Abstract
Impaired ribosome biogenesis is attributed to nucleolar disruption and diffusion of a subset of 60S ribosomal proteins, particularly ribosomal protein (rp)L11, into the nucleoplasm, where they inhibit MDM2, leading to p53 induction and cell-cycle arrest1–4. Previously, we demonstrated that deletion of the 40S rpS6 gene in mouse liver prevents hepatocytes from re-entering the cell cycle after partial hepatectomy5. Here, we show that this response leads to an increase in p53, which is recapitulated in culture by rpS6-siRNA treatment and rescued by the simultaneous depletion of p53. However, disruption of biogenesis of 40S ribosomes had no effect on nucleolar integrity, although p53 induction was mediated by rpL11, leading to the finding that the cell selectively upregulates the translation of mRNAs with a polypyrimidine tract at their 5´-transcriptional start site (5´-TOP mRNAs), including that encoding rpL11, on impairment of 40S ribosome biogenesis. Increased 5´-TOP mRNA translation takes place despite continued 60S ribosome biogenesis and a decrease in global translation. Thus, in proliferative human disorders involving hypomorphic mutations in 40S ribosomal proteins6,7, specific targeting of rpL11 upregulation would spare other stress pathways that mediate the potential benefits of p53 induction8.
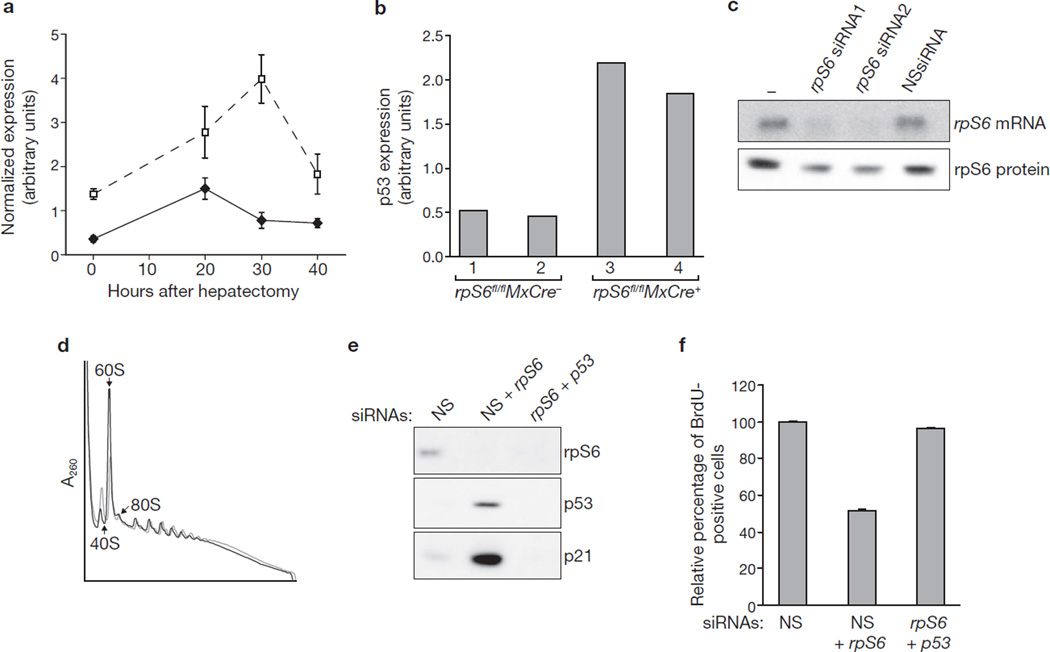
Conditional deletion of the rpS6 gene in mouse liver abrogates 40S ribosome biogenesis and prevents hepatocytes from re-entering the cell cycle after partial hepatectomy5. To understand the mechanism responsible for this response, mRNA microarray analyses were performed after hepatectomy on livers of rpS6fl/flMxCre+ mice, after rpS6 gene deletion, and control rpS6fl/flMxCre− mice. Consistent with the effects on cell-cycle progression, hepatocytes from rpS6fl/flMxCre+ mice, in contrast with those from rpS6fl/flMxCre− mice, failed to induce the expression of genes required for DNA synthesis and cell-cycle progression, including those encoding MCM3, MCM5, CDC6 and CDC20 (Supplementary Information, Fig. S1a; Supplementary Information, Table S1). In parallel, p53 target genes involved in cell-cycle inhibition, including those encoding p21, Bax and MDM2, were upregulated in livers of rpS6fl/flMxCre+ mice (Fig. 1a; Supplementary Information, Fig. S1b). These data are consistent with reports implicating p53 in cell-cycle arrest and apoptosis in response to the deletion of one rpS6 allele in tissues other than the liver9–11. Indeed, analysis of p53 in livers of rpS6fl/flMxCre+ versus rpS6fl/flMxCre− mice showed a significant increase in p53 levels (Fig. 1b). These data indicate that deletion of rpS6 leads to p53 induction, preventing hepatocyte cell-cycle progression.
Figure 1.
Depletion of rpS6 leads to p53-dependent cell-cycle arrest. (a) Expression kinetics of p21 mRNA after partial hepatectomy in the livers of rpS6fl/flMxCre− (filled diamonds) and rpS6fl/flMxCre+ (open squares) mice, as measured by hybridization of DNA microarrays. Results are shown as means ± s.e.m. for three independent experiments. (b) Protein levels of p53 in the livers of rpS6fl/flMxCre− and rpS6fl/flMxCre+ mice at 30 h after partial hepatectomy were measured with an ELISA kit (see Methods). For each genotype the measurements from two different animals are shown. (c) Northern blot (upper panel) and western blot (lower panel) showing the levels of rpS6 mRNA and rpS6 protein, respectively, in non-transfected A549 cells (lane 1), or A549 cells transfected for 24 h with the indicated siRNAs. (d) Polysome profiles from extracts of A549 cells treated for 24 h with either NSsiRNA (grey line) or rpS6 siRNA2 (black line). The positions of 40S and 60S native subunits and 80S monosomes are indicated. (e) Western blots showing the levels of rpS6, p53 and p21 proteins in cells transfected for 24 h with NSsiRNA or rpS6 siRNA2 in combination with either NSsiRNA or p53 siRNA. (f) Quantification of BrdU incorporation in A549 cells transfected for 24 h with the indicated siRNAs. For each treatment, the values are normalized to values for cells transfected with NSsiRNA. Results are shown as means ± s.e.m. for three independent experiments. Uncropped images of blots are shown in Supplementary Information, Fig. S5.
