Abstract
Lrp5 is typically described as a Wnt signaling receptor, albeit a less effective Wnt signaling receptor than the better-studied sister isoform, Lrp6. Here we show that Lrp5 is only a minor player in the response to Wnt3a-type ligands in mammary epithelial cells; instead, Lrp5 is required for glucose uptake, and glucose uptake regulates the growth rate of mammary epithelial cells in culture. Thus, a loss of Lrp5 leads to profound growth suppression, whether growth is induced by serum or by specific growth factors, and this inhibition is not due to a loss of Wnt signaling. Depletion of Lrp5 decreases glucose uptake, lactate secretion, and oxygen consumption rates; inhibition of glucose consumption phenocopies the loss of Lrp5 function. Both Lrp5 knockdown and low external glucose induce mitochondrial stress, as revealed by the accumulation of reactive oxygen species (ROS) and the activation of the ROS-sensitive checkpoint, p38α. In contrast, loss of function of Lrp6 reduces Wnt responsiveness but has little impact on growth. This highlights the distinct functions of these two Lrp receptors and an important Wnt ligand-independent role of Lrp5 in glucose uptake in mammary epithelial cells.
INTRODUCTION
All somatic stem cells tested to date rely on Wnt signaling to maintain their pluripotentiality (1). From the point of view of regenerative medicine, this requirement has some disadvantages, since Wnt signaling can also be highly oncogenic (2). If the molecular regulation of Wnt signaling is better understood, it may be possible to tease apart the positive and negative aspects of the pathway.
Our study focuses on the signals generated at the cell surface by the Lrp5 (low-density lipoprotein receptor-related protein 5) and Lrp6 Wnt signaling receptors. Cell surface presentation of Lrp species is considered to be the limiting factor for Wnt signal generation (3). Mammary epithelial cells in vitro and in vivo grow and divide in response to ectopic Wnt signals. Thus, overexpression of Wnt1 or Wnt10B in mouse mammary glands leads to ductal hyperplasia, inducing cell division in both luminal and basal cells that together comprise the mammary ducts (4). Basal stem cells accumulate as a fraction of the total population (5), and solitary adenocarcinomas arise with a median latency of 7 months, comprising both mammary epithelial cell lineages (6). Thus, as for intestinal cell populations (2), Wnt signaling is a robust growth signal for mammary epithelial cells and acts as an oncogenic stimulus with a relatively low efficiency.
We previously showed that Lrp5 is required to sustain the basal stem cell activity in mammary glands, and also for breast tumor development in response to Wnt1 (5, 7, 8). This was a surprising result, since Lrp5 and Lrp6 are coexpressed by basal mammary epithelial cells and because Lrp6 is known to be a more effective transducer of Wnt ligand activation (9, 10). Lrp5 and Lrp6 share 73% and 64% protein sequence identity in their extracellular and intracellular domains, respectively (11).
Almost all the information we know about the canonical Wnt cell surface signaling complex is based on the analysis of Lrp6, which forms a ternary complex with Frizzled (Fzd) receptors and a Wnt ligand (for experimental purposes, this is usually the soluble Wnt ligand, Wnt3A) (2).
There are clues that the primary function of Lrp5 might be different from that of Lrp6 in vivo. For example, reduced Lrp5 activity has been implicated in metabolic changes in mice and humans, though the molecular explanation for these changes has typically been associated with a canonical Wnt signaling function. For example, Lrp5 was cloned as a candidate gene from the IDDM4 locus, which has been linked to genetic susceptibility to diabetes (12). lrp5−/− mice are leaner than wild-type mice and show higher expression levels of key enzymes required for fatty acid β-oxidation (13). So far, the only Lrp5 activity not linked to Wnt receptor activity is the binding of ApoE by Lrp5. This is a functional interaction leading to hypercholesterolemia, impaired fat tolerance, and atherosclerosis in lrp5−/− mice (14, 15). More specifically, humans with syndromes associated with altered bone density have been shown to have mutations in Lrp5. Gain- and loss-of-function Lrp5 alleles show alterations of osteoblast differentiation that correlate with altered glucose uptake (16–18).
Our study was designed to address the hypothesis that Lrp5 and Lrp6 have nonredundant activities in mammary epithelial cells by knocking their expression down separately and testing their relative impacts on Wnt signaling and other aspects of cell regulation. We discovered that Lrp5 controls growth of mammary epithelial cells, but not because of its role in Wnt signaling. We show instead that Lrp5 is important for normal glucose uptake and that decreased glucose uptake accounts for the phenotype of cells lacking Lrp5 function.
MATERIALS AND METHODS
Ethics statement.
All experiments that included the use of mice were approved by the University of Wisconsin IACUC (protocol number MO1422; institutional assurance number for University of Wisconsin—Madison, A-3368-01). For the studies we describe, there are no suitable alternative approaches, and care is taken to minimize animal distress.
Flow cytometric purification of EP substrains from the HC11 parental strain and assay of their tumorigenicity.
Trypsinized HC11 cells were stained according to the method of Kim et al. (6). Briefly, cells were incubated with fluorescein isothiocyanate (FITC)-conjugated CD49f (clone number GoH3; 30 μl/ml) (BD Biosciences) and phycoerythrin (PE)-conjugated EpCAM (clone number G8.8; 0.5 μg/ml) and then sorted using a FACSVantage cell sorter equipped with DiVa software (Becton Dickson, Franklin Lakes, NJ).
To assess relative tumorigenicity, 4 × 105 EpCAM-positive (EP) or EpCAM-negative (EN) cells were suspended in 2 μl medium containing Matrigel (2 to 5 μg/μl) and transferred to cleared mammary fat pads as described previously (7, 19, 20). Mice were assayed by palpation, and 16 weeks after transplantation, fat pads were dissected, processed, and stained with carmine red.
Cells purified by flow cytometry were stained according to the method of Badders et al. (7) and Kim et al. (6). Briefly, single-cell suspensions were dried briefly on microscope slides and fixed in ice-cold methanol and acetone for 4 and 2 min, respectively. Cell preparations were blocked in 10% nonimmune goat serum and stained using antibodies to K5 and K8 (rabbit and rat primary antibodies, respectively) overnight, followed by a rinse and incubation with fluorescently conjugated secondary antibodies (Alexa Fluor 546–anti-rat and Alexa Fluor 488–anti-rabbit antibodies). DAPI (4′,6-diamidino-2-phenylindole) was used for nuclear DNA counterstaining, and immunofluorescence stains were visualized on a confocal microscope (MRC1024; Bio-Rad).
Cell strains, plasmid constructs, transfection, and transduction.
Mouse mammary epithelial EP cells, derived as described above and described further in Results, were maintained as described for parental HC11 cells (21). Thus, RPMI 1640 culture medium was supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals), 5 μg/ml insulin (Sigma-Aldrich), and 10 ng/ml recombinant human epidermal growth factor (EGF) (R&D Systems). Expression plasmids were obtained as follows: a plasmid expressing constitutively active human β-catenin (with an N-terminal truncation of 47 amino acids; CA-βcat) was purchased from Addgene, a plasmid expressing human Lrp5 was kindly gifted by Brian MacDonald (9), a plasmid expressing mouse Wnt3a was kindly gifted by Bart Williams (Van Andel Research Institute, Grand Rapids, MI), and a plasmid expressing Wnt1 was described in our previous studies (6, 21). Cells were transfected with plasmids or transduced with lentiviral particles expressing short hairpin RNAs (shRNAs) for Lrp5 or Lrp6 as described previously (21). A control plko vector encoding a scrambled shRNA was purchased from Addgene. The Rictor shRNA constructs were provided by the Broad Institute (a kind gift of Dudley Lamming, Department of Medicine) and included the following shRNA sequences (also used for the work in reference 18): CGGTTCATACAAGAGTTATTT and CGAGACTTTGTCTGTCTAATT. Transduced cells were selected for and maintained by addition of 6 μg/ml puromycin to the culture medium. We designed the following additional shRNA sequences targeting Lrp5 mRNA for confirmation of our results: Lrp5 shRNA#2, TCAGATACCAGGATCTTTCGG; and Lrp5 shRNA#3, TCATTGATCTCAGTGTTCACA.
The MDA-MB-468 (basal type), MDA-MB231 and Hs578t (both claudinlow), and SKBr3 and MCF7 (both luminal) human breast cancer cell strains (22) and the mouse breast tumor cell line 4T1 (highly malignant) were obtained from the ATCC and were maintained per ATCC instructions. 4T07 cells were a kind gift from Brett Morris and Patti Keely (University of Wisconsin) and were maintained according to the method of Aslakson and Miller (23). Lrp5 was knocked down in MDA-MB-231 cells maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 5% FBS. Briefly, cells were transduced with a lentivirus containing pGipz shRNA for Lrp5 (clone V3LHS_347020) or a pGipz nonsilencing control (clone RHS4346), purchased from Open Biosystems, as described by Esen et al. (18). Transduced cells were selected for and maintained in medium containing 1.6 μg/ml puromycin.
Quantitative real-time PCR analysis.
RNA isolation, cDNA generation, and amplification by real-time PCR were performed as described previously (7, 21). The PCR primers are listed in Table 1.
TABLE 1.
Primers used in this study
| Target name | Primer sequence (5′–3′) |
|
|---|---|---|
| Forward | Reverse | |
| GLUT1 (slc2a1) | GGCATGTGCTTCCAGTATGT | CCTTGGTCTCAGGGACTTTGAAG |
| GLUT2 (slc2a2) | ACTGGGTCTGCAATTTTGTCA | GAATGTAAACAGGGTGAAGACCAG |
| GLUT4 (slc2a4) | GGTTTCCAGTATGTTGCGGAT | CCTCTGGTTTCAGGCACTTTTAG |
| GLUT6 (slc2a6) | GGCTCCTATCTGTGCTGATTGC | CCTTGGCACAAACTGGACGTA |
| GLUT8 (slc2a8) | CCTTCGTGACTGGCTTTGCTGT | TGGGTAGGCGATTTCCGAGAT |
| GLUT9 (slc2a9) | GCCATCATTGCCTCGTTCTGCA | TACGGCGAAGTTTGAGAGCCAG |
| RICTOR | CAGTGTGAGGTCCTTTCCATCC | GCCATAGATGCTTGCGACTGTG |
| HK1 | GCCTAGACCACCTGAATGTAAC | GGAAGGACACGGTACACTTTG |
| HK2 | CCTCAAGACAAGGGGAATCTTC | CTTCACATTGATGCTGTCGTCA |
| IRS1 | ATGCCAAACCTCCTGTTGAG | CTTCTGGGCCATAGTAGCATTC |
| IRS2 | CTGCCAGCACCTATGCAA | TTTTCAACATGGCGGCGAT |
| IRS4 | AATGGACTTTGCCAGACGAG | GCAGATCTGGAGTAGACAAAGATG |
| LDHA | TCGATCCCATTTCCACCATG | TCTTCCTCAGGAGTCAGTGTC |
| LDHB | CCGTGTCTACCATGGTGAA | TCATCGTCCTTCAGCTTCTGA |
| PDK1 | TTACGGATTGCCCATATCACG | GATTCTGTCGACAGAGCCTTAATG |
| PDK2 | TCATCTATCTGAAGGCCCTGT | CGATACGTCGATGTGTTCTTGG |
| PDK3 | AGTGAACCAAGGGATGCATC | GCTCTCTGGTTGACTTGCAG |
| PKM1 | GCAGCAGCTTGGATAGTTCTCA | GAAGATGCCACGGTACAGATG |
| PKM2 | ATTATCGTGCTCACCAAGTCTG | GAAGATGCCACGGTACAGATG |
Western blotting of cell lysates.
