Abstract
Polycomb chromatin modifiers regulate hematopoietic pluripotent stem and progenitor cell self-renewal and expansion. Polycomb complex redundancy and biochemical heterogeneity complicate the unraveling of the functional contributions of distinct components. We have studied the hematopoietic activity of RYBP, a direct interactor and proposed modulator of RING1A/RING1B-dependent histone H2A monoubiquitylation (H2AUb). Using a mouse model to conditionally inactivate Rybp in adult hematopoiesis, we have found that RYBP deletion results in a reversion of B-1-to-B-2 B-cell progenitor ratios, i.e., of the innate (predominantly fetal) to acquired (mostly adult) immunity precursors. Increased numbers of B-1 progenitors correlated with a loss of pre-proB cells, the B-2 progenitors. RYBP-deficient stem and progenitor cell populations (LKS) and isolated common lymphoid progenitors (CLP) gave rise to increased numbers of B-1 progenitors in vitro. Rybp inactivation, however, did not result in changes of global H2AUb and did not interact genetically with Ring1A or Ring1B deletions. These results show that a sustained regulation of the B-1-to-B-2 switch is needed throughout adult life and that RYBP plays an important role in keeping B-2 dominance, most likely independently of its Polycomb affiliation.
INTRODUCTION
RING1 and YY1 binding protein (RYBP) is a component of a subset of type I Polycomb repressive complexes (PRC1), chromatin regulators endowed with histone H2A monoubiquitylating activity (recently reviewed in references 1 and 2). RYBP contacts the C-terminal region of RING1B and its paralog, RING1A (3), heterodimeric RING-type E3 ubiquitin ligases that modify lysine 119 of histone H2A (H2AUb) (4, 5). Canonical PRC1 assemblies are characterized by the presence of both chromobox-containing subunits and oligomerizing SAM motif subunits, PHC proteins (6, 7). Noncanonical PRC1 complexes, on the other hand, contain RYBP or its paralog, YY1-associated factor (YAF1), instead of chromobox proteins (6, 7), as their association with RING1 proteins is mutually exclusive (8). PRC1 association with chromatin not only modifies H2A but also promotes compaction, a structural alteration that usually correlates with transcriptionally repressed states (6, 9–11). PRC1 is recruited to chromatin through PRC2-induced H3K27me3-dependent and -independent mechanisms (6, 7, 12). In vitro, RYBP-PRC1 is more efficient at histone monoubiquitylation than canonical PRC1 complexes (6), an observation consistent with in vivo evidence showing decreased levels of global H2AUb upon short-hairpin downregulation of RYBP (6, 7, 13). However, the relative contributions to the H2A modification of RYBP- or chromobox-containing PRC1 complexes still are controversial (6, 14), possibly because of cell type variations.
In the mouse, genetic analysis of PRC1 functions in differentiation often has focused on the hematopoietic compartment (summarized in references 15 and 16) as one of the best known hierarchies of related cell lineages (17). Most defects in PRC1 mutant lines pertain to the maintenance of stem/progenitor cells and the subsequent expansion of their differentiated derivatives. This is due in part, but not completely, to the derepression of tumor suppressors encoded by the Ink4a locus, which is associated with PRC1 product loss of function (18–22, 25). As a consequence, these mutant mouse lines develop hypoplastic bone marrow (BM) and secondary hematopoietic organs, i.e., spleen and thymus (16). Less frequently, cell lineage defects, such as skewed differentiation toward lymphoid derivatives of Bmi1/Pcgf4-deficient multipotent progenitors, also have been observed (23, 24). In general, most investigated Polycomb mouse models bear mutations in loci encoding either the RING finger proteins that make the core E3 ligase (16, 25, 26) or subunits specific for canonical PRC1 complexes (16).
Here, we investigated noncanonical PRC1 subunit RYBP function in adult hematopoiesis. Rybp genetic analysis is hampered by the embryonic lethality of the inactive allele (27). Using a mouse line in which the Rybp gene can be conditionally deleted in hematopoietic tissues, we find enlarged numbers of mature (peritoneal B-1 cells) and immature (uncommitted and committed bone marrow progenitors) B-1 cells and a concomitant decrease in numbers for the B-2 cell compartment. The observation is consistent with RYBP acting to maintain the ontogenic switch by which numbers of innate immunity B-1 cells, predominant in fetal and newborn stages, decrease while acquired-immunity B-2 cells prevail.
