proteasome inhibition at the initial stages of presynaptic assembly.
Presynaptic assembly is accompanied by an on-site decrease in proteasome activity
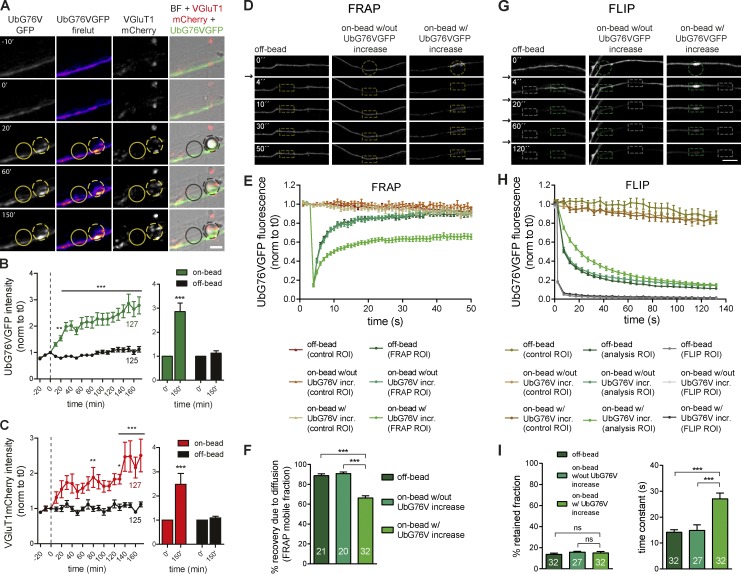
We next asked whether contact with a postsynaptic partner induces changes in the rate of proteasome-mediated degradation along the axon. To address this question, formation of stable presynaptic clusters on beads (Fig. 2) and dendrites (Fig. 3) was monitored using live imaging. Neurons in microfluidic devices coexpressing VGluT1mCherry (presynaptic reporter) and UbG76VGFP (degradation reporter in which GFP bears a signal for rapid ubiquitination and proteasome clearance; Fig. S1 B; Dantuma et al., 2000) extended their axons into the axonal compartment, to which beads were subsequently added (Fig. S2, A and B). Upon contact with a bead, axons responded rapidly (20 min) with an on-bead increased intensity of the degradation reporter that remained elevated, with no detectable changes in adjacent (off-bead) segments (Fig. 2, A and B). This bead-specific enhancement of UbG76VGFP intensity was not due to increases in axonal volume, at least during the first 1 h of contact (Fig. S2, C and D). Moreover, FRAP and fluorescence loss in photobleaching (FLIP) on bead-contacting axons indicated that it was not solely dependent on diffusion from adjacent regions (Fig. 2, D–F) or due to increased retention of the reporter at bead contact sites (Fig. 2, G–I), respectively. Combined, these results indicate that local changes in UbG76VGFP are due to decreased UPS activity at axonal domains contacting beads. Clustering of VGluT1mCherry was later observed at 150 min of bead contact (Fig. 2, A and C). Moreover, poststaining for Bassoon revealed that its clustering was enhanced on beads that were capable of rapidly increasing reporter intensity (Fig. S2, E and F).
Figure 2.
Rapid increase in the degradation reporter intensity on beads precedes clustering. (A) Bead contact (dashed circles) rapidly increased the intensity of UbG76VGFP (firelut and green) with later clustering of VGluT1mCherry (red), not observed in off-bead sites (solid circles). (B and C, left) UbG76VGFP (B) and VGluT1mCherry (C) intensities at on- and off-beads throughout time (normalized to time 0, dashed line). ***, P < 0.001; **, P < 0.01; and *, P < 0.05 for on- versus off-bead (two-way ANOVA). n, beads and equivalent off-sites. (B and C, right) Values at time 0 and 150 min. ***, P < 0.001 (Wilcoxon paired t test). (D and G) Dynamics of UbG76VGFP accumulation on beads (circles) by FRAP (D) and FLIP (G) at off-bead sites and on-bead sites without or with enhanced local intensity of UbG76VGFP (arrows indicate bleaching). (D) Recovery after FRAP (boxes) is not complete at beads with enhanced reporter intensity. (G) Fluorescence loss (green boxes) after FLIP (gray boxes) is almost complete in all conditions. (E and H) Mean changes in UbG76V-GFP fluorescence after FRAP (E) and FLIP (H). incr., increase. Control ROIs, brownish traces. (E) FRAP ROI, greenish traces. Time constant (τ) does not differ, showing unaltered speed of recovery (off-bead, τ = 4.1 ± 0.4; on-bead without UbG76V-GFP, τ = 5.4 ± 1.0; on-bead with UbG76V-GFP, τ = 6.2 ± 0.6). n = 20–32. (H) FLIP ROI, grayish traces; analysis ROI, greenish traces. n = 27–32. (F) Percent UbG76V-GFP recovery after FRAP caused by diffusion from adjacent regions. (I, left) Percentage of UbG76V-GFP–retained fraction after FLIP in adjacent regions. (I, right) Mean time constant (in seconds) shows slower rate of fluorescence loss from beads with enhanced UbG76V-GFP. ns, not significant. (F and I) ***, P < 0.001 (Kruskal-Wallis test followed by the Dunn’s multiple comparison test). n, events. (A–I) Three independent experiments. Results are presented as mean values ± SEM. Bars, 5 µm.
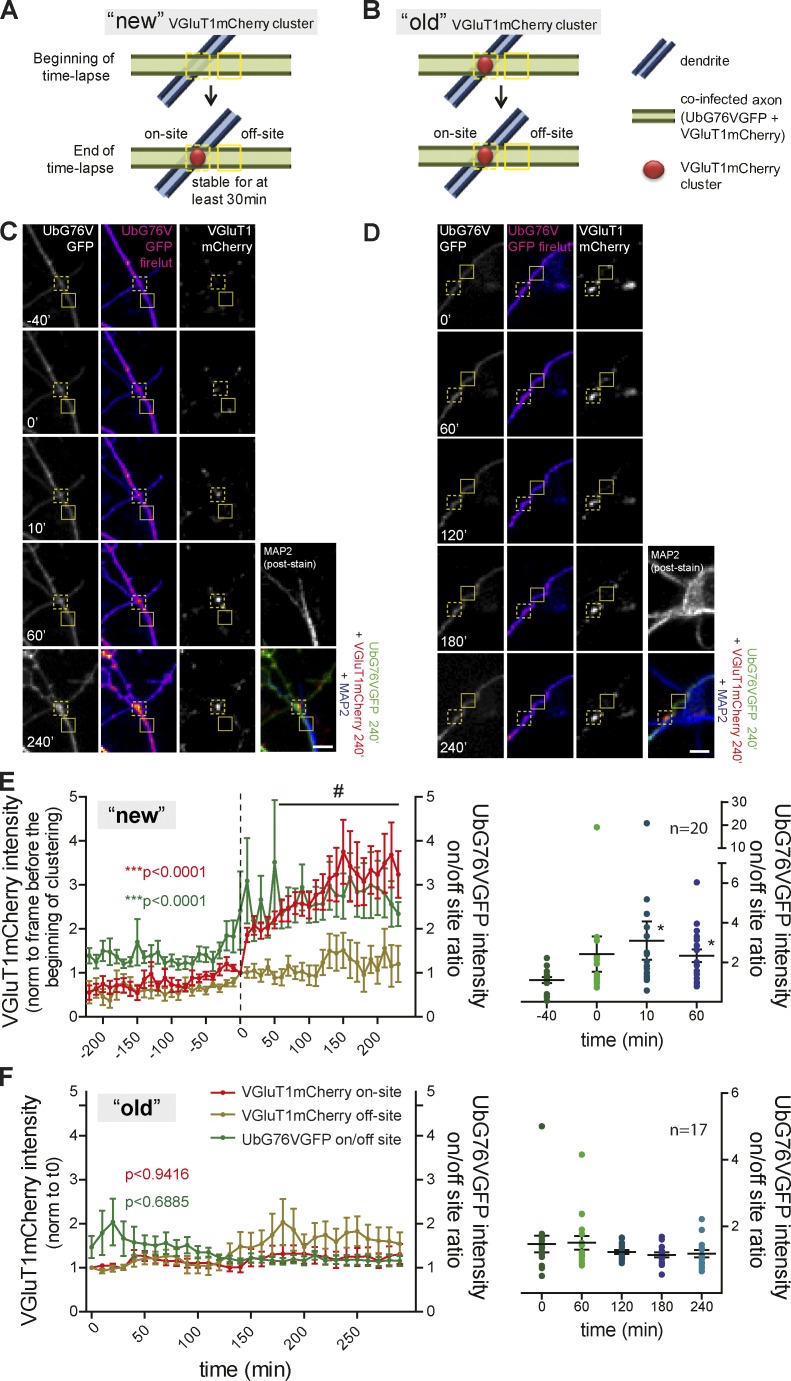
Figure 3.
On-site decreased proteasome activity during formation of stable VGluT1 clusters on dendrites. (A–D) VGluT1mCherry puncta on MAP2+ structures were grouped as “new” (A and C) or “old” (B and D). (C) Increased intensity of UbG76VGFP at the site of clustering (on-site, dashed box) during formation of a new VGluT1mCherry cluster, not observed throughout the lifetime of an old cluster (D). Solid box, off-site. (E and F, left) Time courses of on-site (red line) and off-site (brown line) VGluT1mCherry intensity normalized to time 0 and the on/off-site ratio of UbG76VGFP intensities (green line) for "new" (E) and "old" (F) clusters. For E and F, time 0 corresponds to the frame preceding clustering (dashed line) and the beginning of the time-lapse. VGluT1mCherry (red): #, P < 0.05 for on- versus off-site (***, P < 0.0001, two-way ANOVA). UbG76VGFP ratio (green): ***, P < 0.0001 (Kruskal-Wallis test). (E and F, right) On/off-site ratio of UbG76VGFP intensities at specific time points. *, P < 0.05 in comparison to −40 min (Kruskal-Wallis test followed by the Dunn’s multiple comparison test). n, clusters. Three independent experiments. Results are presented as mean values ± SEM. Bars, 5 µm.
