Abstract
NuA4 histone lysine (K) acetyltransferase (KAT) promotes transcriptional initiation of TATA-binding protein (TBP)-associated factor (TAF)-dependent ribosomal protein genes. TAFs have also been recently found to enhance antisense transcription from the 3′ end of the GAL10 coding sequence. However, it remains unknown whether, like sense transcription of the ribosomal protein genes, TAF-dependent antisense transcription of GAL10 also requires NuA4 KAT. Here, we show that NuA4 KAT associates with the GAL10 antisense transcription initiation site at the 3′ end of the coding sequence. Such association of NuA4 KAT depends on the Reb1p-binding site that recruits Reb1p activator to the GAL10 antisense transcription initiation site. Targeted recruitment of NuA4 KAT to the GAL10 antisense transcription initiation site promotes GAL10 antisense transcription. Like NuA4 KAT, histone H3 K4/36 methyltransferases and histone H2B ubiquitin conjugase facilitate GAL10 antisense transcription, while the Swi/Snf and SAGA chromatin remodeling/modification factors are dispensable for antisense, but not sense, transcription of GAL10. Taken together, our results demonstrate for the first time the roles of NuA4 KAT and other chromatin regulatory factors in controlling antisense transcription, thus illuminating chromatin regulation of antisense transcription.
INTRODUCTION
Noncoding RNAs have been implicated in various cellular processes such as X-chromosome inactivation, genomic imprinting, dosage compensation, heterochromatin formation, metabolism, development, and differentiation (1–5). There are several classes of noncoding RNAs, which include microRNAs, small nuclear RNAs, small interfering RNAs, Piwi-interacting RNAs, and natural antisense transcripts (6). About 72% of genes in human and mouse are associated with antisense transcription (7, 8). Antisense transcripts arise from the strand opposite to the sense strand and play regulatory functions in interfering with the stability of sense transcripts, and hence gene expression. Therefore, a number of studies have been focused on the use of antisense oligonucleotides in regulation of gene expression and treatment of diseases without permanently altering the genes. In fact, antisense oligonucleotides are in various clinical trials for treatment of diseases such as cancers, hypertension, respiratory illness, and HIV infection (9–13).
Despite great potentials of antisense transcripts/transcription in disease pathogenesis and treatment, it is not clearly understood how antisense transcription is initiated. Recently, we have demonstrated that, like in sense transcription, RNA polymerase II is targeted to the 3′ end of the GAL10 coding sequence by an activator Reb1p or Reb1p-binding site and general transcription factors (GTFs) such as transcription factor IID (TFIID) (which is composed of TATA-binding protein [TBP] and a set of TBP-associated factors [TAFs]), TFIIB, and Mediator to initiate antisense transcription (14). Further, we have shown that the Gal4p activator and proteasome that facilitate GAL10 sense transcription are dispensable for GAL10 antisense transcription (14), supporting that GAL10 sense and antisense transcriptions are initiated independently and differently. These recent results shed much light on the initiation of antisense transcription (14). However, the involvement of the chromatin structure/dynamics and the associated factors in regulation of antisense transcription remains poorly understood.
Here, we have carried out experiments to analyze chromatin regulation of antisense transcription. We have taken the advantage of the GAL gene cluster, which consists of three genes, namely, GAL1, GAL7, and GAL10 (Fig. 1A). These genes are induced for sense transcription in galactose-containing growth medium (14). However, long noncoding antisense transcripts are generated from the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium (Fig. 1A), which is repressive to GAL sense transcription (14–16). Therefore, using this GAL gene cluster, the chromatin regulation of GAL10 antisense transcription in dextrose-containing growth medium can be clearly analyzed without any interference from the sense transcription, hence contributing to our understanding of the regulation of antisense transcription by epigenetic factors. We initially focused our studies on NuA4 histone lysine (K) acetyltransferase (KAT), as NuA4 KAT is known to facilitate TAF-dependent sense transcription of the ribosomal protein genes (17–19), while its role in regulation of TAF-dependent GAL10 antisense transcription remains unknown. Here, we find that NuA4 KAT is associated with the 3′ end of the GAL10 coding sequence (i.e., GAL10 antisense transcription initiation site) (Fig. 1A) for histone H4 acetylation and antisense transcription. However, NuA4 KAT-regulated GAL10 antisense transcription is not controlled by TOR (target of rapamycin), while TOR regulates sense transcription of NuA4 KAT-dependent ribosomal protein genes. Like NuA4 KAT, histone H3 K4 methyltransferase (Set1p) and histone H3 K36 methyltransferase (Set2p), which are essential for H3 K4 and K36 methylation, respectively, facilitate GAL10 antisense transcription. Further, we find that histone H2B ubiquitin conjugase Rad6p (which is essential for histone H2B ubiquitylation) promotes GAL10 antisense transcription. However, the Swi/Snf (switch/sucrose nonfermentable) chromatin remodeling factor and SAGA (Spt-Ada-Gcn5-acetyltransferase) chromatin modification factor are dispensable for GAL10 antisense transcription, while these factors have stimulatory functions in GAL10 sense transcription. Collectively, our results demonstrate the roles of different chromatin modification/regulatory factors in controlling GAL10 antisense transcription, thus significantly advancing our knowledge of the chromatin regulation of antisense transcription, as presented below.
FIG 1.
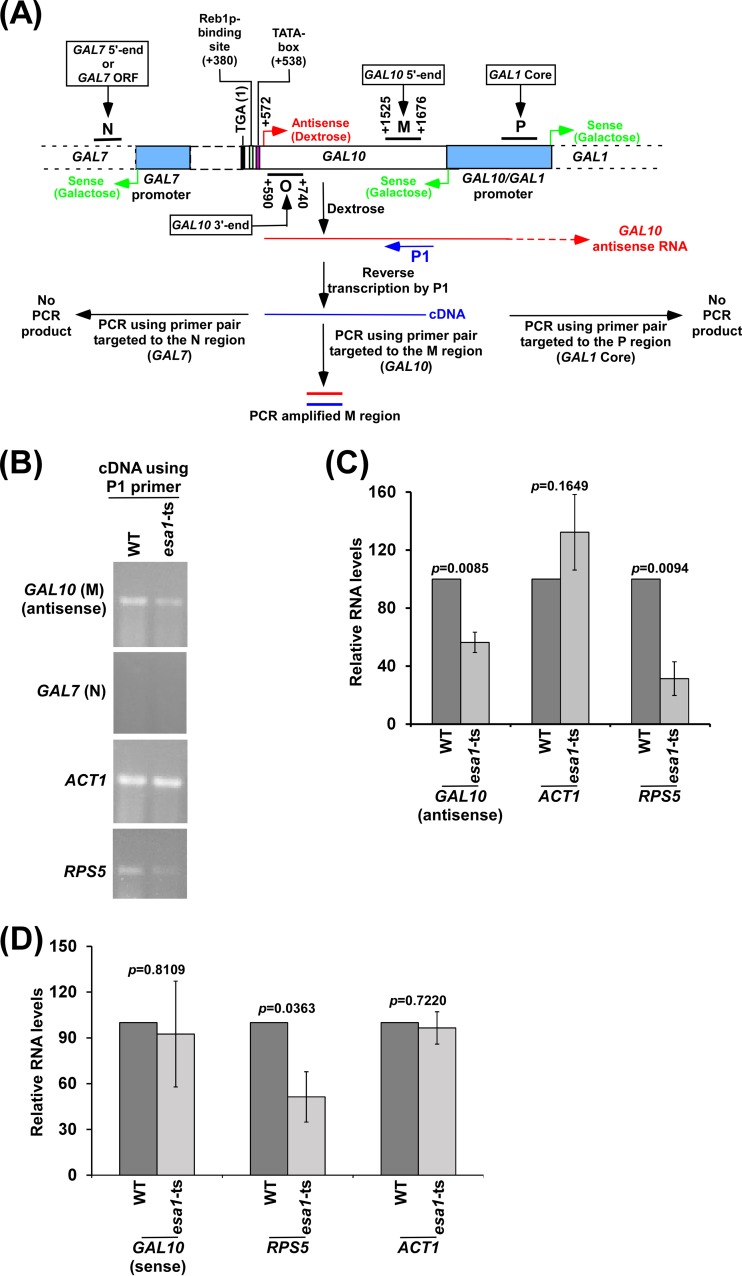
NuA4 KAT promotes GAL10 antisense transcription. (A) Schematic diagram showing the experimental strategy for analysis of the GAL10 antisense transcript. The P1 primer targeted toward the 5′ end of the GAL10 antisense transcript was extended by AMV reverse transcriptase-based reverse transcription at 42°C, and subsequently the extended primer was amplified by primer pairs targeted to the coding regions, M and N, of GAL10 and GAL7, respectively. The numbers are presented with respect to the position of the translational stop codon (TGA) of GAL10. Regions P, M, O, and N represent GAL1 core, GAL10 5′ end, GAL10 3′ end, and GAL7 5′ end (or GAL7 open reading frame [ORF]), respectively, in the ChIP assay. (B and C) Analysis of the GAL10 antisense transcript in the esa1-ts mutant and wild-type (WT) strains in dextrose-containing growth medium. The RNA level in the wild-type strain was set to 100, and the relative RNA level in the mutant strain was plotted in panel C. (D) Sense transcription analysis of GAL10, RPS5, and ACT1 in the esa1-ts and wild-type strains in galactose-containing growth medium by RT-PCR.
MATERIALS AND METHODS
Plasmids.
The plasmid pFA6a-13Myc-KanMX6 (20) was used for genomic tagging of the Esa1p and Eaf5p components of NuA4 KAT, Set1p, and Rad6p. The plasmids PRS406 (21), PRS403 (21), and pFA6a-13Myc-HIS3MX6 (20) were used for PCR-based disruption of SET1, SET2, EAF1, and the RING (really interesting new gene) domain of BRE1.
Strains.