MATERIALS AND METHODS
Rybp conditional inactivation and other mouse lines.
Genomic Rybp sequences to construct a targeting plasmid were isolated from a λ phage obtained from a mouse 129SVJ genomic library. In the targeting vector (Fig. 1), sequences spanning exons 3 to 5 (i.e., all potential coding regions interacting with Polycomb subunits and other proteins [3, 28]) were flanked by loxP sites (recognized by Cre recombinase). A PGK-neo selection cassette, flanked by frt sites (recognized by flippase [FLP] recombinase), was located at intervening sequence 3, and at the end of the short homology arm a PGK-DTA cassette was inserted for positive and negative selection. The linearized plasmid was electroporated into R1 embryonic stem (ES) cells grown on a layer of mitotically inactive mouse embryonic fibroblasts as feeder cells. Colonies surviving the G418/ganciclovir selection were transferred into duplicate 48-well plates. Successfully targeted events were identified by Southern blotting using probes external to the homology arms in the targeting vector. Chimeric mice were generated by aggregation of mutant ES cells with C57BL/6 morulae and subsequently mated to C57BL/6 mice. Heterozygous animals then were backcrossed to C57BL/6 mice. Crosses with a mouse line ubiquitously expressing the FLP recombinase were done to remove the PGK-neo cassette. In vivo Rybp inactivation, in 8- to 12-week-old mice, was done in animals previously mated to the Mx1-Cre transgenic line, in which Cre expression is induced predominantly in the hematopoietic compartment (29) after intraperitoneal (i.p.) administration of polyinosine-polycytidine [poly(I·C); three injections, 250 μg poly(I·C) each, every other day; GE Healthcare]. For ex vivo Rybp inactivation, mice were mated to a mouse line carrying an IRES-Cre-ERT2 cassette knocked in at the 3′ untranslated region (UTR) of the Polr2a locus (30) that expresses a 4′-hydroxytamoxifen (4′-OHT)-inducible Cre recombinase. Ring1A−/−, Ink4a−/−, and Ring1Bf/f lines were described previously (25, 31, 32). Mouse handling procedures were institutionally approved and were in accordance with national and European regulations.
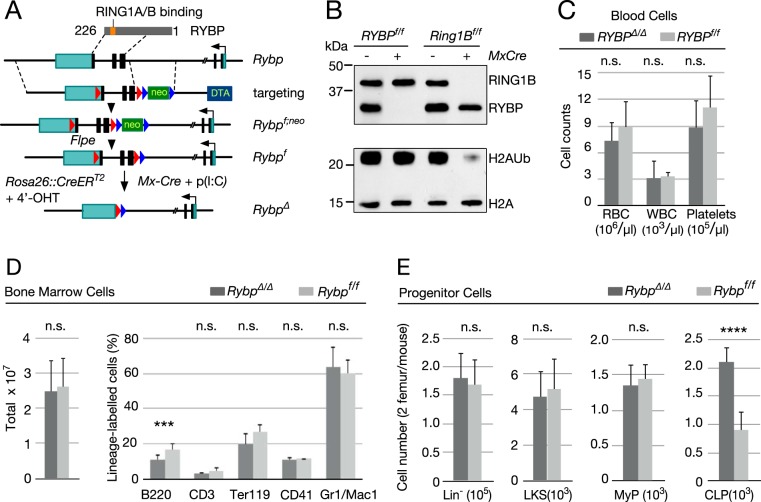
FIG 1.