Primary mammary epithelial cells were isolated from chopped mammary glands as previously described and then directly lysed for protein analysis (7). For analysis of cultured cells, cells were solubilized in either standard lysis buffer (25 mM HEPES, pH 7.4, 300 mM NaCl, 1.5 mM MgCl2, 1 mM EGTA, 50 mM glycerophosphate, 0.5% Triton X-100) or radioimmunoprecipitation assay (RIPA) lysis buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM Na2EDTA, 1 mM EGTA, 1% NP-40, 1% sodium deoxycholate, 2.5 mM sodium pyrophosphate, 1 mM glycerophosphate) with freshly added protease and phosphatase inhibitors (Thermo Fisher Scientific). Western blotting of SDS-PAGE gels was performed as described previously, using the following primary antibodies from Cell Signaling Technology: anti-Lrp5 (5731), anti-Lrp6 (3395), anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) (2118), anti-p-Lrp6 (2568), anti-p-AKT (T308) (2965), anti-p-AKT (S473) (4058), anti-AKT (4685), anti-p-p70 S6 kinase (T389) (9205), anti-p70 S6 kinase (2708), anti-p-4E-BP1 (T37/46) (2855), anti-4E-BP1 (9452), anti-p-S6 ribosomal protein (S235/236) (2211), anti-S6 ribosomal protein (2217), anti-p-SAPK/JNK (T183/Y185) (4668), anti-SAPK/JNK (9252), anti-p-p38 (T180/Y182) (9215), anti-p38 (9212), anti-p-ERK1/2 (T202/Y204) (4377), anti-ERK1/2 (4695), anti-p-LKB1 (S428) (3482), anti-LKB1 (3047), anti-p-AMPKα (T172) (2535), and anti-AMPKα (2603). Anti-β-actin antibody was purchased from Sigma (A5441), and antivinculin antibody was purchased from Millipore (95-386).
Small-molecule inhibitors and growth factors.
EP cells were treated for 48 h with the following small-molecule inhibitors at the indicated doses: 500 μM phenformin (Sigma), 10 nM rotenone (Sigma), 10 μM UO126 (Promega), 10 nM LGK974 (Xcessbio), rapamycin (concentrations as indicated in the figures) (Calbiochem), and 2-deoxy-d-glucose (2-DG) (Sigma). The following growth factors were used at the concentrations indicated for proliferation and glucose uptake assays: 50 ng/ml recombinant mouse Wnt3a (R&D Systems), 2 μg/ml insulin (Sigma), 50 ng/ml recombinant EGF (R&D Systems), and 50 ng/ml recombinant fibroblast growth factor (FGF) (R&D Systems). Growth factors or inhibitors were added to a basal medium comprising 2% FBS, PenStrep antibiotic, and 6 μg/ml puromycin diluted in RPMI 1640 medium.
Assay of cell number.
For assessment of relative cell numbers, cells were plated at a density of 7 × 104 cells/ml in 24-well plates. The next day, cells were changed to test condition medium, and 48 h later, cells were rinsed with phosphate-buffered saline (PBS), fixed with ice-cold methanol for 20 min, and stained with 1% crystal violet (CV) solution (Sigma) for 30 min. Plates were washed three times with deionized water and dried overnight before measurement of the optical density at 595 nm (OD595) (calculated using ImageJ software). This assay was compared with a direct assay of cell numbers by trypsinization and counting (data not shown) to confirm that it could be applied as a surrogate for cell number. Relative differences in cell growth were quantified as percent differences in cell number, i.e., (OD595 for test − OD595 for control)/OD595 for control; the control condition for each assay is indicated. For assays of proliferation in response to glucose deprivation, cell strains were preincubated for 48 h in culture medium made with RPMI 1640 lacking glucose. The fetal bovine serum batch used for these experiments (Atlanta Biologicals) contained 110 mg/dl glucose; therefore, the cultures under “low-glucose” conditions contained 22 mg glucose/liter of medium.
Dual-luciferase assay.
Canonical Wnt signaling activity was measured using the TOPFLASH reporter assay as described by Goel et al. (21), with the following modification: 5 × 104 EP cells in a well of a 24-well plate were transduced with the Super TOPFLASH reporter plasmid (0.25 μg) together with renilla luciferase (0.01 μg).
Glucose uptake and lactate efflux assays.
Cells were seeded in 24-well plates at a density of 7.5 × 104 cells/ml in complete culture medium. The next day, the medium was replaced with phenol red-free RPMI medium supplemented with 2% FBS and antibiotics. The next day, medium containing test compounds was added, and cells were incubated for an additional 24 h. The medium was harvested from cells, and cells were washed, fixed, and stained with crystal violet; glucose and lactate concentrations were determined using colorimetric kits from Eton Biosciences according to the manufacturer's instructions. The percent difference in glucose or lactate uptake/efflux for Lrp5 knockdown cells (Lrp5KD cells) or Lrp6KD cells was calculated relative to the value for control (plko) cells, and all values were normalized to the crystal violet staining intensity (CV/OD595). Briefly, medium samples were removed before and after incubation with a known number of cells (measured by CV/OD595), glucose concentrations were assayed, and the amount of glucose consumed/cell was calculated (Δglucose/CV).
ATP assay.
Cells were plated in 6-cm2 dishes at 7.5 × 104 cells/ml in triplicate and incubated in RPMI medium. Cells were trypsinized and counted, and 1 × 106 cells per replicate were assayed for the amount of ATP per cell according to the manufacturer's instructions (BioVision).
Analysis of OCR and ECAR.
Cells were seeded at a density of 10,000 cells/well in RPMI medium in a 96-well plate. A total of five replicate wells per cell strain were plated. The next day, cells were washed 3 times with test medium, consisting of unbuffered, phenol red-free RPMI medium (Sigma), 2% FBS, 1 mM glutamine, and antibiotics. The oxygen consumption rate (OCR) and the extracellular acidification rate (ECAR) were measured using a Seahorse XFe96 extracellular flux analyzer.
To normalize OCR and ECAR to cell number, standard curves of cells (0 to 20,000) of each strain were plated in parallel tissue culture plates and stained with crystal violet. OCR and ECAR values were calculated using Seahorse Wave software.
Analysis of intracellular reactive oxygen species (ROS) levels.
Cells were plated at 7.5 × 104 cells/ml in 10-cm dishes and incubated in RPMI medium as described previously for glucose and lactate assays. Cells were trypsinized, resuspended in PBS, and stained for 30 min with H2DCFDA in a tissue culture incubator (37°C, 5% CO2) (Life Technologies). Cells were washed once with PBS and resuspended in PBS plus 2% FBS containing 50 nM TOPRO3 (Life Technologies). For a positive control, cells were treated with hydrogen peroxide (200 μM) for 10 min prior to analysis by flow cytometry. Data were collected from 20,000 live cells (per replicate) by using a BD Biosciences LSR II flow cytometer, and the median staining intensity was calculated using FlowJo software.
Statistical analysis.
All experiments were performed with at least three replicates, repeated three times. Statistical significance was determined by performing two-tailed Student's t test, and significance is reported for P values of <0.05.
RESULTS
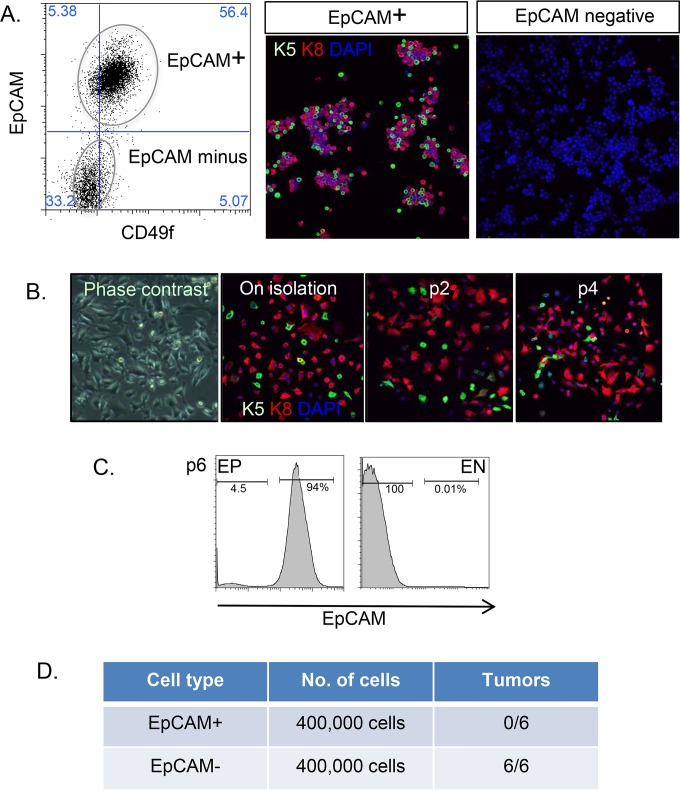
HC11 cells were derived from COMMA-D1 cells, the ancestor of most of the mouse epithelial cell lines in use today (24, 25). Originally described as a nontumorigenic cell line, HC11 cells respond to prodifferentiation protocols with milk secretion and morphological lumen formation. However, this cell line has drifted with passage to become tumorigenic. We rederived a nontumorigenic cell strain from an isolate of HC11 cells kindly provided by Nancy Hynes; thus, using flow cytometry, we separated epithelial cells (expressing the epithelial cell adhesion molecule EpCAM and the mammary epithelial cytokeratin keratin-8 or keratin-5 [K8 or K5]) (Fig. 1A) from nonepithelial cells, which showed a typical epithelial-mesenchymal transition (loss of keratin expression [Fig. 1A], acquisition of vimentin expression, and a transcriptional profile typical of the epithelial-mesenchymal transition [data not shown]). Vimentin-positive cells have been observed in COMMA1D cultures before and were cloned out at a limiting dilution (26). We confirmed that both alleles of p53 were mutant (C138W, Δ123–130), as described by Nancy Hynes and colleagues (27). Upon passage, the epithelial phenotype of these EpCAM-positive cells (EP cells) was stable (Fig. 1B and C). Furthermore, we assayed the tumorigenicity of EP and EpCAM-negative (EN) cells by transfer to mammary fat pads, and we found that this activity partitioned to the EN cell fraction. EP cells are therefore a nontumorigenic mouse epithelial cell line with predominantly luminal characteristics and a minor stable subpopulation expressing some basal epithelial cell markers.
FIG 1.
