Abstract
Prostaglandin (PG) actions on disc metabolism are unclear even though certain PGs are highly expressed by disc cells under inflammatory conditions and nonsteroidal anti-inflammatory drugs (NSAIDs) are frequently used to block PG production to treat back pain. Hence this study aimed to (1) quantify gene expression of arachidonic acid cascade components responsible for PG synthesis and (2) examine the effects of key PGs on disc matrix homeostasis. Microarray analysis revealed that inflammatory stress increases expression of synthases and receptors for prostaglandin E2 (PGE2) and prostaglandin F2α (PGF2α), resulting in elevated PGE2 and PGF2α production in conditioned media of disc cells. PGE2 diminished disc cell proteoglycan synthesis, in a dose-dependent manner. Semiquantitative RT-PCR revealed differential effects of PGE2 and PGF2α on disc cell expression of key matrix structural genes, aggrecan, versican, collagens type I and II. PGE2 and PGF2α also decreased message for the anabolic factor, IGF-1. PGE2 decreased mRNA expression for the anti-catabolic factor TIMP-1 while PGF2α increased mRNAs for catabolic factors MMP-1 and MMP-3. Thus, PGE2 and PGF2α may have an overall negative impact on disc matrix homeostasis, and the use of NSAIDs may impact disc metabolism as well as treat back pain.
Keywords: nucleus pulposus cells, prostaglandins, PGE2, PGF2α, intervertebral disc matrix
Low back pain is often associated with intervertebral disc degeneration (IDD) and remains a significant health problem.1 Etiology of IDD is complex and still not well understood.2 Thus, current treatment options often address symptoms without an understanding of the mechanism of action. For example, nonsteroidal anti-inflammatory drugs (NSAIDs) targeting prostaglandin (PG) production are frequently prescribed for their analgesic and anti-inflammatory effects. However, few studies have addressed potential effects of PG on disc cell metabolism.
Several lines of evidence demonstrate the presence of PGs in discs under stress and implicate the possible involvement of PGs in IDD progression. The pro-inflammatory cytokines IL-1 and TNF-α are increased in degenerated human discs3,4 and have multiple actions that may contribute to disc matrix loss.5,6 Both cytokines induce cyclooxygenase 2 (COX-2), a key enzyme for PG biosynthesis, in disc cells with subsequent synthesis of PGs.7–10 COX-2 expression and PG synthesis in disc cells are also induced by mechanical stress, another predisposing factor that can disrupt disc structure and initiate the degenerative cascade.11,12 PGE2, a major inflammatory PG mediator associated with sensitization of proprioceptive neurons and involved with pain induction, is elevated in herniated lumbar discs.13
The presence of PGs in disc cells suggests they have functional roles in disc metabolism, but current evidence is indirect, limited, and at times contradictory. Chemical inhibition of COX-2 partially dampens the IL-1-mediated induction of catabolic response in human disc cells.14 In this study, COX-2 inhibition also diminishes IL-1-mediated suppression of proteoglycan sysnthesis. In contrast, Yoo et al.15 found that proteoglycan synthesis by canine nucleus pulposus (NP) cells was inhibited by NSAIDs that inhibit both the constitutive COX-1 and inducible COX-2. This suggested that either endogenously produced PGs actually supported matrix proteoglycan synthesis, or that the NSAIDs decrease proteoglycan synthesis through nonspecific actions unrelated to PGs under these experimental conditions. On the other hand, Karppinen et al.16 treated pigs with stab-induced disc degeneration with tiaprofenic acid or indomethacin and reported no negative effects on matrix metabolism from long-term administration of these NSAIDs in this in vivo model. Iwabuchi et al.17 measured the effect of PGE2 on rat disc wet weights and found no significant difference in weight loss, except in those discs concurrently treated with low-intensity pulsed ultrasound where PGE2 diminished disc weight loss, for example, PGE2 was matrix protective. Though indirect, these findings implicate regulatory roles of PGs in disc matrix homeostasis.
The minimal exploration of PG actions on intervertebral disc cells contrasts with the many studies exploring the role of PGs, in particular PGE2, in chondrocyte metabolism, as it relates to the pathophysiology of arthritis (reviewed in Refs.18,19). Depending on the dose, time, and experimental model, PGE2 actions are diverse and include induction of chondrocyte apoptosis, reversal of proteoglycan degradation induced by IL-1, stimulation of aggrecan synthesis, increasing or decreasing collagen synthesis, inhibition of DNA synthesis or increasing growth, and sensitizing chondrocytes to the anabolic actions of IGF-1. Goldring et al. have defined the mechanisms through which PGE2 inhibits type I collagen gene expression and stimulates type II collagen gene expression in chondrocytes and fibroblasts 20,21.
Given the similarities between chondrocytes and cells of the NP, and the frequency with which patients with low back pain are treated with NSAIDs, it is appropriate to determine the potential of human NP (hNP) cells for PG synthesis and response to PGs. The current studies approach this problem through (1) microarray analysis for components of the arachidonic acid cascade, and (2) determining the response of hNP to exogenous PGE2 and PGF2α.
