Abstract
Many high-quality genomes are available for dixenous (two hosts) trypanosomatid species of the genera Trypanosoma, Leishmania, and Phytomonas, but only fragmentary information is available for monoxenous (single-host) trypanosomatids. In trypanosomatids, monoxeny is ancestral to dixeny, thus it is anticipated that the genome sequences of the key monoxenous parasites will be instrumental for both understanding the origin of parasitism and the evolution of dixeny. Here, we present a high-quality genome for Leptomonas pyrrhocoris, which is closely related to the dixenous genus Leishmania. The L. pyrrhocoris genome (30.4 Mbp in 60 scaffolds) encodes 10,148 genes. Using the L. pyrrhocoris genome, we pinpointed genes gained in Leishmania. Among those genes, 20 genes with unknown function had expression patterns in the Leishmania mexicana life cycle suggesting their involvement in virulence. By combining differential expression data for L. mexicana, L. major and Leptomonas seymouri, we have identified several additional proteins potentially involved in virulence, including SpoU methylase and U3 small nucleolar ribonucleoprotein IMP3. The population genetics of L. pyrrhocoris was also addressed by sequencing thirteen strains of different geographic origin, allowing the identification of 1,318 genes under positive selection. This set of genes was significantly enriched in components of the cytoskeleton and the flagellum.
The trypanosomatids (family Trypanosomatidae, class Kinetoplastea) are a group of protists distinguished by a highly specialized mitochondrion and a prominent mitochondrial genome, the kinetoplast. All known trypanosomatids are exclusively parasitic and found primarily in insects1. Three lineages have developed a dixenous life cycle that involves a secondary host, which can either be a vertebrate or a vascular plant2. Members of the genera Trypanosoma and Leishmania cause serious diseases in animals and humans. Species of the genus Leptomonas have a simpler monoxenous (one-host) life cycle, which is confined to insects, where they almost exclusively occupy the intestinal tract3. Monoxenous trypanosomatids are transmitted from one insect to another via food sharing, coprophagy and/or cannibalism4. Together with Crithidia, Lotmaria, and Leishmania, most nominal Leptomonas species belong to the recently established subfamily Leishmaniinae5. Such a phylogenetic position implies that acquisition of the Leishmania dixenous life style occurred within this clade. We propose that direct comparison of the genomes of Leishmania and Leptomonas will reveal specific genes and/or pathways driving evolution of dixeny within this group1.
The border between monoxenous and dixenous species is not impenetrable. Some monoxenous trypanosomatids can withstand the high temperature encountered in warm-blooded vertebrates6,7,8. It has been proposed recently that some monoxenous species are facultatively dixenous if conditions permit and favor such an alteration of their life cycle9,10. One such requirement is the absence of an efficient immunological mechanism for clearing the parasite from the potential host. Indeed, several monoxenous species have been found co-infecting vertebrates along with HIV, Trypanosoma cruzi, or Leishmania spp9,10,11.
Our knowledge of protist genomes is skewed towards parasitic species, with trypanosomatids being one of the most prominent cases12. However, this applies exclusively to dixenous trypanosomatids, as they encompass all medically and veterinary relevant species13. Hence, dozens of species and strains of Leishmania and Trypanosoma have been subjected to genome-wide and transcriptome analyses by next-generation sequencing (reviewed in12,14,15). The genomes of all studied Leishmania species are fairly similar and range in size from 27 Mb in Leishmania enriettii (W. Warren and S. Beverley, pers. commun.) to 33 Mb in L. major and L. infantum16,17. The number of chromosomes ranges between 34 and 36, and the overall genome synteny for these species is remarkably conserved. Genome sizes of characterized trypanosomes vary from 22 Mb in T. b. gambiense to 47.5 Mb in T. vivax18,19,20. The genomes of Leishmania and Trypanosoma are densely packed with almost invariably intron-less genes organized into long polycistronic clusters21.
The only four monoxenous sequenced genomes, albeit in draft quality, are from Lotmaria passim (published as Crithidia mellificae strain SF) parasitizing honey bees (scaffold N50 = 32 kb), Leptomonas seymouri isolated from the cotton stainer (N50 = 70.6 kb), and two endosymbiont-bearing species, Angomonas deanei and Strigomonas culicis described from hemipterid bugs and mosquitoes (N50 = 2.5 and 2.7 kb, respectively)8,22,23. An annotated genome sequence of Crithidia fasciculata encountered in dipteran flies is available in the TriTryp database (N50 = 920 kb, but with 427 unplaced contigs comprising nearly 8.7 Mb)24, in addition to contigs of an unidentified Leptomonas sp. (N50 = 3.4 kb), and Herpetomonas muscarum (N50 = 6.8 kb) in GenBank25,26. Preliminary analyses of the genomic content of these species revealed several genes specific for monoxenous lineages, but none has been examined in detail.
The model organism for the current study is Leptomonas pyrrhocoris (Zotta 1912). This species was isolated from the midgut of the firebug Pyrrhocoris apterus (Heteroptera: Pyrrhocoridae), but the range of suitable hosts is now known to embrace several species of the hemipterid family Pyrrhocoridae, including Pyrrhocoris apterus, P. marginatus and Scantius aegyptius in Europe and the Mediterranean, Dysdercus poecilius in China, and several Dysdercus spp. in the Neotropics and Africa. Our previous work suggested that L. pyrrhocoris originated in the Neotropics3. This is one a few truly globally distributed monoxenous trypanosomatids, the dissemination of which is facilitated by the omnipresence of its Pyrrhocoridae hosts in all regions studied thus far2.
The population genetics and speciation of monoxenous trypanosomatids remains poorly investigated, with information exclusively derived from dixenous Trypanosoma and Leishmania species27. The theory of preponderant clonal evolution states that, in natural populations of pathogens, recombination is infrequent and thus asexual binary division is the main means of reproduction28. Nevertheless, recombination has been documented in many Trypanosomatidae species, including Leishmania major, L. donovani, L. guyanensis, L. infantum, Trypanosoma cruzi, T. brucei, and T. congolense29,30,31. The relative contribution of these competing evolutionary forces remains to be scrutinized further, but some Leishmania hybrids were shown to have enhanced their transmission potential and fitness32,33. The only monoxenous species investigated in this regard are Crithidia bombi and C. fasciculata34,35,36. While recombination was not demonstrated in C. fasciculata, different strains of C. bombi regularly exchange genetic material, with occasional crossing during mixed infections37,38.
Here we describe the first complete, annotated genome of a monoxenous trypanosomatid parasite, L. pyrrhocoris. We also present population analysis of 13 isolates of L. pyrrhocoris collected worldwide, analysis of positive selection signatures in the protein-coding genes, and of their synteny, as compared to model trypanosomatids. We investigated gene family gains and losses in a phylogeny including dixenous species with published genomes and five monoxenous species (L. pyrrhocoris, L. seymouri, C. fasciculata, Blechomonas ayalai, Paratrypanosoma confusum), with a focus on genes gained in Leishmania and Leishmaniinae. In addition, we overlapped differential gene expression data for Leishmania mexicana, Leishmania major, and Leptomonas seymouri with gene family phyletic patterns and identified several novel proteins probably involved in Leishmania virulence.
Results
The genome of L. pyrrhocoris: general features
The L. pyrrhocoris genome was assembled almost to chromosome level (see Supplementary Methods for details) and contains 60 scaffolds (37× average coverage with 454 reads, maximum scaffold length 2,995,728 bp) with scaffold N50 of 910,096 bp and a total assembly length of approximately 30.4 Mb. High degree of synteny with other species (Supplementary Note 1, Supplementary Tables S1, S2 and Supplementary Figs S1–S3) indicates that L. pyrrhocoris genome was assembled correctly. Its current annotation contains 10,148 genes, of which 9,878 are protein-coding (plus rRNAs, tRNAs, snRNAs, snoRNAs, and other non-coding RNAs). This number falls within the range of previously annotated genomes of Leishmania and Trypanosoma (8,400 protein-coding genes for L. major Friedlin and 11,567 for T. brucei TREU927, TriTrypDB, release 25)24 and is significantly higher than that of the streamlined genomes of two plant-infecting Phytomonas spp., with 6,381 and 6,451 protein-coding genes39.