Rederivation of EP mammary epithelial cells. (A) HC11 cells were stained with antibodies to EpCAM and CD49f and sorted by flow cytometry into EpCAM-positive (EP) and EpCAM-negative (EN) fractions. Postsorting, cell aliquots were fixed and stained with epithelial markers, i.e., anti-keratin-5 (K5) and anti-keratin-8 (K8) antibodies (nuclei were counterstained with DAPI [blue]). Lineage specification is indicated as either basal/myoepithelial (K5 positive; green) or luminal (K8 positive; red). Note that EN cells were entirely negative for keratin expression. (B) To test the stability of the EP cell phenotype, cells were passaged, and samples at each passage were analyzed for keratin expression (as described for panel A). (C) After 6 passages, cells were reanalyzed by flow cytometry to show that EP cells maintained EpCAM expression and that EN cells were consistently EpCAM negative. (D) EP or EN cells (4 × 105) were inoculated into mammary fat pads, and tumor formation was measured 12 weeks later.
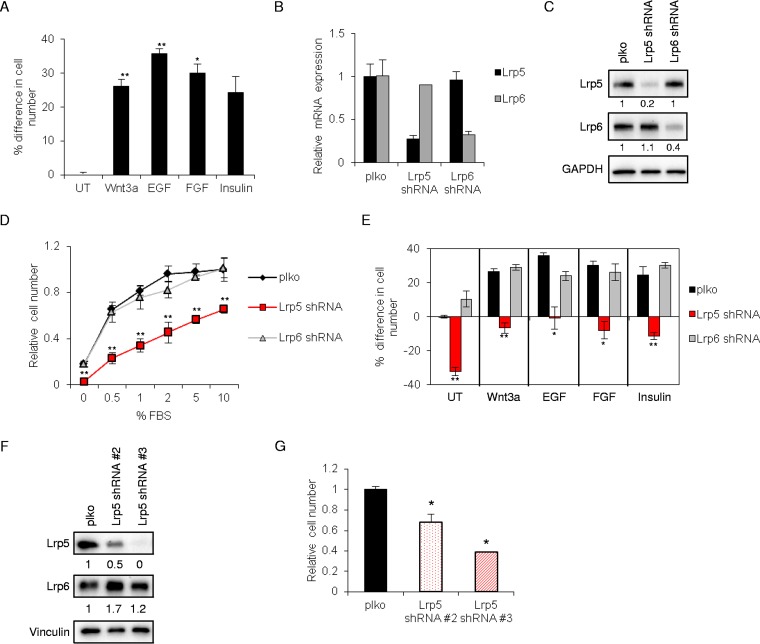
Mammary epithelial cells (EP cells) are predictably responsive to a number of specific growth factor ligands (EGF, FGF, and insulin) (Fig. 2A), including recombinant Wnt3a. To determine the role of Lrp5 in Wnt-induced growth and Wnt-induced transactivation, Lrp5 and Lrp6 were each knocked down by use of shRNA lentiviral expression vectors (Fig. 2B), resulting in 60 to 80% knockdown of each at the protein level (Fig. 2C). Given that these proteins oligomerize (28, 29), we tested the relative stability of each in the absence of the other, and we found no compensatory changes in the amount of Lrp6 in the absence of Lrp5, and vice versa.
FIG 2.
Lrp5 regulates mammary epithelial cell growth. (A) EP mammary epithelial cells were treated with Wnt3a, EGF, FGF, insulin, or no additions (the basal medium [UT] contained 2% fetal bovine serum) for 48 h, and cell number was assayed using crystal violet staining (CV/OD595). Results are reported as % differences in cell number relative to control, plko-transduced cells in basal medium. (B) EP mammary epithelial cells were stably transduced with lentiviruses encoding a scrambled shRNA (plko), Lrp5 shRNA, or Lrp6 shRNA and then selected with puromycin. Knockdown efficiency was assessed by quantitative reverse transcription-PCR (qRT-PCR) of Lrp5 and Lrp6 mRNAs (relative expression of each was normalized to expression in the plko control line). (C) EP cell strains were analyzed by Western blotting for Lrp5 and Lrp6 (the loading control was GAPDH). (D) Growth curves for EP cell strains in response to increasing serum concentrations (0 to 10% FBS) were assayed 48 h after plating (CV/OD595). Results are displayed as cell numbers relative to those of the control (plko cells in 10% FBS). (E) The growth of EP cell strains was determined after 48 h of treatment with specific ligands, namely, Wnt3a (50 ng/ml), insulin (2 μg/ml), EGF (50 ng/ml), and FGF (50 ng/ml), and expressed with respect to plko cells in basal medium (2% serum; untreated [UT]). Percent differences in cell numbers were normalized to control (plko) cells cultured in basal medium. (F) Two more shRNA constructs (shRNA#2 and shRNA#3) for Lrp5 were tested for % knockdown by Western blotting. (G) These Lrp5KD cell strains were assayed for cell number relative to that of control (plko) cells 48 h after plating. Statistical significance is shown for Lrp5KD versus control plko cells (**, P < 0.005; *, P < 0.05).
Surprisingly, Lrp5 knockdown cells showed a dramatically decreased growth rate at all serum levels screened. Cells were counted after 48 h of growth in 0.5 to 10% FBS (Fig. 2D). For cells growing in 1% serum, the number of Lrp5KD cells was reduced by 60%.
To evaluate whether Lrp5 was important for growth induced by specific types of growth factors, EP cells were incubated with serum (2% FBS in basal medium), rWnt3a, EGF, FGF, or insulin for 48 h (Fig. 2E). Surprisingly, although Lrp5KD cells responded to each growth factor, including rWnt3a, the overall cell number was much reduced compared to that of controls. Using two independent shRNA sequences targeting Lrp5, we confirmed the dramatic growth suppression (Fig. 2F), which correlated with the degree of Lrp5 knockdown (Fig. 2G). Interestingly, although Wnt ligands induced growth of EP mammary epithelial cells, canonical Wnt signaling via Lrp6 did not appear to be the driver (Fig. 2E), since there was no effect on growth when Lrp6 was reduced.
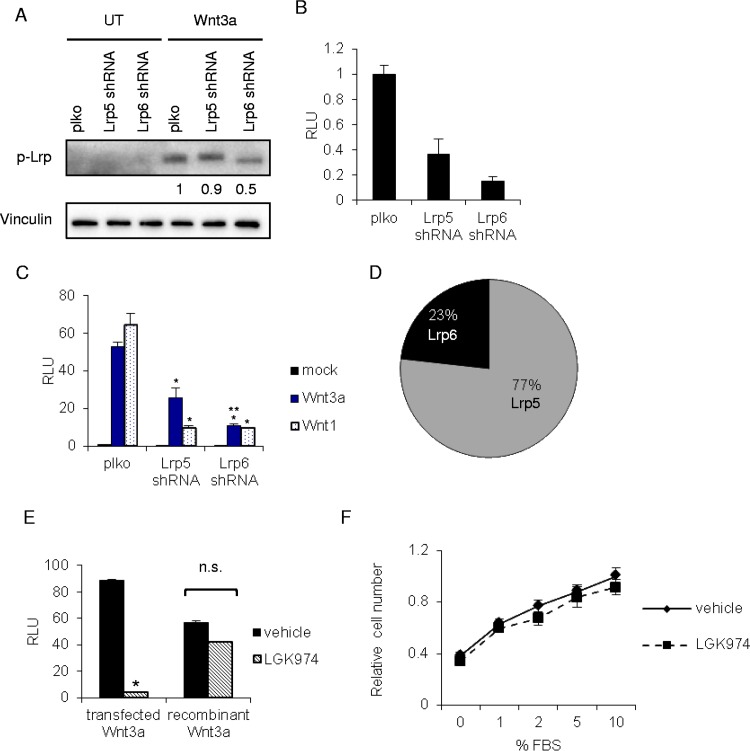
Based on the generality of the growth suppression we observed in Lrp5 knockdown cells, it appeared unlikely that Lrp5 regulated growth by a specific, Wnt-dependent mechanism. However, we considered the possibility that endogenous Wnt signaling could be responsible for maintaining growth of these cells and for promoting growth and survival under all these different growth conditions. Indeed, our analysis of mRNAs expressed by these cells showed significant expression of several Wnt signaling components (5), and at least one breast tumor cell line has been shown to be Wnt dependent for growth and motility, relying on autocrine Wnt signaling (30). To evaluate the effect of each Lrp species on basal and induced Wnt signaling, the relative activation of Lrp receptors (assayed using an anti-phosphoserine-1490 Lrp antibody) was evaluated for control cells (plko), Lrp6KD cells, and Lrp5KD cells and was found to be low/undetectable in the absence of exogenous ligand addition (Fig. 3A). Lrp6KD cells showed a 50% reduction in Lrp activation 60 min after treatment with rWnt3a, whereas Lrp5KD cells showed little change. We also tested the basal (Fig. 3B) and inducible (Fig. 3C) expression of a canonical Wnt reporter (TOPFLASH) in Lrp5KD and Lrp6KD cells, and we found that although knockdown of Lrp5 species reduced Wnt signaling by over 50%, knockdown of the minority Lrp6 protein had a much larger impact. We concluded that despite the fact that Lrp5 comprises the majority of Lrp species in these cells (Fig. 3D) (21), Lrp5 is indeed a much less potent transducer of Wnt signals (9).
FIG 3.
Lrp5 does not signal through the canonical Wnt pathway to regulate cell growth. (A) Relative activation of Lrp receptors in Lrp KD strains was determined after treatment of cells with Wnt3a (50 ng/ml) for 1 h, followed by Western blotting of phospho-Lrp. (B) Basal levels of TCF/β-catenin transactivation were measured in unstimulated EP cell strains by transfection of the Wnt β-catenin/TCF transactivation reporter Super TOPFLASH (and a renilla luciferase normalization control), followed by assay of relative light units (RLU) 48 h later (normalized to control [plko] cells). (C) To examine Wnt-induced canonical TCF/β-catenin transactivation, EP cell strains were transfected with a plasmid encoding Wnt3a, Wnt1, or green fluorescent protein (GFP) (mock), together with Super TOPFLASH/renilla luciferase, and the number of RLU was determined 48 h later (normalized to mock-transfected control [plko] cells). Statistically significant differences between RLU assays are shown for plko compared to Lrp5KD or Lrp6KD cells (*) and between Lrp5KD and Lrp6KD cells (**) (P < 0.05). (D) The relative amounts of Lrp5 and Lrp6 in EP cells were deduced by comparison of myc-tagged Lrp constructs (21). (E) To evaluate whether autocrine Wnt ligands are produced by EP cells, cells were transfected with Super TOPFLASH/renilla luciferase and either dually transfected with a Wnt3a expression plasmid or treated with recombinant Wnt3a (50 ng/ml), in the presence or absence of the PORCN inhibitor LGK974 (10 nM), followed by determination of the number of RLU. n.s., not significant. (F) EP cells were treated with LGK974 in the presence of various serum concentrations, and cell numbers were assayed 48 h later (CV/OD595). The value for control cells (plko cells in 10% serum) was set to 1.