MATERIALS AND METHODS
Nucleus Pulposus Cell Isolation and Culture
The NP was dissected from patient disc surgical specimens (19–59 years, mean = 42.2 years, and average Thompson degeneration grade 2–3.22 All of these NP tissues were isolated from patients underwent surgeries for disc degeneration and not disc herniation, and therefore contact between these tissues and cells outside of the disc, that is, macrophages, was minimal or nonexistent. The experimental protocol was approved by the human subjects Institutional Review Board at the University of Pittsburgh. Cells were isolated and cultured in F-12/D-MEM containing 10% FCS, 1% PS, and 25 μg/ml L-ascorbic acid under standard conditions (37°C, 5% CO2, 95% air, bicarbonate buffering to maintain pH 7.2) as previously described.23 Cell culture materials were purchased from Invitrogen/Gibco (Carlsbad, CA) unless noted otherwise, and the hNP cells were used at passage 1 or 2.
Microarray Analysis of NP Gene Expression
hNP cells (grade 3) at passage 1 were exposed to 0 and 5 ng/ml TNF-α for 24 h. Total RNA was isolated and hybridized to Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays designed for analysis of 47,000 transcripts, following standard procedures recommended by the manufacturer. Array data were collected from three independent experiments and analyzed using RMA24 and Limma25 in the R statistical environment. Expression levels are averages from n = 3 ± 1 SE.
Exposure to Prostaglandins
hNP cells were isolated and expanded in monolayer, and passed to three-dimensional culture in alginate beads as previously described.23 Experiments were begun after the cells had been in three-dimensional culture for at least 1 week and the response of hNP cellsto PGE2 or PGF2α (Sigma–Aldrich, St. Louis, MO) in low serum (1%) media was evaluated after 3 days exposure. The concentrations used (100 ng/ml PGE2 and 75 ng/ml PGF2α) were chosen based on the medium concentrations attained in response to 1 ng/ml IL-1 (104 ± 14 and 62 ± 9 ng/ml for PGE2 and PGF2α, respectively). Cells were also exposed to 10 μM butaprost (a specific PG PTGER2 receptor agonist) and 50 ng/ml fluprostenol (a metabolically stable analog of PGF2α with potent PTGFR receptor agonist activity, Sigma–Aldrich) for 3 days. Total RNA were extracted using the RNeasy Mini Kit (Qiagen, Germantown, MD), and DNA measured by Picogreen (Molecular Probes, Eugene, OR) after digestion in papain buffer.23
Matrix Protein Synthesis
Proteoglycan synthesis was measured on day 3 and total protein/collagenase sensitive incorporation of 3H proline into proteins was done from 48 to 72 h of PG activation. Proteoglycan synthesis was measured from a 6 h pulse labeling with 35S-sulfate at 20 μCi/ml as previously described23 and the results calculated as pmol 35S incorporated/μg DNA and expressed relative to control. Collagenase sensitive and total protein synthesis was measured as 24 h 3H-Proline incorporation as described,26 calculated as dpm/μg DNA, and expressed relative to untreated control.
Gene Expression
Gene expression was analyzed by real-time RT-PCR using Bio-Rad iCycler IQ4 detection system. The reactions were done with the validated primers (Table 1). Real-time RT-PCR reactions were done in duplicate in 96-well plates in a volume of 25 μl using the reagents and protocol per the Bio-Rad iScript One-Step RT-PCR Kit (Hercules, CA). The cycle threshold (Ct) values were obtained, and data normalized to GAPDH expression using the ΔΔCt method to calculate relative mRNA levels compared to untreated samples.
Table 1.
Primers Used in Real-Time RT-PCR Analysis of Gene Expression
| Gene | Forward (5′–3′) | Reverse (5′–3′) |
|---|---|---|
| Aggrecnn | AAGAATCAAGTGGAGCCGTGTGTC | TGAGACCTTGTCCTGATAGGCACT |
| Verslcan | GCGGAGACCAGTGTGAACTTGATT | ACATAACTTGGAAGGCAGAGGCAC |
| Col la 2 | GGAAACAGACAAGCAACCCAAACT | GGTCATGTTCGGTTGGTCAAAGATAA |
| Col 2a 1 | ATGACAATCTGGCTCCCAAC | GAACCTGCTATTGCCCTCTG |
| Col 8a 1 | CCTGGGTCAGCAAGIACCTC | TTGTTCCCCTCGTAAACTGG |
| Col 15 | GCCTTCCTTGACATTGCTGAAGA | CTCCGTTGGACATAGAGAGGGTT |
| MMP-1 | CCCAAAAGCGTGTGACAGTA | GAGCTCAACTTCCGGGTAGA |
| MMP-3 | CAAGGAGGCAGGCAAGACAGC | GCCACGCACAGCAACAGTAGG |
| TIMP-1 | TGGCTTCTGGCATCCTGTTGTTG | CGCTGGTATAAGGTGGTCTGGTTG |
| IGF-1 | AGCTGGTGGATGCTCTTCAGTT | GAAGCAGCACTCATCCACGAT |
| TGF-IS | AAGGGCTACCACGCCAACTT | CCGGGTTGTGCTGGTTGTAC |
Statistical Analysis
Data are given as mean ± standard error, and p-value of Student’s t-test were calculated, and a p-value of <0.05 defined statistically significant differences.