Analysis of Rybp-deficient hematopoietic cells. (A) Schematic representation of strategy used to conditionally inactivate Rybp. (Top) Rybp cDNA denoting Zn finger (ZnF) and C-terminal region that interacts with Polycomb RING1A and RING1B. Rybp coding sequences are shown as black rectangles, and 5′ and 3′-UTR sequences are shown as green rectangles; the broken line in the Rybp locus is due to the large size of intervening sequence that is not represented at the scale used. Dashed lines delineate homology arms contained in the targeting construct, interrupted by a neo cassette flanked by frt sites (blue arrowheads); Rybp coding sequences and the neo cassette are flanked by loxP sites (red arrowheads). The Rybpf allele was generated by mating Rybpf;neo mice to transgenic mice expressing FLPE recombinase. Subsequent mating to MxCre mice or to mice expressing a Cre-ER fusion allowed for in vivo or ex vivo inactivation after poly(I·C) injection or exposure to 4′-hydroxytamoxifen (4′-OHT), respectively. (B) H2AUb levels in Rybp-depleted hematopoietic cells. Shown are representative Western blots of total lysate (top) or histone (bottom) extracts from cKit+ cells from mice of the indicated genotypes. Extracts from Ring1B-deficient hematopoietic cells (on the right) were used as controls. On the left are molecular mass markers. (C) Absolute numbers of circulating cells in Rybpf/f (Cre−) and RybpΔ/Δ (Cre+) mice. RBC, red blood cells; WBC, white blood cells. (D) Bone marrow cells from mice of the indicated genotype. Left, total absolute cell numbers. Right, relative numbers of lineage markers: B and T cell (B220+ and CD3+, respectively), erythroid (Ter119+), megakaryocytic (CD41+), and myeloid (Mac1+/Gr1+). (E) Absolute numbers of immature cell populations from mice of the indicated genotype. Lineage marker-negative bone marrow cells (Lin− population, containing HSCs and all progenitor cells), Lin− ckit+ Sca1+(LKS population, including long- and short-term HSCs), Lin− Sca1− ckit+ (MyP population, corresponding to myeloid progenitors), and Lin− Sca1lo ckit+ IL-7+ (CLP population, corresponding to common lymphoid progenitors) were used. Ten (C and E) or 20 (D) mice were analyzed for each genotype. Data are mean values and standard deviations (SD). ****, P ≤ 0.0001; n.s., not statistically significant (P ≥ 0.05).
Transplantation procedures.
Transplants were performed using at least 6 recipients, with 2 or 3 recipients per donor mouse per genotype. Transplanted animals were provided with acidified sterilized drinking water supplemented with 100 μg/ml neomycin and 5,000 U/ml polymyxin B from 1 week before to 2 weeks after transplant. Animals were irradiated in a Shepherd Mark I-30 gamma irradiator and transplanted within 24 h. Five million CD45.2+ Rybpf/f MxCre+ or Rybpf/f MxCre− BM cells were injected intravenously (i.v.) into lethally irradiated (10-Gy) CD45.1+ recipient mice. At week 6 after transplant, blood was extracted and analyzed for CD45.1/CD45.2 chimerism by flow cytometry. At week 4 to 6 after transplant, only mice with more than 90% CD45.1+ circulating cells were further used for Rybp inactivation.
Hematopoietic cell isolation.
BM cells were flushed out of both femurs and tibiae with phosphate-buffered saline containing 2% fetal calf serum (PBS–2% FCS), and cells were counted in a hemocytometer. Single-cell suspensions of spleens were obtained by homogenization in PBS–2% FCS and gentle straining through nylon mesh. The peritoneal cavity was gently washed with 10 ml PBS, and cells were recovered, counted, and analyzed. Pup livers were homogenized in PBS–2% FCS and equally strained through nylon mesh.
Flow cytometry analysis and isolation.