A yeast (Saccharomyces cerevisiae) strain harboring a temperature-sensitive (ts) mutation in Esa1p (LPY3291) and its isogenic wild-type equivalent (LPY3498) were obtained from the Pillus laboratory (Lorraine Pillus, University of California, San Diego, CA) (22). A yeast strain harboring a null mutation in SWI2 (MSY143) and its wild-type equivalent (FY406) were obtained from the Struhl laboratory (Kevin Struhl, Harvard Medical School, Boston, MA) (23). A yeast strain harboring a null mutation in RAD6 (STY2; Δrad6 in FM392) and its wild-type equivalent (STY1; FM392) were obtained from the Shilatifard laboratory (Ali Shilatifard, Stowers Institute for Medical Research; purchased from Research Genetics). The PAF1 deletion mutant strain (DY7014) in the W303a background was obtained from the Stillman laboratory (David Stillman, University of Utah Health Sciences Center) (24). A yeast strain harboring a null mutation in SPT20 (FY1097) and its isogenic wild-type equivalent (FY67) were obtained from the Winston laboratory (Fred Winston, Harvard Medical School, Boston, MA) (25). A yeast strain carrying mutations in the Reb1p-binding site at the 3′ end of the GAL10 coding sequence and its isogenic wild-type equivalent were obtained from the Tollervey laboratory (David Tollervey, University of Edinburg, United Kingdom) (15). A yeast strain (YKH045) expressing Flag-tagged histone H2B and hemagglutinin (HA)-tagged ubiquitin was obtained from the Osley laboratory (Mary Ann Osley, University of New Mexico School of Medicine) (26). The Δset3, Δrpd3, and wild-type (BY4741) strains were from the Davie laboratory (Judith K. Davie, Southern Illinois University School of Medicine; purchased from Open Biosystems). SET1 was deleted in the wild-type strain (W303a) by the PCR-based gene disruption method to generate ASY16. SET2 was deleted in the wild-type strain (ZDY2; derived from W303a) (27) by the PCR-based gene disruption method to generate SLY7 (28). The RING domain of Bre1p was deleted in YKH045 to generate BUY57 as done previously (29). Multiple Myc epitope tags were added at the original chromosomal loci of SET1 and RAD6 in the wild-type strain (W303a) to generate the PSY4 and PSY2 strains, respectively. Multiple Myc epitope tags were added at the original chromosomal locus of EAF5 in the wild-type strain (W303a) to generate RSY70. Likewise, Eaf5p was tagged with multiple Myc epitopes at its C terminus in the BUY24 strain (which was derived from W303a by deleting EAF1) to generate RSY69. Multiple Myc epitope tags were added at the original chromosomal locus of ESA1 in the wild-type strain (Sc599) (30) to generate BUY12 (17). Multiple Myc epitope tags were added at the original chromosomal locus of ESA1 in the yeast strain carrying mutations in the Reb1p-binding site at the 3′ end of the GAL10 coding sequence and its isogenic wild-type equivalent to generate BUY46 and BUY45, respectively. All these strains with their genotypes are listed in Table S1 in the supplemental material.
Growth media.
Yeast cells were grown in yeast extract-peptone plus 2% dextrose (YPD) up to an optical density at 600 nm (OD600) of 1.0 at 30°C prior to harvesting for analysis of GAL10 antisense transcripts or formaldehyde-based in vivo cross-linking for chromatin immunoprecipitation (ChIP) experiments. For experiments in the wild-type and ts mutant strains, yeast cells were grown in YPD medium at 23°C up to an OD600 of 0.85 and then transferred to 37°C for 1 h before cross-linking or harvesting for RNA analysis. For analysis of rapamycin response, yeast cells were grown in YPD medium up to an OD600 of 1.0 and then treated with 100 nM rapamycin (Sigma) for 30 min prior to harvesting.
Antibodies.
Various antibodies were used in the ChIP and Western blot analyses. These are anti-Rpb1 (8WG16; Covance), anti-Myc (9E10; Santa Cruz Biotechnology, Inc.), anti-HA (F-7; Santa Cruz Biotechnology, Inc.), anti-histone H3 (Ab-1791; Abcam), anti-H3 K4 trimethylation (Ab-8580; Abcam), anti-H3-K36 trimethylation (Ab-9050; Abcam), anti-Flag (F3165; Sigma), and anti-acetylated histone H4 (06866; Millipore) antibodies.
ChIP assay.
The ChIP assay was performed as described previously (17, 28). Briefly, yeast cells were treated with 1% formaldehyde, collected, and resuspended in lysis buffer. Following sonication, cell lysate (400 μl of lysate from 50 ml of yeast culture) was precleared by centrifugation, and then 100 μl of lysate was used for each immunoprecipitation. Immunoprecipitated protein-DNA complexes were treated with proteinase K, the cross-links were reversed, and DNA was purified. Immunoprecipitated DNA was dissolved in 20 μl of TE 8.0 (10 mM Tris-HCl [pH 8.0] and 1 mM EDTA), and 1 μl of immunoprecipitated DNA was analyzed by PCR (a total of 23 cycles). The PCR mixture contained [α-32P]dATP (2.5 μCi for 25 μl of reaction mixture), and the PCR products were detected by autoradiography after separation on a 6% polyacrylamide gel. As a control, “input” DNA was isolated from 5 μl of lysate without going through the immunoprecipitation step and was dissolved in 100 μl of TE 8.0. To compare the PCR signal arising from the immunoprecipitated DNA with that from the input DNA, 1 μl of input DNA was used in the PCR analysis. For ChIP analysis of histone H3, histone H2B, and Myc-tagged Esa1p, Eaf5p, Set1p, Rad6p, and Rpb1p, the protocol described above was modified as described previously (17, 28). Serial dilutions of input and immunoprecipitated DNA samples were used to assess the linear range of PCR amplification as described previously (18). The data presented here are within the linear range of PCR analysis. The primer pairs used for PCR analysis were as follows: GAL10 3′ end, 5′-CTATGTTCAGTTAGTTTGGCTAGC-3′ and 5′-TTGATGCTCTGCATAATAATGCCC-3′; GAL10 5′ end, 5′-CTACGAGATTCCCAAATATGATTCC-3′ and 5′-TAACGCAAGATAGCAAACTTCCAAC-3′; GAL7 5′ end, 5′-AAAGTGCAATCTGTGAGAGGCAATT-3′ and 5′-TTTTCTCTTGCTTCTCTGGAGAGAT-3′; GAL1 core, 5′-ATAGGATGATAATGCGATTAGTTTTTTAGCCTT-3′ and 5′-GAAAATGTTGAAAGTATTAGTTAAAGTGGTTATGCA-3′; RPS5 UAS, 5′-AGAAACAATGAACAGCCTTGAGTTCTC-3′ and 5′-GCAGGGCCATTCTCATCTGA-3′; Chr.-V, 5′-GGCTGTCAGAATATGGGGCCGTAGTA-3′ and 5′-CACCCCGAAGCTGCTTTCACAATAC-3′; ADH1 5′ end, 5′-CTGGTTACACCCACGACGGTTCTT-3′ and 5′-CAGACTTCAAAGCCTTGTAGACG-3′; and 18S ribosomal DNA (rDNA), 5′-GAGTCCTTGTGGCTCTTGGC-3′ and 5′-AATACTGATGCCCCCGACC-3′.
Autoradiograms were scanned and quantitated by the National Institutes of Health ImageJ (version 1.62) program. Immunoprecipitated DNAs were quantitated as the ratio of immunoprecipitate to input in the autoradiogram. The average ChIP signal from the biologically independent experiments is reported with standard deviation (SD) (Microsoft Office Excel 2003). The Student t test in Microsoft Excel 2003 (with tail = 2 and types = 3) was used to determine the P values for statistical significance of the changes in the ChIP signals. The changes were considered to be statistically significant at a P value of <0.05. ChIP signals were determined for the upstream activating sequence (UAS), the core promoter (core), toward the 5′ end of the coding sequence (5′ end), toward the 3′ end of the coding sequence (3′ end), and an inactive region within chromosome V (Chr.-V).
ChDIP assay.
To determine histone H2B ubiquitylation at GAL10, we performed a chromatin double-immunoprecipitation (ChDIP) assay as described previously (31). Briefly, 400 μl lysate from 50 ml yeast culture was first immunoprecipitated using anti-Flag antibody and protein A/G Plus-agarose beads. Following elution of anti-Flag immunoprecipitate by Flag peptide (Sigma), the eluate was immunoprecipitated using an anti-HA antibody, and immunoprecipitated DNA sample was dissolved in 10 μl TE 8.0, of which 1 μl was used for PCR analysis. The “input” DNA was isolated from 5 μl lysate and suspended in 100 μl TE 8.0, of which 1 μl was used for PCR analysis.
RNA preparation.
Total RNA was prepared from yeast cell culture as described by Peterson et al. (32). Briefly, 10 ml of yeast culture was harvested and then suspended in 100 μl of RNA preparation buffer (500 mM NaCl, 200 mM Tris-HCl, 100 mM Na2-EDTA, and 1% SDS) along with 100 μl of phenol-chloroform-isoamyl alcohol and a 100-μl volume-equivalent of glass beads (acid washed) (Sigma). Subsequently, the yeast cell suspension was vortexed at maximum speed (10 in a VWR Mini-vortex mixer) (catalog number 58816-121; VWR) five times (30 s each). The cell suspension was put on ice for 30 s between pulses. After vortexing, 150 μl of RNA preparation buffer and 150 μl of phenol-chloroform-isoamyl alcohol were added to the yeast cell suspension, followed by vortexing for 15 s at maximum speed on a VWR mini-vortex mixer. The aqueous phase was collected following a 5-min centrifugation at maximum speed in a microcentrifuge machine. Total RNA was isolated from the aqueous phase by ethanol precipitation.
Analysis of GAL10 antisense transcript.
Reverse transcription-PCR (RT-PCR) analysis of the GAL10 antisense transcript was performed according to standard protocols (14). Briefly, total RNA was prepared from yeast culture as described above. Equal amounts (15 to 30 μg) of total RNA were used in the reverse transcription assay for both the wild-type and mutant strains. RNA was treated with RNase-free DNase (M610A; Promega), and then reverse transcribed into cDNA using primer P1 (Fig. 1A) targeted to the GAL10 antisense transcript (P1, 5′-CTACGAGATTCCCAAATATGATTCC-3′) as described in the protocol supplied by Promega (A3800; Promega). Reverse transcription was carried out at 42°C using avian myeloblastosis virus (AMV) reverse transcriptase. PCR was performed within the linear range using synthesized first-strand cDNA (or the extended P1 primer shown in Fig. 1A) as the template and primer pairs targeted to the GAL10 coding sequence (represented as region M in Fig. 1A), GAL1 core promoter (marked as region P in Fig. 1A), and GAL7 coding sequence (marked as region N in Fig. 1A). Primer pairs targeted to the ACT1, RPL2B, RPS5, PYK1, and ADH1 coding sequences were also used to amplify the above cDNAs. RT-PCR products were separated by 2.2% agarose gel electrophoresis and visualized by ethidium bromide staining. RT-PCR experiments were carried out three times. These experiments are biologically independent. The average signal from these biologically independent experiments is reported with SD (Microsoft Excel 2003). Student's t test (with tail = 2 and types = 3) was used to determine P values for statistical significances of the changes in the RT-PCR signals. The changes were considered to be statistically significant at a P value of <0.05. The primer pairs used in the PCR analysis of cDNAs were as follows: GAL7 (N), 5′-AAAGTGCAATCTGTGAGAGGCAATT-3′ and 5′-TTTTCTCTTGCTTCTCTGGAGAGAT-3′; GAL10 (M), 5′-CTACGAGATTCCCAAATATGATTCC-3′ and 5′-TAACGCAAGATAGCAAACTTCCAAC-3′; ACT1, 5′-TCCACCACTGCTGAAAGAGAAATTG-3′ and 5′-AATAGTGATGACTTGACCATCTGGA-3′; RPL2B, 5′-GTGCTTTCCACAAGTACAGATTGAA-3′ and 5′-TTTGACCAGAAACGGCACCTCTAGA-3′; RPS5, 5′-AGGCTCAATGTCCAATCATTGAAAG-3′ and 5′-CAACAACTTGGATTGGGTTTTGGTC-3′; ADH1, 5′-CGGTAACAGAGCTGACACCAGAGA-3′ and 5′-ACGTATCTACCAACGATTTGACCC-3′; PYK1, 5′-AAGTTTCCGATGTCGGTAACGCTAT-3′ and 5′-TTGGCAAGTAAGCGATAGCTTGTTC-3′; GAL1 (P), 5′-ATAGGATGATAATGCGATTAGTTTTTTAGCCTT-3′ and 5′-GAAAATGTTGAAAGTATTAGTTAAAGTGGTTATGCA-3′; and 18S rDNA, 5′-GAGTCCTTGTGGCTCTTGGC-3′ and 5′-AATACTGATGCCCCCGACC-3′.