Autocrine signaling depends on the production of myristoylated Wnt ligands, and this process is sensitive to inhibitors of specific acylases (31–34). Thus, LGK974 is a PORCN inhibitor that specifically inhibits Wnt ligand production. If autocrine Wnt ligands maintain growth of EP cells in vitro, growth will be inhibited by LGK974. To test the efficacy and specificity of LGK974 for inhibiting Wnt-dependent responses, we compared TOPFLASH reporter expression levels after transfection of Wnt3a or addition of recombinant Wnt3a protein (Fig. 3E). As anticipated, this inhibitor effectively reduced TOPFLASH expression by >90% in cells synthesizing their own Wnt ligands, whereas it had little effect on the response to recombinant Wnt3a. However, LGK974 had no effect on EP cell growth (Fig. 3F), leading us to conclude that Wnt signaling does not drive growth of these cells (at any of the tested serum concentrations). In sum, the loss of growth observed in Lrp5-deficient cells does not appear to be mediated by canonical Wnt signaling complexes.
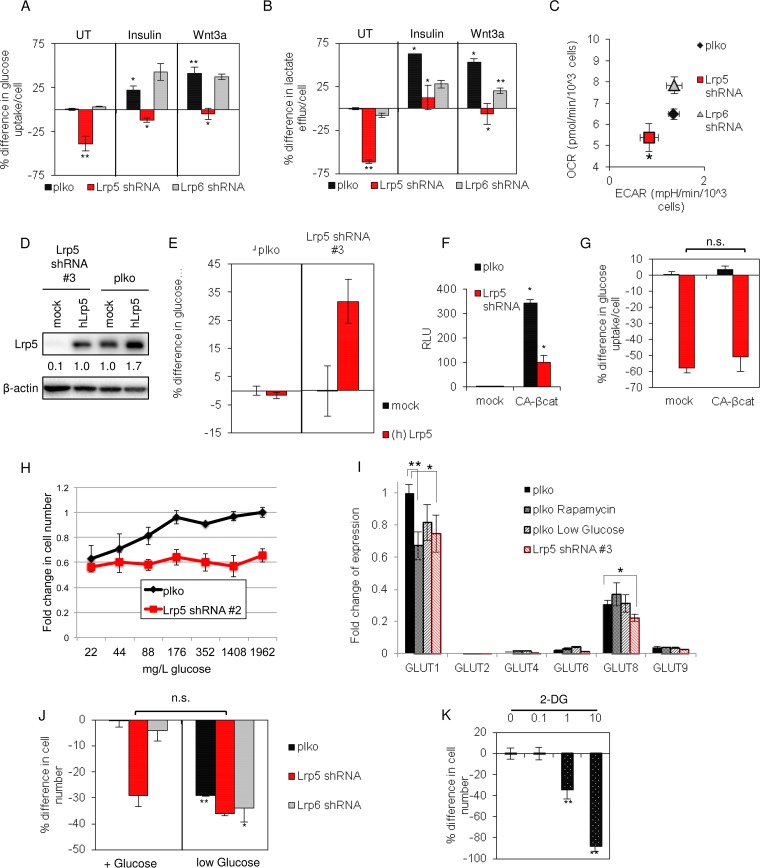
A recent study showed that Lrp5 affects glucose uptake in osteoblast cells (18). Therefore, we evaluated glucose uptake by EP mammary epithelial cells and found that it was reduced by at least 30% by knockdown of Lrp5 (Fig. 4A). Insulin is known to increase glucose uptake by various cell-specific mechanisms; for EP cells, insulin increased glucose uptake by 20%. Lrp5 KD also reduced glucose uptake induced by insulin and Wnt3a. Indeed, the impact of Lrp5 KD on growth directly mirrored the relative glucose uptake, whether it was basal or induced by specific growth factors.
FIG 4.
Lrp5 regulates glucose uptake in mammary epithelial cells. (A) Glucose uptake was measured for EP cell strains over a 48-h period. Results are expressed relative to those for control cells (plko) under basal conditions (UT). (B) Lactate accumulation was measured for media collected in parallel. (C) The oxygen consumption rate (OCR) and extracellular acidification rate (ECAR) for each EP cell strain were analyzed using a Seahorse Biosciences flux analyzer and then normalized (per cell). (D) Human Lrp5 (hLrp5) was overexpressed in Lrp5KD cells (Lrp5 shRNA#3 knockdown) and control (plko) cells, and the amounts of Lrp5 were analyzed by Western blotting. (E) The effect of overexpressed hLrp5 on glucose uptake was assayed over a 24-h period in plko cells (left half) and as a rescue of Lrp expression in Lrp5KD cells (shRNA#3) (right half). (F) To test the effect of a gain of function of canonical TCF/β-catenin transactivation on glucose uptake, EP cells were transfected with a plasmid encoding constitutively active β-catenin (CA-βcat), together with Super TOPFLASH/renilla luciferase. Cell lysates were analyzed 24 h later, and the number of RLU was calculated relative to that for control (plko) cells transfected with a mock plasmid. (G) The cells from panel F were analyzed for their glucose consumption (24 h of consumption). n.s., not significant. (H) Glucose dose-response curves for EP cells and EP Lrp5KD (shRNA#2) cells, grown for 72 h in RPMI medium-2% FBS with the indicated glucose concentrations. Cell numbers were assayed by CV staining. (I) Expression of mRNAs for various GLUT species (GLUT1, -2, -4, -6, -8, and -9; GLUT3 was not detected) was assayed by qPCR analysis of EP cells transduced with Lrp5 shRNA#2 or a control lentiviral vector (plko), either under control conditions or with low glucose (0.12 mM or 22 mg/liter) or 10 nM rapamycin for 24 h. All mRNA amounts are expressed with respect to GLUT1 expression by control EP cells. (J) EP cell strains were grown in medium with glucose (11.1 mM or 2 g/liter) or in low-glucose medium (the glucose concentration in 2% FBS for this serum batch was 0.12 mM or 22 mg/liter), and cell numbers were assayed 48 h later (CV/OD595). The percent difference in cell number was calculated relative to control (plko) cells cultured in the presence of glucose. (K) EP cells were grown with various concentrations of 2-deoxyglucose, an inhibitor of glucose uptake, and cell numbers were assayed and reported relative to that for cells treated with vehicle. Statistical significance is reported as follows: **, P < 0.005; *, P < 0.05.
To determine whether glucose disposition was affected in Lrp5KD cells, we measured lactate efflux under basal and induced culture conditions (Fig. 4B). Smaller glucose amounts taken up by Lrp5KD cells were reflected in smaller amounts of secreted lactate, suggesting that there is no dramatic redirection of glycolysis in these cells. If there is less glucose imported and oxidized in mitochondria, then oxygen consumption is predicted to decrease. Metabolic analysis of O2 consumption confirmed that cells were less metabolically active, consuming less oxygen (OCR), and also confirmed the decreased extracellular acidification rate (ECAR) characteristic of secreted lactate (Fig. 4C). We confirmed the specificity of the shRNA knockdown by rescuing glucose uptake in Lrp5KD cells with expression of the human Lrp5 protein (Fig. 4D and E). We found that overexpression of Lrp5 alone was not sufficient to increase glucose uptake for these cells, and we concluded that the amount of Lrp5 present in EP cells does not solely determine their rate of glucose uptake.
Although we showed that Wnt-induced transactivation is not likely to be implicated in this pathway, other studies have suggested that there are transcriptional targets of Wnt signaling that could affect glucose uptake rates, including IRS1 (35). Therefore, to test whether a gain of function for canonical Wnt signaling is sufficient to increase glucose uptake, constitutively active β-catenin (CA-βcat) was expressed in EP cells, effectively inducing the TOPFLASH reporter (Fig. 4F). (Interestingly, Wnt-dependent transactivation was significantly suppressed in Lrp5KD cells.) However, the expression of CA-βcat did not increase glucose uptake by mammary epithelial cells and did not rescue glucose uptake in Lrp5KD cells (Fig. 4G), ruling out a significant role for β-catenin/T-cell factor (TCF)-induced transcriptional responses.
The rate of glucose uptake by cells is determined by the number, activation status, and type of hexose (GLUT) transporters expressed on the cell surface (36). We incubated EP cells in media with various glucose concentrations and showed that the growth rate of EP cells decreased at glucose concentrations below 176 mg/liter (1 mM) (Fig. 4H). Thus, glucose availability limits growth of EP cells below this threshold concentration. (Note that the glucose concentration of regular RPMI 1640 cell culture medium is 2 g/liter, or 11.1 mM). Lrp5 knockdown cells did not grow in response to increasing glucose in the medium (Fig. 4H).
To investigate whether Lrp5 could be regulating the transcription of GLUT mRNA species, we measured their expression in Lrp5KD cells under two glucose restriction conditions: low (22 mg/liter)-glucose medium and rapamycin treatment (Fig. 4I). In common with the breast cancer cell lines reported by Anderson and colleagues (37), we found that mRNAs for GLUT1 and GLUT8 predominated in EP cells (Fig. 4I). All three glucose restriction conditions, i.e., rapamycin treatment, low-glucose medium, and Lrp5 KD, suppressed mRNA expression for GLUT1 (by approximately 25%), suggesting that this transcriptional response is a component of a compensatory reaction rather than indicating a specific role of Lrp5-mediated transactivation. We also assayed mRNAs for proteins implicated as important for glucose uptake or metabolism in general by other studies (HK1/2, LDHA/B, PKM1/M2, PDK1-3, and IRS1-4), and we found that these were not affected by Lrp5 knockdown (data not shown).
If low glucose uptake by Lrp5KD cells was sufficient to explain why cells grow more slowly, we predicted that the Lrp5 KD phenotype would be mimicked by reducing the glucose concentration in the growth medium. When EP cells were moved to low-glucose medium (22 mg/liter, or 0.12 mM; present in 2% serum), the growth of control cell strains was slowed to approximately the same degree as that of Lrp5KD cells (Fig. 4J). The addition of the nonmetabolized glucose variant 2-deoxyglucose (2-DG) also mimicked the slow-growth phenotype of Lrp5KD cells (Fig. 4K). Together, these data led us to conclude that these cells are highly dependent on Lrp5-regulated glucose uptake for their growth.