Given the difficulties in dissecting cell-cycle checkpoints in vivo, we determined whether these responses could be recapitulated by depleting rpS6 in human A549 cells, which are wild-type for p53 (ref. 12). Transfection of two distinct rpS6-siRNAs led to a sharp decrease in rpS6 transcript levels in comparison with untransfected cells or cells transfected with a non-silencing siRNA (NSsiRNA) (Fig. 1c). In contrast, the effect on rpS6 protein levels was modest because of the long half-life of ribosomes5. Such treatment also resulted in a decrease in free 40S subunits and an increase in free 60S subunits (Fig. 1d), which was paralleled by a decrease in the incorporation of 3H-uridine into nascent 18S, but not 28S, rRNA (Supplementary Information, Fig. S1c). Moreover, as reported for deletion of rpS6, rpS6-siRNA treatment led to the accumulation of a 34S rRNA (ref. 5), which we found, by employing specific probes, to be the 34S precursor of 18S rRNA (Supplementary Information, Fig. S1d). Finally, rpS6-siRNA treatment resulted in an induction of p53 and p21 (Fig. 1e), a 60% decrease in incorporation of bromodeoxyuridine (BrdU) into DNA (Fig. 1f) and an accumulation of cells in G1 (Supplementary Information, Fig. S1e); all these effects were reversed by co-transfection of p53-siRNA (Fig. 1e, f; Supplementary Information, Fig. S1e). Thus, depletion of rpS6 largely phenocopies the responses observed in mouse liver on rpS6 deletion.
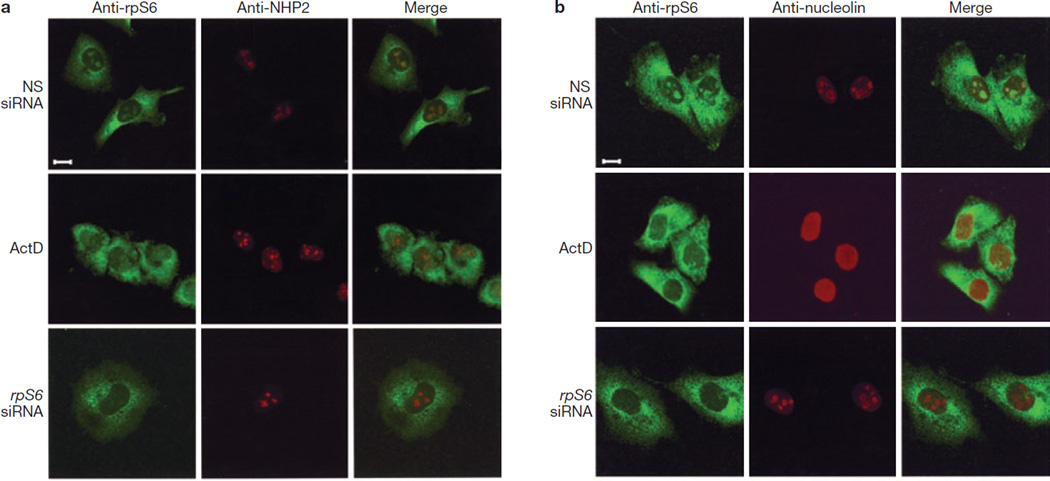
Induction of p53 in response to inhibition of ribosome biogenesis is attributed to nucleolar disruption13, which releases 60S rpL11 into the nucleoplasm, where it binds and inhibits MDM2 (refs 1, 4). To test this possibility, we analysed the nucleolar status of cells treated with NSsiRNA, actinomycin D, or rpS6-siRNA. RpS6 immunostaining of NSsiRNA-treated A549 cells revealed diffuse cytoplasmic, but discrete nucleolar, staining (Fig. 2a), presumably representing mature and nascent 40S ribosomes, respectively. Nucleolar rpS6 staining co-localized with that of nuclear high-mobility group-like protein 2 (NHP2), which localizes to dense fibrillar centres14. Treatment with actinomycin D led to a loss of nucleolar rpS6 staining and dispersion of NHP2 into nuclear clusters, which is consistent with its ability to inhibit rRNA transcription and to disrupt nucleoli13. In rpS6-siRNA-depleted cells, nucleolar rpS6 staining was absent, whereas it was largely unaffected in mature ribosomes of the cytoplasm (Fig. 2a). However, rpS6 depletion did not disrupt nucleoli, as demonstrated by the maintenance of NHP2 staining (Fig. 2a). Similarly, the localization of nucleolin, which is enriched at the periphery of nucleoli, was unaffected by rpS6 depletion, whereas it was drastically altered by actinomycin D (Fig. 2b). Consistent with rpS6 depletion having no effect on the integrity of the nucleolus (Fig. 2), it did not affect the localization of nascent 60S ribosomal proteins, as judged by immunostaining of 60S rpL7a (Supplementary Information, Fig. S2). Thus, nucleolar disruption is not a prerequisite for p53 induction after inhibition of 40S ribosome biogenesis13.
Figure 2.
Depletion of rpS6 does not result in nucleolar disruption. (a, b) A549 cells treated with either NSsiRNA or rpS6 siRNA2 for 24 h or with 5 ng ml−1 actinomycin D (ActD) for 8 h were fixed and stained with anti-rpS6 and anti-NHP2 (a) or anti-rpS6 and anti-nucleolin (b) antibodies and analysed by confocal microscopy with a Plan-Apochromat 100×/1.4 numerical aperture Oil Dic objective (Zeiss; see Methods). Scale bar, 10 µm.