Cell separation was performed by immunodepletion followed by cell sorting in a FACSVantageSE-Diva flow cytometer (Becton Dickinson). Lineage-negative (Lin−) or Lin−/B220 cells from bone marrow and perinatal livers were prepared by using a lineage cell depletion kit (Miltenyi Biotec) or the individual biotin-labeled monoclonal antibodies against mature hematopoietic cells except for B220, respectively. LD columns together with a QuadroMACS separation unit from Miltenyi Biotec were used. Lin− cells were stained with antibodies against CD117 (cKit, 2B8)-fluorescein isothiocyanate (FITC), phycoerythrin (PE), allophycocyanin (APC), or biotin; Sca1 (D7)-FITC, peridinin chlorophyll protein (PerCP)-Cy5.5, or biotin (E13-161.7); or CD127 (IL7R, B12.1)-PE or biotin for the identification of most immature (LKS; Lin− ckit+ Sca1+), myeloid progenitor (MyP; Lin− ckit+ Sca1−), or common lymphoid progenitor (CLP; Lin− ckit+Sca1int CD127+) cells. For the analysis of mature differentiated cell types, total BM, spleen, peritoneal cavity, or perinatal liver cells were resuspended in staining buffer (PBS with 2% FCS and 0.1% sodium azide) and blocked for 5 min at 4°C with anti-mouse BD Fc block (Becton Dickinson) and further incubated for 20 min with the appropriate antibodies: anti-CD11b (Mac-1, M1/70)-PerCP-Cy5.5 or biotin; Gr1 (RB6-8C5)-FITC or biotin; CD3e (500A2)-biotin, CD45R (B220, RA3-6B2)-FITC, biotin, APC-Cy7, or APC-Alexa Fluor 750 (AF750); CD19 (1D3)-PE, biotin, PerCP-Cy5.5, or PE-Cy7; Ter119-PE or biotin; CD45.1 (A20)-PE; CD45.2 (104)-FITC; CD21 (7G6)-FITC; CD23 (B3B4)-PE; IgD (11-26c.2a)-FITC; IgM (R6-60.2)-PE or biotin; CD93 (AA4.1, C1qRp)-APC; or CD5 (53-7.3)-FITC. Streptavidin conjugates used were PE, PerCP-Cy5.5, APC, APC-Cy7, PE-Cy5, and Pacific Blue. All monoclonal antibody conjugates were purchased from Becton Dickinson, except for CD19-PE (from Miltenyi), B220-AF750 and CD93-APC (from eBioscience), and CD19-PE-Cy7, IgM-biotin, and streptavidin-APC (from BioLegend). Propidium iodide or 4′,6-diamidino-2-phenylindole (DAPI) was used for live-dead cell exclusion. Fluorescence-activated cell sorter (FACS) analysis was performed on a FACScan or FACSCanto instrument (Becton Dickinson) when four to seven colors were used. Mature bone marrow cells were defined as B and T cell (B220+ and CD3+, respectively), erythroid (Ter119+), megakaryocytic (CD41+), and myeloid (Mac1+/Gr1+). Spleen and peritoneal cavity (PerC) populations are described below. B-1 and B-2 progenitors were defined as Lin− CD19+ B220− ckit+ AA4.1+ and Lin− CD19− B220int ckitlo AA4.1+, respectively. Relevant gates used to analyze the different populations shown in the figures are indicated in Fig. S1 to S3, which are available at https://copy.com/EJdQJFI0BYvq4DAo. Data were analyzed with FlowJo software v.8.8.6 (Tree Star, Ashland, OR) using the bi-exponential scaling (“logicle” transformation) (33). Five percentiles, including outliers, and high-resolution contour plots from 5,000 to 10,000 cells are shown.
B-1P culture.
Medium for B-1 progenitor (B-1P) culture was RPMI medium supplemented with 10% FCS, 50 μM ß-mercaptoethanol (Merck), 1 mM l-glutamine, 100 U/ml streptomycin, 100 μg/ml penicillin, and 20 ng/ml interleukin-3 (IL-3) or 10% WEHI conditioned medium, 20 ng/ml IL-6, 20 ng/ml SCF, 10 ng/ml Flt3 ligand, and 10 ng/ml human thymic stromal lymphopoietin (TSLP) (all cytokines were from Peprotech). S17 cells were maintained in alpha-minimum essential medium with 5% FCS, 1 mM l-glutamine, 100 U/ml streptomycin, and 100 μg/ml penicillin. When subconfluent, cells were trypsinized and seeded at 0.6 × 105 cells per cm2 into 0.4-μm Transwell inserts (Falcon) the day before coculture. The original protocol (34) was followed. Sorted pools enriched for B-1 progenitors (Lin− B220− CD19+) were seeded at a concentration of 1,500 cells per ml of B-1P medium with human TSLP at 10 ng/ml or at different concentrations as indicated. The inserts with S17 cell confluent monolayers then were inserted into 6- or 12-well plates containing seeded progenitors at 3 or 1.5 ml per well, respectively. Cultures were incubated at 37°C in 5% CO2, one-third of the medium was changed twice weekly, and the generated cells were analyzed at the indicated times. Newly S17-seeded Transwells were changed after 8 to 10 days of culture or when signs of disrupted monolayer were observed. All media and supplements were from Life Technologies.