We then addressed this issue in axon–dendrite contacts by monitoring presynaptic clustering on axons contacting MAP2+ structures (Fig. S2, G–I). We quantified changes in the axonal degradation rate occurring at the site of a newly formed VGluT1-mCherry cluster (“new”) in comparison to a preexisting cluster (“old”) on dendrites (Fig. 3, A and B). During formation of a new stable VGluT1mCherry cluster, there was a significant increase in the ratio of UbG76VGFP intensity between the site of clustering and a nonsynaptic adjacent site (UbG76VGFP intensity on/off-site ratio; Fig. 3, C and E). Occasional UbG76VGFP accumulation events accompanied by VGluT1mCherry clustering occurred fleetingly but did not form stable clusters. On the contrary, no local changes in the degradation reporter signal were observed throughout the lifetime of an old puncta (Fig. 3, D and F). In conclusion, assembly of an excitatory presynaptic terminal onto a postsynaptic partner is preceded and accompanied by a localized reduction in the activity of the proteasome.
Proteasome inhibition-induced presynaptic accumulation of ubiquitinated conjugates as the trigger for presynaptic differentiation
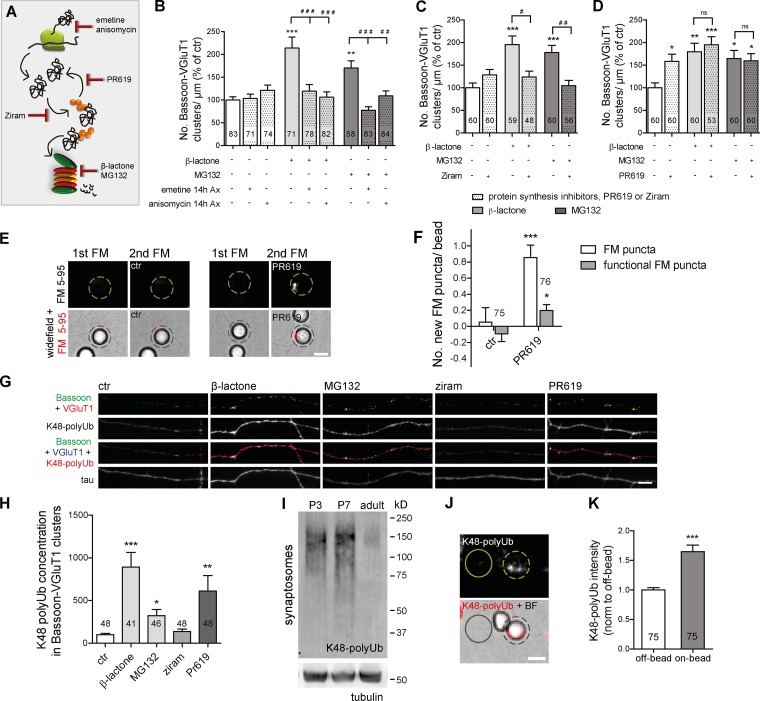
To unmask the local mechanism generating new presynaptic clusters, we first assessed its dependence on protein synthesis (Fig. 4 A). The synaptogenic effect of axonal proteasome inhibition relied on axonal protein synthesis (Fig. 4 B and Fig. S3 A) but not on soma synthesized proteins (Fig. S3, D and E). The fact that presynaptic assembly was only reverted when preceded by long-term (14 h) axonal protein synthesis inhibition (Fig. 4 B and Fig. S3 A), in opposition to an acute (1 h) treatment (Fig. S3, B and C), indicated that rapid accumulation of no longer degraded proteins from a pool of previously synthesized ones underlies the presynaptogenic effect. Interestingly, formation of sensory-motor synapses in Aplysia californica has also been suggested to require local protein synthesis (Lyles et al., 2006). Nonetheless, the difference in terms of synapse type and organisms’ complexity between this study and ours points to the possibility of different mechanisms controlling presynaptic assembly through local translation.
Figure 4.
Presynaptic accumulation of ubiquitinated conjugates is required and sufficient to induce presynaptic assembly. (A) Selective targeting of UPS and protein synthesis. (B–D) Density of presynaptic clusters (percentage of control) after axonal proteasome inhibition in the absence or presence of protein synthesis inhibitors emetine (10 µM) or anisomycin (10 µM; B), ziram (1 µM; C), or PR619 (1 µM; D). Long-term (14 h) inhibition of axonal protein synthesis (B) and inhibition of E1-mediated ubiquitination (1 h; C) abolished the synaptogenic effect of proteasome inhibition. (D) PR619 locally boosted formation of clusters. Five independent experiments. ns, not significant. (E) PR619 led to an increase in the formation of FM puncta on beads (dashed circles; protocol; Fig. 1, D and E). (F) Number of new total and active FM puncta per bead. ***, P < 0.001; *, P < 0.05 (Mann-Whitney test). (G) Accumulation of K48 polyUb (red) signal on presynaptic clusters by axonal proteasome and deubiquitinases inhibition. (H) Quantification of G (percentage of control). Four independent experiments. (I) Representative immunoblot of synaptosomal K48 Ub chains shows up-regulation at early postnatal development. Tubulin, loading control. (J) Axonal accumulation of K48 polyUb conjugates on beads (dashed circles; 4–5 h on the axonal side). Solid circles indicate off-bead. (K) K48 Ub intensity (normalized to off-bead). ***, P < 0.001 (Wilcoxon paired t test). (B, C, D, and H) ***, P < 0.001; **, P < 0.01; and *, P < 0.05 compared with control and ###, P < 0.001; ##, P < 0.01; and #, P < 0.05 (Kruskal-Wallis test followed by the Dunn´s multiple comparison test). n, microscope FOVs. (F and K) n, beads. Three independent experiments. Results are presented as mean values ± SEM. Bars, 5 µm.
Because proteasome blocking triggers an extensive accumulation of ubiquitinated proteins (Fig. S3, J and K) while reducing free Ub levels (Fig. S3, H and I), we next wondered whether the effect would be dependent on de novo ubiquitination. We used an inhibitor of the E1 Ub-activating enzyme, ziram (Fig. 4 A), which prevents formation of E1–Ub conjugates (Rinetti and Schweizer, 2010), and so no degradation of nonubiquitinated proteins occurs. Treatment of isolated axons with ziram alone had no effect on the number of presynaptic clusters, but when in combination with proteasome inhibitors, it led to a complete reversion of their clustering effect (Fig. 4 C and Fig. S3 F). This result demonstrated that accumulation of proteins in their ubiquitinated state is required for local proteasome inhibition-induced presynaptic assembly. To further validate this hypothesis, we used a broad-range inhibitor of deubiquitinases, PR619 (Fig. 4 A; Altun et al., 2011), which expectedly led to an accumulation of ubiquitinated conjugates (Fig. S3, J and K). Similar to proteasome inhibitors, PR619 also increased the number of presynaptic clusters along axons, yet no cumulative effect was observed (Fig. 4 D and Fig. S3 G), suggesting that both inhibitors act through the same mechanism. Furthermore, local inhibition of deubiquitination resulted in a significant increase in the number of new FM puncta on beads (Fig. 4, E and F).
Considering that contact with a postsynaptic partner induced an on-site decreased activity of the proteasome coincident with the moment of presynaptic assembly (Figs. 2, 3, and S2), we asked whether ubiquitinated conjugates accumulate at sites of newly formed presynaptic clusters. Lysine 48 (K48)–linked polyUb chains on substrates function as tags for proteasomal degradation (Thrower et al., 2000) and as such, undergo robust up-regulation upon proteasome inhibition (Fig. S3, L and M; Xu et al., 2009). Staining cultures with an antibody specific for K48 polyUb chains (Apu2; Newton et al., 2008) revealed a pronounced concentration of K48 Ub signals in newly formed presynaptic clusters as opposed to a diffuse pattern in control conditions (Fig. 4, G and H). Interestingly, synaptic expression of K48 Ub chains is higher at developmental stages coincident with the peak of synaptogenesis in the hippocampus (Fig. 4 I; Mody et al., 2001), which recapitulates the pattern observed in whole brain extracts (Chen et al., 2011). Moreover, contact with beads induces accumulation of K48 polyubiquitinated proteins in the contacting axon (Fig. 4, J and K). Altogether, these results demonstrate that polyubiquitinated conjugates bearing the K48 polyUb tag accumulate at nascent presynaptic sites.
Collectively, this set of data suggests that accumulation of polyubiquitinated conjugates at the site of a nascent presynapse, in response to a local halt in proteasome degradation, functions as the trigger for presynaptic clustering.