RESULTS
NuA4 KAT facilitates antisense transcription from the 3′ end of the GAL10 coding sequence.
NuA4 KAT has been previously shown to be required for recruitment of TAFs (or TFIID) to the promoters of the ribosomal protein genes for transcriptional initiation (17). However, the role of NuA4 KAT in antisense transcriptional initiation remains unknown. We have recently demonstrated the roles of TAFs in antisense transcriptional initiation from the 3′ end of the GAL10 coding sequence (14). Like sense transcription of the ribosomal protein genes, the TAF-dependent antisense transcription from GAL10 may also require NuA4 KAT. To test this, we analyzed the role of NuA4 KAT in antisense transcription from the 3′ end of the GAL10 coding sequence. In this direction, the ESA1 (which is an integral component of NuA4 with KAT activity) wild-type and ts strains were grown in dextrose-containing medium (which induces GAL10 antisense transcription and represses GAL10 sense transcription [14]) at 23°C up to an OD600 of 0.85 and then switched to 37°C for 1 h for ts inactivation of Esa1p prior to harvesting for GAL10 antisense transcript analysis, as done previously (17). Total RNAs from the wild-type and ts mutant strains were isolated and analyzed as described in our recent publication (14) and schematically shown in Fig. 1A. Briefly, a primer (P1) targeted toward the 5′ end of the GAL10 antisense transcript was used for synthesis of cDNA, and subsequently, cDNA was amplified using a primer pair encompassing region M in the GAL10 coding sequence. Such analysis would generate the GAL10 antisense transcript but not sense transcript in dextrose-containing growth medium. However, the use of the P1 primer in the cDNA synthesis can also detect sense transcripts of other genes (e.g., ACT1, ADH1, RPS5, and RPL2B) that are expressed in dextrose-containing growth medium, since the P1 primer can randomly hybridize to RNAs (including mRNAs and rRNAs) at 42°C during cDNA synthesis via matched (A·T and G·C) as well as mismatched (G·T, G·A, A·C, T·C, G·G, A·A, T·T, and C·C) base pairs (33–35), as discussed previously (14). We found that GAL10 antisense transcription was significantly decreased in the esa1-ts mutant strain in comparison to the wild-type equivalent (Fig. 1B and C; see Fig. S1A and B in the supplemental material). Likewise, Esa1p promoted sense transcription of the NuA4-dependent RPS5 gene (Fig. 1B and C). However, sense transcription of the NuA4-independent ACT1 gene (19) was not changed in the esa1-ts mutant strain (Fig. 1B and C). Further, as a control, we used a primer pair targeted to the GAL7 coding sequence (region N) in the above PCR analysis. We did not observe a PCR signal (Fig. 1B), as the GAL7 primer pair (which was successfully used in our previous studies and generated a PCR signal when genomic DNA was used as a template [see Fig. S1C in the supplemental material]) (14) was located upstream of the GAL10 antisense initiation site (Fig. 1A) (14–16) and GAL7 sense transcription does not occur in dextrose-containing growth medium (14). Likewise, using a primer pair (at region P) upstream of the P1 primer (Fig. 1A) in the above RT-PCR analysis, we did not observe a PCR signal (see Fig. S1D in the supplemental material), as the primer pair (which amplified the P region when genomic DNA was used as a template [see Fig. S1C in the supplemental material]) at region P is located outside the P1-generated cDNA of the GAL10 antisense transcript. The absence of the PCR signals at the N region of GAL7 or the P region (Fig. 1A and B; see Fig. S1D in the supplemental material) supports that there was no residual DNA contamination in the above RT-PCR analysis. Collectively, our results support that NuA4 KAT facilitates GAL10 antisense transcription. However, GAL1/10 sense transcription is not regulated by NuA4 KAT (19) (Fig. 1D), while RPS5 and other ribosomal protein genes are dependent on NuA4 KAT for sense transcription (17–19) (Fig. 1D). Consistent with these results, NuA4 KAT was not found to be associated with the GAL1/10 promoter (19) but was associated with RPS5 and other ribosomal protein genes (17–19).
NuA4 KAT associates with the 3′ end of the GAL10 coding sequence.
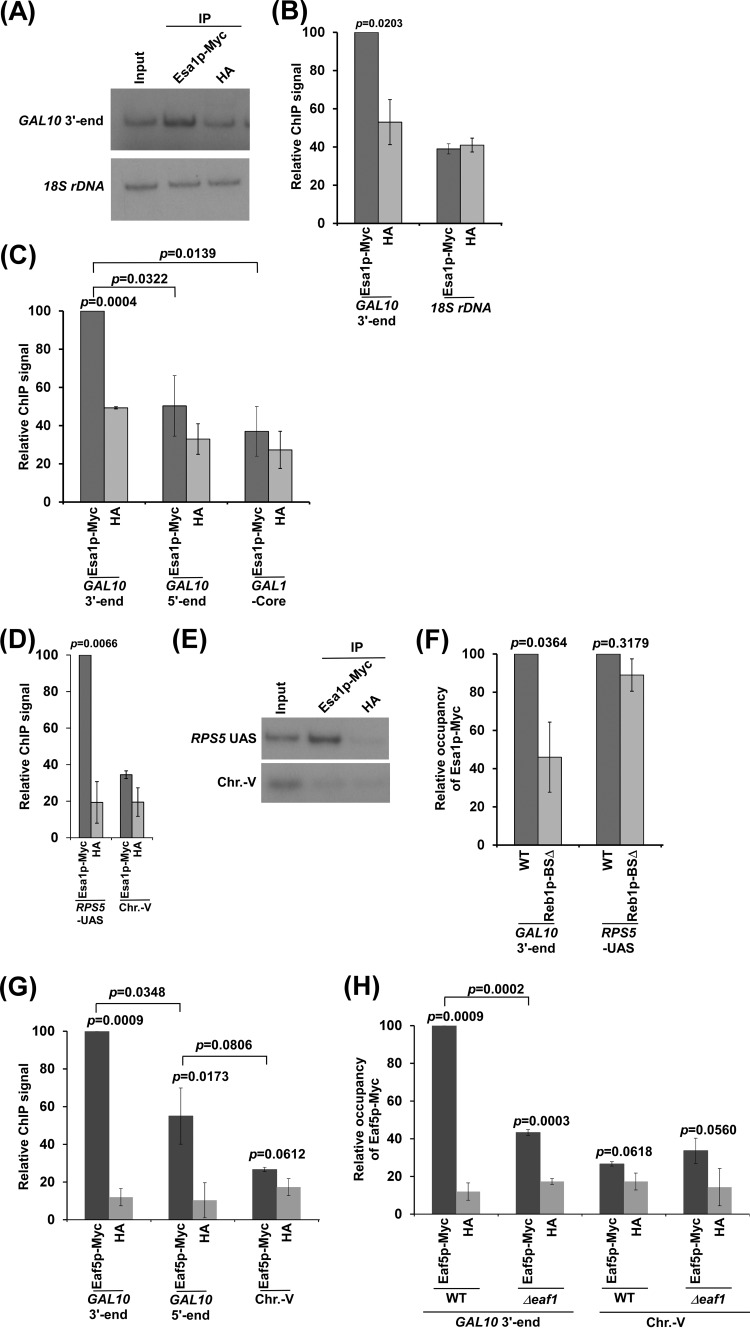
To determine whether NuA4 KAT plays a direct role in facilitating antisense transcription from the 3′ end of the GAL10 coding sequence, we analyzed the association of Myc epitope-tagged Esa1p with the GAL10 3′ end in dextrose-containing growth medium. We found significant association of Myc-tagged Esa1p with the 3′ end of the GAL10 coding sequence (region O in Fig. 1A) in comparison to the nonspecific anti-HA antibody (or background signal) (Fig. 2A and B). As a negative control, we also analyzed the association of Myc-tagged Esa1p with 18S rDNA (or the gene transcribed by RNA polymerase I) and found that the Myc-tagged Esa1p ChIP signal was same as that of anti-HA (or background) (Fig. 2A and B). Further, the PCR analysis for 18S rDNA was carried out within the linear range, as the ratio of the Myc-tagged Esa1p and HA signals was not significantly altered at various dilutions (see Fig. S2 in the supplemental material). Thus, our results support that Esa1p is associated with the GAL10 3′ end where antisense transcription is initiated. However, similar recruitment of Esa1p was not observed at the 5′ end of the GAL10 coding sequence (region M in Fig. 1A) or GAL1 core promoter (region P in Fig. 1A) in dextrose-containing growth medium (Fig. 2C). NuA4 KAT associates with the UASs of the NuA4-dependent ribosomal protein genes for TAF-dependent sense transcription (17–19). Thus, as a positive control, we showed that Esa1p is associated with the UAS of a ribosomal protein gene, RPS5, but not with an inactive region within chromosome V (Chr.-V) (Fig. 2D and E), consistent with previous studies (17, 18). Collectively, our results demonstrate that Esa1p associates with the GAL10 antisense transcription initiation site. Such association of NuA4 KAT with GAL10 is dependent on the Reb1p-binding site (Fig. 2F) that recruits Reb1p to the GAL10 antisense transcription initiation site to trigger transcription (15). As a control, we showed that the targeting of NuA4 KAT to the RPS5 UAS is not altered in the absence of the Reb1p-binding site (Fig. 2F), since the Reb1p-binding site is absent at the RPS5 promoter/UAS. At the RPS5 promoter/UAS, recruitment of NuA4 KAT depends on the RPG box that targets the activator Rap1p (17–19). Thus, our results support that NuA4 KAT is targeted by Reb1p at the GAL10 antisense transcription initiation site. Consistently, the Eaf5p component of NuA4 KAT is predominantly recruited to the 3′ end of the GAL10 coding sequence (Fig. 2G). Further, NuA4 KAT's overall structural integrity is maintained by its Eaf1p component (18, 36, 37). In agreement with this fact, we find that the recruitment of the Eaf5p component of NuA4 KAT to the 3′ end of the GAL10 coding sequence is impaired in the Δeaf1 strain (Fig. 2H). Similarly, NuA4 KAT components are not recruited to the promoters of the ribosomal protein genes in the absence of Eaf1p (18).
FIG 2.