Proliferation and apoptosis analysis.
In vivo 5-bromodeoxyuridine (BrdU; Becton Dickinson) labeling was performed by i.p. injection of 0.1 ml solution (10 mg/ml) per mouse. Sixteen hours later, myeloid and B1 progenitors were isolated, fixed, permeabilized, and stained using an FITC BrdU flow kit (Becton Dickinson). Early apoptosis was assessed by staining with an annexin V/FITC kit (Bender MedSystems).
Western blotting.
Cells were lysed in Laemmli buffer with protease inhibitors for 5 min at 95°C and sonicated 3 times (30-s on/off cycles) with a Bioruptor Diagenode. Cell lysates were cleared of debris by centrifugation at 15,000 × g for 15 min. For monoubiquitylated histone H2A analysis, cell pellets were lysed in PBS containing 0.5% Triton X-100, 2 mM phenylmethylsulfonyl fluoride, 10 mM iodoacetamide, and protein inhibitors (Complete; Roche). Histones then were extracted in 0.2N HCl. For protein and histone analysis, a volume equivalent to 105 cells was subjected to SDS-PAGE and transferred to nitrocellulose membrane (Amersham Protran; GE Healthcare) for 1 h at 2 mA/cm2 on a wet transfer apparatus (Bio-Rad). After being blocked in Tris-buffered saline containing 0.1% Tween 20 and 5% nonfat dry milk, filters were incubated overnight at 4°C with the following antibodies: anti-RING1B and anti-RYBP (3), anti-histone H2A (Millipore), anti-histone H2AK119Ub1 (D2754; Cell Signaling), and anti-α-tubulin (clone B-5-1-2; Sigma). After a washing step and incubation with horseradish peroxidase-conjugated secondary antibodies (Dako), signals were detected using an enhanced chemiluminescence system (GE Healthcare).
Kidney histology and pathology assessment.
Kidney fixation was carried out for 24 h in 10% neutral buffered formalin, and samples were further dehydrated and embedded in paraffin wax. Sections (3 μm through the center of the longitudinal axis of each whole kidney) were stained with hematoxylin and eosin (HE), Periodic acid-Schiff (PAS), and methenamine silver stains. Images were acquired using an Olympus PRovis AX70R microscope (Tokyo, Japan) with a Nikon DXM 1200W digital camera. At least 60 glomeruli per animal were evaluated in fixed and stained kidney samples for cellular hypertrophy and hyperplasia, adhesions, sclerosis, or hyalinosis. Lesions were scored according to the number of affected glomeruli and degree of damage: 0 (<5%), I (5 to 25% mild or 5 to 10% moderate to severe), II (25 to 50% mild or 10 to 25% moderate to severe), and III (>50% mild or >25% moderate to severe). Interstitial inflammation and tubular injury also were graded semiquantitatively on a scale of 0 to 3+. Lesion evaluation and grading were assessed according to previously described methods (35–37).
Statistical analysis.
Data sets were compared using two-tailed Student's t tests, except for kidney lesions, for which a Mann-Whitney test was used. Statistically significant differences were considered and are indicated for P values below 0.05 in each figure legend.
RESULTS
Altered bone marrow B-cell differentiation in Rybp-deficient hematopoiesis.
Poly(I·C)-induced deletion of floxed Rybp alleles (Rybpf) removes coding sequences (Fig. 1A) downstream of the zinc finger (RybpΔ/Δ allele). Efficient Rybp inactivation in bone marrow cells was assessed by Western blotting (Fig. 1B, top) but did not affect global H2AUb levels, in contrast to the observed decrease in Ring1B-depleted bone marrow cells (Fig. 1B, bottom).
Circulating or total bone marrow cells (Fig. 1C and D) did not show gross alterations between mutant and control mice. The relative proportion of erythroid, megakaryocytic, myeloid, and T-cell precursors was not affected by Rybp depletion; however, the B-cell pool significantly decreased by one-third in mutant mice (from 17.9% to 11.2%, as shown in Fig. 1D, right). Immature populations as a whole lineage-negative (Lin−) Sca1+ ckit+ (LKS) pool or containing hematopoietic stem cell (HSC) or myeloid progenitor (MyP) subpopulations showed no alteration (Fig. 1E). Only the lymphoid-committed progenitor pool (CLP) was enlarged in Rybp-deficient mice (Fig. 1E, right).