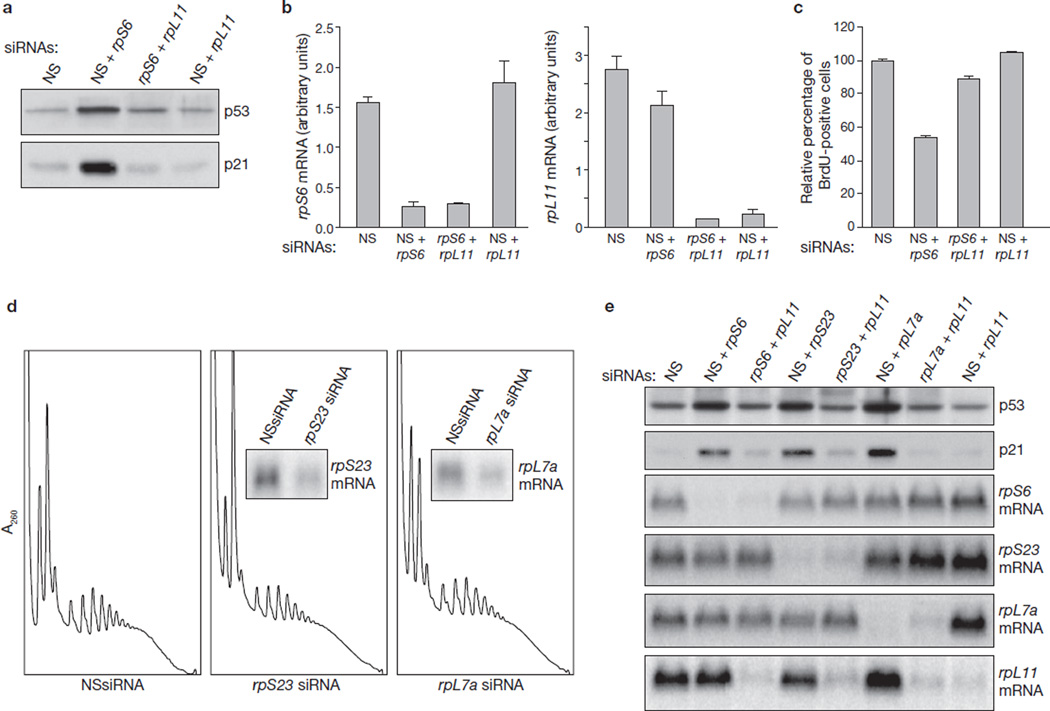
Because rpS6 depletion neither blunts 60S ribosome biogenesis (Fig. 1d; Supplementary Information, Fig. S1c) nor causes nucleolar disruption (Fig. 2), it seemed unlikely that p53 induction would be mediated by rpL11 (refs 1, 4). However, the induction of p53 and p21 by rpS6 depletion was abolished by the simultaneous depletion of rpL11 (Fig. 3a). As with rpS6 depletion, depletion of rpL11 had only a modest effect on protein levels (data not shown), although it clearly decreased mRNA levels (Fig. 3b). Moreover, rpL11 depletion in rpS6-siRNA-treated cells rescued S-phase entry (Fig. 3c). To ascertain the specificity of this response, we also analysed cells depleted of 40S rpS23 and 60S rpL7a. As judged from polysome profiles and incorporation of 3H-uridine into nascent rRNA, depletion of rpS23 or rpL7a resulted in the inhibition of 40S or 60S ribosome biogenesis, respectively (Fig. 3d; Supplementary Information, Fig. S3a). In a similar manner to rpS6 depletion (Fig. 1e), a decrease in either protein led to p53 and p21 induction, which was suppressed by the simultaneous depletion of rpL11 (Fig. 3e). Equivalent results were obtained with a second rpL11-siRNA targeting a distinct region of the transcript (Supplementary Information, Fig. 3b, c). These findings indicate that rpL11-mediated p53 induction is a general response to inhibition of 40S or 60S ribosome biogenesis.
Figure 3.
Upregulation of p53 by depletion of ribosomal proteins is mediated by rpL11. (a) Western blots showing the levels of p53 and p21 in A549 cells transfected for 24 h with the indicated siRNAs. (b) Total RNA extracted from the indicated transfections, performed in parallel with those in a, were used to measure the levels of the rpS6 and rpL11 mRNAs by quantitative real-time PCR. Each bar represents the ratio of the indicated mRNA to that of β-actin mRNA, and results are shown as means ± s.e.m. for three independent transfection experiments. (c) BrdU incorporation in A549 cells transfected for 24 h with the indicated siRNAs. For each treatment the values are normalized to values for cells transfected with NSsiRNA. Results are shown as means ± s.e.m. for three independent experiments. (d) Polysome profiles from extracts of A549 cells treated for 24 h with either NSsiRNA (left panel) or siRNAs specific for rpS23 or rpL7a mRNAs. Insets: northern blots hybridized to probes specific for rpS23 and rpL7a mRNAs. (e) Western blots showing the levels of p53 and p21 proteins, and northern blots showing the levels of rpS6, rpS23, rpL7a and rpL11 mRNAs in A549 cells treated for 24 h with the indicated siRNAs. Uncropped images of blots are shown in Supplementary Information, Fig. S5.
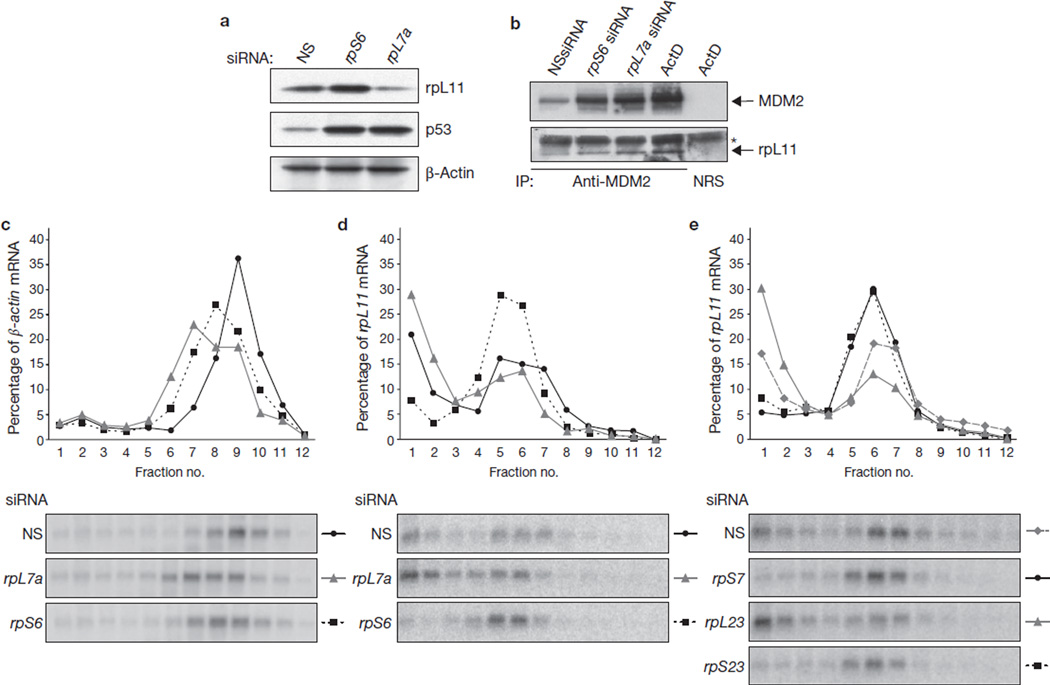
If p53 induction, after rpS6 depletion, is dependent on rpL11, this would require increased amounts of rpL11 protein in the face of 60S ribosome biogenesis. Treatment with either rpS6-siRNA or rpL7a-siRNA leads to p53 induction, in contrast with the NSsiRNA-treated control cells (Fig. 4a). However, whereas depletion of rpS6 led to an increase in total cellular L11 protein, depletion of rpL7a had the opposite effect (Fig. 4a), which is consistent with the decrease in total native 60S ribosomes (Fig. 3d). The prediction from these findings is that there is more ribosome-free rpL11 in rpS6-siRNA-treated cells than in NSsiRNA-treated cells. To examine this possibility, the distributions of rpL11 and β-actin, the latter as a loading control, were examined on western blots after centrifugation of cellular extracts on shallow sucrose gradients12, in which large polysomes pellet to the bottom of the tube. In samples from NSsiRNA-treated cells, most rpL11 protein sedimented with native 60S ribosomes and polysomes, although ribosome-free rpL11 could be detected at the top of the gradient (Supplementary Information, Fig. S4a). Similarly, in samples from rpS6-siRNA-treated cells, most rpL11 was associated with free 60S ribosomes and polysomes, although the amount that sedimented with native 60S ribosomes was significantly increased (Supplementary Information, Fig. S4a), which is consistent with the increase in native 60S ribosomes (Fig. 1d). However, despite continued 60S ribosome biogenesis, the amount of free rpL11 at the top of the gradient was clearly increased over that detected in NSsiRNA-treated cells, with comparable levels of cytosolic β-actin present in each sample (Supplementary Information, Fig. S4a). An increase in ribosome-free rpL11 was also observed in rpL7a-depleted cells (data not shown), most probably due to inhibition of 60S ribosome biogenesis (Fig. 3d). In rpS6-depleted and rpL7a-depleted cells, we found an increase in the amount of rpL11 that co-immunoprecipitated with enhanced levels of p53-induced MDM2, which were comparable to those observed in cells treated with actinomycin D (Fig. 4b). Thus, inhibition of 40S ribosome biogenesis leads to an increase in ribosome-free rpL11, which can bind and inhibit MDM2.