Many genes in the L. pyrrhocoris resulted from recent duplications. Using an E-value cut-off of 10−10 combined with a filter on percent identity (>70%) and the ratio of BLAST alignment length to query length (>0.8), the percentage of genes present as two or more homologous copies is estimated at ~14% (1,382 of all protein-coding genes). Percentages of duplicated protein-coding genes for Phytomonas sp. EM1, Phytomonas sp. HART1, L. seymouri, L. major, T. brucei TREU927, and C. fasciculata are 2.8%, 4.1%, 0.8%, 9.8%, 31.1%, and 33.0%, respectively. Notably, most groups of paralogs analyzed here are composed of exact duplicates with 99–100% (in T. brucei TREU927, L. pyrrhocoris, and L. major) or 95–100% in (C. fasciculata) identity at the amino acid level (Supplementary Fig. S4). The same picture was revealed by mapping gene family expansions on the tree of trypanosomatids: most striking expansions were mapped on the C. fasciculata, T. brucei, and T. congolense branches (Supplementary Fig. S5). Gene duplication represents a major genome evolution mechanism among trypanosomatids, given the general lack of transcription regulation in this group40. For example, 102 copies of a putative autophagy gene ATG8, 21 copies of a Leishmania GP46 surface-antigen homolog and 13 folate/biopterin transporters were found in the L. pyrrhocoris genome, probably reflecting the need for higher gene expression levels. On the other hand, certain lineages demonstrate a trend for streamlining of gene families, as exemplified by Phytomonas spp. (the percentage of duplicated genes in Phytomonas sp. EM1 was estimated at 2.8%).
Metabolism and other functional modules in L. pyrrhocoris
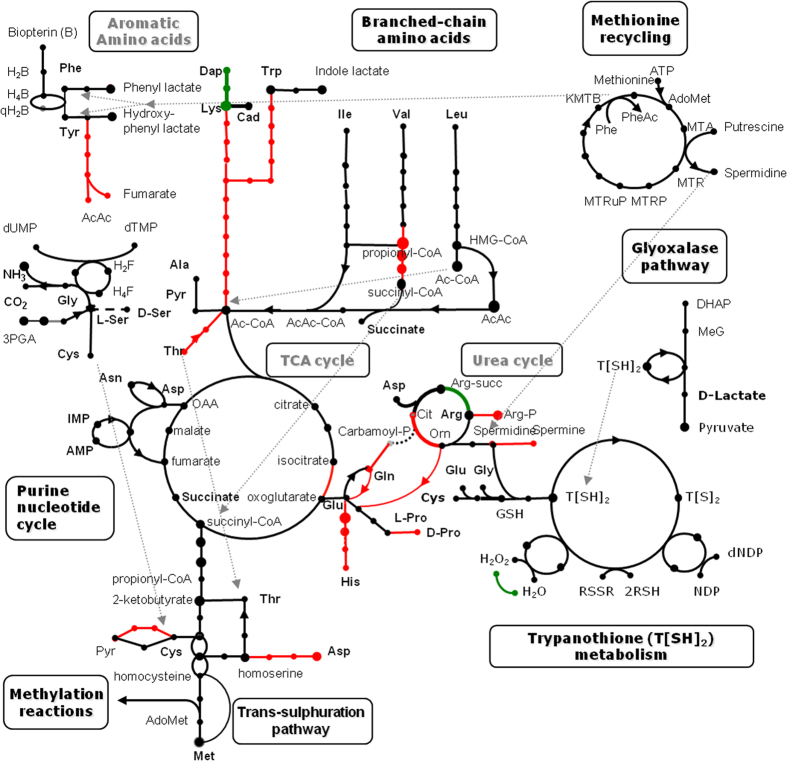
All predicted protein sequences of L. pyrrhocoris were compared with an in-house database of proteins representing over 550 metabolic enzymes from six previously sequenced trypanosomatid genomes as described previously8. The genome of L. pyrrhocoris contains genes coding for a fully functional mitochondrion with respiratory chain and functional glycosomes. A classic glycolytic pathway responsible for the metabolism of various exogenous sugars is partly located inside glycosomes (Supplementary Table S3). Carbohydrate metabolism is characterized by an incomplete aerobic oxidation because one of the classic mitochondrial TCA cycle enzymes, NAD-linked isocitrate dehydrogenase, has been replaced by an NADP-linked isoenzyme, making the cycle non-functional (Supplementary Note 2). This is a general feature of all trypanosomatids studied thus far41. However, all other TCA-cycle enzymes are encoded in the genome (Supplementary Table S4). The enzymes involved in the β-oxidation of fatty acids are also present. Similarly to Leishmania spp., L. pyrrhocoris is able to synthesize its own pyrimidines, but depends on a supply of external purines. Other metabolic features shared with Leishmania include the inability to oxidize aromatic amino acids and requirement for an external supply of most of the essential amino acids, cofactors, and vitamins for growth (Fig. 1, Supplementary Note 2, and Supplementary Table S5). A unique property amongst trypanosomatids, only shared with C. fasciculata, is the capacity to convert diaminopimelic acid, an amino acid of bacterial cell walls, into lysine. Diaminopimelate epimerase, one of the enzymes involved in this process, has apparently been acquired by the clade of C. fasciculata and L. pyrrhocoris, probably via horizontal gene transfer from bacteria, and then lost in L. seymouri. Oxidative stress protection in L. pyrrhocoris, L. seymouri, and C. fasciculata also differs from the other trypanosomatids analyzed thus far - in addition to the trypanothione system and many homologs of tryparedoxins and peroxiredoxins, the ancestor of these species has acquired a bacterial-type catalase by lateral gene transfer (Supplementary Table S6).
Figure 1. Schematic representation of Leptomonas pyrrhocoris amino-acid metabolism and some related pathways.
Each dot represents a metabolite and each line represents the presence or absence of a predicted enzyme. Color coding: black, enzyme present; red, enzyme present in most free-living eukaryotic organisms, but lost in L. pyrrhocoris; green, enzyme present in L. pyrrhocoris and Crithidia fasciculata, but absent in Leishmania.
To illuminate specific features of L. pyrrhocoris, we also focused on other selected pathways and functional modules previously shown to be critical for the trypanosomatid biology, including membrane trafficking and small GTPases (Supplementary Note 3, Supplementary Tables S7, S8, and Supplementary Fig. S6), cell surface proteins (Supplementary Note 4 and Supplementary Figs S7–S9), and protein kinases (Supplementary Note 5 and Supplementary Table S9). Of particular interest is the presence of genes encoding Argonaute, Dicer1 and PIWI-like proteins (Supplementary Table S10). This suggests that L. pyrrhocoris is endowed with RNA interference capacity, similar to the African trypanosomes, Leishmania braziliensis, and C. fasciculata. Interestingly, genes for RNAi are lost in the close relative of L. pyrrhocoris, L. seymouri8.
Gene gains and losses in trypanosomatids
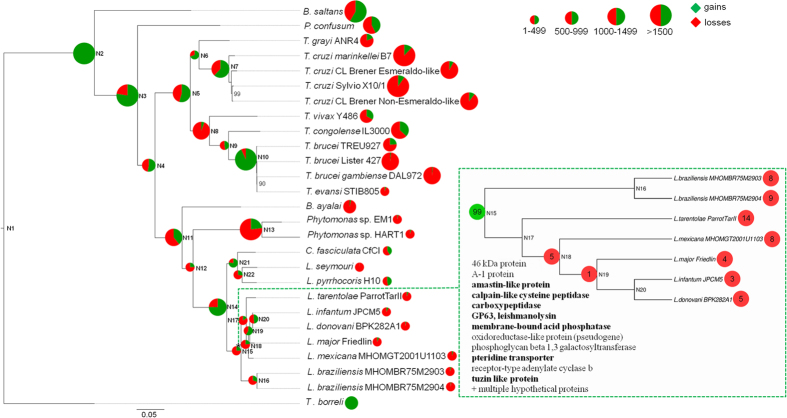
For unraveling evolution of the dixenous life style in Leishmania we focused on lineage-specific gene family gains and losses in the sister lineages Leptomonas/Crithidia and Leishmania, forming the clade Leishmaniinae5. We performed OrthoMCL analysis (see Methods for details) on a dataset of 27 annotated trypanosomatid genomes (Supplementary Table S1). This resulted in 19,866 orthologous groups (OGs), 8,318 of which contained proteins of L. pyrrhocoris. Next, we mapped gene family (i.e., OG) gains and losses on the tree of trypanosomatids with Dollo and Wagner (to infer OG expansions/contractions) parsimony algorithms (Fig. 2, Supplementary Figs S5, S10, S11)42. Gene gains clearly dominate at the basal node of trypanosomatids, and at the basal nodes of Leishmaniinae, Leptomonas/Crithidia, American trypanosomes, T. cruzi, and T. brucei. The other internal nodes and leafs are either dominated by losses or have almost equal counts of gains and losses (Fig. 2; Supplementary Fig. S11).