Analysis of NuA4 KAT recruitment to the 3′ end of the GAL10 coding sequence. (A and B) Esa1p is associated with the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium. Immunoprecipitation (IP) was carried out using an anti-Myc antibody against Myc-tagged Esa1p. Immunoprecipitated DNA was analyzed by PCR using the primer pairs encompassing the 3′ end of the GAL10 coding sequence (region O in Fig. 1A) and a region with 18S rDNA. The ratio of the immunoprecipitate over the input in the autoradiogram (termed a ChIP signal) was measured. The maximum ChIP signal was set to 100, and other ChIP signals relative to the maximum ChIP signal (represented as relative ChIP signal or relative occupancy) were plotted. (C, D, and E) ChIP analysis of Myc-tagged Esa1p at the 3′ and 5′ ends (regions O and M, respectively, in Fig. 1A) of the GAL10 coding sequence, the GAL1 core promoter (region P in Fig. 1A), the RPS5 UAS, and an inactive region within chromosome V (Chr.-V). (F) ChIP analysis of Myc-tagged Esa1p at the 3′ end of the GAL10 coding sequence and RPS5 UAS in the presence and absence of a Reb1p-binding site at the 3′ end of the GAL10 coding sequence. (G) ChIP analysis of Myc-tagged Eaf5p at the 3′ and 5′ ends of the GAL10 coding sequence and Chr.-V in dextrose-containing growth medium. (H) ChIP analysis of Myc-tagged Eaf5p at the 3′ end of the GAL10 coding sequence in the wild-type and Δeaf1 strains.
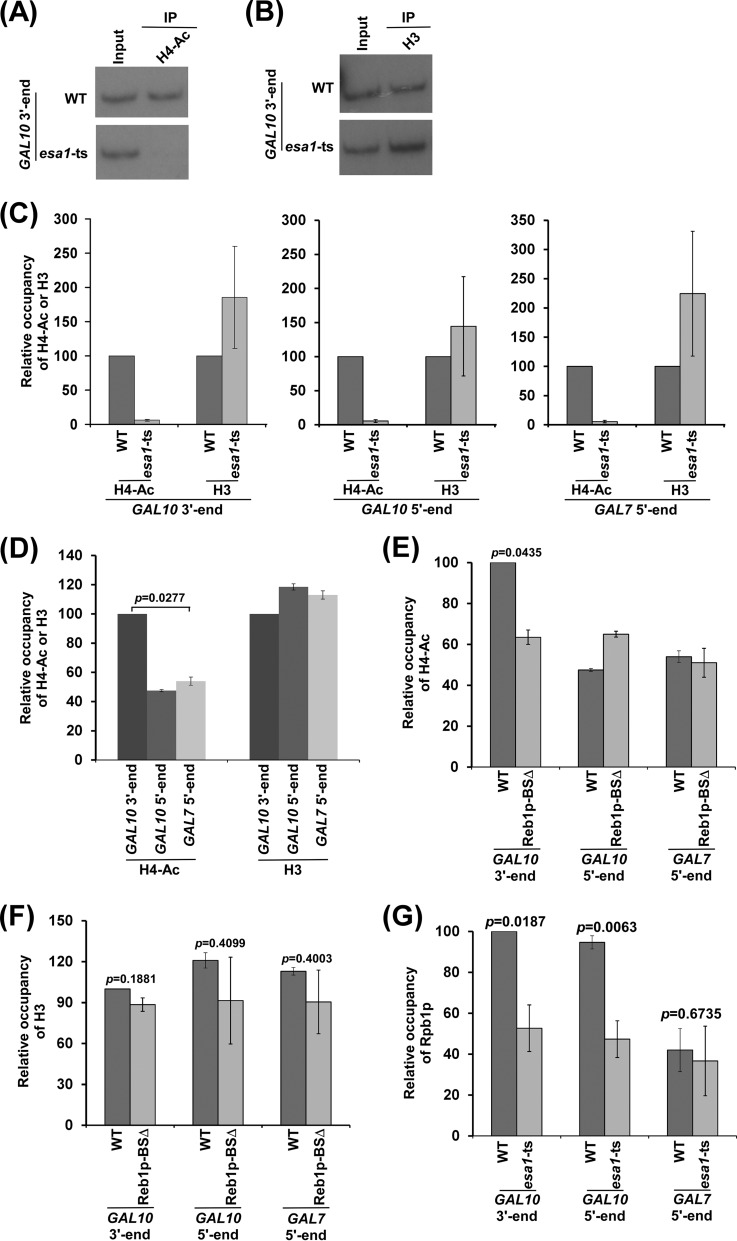
Since NuA4 KAT is associated with the GAL10 antisense transcription initiation site, it is likely to regulate targeted histone H4 acetylation. To test this, we analyzed the level of histone H4 acetylation at the 3′ end of the GAL10 coding sequence in the wild-type and esa1-ts mutant strains at the nonpermissive temperature. We found that histone H4 acetylation at the 3′ end of the GAL10 coding sequence is impaired in the esa1-ts mutant strain in comparison to the wild-type equivalent (Fig. 3A to C). Such a decrease of histone H4 acetylation is not due to the eviction of histone H3/H4 tetramer, since the level of histone H3 (a representative component of histone H3/H4 tetramer) at the GAL10 antisense transcription initiation site is not reduced in the esa1-ts mutant strain (Fig. 3B and C). Further, we found that the 5′ end of the GAL10 coding sequence is also acetylated at histone H4 in an Esa1p-dependent manner (Fig. 3C). Similarly, Esa1p-dependent histone H4 acetylation is also observed at the GAL7 coding sequence (Fig. 3C) and inactive region within Chr.-V (18) in dextrose-containing growth medium. The presence of such histone H4 acetylation at the 5′ ends of the GAL10 and GAL7 coding sequences or Chr.-V is likely due to nontargeted global histone H4 acetylation by piccolo-NuA4 (picNuA4) (which is composed of Esa1p, Yng2p, and Epl1p [38, 39]), as previous studies (38, 39) demonstrated the role of picNuA4 in global histone H4 acetylation in a nontargeted fashion. Consistently, we also observed nontargeted histone H4 acetylation at the promoters of the NuA4-dependent ribosomal protein genes in the absence of NuA4 or Eaf1p (18), and such acetylation is impaired in the esa1-ts mutant (18), hence supporting the role of picNuA4 in nontargeted histone H4 acetylation in the absence of NuA4. Here, we observed a high level of histone H4 acetylation predominantly at the 3′ end of the GAL10 coding sequence (Fig. 3D), and such acetylation was impaired in the absence of the Reb1p (or activator)-binding site (Fig. 3E and F). These results indicate nontargeted histone H4 acetylation at GAL10 in the absence of NuA4 KAT, similar to the results for the ribosomal protein gene and other genes (18). Although picNuA4 is involved in global genome-wide histone H4 acetylation, its nontargeted association with chromatin is not generally observed by the ChIP assay (18, 19), possibly due to weak/transient interaction. Nontargeted histone H4 acetylation is not associated with transcription (19). For example, many genes/promoters (e.g., ADH1, ACT1, and GAL1) are globally acetylated at histone H4 by picNuA4, and such nontargeted histone modification does not modulate their transcription (19). However, targeted histone H4 acetylation by NuA4 KAT regulates transcription. For example, the promoters of the ribosomal protein genes are acetylated at histone H4 by targeted recruitment of NuA4 KAT for stimulation of transcription (17–19). Likewise, targeted histone H4 acetylation at the 3′ end of the GAL10 coding sequence facilitates antisense transcription (Fig. 1B and C; see Fig. S1A and B in the supplemental material) via enhanced recruitment of RNA polymerase II (Fig. 3G).
FIG 3.
Analysis of histone H4 acetylation at the 3′ end of the GAL10 coding sequence. (A to C) Analysis of the levels of histone H4 acetylation and histone H3/H4 tetramer at the 3′ and 5′ ends of the GAL10 coding sequence and GAL7 ORF (or GAL7 5′ end) in the esa1-ts mutant and wild-type strains. Immunoprecipitation was carried out using an anti-histone H4 acetylation antibody against acetylated histone H4 or an anti-histone H3 antibody against histone H3 of the histone H3/H4 tetramer. (D) Analysis of histone H4 acetylation and histone H3 at the 5′ and 3′ ends of the GAL10 coding sequence and 5′ end of the GAL7 coding sequence. (E and F) The Reb1p-binding site regulates histone H4 acetylation at the 3′ end of the GAL10 coding sequence. (G) Analysis of Rpb1p association with GAL10 in dextrose-containing growth medium in the esa1-ts mutant and wild-type strains.
NuA4 KAT-dependent GAL10 antisense transcription is not regulated by TOR, while NuA4 KAT-mediated sense transcription of the ribosomal protein genes is impaired following inhibition of TOR.
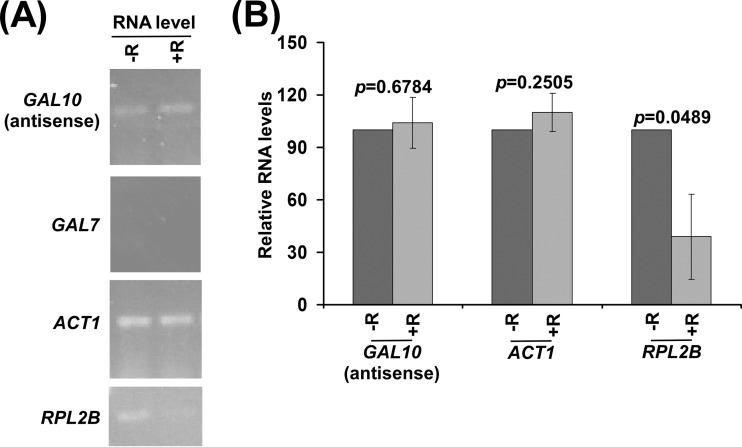
Previous studies (17, 40) demonstrated that the association of NuA4 KAT with the ribosomal protein genes is impaired by rapamycin-mediated inhibition of the TOR pathway. Consequently, sense transcription of the ribosomal protein genes is decreased following rapamycin treatment (40). To determine whether TOR similarly regulates antisense transcription, we analyzed the levels of GAL10 antisense transcripts with or without rapamycin treatment. We found that GAL10 antisense transcription is not altered in response to rapamycin treatment (Fig. 4). As a control, we showed that sense transcription of the NuA4-independent ACT1 gene (17, 19) is not altered following rapamycin treatment (Fig. 4). However, sense transcription of a ribosomal protein gene, RPL2B, is impaired in response to rapamycin treatment (Fig. 4), consistent with previous studies (17, 40). Thus, rapamycin (or the TOR pathway) differentially regulates NuA4 KAT-dependent sense transcription at the ribosomal protein genes (e.g., RPL2B) and antisense transcription at GAL10.
FIG 4.
NuA4 KAT-dependent GAL10 antisense transcription is not altered in response to rapamycin treatment. (A) RT-PCR analysis of the GAL10 antisense transcript with (+R) or without (−R) rapamycin treatment. (B) The transcription data shown in panel A were plotted.
Histone H3 K4 and K36 methyltransferases facilitate GAL10 antisense transcription.