A role for proteasome-related polyUb chains in presynaptic assembly
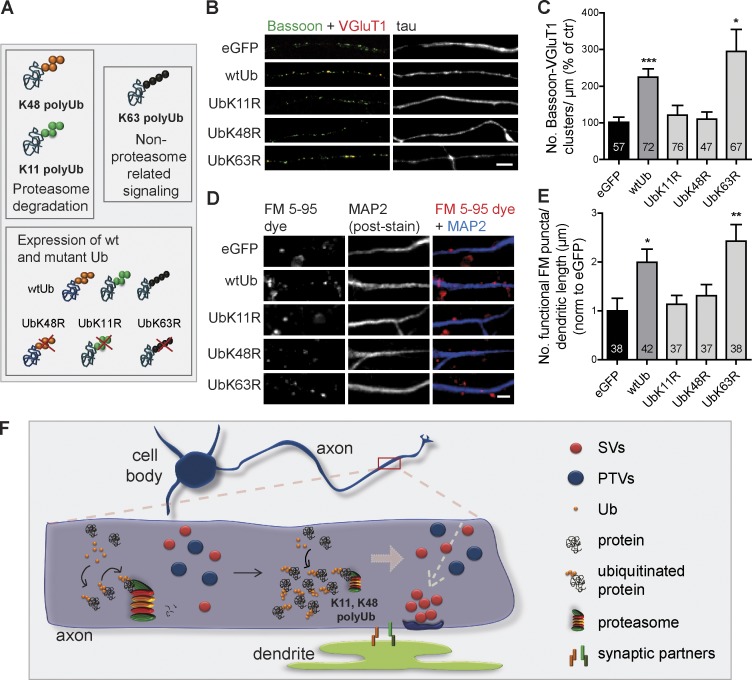
Polyubiquitination can occur at any of the lysines present in Ub 6, 11, 27, 29, 33, 48, and 63 (Peng et al., 2003), with distinct biological outcomes (Sadowski et al., 2012). Although K48 and K11 polyubiquitination triggers proteasome degradation, K63 Ub chains have a role in nondegradative pathways in cells (Fig. 5 A; Sadowski et al., 2012). We further tested our working hypothesis by generating Sindbis virus expressing wild-type (wt) and mutant forms of Ub that prevent polyubiquitination onto specific lysines by their mutation into arginine (UbK11R, UbK48R, and UbK63R; Fig. 5 A and Fig. S3, N–S). Expression of wtUb in neurons cultured in microfluidic devices strongly increased the number of presynaptic clusters (Fig. 5, B and C; and Fig. S3, T–V) and the number of total and functional FM puncta along dendrites (Fig. 5, D and E; and Fig. S3 W), which were suppressed by prevention of K11 and K48 polyubiquitination, as opposed to K63. We thus concluded that up-regulation of polyubiquitinated conjugates upon expression of Ub triggers presynaptic assembly. Interestingly, Ub chains with expected roles in proteasome degradation (K11 and K48) were required. Both proteasome inhibition and prevention of protein polyubiquitination reduce global protein degradation. The difference resides in the state of the substrates being accumulated: polyubiquitinated versus mono- or nonubiquitinated. Accordingly, the lack of synaptogenic effect of UbK48R (or UbK11R) further supports the hypothesis that the effect of proteasome inhibitors is attributed to the remaining pool of polyubiquitinated substrates. We hereby demonstrate that K11- and K48-polyubiquitinated proteins are the triggers for presynaptic formation.
Figure 5.
K48 and K11 polyubiquitination triggers formation of presynaptic sites. (A) Altering levels of endogenous polyUb chains by overexpressing wt and mutant Ub. (B and D) wtUb led to increased presynaptic clusters in isolated axons (B) and FM puncta along dendrites (MAP2, blue; D), which did not occur for UbK11R and UbK48R mutants. eGFP, control. Bars, 5 µm. (C) Number of presynaptic clusters per axonal length (percentage of eGFP). n, microscope FOVs. Five independent experiments. (E) Number of functional FM puncta per dendritic length (normalized to eGFP). n, dendrites. Two individual experiments. (F) Current model: a proteasome inhibition-derived polyUb nest for presynaptic assembly. PTV, piccolo-Bassoon transport vesicle. ***, P < 0.001; **, P < 0.01; and *, P < 0.05 compared with eGFP (Kruskal-Wallis test followed by the Dunn’s multiple comparison test). Results are presented as mean values ± SEM.
A proteasome inhibition-derived polyUb nest for presynaptic assembly
In this study, we unmask a new on-site UPS-related mechanism controlling formation of presynaptic sites. Our results suggest a model in which transient and local reduction of proteasome activity after contact with a postsynaptic partner leads to an on-site accumulation of proteins in their polyubiquitinated state (K48 and K11), which in turn functions as a nesting platform for the clustering of presynaptic material and subsequently, presynaptic differentiation (Fig. 5 F).
Developmental regulation of the UPS controls presynaptic formation. In the rat brain, activity of proteasome catalytic subunits is high at the first postnatal week with subsequent reduction (Petersen et al., 2010). Also, UPS components are up-regulated at this developmental stage, and high concentrations of K48-tagged proteins are observed (Chen et al., 2011). Moreover, in the Drosophila embryo, K48 and K11 Ub chains represent 57% and 11%, respectively, of the neuronal ubiquitome (Franco et al., 2011). These studies show that at the peak of synaptogenesis, the UPS is highly active. Somewhat similar to the effect on sensory synaptic inputs in A. californica motoneurons (Zhao et al., 2003), we show enhanced presynaptic assembly upon local proteasome inhibition. Hence, one can speculate that in the young brain, high proteasome activity acts as a brake on the formation of presynaptic sites, which will be relieved upon spatiotemporal reduction. The idea that local decreased proteasome activity dictates the onset of synaptogenesis implies that the developing axon has ways of promoting endogenous proteasome inhibition. Indeed, dynamic modifications can alter proteasome activity in neurons (Upadhya et al., 2006; Djakovic et al., 2009; Tai et al., 2010; Caldeira et al., 2013; Huang et al., 2013); however, this has yet to be linked to presynaptogenic cues. Contradictorily, in Caenorhabditis elegans axons, the extent of presynaptic differentiation is controlled by localized function of E3 ligases, either by removing kinases that suppress synaptogenesis (Liao et al., 2004; Nakata et al., 2005) or by eliminating unwanted terminals (Ding et al., 2007). These studies reveal a role for enhanced proteasome clearance in the control of nascent presynapses, which together with our results indicate that UPS may govern synapse formation in diverse ways.
In this study, we gathered a substantial set of data demonstrating that formation of presynaptic clusters is triggered by an on-site accumulation of polyubiquitinated proteins. Interestingly, the two types of chains known to target proteins for degradation (K11 and K48; Thrower et al., 2000; Kravtsova-Ivantsiv and Ciechanover, 2012) were identified as the prime signals. So far, few studies have revealed roles for K48 and K11 Ub chains other than this classical labeling (Ye et al., 2003; Flick et al., 2004; Dynek et al., 2010; Goto et al., 2010; Hay-Koren et al., 2011). Herein, we propose a novel role for these two types of Ub linkages in signaling presynaptic assembly. In agreement, various presynaptic proteins are known to be ubiquitinated (Segref and Hoppe, 2009; Bingol and Sheng, 2011; Franco et al., 2011; Na et al., 2012). Future efforts should be directed at identifying the polyubiquitinated local players instructing presynaptic formation. We predict these might be presynaptic scaffolds or elements of the axonal cytoskeleton machinery, which are currently believed to scaffold nascent presynapses (Chia et al., 2013). We further hypothesize that transient enrichment of proteasome-related polyubiquitinated proteins may act as a hub for the recruitment of SV protein transport vesicles and piccolo-Bassoon transport vesicles and formation of en passant boutons (Fig. 5 F). Analogously, some strategies for DNA repair involve recruitment of repair machinery to the site of DNA lesion by the recognition of loci of extensively ubiquitinated histones (Jackson and Durocher, 2013). Also, activation of the NF-κB pathway requires recruitment of kinases to a highly polyubiquitinated signaling complex formed by activation of transmembrane receptors (Kulathu and Komander, 2012). This study opens the exciting question of how a pool of polyubiquitinated conjugates functions as an intracellular signal to presynaptic assembly. Furthermore, we anticipate the need to identify the axonal machinery responsible for the recognition and decoding of these polyUb signals.
Materials and methods
Constructs
F(syn)WRBN-VGluT1mCherry, a vector for lentiviral expression of a fusion version of VGluT1 to mCherry, was provided by E. Herzog (Interdisciplinary Institute for Neuroscience, Bordeaux, France; Herzog et al., 2011). The VGluT1mCherry coding sequence was amplified by PCR (primers 5′-CCACCTCTAGACGCGTGCCGCCGCCATGGAGTTCCGGCAGGAGGAGTT-3′ and 5′-CTGGTCGGATCATTGGGCCCTTACTTGTACAGCTCGTCCATGCCG-3′) and cloned into pSinRep5 vector (Invitrogen) through ApaI and MluI sites by a cloning kit (639648; In-Fusion HD; Takara Bio Inc.), so that a Sindbis viral expression version of this fusion protein was generated. The degradation reporter UbG76VGFP (plasmid 11941; Addgene; Dantuma et al., 2000) and GFP-Ub (plasmid 11928; Addgene) were provided by C. Duarte (University of Coimbra, Coimbra, Portugal). To generate a plasmid for Sindbis viral–mediated expression of the degradation reporter, the UbG76V-GFP coding sequence was amplified by PCR (primers 5′-CCACCTCTAGACGCGTGCCGCCGCCATGCAGATCTTCGTGAAGACTCTG-3′ and 5′-CTGGTCGGATCATTGGGCCCTTACTTGTACAGCTCGTCCATGCCGAGAGT-3′) and cloned into pSinRep5 vector through ApaI and MluI sites by the cloning kit. To generate a Sindbis viral construct for the expression of Ub, the coding sequence of the wtUb was amplified from GFP-Ub by PCR (primers 5′-CACCACCACCTCTAGACGGGCCGCATGCAGATCTTCGTGAA-3′ and 5′-AGGGGCGGAATCTAGATCACCCACCTCTGAGACGGAGTAC-3′) and cloned into pSinRep-IRES-eGFP vector (provided by U. Hengst, Columbia University, New York, NY; Wu et al., 2005) through the XbaI site by the cloning kit. In the resulting plasmid, pSinRep-wtUb-IRES-eGFP, the expression of Ub is under control of the subgenomic promoter, whereas eGFP expression is controlled by a ribosome entry site (IRES). The empty vector pSinRep-IRES-eGFP was used as the control. Site-directed mutagenesis to the coding sequence of Ub inserted into the pSinRep-wtUb-IRES-eGFP plasmid was performed by a site-directed mutagenesis kit (200521; QuikChange II XL; Agilent Technologies). Lysines in positions 11, 48, and 63 of Ub sequence were mutated to arginine to generate the Ub mutant forms UbK11R (primers 5′-GAAGACTCTGACTGGTAGGACCATCACCCTCGA-3′ and 5′-TCGAGGGTGATGGTCCTACCAGTCAGAGTCTTC-3′), UbK48R (primers 5′-AGAGGCTGATCTTTGCTGGAAGACAGCTGGAAGA-3′ and 5′-TCTTCCAGCTGTCTTCCAGCAAAGATCAGCCTCT-3′), and UbK63R (primers 5′-CCTGTCTGACTACAACATCCAGAGAGAGTCCACCCT-3′ and 5′-AGGGTGGACTCTCTCTGGATGTTGTAGTCAGACAGG-3′), respectively.