Figure 4.
Depletion of 40S ribosomal proteins results in translational upregulation of the rpL11 mRNA. (a) Western blots showing the levels of rpL11, p53 and β-actin proteins in cells transfected for 48 h with NSsiRNA, rpS6 siRNA2 or rpL7a siRNA. (b) Western blots showing the levels of rpL11 and MDM2 present in immunoprecipitates (IP) by using either an anti-MDM2 antibody or normal rabbit serum (NRS). Extracts used in the immunoprecipitation are from cells transfected for 24 h with NSsiRNA or with rpS6 or rpL7a siRNAs, or treated with actinomycin D (ActD) at 10 ng ml−1 for 10 h. The asterisk indicates the light chain of the anti-MDM2 antibody, which reacts with the secondary antibody. (c, d) Northern blot analysis of β-actin mRNA (c) and rpL11 mRNA (d) distribution on polysomes of A549 cells transfected with the indicated siRNAs for 30 h. (e) Northern blot analysis of rpL11 mRNA distribution on polysomes of A549 cells transfected with the indicated siRNAs. Fractions 4–12 in c–e contained translating polysomes. Uncropped images of blots are shown in Supplementary Information, Fig. S5.
Several mechanisms could explain the increase in ribosome-free rpL11 protein levels after disruption of 40S ribosome biogenesis. However, because ribosomal proteins have long half-lives and rpL11 mRNA levels are not significantly changed in rpS6-siRNA-treated cells from those in NSsiRNA-treated cells (Fig. 3b; Supplementary Information, Fig. 3c), we focused on translation. Consistent with rpS6-siRNA-treated cells and rpL7a-siRNA-treated cells having fewer polysomes than NSsiRNA-treated cells (Figs 1d and 3d), β-actin mRNA was distributed to smaller polysomes in these cells (Fig. 4c). Similarly, more rpL11 transcripts were found in the non-polysome fraction in cells treated with rpL7a-siRNA than in those treated with NSsiRNA (Fig. 4d). However, despite comparable inhibitory effects of rpS6-siRNA and rpL7a-siRNA on β-actin mRNA translation (Fig. 4c) and global protein synthesis (Fig. 5b), most of the rpL11 transcripts were recruited to actively translating polysomes in rpS6-siRNA-treated cells (Fig. 4d). Similarly, depletion of rpS23 or rpS7 resulted in inhibition of 40S ribosome biogenesis and the recruitment of rpL11 mRNAs to polysomes, whereas rpL23 depletion led to inhibition of 60S ribosome biogenesis and the accumulation of rpL11 mRNAs in the non-polysome fraction (Fig. 4e, and data not shown). During liver regeneration, the translation of mRNAs encoding ribosomal proteins is upregulated, whereas in non-proliferating liver they reside largely in the non-polysome portion of the gradient15. Consistent with this finding was our observation, in the regenerating livers of rpS6fl/flMxCre− mice, that most of the rpL11 mRNA was recruited to polysomes, with a small amount present in the non-polysome fraction (Supplementary Information, Fig. S4b). However, in the non-regenerating livers of rpS6fl/flMxCre+ mice, we found that all rpL11 mRNA shifted to polysomes, despite a decrease in the mean polysome size of rpL11 mRNA transcripts (Supplementary Information, Fig. S4b). Thus, translational upregulation of rpL11 mRNA seems to be a general response to inhibition of 40S ribosome biogenesis.
Figure 5.
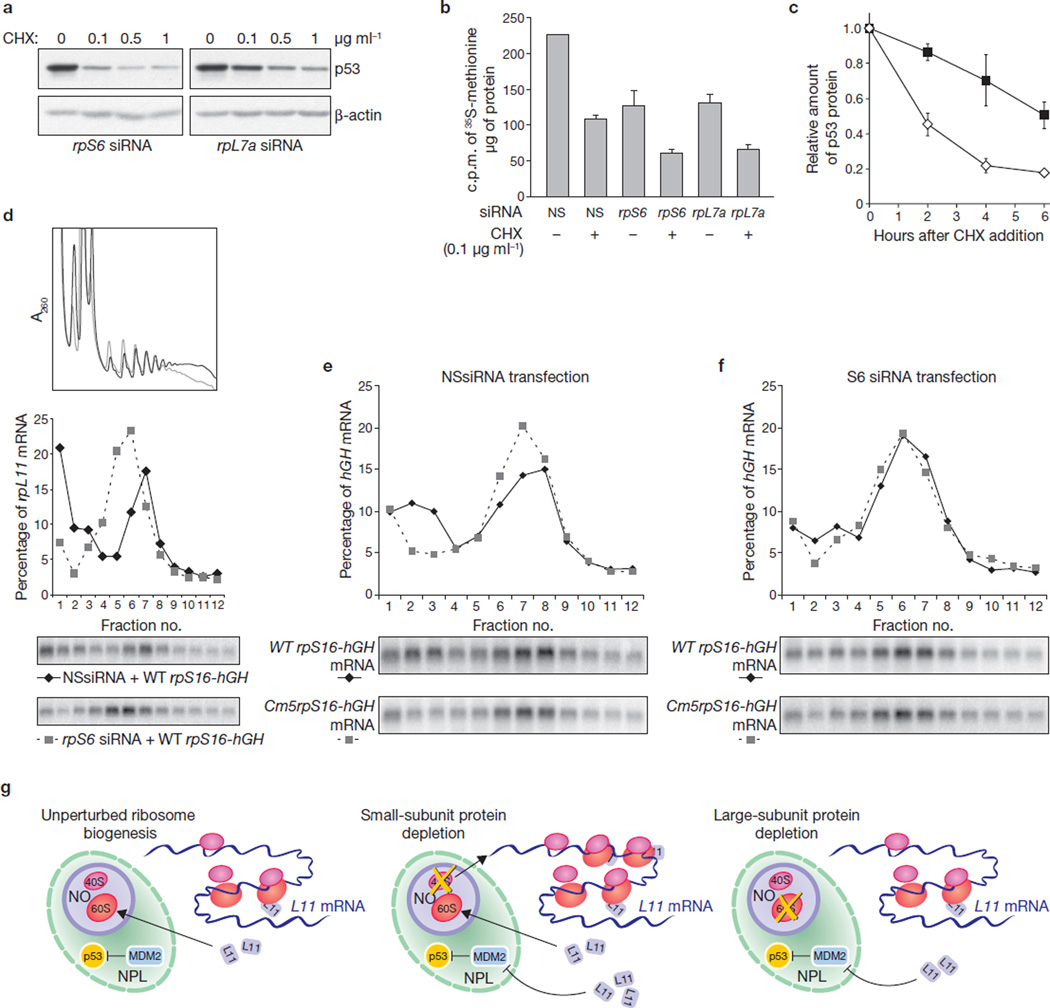
Depletion of rpS6 upregulates 5´-TOP mediated translation. (a) Western blots showing levels of p53 and β-actin in A549 cells transfected with either rpS6 or rpL7a siRNA and treated for 6 h with the indicated doses of cycloheximide (CHX). (b) Measurement of [35S]methionine incorporation in cells transfected with the indicated siRNAs and treated, or left untreated, for 4 h with 0.1 µg ml−1 cycloheximide. Results are shown as means ± s.e.m. for three independent experiments. (c) Quantification of the ratio of p53 to β-actin (measured by densitometry on western blots) in cells transfected for 24 h with either rpS6 siRNA (open diamonds) or rpL7a siRNA (filled squares) and treated for the indicated times with 0.1 µg ml−1 cycloheximide. Results are shown as means ± s.e.m. for three independent experiments. A representative western blot used for this analysis is shown in