RESULTS
Microarray Analysis
Microarray analysis was done to assess the expression of the components of the arachidonic acid cascade (Table 2). Typically considered a constitutively expressed gene, COX-1 mRNA (67 ± 25) is low compared to inducible COX-2 (206 ± 37) in hNP cells under basal and TNFα-induced conditions. mRNA levels of prostaglandin E synthase genes, all three isoforms, are also high, especially PTGES3 which is induced by TNFα ~3-fold. Synthases for PGF2α and prostacyclin synthase (PTGIS) are also highly expressed. Receptors for PGE2, in particular PTGER2 and 4 are expressed at high basal levels and are upregulated ~2-fold by TNFα. Receptor for PGF2α is also expressed at a moderate level and upregulated by TNFα. Synthetases responsible for synthesis of PGE2, PGF2α, and their receptors were upregulated by TNF-α, suggesting that PGE2 and PGF2α are important PGs in disc metabolism. These results are consistent with previous findings of high levels of PGE2 in herniated lumbar discs13 and high PGF2α production by hNP cells14 as noted in the Materials and Methods Section. The high expression of synthases and receptors for PGE2 and PGF2α, and their documented secretion by hNP in response to both IL-1 and TNF-α directed our choice to evaluate hNP cell response to these two PGs in this study.
Table 2.
Gene Expression of Components of the Arachidonic Acid Cascade in Human NP Cells
| Gene Symbol | Gene Title | Known Function | Basal (−TNFα) | Induced (+TNFα) |
|---|---|---|---|---|
| Cyclooxygenases | ||||
| PTGS1 | Cyclooxygenase 1 (COX-1) | Constitutive COX. PTG biosynthesis | 67.2 ± 25.3 | 51.3 ± 24.4 |
| PTGS2 | Cyclooxygenase 2 (COX-2) | Inducible COX. PTG biosynthesis | 206.3 ± 37.4 | 401.3 ± 37.1 |
| Isomerases | ||||
| PTGES | Prostaglandin E synthase | Microsomal glutathione-dependent PGE synthase. P53-induced gene |
413.1 ± 189.5 | 1150.2 ± 454.3 |
| PTGES2 | Prostaglandin E synthase 2 | Membrane-associated PGE synthase Converts PGH2 to PGE2 |
264.3 ± 81.1 | 304.1 ± 116.1 |
| PTGES3 | Prostaglandin E synthase 3 | Constitutive cytosolic PGE synthase Chaperone for steroid receptor |
8542 ± 2302 | 9988.9 ± 3535.3 |
| PTGDS | Prostaglandin D2 synthase | Converts PGH2 to PGD2 | 177.7 ± 46.8 | 186.8 ± 47.7 |
| AKR1C3 (PTGFS) | Prostaglandin F synthase | Converts PGH2 to PGF2α | 730 ± 298 | 1001 ± 392.6 |
| PTGIS | Prostaglandin 12 synthase (prostacyclin synthase) | Converts PGH2 to PGI2 | 1320 ± 215.5 | 407.9 ± 16 |
| PTG receptors | ||||
| PTGER1 | Prostaglandin E receptor 1 | One of four receptor for PGE2 | 32.4 ± 10.2 | 26.8 ± 8.5 |
| PTGER2 | Prostaglandin E receptor 2 | One of four receptor for PGE2 | 889.6 ± 160.6 | 2278.5 ± 173 |
| PTGER3 | Prostaglandin E receptor 3 | One of four receptor for PGE2 | 25.2 ± 3.2 | 18.4 ± 4.5 |
| PTGER4 | Prostaglandin E receptor 4 | One of four receptor for PGE2 | 417 ± 236 | 752.9 ± 603.7 |
| PTGFR | Prostaglandin F receptor | Receptor for PGF2α | 266.9 ± 73.7 | 719.6 ± 118.9 |
| PTGIR | Prostaglandin 12 receptor | Receptor for prostacyclin (PGI2) | 58.3 ± 10.7 | 67.2 ± 17.1 |
Microarray analysis of gene expression of components of the arachidonic acid cascade. RNAs from hNP cells exposed to 0 and 5 ng/ml TNF-α for 24 h were hybridized to Affymetrix GeneChip Human Genome U133 Plus 2.0 Arrays mRNA expression levels of genes involved in the arachidonic acid pathway were analyzed. Basal = untreated control. Values are mean ± SE of n = 3. Genes expressed highly at basal level or inducible by TNF-α are highlighted in bold face.
Response of NP Cells to PGE2 and PGF2α
PGE2 but Not PGF2α Decreases Proteoglycan Synthesis
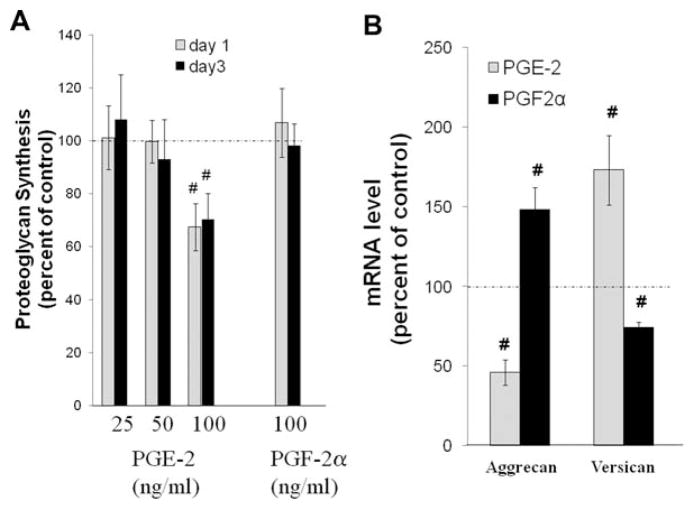
PGE2 at 100 ng/ml significantly decreased total proteoglycan synthesis by ~30% after 24 or 72 h exposure; in contrast, PGF2α did not affect total proteoglycan synthesis (Fig. 1A). No effects were seen at lower concentrations of PGF2α (data not shown). mRNA for two major aggregated proteoglycans, aggrecan core protein and versican, were also determined. As seen in Figure 1B, PGE2 decreased aggrecan mRNA to 46% while increasing versican mRNA to 173% of control. In contrast, PGF2α increased aggrecan message to 148% while decreasing versican mRNA to 75% of control.