Antibodies
We used the following primary antibodies: mouse monoclonal Bassoon (1:400, immunocytochemistry [ICC]; 1:1,000, Western blot [WB]; ADI-VAM-PS003; Enzo Life Sciences), rabbit monoclonal K48 polyUb (Apu2; 1:500, ICC; 1:1,000, WB; 05-1307; EMD Millipore), chicken polyclonal MAP2 (1:5,000, ICC; AB5543; EMD Millipore), chicken polyclonal tau (1:1,000, ICC; ab75714; Abcam), mouse monoclonal tubulin (1:300,000, WB; T7816; Sigma-Aldrich), mouse monoclonal tuj1 (1:1,000, ICC; MMS-435P; Covance), rabbit polyclonal Ub (1:200, ICC; 1:1,000, WB; Z0458; Dako), guinea pig polyclonal VGluT1 (1:1,500, ICC; AB5905; EMD Millipore), and rabbit polyclonal VGluT1 (1:5,000, WB; 135503; Synaptic Systems). As for secondary antibodies, alkaline phosphatase–conjugated antibodies (Jackson ImmunoResearch Laboratories, Inc.) were used for blotting, and Alexa Fluor 350–, 488–, 568–, and 647–conjugated antibodies (1:1,000; Thermo Fisher Scientific) were used for immunocytochemistry.
Microfluidic devices for neuron culture
Microfluidic devices consist of a molded polydimethylsiloxane (PDMS) chamber assembled in a glass coverslip (Taylor et al., 2003). The molds for the PDMS devices used in this study were fabricated by N.L. Jeon. Microfluidic devices were prepared and assembled similarly to those previously described (Taylor et al., 2005). In brief, PDMS was prepared from a silicon elastomer kit (Sylgard 184; Dow Corning) and cured in molds, and each device was properly cleaned, sterilized with 75% ethanol, and air dried in the culture hood. Glass coverslips (Assistent) were cleaned in 65% nitric acid and extensively washed with mQH2O (five washes for 30 min each), rinsed twice in 100% ethanol, dried, and sterilized under UV radiation for at least 15 min. The PDMS mold was properly assembled on the glass coverslip under sterile conditions. Coverslips were double coated with PDL (0.1 mg/ml overnight at 37°C) and laminin (2 µg/ml for 2 h at 37°C). Before plating cells, devices were washed once with plating medium (MEM supplemented with 0.026 M NaHCO3, 0.025 M glucose, 1 mM sodium pyruvate, and 10% FBS).
Primary culture of hippocampal neurons
Primary cultures of rat hippocampal neurons were prepared as previously described (Baptista et al., 2010; Baeza et al., 2012), with minor changes. After dissection, hippocampi from E18 Wistar rat embryos were dissociated in 0.045% trypsin/0.01% vol/vol deoxyribonuclease in HBSS for 15 min at 37°C. Hippocampi were then washed once in plating medium containing 10% FBS, thus stopping trypsin activity, and mechanically dissociated in fresh plating medium, and cell density was determined. Cells were plated in plating medium: in 450-µm microfluidic devices, 7 × 104 cells were plated in the somal compartment; in synapse formation chambers, 7 × 104 and 105 cells were plated in the presynaptic and postsynaptic compartment, respectively. Neurons were allowed to attach for 2–4 h, and then plating medium was replaced with culture medium (neurobasal medium supplemented with 2% B27, 25 µM glutamate, 0.5 mM glutamine, and 1:400 penicillin/streptomycin). In microfluidic devices, as a way of reducing glutamate excitotoxicity in growing axons, glutamate-free culture medium was added to the axonal compartment of 450-µm microfluidic devices and to the synaptic compartment of synapse formation chambers. Cells were maintained in a humidified incubator with 5% CO2/95% air at 37°C. At day in vitro (DIV) 3/4, the mitotic inhibitor 5-fluorodeoxyuridine (10-µM final concentration) was added to reduce contamination with glia cells. Cells were allowed to grow, and, unless otherwise indicated, experiments were performed at DIV 7/8, which corresponds to the peak of synaptogenesis in primary hippocampal cultures (Fletcher et al., 1994).
Synaptosome preparation
Purification of synaptosomes was performed as previously described (Ribeiro et al., 2014). Hippocampi from Wistar rats (P3, P7, and adult) were dissected and homogenized in a motor-driven glass Teflon homogenizer (30 stokes and 900 rpm at 4°C) in Hepes-buffered sucrose buffer (0.32 M sucrose and 4 mM Hepes, pH 7.4) supplemented with protease and phosphatase inhibitors (0.2 mM PMSF, 1 µg/ml chymostatin/leupeptin/antipain/pepstatin, 0.1 mM sodium ortovanadate [Na3VO4], and 50 mM NaF). The homogenate was then centrifuged at 900 g for 15 min at 4°C, and the supernatant was collected and centrifuged at 18,000 g for 15 min at 4°C to yield the synaptosomal fraction. It was further washed by resuspension in Hepes-buffered sucrose buffer and centrifugation at 18,000 g for 15 min at 4°C. Quantification of protein was performed by the bicinchoninic acid assay, and 40-µg samples were denatured with denaturating buffer (62.5 mM Tris-HCl, pH 6.8, 10% vol/vol glycerol, 2% vol/vol SDS, 0.01% weight/vol bromophenol blue, and 5% vol/vol β-mercaptoethanol, added fresh) and boiled at 95°C for 5 min before running the WB.
Generation of Sindbis and lentivirus
For the generation of Sindbis virus, the pSinRep construct expressing the desired gene of interest and the helper plasmid DH26S were linearized with either XhoI, PacI, or NotI and properly treated for the removal of RNase contamination. Synthesis of RNA from linearized DNAs was performed by in vitro transcription using the mMESSAGE mMACHINE SP6 kit (1340; Ambion). BHK-1 cells were electroporated with 12 µg DH26S RNA and 12 µg of the desired pSinRep RNA, and production of virus was allowed to occur for 24–36 h. Supernatant was then collected, and virus particles were purified by centrifugation at 60,000 g for 2 h and 20 min at 15°C. The viral pellet was then resuspended in PBS (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, and 1.8 mM KH2PO4, pH 7.4) with 0.1% BSA and stored at −80°C. The virus titer was determined in BHK-1 cells, and the volume of virus for infection was adjusted so that >85% of neurons were transduced. For expression of Ub and its mutant forms in microfluidic devices, expression was allowed to occur for 18–20 h. For live-imaging experiments, cells were incubated with virus for 6–8 h before imaging.
For the generation of lentivirus, HEK293T cells were transfected using calcium phosphate transfection with the lentiviral expression vector F(syn)WRBN-VGluT1mCherry and three lentiviral packaging vectors, pLP1, pLP2, and pLP-VSVG, for the expression of gag/pol genes, rev gene, and VSVG envelope glycoprotein gene, respectively. The supernatant containing virus particles was collected 48–60 h after transfection and concentrated by centrifugation at 60,000 g for 2 h at 22°C. The viral pellet was then resuspended in PBS with 0.1% BSA and stored at −80°C. After infection, expression of protein occurred for 48–60 h.
PDL-coated beads
Aliphatic amine latex beads were prepared as previously described (Taylor et al., 2013). They were incubated with PDL for 30 min at 37°C, centrifuged to remove supernatant, washed twice in sterile mQH2O, and diluted in culture medium. 30 µl of bead suspension was added to the axonal compartment of microfluidic devices and incubated at 37°C for the indicated period of time. For live-imaging experiments, beads were diluted in Hepes-buffered solution imaging medium (119 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 30 mM glucose, and 10 mM Hepes, pH 7.4).
Drug treatment
Treatment of cells with UPS or protein synthesis inhibitors was performed in conditioned medium. For proteasome inhibitors, a preincubation of 30 min and 15 min was performed with β-lactone and MG132, respectively. When cells were cotreated with proteasome inhibitors and protein synthesis inhibitors PR619 or ziram, both inhibitors were added simultaneously. All inhibitors were diluted in conditioned medium from a 1,000× stock in DMSO and added to cells, except for the protein synthesis inhibitors emetine and anisomycin, which were diluted in mQH2O. Equal amounts of DMSO or water were added to the control. β-Lactone, emetine, and anisomycin were applied at 10 µM. As for MG132, PR619, and ziram, a concentration of 1 µM was used.