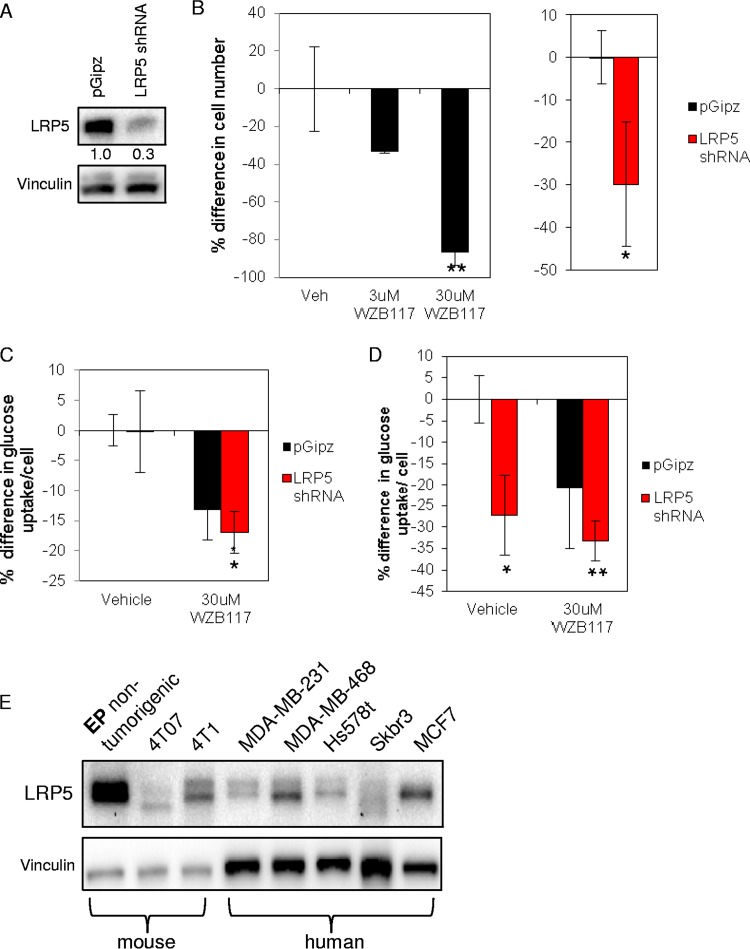
Typically, cancer cells consume more glucose than normal cells (22) (secreting partially oxidized glucose as lactate, per the “Warburg” effect). We therefore tested whether the breast cancer cell line MDA-MB-231 showed Lrp5-regulated glucose uptake (Fig. 5). We confirmed that these breast cancer cells showed profound growth inhibition in response to the selective GLUT1 inhibitor WZB117 (38) (Fig. 5B). Reducing the level of Lrp5 mRNA by 70% by use of shRNA lentiviral expression constructs reduced the cell number by 30%. Furthermore, MDA-MB-231 cells showed a 30% decrease in glucose uptake in response to the specific GLUT1 inhibitor WZB117 (Fig. 5C), and a similar decrease in glucose uptake was seen for Lrp5KD cells, but only when glutamine was omitted from the culture medium (Fig. 5D). MDA-MB-231 cells are highly glutamine dependent (22), so Lrp5-regulated glucose uptake may become growth limiting only when glutamine is absent. Lrp5 is expressed by all the breast cancer and nontumorigenic breast epithelial cells that we tested, and interestingly, there were significant differences in the number and size of bands identified on high-resolution Western blots (Fig. 1E), implying the regulation of Lrp5 by posttranslational modification. (Note that we could not detect an alternatively spliced Lrp5 variant described as characteristic of breast cancer cell lines [39; data not shown].)
FIG 5.
Lrp5 controls glucose uptake and growth of MDA-MB-231 human breast cancer cells. (A) MDA-MB-231 cells transduced with lentiviruses containing either a pGipz nonsilencing control or Lrp5 shRNA were analyzed by Western blotting to assess the efficiency of knockdown. (B) MDA-MB-231 cells were plated at 3.0 × 104 cells/ml in 24-well plates and treated with the GLUT1 inhibitor WZB117; changes in cell number were assessed 48 h after administering the inhibitor. Growth of Lrp5KD and pGipZ control cells was likewise assessed after 48 h (in low-glucose medium [22 mg/liter]). (C) Glucose uptake was assessed in vehicle (pGipZ-transduced)-treated and Lrp5KD MDA-MB-231 cells in full medium (with 2 g/liter glucose); cells were plated at 6.0 × 104 cells/ml. As a control, cells were treated with WZB117 for 24 h. (D) In parallel, glucose uptake was assessed for MDA-MD-231 cells cultured in RPMI medium without glutamine for 24 h. (E) Lrp5 expression in a panel of normal and breast cancer cell lines from mouse and human sources was analyzed by Western blotting. **, P < 0.005; *, P < 0.05.
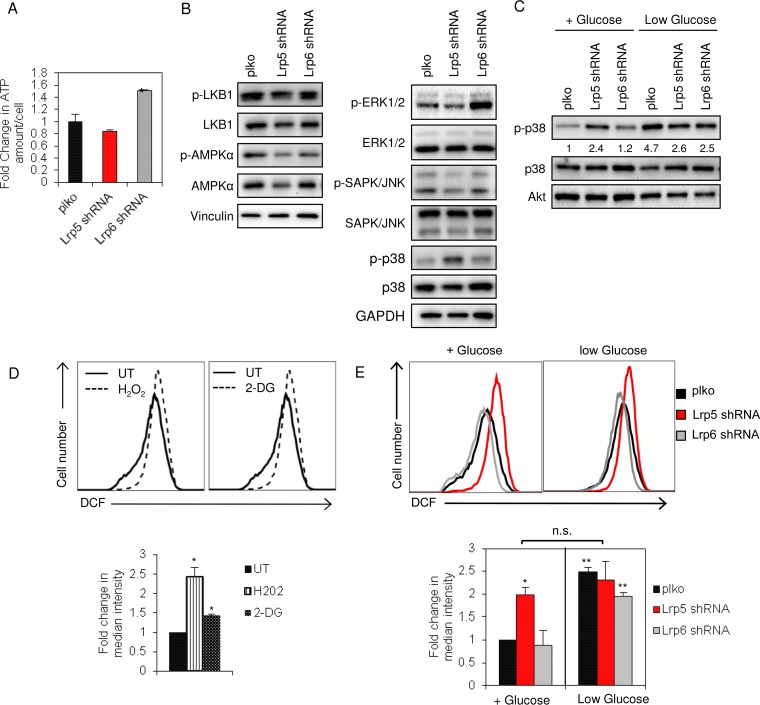
A well-characterized response to energetic stress is the activation of AMPK. Indeed, this molecule is considered to be one of the principal brakes on cell growth under conditions with limiting nutrients (40). We measured ATP concentrations of the cell strains under basal culture conditions and found that Lrp5KD cells were not statistically different from control cells (Fig. 6A). Not surprisingly, then, activation of AMPK was not increased in Lrp5KD cells (Fig. 6B). The activation of other signaling pathways known to determine growth rate (LKB and extracellular signal-regulated kinase [ERK] pathways) was also unchanged, including the c-Jun N-terminal kinase (JNK) pathway, known to be a noncanonical Wnt signaling output. However, there was a significant increase in activation of the stress checkpoint protein p38α/MAPK14 in Lrp5KD cells but not Lrp6KD cells. Activation of p38α was also observed in control cells cultured in low-glucose medium (Fig. 6C).
FIG 6.
Lower glucose uptake by EP cells increases ROS levels and p38α activation. (A) ATP levels were measured for EP cell strains and normalized to the cell number. Fold changes in the amount of ATP per cell were normalized to control (plko) cells cultured in basal medium. (B) Lysates of EP cell strains were analyzed by Western blotting for activation of several metabolic sensors and regulators, together with controls (vinculin and GAPDH). (C) EP cell strains were incubated with or without glucose (48 h), and cell lysates were analyzed by Western blotting for relative activation of p38α. (D) EP cells were treated with 2-DG (1 mM) (right panel) or with H2O2 (0.2 mM) as a positive control (left panel), and ROS levels were measured by staining cells with intracellular DCF dye followed by flow cytometry (quantified in the bottom panel). Fold changes in the median intensity of DCF staining were normalized to control cells cultured in basal medium. (E) ROS levels of EP cells were determined after 48 h of culture in the presence or absence of glucose. Statistical comparisons are shown with respect to control plko cells grown in glucose-containing medium. **, P < 0.005; *, P < 0.05; n.s., not significant.
Mitochondrial stress associated with dysregulation of the electron transport chain (ETC) is a source of reactive oxygen species (ROS), which in turn stimulate the activation of the stress checkpoint p38α (41, 42). Indeed, inhibition of glucose uptake is typically sufficient to stress the ETC and to increase the steady-state levels of ROS (42); we confirmed that 2-DG-treated EP cells showed an increased ROS level (Fig. 6D). Similarly, EP cells growing in low-glucose medium and Lrp5KD EP cells also showed elevated (2 to 3 times) ROS levels (Fig. 6E). Therefore, the loss of glucose uptake observed in Lrp5KD cells is sufficient to account for increased ROS levels, p38α activation, and cell growth inhibition.
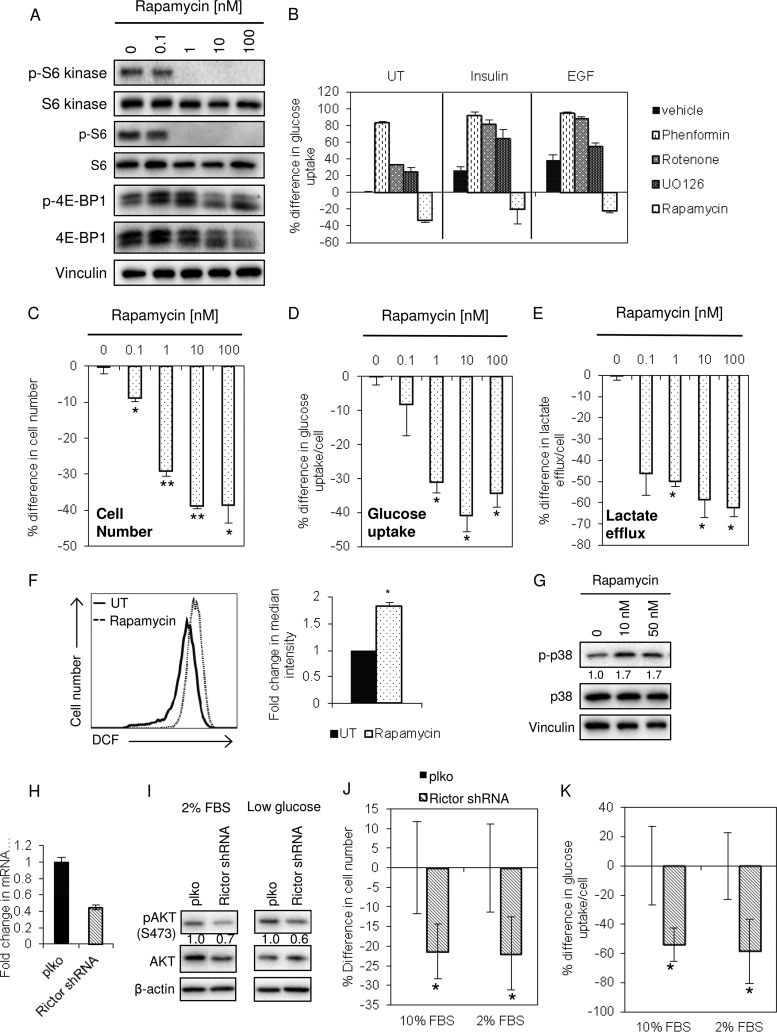
Given the importance of mTOR signaling as a regulator of cell growth and glucose uptake (43), we tested whether the mTORC1 inhibitor rapamycin would reduce glucose uptake in EP cells and phenocopy other aspects of Lrp5KD cells. Rapamycin abrogated the activation of mTOR signaling targets that affect translation, including S6 kinase, S6, and (less so) 4E-BP1 (Fig. 7A). Inhibiting mTOR signaling effectively inhibited glucose uptake (Fig. 7B). To provide a comparison, cells were also treated with inhibitors of various aspects of metabolism, including the biguanide drug phenformin (by analogy to metformin, the likely target is mitochondrial GPDH [44]), rotenone (a classic complex 1 inhibitor), and UO126 (an ERK inhibitor). Interestingly, and in contrast to rapamycin, these inhibitors tended to promote glucose uptake (Fig. 7B). Rapamycin reduced growth (Fig. 7C), glucose uptake (Fig. 7D), and lactate efflux (Fig. 7E), to degrees similar to those observed with the knockdown of Lrp5. Rapamycin also replicated other aspects of the Lrp5KD phenotype, inducing ROS and p38α activation (Fig. 7F and G).