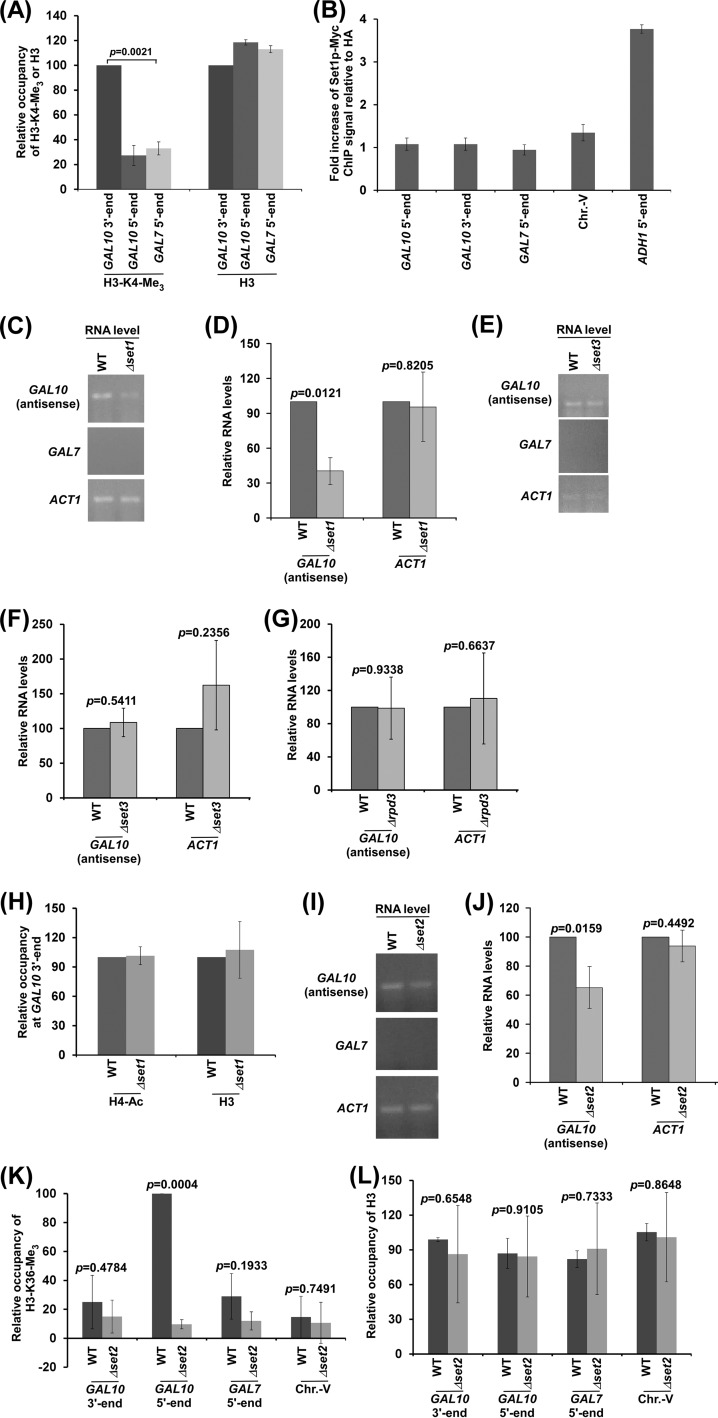
Like histone H4 acetylation, histone H3 K4 trimethylation is also found at the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium (15, 16) (Fig. 5A). Such modification is targeted by Reb1p (15). However, Set1p, which methylates K4 of histone H3 (41), is not found to be associated with the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium (15). Likewise, we also did not observe the association of Set1p with the 3′ end of the GAL10 coding sequence in dextrose-containing growth medium (Fig. 5B), while Set1p is associated with the 5′ end of the coding sequence of ADH1, which is constitutively engaged in sense transcription (Fig. 5B), consistent with previous studies (42). This could be due to weak/transient interaction (and/or cross-linking) of Set1p with the GAL10 antisense transcription initiation site during less frequent antisense transcription, as also suggested in previous studies (15). Nonetheless, Set1p-mediated H3 K4 methylation is observed at the GAL10 antisense transcription initiation site (15) (Fig. 5A). To determine the role of histone H3 K4 methylation in antisense transcription, we analyzed the levels of GAL10 antisense transcripts in the wild-type and deletion mutant strains of SET1. We found that GAL10 antisense transcription is impaired in the Δset1 strain in comparison to the wild-type equivalent (Fig. 5C and D). As a control, we demonstrated that sense transcription of the Set1p-independent ACT1 gene (43, 44) is not altered in the absence of Set1p or histone H3 K4 methylation (Fig. 5C and D). Thus, our results support that histone H3 K4 methylation is associated with GAL10 antisense transcription. Histone H3 K4 methylation has been previously implicated in controlling sense transcription via Rpd3S (histone deacetylase complex small or Rpd3 small) and the Set3p-containing histone deacetylase complex (15, 16, 45). However, we found that Set3p or Rpd3p does not regulate GAL10 antisense transcription (Fig. 5E to G). Thus, Set1p regulates GAL10 antisense transcription independently of Set3p and Rpd3p. Further, such function of Set1p is not mediated via histone H4 acetylation, as we did not observe an alteration of histone H4 acetylation at the GAL10 antisense transcription initiation site in the Δset1 strain in comparison to the wild-type equivalent (Fig. 5H).
FIG 5.
GAL10 antisense transcription is regulated by histone H3 K4 and K36 methyltransferases. (A) Analysis of histone H3 K4 trimethylation (H3-K4-Me3) and histone H3 levels at the 5′ and 3′ ends of the GAL10 coding sequence and the 5′ end of the GAL7 coding sequence. (B) ChIP analysis of Myc-tagged Set1p at the 5′ and 3′ ends of the GAL10 coding sequence, Chr.-V, and 5′ ends of the ADH1 and GAL7 coding sequence. The fold increase of Set1p ChIP signal relative to HA is plotted. (C and D) RT-PCR analysis of GAL10 antisense RNA in the Δset1 and wild-type strains. The transcription data shown in panel C were plotted in panel D. (E and F) RT-PCR analysis of GAL10 antisense RNA in the Δset3 and wild-type strains. (G) RT-PCR analysis of GAL10 antisense RNA in the Δrpd3 and wild-type strains. (H) ChIP analysis of histone H4 acetylation and histone H3 levels at GAL10 in the wild-type and Δset1 strains. (I and J) RT-PCR analysis of GAL10 antisense RNA in the Δset2 and wild-type strains. (K and L) ChIP analysis of histone H3 K36 trimethylation (H3 K36-Me3) and histone H3 at GAL10, GAL7, and Chr.-V in dextrose-containing growth medium. The maximum ChIP signal was set to 100, and other ChIP signals relative to the maximum ChIP signal were plotted.
Previous studies (15) demonstrated Set2p-mediated histone H3 K36 trimethylation at GAL10 under growth conditions permissive to GAL10 antisense transcription. Such covalent modification is dependent on Reb1p-mediated GAL10 antisense transcription (15). Histone H3 K36 methylation has been implicated in repression of sense transcription via the recruitment of Rpd3S (46, 47). In addition, histone H3 K36 methylation may also be involved in regulation of GAL10 antisense transcription. To test this, we analyzed the levels of GAL10 antisense transcripts in the wild-type and deletion mutant strains of SET2. We found that the level of GAL10 antisense transcript is decreased in the Δset2 strain in comparison to the wild-type equivalent (Fig. 5I and J). However, the level of ACT1 sense transcript is not altered in the Δset2 strain (Fig. 5I and J), consistent with previous studies (48). Thus, our results support that histone H3 K36 methylation favors antisense transcription. Further, the deletion of Rpd3p does not alter GAL10 antisense transcription (Fig. 5G). Thus, Set2p regulates GAL10 antisense transcription independently of Rpd3p. Moreover, we found that the GAL10 antisense coding sequence (i.e., the GAL10 5′ end) (Fig. 1A), but not the initiation site (i.e., GAL10 3′ end) (Fig. 1A), is methylated predominantly at K36 of histone H3 (Fig. 5K and L), consistent with previous studies (15), suggesting a role of histone H3 K36 methylation in regulation of GAL10 antisense transcription elongation.
GAL10 antisense transcription is facilitated by histone H2B ubiquitylation.
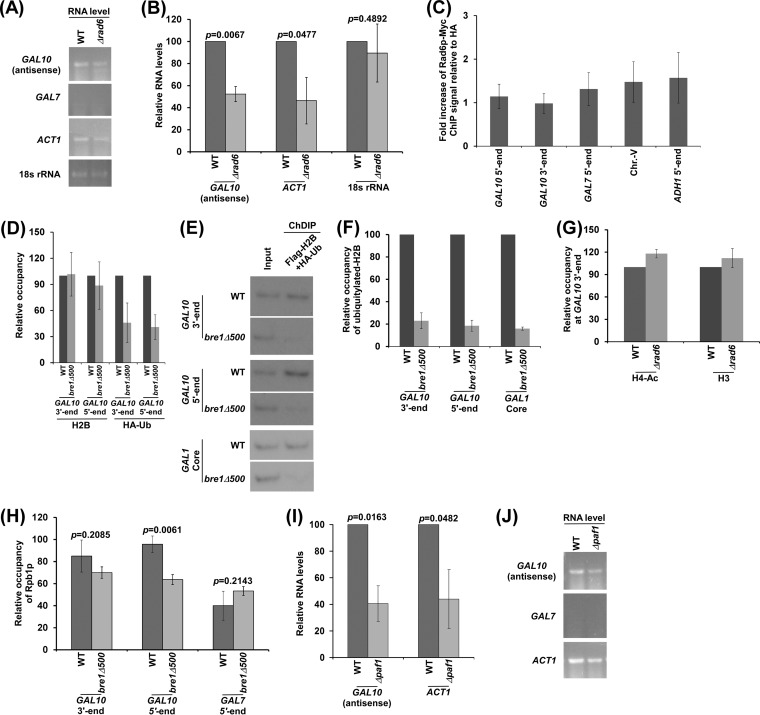
Histone H3 K4 methylation is promoted by histone H2B ubiquitylation during sense transcription (49). Further, histone H2B ubiquitylation has been shown to enhance sense transcription (50, 51). Similar to the regulation of sense transcription, histone H2B ubiquitylation may also control antisense transcription. To test this, we analyzed the levels of GAL10 antisense transcripts in the RAD6 (which has histone H2B ubiquitin conjugase activity for histone H2B ubiquitylation via Bre1p ubiquitin ligase [49]) wild-type and deletion mutant strains. We found that the level of GAL10 antisense transcript was decreased in the Δrad6 strain in comparison to the wild-type equivalent (Fig. 6A and B). Likewise, Rad6p promoted sense transcription of ACT1 (Fig. 6A and B), consistent with previous studies (26, 29). As a loading control, we showed that the 18S rRNA level was not altered in the Δrad6 strain (Fig. 6A and B). Thus, our results support that GAL10 antisense transcription is facilitated by Rad6p (or histone H2B ubiquitylation). However, we could not detect Rad6p ubiquitin conjugase at GAL10 in dextrose-containing growth medium (Fig. 6C). Similarly, Rad6p, which ubiquitylates histone H2B during sense transcription, was also not found to be associated with the active ADH1 gene (Fig. 6C). This could be due to weak/transient association (or poor cross-linking) of Rad6p with the active gene, analogous to the fact that Mediator is not found to be associated with the promoters of the ribosomal protein genes which require the Mediator complex for their transcription (52). Nonetheless, we found histone H2B ubiquitylation at GAL10, and such modification was impaired in the absence of the RING domain (which is essential for histone H2B ubiquitylation) of Bre1p ubiquitin ligase (Fig. 6 to F), consistent with previous studies (53). Further, histone H2B ubiquitylation promotes GAL10 antisense transcription (Fig. 6A and B). Such function of histone H2B ubiquitylation is not mediated via histone H4 acetylation, as we did not observe an alteration of histone H4 acetylation in the Δrad6 strain in comparison to the wild-type equivalent (Fig. 6G).