Biochemistry
Biochemical analysis of protein levels was performed by WB as previously described (Shin et al., 2010; Baptista et al., 2014), with minor changes. Cells were first washed twice in cold PBS. Protein extracts were then prepared by scrapping cells in radioimmunoprecipitation assay lysis buffer (150 mM NaCl, 50 mM Tris-HCl, pH 7.4, 5 mM EGTA, 1% Triton X-100, 0.5% deoxycholate, and 0.1% SDS, pH 7.5, freshly supplemented with 50 mM NaF, 1.5 mM Na3VO4, 0.1 mM PMSF, and 1 µg/ml chymostatin/leupeptin/antipain/pepstatin). Lysates were sonicated and centrifuged at 16,100 g for 10 min at 4°C, and the supernatant was collected. Quantification of protein was performed by the bicinchoninic acid assay, and samples (40 µg in 40 µl) were denatured with denaturating buffer and boiled at 95°C for 5 min. Protein extracts were electrophoresed in a Tris-glycine-SDS buffer (25 mM Tris, 192 mM glycine, and 0.1% weight/vol SDS, pH 8.3) in 7.5% (Bassoon and VGluT1), 15% (free Ub), or 4–15% gradient (ubiquitinated conjugates with Ub and Apu2 antibodies) polyacrylamide gel 1.5 mm thick. Electrotransfer onto a polyvinylidene difluoride membrane was performed either overnight at 40 V at 4°C or by using an equivalent protocol for rapid transfer (250 mA for 4 h or 250 mA for 6 h at 4°C depending on the protein of interest weight). Membranes were washed once with TBS (20 mM Tris and 137 mM NaCl) with 0.1% vol/vol Tween 20 (TBS-T) and blocked for 1 h at RT in TBS-T with 5% nonfat dry milk or 3% BSA. Membranes were again washed for three times with TBS-T and incubated with the primary antibody diluted in TBS-T containing 5% or 0.5% weight/vol nonfat dry milk or 3% BSA. Incubation was performed either overnight at 4°C or at RT for 1 h. After three washes, membranes were incubated for 1 h with alkaline phosphatase–conjugated secondary antibodies (anti-mouse or anti-rabbit, depending on the primary antibody host species) at RT, washed again for three times, and resolved with enhanced chemifluorescence substrate for a maximum of 5 min. Membranes were scanned with a gel and blot imaging system (Storm 860; GE Healthcare), and quantification was performed using ImageQuant software under linear exposure conditions. Whenever necessary, membranes were stripped with 0.2 M NaOH for 20 min and reprobed.
Live-cell imaging
Live-imaging experiments for fluorescently tagged proteins (eGFP and mCherry; excitation wavelength of 488 and 561 nm) and for the FM 5-95 lypophilic styryl dye (T23360; Thermo Fisher Scientific; excitation wavelength of 560 nm) were performed at RT (∼20°C) in imaging medium (119 mM NaCl, 5 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 30 mM glucose, and 10 mM Hepes, pH 7.4). Acquisition was performed using a spinning disk confocal imaging system (CSU-X1-M1N-E; Yokogawa Electric Corporation) configured for a motorized inverted microscope (IX81; Olympus) driven by iQ 3.1 software (Andor Technology). Images were collected with a 60× water objective (1.2 NA; UPLSAPO; Olympus) and an electron-multiplying charged-coupled device (CCD) camera (IXON-X3; Andor Technology).
Before imaging, culture medium was replaced with prewarmed imaging medium, and cells were allowed to recover for 30 min at 37°C. For the time-lapse imaging experiments, the device was mounted on the microscope stage, and positions of interest (in which dually infected axons, UbG76V-GFP+ and VGluT1mCherry+, have crossed the microgrooves into the axonal compartment) were selected. Data were collected sequentially from several defined positions (a maximum of 15 per device). Focal drift during the experiment was corrected automatically using the autofocus feature of the Olympus system. Laser intensities were kept as low as possible, and acquisition at 2 × 2 binning was performed. In experiments involving beads, three frames were captured of the selected positions on the axonal compartment before addition of beads, and then beads were added to both compartments (the same volume was added to both sides to prevent focus drift caused by unbalanced preparation). Time-lapse was resumed 10 min after addition of beads. Frames were acquired as z stacks (35–40 slides, 8–10-µm range) every 10 min.
For the FM 5-95 dye experiments, each cycle of FM dye loading/unloading comprised the following steps: FM 5-95 dye loading solution containing a high KCl concentration (imaging medium with 90 mM KCl supplemented with 10 µM FM 5-95 dye, 20 µM 6-cyano-7-nitroquinoxaline-2,3-dione [CNQX], and 50 µM D-AP5) to promote depolarization was added to microfluidic devices for 1 min and washed once with 10 µM FM 5-95 dye in imaging medium for 1 min. An additional three washes with 1 mM Advasep-7 in imaging medium, for 1 min each, were performed for optimal removal of FM dye excess. This medium was then replaced by imaging medium containing 20 µM CNQX and 50 µM D-AP5, and the device was placed on the microscope stage. CNQX and D-AP5, which are α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid (AMPA)/kainate and N-methyl-d-aspartate (NMDA) receptor antagonists, respectively, were added to the medium to block recurrent excitation. Positive and negative electrodes were placed on each well of either the somal or axonal compartment, and unloading of the FM dye was induced by electrical stimulation, which was performed by a two-channel stimulus generator (model STG4002; Multi Channel Systems) in current mode with an asymmetric waveform (−480 µA for 1 ms and 1,600 µA for 300 µs) at 20 Hz for 1,200 pulses for 1 min. At least four frames were acquired before stimulation. The frames were acquired as z stacks (35–40 slides, 8–10-µm range) every 15 s for 5 min in a 2 × 2 binning mode. FM puncta that could unload >5% of their content after 1 min of electrical stimulation were considered functional presynaptic clusters (Taylor et al., 2013). For detection of functional FM puncta along dendrites (Fig. 5, D and E), one cycle of FM dye loading/unloading protocol was performed on the somal side of microfluidic devices after 18–20-h viral infection with the Ub constructs. Under the brightfield light, positions to image were carefully chosen to include nearly the same number of cell bodies. Dendrites were later detected by retrospective immunocytochemistry for MAP2.
For the experiments in which we were interested in monitoring the appearance of new FM puncta on beads (Fig. 1, D–G; and Fig. 4, E and F), beads were first added to the axonal compartment of microfluidic devices and incubated for 3 h. Then, a first cycle of FM 5-95 dye was performed, followed by complete destaining with high KCl imaging medium for 1 min and two washes with imaging medium containing 20 µM CNQX and 50 µM D-AP5. An image after FM dye destaining was acquired for later subtraction from the initial image of the second FM dye cycle. Cultures were then treated with proteasome inhibitors, PR619 or DMSO, for 1 h. A second cycle of FM dye with live-monitored unloading at exactly the same position was then performed to look for the formation of new functional FM puncta. The position of the beads and the microgrooves were instrumental for localizing the imaging position between the first and second FM dye cycles.
FRAP/FLIP experiments were performed at 37°C with a confocal microscope (LSM 710; ZEISS) driven by Zen Black 2012 software (ZEISS) with a 63× oil differential interference contrast (DIC) objective (M17n; NA 1.40; Plan-Apochromat) at a magnification of 2×. Neurons in microfluidic devices expressing UbG76V-GFP for 12 h were used, and imaging was performed on the axonal compartment. Medium was replaced for imaging medium, and beads were added to the axonal compartment for 20 min before FRAP/FLIP imaging. Imaging was confined to the time interval between 20 min and 1 h after bead contact, and acquisition was done in a single z-slice manner. Bleach was accomplished by performing 10 bleach iterations with the 488-nm laser power set to 100% (100× greater than for acquisition) in a total bleach time of 0.45 s at 5 × 2–µm regions of interest (ROIs) carefully placed along axons. Before bleaching, three baseline images were taken. For FRAP, regions to bleach were chosen at off-bead and on-bead axonal sites. For the latter, a distinction was made between beads causing an increased intensity of UbG76V-GFP (on-bead with UbG76V-GFP increase) and the ones that did not alter its fluorescence along axons (on-bead without UbG76V-GFP increase). A minimal distance of 45 µm was maintained between bleached and control ROIs. After a single bleaching event, changes in fluorescence were analyzed at bleached and control ROIs every 1 s for a total of 50 s. For FLIP, repeated bleaching was performed at a 5-µm distance from the measurement site (off-bead, on-bead with UbG76V-GFP, or on-bead without UbG76V-GFP). An image was taken after each bleaching episode in a total experiment time of 130 s. Changes in fluorescence were analyzed at the bleaching region (FLIP ROI), at the measurement region (analysis ROI), and at regions of nonbleached axons (control ROI) to control for fluorescence decay as a result of bleaching throughout the experiment.
Immunocytochemistry
Immunocytochemistry was performed similarly to that previously described (Baptista et al., 2013). Cells were fixed in prewarmed 4% paraformaldehyde (in PBS with 4% sucrose) for 10 min at RT. A prefixation of 5 min in 1% paraformaldehyde was performed to prevent damaging the population of isolated axons. Cultures were washed three times in PBS and then permeabilized in PBS with 0.25% Triton X-100 for 5 min at RT and washed once in PBS before blocking for 30 min in PBS with 3% BSA. Preparations were incubated with the mix of primary antibodies in 3% BSA either overnight at 4°C or for 2 h at 37°C, washed three times in PBS, and incubated with the mix of secondary antibodies for 1 h at RT in 3% BSA. Cultures were again washed, this time twice in PBS with 0.1% Triton X-100 and once in PBS, and the coverslip was rinsed in mQH2O and mounted in prolong mounting media with or without DAPI. For microfluidic chambers, the PDMS device was disassembled from the coverslip only before mounting on the microscope glass.