Supplementary Information, Fig. S4c. (d) Polysome profiles (top) and northern blot analysis of the distribution of rpL11 mRNA on polysomes (bottom) in HEK 293 cells co-transfected with wild-type (WT) rpS16-hGH plasmid DNA in combination with either NSsiRNA (black line) or rpS6 siRNA (grey line). (e) Northern blot analysis of the distribution on polysomes of hGH reporter mRNA in HEK 293 cells co-transfected with NSsiRNA in combination with either wild-type rpS16-hGH or Cm5rpS16-hGH plasmid DNA. (f) Northern blot analysis of the distribution on polysomes of hGH reporter mRNA in HEK 293 cells co-transfected with rpS6 siRNA2 in combination with either wild-type rpS16-hGH or Cm5rpS16-hGH plasmid DNA. In the northern blot analysis of all three panels, fractions 4–12 contained translating polysomes. (g) Model of p53 stabilization in response to impaired 40S ribosome biogenesis. See the text for details. NPL, nucleoplasm; NO, nucleolus. Uncropped images of blots are shown in Supplementary Information, Fig. S5.
The increased rpL11 translation induced by depletion of 40S ribosomal proteins was unexpected, because decreased ribosomal protein mRNA translation is associated with global inhibition of protein synthesis16. Given that translational upregulation of rpL11 is responsible for the increase in p53 levels in cells in which 40S, but not 60S, ribosome biogenesis has been disrupted, this response should be more sensitive to global translational inhibition. We found that cycloheximide, a general translational inhibitor, blunted p53 expression in a dose-dependent manner in either rpS6-siRNA-transfected or rpL7a-siRNA-transfected cells, with the effect more pronounced in rpS6-depleted cells (Fig. 5a). Depletion of rpS6 or rpL7a decreased the incorporation of [35S]methionine into nascent protein by 50%, and in both cases further treatment with 0.1 µg ml−1 cycloheximide decreased translation a further 50% (Fig. 5b). However, treatment with cycloheximide impaired p53 accumulation to a larger extent in cells transfected with rpS6-siRNA than in those transfected with rpL7a-siRNA (Fig. 5c; Supplementary Information, Fig. S4c). These findings agree with rpL11 translational upregulation being responsible for p53 induction in cells with impaired 40S ribosome biogenesis.
Ribosomal protein mRNA translation is negatively controlled by a 5´-TOP (refs 17, 18): replacement of pyrimidines with purines at this site leads to their constitutive translation19,20. The fact that other 5´-TOP mRNAs, including those encoding rpS8, rpS16 and rpL26, were also upregulated in rpS6-depleted cells (Supplementary Information, Fig. S4d) suggested that selective translation of rpL11 is mediated by the 5´-TOP. We tested this possibility by transiently expressing two reporter constructs containing the coding region of the mRNA encoding human growth hormone (hGH) fused to either the first 29 nucleotides of the rpS16 5´-untranslated region, including the 5´-TOP (wtrpS16-hGH), or to a mutant in which five of the eight 5´-TOP pyrimidines were replaced by purines (Cm5rpS16-hGH; refs 19, 20). RpS6 depletion, in comparison with controls, led to a decrease in the number of 40S ribosomes, a decrease in the mean polysome size and an increase in the number of free 60S ribosomes (Fig. 5d). This also resulted in the almost complete shift of rpL11 transcripts onto actively translating polysomes, despite decreased global translation, as reflected by the decreased mean polysome size of rpL11 mRNA (Fig. 5d). In a similar manner to rpL11, wtrpS16-hGH transcripts, in NSsiRNA-treated cells, were distributed between non-polysomes and polysomes (Fig. 5e), whereas the Cm5rpS16-hGH transcript was present almost exclusively in polysomes (Fig. 5e). After rpS6 depletion, wtrpS16-hGH transcript, as with rpL11, largely shifted to polysomes, behaving as the Cm5rpS16-hGH transcript (Fig. 5f), despite the inhibition of global translation, as reflected by the shift of wtrpS16-hGH and Cm5rpS16-hGH transcripts to polysomes with a smaller mean size. These data show that translational upregulation of rpL11 mRNAs is mediated through derepression of the 5´-TOP.