Figure 2. Gene family gains/losses mapped on the tree of kinetoplastids using Dollo parsimony algorithm.
The maximum likelihood tree is based on the alignment of 57 conserved proteins and inferred using the LG + Г + F model and 1,000 bootstrap replicates. Only bootstrap support values lower than 100% are shown. The horizontal bar represents 0.05 substitutions per site. Gene gains dominate at the basal node of trypanosomatids, and at the basal nodes of Leishmaniinae, Leptomonas/Crithidia, American trypanosomes, T. cruzi, and T. brucei. The other internal nodes and leafs are either dominated by losses or have almost equal counts of gains and losses. An inset figure on the right depicts the losses for 99 OGs gained at the Leishmania node. Annotations for proteins within these OGs are shown at the bottom left corner of the inset. Annotations of the known proteins implicated in virulence are in bold.
The OG gains and losses at the Leishmaniinae, Leishmania, and Leptomonas/Crithidia nodes are of primary interest for identification of novel genes involved in Leishmania virulence (Fig. 2, Supplementary Note 6, and Supplementary Figs S12–S22). Using genome sequences of three monoxenous Leishmaniinae species (L. pyrrhocoris, L. seymouri, and C. fasciculata) as a robust reference, we have delineated a group of 99 OGs gained at the basal node of Leishmania (Fig. 2). This group of proteins includes several known virulence factors, but a great majority, 87 of 99 OGs, is represented by proteins of unknown function, which highlights the need of future gene-focused functional studies. However, in the next section we will use differential expression data to zoom in on a group of most promising virulence factor candidates among Leishmania-specific proteins. Functions of genes gained at the Leishmania and Leishmaniinae nodes and their possible implications for the evolution of dixenous parasitism are discussed in Supplementary Note 6.
Identification of novel Leishmania virulence factor candidates
In our search for new candidates for Leishmania virulence factors we used OG gains/losses mapped on the tree of trypanosomatids and differential gene expression data generated by our team for Leptomonas seymouri (genes up-regulated at an elevated temperature8), Leishmania major LV561 (genes up-regulated in a virulent isolate compared to an avirulent one) and L. mexicana M379 (genes overexpressed at the virulent stages of the life cycle: metacyclic promastigotes and amastigotes) (Fig. 3, Supplementary Figs S23–S25). At least some of Leishmania-specific virulence factors are expected to be gained either at the Leishmaniinae node N14 or at the basal Leishmania node N15 (Fig. 2). In order to verify this assumption, we first focused on 47 proteins involved in virulence and confirmed by experimental studies (Supplementary Table S11), identified corresponding OGs and mapped their gains and losses on the tree (Supplementary Fig. S26). Five OGs were gained at the Leishmaniinae node, four at nodes within Leishmania, and the rest at more basal nodes of the tree.
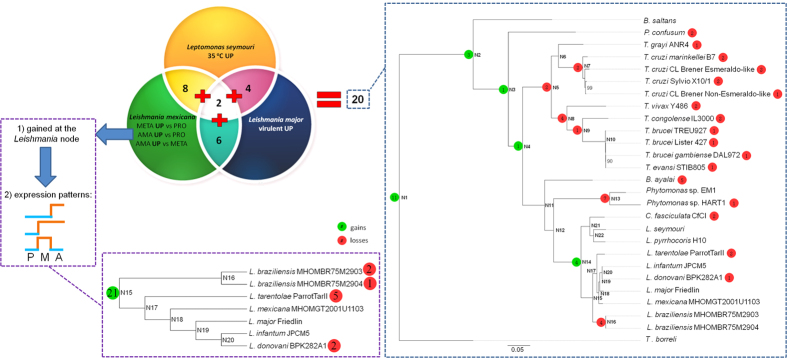
Figure 3. Approaches used for identification of novel Leishmania virulence factors.
The Venn diagram represents two- and three-way intersections between differential expression datasets (A) (Leptomonas seymouri genes up-regulated at elevated temperature of 35 °C compared to 23 °C), (B) (genes up-regulated in a Leishmania major LV561 virulent isolate compared to an avirulent one), and (C) (differential expression data for L. mexicana developmental stages). Two phylogenetic trees of kinetoplastids with gene family gains/losses mapped using Dollo parsimony algorithm are shown. Gain and loss counts for 20 OGs obtained through overlapping the differential expression datasets are depicted in the tree on the right. The Leishmania cladogram at the bottom shows gains and losses for 21 OGs containing 40 L. mexicana genes gained at the Leishmania node and having differential expression patterns suggesting a potential role in virulence. Leishmania life cycle stages are abbreviated as follows: PRO or P, procyclics; META or M, metacyclics; AMA or A, amastigotes.
Then, we took only those OGs gained at the Leishmania node which included L. mexicana M379 genes with relevant expression profiles in the life cycle, 21 OGs in total (Fig. 3, and Supplementary Table S12). The first group includes 35 genes (in 16 OGs) up-regulated in L. mexicana amastigotes as compared to the metacyclic and procyclic stages. A majority of these OGs in L. mexicana is annotated as proteins with unknown functions, with only one OG including amastin-like and A1 proteins (amastin is a known virulence-associated protein)43,44. The second group of interest contained two hypothetical proteins (two OGs) up-regulated in amastigotes and metacyclics in comparison to procyclics. Finally, the third group brought together three hypothetical proteins (three OGs) up-regulated in metacyclics only. Although, to the best of our knowledge, any functional data that might link these proteins to Leishmania virulence are lacking, the combination of two properties makes them a reasonably good starting group for future experimental studies: i/the gene families originated at the basal Leishmania node; ii/expression of the genes is up-regulated only at the virulent stages of the Leishmania life cycle. Excluding the OG annotated as amastin-like proteins leaves 20 novel OGs potentially involved in virulence.
By the second approach, we identified OG overlaps between the three differential expression datasets. As a result, 20 OGs were found to be shared by at least two datasets, and only two OGs were shared by all three datasets (Fig. 3 and Supplementary Table S13). Eleven of the 20 OGs had annotations of known virulence factors (Supplementary Table S13), while the remaining nine OGs represented proteins, the involvement of which in Leishmania virulence remains to be elucidated. We also inferred phyletic patterns for the OGs shared by the differential expression datasets (Fig. 3). The situation resembles that observed for the known Leishmania virulence factors (Supplementary Fig. S26) - a majority of the OGs were gained at the basal nodes, a high proportion of them was later lost in Trypanosoma and Phytomonas spp. and retained within the Leishmaniinae clade, and no OG was gained at the basal Leishmania node. Detailed information about four OGs gained at the Leishmaniinae node is presented in Table 1. Three of them have annotations of known virulence-associated proteins, while one of the OGs contains proteins with unknown function. Importantly, hypothetical proteins found within the latter OG have relevant differential expression patterns (up-regulated in L. seymouri at 35 °C and in L. mexicana amastigotes, Table 1). All proteins belonging to this OG have no known domains, are about 380 amino acids long, and are present in the Leishmania genomes (L. donovani, L. infantum, L. major, L. mexicana, and L. tarentolae) as single copies. They represent primary targets for gene ablation experiments in order to shed light on their function.