FIG 6.
GAL10 antisense transcription is regulated by histone H2B ubiquitin conjugase and Paf1p. (A and B) Analysis of GAL10 antisense RNA in the Δrad6 and wild-type strains. (C) ChIP analysis of Myc-tagged Rad6p at the 5′ and 3′ ends of the GAL10 coding sequence, Chr.-V, and 5′ ends of the ADH1 and GAL7 coding sequences. (D) ChIP analysis of Flag-tagged histone H2B and HA-tagged ubiquitin at the 5′ and 3′ ends of the GAL10 coding sequence in the absence of the RING domain of Bre1p (the bre1Δ500 strain without 500 amino acids at Bre1p's C terminus that contain the RING domain). Immunoprecipitation was carried out using anti-Flag and anti-HA antibodies against Flag-tagged histone H2B and HA-tagged ubiquitin as described previously (31). (E and F) ChDIP analysis for the levels of ubiquitylated histone H2B in the wild-type (YKH045) and bre1Δ500 strains expressing Flag-tagged H2B and HA-tagged ubiquitin. (G) Analysis of histone H4 acetylation and histone H3 levels at GAL10 in the wild-type and Δrad6 strains. (H) Analysis of Rpb1p association with the 3′ and 5′ ends of the GAL10 coding sequence in the wild-type and bre1Δ500 strains. The maximum ChIP signal was set to 100, and the other ChIP signals relative to maximum ChIP signal were plotted. (I and J) Analysis of GAL10 antisense RNA in the Δpaf1 and wild-type strains.
The fact that histone H2B is ubiquitylated at the GAL10 5′ and 3′ ends in dextrose-containing growth medium (Fig. 6 to F) indicates that histone H2B ubiquitylation may regulate both GAL10 antisense transcription initiation and elongation. To test this, we analyzed the association of RNA polymerase II with the GAL10 antisense transcription initiation site (i.e., GAL10 3′ end) (Fig. 1A) and coding sequence (i.e., GAL10 5′ end) (Fig. 1A) in dextrose-containing growth medium in the wild-type and bre1Δ500 strains. We found that the loss of histone H2B ubiquitylation in the bre1Δ500 strain (Fig. 6 to F) did not significantly alter the association of RNA polymerase II with the GAL10 antisense transcription initiation site (Fig. 6H). However, RNA polymerase II association with the GAL10 antisense coding sequence was significantly decreased in the bre1Δ500 strain (Fig. 6H). These observations indicate the role of histone H2B ubiquitylation in facilitation of GAL10 antisense transcriptional elongation. Likewise, histone H2B ubiquitylation promotes sense transcriptional elongation (50, 51).
Histone H2B ubiquitylation is regulated by Paf1p or the Paf1p-containing complex (53, 54). Further, Paf1p interacts with RNA polymerase II and promotes sense transcription. We have recently demonstrated that Reb1p targets RNA polymerase II to GAL10 for antisense transcription (14). These observations indicate that Paf1p may regulate GAL10 antisense transcription. To test this, we analyzed the levels of GAL10 antisense transcripts in the wild-type and Δpaf1 strains. We found that GAL10 antisense transcription is significantly decreased in the Δpaf1 strain in comparison to the wild-type equivalent (Fig. 6I and J). Likewise, the loss of Paf1p impairs sense transcription of ACT1 (Fig. 6I and J), consistent with previous studies (29). Thus, our results demonstrate that Paf1p facilitates GAL10 antisense transcription. Similarly, Paf1p also regulates GAL sense transcription (29).
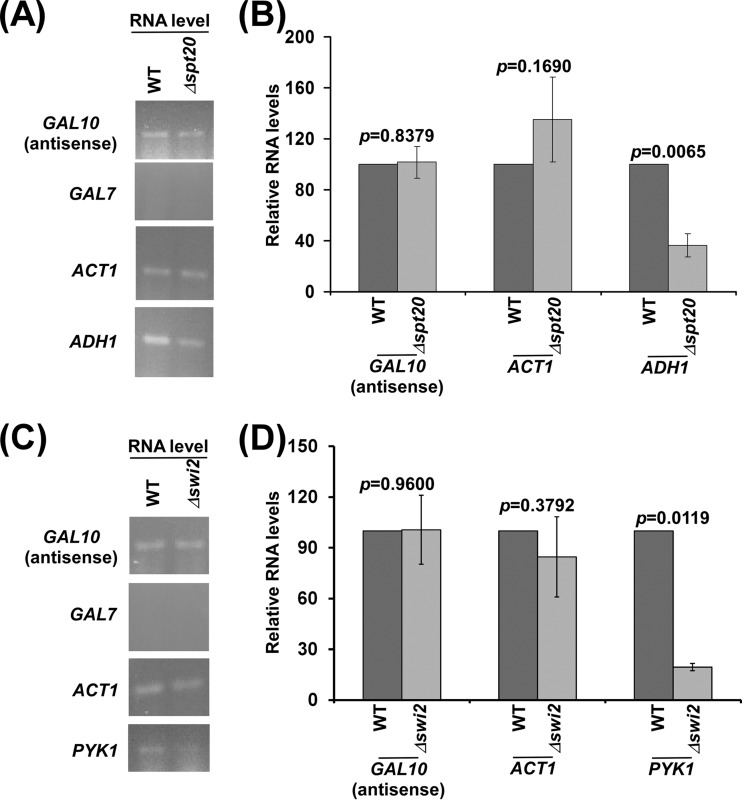
SAGA does not regulate GAL10 antisense transcription but rather promotes sense transcription.
We found that histone H2B ubiquitylation facilitates GAL10 antisense transcription. Histone H2B ubiquitylation is regulated by the histone H2B deubiquitylation activity of SAGA (31, 49). Thus, SAGA may regulate GAL10 antisense transcription. To test this, we analyzed GAL10 antisense transcription in the SPT20 (which impairs the structural and functional integrity of SAGA) wild-type and deletion mutant strains (31, 55). We found that GAL10 antisense transcription is not altered in the absence of Spt20p (Fig. 7A and B). Likewise, transcription of the SAGA-independent ACT1 gene (56, 57) but not the SAGA-dependent ADH1 gene (31) was not altered in the absence of Spt20p (Fig. 7A and B). These results support that SAGA does not regulate GAL10 antisense transcription. However, SAGA promotes GAL1/10 sense transcription (58, 59). Hence, SAGA differentially regulates sense and antisense transcription of GAL10. The fact that SAGA or its histone H2B deubiquitylation activity is dispensable for GAL10 antisense transcription indicates that SAGA may not be targeted to GAL10 under the growth conditions (or dextrose-containing growth medium) permissive to GAL10 antisense transcription. Indeed, SAGA is not targeted to GAL1/10 in dextrose-containing growth medium (59) but is recruited to these genes in galactose-containing growth medium by an activator, Gal4p, for sense transcription (58, 59).
FIG 7.
GAL10 antisense transcription is not regulated by SAGA and Swi/Snf. (A and B) Analysis of GAL10 antisense RNA in the Δspt20 and wild-type strains. (C and D) Analysis of GAL10 antisense RNA in the Δswi2 and wild-type strains.
Swi/Snf does not regulate GAL10 antisense transcription, while it promotes sense transcription.
So far, we had found that GAL10 antisense transcription is facilitated by NuA4 KAT, histone H3 K4 and K36 methyltransferases, and histone H2B ubiquitin conjugase, thus indicating the roles of different histone covalent modifications in regulation of antisense transcription. However, the functions of chromatin remodeling factors in regulation of antisense transcription remain largely unknown. In view of this, we analyzed the role of an important ATP-dependent chromatin remodeling factor, Swi/Snf, that is known to enhance GAL1/10 sense transcription by remodeling chromatin and/or nucleosomal disassembly (23), in regulation of GAL10 antisense transcription. In this direction, we determined the levels of GAL10 antisense transcripts in the SWI2 (which has an ATPase activity) wild-type and deletion mutant strains. We found that the level of the GAL10 antisense transcript was not altered in the Δswi2 strain in comparison to the wild-type equivalent (Fig. 7C and D). Likewise, Swi/Snf did not regulate the sense transcription of ACT1 (Fig. 7C and D), consistent with previous studies (60). However, sense transcription of PYK1 was impaired in the Δswi2 strain (Fig. 7C and D), consistent with previous studies (23). Therefore, our results support that GAL10 antisense transcription is not regulated by Swi/Snf. On the other hand, GAL1/10 sense transcription is promoted by Swi/Snf (61). Thus, GAL10 sense and antisense transcriptions are differentially controlled by Swi/Snf. However, another ATP-dependent chromatin remodeling factor may regulate GAL10 antisense transcription, which remains to be further elucidated.
DISCUSSION
Although antisense transcripts/transcription is involved in regulation of sense transcription and cellular functions, it is not clearly understood how antisense transcription is initiated. We have recently demonstrated that an activator (Reb1p) and a set of GTFs target RNA polymerase II to initiate antisense transcription from the 3′ end of the GAL10 coding sequence (14). Here, our results reveal that NuA4 KAT is targeted to the 3′ end of the GAL10 coding sequence by Reb1p for histone H4 acetylation (Fig. 2 and 3). The loss of NuA4 KAT-mediated histone H4 acetylation is associated with impaired GAL10 antisense transcription (Fig. 1, 2, and 3). Thus, our results support the role of NuA4 KAT in facilitation of GAL10 antisense transcription.
Like its role in stimulation of GAL10 antisense transcription, Reb1p has been shown to promote sense transcription of many housekeeping genes (15, 62–64). Further, intergenic Reb1p-binding sites have been detected in genome-wide ChIP-on-chip studies (65). Moreover, 215 perfect matches to the Reb1p-binding sequences within coding sequences in Saccharomyces cerevisiae have been identified by BLAST analysis (15). Thus, Reb1p is likely to act in these regions to initiate antisense transcription, similar to the results for GAL10. Further, it is yet unknown how Reb1p promotes transcriptional initiation. The results presented here demonstrate that Reb1p targets NuA4 KAT for histone H4 acetylation at the GAL10 antisense transcription initiation site for recruitment of RNA polymerase II and hence GAL10 antisense transcription (Fig. 1, 2, and 3). Reb1p might similarly regulate antisense transcription and/or sense transcription at other genes via NuA4 KAT, which remains to be further elucidated.
Like its recruitment at the GAL10 antisense transcription initiation site, NuA4 KAT is targeted to the promoters of the TAF-dependent ribosomal protein genes by an activator, Rap1p, to enhance their sense transcription (17–19). Transcription of the ribosomal protein genes depends on the TOR pathway via a mechanism that acts on Ifh1p and Fhl1p, which are recruited to the ribosomal protein genes by Rap1p. Although both sense transcription of the ribosomal protein genes and GAL10 antisense transcription depend on NuA4 KAT, the latter requires Reb1p rather than Rap1p and thus may not be regulated by the TOR pathway. We tested this by using rapamycin to inhibit the TOR pathway and indeed found that GAL10 antisense transcription was not affected (Fig. 4).