Figure 1.
Effects of PGE2 and PGF2α on proteoglycan gene expression and synthesis. (A) Effects of PGE2 and PGF2α on total proteoglycan synthesis. hNP in alginate beads were treated with a range of PGE2 concentrations or 75 ng/ml PGF2α as described in the Materials and Methods Section and pulse labeled with 35S-Sulfate to determine relative rates of total proteoglycan synthesis. Values are mean ± SE of n = 4–8. #p < 0.05 versus Control. (B) Effects of PGE2 and PGF2α on mRNA expression of two major proteoglycans, aggrecan and versican. HNP in alginate beads were exposed to 100 ng/ml PGE2 or 75 ng/ml PGF2α for 72 h, RNA isolated, and rtPCR done to evaluate mRNA for aggrecan and versican as described in the Materials and Methods Section. Values are mean ± SE of n = 8–10. #p < 0.05 versus Control. Results demonstrated that PGE2 and PGF2α differentially increase and decrease mRNA for aggrecan and versican.
PGE2 but Not PGF2α Increase hNP Collagen Synthesis
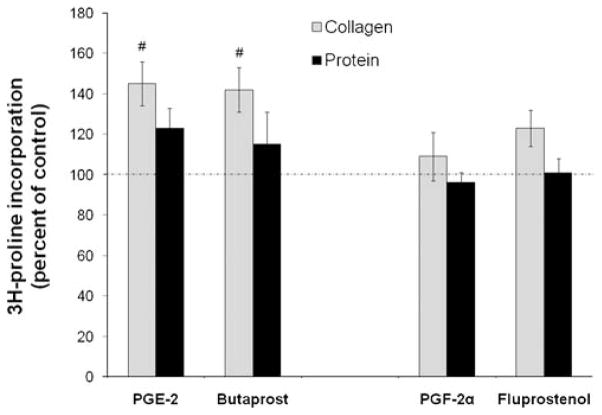
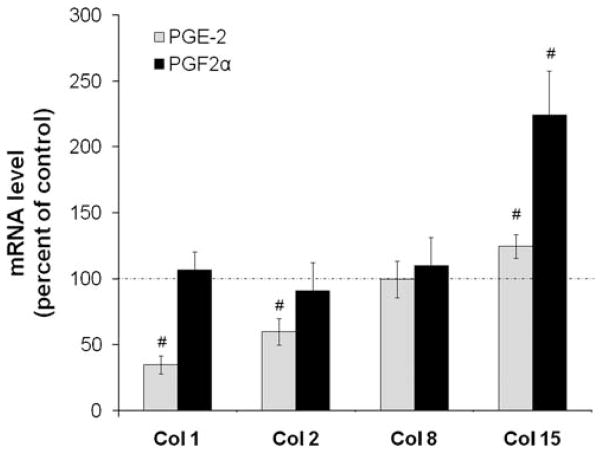
As seen in Figure 2, neither PGE2 nor PGF2α significantly changed 3H proline incorporation into total proteins. However, there were modest increases in collagen synthesis in the presence of PGE2 or 10 μM butaprost, a specific PTGER2 receptor agonist. Neither PGF2α nor 50 ng/ml fluprostenol, a metabolically stable analog of PGF2α with potent PTGFR receptor agonist activity, significantly affected collagen or total protein synthesis. Figure 3 shows that, in contrast with its effects on total collagen synthesis, PGE2 actually decreased message for both collagens type I and II by ~ 65% and 40%, respectively. However, it did modestly increase mRNA for collagen XV by 25%. 100 ng/ml PGE2 also exerted similar control on gene expression of these collagens after 24 h exposure (0.35 ± 0.013 for collagen I, 0.55 ± 0.05 for collagen II, 0.91 ± 0.32 for collagen VIII, and 1.85 ± 0.41 for collagen XV). The only significant effect of PGF2α on message for these four types of collagen was to increase mRNA for Collagen XV by twofold. Thus, PGE2 and PGF2α exert distinct actions on gene expression and synthesis of disc matrix collagen and proteoglycans.
Figure 2.
PGE2 and the PTGER2 receptor agonist butaprost increase hNP collagen, but not total protein synthesis. HNP were exposed to 100 ng/ml PGE2, 10 μM butaprost, 75 ng/ml PGF2α, or 50 ng/ml fluprostenol for 72 h with 3H proline added for the final 24 h of culture. Values are mean ± SE of n = 4–6. #p < 0.05 versus Control.