Microscopy of antibody-labeled cultures
Fixed preparations were imaged using a microscope (Observer Z.1; ZEISS) equipped with a 63× oil objective (1.4 NA; Plan-NeoFluar), a CCD digital camera (AxioCam HRm), and the Zen Blue 2011 software (ZEISS). In microfluidic devices, unless otherwise indicated, images were taken of the axonal compartment. XY reconstructions of microfluidic devices (with both somal and axonal compartments) were performed either with a confocal microscope (LSM 510 Meta; ZEISS) with a 40× objective (1.3 NA; DIC M27; EC-PlanNeofluar) driven by the LSM 510 software or a spinning disk confocal imaging system (CSU-X1-M1N-E; Yokogawa Electric Corporation) configured for a microscope (IX81; Olympus) driven by Andor iQ 3.1 software with a 60× water objective (1.2 NA; UPLSAPO; Olympus) and an electron-multiplying CCD camera (IXON-X3; Andor Technology). Posthoc immunocytochemistry after live experiments was performed using the spinning disk system. In these preparations, the device was not disassembled after immunocytochemistry, and retrospective imaging was performed in PBS using the 60× water objective. This allowed us to easily locate the multiple positions where live data were collected by using the microgrooves as coordinates. In brief, microgrooves were numbered according to the orientation of the device on the microscope stage, and at the end of acquisition, each position of the microgroove nearer to its center was marked down. The position was then corrected manually by comparing the DIC image acquired at the end of the live experiment with that of the fixed preparation. Moreover, the plug-in Align images by line ROI in ImageJ was further used to guarantee perfect alignment.
Quantitative imaging analysis
Quantification of fluorescence images was performed using ImageJ software (National Institutes of Health). For fixed cells, samples within an experiment were simultaneously stained and imaged with identical settings (exposure time and fluorescence light intensity were kept constant throughout acquisition). Images of random fields of view (FOVs) of either isolated axons or cell bodies were taken. Selection of ROIs to acquire fluorescent images was performed either on the axonal or somal marker to avoid bias acquisition. All images were converted to 8 bit for quantification purposes.
To quantify the number of presynaptic puncta along axons, the axonal marker image was used to select populations of axons to quantify. Axonal length was determined by performing analysis (ImageJ plug-in Analyze skeleton) of a “skeletonized” version of the axonal marker. The sum of the length of all the axonal branches identified in an image was used as the axonal length. Correspondent images of synaptic markers were thresholded (threshold values conserved in individual experiments), and particle analysis was performed to calculate number and area of puncta. To quantify the number of presynaptic clusters (Bassoon-VGluT1 clusters), the presence or absence of VGluT1 puncta within Bassoon puncta ROIs was determined, and the total number of Bassoon ROIs containing VGluT1 puncta was divided by the axonal length. Analysis was limited to Bassoon puncta bigger than the smaller quantifiable object (0.05 µm2) in accordance with our imaging settings (Waters, 2009) in order to exclude the possible contribution of mobile piccolo-Bassoon transport vesicles (0.02 µm2; Zhai et al., 2001; Shapira et al., 2003), thus narrowing down analysis to nascent presynaptic sites. A minimum of 12 microscope FOVs of the axonal side were analyzed per individual experiment in each condition.
For quantification of live experiments, confocal slices were sum projected in ImageJ, and alignment of frames within each video was performed by TurboReg or StackReg plug-ins and converted to 8 bit. In experiments involving accumulation of synaptic material on beads, analysis was performed similarly to a study by Lucido et al. (2009). The brightfield image was used to locate beads in contact with axons, and ROIs for individual beads and adjacent sites along the axon were created (on-bead and off-bead, respectively). Change in signal was quantified by measuring the fluorescence intensity at each bead and correspondent off-bead site in each individual frame of the time-lapse video. Individual values were then normalized to the fluorescence intensity in that site at the frame preceding addition of beads (time 0, 0 min) both for on-bead and off-bead sites.
For the experiment in which formation of presynaptic clusters on dendrites was monitored (Fig. 3, A–F), digital videos of the time-lapse sequence for each position were prepared and carefully analyzed to detect formation of stable VGluT1mCherry clusters in sites not detected at prior time points (“new”) or clusters stable at approximately the same location throughout the entire time-lapse (“old”). Puncta were only considered new if stable until the end of the time-lapse for at least 30 min. After identification of new and old puncta, their locations were overlaid on the corresponding posthoc MAP2 immunostained images (see Microscopy of antibody-labeled cultures for further details on acquisition of images after retrospective labeling). Alignment of VGluT1mCherry and UbG76V-GFP videos with MAP2 retrospective ones was done according to the brightfield images of the same region taken at the end of the time-lapse and the one after immunostaining by the ImageJ plug-in Align images by line ROI. The alignment correction used for DIC images was applied to VGluT1mCherry and UbG76V-GFP, and new and old VGluT1mCherry clusters formed onto dendrites were considered. ROIs encompassing the whole VGluT1mCherry cluster were created at the site of clustering (on-site) and equal-sized ROIs at adjacent axonal sites (off-site; Fig. 3, A and B, on-site and off-site correspond to dashed and solid boxes, respectively). Change in signal was quantified by measuring the fluorescence intensity at each ROI in each individual frame of both VGluT1mCherry and UbG76V-GFP time-lapse videos and used to generate the dataset presented in Fig. 3 (E and F). For VGluT1mCherry, individual values were normalized to the fluorescence intensity in that site at time 0. For new and old puncta, time 0 is considered to be the frame before clustering was initiated or the first frame of the time-lapse, respectively. For new puncta, we considered the beginning of clustering as the frame at which the stable puncta first appeared, recognized as a high increase in fluorescent signal (at least a 30% increase in signal intensity in relation to the previous frame). For UbG76V-GFP, the ratio of its intensities between the on- and off-site was calculated.
For the FM dye experiments, the brightfield image was used to create ROIs encompassing beads or dendrites that contact with axons. Then, the number of FM puncta on each bead or along each dendritic segment was quantified by performing particle analysis in ImageJ. To evaluate the unloading capacity of FM puncta, quantification was adapted from previous work (Taylor et al., 2013). Each puncta intensity is measured throughout the registered sequence of images (frames every 15 s for 5 min), normalized to the frame before stimulation, and corrected for the baseline slope (calculated from the change in intensity in the three frames preceding stimulation). Puncta that unloaded >5% of their FM dye content after 1 min of stimulation were considered as functional (Taylor et al., 2013). For the experiment with a double FM dye cycle (Fig. 1, D–G; and Fig. 4, E and F), the net gain in functional and total FM puncta was calculated by subtracting the number of puncta after and before treatment.
For quantification of axonal volume, 0.23-µm–spaced z stacks were acquired from mApple-expressing axons in the axonal compartment. Volume at designated ROIs was quantified with the ImageJ plug-in 3D object counter (Bolte and Cordelières, 2006), which determines the number of voxels in an object and calibrates it into xyz dimensions, with the threshold kept constant between on-bead and the adjacent ROI. 3D surface reconstructions were obtained in ImageJ with the plug-in ImageJ 3D viewer.
For FRAP and FLIP, change in fluorescence at designated ROIs was obtained from the sequence of images, and values were normalized to the frame preceding bleaching. For quantification of FRAP experiments, data for each FRAP event were fitted to a one-phase exponential association function (y = y0 + [plateau − y0] × [1 − exp(−x/τ)], with y0 constrained to between 0 and 1) in Prism 5 (GraphPad Software). Individual values of the plateau and time constant (τ) were obtained for each curve fitting and compared. The recovery fraction was calculated as equal to the plateau × 100 (Fig. 2 F). For FLIP experiments, data for each FLIP event were fitted to a one-phase exponential decay function (y = [y0 − plateau] × exp[−x/τ] + plateau, with y0 constrained to between 0 and 1) in Prism 5 software. Individual values of the plateau and time constant (τ) were obtained for each curve fitting. The retained fraction was calculated as equal to (1 − plateau) × 100 (Fig. 2 I, left).
All images were processed and prepared for presentation using Photoshop (Adobe). Figures were created in Illustrator (Adobe).
Statistical analysis
Results are presented as mean values ± SEM. Graphs and statistical analysis were performed in Prism 5 software. Statistical significance was assessed by nonparametric tests. Mann-Whitney test or Wilcoxon paired t test was performed for comparisons of changes between two groups. For comparisons between multiple groups, we used the Kruskal-Wallis test followed by the Dunn’s multiple comparison test. For the live-imaging data, we performed two-way analysis of variance (ANOVA) with time as the repeated measure to assess changes between two groups throughout time.
Online supplemental material
Fig. S1 provides control experiments validating the specific inhibition of the proteasome in isolated axons and its synaptogenic effect. Fig. S2 gathers additional information that supports a decreased proteasome activity at sites of newly formed presynaptic clusters. Fig. S3 provides supporting information for the triggering of presynaptic assembly by proteasome-related ubiquitinated conjugates. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201509039/DC1.
Supplementary Material
Acknowledgments
We thank Dr. Etienne Herzog for plasmids and Drs. Carlos Duarte and Ulrich Hengst for plasmids and comment on the manuscript. We thank Dr. Luísa Cortes for technical microscope support and Drs. Ana Luísa Carvalho, Carlos Duarte, and João Peça for helpful discussions.
This work was supported by Fundação para a Ciência e Tecnologia (FCT) with grant SFRH/BD/51196/2010; by Federación Española de Enfermedades Raras through Programa Operacional Factores de Competitividade; by national funds through FCT (PTDC/SAU-NEU/104100/2008, EXPL/NEU-NMC/0541/2012, and UID/NEU/04539/2013); and by a Marie Curie Actions International reintegration grant (249288), Seventh Framework program.