During growth, rpL11 is assembled into nascent 60S ribosomes; however, impairment of this process, or of rRNA transcription, allows excess rpL11 to inhibit MDM2 and stabilize p53 (Fig. 5g). In contrast, when 40S ribosome biogenesis is impaired, 60S ribosome biogenesis continues, leading to the translational upregulation of 5´-TOP mRNAs, despite inhibition of global protein synthesis (Fig. 5g). Apparently, the translational upregulation of 5´-TOP mRNAs is not highly energy-consuming, because the rate of translation in rpS6-depleted cells is equivalent to that in rpL7a-depleted cells (Fig. 5b), in which 5´-TOP mRNA translation is suppressed (Fig. 4d; Supplementary Information, Fig. S4d). The importance of the cellular response to disrupted ribosome biogenesis is underscored by the finding that hypomorphic mutations in 40S ribosomal proteins rpS19, rpS24, and rpS14 are causal agents in Diamond–Blackfan anaemia6,21 and 5q− syndrome7, which are disorders characterized by erythroid hypoplasia and increased susceptibility to leukemia. Depletion of ribosomal proteins results in alterations in the ratio of mRNA to ribosomes, potentially changing the pattern of translation and the genetic program5. Such deficiencies lead to p53 induction, which is also associated with other pathological conditions such as Treacher–Collins syndrome (TCS). TCS is a congenital disorder characterized by hypoplasia of craniofacial elements caused by mutations in the nucleolar protein Treacle that impair ribosome biogenesis and result in p53 induction22. These effects are reversed in a p53 heterozygous background, suggesting that suppression of p53 could be used in the treatment of TCS22. However, given that ribosomal proteins are haploinsufficient tumour suppressors in humans6,7,21, inactivation of p53 in conditions under which ribosome biogenesis is impaired could induce leukaemia in Diamond–Blackfan anaemia6,21 and 5q− syndrome7, as recently described in zebrafish23. In contrast, our studies suggest that signalling components involved in rpL11 upregulation could be clinically targeted without interfering with other pathways involved in p53 induction8. It will therefore be important to elucidate the mechanisms by which translation of 5´-TOP mRNAs is controlled in 40S ribosomal-protein-depleted cells, including those regulated by lupus antigen and mTOR. Lupus antigen has been shown to regulate the translation of 5´-TOP mRNA; however, it is unclear whether it is a positive or negative effector24,25. An inhibitor of mTOR function, rapamycin, has been shown to selectively attenuate the translation of 5´-TOP mRNAs20; however, we found the inhibitory effect of rapamycin on the translation of rpL11 mRNA in either rpS6-depleted or rpS23-depleted cells to be much less pronounced than in NSsiRNA-treated cells (S.F. and G.T., unpublished observations). Unravelling the signalling pathways that regulate 5´-TOP mRNA translation will be a key step in developing specific therapies for diseases associated with hypomorphic lesions in ribosomal protein expression.
METHODS
Cell culture and transfection
A549 and HEK 293 cells were cultured and transfected with siRNAs as described10,19, with the final concentration of each siRNA being 16.7 nM. For co-transfection, cells in 10-cm dishes were transfected with calcium phosphate26 and 2 µg of plasmid DNA, followed 24 h later with 6 nM siRNA. The siRNAs employed targeted mRNAs encoding human rpS6 (5´-AAGAAAGCCCTTAAATAAAGA-3´ (siRNA1) and 5´-TTGTAAGAAAGCCCTTAAATA-3´ (siRNA2)), rpS7 (5´-TAGATGGCAGCCGGCTCATAA-3´), rpS23 (5´-CAGCCCTAAAGGCCAACCCTT-3´), rpL7a (5´-CACCACCTTGGTGGAGAACAA-3´), rpL11 (5´-AAGGTGCGGGAGTATGAGTTA-3´), rpL23 (5´-CTGGTGCGAAATTCCGGATTT-3´), p53 (ref. 12) and Nonsilencing siRNA (Qiagen).
Mice
The floxed S6 mice and the rpS6 gene deletion have been described (Sigma)5. Three days after rpS6 gene deletion, partial hepatectomy was performed27, after injection of 0.08% Narketan (Vetoquinol AG)/0.24% Rompun (Bayer) at 10 µl g−1 body weight. Livers of mice killed at 20, 30 and 40 h after surgery were quickly frozen and stored at −80 °C. The portion of liver resected was used for the 0 h time point.
Measurement of [35S]methionine incorporation
Cells were labelled for 30 min with a 35SMeth/Cys mix (10 µCi ml−1; NEG072; Perkin Elmer); proteins were extracted and precipitated and the amount of radioactivity incorporated was determined28.
Protein extraction
For western blot analysis of total extracts, cells were washed with ice-cold PBS and lysed in 50 mM Tris-HCl pH 8, 250 mM NaCl, 1% Triton X-100, 0.25% sodium deoxycholate, 0.05% SDS, 1 mM dithiothreitol (DTT). The lysates were cleared by centrifugation. For co-immunoprecipitation of MDM2–rpL11 complexes, cells were lysed in 50 mM Tris-HCl pH 8, 250 mM NaCl, 0.5% Nonidet P40, 1 mM DTT, and the lysates were cleared by centrifugation. For the analysis and distribution of rpL11 protein on polysome profiles, cells were washed with ice-cold buffer A (20 mM Tris-HCl pH 7.5, 250 mM KCl, 10 mM MgCl2) and lysed in buffer A containing 0.5% Triton X-100, 0.5% sodium deoxycholate, 120 U ml−1 RNAsin (Promega), 3 mM DTT. The extracts were incubated on ice for 30 min and cleared by centrifugation. All extraction buffers contained a cocktail of protease inhibitors (Roche).
Immunoprecipitation
Samples (1.6 mg of extract) were precleared for 1 h with protein A-Sepharose and then incubated for 4 h at 4 °C with anti-MDM2 antibody (H-221; Santa Cruz Biotechnology). After 1 h with protein A-Sepharose, immunoprecipitates were washed three times in extraction buffer and resuspended in protein sample buffer and analysed as described29.
Western blotting and enzyme-linked immunosorbent assay (ELISA)
Protein samples were resolved by SDS–PAGE and transferred to poly(vinylidene difluoride) membranes (Millipore), with a semi-dry transfer apparatus (LTF-Labortechnik). The membranes were blocked in Tris-buffered saline (TBS) containing 0.2% Tween 20 and 5% milk. Both primary and horseradish peroxidase-conjugated secondary antibodies (Amersham Biosciences) were added in TBS containing 0.2% Tween 20 and 3% BSA (Sigma). After incubation with an enhanced chemiluminescence (ECL) reagent (Amersham Biosciences), blots were exposed to X-ray films (Kodak). Primary antibodies used were as follows: mouse anti-rpS6 (Novartis), rabbit anti-rpL11 (from Karen Vousden), anti-rpL11 (Zymed Laboratories) antip53 (Santa Cruz Biotechnology), anti-p21 (Pharmingen), anti-MDM2 (Santa Cruz Biotechnology), and anti-β-actin (Cell Signaling). Quantification of western blots by densitometry was performed with ImageQuant software (Molecular Dynamics). For the determination of liver p53 levels, an ELISA kit was used as described by the manufacturer (Roche), with duplicate measurements for each sample.