Table 1. Expression patterns of putative virulence factors gained at the Leishmaniinae node.
| OG | Gene IDs | Annotation | Upregulated in |
||
|---|---|---|---|---|---|
| L. seymouri | L. mexicana | L. majorFriedlin | |||
| 06971 | LmjF.19.0840, LmjF.19.0844, LmjF.19.0848, LbrM2903_19_0920, LbrM.19.1140, LdBPK_190840.1, LinJ.19.0840, LmxM.19.0870, LtaP19.0810 | ATG8/AUT7/APG8/PAZ2 (ATG8A.1) | 35 °C | META | VIRULENT |
| 09307 | LdBPK_051210.1, LinJ.05.1210, LmjF.05.1215, LmxM.05.1215, LbrM2903_05_1260, LbrM.05.1210, LtaP05.1300 | surface antigen-like protein | 35 °C | PRO and META | VIRULENT |
| 09635 | LbrM2903_27_2010, LbrM.27.1900, LdBPK_271680.1, LinJ.27.1680, LmjF.27.1780, LmxM.27.1780, LtaP27.1830 | casein kinase I-like protein | no significant changes | META and AMA | VIRULENT |
| 10354 | LdBPK_312140.1, LinJ.31.2140, LmjF.31.2090, LmxM.30.2090, LtaP31.2520 | protein with unknown function | 35 °C | AMA | no significant changes |
Only Leishmania spp. gene IDs are shown. Leishmania life cycle stages are abbreviated as follows: PRO, procyclics; META, metacyclics; AMA, amastigotes.
Sequence polymorphism in the L. pyrrhocoris genome
Whole genome sequencing of clonal cultures of 13 L. pyrrhocoris lineages allowed us to study ploidy and the patterns of sequence variation across the genome. We sampled as widely as possible, including isolates from Europe, Central America and Africa (Supplementary Table S14). Despite this, the overall level of sequence variation was relatively modest, with an average number of sequence differences per nucleotide of 0.0058, comparable to Arabidopsis thaliana and about 5 times higher than in humans45,46. Total number of homozygous and heterozygous single nucleotide polymorphic (SNP) sites (inferred using L. pyrrhocoris H10 as a reference) varied from 18,654 in the F165 lineage to 219,139 in G58. In total, 907,015 SNP sites were identified. The majority of SNPs (892,221; 98.37%) were biallelic. The percentage of nonsynonimous SNPs ranged from 27% in the G58 lineage to 31% in P59, with 29% on average across all lineages. The average percentage of heterozygous sites was 2.6%, with the lowest value (1.1%) observed in the K06 lineage and the highest (6.2%) in CH278. Sequence variation was significantly lowered at non-synonymous, compared to synonymous sites in protein-coding genes (Supplementary Fig. S27), reflecting the action of purifying selection against deleterious amino acid replacements. The distribution of polymorphisms was uniform across the genome (Supplementary Fig. S28) with no apparent peaks or valleys typically seen around centromeres or telomeres in actively recombining genomes. Indeed, we found no evidence for the presence of recombination in our sequence polymorphism data using a coalescent-based approach47, indicating that this species is entirely clonal (see Methods for details). Interestingly, the L. pyrrhocoris genome harbors the same complement of meiosis-associated genes as L. major and T. brucei and also codes for homologs of proteins involved in gamete membrane fusion (HAP2/GCS1) and karyogamy (GEX1) in other eukaryotes48,49, suggesting that the cellular machinery for executing a (para)sexual cycle is present in this species, but it is not functional. An alternative explanation for the apparent lack of recombination may be the lack of genetic exchange between geographically remote populations of L. pyrrhocoris. Either way, the 13 L. pyrrhocoris isolates with sequenced genomes represent independent lineages that do not appear to have recombined with each other.
The ploidy of L. pyrrhocoris lineages was inferred using average read coverage for scaffolds under the assumption that most of them are diploid (which was supported by heterozygosity patterns). Twenty L. pyrrhocoris scaffolds (LpyrH10_01–02, 04–11, 13, 15, 17, 19, 21–22, 25, 31, 35, and 48), which constitute ~66% of the genome, were stably diploid in all 14 lineages investigated. Eleven scaffolds were found to be diploid in all lineages except one or two, while four scaffolds (LpyrH10_12, 29, 32, 42) are almost invariably tetraploid, scaffold 53 is almost invariably haploid, and five scaffolds (44, 45, 49, 56, 60) are polyploid or, more likely, represent unplaced repeats. Such ploidy pattern in L. pyrrhocoris strains is not surprising given the variable ploidy of Leishmania and Trypanosoma strains50,51,52.
Adaptive evolution in the L. pyrrhocoris genome
To test whether any L. pyrrhocoris genes show evidence of molecular adaptation in the recent past (since the divergence of L. pyrrhocoris strains from their common ancestor), we employed a maximum likelihood phylogeny-based approach that is appropriate given the lack of recombination53. This approach is based on comparing the fit of nested models to data, with a simpler model (‘M7’) allowing only for purifying selection, and the more general model (‘M8’) accommodating both purifying and positive selection54. Likelihood ratio tests comparing these models demonstrated a significantly better fit of the M8 model for 1,318 L. pyrrhocoris genes, corresponding to 1,253 OGs (see Methods for details of the analysis). Thus, genes that putatively evolved under positive selection account for 13.3% of all protein-coding genes in L. pyrrhocoris. All three GO term categories (biological process, molecular function, and cell compartment) point to the fact that cytoskeleton- and flagellum-related functions occur more frequently among the positively selected genes as compared to the whole genome (Supplementary Table S15). Flagellum in trypanosomatids participates in cell mobility, cell division, influences cell size and organelle positioning, and may even function as a sensory platform for host-parasite interactions55.
Mapping the 1,253 OGs containing positively selected genes on the trypanosomatid phylogeny (Supplementary Fig. S29) revealed that the highest numbers of OGs were reconstructed to the basal nodes (N1–N3) and to the Leishmaniinae node N14: 493, 245, 143, and 162 OGs, respectively. Sixty one percent of the OGs containing genes under positive selection were gained within the studied phylogeny (i.e. cannot be reconstructed to the most basal node N1), while 44% were never lost, and 17% of OGs were never gained or lost within the studied phylogeny. A noticeable fraction of positively selected OGs (70 or 5.6%) was gained at the basal node of monoxenous species C. fasciculata, L. pyrrhocoris, and L. seymouri, and a substantial share was lost in both Phytomonas spp. (249 OGs, 19.9%) and in all Leishmania spp. (42 OGs, 3.4%). These observations highlight the functional significance of genes under positive selection for the monoxenous species of the Leptomonas/Crithidia clade. Indeed, the percentage of monoxenous-specific genes (i.e. genes gained at the Crithidia/Leptomonas node) under positive selection (16.8% or 70 out of 416, see Supplementary Table S16) is significantly higher than that across the genome (13.3%), with a 2 × 2 contingency test p-value of 0.0494. A great majority of those genes (52 of 70) have no functional annotation, and the remaining handful of genes are involved in carbohydrate and amino acid metabolism, glycolysis, cAMP signaling, protection against ROS and phenolic acid metabolism. These findings highlight the need for experimental studies to unravel functions of the proteins apparently important for the lifestyle of monoxenous trypanosomatids.
Five OGs containing positively selected genes were unique to L. pyrrhocoris (Supplementary Table S17). Two genes within these groups are annotated as proteins with unknown function; two other genes showed homology to L. braziliensis TATE transposable elements, and one gene was annotated as a putative surface antigen protein. For each of these genes positions of codons under positive selection were determined using the empirical Bayes method (Supplementary Table S17). The presence of mobile elements with a telomeric-repeat site specificity (TATE) resembling those found in the L. braziliensis genome correlates well with our finding that the L. pyrrhocoris genome encodes the full set of RNAi pathway genes with a potential role in control of transposable element mobilization56.