A.M. Taylor and N.L. Jeon are inventors of the microfluidic chambers to compartmentalize neurons (US 7419822 B2), and both have financial interest in Xona Microfluidics, LLC. The authors declare no further competing financial interests.
Footnotes
Abbreviations used in this paper:
- ANOVA
- analysis of variance
- CCD
- charged-coupled device
- CNQX
- 6-cyano-7-nitroquinoxaline-2,3-dione
- DIC
- differential interference contrast
- DIV
- day in vitro
- FLIP
- fluorescence loss in photobleaching
- FOV
- field of view
- ICC
- immunocytochemistry
- NMJ
- neuromuscular junction
- PDL
- poly-d-lysine
- PDMS
- polydimethylsiloxane
- ROI
- region of interest
- SV
- synaptic vesicle
- Ub
- ubiquitin
- UPS
- ubiquitin–proteasome system
- WB
- Western blot
- wt
- wild type
References
- Altun M., Kramer H.B., Willems L.I., McDermott J.L., Leach C.A., Goldenberg S.J., Kumar K.G.S., Konietzny R., Fischer R., Kogan E., et al. . 2011. Activity-based chemical proteomics accelerates inhibitor development for deubiquitylating enzymes. Chem. Biol. 18:1401–1412. 10.1016/j.chembiol.2011.08.018 [DOI] [PubMed] [Google Scholar]
- Baeza J.L., de la Torre B.G., Santiveri C.M., Almeida R.D., García-López M.T., Gerona-Navarro G., Jaffrey S.R., Jiménez M.Á., Andreu D., González-Muñiz R., and Martín-Martínez M.. 2012. Cyclic amino acid linkers stabilizing key loops of brain derived neurotrophic factor. Bioorg. Med. Chem. Lett. 22:444–448. 10.1016/j.bmcl.2011.10.107 [DOI] [PubMed] [Google Scholar]
- Baptista F.I., Pinto M.J., Elvas F., Almeida R.D., and Ambrósio A.F.. 2013. Diabetes alters KIF1A and KIF5B motor proteins in the hippocampus. PLoS One. 8:e65515 10.1371/journal.pone.0065515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptista F.I., Pinto M.J., Elvas F., Martins T., Almeida R.D., and Ambrósio A.F.. 2014. Diabetes induces changes in KIF1A, KIF5B and dynein distribution in the rat retina: implications for axonal transport. Exp. Eye Res. 127:91–103. 10.1016/j.exer.2014.07.011 [DOI] [PubMed] [Google Scholar]
- Baptista M.S., Melo C.V., Armelão M., Herrmann D., Pimentel D.O., Leal G., Caldeira M.V., Bahr B.A., Bengtson M., Almeida R.D., and Duarte C.B.. 2010. Role of the proteasome in excitotoxicity-induced cleavage of glutamic acid decarboxylase in cultured hippocampal neurons. PLoS One. 5:e10139 10.1371/journal.pone.0010139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bingol B., and Sheng M.. 2011. Deconstruction for reconstruction: the role of proteolysis in neural plasticity and disease. Neuron. 69:22–32. 10.1016/j.neuron.2010.11.006 [DOI] [PubMed] [Google Scholar]
- Bolte S., and Cordelières F.P.. 2006. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224:213–232. 10.1111/j.1365-2818.2006.01706.x [DOI] [PubMed] [Google Scholar]
- Caldeira M.V., Curcio M., Leal G., Salazar I.L., Mele M., Santos A.R.A., Melo C.V., Pereira P., Canzoniero L.M.T., and Duarte C.B.. 2013. Excitotoxic stimulation downregulates the ubiquitin-proteasome system through activation of NMDA receptors in cultured hippocampal neurons. Biochim. Biophys. Acta. 1832:263–274. 10.1016/j.bbadis.2012.10.009 [DOI] [PubMed] [Google Scholar]
- Chen P.C., Qin L.N., Li X.M., Walters B.J., Wilson J.A., Mei L., and Wilson S.M.. 2009. The proteasome-associated deubiquitinating enzyme Usp14 is essential for the maintenance of synaptic ubiquitin levels and the development of neuromuscular junctions. J. Neurosci. 29:10909–10919. 10.1523/JNEUROSCI.2635-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen P.C., Bhattacharyya B.J., Hanna J., Minkel H., Wilson J.A., Finley D., Miller R.J., and Wilson S.M.. 2011. Ubiquitin homeostasis is critical for synaptic development and function. J. Neurosci. 31:17505–17513. 10.1523/JNEUROSCI.2922-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chia P.H., Li P., and Shen K.. 2013. Cellular and molecular mechanisms underlying presynapse formation. J. Cell Biol. 203:11–22. 10.1083/jcb.201307020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cristovão G., Pinto M.J., Cunha R.A., Almeida R.D., and Gomes C.A.. 2014. Activation of microglia bolsters synapse formation. Front. Cell. Neurosci. 8:153 10.3389/fncel.2014.00153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantuma N.P., Lindsten K., Glas R., Jellne M., and Masucci M.G.. 2000. Short-lived green fluorescent proteins for quantifying ubiquitin/proteasome-dependent proteolysis in living cells. Nat. Biotechnol. 18:538–543. 10.1038/75406 [DOI] [PubMed] [Google Scholar]
- DiAntonio A., Haghighi A.P., Portman S.L., Lee J.D., Amaranto A.M., and Goodman C.S.. 2001. Ubiquitination-dependent mechanisms regulate synaptic growth and function. Nature. 412:449–452. 10.1038/35086595 [DOI] [PubMed] [Google Scholar]
- Ding M., Chao D., Wang G., and Shen K.. 2007. Spatial regulation of an E3 ubiquitin ligase directs selective synapse elimination. Science. 317:947–951. 10.1126/science.1145727 [DOI] [PubMed] [Google Scholar]
- Djakovic S.N., Schwarz L.A., Barylko B., DeMartino G.N., and Patrick G.N.. 2009. Regulation of the proteasome by neuronal activity and calcium/calmodulin-dependent protein kinase II. J. Biol. Chem. 284:26655–26665. 10.1074/jbc.M109.021956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dynek J.N., Goncharov T., Dueber E.C., Fedorova A.V., Izrael-Tomasevic A., Phu L., Helgason E., Fairbrother W.J., Deshayes K., Kirkpatrick D.S., and Vucic D.. 2010. c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J. 29:4198–4209. 10.1038/emboj.2010.300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fletcher T.L., De Camilli P., and Banker G.. 1994. Synaptogenesis in hippocampal cultures: evidence indicating that axons and dendrites become competent to form synapses at different stages of neuronal development. J. Neurosci. 14:6695–6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flick K., Ouni I., Wohlschlegel J.A., Capati C., McDonald W.H., Yates J.R., and Kaiser P.. 2004. Proteolysis-independent regulation of the transcription factor Met4 by a single Lys 48-linked ubiquitin chain. Nat. Cell Biol. 6:634–641. 10.1038/ncb1143 [DOI] [PubMed] [Google Scholar]
- Franco M., Seyfried N.T., Brand A.H., Peng J., and Mayor U.. 2011. A novel strategy to isolate ubiquitin conjugates reveals wide role for ubiquitination during neural development. Mol. Cell. Proteomics. 10 10.1074/mcp.M110.002188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman H.V., Bresler T., Garner C.C., and Ziv N.E.. 2000. Assembly of new individual excitatory synapses: time course and temporal order of synaptic molecule recruitment. Neuron. 27:57–69. 10.1016/S0896-6273(00)00009-X [DOI] [PubMed] [Google Scholar]
- Goto E., Yamanaka Y., Ishikawa A., Aoki-Kawasumi M., Mito-Yoshida M., Ohmura-Hoshino M., Matsuki Y., Kajikawa M., Hirano H., and Ishido S.. 2010. Contribution of lysine 11-linked ubiquitination to MIR2-mediated major histocompatibility complex class I internalization. J. Biol. Chem. 285:35311–35319. 10.1074/jbc.M110.112763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay-Koren A., Caspi M., Zilberberg A., and Rosin-Arbesfeld R.. 2011. The EDD E3 ubiquitin ligase ubiquitinates and up-regulates β-catenin. Mol. Biol. Cell. 22:399–411. 10.1091/mbc.E10-05-0440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herzog E., Nadrigny F., Silm K., Biesemann C., Helling I., Bersot T., Steffens H., Schwartzmann R., Nägerl U.V., El Mestikawy S., et al. . 2011. In vivo imaging of intersynaptic vesicle exchange using VGLUT1Venus knock-in mice. J. Neurosci. 31:15544–15559. 10.1523/JNEUROSCI.2073-11.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Wang H., Perry S.W., and Figueiredo-Pereira M.E.. 2013. Negative regulation of 26S proteasome stability via calpain-mediated cleavage of Rpn10 subunit upon mitochondrial dysfunction in neurons. J. Biol. Chem. 288:12161–12174. 10.1074/jbc.M113.464552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson S.P., and Durocher D.. 2013. Regulation of DNA damage responses by ubiquitin and SUMO. Mol. Cell. 49:795–807. 10.1016/j.molcel.2013.01.017 [DOI] [PubMed] [Google Scholar]
- Jin Y., and Garner C.C.. 2008. Molecular mechanisms of presynaptic differentiation. Annu. Rev. Cell Dev. Biol. 24:237–262. 10.1146/annurev.cellbio.23.090506.123417 [DOI] [PubMed] [Google Scholar]
- Kravtsova-Ivantsiv Y., and Ciechanover A.. 2012. Non-canonical ubiquitin-based signals for proteasomal degradation. J. Cell Sci. 125:539–548. 10.1242/jcs.093567 [DOI] [PubMed] [Google Scholar]
- Kulathu Y., and Komander D.. 2012. Atypical ubiquitylation - the unexplored world of polyubiquitin beyond Lys48 and Lys63 linkages. Nat. Rev. Mol. Cell Biol. 13:508–523. 10.1038/nrm3394 [DOI] [PubMed] [Google Scholar]