FIG 7.
Both rapamycin and RICTOR/mTORC2 knockdown phenocopy Lrp5 knockdown. (A) EP cells were treated for 48 h with rapamycin at the indicated doses, and lysates were analyzed by Western blotting for the mTORC1 target proteins S6, S6 kinase, and 4E-BP1 (vinculin was used as a loading control). (B) EP cells were treated with the indicated metabolic inhibitors, and glucose uptake was assayed after 48 h. (C to E) The effects of 48 h of rapamycin treatment on cell number, glucose uptake, and lactate efflux were assayed as described in the legend to Fig. 4. (F) ROS levels were assayed by DCF staining and flow cytometric analysis. All results were normalized to EP cells treated with vehicle. (G) Relative activation of p38α was measured by Western blotting. (H) EP cells were transduced with shRNA species for RICTOR, a component of the mTORC2 complex. (I) Lysates of EP cells (control and RICTOR shRNA-treated cells), cultured in 2% serum or low-glucose (22 mg/liter) medium for 24 h, were analyzed by Western blotting for their relative levels of pAkt activation (at S473), together with total Akt and a β-actin loading control. (J) RICTOR KD cells were grown in 2 or 10% serum (as indicated) for 48 h, and cell numbers were assayed. (K) Glucose uptake over 24 h was assessed for the cells from panel J. **, P < 0.005; *, P < 0.05.
mTORC2 is frequently implicated as a key component in glucose sensing and uptake, as opposed to mTORC1, which detects and reacts to amino acids (45). Since rapamycin inhibits both mTORC1 and mTORC2, depending on the time course of exposure, cell type, and drug dose, we asked whether a specific ablation of mTORC2 function could phenocopy Lrp5 knockdown in mammary epithelial cells. RICTOR is an mTORC2-specific subunit (as opposed to mTORC1) (46); approximately 50% knockdown significantly inhibited the phosphorylation of Akt S473, a known target of mTORC2. Like Lrp5 knockdown, RICTOR knockdown inhibited glucose uptake and EP cell growth (Fig. 7H to K).
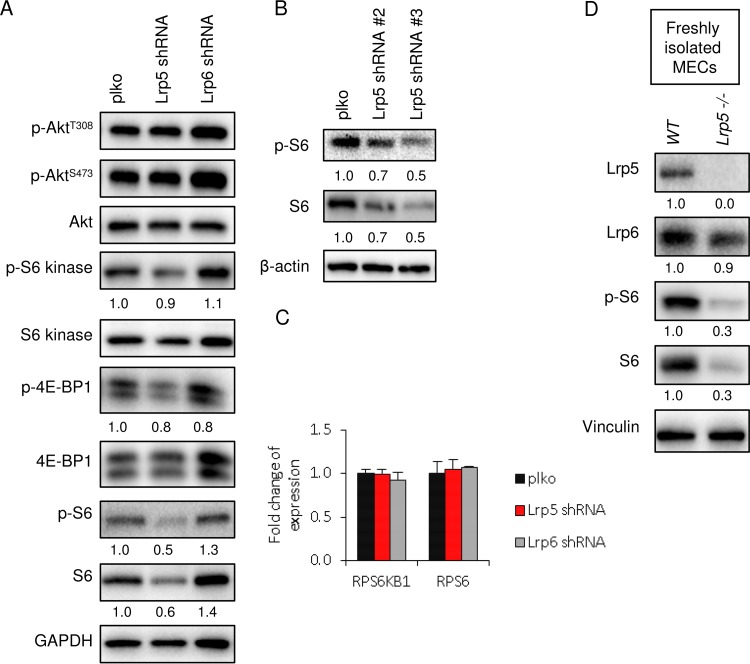
We tested whether other mTOR signaling outputs were inhibited in the absence of Lrp5, and we found reduced activation of S6, S6 kinase, and 4E-BP (Fig. 8A). S6 is a highly conserved mTOR target which is also a metabolic regulator in yeast (47). Interestingly, there was no impact of decreased Lrp5 on the activation of Akt (S473 or T308) or on other signaling hubs (such as ERK) (Fig. 6B). Thus, Lrp5KD cells show some phenotypes in common with cells with a loss of function of mTORC2 (reduced glucose uptake and growth), as well as others that are not shared, including reduced pS473-Akt. One phenotype of Lrp5KD cells that may be a unique signature is that the total extractable amount of the translation-associated protein S6 is much smaller (Fig. 8A and B), despite expression of S6 (and S6 kinase) mRNAs at wild-type levels (Fig. 8C). A much smaller amount of extractable S6 was also observed in primary mammary epithelial cells isolated from lrp5−/− mammary glands (Fig. 8D). Studies of S6 function have focused on a nonphosphorylatable knock-in form (rpS6P−/−); therefore, the predicted cause and effects of absent S6 are not well understood.
FIG 8.
Lrp5 knockdown cells show a specific S6 signature. (A) The relative activation of Akt, S6, S6 kinase, and 4E-BP1 was analyzed by Western blotting of lysates of EP cell strains. (B) A similar analysis was performed with EP cell strains knocked down by use of shRNA#2 and shRNA#3. (C) Assay of expression of S6 and S6 kinase mRNAs in Lrp5KD EP cells by qPCR. (D) Assay of lysates of primary mammary epithelial cells (MECs) isolated from lrp5−/− or control (C57BL/6) mammary glands by Western blotting.
DISCUSSION
This study has revealed that Lrp5 is a minor player in the response to Wnt3a-type ligands; instead, Lrp5 serves to regulate glucose uptake in mammary epithelial cells. This is key, since glucose uptake determines the rate of mammary epithelial cell growth in vitro. Decreased glucose uptake inhibits the growth of mammary epithelial cells in general, whether growth is induced by serum, by specific growth factors (including FGF and EGF), or by Wnt ligands. In contrast, loss of function of Lrp6, the dominant Wnt responder, leads to a predictable loss of Wnt reporter activity but little change in the growth rate. It has been shown that Lrp5 is not optimized for stimulating β-catenin/TCF signaling (9), as the carboxy-terminal domain is not easily activated. Our results suggest that Lrp5 structure and function may be optimized to perform a different function.
Since Lrp5 and Lrp6 proteins form heteromers (21, 28, 29), there is great potential for a coordinated response to Wnt ligands that includes an Lrp6-dependent β-catenin/TCF transcriptional response together with an Lrp5-dependent glucose metabolic response. Our previous studies have shown that canonical Wnt signaling, and therefore mammary stem cell activity, is constrained to cells expressing both Lrp5 and Lrp6 (21, 48, 49). Thus, all normal mammary stem cells express both Lrp species, and presumably both Lrp-associated activities. The heteromerization of Lrp5 and Lrp6 may explain why Wnt signaling outputs often include metabolic endpoints. For example, Inoki et al. (50) reported the indirect activation of mTOR signaling downstream of Wnt-signaling breast tumor cell lines. The growth of these cells was inhibited by rapamycin both in vitro and in vivo. For our study, loss of function of Lrp6 induced a gain of function for some signaling and metabolic parameters, suggesting that Lrp6 has suppressor activity. Thus, ERK and mTOR signaling increased (Fig. 4B and 6A), oxygen consumption and ATP amounts increased (Fig. 3C and 4A), lactate efflux declined (Fig. 3B), and cells became less stressed under low-glucose conditions (Fig. 4E). Thus, these two receptors may have interactive functionality.
Recent studies have begun to implicate Lrp5 as a metabolic regulator, both systemically and locally. However, many of these studies have focused on Wnt ligand-regulated changes in metabolism and canonical Wnt signaling outputs to explain their phenotypes. For example, in humans, rare mutations in Lrp5 control bone and cardio-metabolic disorders, such that gain-of-function mutations lead to high bone mass and enhanced lower body fat accumulation, whereas loss-of-function mutations give low bone mineral density and increased abdominal adiposity (51, 52). Other investigators have shown that Lrp5 knockout mice show normal glucose levels but impaired glucose tolerance, thus showing less insulin secretion in response to glucose administration (14).
Dissection of the molecular basis for Lrp5 activity in osteoblasts has shown that Lrp5 controls glucose uptake for the bone marrow stromal cell line ST2 during Wnt-induced osteoblastic differentiation (18). This study showed that loss of Lrp5 function decreased Wnt-induced differentiation for these cells but did not inhibit growth. In contrast, our study showed that the growth rate of mammary epithelial cells strictly depends upon Lrp5; indeed, cell viability is severely reduced when knockdown is more complete (J. A. Martin and C. M. Alexander, unpublished data). For mammary epithelial cells, Lrp5 is present in great excess compared to Lrp6 (3-fold), and there is no simultaneous, Wnt-induced differentiation process; thus, it can be demonstrated unambiguously that Lrp5 plays a role in glucose uptake. Indeed, we showed that Lrp5-dependent regulation of glucose uptake in mammary epithelial cells occurs regardless of the presence of Wnt ligands.
Although Lrp5-deficient mammary epithelial cells showed some decrease in Wnt transactivation responses (measured using the TOPFLASH reporter), the loss of glucose uptake was not related to canonical β-catenin/TCF signaling. This conclusion is supported by the following results: (i) gain of function for β-catenin/TCF signaling did not induce an increase of glucose uptake and (ii) loss of function for β-catenin/TCF signaling (Lrp6 KD or LGK974 treatment) did not decrease glucose uptake. Esen and colleagues also concluded that Wnt-mediated enhancement of glucose uptake was independent of canonical β-catenin/TCF signaling (using a glycogen synthase kinase [GSK] inhibitor, shRNA for β-catenin, and the tankyrase inhibitor XAV939) (18). They confirmed that β-catenin/TCF signaling induced transcription of IRS1, known to be important to insulin-induced glycogen synthesis in muscle cells (35), but showed that this was not important to glucose uptake in osteoblasts.
It is increasingly clear that glucose uptake is a highly regulated process that can determine growth and differentiation. Thus, glucose uptake increases in response to intracellular signals and extracellular cues, and a larger glucose supply feeds forward to determine signaling activation and establish homeostasis. Recently, a study of a panel of 46 breast cancer cell lines revealed their remarkable range of glucose uptake rates and glucose dependences (22). Glucose uptake has been shown to regulate the growth of human breast cancer cell lines via GLUT1 (37), as well as the malignancy of mouse breast cancer cell lines (via GLUT3 amplification) (53).