Further, we have recently demonstrated that the 19S regulatory particle (RP) facilitates the targeting of NuA4 KAT to the promoters of the ribosomal protein genes to enhance sense transcription (17). Consistently, the 19S RP has been shown to promote sense transcription of the NuA4 KAT-dependent ribosomal protein genes (17). It is not clearly known whether other genes that are regulated by NuA4 KAT also require the 19S RP. Our results reveal that NuA4 KAT-regulated GAL10 antisense transcription is independent of the 19S RP (14). Thus, NuA4 KAT-mediated transcription is differentially regulated by the 19S RP. Such a differential requirement of the 19S RP could be dictated by activators, as sense transcription of the ribosomal protein genes and GAL10 antisense transcription are activated by the Rap1p and Reb1p activators, respectively.
Like NuA4 KAT, histone H3 K4 methyltransferase (Set1p) also favors GAL10 antisense transcription (Fig. 5A to D). Histone H3 K4 methylation associated with GAL10 antisense transcription has been implicated in repression of sense transcription (15, 16), thus deciphering the role of antisense transcription in regulation of sense transcription. Our results demonstrate that, in addition to repressing sense transcription, histone H3 K4 methylation also promotes antisense transcription. Like histone H3 K4 methylation (or Set1p), histone H3 K36 methylation or methyltransferase (Set2p) facilitates GAL10 antisense transcription (Fig. 5I and J). Histone H3 K36 methylation has been previously shown to repress cryptic transcription via the recruitment of the Rpd3S complex at the coding sequence during sense transcription (46, 47). Further, previous studies (15) at the GAL10 locus revealed a high level of histone H3 K36 methylation under the growth conditions permissive to GAL10 antisense transcription. Such methylation of histone H3 at the GAL10 locus has been implicated in repression of sense transcription via Rpd3S and hence provided a mechanism for regulation of sense transcription by antisense transcription. However, the role of histone H3 K36 methylation in antisense transcription was not clear. Here, we show that Set2p (or histone H3 K36 methylation) promotes antisense transcription (Fig. 5I and J). Thus, in addition to repressing sense/cryptic transcription, histone H3 K36 methylation also promotes antisense transcription.
Like histone H3 K4/36 methylation, histone H2B ubiquitylation also facilitates GAL10 antisense transcription (Fig. 6A, B, D to F and H). Since the level of histone H2B ubiquitylation is regulated by ubiquitin protease (or deubiquitylation), histone H2B ubiquitin protease or deubiquitylation activity of SAGA is likely to alter the level of GAL10 antisense transcription. To test this, we analyzed the role of SAGA in regulation of GAL10 antisense transcription. SAGA has histone H3 acetylation activity in addition to its histone H2B deubiquitylation activity. Moreover, SAGA functions to stimulate the preinitiation complex formation during sense transcription (58, 59). We found here that SAGA is dispensable for GAL10 antisense transcription (Fig. 7A and B). However, SAGA is essential for GAL1/10 sense transcription (58, 59). Thus, SAGA is differentially required for GAL10 sense and antisense transcription. Like SAGA, the activator Gal4p (which is essential for GAL10 sense transcription) is dispensable for GAL10 antisense transcription (14). These results support that both GAL10 sense and antisense transcriptions are regulated independently and differently. Further, the fact that we did not observe the effect of SAGA on GAL10 antisense transcription is due to the failure of SAGA association with GAL1/10 in the absence of active Gal4p in dextrose-containing growth medium (58, 59). However, some other ubiquitin protease(s) may be involved in regulation of histone H2B ubiquitylation associated with antisense transcription.
Previous studies (23, 66) have demonstrated that histones are evicted during sense transcription. Such nucleosomal disassembly promotes transcription. The Swi/Snf chromatin remodeling complex participates in histone eviction during sense transcription and facilitates transcription (23). We found here that Swi/Snf is not required for GAL10 antisense transcription (Fig. 7C and D), while it promotes GAL1/10 sense transcription (23, 61). Thus, another factor may be involved in altering chromatin structure/disassembly during antisense transcription, which needs to be further elucidated.
In summary, we demonstrate here that NuA4 KAT is associated with the 3′ end of the GAL10 coding sequence for histone H4 acetylation and antisense transcription initiation. However, such a function of NuA4 KAT in stimulation of GAL10 antisense transcription is independent of the 19S RP and TOR pathway. On the other hand, the 19S RP and TOR play important roles in recruiting NuA4 KAT to the promoters of the ribosomal protein genes during sense transcription (17, 40). Thus, NuA4 KAT recruitment is differentially regulated at the Rap1p-dependent ribosomal protein genes and Reb1p-dependent antisense GAL10 to control sense and antisense transcription. Like NuA4 KAT, histone H3 K4 and K36 methyltransferases also facilitate antisense transcription. Similarly, histone H2B ubiquitylation promotes antisense transcription. Collectively, our results illuminate for the first time the chromatin regulation of antisense transcription. Since the aforementioned histone covalent modifications or associated factors are conserved among eukaryotes, similar chromatin regulation of antisense transcription is likely to exist in humans and other eukaryotes.
Supplementary Material
ACKNOWLEDGMENTS
We thank Kevin Struhl, Mary Ann Osley, David Tollervey, Lorraine Pillus, Fred Winston, Ali Shilatifard, and David Stillman for yeast strains and Judy Davie for yeast strains and technical assistance.
Funding Statement
The work in the Bhaumik laboratory was supported by grants from the National Institutes of Health (1R15GM088798-01 and 2R15GM088798-02) and the American Heart Association (15GRNT25700298).
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00808-15.
REFERENCES
- 1.Lapidot M, Pilpel Y. 2006. Genome-wide natural antisense transcription: coupling its regulation to its different regulatory mechanisms. EMBO Rep 7:1216–1222. doi: 10.1038/sj.embor.7400857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lavorgna G, Dahary D, Lehner B, Sorek R, Sanderson CM, Casari G. 2004. In search of antisense. Trends Biochem Sci 29:88–94. doi: 10.1016/j.tibs.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 3.Lee JT. 2009. Lessons from X-chromosome inactivation: long ncRNA as guides and tethers to the epigenome. Genes Dev 23:1831–1842. doi: 10.1101/gad.1811209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR. 2003. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat Genet 34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
- 5.Ohhata T, Hoki Y, Sasaki H, Sado T. 2008. Crucial role of antisense transcription across the Xist promoter in Tsix-mediated Xist chromatin modification. Development 135:227–235. [DOI] [PubMed] [Google Scholar]
- 6.Tisseur M, Kwapisz M, Morillon A. 2011. Pervasive transcription: lessons from yeast. Biochimie 93:1889–18896. doi: 10.1016/j.biochi.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 7.Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, Kawai J. 2005. Antisense transcription in the mammalian transcriptome. Science 309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 8.He Y, Vogelstein B, Velculescu VE, Papadopoulos N, Kinzler KW. 2008. The antisense transcriptomes of human cells. Science 322:1855–1857. doi: 10.1126/science.1163853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrari N, Seguin R, Renzi P. 2011. Oligonucleotides: a multi-targeted approach for the treatment of respiratory diseases. Future Med Chem 3:1647–1662. doi: 10.4155/fmc.11.108. [DOI] [PubMed] [Google Scholar]
- 10.Lu X, Yu Q, Binder GK, Chen Z, Slepushkina T, Rossi J, Dropulic B. 2004. Antisense-mediated inhibition of human immunodeficiency virus (HIV) replication by use of an HIV type 1-based vector results in severely attenuated mutants incapable of developing resistance. J Virol 78:7079–7088. doi: 10.1128/JVI.78.13.7079-7088.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeVos SL, Miller TM. 2013. Antisense oligonucleotides: treating neurodegeneration at the level of RNA. Neurotherapeutics 10:486–497. doi: 10.1007/s13311-013-0194-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gao Z, Cooper TA. 2013. Antisense oligonucleotides: rising stars in eliminating RNA toxicity in myotonic dystrophy. Hum Gene Ther 24:499–507. doi: 10.1089/hum.2012.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Visser ME, Witztum JL, Stroes ES, Kastelein JJ. 2012. Antisense oligonucleotides for the treatment of dyslipidaemia. Eur Heart J 33:1451–1458. doi: 10.1093/eurheartj/ehs084. [DOI] [PubMed] [Google Scholar]
- 14.Malik S, Durairaj G, Bhaumik SR. 2013. Mechanisms of antisense transcription initiation from the 3′ end of the GAL10 coding sequence in vivo. Mol Cell Biol 33:3549–3567. doi: 10.1128/MCB.01715-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Houseley J, Rubbi L, Grunstein M, Tollervey D, Vogelauer M. 2008. A ncRNA modulates histone modification and mRNA induction in the yeast GAL gene cluster. Mol Cell 32:685–695. doi: 10.1016/j.molcel.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinskaya M, Gourvennec S, Morillon A. 2009. H3 lysine 4 di- and tri-methylation deposited by cryptic transcription attenuates promoter activation. EMBO J 28:1697–1707. doi: 10.1038/emboj.2009.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uprety B, Lahudkar S, Malik S, Bhaumik SR. 2012. The 19S proteasome subcomplex promotes the targeting of NuA4 HAT to the promoters of ribosomal protein genes to facilitate the recruitment of TFIID for transcriptional initiation in vivo. Nucleic Acids Res 40:1969–1983. doi: 10.1093/nar/gkr977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uprety B, Sen R, Bhaumik SR. 2015. Eaf1p is required for recruitment of NuA4 in targeting TFIID to the promoters of the ribosomal protein genes for transcriptional initiation in vivo. Mol Cell Biol 35:2947–2964. doi: 10.1128/MCB.01524-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reid JL, Iyer VR, Brown PO, Struhl K. 2000. Coordinate regulation of yeast ribosomal protein genes is associated with targeted recruitment of Esa1 histone acetylase. Mol Cell 6:1297–1307. doi: 10.1016/S1097-2765(00)00128-3. [DOI] [PubMed] [Google Scholar]
- 20.Longtine MS, McKenzie A III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pingle JR. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14:953–961. [DOI] [PubMed] [Google Scholar]