Discussion
Kinetoplastid flagellates are a well-defined, clearly monophyletic lineage with a range of dramatically different life strategies, ranging from a free-living one, to obligatory endosymbiosis in an amoeba57 or complex parasitic life cycles involving invertebrate and vertebrate hosts or plants1. Due to their medical importance, virtually all attention was focused on trypanosomatids parasitizing humans, which resulted in high-quality nuclear genome sequences for T. brucei, T. cruzi, and L. major58,59,60. However, a decade later, when dozens of genomes of Trypanosoma and Leishmania have been published or are in progress, our information about the genomes of monoxenous trypanosomatids remains fragmentary. Since several genomes of draft quality available for these highly diverse parasites of insects8,22,23 are insufficient for a detailed comparison with dixenous genomes, we generated a high-quality genome sequence of a globally distributed parasite of firebugs, L. pyrrhocoris.
Both the size of the Leptomonas genome and its coding capacity are similar to the dixenous kins. The levels of synteny, ranging from 22% to 90% with T. brucei and L. seymouri, respectively, are not surprising and correlate with evolutionary distances between the compared trypanosomatids (Supplementary Note 1). The studied parasite possesses all core components of the RNAi pathway making L. pyrrhocoris particularly interesting from the perspective of functional studies.
The transition from the free-living style to parasitism in kinetoplastids was apparently followed by a massive gene loss, as suggested by comparing the ~18,943 gene content of Bodo saltans61, a representative of the free-living kinetoplastid group most closely related to parasitic trypanosomatids, with a mere 6,000 to 12,000 gene complement encountered in all sequenced trypanosomatids24,39,62. Detailed analysis has shown that in the process of streamlining its genome the ancestral trypanosomatid have lost genes that functioned in macromolecular digestion and assimilation, as well as many intracellular membrane pumps and ABC transporters, while expanded the families of membrane transporters for scavenging amino acids, nucleosides, and other metabolites from the host61. Our analysis also revealed gene losses in trypanosomatid amino acid metabolism (Fig. 1), as well as the loss of murein- and glycogen-degrading capabilities (Supplementary Note 2). A similarly drastic gene reduction occurred in the evolutionary history of apicomplexans, another highly successful and diverse parasitic lineage63, and genome reduction is thought to be a major evolutionary trend for parasites64.
Based on the patterns of gene family gains and losses in trypanosomatids (Fig. 2, Supplementary Fig. S11), we speculate that not only losses, but also massive gene gains at certain nodes, in particular the basal node of trypanosomatids, the Leishmaniinae node, and the T. brucei and T. cruzi nodes, have driven expansion into novel niches. This was followed by rather limited lineage-specific losses as a result of adaptation to specific hosts. Phytomonas, with its minimized gene repertoire, seems to be an extreme case of a prevailing OG loss trend39.
Emergence of novel gene families of transmembrane transporters and proteins participating in fatty acid biosynthesis and amino acid metabolism at the Leishmaniinae node clearly reflects continuing adaptation to the insect host, where sugars and amino acids serve as important energy substrates65. Previously reported presence of additional desaturases and elongases in Leishmania spp. and their absence in T. brucei and T. cruzi66 correlates with our OG analysis. Of importance is also the presence in Leptomonas spp. of at least a partial LPG (lipophosphoglycan) pathway, highly expressed in the Leishmania insect stages, indicating that both protists utilize similar molecules in their interaction with the insect hosts67,68. A detailed analysis of the amastin family in L. pyrrhocoris (Supplementary Note 4) further confirms that a substantial elaboration of amastin transmembrane glycoproteins occurred after the origin of Leishmania43.
Analyses of the protein machinery involved in vesicle trafficking in L. pyrrhocoris revealed a set of genes highly similar to the previously studied dixenous trypanosomatids (Supplementary Note 3, Supplementary Table S7). Notably, L. pyrrhocoris has retained all four adaptor complexes (AP-1 to AP-4) inherited from free-living kinetoplastids ancestors, whereas some of the complexes are missing in Leishmania, Phytomonas and African trypanosomes. However, T. cruzi also exhibits all four adaptor complexes, so their reduction is not directly linked to the transition from monoxeny to dixeny. The comparison of the small GTPase genes in different trypanosomatids (Supplementary Note 3, Supplementary Table S8) showed that gene loss is a much more significant trend in the evolution of this group of genes than innovation by gene duplications. Given the association of small GTPases with various basic cellular processes, this finding suggests the continuing simplification of the general cellular physiology in the trypanosomatid evolution. In terms of the number of ancestral small GTPases retained, L. pyrrhocorris stands between T. cruzi exhibiting a complete set and other dixenous trypanosomatids that show some secondary simplification of the ancestral small GTPase complement. Again, no straightforward correlation between the lifestyle and the complexity of this gene cohort is apparent. The analysis of small GTPases also revealed minimization of their ARF clade in L. pyrrhocoris (Supplementary Note 3). Overall, the studied trypanosomatid has an endomembrane system similar to that of its dixenous relatives with no evidence for massive remodeling accompanying the dixenous life style. As ARFs/ARLs tend to control vesicle coat assembly and other cytoskeletal-related functions, this may also reflect a decreased requirement for regulation of pathways accompanying switches between hosts. However, the stability of the Ras-like GTPase cohort between distantly related L. pyrrhocoris and T. brucei69 suggests that the system is unlikely to play a major role in life stage transition, and that these proteins rather play roles in general cellular physiology.
This study revealed that 38 OGs corresponding to known Leishmania virulence factors were gained at the basal nodes, 5 OGs were gained at the Leishmaniinae node, and only 4 OGs emerged at the nodes within the genus Leishmania. Notably, a high proportion of the putative virulence factors (from the 38 OG mentioned above) were lost in the streamlined genomes of Phytomonas spp. We consider two alternative explanations for the rather basal acquisition of most virulence factors, with only a small proportion gained at the Leishmaniinae node. Firstly, it is possible that not the genes themselves, but their coordinated expression is required for virulence. The second explanation relies on the fact that, due to the unavailability of monoxenous outgroups, previous studies lacked defined sets of OGs gained at the Leishmaniinae and Leishmania nodes and, consequently, these OGs were never a focus of Leishmania virulence factors studies. This could also explain the fact that none of the virulence factors in our dataset was gained at the Leishmania node. However, this does not imply that this node does not deserve attention in further studies specifically targeting Leishmania virulence factors. The inclusion of a high-quality monoxenous trypanosomatid genome and differential gene expression data in the OG analysis across all trypanosomatids has the added value of identifying additional virulence factors confined to Leishmania. Indeed, in this study we have identified 29 orthologous groups representing novel Leishmania virulence factor candidates.
Methods
Parasites and establishment of the clonal lines
L. pyrrhocoris isolate H10 isolated from the firebug, Pyrrhocoris apterus, in the Czech Republic3 was used for initial genome and transcriptome sequencing. Cultures were routinely maintained in Brain Heart Infusion (BHI) medium (Sigma-Aldrich, St. Louis, USA) supplemented with 10% Fetal Bovine Serum (Thermo Fisher Scientific, Waltham, USA), 50 units/ml of penicillin, 50 μg/ml of streptomycin (both from Sigma-Aldrich), and 10 μg/ml of hemin (Jena Bioscience GmbH, Jena, Germany) at +23 °C. Origin of other isolates used in this work is summarized in Supplementary Table S14. Two or three independent clonal lines for each of the of L. pyrrhocoris isolates 10VL, 121AL, 14BT, 25EC, 324RV, 329MV, CH278, F165, F19, G58, K06, P59, and SERG were established as described before70,71. In brief, parasite primary cultures were plated in multiple serial dilutions onto a 1% agar medium supplemented with BHI and antibiotics as described earlier72. The identities of clonal lines used in downstream analyses were confirmed by sequencing their spliced leader (SL) RNA gene repeats (see below). Cultures and clonal lines obtained were deposited in the collection of the Life Science Research Centre of the University of Ostrava and are available upon request.
SL RNA gene repeats were amplified using primers M167 and M168, and the PCR products were cloned and sequenced as described previously73,74,75. Obtained sequences were compared to published data3,13, and one clonal line from each isolate was selected for whole genome sequencing. SL sequences determined in the course of this work are deposited in GenBank under accession numbers KT012485–KT012505.