One of the intracellular pathways that controls glucose uptake is the mTOR pathway; thus, inhibition of mTORC1 with the specific inhibitor rapamycin inhibits glucose uptake in most cell types. We found that rapamycin-treated cells shared symptoms of mitochondrial stress with Lrp5-deficient cells, including higher ROS levels and p38α activation. Lrp5-deficient cells showed decreased activation of mTOR signaling, as assayed by phosphorylation of the ribosome-associated proteins S6 and S6 kinase. However, we could detect no differences in protein amount per cell (measured as the amount of protein per cell counted), typically used as an index of relative translation rates, nor did we observe a change in cell size (measured by flow cytometry), typically used as a measure of mTOR activation (54; data not shown). Interestingly, Lrp5-deficient cells showed a profound deficiency of S6 protein in lysates analyzed by Western blotting, despite normal levels of S6 mRNA (Fig. 8C). This is an unusual and possibly unique phenotype, not shared by rapamycin-treated or RICTOR knockdown (mTORC2 loss of function) cells. The function of the S6 protein still remains obscure, though it is poised at the interface of the 40S and 60S subunits of the mature ribosome, suggesting that it could modify translation depending upon signaling inputs (47, 55). Mice with a nonactivatable version of S6 (rpS6P−/−) were reported to be viable and grossly normal, though they showed subtle alterations in glucose homeostasis, including reduced pancreatic islet function but enhanced insulin sensitivity in peripheral tissues (56).
Glucose uptake is controlled by a series of GLUT transporter proteins, which facilitate glucose transport at the plasma membrane (57, 58). These channel proteins show different mRNA expression patterns and highly regulated cell surface presentation. Several studies have shown that the rate of GLUT-dependent glucose uptake can determine mammary epithelial cell growth and the invasive behavior of breast tumor cell lines (37, 53, 59). Specifically, GLUT1 is the most abundant glucose transporter species in mammary epithelial cells, and knockdown of GLUT1 in mouse mammary tumor cell lines reduces glucose uptake, with a proportional loss of lactate secretion and a corresponding decrease in growth rates, both in tissue culture and in vivo (37). Our study shows that both glucose-limited and Lrp5KD mammary epithelial cells show proportionally less glucose consumption, lactate secretion, and oxygen consumption. All three glucose-inhibited conditions (rapamycin treatment, Lrp5 KD, and low-glucose medium) showed significantly decreased GLUT1 mRNA expression (approximately 25%), which we assume reflects a compensatory switch to alternate calorie acquisition strategies. Since losses of function of Lrp5 and GLUT1 generate broadly similar phenotypes, we suggest that these two proteins are likely to be functionally interactive.
We conclude that Lrp5 is required for glucose uptake, whether by mouse mammary epithelial cells or human breast cancer cells, and that, in this role, it serves to limit and determine the cellular growth rate in a Wnt-independent manner. We propose that since the amount of Lrp5 determines metabolism, this in turn determines how cells respond to growth factors. We suggest that since glucose uptake is typically important to breast tumor cells (37, 53), Lrp5-associated glucose processing may become highly significant during breast epithelial cell transformation.
ACKNOWLEDGMENTS
We thank Dudley Lamming (Department of Medicine, University of Wisconsin—Madison) for his kind gift of RICTOR shRNA expression vectors and Brett Morris and Patti Keely (Department of Cell and Regenerative Biology, University of Wisconsin-Madison) for consultation on the analysis of mouse breast tumor cell lines. We give many thanks to those who provided critical reviews and interpretations of the manuscript, including Avtar Roopra, Dudley Lamming, and Bart Williams.
We are also grateful to our funding sources, i.e., grant T32 CA009135 to E.N.C. and J.A.M., Era of Hope scholar award W81XWH 06-1-0491 to C.M.A. and S.K., and Cancer Center Core Support for the University of Wisconsin (grant P30 CA014520 to C.M.A.).
We have no conflicts of interest to declare.
REFERENCES
- 1.Schuijers J, Clevers H. 2012. Adult mammalian stem cells: the role of Wnt, Lgr5 and R-spondins. EMBO J 31:2685–2696. doi: 10.1038/emboj.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Clevers H, Nusse R. 2012. Wnt/beta-catenin signaling and disease. Cell 149:1192–1205. doi: 10.1016/j.cell.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Brennan K, Gonzalez-Sancho JM, Castelo-Soccio LA, Howe LR, Brown AM. 2004. Truncated mutants of the putative Wnt receptor LRP6/Arrow can stabilize beta-catenin independently of Frizzled proteins. Oncogene 23:4873–4884. doi: 10.1038/sj.onc.1207642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mastroianni M, Kim S, Kim YC, Esch A, Wagner C, Alexander CM. 2010. Wnt signaling can substitute for estrogen to induce division of ERalpha-positive cells in a mouse mammary tumor model. Cancer Lett 289:23–31. doi: 10.1016/j.canlet.2009.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Alexander CM, Goel S, Fakhraldeen SA, Kim S. 2012. Wnt signaling in mammary glands: plastic cell fates and combinatorial signaling. Cold Spring Harb Perspect Biol 4:a008037. doi: 10.1101/cshperspect.a008037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim S, Goel S, Alexander CM. 2011. Differentiation generates paracrine cell pairs that maintain basaloid mouse mammary tumors: proof of concept. PLoS One 6:e19310. doi: 10.1371/journal.pone.0019310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Badders NM, Goel S, Clark RJ, Klos KS, Kim S, Bafico A, Lindvall C, Williams BO, Alexander CM. 2009. The Wnt receptor, Lrp5, is expressed by mouse mammary stem cells and is required to maintain the basal lineage. PLoS One 4:e6594. doi: 10.1371/journal.pone.0006594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindvall C, Evans NC, Zylstra CR, Li Y, Alexander CM, Williams BO. 2006. The Wnt signaling receptor Lrp5 is required for mammary ductal stem cell activity and Wnt1-induced tumorigenesis. J Biol Chem 281:35081–35087. doi: 10.1074/jbc.M607571200. [DOI] [PubMed] [Google Scholar]
- 9.Macdonald BT, Semenov MV, Huang H, He X. 2011. Dissecting molecular differences between Wnt coreceptors LRP5 and LRP6. PLoS One 6:e23537. doi: 10.1371/journal.pone.0023537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.MacDonald BT, Tamai K, He X. 2009. Wnt/beta-catenin signaling: components, mechanisms, and diseases. Dev Cell 17:9–26. doi: 10.1016/j.devcel.2009.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.He X, Semenov M, Tamai K, Zeng X. 2004. LDL receptor-related proteins 5 and 6 in Wnt/beta-catenin signaling: arrows point the way. Development 131:1663–1677. doi: 10.1242/dev.01117. [DOI] [PubMed] [Google Scholar]
- 12.Hey PJ, Twells RC, Phillips MS, Yusuke N, Brown SD, Kawaguchi Y, Cox R, Guochun X, Dugan V, Hammond H, Metzker ML, Todd JA, Hess JF. 1998. Cloning of a novel member of the low-density lipoprotein receptor family. Gene 216:103–111. doi: 10.1016/S0378-1119(98)00311-4. [DOI] [PubMed] [Google Scholar]
- 13.Frey JL, Li Z, Ellis JM, Zhang Q, Farber CR, Aja S, Wolfgang MJ, Clemens TL, Riddle RC. 2015. Wnt-Lrp5 signaling regulates fatty acid metabolism in the osteoblast. Mol Cell Biol 35:1979–1991. doi: 10.1128/MCB.01343-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujino T, Asaba H, Kang MJ, Ikeda Y, Sone H, Takada S, Kim DH, Ioka RX, Ono M, Tomoyori H, Okubo M, Murase T, Kamataki A, Yamamoto J, Magoori K, Takahashi S, Miyamoto Y, Oishi H, Nose M, Okazaki M, Usui S, Imaizumi K, Yanagisawa M, Sakai J, Yamamoto TT. 2003. Low-density lipoprotein receptor-related protein 5 (LRP5) is essential for normal cholesterol metabolism and glucose-induced insulin secretion. Proc Natl Acad Sci U S A 100:229–234. doi: 10.1073/pnas.0133792100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magoori K, Kang MJ, Ito MR, Kakuuchi H, Ioka RX, Kamataki A, Kim DH, Asaba H, Iwasaki S, Takei YA, Sasaki M, Usui S, Okazaki M, Takahashi S, Ono M, Nose M, Sakai J, Fujino T, Yamamoto TT. 2003. Severe hypercholesterolemia, impaired fat tolerance, and advanced atherosclerosis in mice lacking both low density lipoprotein receptor-related protein 5 and apolipoprotein E. J Biol Chem 278:11331–11336. doi: 10.1074/jbc.M211987200. [DOI] [PubMed] [Google Scholar]
- 16.Monroe DG, McGee-Lawrence ME, Oursler MJ, Westendorf JJ. 2012. Update on Wnt signaling in bone cell biology and bone disease. Gene 492:1–18. doi: 10.1016/j.gene.2011.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cui Y, Niziolek PJ, MacDonald BT, Zylstra CR, Alenina N, Robinson DR, Zhong Z, Matthes S, Jacobsen CM, Conlon RA, Brommage R, Liu Q, Mseeh F, Powell DR, Yang QM, Zambrowicz B, Gerrits H, Gossen JA, He X, Bader M, Williams BO, Warman ML, Robling AG. 2011. Lrp5 functions in bone to regulate bone mass. Nat Med 17:684–691. doi: 10.1038/nm.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esen E, Chen J, Karner CM, Okunade AL, Patterson BW, Long F. 2013. WNT-LRP5 signaling induces Warburg effect through mTORC2 activation during osteoblast differentiation. Cell Metab 17:745–755. doi: 10.1016/j.cmet.2013.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu BY, McDermott SP, Khwaja SS, Alexander CM. 2004. The transforming activity of Wnt effectors correlates with their ability to induce the accumulation of mammary progenitor cells. Proc Natl Acad Sci U S A 101:4158–4163. doi: 10.1073/pnas.0400699101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S, Roopra A, Alexander CM. 2012. A phenotypic mouse model of basaloid breast tumors. PLoS One 7:e30979. doi: 10.1371/journal.pone.0030979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goel S, Chin EN, Fakhraldeen SA, Berry SM, Beebe DJ, Alexander CM. 2012. Both LRP5 and LRP6 receptors are required to respond to physiological Wnt ligands in mammary epithelial cells and fibroblasts. J Biol Chem 287:16454–16466. doi: 10.1074/jbc.M112.362137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Timmerman LA, Holton T, Yuneva M, Louie RJ, Padro M, Daemen A, Hu M, Chan DA, Ethier SP, van't Veer LJ, Polyak K, McCormick F, Gray JW. 2013. Glutamine sensitivity analysis identifies the xCT antiporter as a common triple-negative breast tumor therapeutic target. Cancer Cell 24:450–465. doi: 10.1016/j.ccr.2013.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aslakson CJ, Miller FR. 1992. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res 52:1399–1405. [PubMed] [Google Scholar]