Figure 3.
PGE2 and PGF2α differentially increase and decrease mRNA for collagen I, II, VIII, and XV. HNP in alginate beads were exposed to 100 ng/ml PGE2 or 75 ng/ml PGF2α for 72 h, RNA isolated, and rtPCR done to evaluate expression of collagens as described in the Materials and Methods Section. Values are mean ± SE of n = 6–10. #p < 0.05 versus Control.
PGE2 and PGF2α Differentially Regulate Expression of Growth Factors and Catabolic Genes
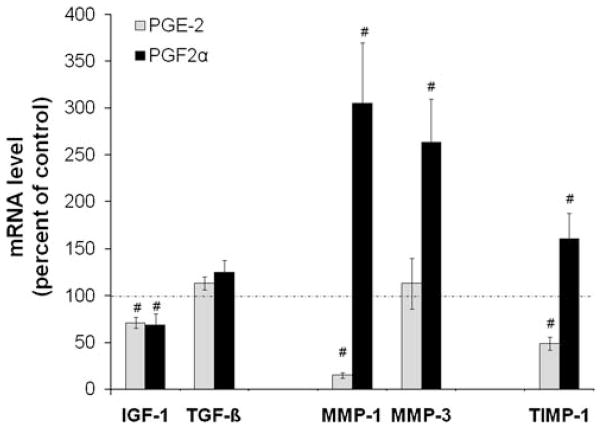
The effects of these PGs on the expression of growth factors known to modulate matrix protein synthesis and some of the TIMPs and MMPs involved in matrix protein degradation are shown in Figure 4. Neither PGE2 nor PGF2α affected mRNA for TGF-β but had similar effects on IGF-1 mRNA (decrease by ~30%). MMP-3 and MMP-1 mRNA were increased two- to threefold by PGF2α while PGE2 decreased mRNA for MMP-1 to ~15% of control values. PGE2 decreased (to 50% of control) while PGF2α increased (to 161% of control) mRNA of the anti-catabolic gene, TIMP-1.
Figure 4.
PGE2 and PGF2α variably increase and decrease mRNA for factors that modulate disc matrix synthesis and degradation. hNP in alginate beads were treated as described for Figure 2, RNA isolated, and rtPCR done as described in the Materials and Methods Section. Values are mean ± SE of n = 6–10. #p < 0.05 versus Control
DISCUSSION
This study evaluated the induction of constituents of the arachidonic acid pathway in response to inflammatory cytokines in order to identify key control points. The microsomal prostaglandin E synthase 1 (PTGES) is induced (~3-fold) in hNP by TNF-α, as it is in articular chondrocytes by this and other inflammatory cytokines27 as well as by mechanical stress.28 PTGES has been implicated in the development of arthritis and suggested as a therapeutic target in this joint disease. However, given the much higher, albeit stable amount of message of the constitutively expressed cytosolic PTGES3 in these disc cells, further studies are needed to determine the actual synthase protein and activity responsible for PGE2 synthesis in the disc and to conclude which synthase could potentially be targeted to diminish PG production in IDD.
PTGFS gene expression is actually greater than PTGES under basal conditions and is also increased (~2-fold) to a similar level as that of PTGES in TNF-α-activated hNP cells (Table 2). Thus, cytokine activation of PG production works through increasing both COX-2 and the downstream PG synthases. Aside from a study performed by Jacob et al.29 to test the ability of PGF2α to alter chondrocyte matrix protein production, this is the first report, to the best of our knowledge, of the expression of genes responsible for the production of PGF2α as well as response of NP cells to this PG.
PTGIS converts PGH2 into prostacyclin (PGI2), a potent vasodilator and inhibitor of platelet aggregation with roles in acute and chronic inflammation. mRNA of PTGIS is present in hNP, but in contrast with the synthases for PGE2 and PGF2α, it is actually decreased to 1/3 of basal levels in TNF-α-activated cells. The relevance of this change in hNP cell function remains to be tested, as very low levels of receptor mRNA for prostacyclin are identified. mRNA for receptors of both PGE2 and PGF2α are present and increase two- to threefold in cytokine-activated cells, thus, suggesting that PGE2 and PGF2α play prominent roles in discs.
This study investigated the action of specific PGs, that is, PGE2 and PGF2α, on disc cells that show potential to alter the matrix composition in ways that could affect both structure and function. PGE2 decreased aggrecan mRNA to 46% of control while decreasing proteoglycan synthesis to ~70% of control. PGF2α, while not affecting total proteoglycan synthesis, also changed the relative gene expression of aggrecan and versican, but in the opposite direction as compared to PGE2, that is, increased aggrecan while decreasing versican. Our results with disc cells differ from those of Jakob et al.29 who evaluated the actions of specific PGs after longer exposure (2 weeks) on adult articular chondrocyte pellets and found no effect of PGE2 but a twofold increase in GAG/DNA content in PGF2α-treated cells. This is consistent with evidence of other phenotypic differences between disc cells and articular chondrocytes. These data suggest that the predominant type of disc proteoglycans may very well be affected by PG exposure, but the consequences for disc matrix structure and function remain to be explored.