Genome assembly and annotation, transcriptomic data processing and analysis
Gene family analysis using the OrthoMCL approach
Orthologous groups (OGs) for the Leptomonas proteins were inferred using the OrthoMCL v.2.0 software76. Full lists of annotated proteins for 23 trypanosomatid species were downloaded from the TriTrypDB v.7.0 and combined with newly annotated proteins from L. pyrrhocoris and three other trypanosomatid species: L. seymouri, Blechomonas ayalai and Paratrypanosoma confusum (Supplementary Table S1). The reference protein dataset was subjected to removal of poor quality proteins (based on sequence length and percent of in-frame stop codons), all vs. all BLAST search (E-value cut-off 10−10) and a clustering procedure implemented in the OrthoMCL algorithm.
Gene family gains and losses were mapped on the reference species tree using the COUNT software42. Gains and losses pattern was inferred using the Dollo and Wagner parsimony algorithms implemented in COUNT (with gain penalty value set to 3 for Wagner algorithm). Dollo parsimony allows a gene family to be gained only once, while Wagner parsimony allows multiple gains with a certain gain penalty and, in addition, inferring gene family expansions and contractions.
Phylogenetic analysis
For the phylogenetic tree construction 644 OGs containing only one protein for each of 27 kinetoplastid species were taken. The protein sequences were aligned using MUSCLE77 with default parameters. The alistat script from the HMMER package v.3.1 was used to calculate average percent identity within each OG. Fifty seven OGs having average percent identity within the group >75% were used for constructing a multi-gene phylogeny. Protein alignments were individually trimmed in Gblocks v.0.91b with relaxed parameters (−b3 = 10 −b4 = 5 −b5 = h) and concatenated, producing an alignment of 22,544 characters. Maximum likelihood phylogenetic analysis was performed using RA × ML v.8.0.24 with the LG + Г phylogenetic model and with amino acid equilibrium frequencies inferred from the data78. One thousand bootstrap replicates and 200 iterations of the maximum likelihood algorithm were performed, and the best tree was visualized in FigTree v.1.4.1.
Gene ontology analysis
Gene ontology (GO) annotation was performed for L. pyrrhocoris gene families gained/lost at certain nodes in the trypanosomatid tree using the Blast2GO PRO software79 with the following settings: BLAST E-value cut-off, 10−10; maximum number of hits per sequence, 100; with filtering of low complexity regions turned on. A BLASTP search was run against a local NCBI nr database (download date: 13.02.2015). Subsequently, mapping GO terms associated with BLAST hits onto query sequences was performed, followed by GO annotation, i.e. selection of most specific GO terms. Annotation was conducted with an E-value cut-off of 10−10 and an annotation cutoff of 55. GO term graphs were generated, and multi-level pie charts were created for each GO term category: cellular component, biological process, and molecular function. Combined graphs and pie charts were plotted with a sequence filter, i.e. a minimal number of sequences a GO node must contain in order to be displayed on the graph or chart, set to 5 in the case of gains/losses and to 10 for expansions/contractions. Analysis of GO terms enrichment including all the standard steps (BLAST search, GO terms mapping and annotation) was also performed for L. pyrrhocoris protein-coding genes showing signs of positive selection vs. all protein-coding genes using Fisher’s exact test with a p-value cut-off of 0.01.
Identification of novel Leishmania virulence factor candidates
In order to identify novel proteins involved in Leishmania virulence, phyletic patterns for OGs were overlapped with three gene expression datasets generated with the Illumina HiSeq technology. The first dataset contained 189 genes of L. seymouri up-regulated in an axenic culture at an elevated temperature (35 °C) compared to 23 °C8. The second dataset was comprised of 53 genes up-regulated in a virulent isolate of L. major LV561 compared to its avirulent counterpart (our unpublished data). The third dataset included differential expression data for L. mexicana M379 developmental stages in axenic cultures (our unpublished data). This dataset, in turn, consisted of four subsets: i/genes up-regulated in metacyclics compared to procyclics (12 genes); ii/genes up-regulated in amastigotes compared to procyclics (433 genes); iii/genes up-regulated in metacyclics compared to amastigotes (713 genes); iv/genes up-regulated in amastigotes compared to metacyclics (358 genes). Subset iii/was omitted since it showed considerable overlap with subset ii/and probably included a lot of genes functioning in the insect host environment and not involved in virulence directly. In all datasets mentioned above, genes with expression fold change ≥1.5 and an FDR-corrected p-value ≤0.05 were chosen for further analyses. First, differential expression patterns of L. mexicana M379 genes gained at the Leishmania node were investigated. In brief, L. mexicana gene IDs were identified within OGs gained at the Leishmania node and genes (showing at least a 1.5-fold change in expression) with the following expression profiles in the life cycle were selected: i/up-regulated in amastigotes only; ii/up-regulated in metacyclics only; iii/up-regulated in amastigotes and metacyclics. Second, for each gene within the three differential expression datasets an OG number was inferred. All OGs overlapping between at least two expression datasets were identified. Both approaches combined resulted in identification of 29 OGs containing novel virulence factor candidates.
Analysis of positive selection in Leptomonas
Paired-end 100 nt genomic reads for 13 clonal lines of L. pyrrhocoris (10VL, 121AL, 14BT, 25EC, 324RV, 329MV, CH278, F165, F19, G58, K06, P59, SERG) were generated using the Illumina HiSeq platform (Wellcome Trust Centre for Human Genetics, Oxford, UK). An average read count per strain was 12,980,951 (with 43× average coverage), ranging from 11,505,714 reads for the 10 VL isolate (~38× coverage) to 15,457,054 reads for F19 (~51× coverage). Prior to further analysis, reads were subjected to trimming and quality filtering using CLC Genomics Workbench v. 7.0 with the following settings: regions with Phred quality <20 were trimmed, no more than one N was allowed in the remaining sequence, then TruSeq adapter trimming and a minimum length threshold of 75 nt were applied. Filtered reads were mapped to the reference genome of L. pyrrhocoris isolate H10 using CLC Genomics Workbench v. 7.0. The mapping parameters were as follows: mismatch cost, 2; insertion cost, 2; deletion cost, 2; length fraction, 0.75; and similarity fraction, 0.95. Variant calling procedure implemented in Platypus80 produced 907,015 single nucleotide variants (SNVs) and 261,475 insertions or deletions (indels), 1,168,490 variants in total. Consensus coding sequences (9,878 within the L. pyrrhocoris genome) were extracted for each strain, taking into account the observed SNVs, and aligned using MUSCLE with default parameters.
The recombination analysis was performed using Ldhat v.2.247 and suggested the absence of recombination between populations. Selection analysis was performed using the CodeML script from the PAML package v.4.854. We compared the M7 (β) and M8 (β + ω) models for inferring positive selection. Both models are codon-based and allow ω, i.e. the dN/dS ratio (the rate of nonsynonymous substitutions divided by the rate of synonymous substitutions per site), to vary between sites. Model M7 splits codons of analyzed sequence into 9 classes with 0 <ω < 1, while M8 allows one of the codon classes to have ω > 1 (estimated from the data). Comparison between the two models was performed using the likelihood ratio test (LRT). LRT allows determining which of the opposing models, M7 or M8, fits the data better. If the M8 model, which allows ω values >1, fitted the data better than model M7, then positive selection was inferred. It was considered that model M8 fitted the data significantly better than M7 if 2 × (lnLModel8 − lnLModel7) > 4.61 (10% χ2 critical value with 2 degrees of freedom). If the conditions above were satisfied, a gene was considered to be under positive selection. For genes showing a signature of positive selection, an attempt to determine particular codons clustering into the codon class with ω > 1 was made using the empirical Bayes method81. A codon was considered to be under positive selection if a p-value under model M8 was >0.95.
Additional Information
How to cite this article: Flegontov, P. et al. Genome of Leptomonas pyrrhocoris: a high-quality reference for monoxenous trypanosomatids and new insights into evolution of Leishmania. Sci. Rep. 6, 23704; doi: 10.1038/srep23704 (2016).