Analysis of mRNA and protein distribution on polysome profiles
Analysis of mRNA distribution on polysome profiles was performed as described29. To analyse the distribution of rpL11 protein on polysome profiles, cell extracts were centrifuged on a shallow sucrose gradient12 and fractionated. Proteins from each fraction were precipitated with 20% trichloroacetic acid. After centrifugation, the pellets were washed with 80% ethanol/20% diethyl ether, resuspended in protein sample buffer and processed for western blot analysis.
RNA extraction, northern blot analysis, and probes
Isolation of total RNA and northern blot analyses were largely performed as described previously12. The probes used were as follows: rpS6, 5´-GAGATGTTCAGCTTCATCTTGAAGCAGCTGAACGCCTCCGACGCCACGGAAAAGAGG-3´; rpL7a, 5´-GGCGATACGAGCCACAGACTTAGGACCCAGGACATTGCCACCCCAGTGACGGCGGATC-3´; rpL11 (human), 5´-GTTTGGCCCCAATGCAGCCTGTCCTGCGCTTCTTGTCTGCGATGCTGAAACCTGGCC-3´ ; rpS23, 5´-CCACTTCTGGTCTCGTGCGTGACTACGGAGCTTCCTAGCAGTACGAAGTCCACGACAC-3´; β-actin, 5´-CAGATTTTCTCCATGTCGTCCCAGTTGGTGACGATGCCGTGCTCGATGGGGTACTTCAG-3 ´ ; r pL1 1 (mouse), 5´-GTGTATCTAGCTTTGGAGAACACCGGGGTCTGGCCTGTGAGCTGCTCCAACACCTTGGC-3´; rRNA, 5´-GGCGAGCGACCGGCAAGGCGGAGG TCGACCCACGCCACACGTCGCACGAACGCCTGTC-3´. The hGH probe has been described20. The blots were developed with a Storm 840 phosphorimager and quantified with ImageQuant software.
Quantitative real-time PCR
Total RNAs were reverse-transcribed with a Superscript III kit (Invitrogen). The complementary DNAs were used in PCR reactions containing Absolute BlueSYBR Green Fluorescein mix (Thermo Scientific). The reactions were analysed in a Realplex S apparatus (Eppendorf). A standard curve was generated for each assay. Reactions were performed as follows: 95^°C for 15^min followed by 40 cycles of 95^°C for 15^s, 60^°C for 20^s, and 72^°C for 20^s. Melting-curve analyses were performed to verify the synthesis of single products. The primers used were as follows: rpS6, 5´-TCTTGACCCATGGCCGTGTC-3´ (forward) and 5´-GCGGCGAGGCACTGTAGTAT-3´ (reverse); rpL7a, 5´-GCTGAAAGTGCCTCCTGCGA-3´ (forward) and 5´-CACCAAGGTGGTGACGGTGT-3´ (reverse); rpL11, 5´-TCCACTGCACAGTTCGAGGG-3´ (forward) and 5´-AAACCTGGCCTACCCAGCAC-5´ (reverse). The β-actin primers have been described30.
Immunofluorescence
Cells seeded on coverslips were washed in PBS, fixed for 10 min at room temperature 20–22 °C with 4% paraformaldehyde, permeabilized for 5 min in PBS containing 0.5% Triton X-100 and washed in PBS. After blocking for 30 min in PBS containing 1% BSA at room temperature, the preparations were incubated overnight at 4 °C with the following antibodies diluted in PBS containing 1% BSA: mouse anti-rpS6, rabbit anti-NHP2 (ref. 14), anti-nucleolin (H-250; Santa Cruz) or rabbit anti-rpL7a. The coverslips were then washed in PBS and incubated for 30 min at room temperature with the secondary antibodies Alexa Fluor 488 goat anti-mouse and Alexa Fluor 555 donkey anti-rabbit (Molecular Probes) diluted in PBS containing 1% BSA and then washed in PBS. The coverslips were mounted on slides and analysed by fluorescence imaging with a Zeiss Axioplan II confocal microscope.
Microarray analysis
RNAs were extracted from liver portions, with the guanidinium thiocyanate–phenol–chloroform extraction protocol31, and further purified with an RNeasy Kit (Qiagen). Total RNA (10 µg) was reverse transcribed with the Affymetrix cDNA synthesis kit. The cDNA was then used in an in vitro transcription reaction using the in vitro transcription kit from Affymetrix. The resulting cRNAs were hybridized to MGU-74A GeneChips (Affymetrix) in accordance with the protocols provided by Affymetrix. Expression values were estimated with the robust multichip analysis summary algorithm (RMA)32 implemented in RMAExpress (http://rmaexpress.bmbolstad.com/). Expression analysis was performed with GeneSpring 6.2 (Silicon Genetics). For each time point we produced a list of genes that were increased and a list of genes that were decreased in S6fl/flMxCre− mice in comparison with S6fl/flMxCre+ mice. Consideration was limited to those genes whose change in expression was at least 1.5-fold relative to the control genotype at one or more time points and that passed a one-way analysis of variance (cutoff P < 0.01). Microarray data can be accessed at http:// www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE13864.
Flow cytometry
Cells pulsed for 10 min with 10 µM BrdU (Sigma) were collected and fixed in 70% ethanol. Staining with a fluorescein isothiocyanate-conjugated anti-BrdU antibody (Becton Dickinson) was performed in accordance with the manufacturer’s instructions. A propidium iodide/RNAse solution (Phoenix Flow Systems) was added to the cell suspension before the analysis. Flow cytometry data were acquired on a Coulter Epics XL (Beckman Coulter) with a 488-nm argon-ion laser. Data acquisition was performed with System II software (Beckman Coulter). Doublet discrimination was performed by gating on peak versus integral signals. Cell-cycle analysis was performed with ModFit LT version 2.0 (Verity Software House, Inc.). Histograms were made with FlowJo software version 6.3.2 (Tree Star, Inc.). In all experiments, 10,000 gated events were collected.
Supplementary Material
Acknowledgments
We are indebted to P. D. Plas, A. Selvaraj and P. B. Dennis for their critical reading of the manuscript, as well as members of the laboratory for numerous discussions. We are also thankful to K. Vousden for the L11 polyclonal antibody. Finally we thank G. Doerman and M. Daston for computer graphics and the editing of the manuscript, respectively. A.D.C. was supported by fellowship from the Boehringer Ingelheim Fund. These studies were supported by the Mouse Models in Human Cancer Consortium grant U01 CA-84292 to P.P.P. and G.T., and G.T. is also supported by the Gladys and John Strauss Chair in Cancer Research and start-up funds from the University of Cincinnati.
Footnotes
Note: Supplementary Information is available on the Nature Cell Biology website.
COMPETING FINANCIAL INTERESTS
The authors declare that they have no competing financial interests.