Neither PG affected total protein synthesis, however, PGE2 and the PTGER2 agonist butaprost increased collagen synthesis, suggesting this action of PGE2 may be through the PTGER2 receptor. PGE2 decreased message for both collagen I and II and only modestly increased that for collagen XV. Because collagen synthesis represents all collagen gene products, it is not surprising that increased or decreased collagen synthesis in response to PGE did not reflect the same changes in expression of a few collagen types such as type I and II. Indeed, regulation of collagen synthesis is enormously complex which is often involved the coordination of many collagens, each of which is under multiple steps of gene expression control in a temporal manner.30,31 Our findings suggest that either additional collagen types are being induced that were not identified in our RT-PCR analysis or additional posttranscriptional regulations of collagen I and II by PGE2 exist within the disc cells. The decrease in collagen I is consistent with that documented in chondrocytes19,20 treated with PGE2; the decrease in message for collagen II is not, as PGE2 is reported to have no effect29 or to actually increase20 collagen II mRNA. However, the actions of PGE2 on chondrocyte collagen metabolism can be dose dependent with low concentrations increasing and high concentrations decreasing synthesis.32
The lack of any significant effect of PGF2α or its nonmetabolizable analog fluprostenol on total protein and collagen synthesis was surprising, as it is frequently associated with anabolic processes in muscle cells.33,34 Furthermore, it is reported that while PGF2α had no effect on collagen I, it did enhance collagen II expression by ~20× in articular chondrocytes.29 Of the parameters evaluated in the current study of hNP, only message for collagen XV was significantly affected by PGF2α. Collagen XV, a nonfibrillar collagen, exhibits diverse conformations and supramolecular assemblies and has many sites of attachment for glycosaminoglycan chains. In addition to its structural role, this collagen has been implicated in migration, proliferation, apoptosis, or survival of different cell types.35,36 If the changes we observed in expression of collagens are translated into changes in the collagen composition of disc matrix proteins, PGs show potential to alter the matrix composition of the disc in ways that could affect both structure and function. Therefore, future studies should address the substrates of collagenases synthesized in response to PG exposure.
In addition to their influence on matrix gene expression and synthesis, these PGs also modulated the expression of factors that can affect disc cell matrix metabolism. Both PGE2 and PGF2α decreased message for IGF-1 by 29% and 32%, respectively. As IGF-1 mediates chondrocyte cell survival,37 rescues annulus cells of the disc from senescence,38 and increases disc matrix synthesis,39 one could postulate that chronic exposure to PGs might have a role in the loss of disc cell viability and function through their downregulation of this growth factor. Prostaglandin E1 was recently reported to induce the production of epidermal growth factor, a factor important in cell proliferation and cell survival, in cultured human annulus cells.40 Thus, PGs appear to have different influences on disc cell metabolism, including matrix metabolism as well as cell growth and survival.
PGF2α and PGE2 also modulate disc matrix catabolic balance through their influence on MMP and TIMP gene expression. Although PGE2 decreases TIMP-1, it decreases message for MMP-1 to an even greater extent (51% vs. 85% decrease from Control) while not affecting message for MMP-3. PGF2α, in contrast, increases both MMP-1 and MMP-3 (~3-fold) while increasing TIMP-1 only 60%. We found no effect of either PG on expression of TIMP-3, however, the effects, if any, on other MMPs and/or aggrecanase expression as well as activity would need to be evaluated before concluding which PG would facilitate the greatest net matrix degradation.
In conclusion, defining effects of specific PGs on disc cell metabolic activities could facilitate targeting specific synthases and/or receptors to protect the disc from degeneration. Based on the chosen endpoint measurements, we conclude that during clinical use of NSAIDs, PGE2 might have greater net effect than PGF2α on anabolic processes while PGF2α primarily increases catabolic activities. As both modulate expression of matrix proteins in a manner that could alter the ratios of the various proteoglycans and collagens in the matrix, the consequences for disc structure and function remain to be defined. Studies evaluating the ability of COX-2 inhibitors and/or NSAIDs to modulate hNP response to inflammatory cytokines would help to predict the changes in matrix homeostasis when these agents are used.
Acknowledgments
This work supported by VA Rehabilitation Research and Development Grant B4-3894RA and the Albert B. Ferguson, Jr., MD. Orthopaedic Fund of the Pittsburgh Foundation. The work was approved by the IRB of the University of Pittsburgh. The authors gratefully acknowledge the technical support of SuLan Huang, Elizabeth Christian, Geoffrey Sirockman, and the staff of the Ferguson Laboratory.