- 24.Danielson KG, Oborn CJ, Durban EM, Butel JS, Medina D. 1984. Epithelial mouse mammary cell line exhibiting normal morphogenesis in vivo and functional differentiation in vitro. Proc Natl Acad Sci U S A 81:3756–3760. doi: 10.1073/pnas.81.12.3756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ball RK, Friis RR, Schoenenberger CA, Doppler W, Groner B. 1988. Prolactin regulation of beta-casein gene expression and of a cytosolic 120-kd protein in a cloned mouse mammary epithelial cell line. EMBO J 7:2089–2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell SM, Taha MM, Medina D, Rosen JM. 1988. A clonal derivative of mammary epithelial cell line COMMA-D retains stem cell characteristics of unique morphological and functional heterogeneity. Exp Cell Res 177:109–121. doi: 10.1016/0014-4827(88)90029-8. [DOI] [PubMed] [Google Scholar]
- 27.Merlo GR, Venesio T, Taverna D, Marte BM, Callahan R, Hynes NE. 1994. Growth suppression of normal mammary epithelial cells by wild-type p53. Oncogene 9:443–453. [PubMed] [Google Scholar]
- 28.Cong F, Schweizer L, Varmus H. 2004. Wnt signals across the plasma membrane to activate the beta-catenin pathway by forming oligomers containing its receptors, Frizzled and LRP. Development 131:5103–5115. doi: 10.1242/dev.01318. [DOI] [PubMed] [Google Scholar]
- 29.Bilic J, Huang YL, Davidson G, Zimmermann T, Cruciat CM, Bienz M, Niehrs C. 2007. Wnt induces LRP6 signalosomes and promotes dishevelled-dependent LRP6 phosphorylation. Science 316:1619–1622. doi: 10.1126/science.1137065. [DOI] [PubMed] [Google Scholar]
- 30.Matsuda Y, Schlange T, Oakeley EJ, Boulay A, Hynes NE. 2009. WNT signaling enhances breast cancer cell motility and blockade of the WNT pathway by sFRP1 suppresses MDA-MB-231 xenograft growth. Breast Cancer Res 11:R32. doi: 10.1186/bcr2317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lum L, Clevers H. 2012. Cell biology. The unusual case of Porcupine. Science 337:922–923. doi: 10.1126/science.1228179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu J, Pan S, Hsieh MH, Ng N, Sun F, Wang T, Kasibhatla S, Schuller AG, Li AG, Cheng D, Li J, Tompkins C, Pferdekamper A, Steffy A, Cheng J, Kowal C, Phung V, Guo G, Wang Y, Graham MP, Flynn S, Brenner JC, Li C, Villarroel MC, Schultz PG, Wu X, McNamara P, Sellers WR, Petruzzelli L, Boral AL, Seidel HM, McLaughlin ME, Che J, Carey TE, Vanasse G, Harris JL. 2013. Targeting Wnt-driven cancer through the inhibition of Porcupine by LGK974. Proc Natl Acad Sci U S A 110:20224–20229. doi: 10.1073/pnas.1314239110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Proffitt KD, Madan B, Ke Z, Pendharkar V, Ding L, Lee MA, Hannoush RN, Virshup DM. 2013. Pharmacological inhibition of the Wnt acyltransferase PORCN prevents growth of WNT-driven mammary cancer. Cancer Res 73:502–507. doi: 10.1158/0008-5472.CAN-12-2258. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, Moon J, Dodge ME, Pan X, Zhang L, Hanson JM, Tuladhar R, Ma Z, Shi H, Williams NS, Amatruda JF, Carroll TJ, Lum L, Chen C. 2013. The development of highly potent inhibitors for porcupine. J Med Chem 56:2700–2704. doi: 10.1021/jm400159c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yoon JC, Ng A, Kim BH, Bianco A, Xavier RJ, Elledge SJ. 2010. Wnt signaling regulates mitochondrial physiology and insulin sensitivity. Genes Dev 24:1507–1518. doi: 10.1101/gad.1924910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bogan JS. 2012. Regulation of glucose transporter translocation in health and diabetes. Annu Rev Biochem 81:507–532. doi: 10.1146/annurev-biochem-060109-094246. [DOI] [PubMed] [Google Scholar]
- 37.Young CD, Lewis AS, Rudolph MC, Ruehle MD, Jackman MR, Yun UJ, Ilkun O, Pereira R, Abel ED, Anderson SM. 2011. Modulation of glucose transporter 1 (GLUT1) expression levels alters mouse mammary tumor cell growth in vitro and in vivo. PLoS One 6:e23205. doi: 10.1371/journal.pone.0023205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liu Y, Cao Y, Zhang W, Bergmeier S, Qian Y, Akbar H, Colvin R, Ding J, Tong L, Wu S, Hines J, Chen X. 2012. A small-molecule inhibitor of glucose transporter 1 downregulates glycolysis, induces cell-cycle arrest, and inhibits cancer cell growth in vitro and in vivo. Mol Cancer Ther 11:1672–1682. doi: 10.1158/1535-7163.MCT-12-0131. [DOI] [PubMed] [Google Scholar]
- 39.Bjorklund P, Svedlund J, Olsson AK, Akerstrom G, Westin G. 2009. The internally truncated LRP5 receptor presents a therapeutic target in breast cancer. PLoS One 4:e4243. doi: 10.1371/journal.pone.0004243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hardie DG. 2015. Molecular pathways: is AMPK a friend or a foe in cancer? Clin Cancer Res 21:3836–3840. doi: 10.1158/1078-0432.CCR-14-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wagner EF, Nebreda AR. 2009. Signal integration by JNK and p38 MAPK pathways in cancer development. Nat Rev Cancer 9:537–549. doi: 10.1038/nrc2694. [DOI] [PubMed] [Google Scholar]
- 42.Wallace DC. 2012. Mitochondria and cancer. Nat Rev Cancer 12:685–698. doi: 10.1038/nrc3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yecies JL, Manning BD. 2011. mTOR links oncogenic signaling to tumor cell metabolism. J Mol Med 89:221–228. doi: 10.1007/s00109-011-0726-6. [DOI] [PubMed] [Google Scholar]
- 44.Madiraju AK, Erion DM, Rahimi Y, Zhang XM, Braddock DT, Albright RA, Prigaro BJ, Wood JL, Bhanot S, MacDonald MJ, Jurczak MJ, Camporez JP, Lee HY, Cline GW, Samuel VT, Kibbey RG, Shulman GI. 2014. Metformin suppresses gluconeogenesis by inhibiting mitochondrial glycerophosphate dehydrogenase. Nature 510:542–546. doi: 10.1038/nature13270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zoncu R, Efeyan A, Sabatini DM. 2011. mTOR: from growth signal integration to cancer, diabetes and ageing. Nat Rev Mol Cell Biol 12:21–35. doi: 10.1038/nrm3025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oh WJ, Jacinto E. 2011. mTOR complex 2 signaling and functions. Cell Cycle 10:2305–2316. doi: 10.4161/cc.10.14.16586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ruvinsky I, Meyuhas O. 2006. Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem Sci 31:342–348. doi: 10.1016/j.tibs.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 48.Gong Y, Bourhis E, Chiu C, Stawicki S, DeAlmeida VI, Liu BY, Phamluong K, Cao TC, Carano RA, Ernst JA, Solloway M, Rubinfeld B, Hannoush RN, Wu Y, Polakis P, Costa M. 2010. Wnt isoform-specific interactions with coreceptor specify inhibition or potentiation of signaling by LRP6 antibodies. PLoS One 5:e12682. doi: 10.1371/journal.pone.0012682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ettenberg SA, Charlat O, Daley MP, Liu S, Vincent KJ, Stuart DD, Schuller AG, Yuan J, Ospina B, Green J, Yu Q, Walsh R, Li S, Schmitz R, Heine H, Bilic S, Ostrom L, Mosher R, Hartlepp KF, Zhu Z, Fawell S, Yao YM, Stover D, Finan PM, Porter JA, Sellers WR, Klagge IM, Cong F. 2010. Inhibition of tumorigenesis driven by different Wnt proteins requires blockade of distinct ligand-binding regions by LRP6 antibodies. Proc Natl Acad Sci U S A 107:15473–15478. doi: 10.1073/pnas.1007428107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Inoki K, Ouyang H, Zhu T, Lindvall C, Wang Y, Zhang X, Yang Q, Bennett C, Harada Y, Stankunas K, Wang CY, He X, MacDougald OA, You M, Williams BO, Guan KL. 2006. TSC2 integrates Wnt and energy signals via a coordinated phosphorylation by AMPK and GSK3 to regulate cell growth. Cell 126:955–968. doi: 10.1016/j.cell.2006.06.055. [DOI] [PubMed] [Google Scholar]
- 51.Loh NY, Neville MJ, Marinou K, Hardcastle SA, Fielding BA, Duncan EL, McCarthy MI, Tobias JH, Gregson CL, Karpe F, Christodoulides C. 2015. LRP5 regulates human body fat distribution by modulating adipose progenitor biology in a dose- and depot-specific fashion. Cell Metab 21:262–272. doi: 10.1016/j.cmet.2015.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Krishnan V, Bryant HU, Macdougald OA. 2006. Regulation of bone mass by Wnt signaling. J Clin Invest 116:1202–1209. doi: 10.1172/JCI28551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Onodera Y, Nam JM, Bissell MJ. 2014. Increased sugar uptake promotes oncogenesis via EPAC/RAP1 and O-GlcNAc pathways. J Clin Invest 124:367–384. doi: 10.1172/JCI63146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu L, Derynck R. 2009. Essential role of TGF-beta signaling in glucose-induced cell hypertrophy. Dev Cell 17:35–48. doi: 10.1016/j.devcel.2009.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Magnuson B, Ekim B, Fingar DC. 2012. Regulation and function of ribosomal protein S6 kinase (S6K) within mTOR signalling networks. Biochem J 441:1–21. doi: 10.1042/BJ20110892. [DOI] [PubMed] [Google Scholar]
- 56.Ruvinsky I, Sharon N, Lerer T, Cohen H, Stolovich-Rain M, Nir T, Dor Y, Zisman P, Meyuhas O. 2005. Ribosomal protein S6 phosphorylation is a determinant of cell size and glucose homeostasis. Genes Dev 19:2199–2211. doi: 10.1101/gad.351605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhao FQ. 2014. Biology of glucose transport in the mammary gland. J Mammary Gland Biol Neoplasia 19:3–17. doi: 10.1007/s10911-013-9310-8. [DOI] [PubMed] [Google Scholar]
- 58.Mueckler M, Thorens B. 2013. The SLC2 (GLUT) family of membrane transporters. Mol Aspects Med 34:121–138. doi: 10.1016/j.mam.2012.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wahdan-Alaswad R, Fan Z, Edgerton SM, Liu B, Deng XS, Arnadottir SS, Richer JK, Anderson SM, Thor AD. 2013. Glucose promotes breast cancer aggression and reduces metformin efficacy. Cell Cycle 12:3759–3769. doi: 10.4161/cc.26641. [DOI] [PMC free article] [PubMed] [Google Scholar]