Supplementary Material
Acknowledgments
We would like to thank members of our laboratories for helpful discussions and technical assistance. We also thank Dr. S. Beverley and The Genome Institute, Washington University School of Medicine for permission to use genomic data of C. fasciculata. We acknowledge the use of research infrastructure that has received funding from the EU 7th Framework Programme under grant agreement No. 316304. The financial support through the project LO1208 of the National Feasibility Programme I of the Czech Republic is gratefully appreciated. This work was supported by the following grants: Czech Science Foundation (14-23986S to J.L., 13-24983S and 15-16406S to M.E., and 16-18699S to J.L. and V.Y.), the Moravskoslezský Kraj research initiatives DT1/RRC/2014 (to V.Y., A.K., P.F.) and 00955/RRC/2015 (to V.Y.), the Russian Science Foundation (project 14-50-00029 to M.D.L.), and the Russian Foundation for Basic Research (project 14-04-31936 to P.F. and E.G.).
Footnotes
Author Contributions Study conceiving and planning – J.L., V.Y. and P.F. Genome and transcriptome sequencing – Č.V., M.D.L., D.F. and P.F. Genome and transcriptome assembly and annotation – S.F., P.F., A.B., O.F., E.S.G., N.K. and S.K. Metabolic pathways analysis – F.O., A.B. and P.F. Annotation of functional pathways – P.F., A.B., N.K., M.E., M.C.F., A.P.J., A.O.R. and D.V. OrthoMCL analysis – A.B., P.F. Leishmania virulence factors – A.B., P.F., J.H., A.I., A.K., J.S., P.V., J.V. and V.Y. L. pyrrhocoris populations sampling – D.A.M., J.V., V.Y. and J.L. Population phylogenomics – A.B., P.F., D.F., T.S. and K.Z. Manuscript preparation – P.F., A.B., V.Y. and J.L.
References
- Lukeš J., Skalický T., Týč J., Votýpka J. & Yurchenko V. Evolution of parasitism in kinetoplastid flagellates. Mol Biochem Parasitol 195, 115–122 (2014). [DOI] [PubMed] [Google Scholar]
- Maslov D. A., Votýpka J., Yurchenko V. & Lukeš J. Diversity and phylogeny of insect trypanosomatids: all that is hidden shall be revealed. Trends Parasitol 29, 43–52 (2013). [DOI] [PubMed] [Google Scholar]
- Votýpka J. et al. Cosmopolitan distribution of a trypanosomatid Leptomonas pyrrhocoris. Protist 163, 616–631 (2012). [DOI] [PubMed] [Google Scholar]
- McGhee R. B. & Cosgrove W. B. Biology and physiology of the lower Trypanosomatidae. Microbiol Rev. 44, 140–173 (1980). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jirků M., Yurchenko V. Y., Lukeš J. & Maslov D. A. New species of insect trypanosomatids from Costa Rica and the proposal for a new subfamily within the Trypanosomatidae. J Eukaryot Microbiol. 59, 537–547 (2012). [DOI] [PubMed] [Google Scholar]
- De Sa M. F., De Sa C. M., Veronese M. A., Filho S. A. & Gander E. S. Morphologic and biochemical characterization of Crithidia brasiliensis sp. n. J Protozool 27, 253–257 (1980). [DOI] [PubMed] [Google Scholar]
- Roitman I., Mundim M. H., De Azevedo H. P. & Kitajima E. W. Growth of Crithidia at high temperature: Crithidia hutneri sp. n. and Crithidia luciliae thermophila s. sp. n. J Protozool 24, 553–556 (1977). [Google Scholar]
- Kraeva N. et al. Leptomonas seymouri: adaptations to the dixenous life cycle analyzed by genome sequencing, transcriptome profiling and co-infection with Leishmania donovani PLoS Pathog 11, e1005127 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira M. S. & Borges A. S. Some aspects of protozoan infections in immunocompromised patients - a review. Mem Inst Oswaldo Cruz 97, 443–457 (2002). [DOI] [PubMed] [Google Scholar]
- Dedet J. P. & Pratlong F. Leishmania, Trypanosoma and monoxenous trypanosomatids as emerging opportunistic agents. J Eukaryot Microbiol. 47, 37–39 (2000). [DOI] [PubMed] [Google Scholar]
- Rosenthal E. et al. HIV and Leishmania coinfection: a review of 91 cases with focus on atypical locations of Leishmania. Clin Infect Dis. 31, 1093–1095 (2000). [DOI] [PubMed] [Google Scholar]
- del Campo J. et al. The others: our biased perspective of eukaryotic genomes. Trends Ecol Evol 29, 252–259 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votýpka J. et al. New approaches to systematics of Trypanosomatidae: criteria for taxonomic (re)description. Trends Parasitol 31, 460–469 (2015). [DOI] [PubMed] [Google Scholar]
- Cantacessi C., Dantas-Torres F., Nolan M. J. & Otranto D. The past, present, and future of Leishmania genomics and transcriptomics. Trends Parasitol 31, 100–108 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. P. Genome evolution in trypanosomatid parasites. Parasitology 142 Suppl 1, S40–56 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real F. et al. The genome sequence of Leishmania (Leishmania) amazonensis: functional annotation and extended analysis of gene models. DNA Res 20, 567–581 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peacock C. S. et al. Comparative genomic analysis of three Leishmania species that cause diverse human disease. Nat Genet 39, 839–847 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sistrom M. et al. Comparative genomics reveals multiple genetic backgrounds of human pathogenicity in the Trypanosoma brucei complex. Genome Biol Evol 6, 2811–2819 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson W. The origins of the trypanosome genome strains Trypanosoma brucei brucei TREU 927, T. b. gambiense DAL 972, T. vivax Y486 and T. congolense IL3000. Parasit Vectors 5, 71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greif G. et al. Transcriptome analysis of the bloodstream stage from the parasite Trypanosoma vivax. BMC Genomics 14, 149 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myler P. J. In Leishmania: after the genome (eds P. J. Myler & N. Fasel) Ch. 2, 15–28 (Caister Academic Press, 2008). [Google Scholar]
- Runckel C., DeRisi J. & Flenniken M. L. A draft genome of the honey bee trypanosomatid parasite Crithidia mellificae. PLoS One 9, e95057 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motta M. C. et al. Predicting the proteins of Angomonas deanei, Strigomonas culicis and their respective endosymbionts reveals new aspects of the trypanosomatidae family. PLoS One 8, e60209 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aslett M. et al. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res 38, D457–462 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh N., Chikara S. & Sundar S. SOLiD sequencing of genomes of clinical isolates of Leishmania donovani from India confirm Leptomonas co-infection and raise some key questions. PLOS One 8, e55738 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alves J. M. et al. Endosymbiosis in trypanosomatids: the genomic cooperation between bacterium and host in the synthesis of essential amino acids is heavily influenced by multiple horizontal gene transfers. BMC Evol Biol 13, 190 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tibayrenc M. & Ayala F. J. How clonal are Trypanosoma and Leishmania? Trends Parasitol 29, 264–269 (2013). [DOI] [PubMed] [Google Scholar]
- Tibayrenc M. & Ayala F. J. Reproductive clonality of pathogens: a perspective on pathogenic viruses, bacteria, fungi, and parasitic protozoa. Proc Natl Acad Sci. USA 109, E3305–3313 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akopyants N. S. et al. Demonstration of genetic exchange during cyclical development of Leishmania in the sand fly vector. Science 324, 265–268 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sádlová J. et al. Visualisation of Leishmania donovani fluorescent hybrids during early stage development in the sand fly vector. PLoS One 6, e19851 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez J. D. et al. Contemporary cryptic sexuality in Trypanosoma cruzi. Mol Ecol 21, 4216–4226 (2012). [DOI] [PubMed] [Google Scholar]
- Rogers M. B. et al. Genomic confirmation of hybridisation and recent inbreeding in a vector-isolated Leishmania population. PLoS Genet 10, e1004092 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volf P. et al. Increased transmission potential of Leishmania major/Leishmania infantum hybrids. Int J Parasitol 37, 589–593 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp M., Erler S. & Lattorff H. M. Seasonal variability of prevalence and occurrence of multiple infections shape the population structure of Crithidia bombi, an intestinal parasite of bumblebees (Bombus spp.). MicrobiologyOpen 1, 362–372 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Votýpka J., Ray D. S. & Lukeš J. Crithidia fasciculata: a test for genetic exchange. Exp Parasitol 99, 104–107 (2001). [DOI] [PubMed] [Google Scholar]