References
- 1.Lohrum MA, Ludwig RL, Kubbutat MH, Hanlon M, Vousden KH. Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell. 2003;3:577–587. doi: 10.1016/s1535-6108(03)00134-x. [DOI] [PubMed] [Google Scholar]
- 2.Dai MS, Lu H. Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 2004;279:44475–44482. doi: 10.1074/jbc.M403722200. [DOI] [PubMed] [Google Scholar]
- 3.Dai MS, et al. Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell. Biol. 2004;24:7654–7668. doi: 10.1128/MCB.24.17.7654-7668.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yuan X, et al. Genetic inactivation of the transcription factor TIF-IA leads to nucleolar disruption, cell cycle arrest, and p53-mediated apoptosis. Mol. Cell. 2005;19:77–87. doi: 10.1016/j.molcel.2005.05.023. [DOI] [PubMed] [Google Scholar]
- 5.Volarevic S, et al. Proliferation, but not growth, blocked by conditional deletion of 40S ribosomal protein S6. Science. 2000;288:2045–2047. doi: 10.1126/science.288.5473.2045. [DOI] [PubMed] [Google Scholar]
- 6.Draptchinskaia N, et al. The gene encoding ribosomal protein S19 is mutated in Diamond–Blackfan anaemia. Nature Genet. 1999;21:169–175. doi: 10.1038/5951. [DOI] [PubMed] [Google Scholar]
- 7.Ebert BL, et al. Identification of RPS14 as a 5q− syndrome gene by RNA interference screen. Nature. 2008;451:335–339. doi: 10.1038/nature06494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vousden KH, Lane DP. p53 in health and disease. Nature Rev. Mol. Cell Biol. 2007;8:275–283. doi: 10.1038/nrm2147. [DOI] [PubMed] [Google Scholar]
- 9.Sulic S, et al. Inactivation of S6 ribosomal protein gene in T lymphocytes activates a p53-dependent checkpoint response. Genes Dev. 2005;19:3070–3082. doi: 10.1101/gad.359305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panic L, et al. Ribosomal protein S6 gene haploinsufficiency is associated with activation of a p53-dependent checkpoint during gastrulation. Mol. Cell. Biol. 2006;26:8880–8891. doi: 10.1128/MCB.00751-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McGowan KA, et al. Ribosomal mutations cause p53-mediated dark skin and pleiotropic effects. Nature Genet. 2008;40:963–970. doi: 10.1038/ng.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beuvink I, et al. The mTOR inhibitor RAD001 sensitizes tumor cells to DNA-damaged induced apoptosis through inhibition of p21 translation. Cell. 2005;120:747–759. doi: 10.1016/j.cell.2004.12.040. [DOI] [PubMed] [Google Scholar]
- 13.Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–6077. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pogacic V, Dragon F, Filipowicz W. Human H/ACA small nucleolar RNPs and telomerase share evolutionarily conserved proteins NHP2 and NOP10. Mol. Cell. Biol. 2000;20:9028–9040. doi: 10.1128/mcb.20.23.9028-9040.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aloni R, Peleg D, Meyuhas O. Selective translational control and nonspecific posttranscriptional regulation of ribosomal protein gene expression during development and regeneration of rat liver. Mol. Cell. Biol. 1992;12:2203–2212. doi: 10.1128/mcb.12.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyuhas O, Avni D, Shama S. In: Translational Control. Hershey JWB, Mathews MB, Sonenberg N, editors. Cold Spring Harbor Laboratory Press; 1996. pp. 363–388. [Google Scholar]
- 17.Meyuhas O. Synthesis of the translational apparatus is regulated at the translational level. Eur. J. Biochem. 2000;267:6321–6330. doi: 10.1046/j.1432-1327.2000.01719.x. [DOI] [PubMed] [Google Scholar]
- 18.Fumagalli S, Thomas G. In: Translational Control of Gene Expression. Sonenberg N, Hershey JWB, Mathews M, editors. Cold Spring Harbor Laboratory Press; 2000. pp. 695–717. [Google Scholar]
- 19.Levy S, Avni D, Hariharan N, Perry RP, Meyuhas O. Oligopyrimidine tract at the 5´ end of mammalian ribosomal protein mRNAs is required for their translational control. Proc. Natl Acad. Sci. USA. 1991;88:3319–3323. doi: 10.1073/pnas.88.8.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jefferies HBJ, et al. Rapamycin suppresses 5´TOP mRNA translation through inhibition of p70s6k . EMBO J. 1997;12:3693–3704. doi: 10.1093/emboj/16.12.3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gazda HT, et al. Ribosomal protein S24 gene is mutated in Diamond–Blackfan anemia. Am. J. Hum. Genet. 2006;79:1110–1118. doi: 10.1086/510020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones NC, et al. Prevention of the neurocristopathy Treacher Collins syndrome through inhibition of p53 function. Nature Med. 2008;14:125–133. doi: 10.1038/nm1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.MacInnes AW, Amsterdam A, Whittaker CA, Hopkins N, Lees JA. Loss of p53 synthesis in zebrafish tumors with ribosomal protein gene mutations. Proc. Natl Acad. Sci. USA. 2008;105:10408–10413. doi: 10.1073/pnas.0805036105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Crosio C, Boyl PP, Loreni F, Pierandrei-Amaldi P, Amaldi F. La protein has a positive effect on the translation of TOP mRNAs in vivo. Nucleic Acids Res. 2000;28:2927–2934. doi: 10.1093/nar/28.15.2927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schwartz EI, Intine RV, Maraia RJ. CK2 is responsible for phosphorylation of human La protein serine-366 and can modulate rpL37 5´-terminal oligopyrimidine mRNA metabolism. Mol. Cell. Biol. 2004;24:9580–9591. doi: 10.1128/MCB.24.21.9580-9591.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen CA, Okayama H. Calcium phosphate-mediated gene transfer: a highly efficient transfection system for stably transforming cells with plasmid DNA. Biotechniques. 1988;6:632–638. [PubMed] [Google Scholar]
- 27.Blindenbacher A, et al. Interleukin 6 is important for survival after partial hepatectomy in mice. Hepatology. 2003;38:674–682. doi: 10.1053/jhep.2003.50378. [DOI] [PubMed] [Google Scholar]
- 28.Iordanov MS, et al. Ribotoxic stress response: activation of the stress-activated protein kinase JNK1 by inhibitors of the peptidyl transferase reaction and by sequence-specific RNA damage to the α-sarcin/ricin loop in the 28S rRNA. Mol. Cell. Biol. 1997;17:3373–3381. doi: 10.1128/mcb.17.6.3373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pende M, et al. S6K1−/−/S6K2−/− mice exhibit perinatal lethality and rapamycin-sensitive 5´-terminal oligopyrimidine mRNA translation and reveal a mitogen-activated protein kinase-dependent S6 kinase pathway. Mol. Cell. Biol. 2004;24:3112–3124. doi: 10.1128/MCB.24.8.3112-3124.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young AR, et al. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. J. Cell Sci. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 31.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 32.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.