References
- 1.Freemont AJ. The cellular pathobiology of the degenerate intervertebral disc and discogenic back pain. Rheumatology. 2009;48(1):5–10. doi: 10.1093/rheumatology/ken396. [DOI] [PubMed] [Google Scholar]
- 2.Masuda K. Biological repair of the degenerated intervertebral disc by the injection of growth factors. Eur Spine J. 2008;17(Suppl 4):441–451. doi: 10.1007/s00586-008-0749-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.LeMaitre CL, Pockert A, Buttle DJ, et al. Matrix synthesis and degradation in human intervertebral disc degeneration. Biochem Soc Trans. 2007;35:652–655. doi: 10.1042/BST0350652. [DOI] [PubMed] [Google Scholar]
- 4.LeMaitre CL, Hoyland JA, Freemont AJ. Catabolic cytokine expression in degenerate and herniated human intervertebral discs: IL-1 beta and TNF alpha expression profile. Arthritis Res Ther. 2007;9:R77. doi: 10.1186/ar2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Weiler C, Nerlich AG, Bachmeier BE, et al. Expression and distribution of tumor necrosis factor alpha in human lumbar intervertebral discs: a study in surgical specimen and autopsy controls. Spine. 2004;30:44–54. doi: 10.1097/01.brs.0000149186.63457.20. [DOI] [PubMed] [Google Scholar]
- 6.Goupille P, Mulleman D, Chevalier X. Is Interleukin-1 a good taraget for therapeutic intervention in intervertebral disc degeneration: lessons from the osteoarthritic experience. Arthritis Res Ther. 2007;9:110. doi: 10.1186/ar2324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Seguin CA, Pilliar RM, Roughley PJ, et al. Tumor necrosis factor-alpha modulates matrix production and catabolism in nucleus pulposus tissue. Spine. 2005;30:1940–1948. doi: 10.1097/01.brs.0000176188.40263.f9. [DOI] [PubMed] [Google Scholar]
- 8.Kang JD, Stefanovic-Racic M, McIntyre LA, et al. Toward a biochemical understanding of human intervertebral disc degeneration and herniation. Spine. 1997;22:1065–1073. doi: 10.1097/00007632-199705150-00003. [DOI] [PubMed] [Google Scholar]
- 9.Miyamoto H, Saura R, Coita M, et al. The role of cyclooxygenase-2 in lumbar disc herniation. Spine. 2002;27:2477–2483. doi: 10.1097/00007632-200211150-00011. [DOI] [PubMed] [Google Scholar]
- 10.Jimbo K, Park JS, Yokosuka K, et al. Positive feedback loop of IL-1beta upregulating production of inflammatory mediators in human intervertebral disc cells in vitro. J Neurosurgery. 2005;2:589–595. doi: 10.3171/spi.2005.2.5.0589. [DOI] [PubMed] [Google Scholar]
- 11.Miyamoto H, Doita M, Nishida K, et al. Effects of cyclic mechanical stress on the production of inflammatory agents by nucleus pulposus and anulus fibrosus derived cells in vitro. Spine. 2006;31:4–9. doi: 10.1097/01.brs.0000192682.87267.2a. [DOI] [PubMed] [Google Scholar]
- 12.Wang Y-J, Shi A, Lu W, et al. Cervical intervertebral disc degeneration induced by unbalanced dynamic and static forces: a novel in vivo rat model. Spine. 2006;31:1532–1538. doi: 10.1097/01.brs.0000222019.84095.23. [DOI] [PubMed] [Google Scholar]
- 13.O’Donnell JL, O’Donnell AL. Prostaglandin E2 content in herniated lumbar disc disease. Spine. 1996;21:1653–1655. doi: 10.1097/00007632-199607150-00007. [DOI] [PubMed] [Google Scholar]
- 14.Studer R, Vo N, Sowa G, et al. COX-2 inhibition modulates human nucleus pulposus cell response to cytokines. Geneva, Switzerland: International Society for the Study of the lumbar spines; 2008. [Google Scholar]
- 15.Yoo JU, Papay RS, Malemud CJ. Suppression of proteoglycan synthesis in chondrocyte cultures derived from canine intervertebral disc. Spine. 1990;17:221–224. doi: 10.1097/00007632-199202000-00017. [DOI] [PubMed] [Google Scholar]
- 16.Karppinen J, Inkinen RI, Kaapa E, et al. Effects of tiaprofenic acid and indomethacin on proteoglycans in the degenerating porciine intervertebral disc. Spine. 1995;20:1170–1177. doi: 10.1097/00007632-199505150-00012. [DOI] [PubMed] [Google Scholar]
- 17.Iwabuchi S, Ito M, Chikanishi T, et al. Role of the tumor necrosis factor-α, cyclooxygenase-2, prostaglandin E2, and effect of low-intensity pulsed ultrasound in an in vitro herniated disc resorption model. J Orthop Res. 2008;26:1274–1278. doi: 10.1002/jor.20525. [DOI] [PubMed] [Google Scholar]
- 18.Amin AR, Dave M, Attur M, et al. COX-2, NO, and cartilage damage and repair. Curr Rheumatol Rep. 2000;2:447–453. doi: 10.1007/s11926-000-0019-5. [DOI] [PubMed] [Google Scholar]
- 19.Goldring MB, Berenbaum R. The regulation of chondrocyte function by proinflammatory mediators; prostaglandins and nitric oxide. Clin Orthop Relat Res. 2004;427S:S37–S46. doi: 10.1097/01.blo.0000144484.69656.e4. [DOI] [PubMed] [Google Scholar]
- 20.Goldring MB, Suen LF, Yamin R, et al. Regulation of collagen gene expression by prostaglandins and IL-1beta in cultured chondrocytes and fibroblasts. Am J Ther. 1996;3:9–16. doi: 10.1097/00045391-199601000-00003. [DOI] [PubMed] [Google Scholar]