- 21.Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clarke AS, Lowell JE, Jacobson SJ, Pillus L. 1999. Esa1p is an essential histone acetyltransferase required for cell cycle progression. Mol Cell Biol 19:2515–2526. doi: 10.1128/MCB.19.4.2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwabish MA, Struhl K. 2007. The Swi/Snf complex is important for histone eviction during transcriptional activation and RNA polymerase II elongation in vivo. Mol Cell Biol 27:6987–6995. doi: 10.1128/MCB.00717-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Biswas D, Dutta-Biswas R, Mitra D, Shibata Y, Strahl BD, Formosa T, Stillman DJ. 2006. Opposing roles for Set2 and yFACT in regulating TBP binding at promoters. EMBO J 25:4479–4489. doi: 10.1038/sj.emboj.7601333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Roberts SM, Winston F. 1996. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol Cell Biol 16:3206–3213. doi: 10.1128/MCB.16.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kao CF, Hillyer C, Tsukuda T, Henry K, Berger S, Osley MA. 2004. Rad6 plays a role in transcriptional activation through ubiquitylation of histone H2B. Genes Dev 18:184–195. doi: 10.1101/gad.1149604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Malik S, Bagla S, Chaurasia P, Duan Z, Bhaumik SR. 2008. Elongating RNA polymerase II is disassembled through specific degradation of its largest but not other subunits in response to DNA damage in vivo. J Biol Chem 283:6897–6905. doi: 10.1074/jbc.M707649200. [DOI] [PubMed] [Google Scholar]
- 28.Malik S, Chaurasia P, Lahudkar S, Durairaj G, Shukla A, Bhaumik SR. 2010. Rad26p, a transcription-coupled repair factor, is recruited to the site of DNA lesion in an elongating RNA polymerase II-dependent manner in vivo. Nucleic Acids Res 38:1461–1477. doi: 10.1093/nar/gkp1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sen R, Lahudkar S, Durairaj G, Bhaumik SR. 2013. Functional analysis of Bre1p, an E3 ligase for histone H2B ubiquitylation, in regulation of RNA polymerase II association with active genes and transcription in vivo. J Biol Chem 288:9619–9633. doi: 10.1074/jbc.M113.450403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Russell SJ, Johnston SA. 2001. Evidence that proteolysis of Gal4 cannot explain the transcriptional effects of proteasome ATPase mutations. J Biol Chem 276:9825–9831. doi: 10.1074/jbc.M010889200. [DOI] [PubMed] [Google Scholar]
- 31.Shukla A, Stanojevic N, Duan Z, Sen P, Bhaumik SR. 2006. Ubp8p, a histone deubiquitinase whose association with SAGA is mediated by Sgf11p, differentially regulates lysine 4 methylation of histone H3 in vivo. Mol Cell Biol 26:3339–3352. doi: 10.1128/MCB.26.9.3339-3352.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peterson CL, Kruger W, Herskowitz I. 1991. A functional interaction between the C-terminal domain of RNA polymerase II and the negative regulator SIN1. Cell 64:1135–1143. doi: 10.1016/0092-8674(91)90268-4. [DOI] [PubMed] [Google Scholar]
- 33.Bhaumik SR, Chary KV, Govil G, Liu K, Miles HT. 1997. Homopurine and homopyrimidine strands complementary in parallel orientation form an antiparallel duplex at neutral pH with A-C, G-T, and T-C mismatched base pairs. Biopolymers 41:773–784. [DOI] [PubMed] [Google Scholar]
- 34.Tikhomirova A, Beletskaya IV, Chalikian TV. 2006. Stability of DNA duplexes containing GG, CC, AA, and TT mismatches. Biochemistry 45:10563–10571. doi: 10.1021/bi060304j. [DOI] [PubMed] [Google Scholar]
- 35.Cisse II, Kim H, Ha T. 2012. A rule of seven in Watson-Crick basepairing of mismatched sequences. Nat Struct Mol Biol 19:623–627. doi: 10.1038/nsmb.2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mitchell L, Lambert JP, Gerdes M, Al-Madhoun AS, Skerjanc IS, Figeys D, Baetz K. 2008. Functional dissection of the NuA4 histone acetyltransferase reveals its role as a genetic hub and that Eaf1 is essential for complex integrity. Mol Cell Biol 28:2244–2256. doi: 10.1128/MCB.01653-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Auger A, Galarneau L, Altaf M, Nourani A, Doyon Y, Utley RT, Cronier D, Allard S, Côté J. 2008. Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants. Mol Cell Biol 28:2257–2270. doi: 10.1128/MCB.01755-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Friis RM, Wu BP, Reinke SN, Hockman DJ, Sykes BD, Schultz MC. 2009. A glycolytic burst drives glucose induction of global histone acetylation by picNuA4 and SAGA. Nucleic Acids Res 37:3969–3980. doi: 10.1093/nar/gkp270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Selleck W, Fortin I, Sermwittayawong D, Côté J, Tan S. 2005. The Saccharomyces cerevisiae Piccolo NuA4 histone acetyltransferase complex requires the Enhancer of Polycomb A domain and chromodomain to acetylate nucleosomes. Mol Cell Biol 25:5535–5542. doi: 10.1128/MCB.25.13.5535-5542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rohde J, Cardenas ME. 2003. The tor pathway regulates gene expression by linking nutrient sensing to histone acetylation. Mol Cell Biol 23:629–635. doi: 10.1128/MCB.23.2.629-635.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bryk M, Briggs SD, Strahl BD, Curcio MJ, Allis CD, Winston F. 2002. Evidence that Set1, a factor required for methylation of histone H3, regulates rDNA silencing in S. cerevisiae by a Sir2-independent mechanism. Curr Biol 12:165–170. doi: 10.1016/S0960-9822(01)00652-2. [DOI] [PubMed] [Google Scholar]
- 42.Lee JS, Shukla A, Schneider J, Swanson SK, Washburn MP, Florens L, Bhaumik SR, Shilatifard A. 2007. Histone crosstalk between H2B monoubiquitination and H3 methylation mediated by COMPASS. Cell 131:1084–1096. doi: 10.1016/j.cell.2007.09.046. [DOI] [PubMed] [Google Scholar]
- 43.Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T. 2002. Active genes are tri-methylated at K4 of histone H3. Nature 419:407–411. doi: 10.1038/nature01080. [DOI] [PubMed] [Google Scholar]
- 44.South PF, Harmeyer KM, Serratore ND, Briggs SD. 2013. H3K4 methyltransferase Set1 is involved in maintenance of ergosterol homeostasis and resistance to Brefeldin A. Proc Natl Acad Sci U S A 110:E1016–E1025. doi: 10.1073/pnas.1215768110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim T, Xu Z, Clauder-Münster S, Steinmetz LM, Buratowski S. 2012. Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell 150:1158–1169. doi: 10.1016/j.cell.2012.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Keogh MC, Kurdistani SK, Morris SA, Ahn SH, Podolny V, Collins SR, Schuldiner M, Chin K, Punna T, Thompson NJ, Boone C, Emili A, Weissman JS, Hughes TR, Strahl BD, Grunstein M, Greenblatt JF, Buratowski S, Krogan NJ. 2005. Cotranscriptional set2 methylation of histone H3 lysine 36 recruits a repressive Rpd3 complex. Cell 123:593–605. doi: 10.1016/j.cell.2005.10.025. [DOI] [PubMed] [Google Scholar]
- 47.Carrozza MJ, Li B, Florens L, Suganuma T, Swanson SK, Lee KK, Shia WJ, Anderson S, Yates J, Washburn MP, Workman JL. 2005. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123:581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 48.Smolle M, Venkatesh S, Gogol MM, Li H, Zhang Y, Florens L, Washburn MP, Workman JL. 2012. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat Struct Mol Biol 19:884–892. doi: 10.1038/nsmb.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shukla A, Chaurasia P, Bhaumik SR. 2009. Histone methylation and ubiquitination with their cross-talk and roles in gene expression and stability. Cell Mol Life Sci 66:1419–1433. doi: 10.1007/s00018-008-8605-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shukla A, Bhaumik SR. 2007. H2B-K123 ubiquitination stimulates RNAPII elongation independent of H3-K4 methylation. Biochem Biophys Res Commun 359:214–220. doi: 10.1016/j.bbrc.2007.05.105. [DOI] [PubMed] [Google Scholar]
- 51.Fleming AB, Kao CF, Hillyer C, Pikaart M, Osley MA. 2008. H2B ubiquitylation plays a role in nucleosome dynamics during transcription elongation. Mol Cell 31:57–66. doi: 10.1016/j.molcel.2008.04.025. [DOI] [PubMed] [Google Scholar]
- 52.Fan X, Chou DM, Struhl K. 2006. Activator-specific recruitment of Mediator in vivo. Nat Struct Mol Biol 13:117–120. doi: 10.1038/nsmb1049. [DOI] [PubMed] [Google Scholar]
- 53.Kim J, Roeder RG. 2009. Direct Bre1-Paf1 complex interactions and RING finger-independent Bre1-Rad6 interactions mediate histone H2B ubiquitylation in yeast. J Biol Chem 284:20582–20592. doi: 10.1074/jbc.M109.017442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ng HH, Dole S, Struhl K. 2003. The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J Biol Chem 278:33625–33628. doi: 10.1074/jbc.C300270200. [DOI] [PubMed] [Google Scholar]
- 55.Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. 1997. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes Dev 11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- 56.Milgrom E, West RW Jr, Gao C, Shen WC. 2005. TFIID and Spt-Ada-Gcn5-acetyltransferase functions probed by genome-wide synthetic genetic array analysis using a Saccharomyces cerevisiae taf9-ts allele. Genetics 171:959–973. doi: 10.1534/genetics.105.046557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li XY, Bhaumik SR, Green MR. 2000. Distinct classes of yeast promoters revealed by differential TAF recruitment. Science 288:1242–1244. doi: 10.1126/science.288.5469.1242. [DOI] [PubMed] [Google Scholar]
- 58.Larschan E, Winston F. 2001. The S. cerevisiae SAGA complex functions in vivo as a coactivator for transcriptional activation by Gal4. Genes Dev 15:1946–1956. doi: 10.1101/gad.911501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Bhaumik SR, Green MR. 2001. SAGA is an essential in vivo target of the yeast acidic activator Gal4p. Genes Dev 15:1935–1945. doi: 10.1101/gad.911401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Biggar SR, Crabtree GR. 1999. Continuous and widespread roles for the Swi-Snf complex in transcription. EMBO J 18:2254–2264. doi: 10.1093/emboj/18.8.2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Malik S, Chaurasia P, Lahudkar S, Uprety B, Bhaumik SR. 2012. Rad26p regulates the occupancy of histone H2A-H2B dimer at the active genes in vivo. Nucleic Acids Res 40:3348–3363. doi: 10.1093/nar/gkr1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Packham EA, Graham IR, Chambers A. 1996. The multifunctional transcription factors Abf1p, Rap1p and Reb1p are required for full transcriptional activation of the chromosomal PGK gene in Saccharomyces cerevisiae. Mol Gen Genet 250:348–356. [DOI] [PubMed] [Google Scholar]
- 63.Schuller HJ, Schutz A, Knab S, Hoffmann B, Schweizer E. 1994. Importance of general regulatory factors Rap1p, Abf1p and Reb1p for the activation of yeast fatty acid synthase genes FAS1 and FAS2. Eur J Biochem 225:213–222. doi: 10.1111/j.1432-1033.1994.00213.x. [DOI] [PubMed] [Google Scholar]
- 64.Remacle JE, Holmberg S. 1992. A REB1-binding site is required for GCN4-independent ILV1 basal level transcription and can be functionally replaced by an ABF1-binding site. Mol Cell Biol 12:5516–5526. doi: 10.1128/MCB.12.12.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harbison CT, Gordon DB, Lee TI, Rinaldi NJ, Macisaac KD, Danford TW, Hannett NM, Tagne JB, Reynolds DB, Yoo J, Jennings EG, Zeitlinger J, Pokholok DK, Kellis M, Rolfe PA, Takusagawa KT, Lander ES, Gifford DK, Fraenkel E, Young RA. 2004. Transcriptional regulatory code of a eukaryotic genome. Nature 431:99–104. doi: 10.1038/nature02800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Durairaj G, Chaurasia P, Lahudkar S, Malik S, Shukla A, Bhaumik SR. 2010. Regulation of chromatin assembly/disassembly by Rtt109p, a histone H3 Lys56-specific acetyltransferase, in vivo. J Biol Chem 285:30472–30479. doi: 10.1074/jbc.M110.113225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.