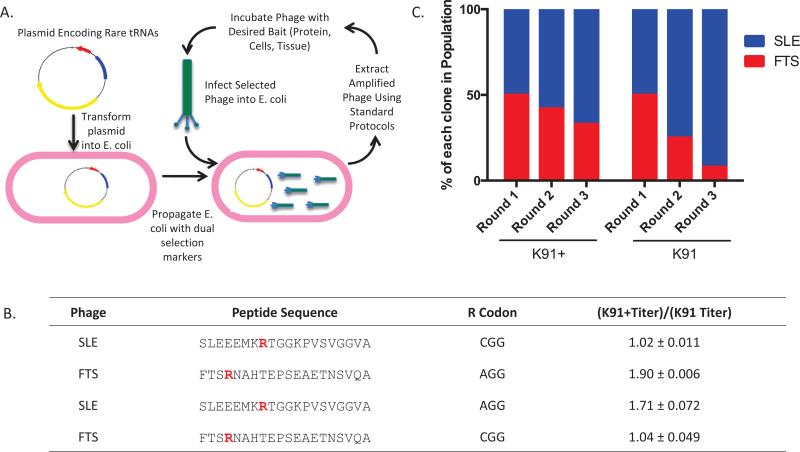
Figure 1. Transformation of K91 cells with the pRARE plasmid increases amplification of a phage clone containing a rare codon.
(A) Scheme outlining the method to reduce phage amplification bias. K91 E. coli cells are transformed with pRARE, which encodes for expression of 6 rare tRNAs and imparts chloramphenicol resistance. Once the cells are transformed, phage amplification occurs by standard protocols using chloramphenicol and tetracycline as section markers. (B) Amino acid sequences of 20-mer phage clones SLE and FTS. Arg residues (R) are highlighted in red to indicate the nucleotide sequence that encodes for the amino acid. The coding sequence for the Arg was switched for each phage clone using site-directed mutagenesis. Presented is quantification of fold changes in titer difference between phage clones amplified in K91+ compared with K91 cells [(K91+ titer)/(K91 titer)]. Titering was performed by inoculating 100-fold phage dilutions into E. coli cultures in log phase growth (0.4–0.6 OD600), incubating at 37°C without shaking, and then plating on YT-tet or YT-tet+cam plates followed by overnight incubation at 37°C. Colonies were counted and CFU/mL was calculated using a standard formula (15). Mean fold change ± SD is presented (n = 3, P < 0.05). (C) Percentage of FTS clones (Arg encoded by AGG) in iterative rounds of amplification in competition with SLE (Arg encoded by CGG) (n = 2). Briefly, 250 phage from each group were incubated with E. coli at 37°C and then plated on 150 cm YT-Tet or YT-Tet+Cam plates. The following day, 12 colonies were picked to monitor phage population, and then phage were extracted using a standard protocol (15). The same procedure was repeated with 500 phage particles from the previous round of amplification.