- Liao E.H., Hung W., Abrams B., and Zhen M.. 2004. An SCF-like ubiquitin ligase complex that controls presynaptic differentiation. Nature. 430:345–350. 10.1038/nature02647 [DOI] [PubMed] [Google Scholar]
- Lucido A.L., Suarez Sanchez F., Thostrup P., Kwiatkowski A.V., Leal-Ortiz S., Gopalakrishnan G., Liazoghli D., Belkaid W., Lennox R.B., Grutter P., et al. . 2009. Rapid assembly of functional presynaptic boutons triggered by adhesive contacts. J. Neurosci. 29:12449–12466. 10.1523/JNEUROSCI.1381-09.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyles V., Zhao Y., and Martin K.C.. 2006. Synapse formation and mRNA localization in cultured Aplysia neurons. Neuron. 49:349–356. 10.1016/j.neuron.2005.12.029 [DOI] [PubMed] [Google Scholar]
- Mody M., Cao Y., Cui Z., Tay K.Y., Shyong A., Shimizu E., Pham K., Schultz P., Welsh D., and Tsien J.Z.. 2001. Genome-wide gene expression profiles of the developing mouse hippocampus. Proc. Natl. Acad. Sci. USA. 98:8862–8867. 10.1073/pnas.141244998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Na C.H., Jones D.R., Yang Y., Wang X., Xu Y., and Peng J.. 2012. Synaptic protein ubiquitination in rat brain revealed by antibody-based ubiquitome analysis. J. Proteome Res. 11:4722–4732. 10.1021/pr300536k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata K., Abrams B., Grill B., Goncharov A., Huang X., Chisholm A.D., and Jin Y.. 2005. Regulation of a DLK-1 and p38 MAP kinase pathway by the ubiquitin ligase RPM-1 is required for presynaptic development. Cell. 120:407–420. 10.1016/j.cell.2004.12.017 [DOI] [PubMed] [Google Scholar]
- Neto E., Alves C.J., Sousa D.M., Alencastre I.S., Lourenço A.H., Leitão L., Ryu H.R., Jeon N.L., Fernandes R., Aguiar P., et al. . 2014. Sensory neurons and osteoblasts: close partners in a microfluidic platform. Integr Biol (Camb). 6:586–595. 10.1039/c4ib00035h [DOI] [PubMed] [Google Scholar]
- Newton K., Matsumoto M.L., Wertz I.E., Kirkpatrick D.S., Lill J.R., Tan J., Dugger D., Gordon N., Sidhu S.S., Fellouse F.A., et al. . 2008. Ubiquitin chain editing revealed by polyubiquitin linkage-specific antibodies. Cell. 134:668–678. 10.1016/j.cell.2008.07.039 [DOI] [PubMed] [Google Scholar]
- Peng J., Schwartz D., Elias J.E., Thoreen C.C., Cheng D., Marsischky G., Roelofs J., Finley D., and Gygi S.P.. 2003. A proteomics approach to understanding protein ubiquitination. Nat. Biotechnol. 21:921–926. 10.1038/nbt849 [DOI] [PubMed] [Google Scholar]
- Petersen A., Honarvar A., and Zetterberg M.. 2010. Changes in activity and kinetic properties of the proteasome in different rat organs during development and maturation. Curr. Gerontol. Geriatr. Res. 2010 10.1155/2010/230697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro L.F., Catarino T., Santos S.D., Benoist M., van Leeuwen J.F., Esteban J.A., and Carvalho A.L.. 2014. Ghrelin triggers the synaptic incorporation of AMPA receptors in the hippocampus. Proc. Natl. Acad. Sci. USA. 111:E149–E158. 10.1073/pnas.1313798111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinetti G.V., and Schweizer F.E.. 2010. Ubiquitination acutely regulates presynaptic neurotransmitter release in mammalian neurons. J. Neurosci. 30:3157–3166. 10.1523/JNEUROSCI.3712-09.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadowski M., Suryadinata R., Tan A.R., Roesley S.N.A., and Sarcevic B.. 2012. Protein monoubiquitination and polyubiquitination generate structural diversity to control distinct biological processes. IUBMB Life. 64:136–142. 10.1002/iub.589 [DOI] [PubMed] [Google Scholar]
- Segref A., and Hoppe T.. 2009. Think locally: control of ubiquitin-dependent protein degradation in neurons. EMBO Rep. 10:44–50. 10.1038/embor.2008.229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapira M., Zhai R.G., Dresbach T., Bresler T., Torres V.I., Gundelfinger E.D., Ziv N.E., and Garner C.C.. 2003. Unitary assembly of presynaptic active zones from Piccolo-Bassoon transport vesicles. Neuron. 38:237–252. 10.1016/S0896-6273(03)00207-1 [DOI] [PubMed] [Google Scholar]
- Shen W., Wu B., Zhang Z., Dou Y., Rao Z.R., Chen Y.R., and Duan S.. 2006. Activity-induced rapid synaptic maturation mediated by presynaptic cdc42 signaling. Neuron. 50:401–414. 10.1016/j.neuron.2006.03.017 [DOI] [PubMed] [Google Scholar]
- Shin S.B.Y., Almeida R.D., Gerona-Navarro G., Bracken C., and Jaffrey S.R.. 2010. Assembling ligands in situ using bioorthogonal boronate ester synthesis. Chem. Biol. 17:1171–1176. 10.1016/j.chembiol.2010.09.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai H.C., Besche H., Goldberg A.L., and Schuman E.M.. 2010. Characterization of the brain 26S proteasome and its interacting proteins. Front. Mol. Neurosci. 3:1–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A.M., Rhee S.W., Tu C.H., Cribbs D.H., Cotman C.W., and Jeon N.L.. 2003. Microfluidic multicompartment device for neuroscience research. Langmuir. 19:1551–1556. 10.1021/la026417v [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A.M., Blurton-Jones M., Rhee S.W., Cribbs D.H., Cotman C.W., and Jeon N.L.. 2005. A microfluidic culture platform for CNS axonal injury, regeneration and transport. Nat. Methods. 2:599–605. 10.1038/nmeth777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A.M., Berchtold N.C., Perreau V.M., Tu C.H., Li Jeon N., and Cotman C.W.. 2009. Axonal mRNA in uninjured and regenerating cortical mammalian axons. J. Neurosci. 29:4697–4707. 10.1523/JNEUROSCI.6130-08.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A.M., Dieterich D.C., Ito H.T., Kim S.A., and Schuman E.M.. 2010. Microfluidic local perfusion chambers for the visualization and manipulation of synapses. Neuron. 66:57–68. 10.1016/j.neuron.2010.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A.M., Wu J., Tai H.C., and Schuman E.M.. 2013. Axonal translation of β-catenin regulates synaptic vesicle dynamics. J. Neurosci. 33:5584–5589. 10.1523/JNEUROSCI.2944-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thrower J.S., Hoffman L., Rechsteiner M., and Pickart C.M.. 2000. Recognition of the polyubiquitin proteolytic signal. EMBO J. 19:94–102. 10.1093/emboj/19.1.94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian X., and Wu C.. 2013. The role of ubiquitin-mediated pathways in regulating synaptic development, axonal degeneration and regeneration: insights from fly and worm. J. Physiol. 591:3133–3143. 10.1113/jphysiol.2012.247940 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upadhya S.C., Ding L., Smith T.K., and Hegde A.N.. 2006. Differential regulation of proteasome activity in the nucleus and the synaptic terminals. Neurochem. Int. 48:296–305. 10.1016/j.neuint.2005.11.003 [DOI] [PubMed] [Google Scholar]
- Waters J.C. 2009. Accuracy and precision in quantitative fluorescence microscopy. J. Cell Biol. 185:1135–1148. 10.1083/jcb.200903097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K.Y., Hengst U., Cox L.J., Macosko E.Z., Jeromin A., Urquhart E.R., and Jaffrey S.R.. 2005. Local translation of RhoA regulates growth cone collapse. Nature. 436:1020–1024. 10.1038/nature03885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu P., Duong D.M., Seyfried N.T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., and Peng J.. 2009. Quantitative proteomics reveals the function of unconventional ubiquitin chains in proteasomal degradation. Cell. 137:133–145. 10.1016/j.cell.2009.01.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada T., Yang Y., Huang J., Coppola G., Geschwind D.H., and Bonni A.. 2013. Sumoylated MEF2A coordinately eliminates orphan presynaptic sites and promotes maturation of presynaptic boutons. J. Neurosci. 33:4726–4740. 10.1523/JNEUROSCI.4191-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Meyer H.H., and Rapoport T.A.. 2003. Function of the p97-Ufd1-Npl4 complex in retrotranslocation from the ER to the cytosol: dual recognition of nonubiquitinated polypeptide segments and polyubiquitin chains. J. Cell Biol. 162:71–84. 10.1083/jcb.200302169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhai R.G., Vardinon-Friedman H., Cases-Langhoff C., Becker B., Gundelfinger E.D., Ziv N.E., and Garner C.C.. 2001. Assembling the presynaptic active zone: a characterization of an active one precursor vesicle. Neuron. 29:131–143. 10.1016/S0896-6273(01)00185-4 [DOI] [PubMed] [Google Scholar]
- Zhao Y., Hegde A.N., and Martin K.C.. 2003. The ubiquitin proteasome system functions as an inhibitory constraint on synaptic strengthening. Curr. Biol. 13:887–898. 10.1016/S0960-9822(03)00332-4 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.