- 21.Riquet FB, Lai W-FT, Birkhead JR, et al. Suppression of type I collagen gene expression by prostaglandins in fibroblasts is mediated at the transcriptional level. Mol Med. 2000;6:705–719. [PMC free article] [PubMed] [Google Scholar]
- 22.Thompson JP, Pearce RH, Schechter MT, et al. Preliminary evaluation of a scheme for grading the gross morphology of the human intervertebral disc. Spine. 1990;15:411–415. doi: 10.1097/00007632-199005000-00012. [DOI] [PubMed] [Google Scholar]
- 23.Studer RK, Aboka AM, Gilbertson LG, et al. p38 MAPK inhibition in nucleus pulposus cells: a potential target for treating intervertebral disc degeneration. Spine. 2007;32:2827–2833. doi: 10.1097/BRS.0b013e31815b757a. [DOI] [PubMed] [Google Scholar]
- 24.Irizarry RA, Hobbs B, Collin F, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 25.Smyth GK. Linear models and empirical Bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3(1):Article 3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 26.Gilberston L, Ahn S-H, Teng P-N, et al. The effects of rhBMP-2, rhBMP-12, and Ad-BMP-12 on matrix synthesis in human annulus fibrosus and nucleus pulposus cells. Spine J. 2008;8(3):449–456. doi: 10.1016/j.spinee.2006.11.006. [DOI] [PubMed] [Google Scholar]
- 27.Fahmi H. mPGES-1 as a novel target for arthritis. Curr Opin Rheumatol. 2004;16:623–627. doi: 10.1097/01.bor.0000129664.81052.8e. [DOI] [PubMed] [Google Scholar]
- 28.Gosset M, Berenbaum R, Levy A, et al. Prostaglandin E2 synthesis in cartilage explants under compression: mPGES-1 is a mechanosensitive gene. Arthritis Res Ther. 2006;8:R135. doi: 10.1186/ar2024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jakob M, Demarteau O, Suetterlin R, et al. Chondrogenesis of expanded adult human articular chondrocytes is enhanced by specific prostaglandins. Rheumatology. 2004;43:852–857. doi: 10.1093/rheumatology/keh197. [DOI] [PubMed] [Google Scholar]
- 30.Kohn DH, Sarmadi M, Helman JI, et al. Effects of pH on human bone marrow stromal cells in vitro: implications for tissue engineering of bone. J Biomed Mater Res. 2002;60(2):292–299. doi: 10.1002/jbm.10050. [DOI] [PubMed] [Google Scholar]
- 31.Wei-Dong D, Zhang YE, Zhai WR, et al. Dynamic changes of type I, III and IV collagen synthesis and distribution of collagen-producing cells in carbon tetrachloride-induced rat liver fibrosis. World J Gastroenterol. 1999;5(5):397–403. doi: 10.3748/wjg.v5.i5.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DiBattista JA, Dore S, Martel-Pelletier J, et al. Prostaglandin E2 stimulates incorporation of proline into collagenase digestible proteins in human articular chondrocytes; identification of an effector autocrine loop involving insulin-like growth factor I. Mol Cell Endocrinol. 1996;123:27–35. doi: 10.1016/0303-7207(96)03887-7. [DOI] [PubMed] [Google Scholar]
- 33.Xu W, Chou C-L, Sun H, et al. FP prostanoid receptor-mediated induction of the expression of early growth response factor-1 by activation of a ras/raf/mitogen-activated protein kinase signaling cascade. Mol Pharmacol. 2008;73:111–118. doi: 10.1124/mol.107.038778. [DOI] [PubMed] [Google Scholar]
- 34.Rice KM, Uddemarri S, Desai DH, et al. PGF2α—associated vascular smooth muscle hypertrophy is ROS dependent and involves the activation of mTOR, p70S6k, and PTEN. Prostaglandins Other Lipid Mediat. 2008;85:49–57. doi: 10.1016/j.prostaglandins.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyers JC, Amenta PS, Dion AS, et al. The molecular structure of human tissue type XV presents a unique conformation among the collagens. Biochem J. 2007;404:535–544. doi: 10.1042/BJ20070201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ortega N, Werb Z. New functional roles for non-collagenous domains of basement membrane collagens. J cell Sci. 2002;115:4201–4214. doi: 10.1242/jcs.00106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cavero JD, Carlson CS, Im HJ, et al. Increased expression of the Akt/PKB inhibitor TRB3 in osteoarthritic chondrocytes inhibits insulin-like growth factor 1-mediated cell survival and proteoglycan synthesis. Arthritis Rheum. 2009;60:492–500. doi: 10.1002/art.24225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gruber HE, Hoelscher GL, Ingram JA, et al. IGF-1 rescues human intervertebral annulus cells from in vitro stress-induced premature senescence. Growth Factors. 2008;26:220–225. doi: 10.1080/08977190802273814. [DOI] [PubMed] [Google Scholar]
- 39.Evans C. Potential biologic therapies for the intervertebral disc. J Bone Joint Surg. 2006;88:95–98. doi: 10.2106/JBJS.E.01328. [DOI] [PubMed] [Google Scholar]
- 40.Gruber HE, Hoelscher G, Loeffler B, et al. Prostaglandin E1 and misoprostol increase epidermal growth factor production in 3D-cultured human annulus cells. Spine J. 2009;9:760–766. doi: 10.1016/j.spinee.2009.04.024. [DOI] [PubMed] [Google Scholar]