- Cisarovsky G. & Schmid-Hempel P. Few colonies of the host Bombus terrestris disproportionately affect the genetic diversity of its parasite, Crithidia bombi. Infect Genet Evol 21, 192–197 (2014). [DOI] [PubMed] [Google Scholar]
- Schmid-Hempel R., Salathe R., Tognazzo M. & Schmid-Hempel P. Genetic exchange and emergence of novel strains in directly transmitted trypanosomatids. Infect Genet Evol 11, 564–571 (2011). [DOI] [PubMed] [Google Scholar]
- Tognazzo M., Schmid-Hempel R. & Schmid-Hempel P. Probing mixed-genotype infections II: high multiplicity in natural infections of the trypanosomatid, Crithidia bombi, in its host, Bombus spp. PLoS One 7, e49137 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porcel B. M. et al. The streamlined genome of Phytomonas spp. relative to human pathogenic kinetoplastids reveals a parasite tailored for plants. PLOS Genet 10, e1004007 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell D. A., Thomas S. & Sturm N. R. Transcription in kinetoplastid protozoa: why be normal? Microbes Infect 5, 1231–1240 (2003). [DOI] [PubMed] [Google Scholar]
- Leroux A. E., Maugeri D. A., Cazzulo J. J. & Nowicki C. Functional characterization of NADP-dependent isocitrate dehydrogenase isozymes from Trypanosoma cruzi. Mol Biochem Parasitol 177, 61–64 (2011). [DOI] [PubMed] [Google Scholar]
- Csuros M. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 26, 1910–1912 (2010). [DOI] [PubMed] [Google Scholar]
- Jackson A. P. The evolution of amastin surface glycoproteins in trypanosomatid parasites. Mol Biol Evol 27, 33–45 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall L. I. & McKerrow J. H. Determinants of disease phenotype in trypanosomatid parasites. Trends Parasitol 30, 342–349 (2014). [DOI] [PubMed] [Google Scholar]
- Sachidanandam R. et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature 409, 928–933 (2001). [DOI] [PubMed] [Google Scholar]
- Clark R. M. et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science 317, 338–342 (2007). [DOI] [PubMed] [Google Scholar]
- McVean G., Awadalla P. & Fearnhead P. A coalescent-based method for detecting and estimating recombination from gene sequences. Genetics 160, 1231–1241 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ning J. et al. Comparative genomics in Chlamydomonas and Plasmodium identifies an ancient nuclear envelope protein family essential for sexual reproduction in protists, fungi, plants, and vertebrates. Genes Dev 27, 1198–1215 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J. L. & Johnson M. A. Is HAP2-GCS1 an ancestral gamete fusogen? Trends Cell Biol 20, 134–141 (2010). [DOI] [PubMed] [Google Scholar]
- Rogers M. B. et al. Chromosome and gene copy number variation allow major structural change between species and strains of Leishmania. Genome Res 21, 2129–2142 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterkers Y., Lachaud L., Crobu L., Bastien P. & Pages M. FISH analysis reveals aneuploidy and continual generation of chromosomal mosaicism in Leishmania major. Cell Microbiol 13, 274–283 (2011). [DOI] [PubMed] [Google Scholar]
- Reis-Cunha J. L. et al. Chromosomal copy number variation reveals differential levels of genomic plasticity in distinct Trypanosoma cruzi strains. BMC Genomics 16, 499 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anisimova M., Nielsen R. & Yang Z. Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics 164, 1229–1236 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- Langousis G. & Hill K. L. Motility and more: the flagellum of Trypanosoma brucei. Nat Rev Microbiol 12, 505–518 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bringaud F., Ghedin E., El-Sayed N. M. & Papadopoulou B. Role of transposable elements in trypanosomatids. Microbes Infect 10, 575–581 (2008). [DOI] [PubMed] [Google Scholar]
- Tanifuji G. et al. Genomic characterization of Neoparamoeba pemaquidensis (Amoebozoa) and its kinetoplastid endosymbiont. Eukaryot Cell 10, 1143–1146 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berriman M. et al. The genome of the African trypanosome Trypanosoma brucei. Science 309, 416–422 (2005). [DOI] [PubMed] [Google Scholar]
- El-Sayed N. M. et al. The genome sequence of Trypanosoma cruzi, etiologic agent of Chagas disease. Science 309, 409–415 (2005). [DOI] [PubMed] [Google Scholar]
- Ivens A. C. et al. The genome of the kinetoplastid parasite, Leishmania major. Science 309, 436–442 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson A. P. et al. Kinetoplastid phylogenomics reveals the evolutionary innovations associated with the origins of parasitism. Curr Biol 26, 161–172 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J. & El-Sayed N. M. Functional genomics of trypanosomatids. Parasite Immunol 34, 72–79 (2012). [DOI] [PubMed] [Google Scholar]
- Woo Y. H. et al. Chromerid genomes reveal the evolutionary path from photosynthetic algae to obligate intracellular parasites. eLife 4, e06974 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf Y. I. & Koonin E. V. Genome reduction as the dominant mode of evolution. Bioessays 35, 829–837 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Opperdoes F. & Michels P. A. In Leishmania: after the genome (eds , Myler P. & Fasel N.) Ch. 7, 123–158 (Caister Academic Press, 2008). [Google Scholar]
- Lee S. H., Stephens J. L. & Englund P. T. A fatty-acid synthesis mechanism specialized for parasitism. Nat Rev Microbiol 5, 287–297 (2007). [DOI] [PubMed] [Google Scholar]
- Kamhawi S. et al. A role for insect galectins in parasite survival. Cell 119, 329–341 (2004). [DOI] [PubMed] [Google Scholar]
- Dostálová A. & Volf P. Leishmania development in sand flies: parasite-vector interactions overview. Parasit Vectors 5, 276 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field M. C. Signalling the genome: the Ras-like small GTPase family of trypanosomatids. Trends Parasitol 21, 447–450 (2005). [DOI] [PubMed] [Google Scholar]
- Votýpka J. et al. Kentomonas gen. n., a new genus of endosymbiont-containing trypanosomatids of Strigomonadinae subfam. n. Protist 165, 825–838 (2014). [DOI] [PubMed] [Google Scholar]
- Hamilton P. T. et al. Infection dynamics and immune response in a newly described Drosophila-trypanosomatid association. MBio 6, e01356–01315 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popp M. & Lattorff H. M. A quantitative in vitro cultivation technique to determine cell number and growth rates in strains of Crithidia bombi (Trypanosomatidae), a parasite of bumblebees. J Eukaryot Microbiol 58, 7–10 (2011). [DOI] [PubMed] [Google Scholar]
- Maslov D. A., Yurchenko V. Y., Jirků M. & Lukeš J. Two new species of trypanosomatid parasites isolated from Heteroptera in Costa Rica. J Eukaryot Microbiol 57, 177–188 (2010). [DOI] [PubMed] [Google Scholar]
- Maslov D. A., Westenberger S. J., Xu X., Campbell D. A. & Sturm N. R. Discovery and barcoding by analysis of spliced leader RNA gene sequences of new isolates of Trypanosomatidae from Heteroptera in Costa Rica and Ecuador. J Eukaryot Microbiol 54, 57–65 (2007). [DOI] [PubMed] [Google Scholar]
- Westenberger S. J. et al. Trypanosomatid biodiversity in Costa Rica: genotyping of parasites from Heteroptera using the spliced leader RNA gene. Parasitology 129, 537–547 (2004). [DOI] [PubMed] [Google Scholar]
- Li L., Stoeckert C. J. Jr. & Roos D. S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13, 2178–2189 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R. C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conesa A. et al. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676 (2005). [DOI] [PubMed] [Google Scholar]
- Rimmer A. et al. Integrating mapping-, assembly- and haplotype-based approaches for calling variants in clinical sequencing applications. Nat Genet 46, 912–918 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., Wong W. S. & Nielsen R. Bayes empirical bayes inference of amino acid sites under positive selection. Mol Biol Evol 22, 1107–1118 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.