To determine if the decreased B-cell pool (B220+ cells) involved all maturation stages or only a given subpopulation, we focused on B-cell markers B220 and CD19. As seen in Fig. 2A, cell distribution in RybpΔ/Δ total bone marrow cells departed from the canonical pattern observed with Rybpf/f cells. Of the two canonical CD19+ cells, the population with the brightest B220+ staining had similar labeling in both mutant and wild-type bone marrow; however, the dimmer B220+ pool appeared systematically shifted toward a lower, even negative fluorescence, as shown in representative plots (Fig. 2A, left). Mutant B cells contained a substantial fraction of CD19+ cells expressing little (B220lo) or no B220 (even below the classical quadrant boundaries drawn according to single-labeled controls), whereas the content of CD19− B220+ cells was significantly decreased. These two populations contain progenitors for B-1 (B-1 progenitors [B-1P]) and for B-2 (pre-proB cells or B-2 progenitors [B-2P]) lymphocytes (38), respectively.
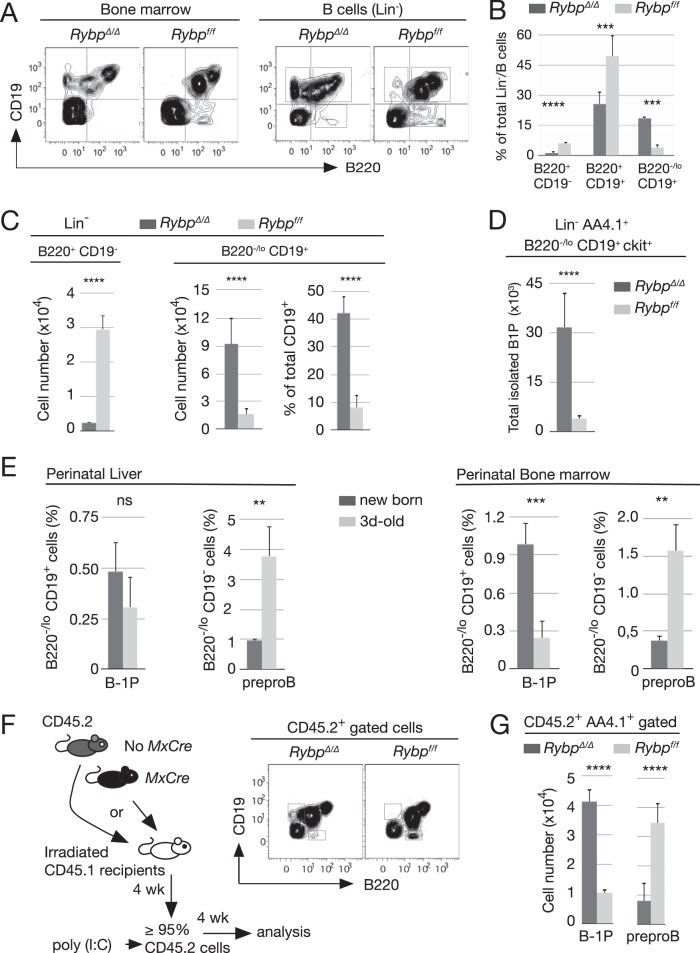
FIG 2.
Cell-autonomous reversal of the B-1 progenitors to pre-proB cell ratio in Rybp-deficient bone marrow. (A) Representative flow cytometry plots of total bone marrow cells or of Lin−/B cells (lineage-depleted cells, except for B220) labeled with antibodies against B220 and CD19 markers. (B) Relative size of B-cell subpopulations (gated as indicated in the corresponding plots) from control (Rybpf/f) and mutant (RybpΔ/Δ) Lin−/B220 cells. (C) Absolute and relative cell numbers of B220−/lo CD19+ and B220+ CD19− subsets within Lin− CD19+ (left) or Lin− B220+ (right) populations in mice of the indicated genotypes. (D) Increased cell numbers of mutant B1 progenitors (Lin− AA4.1+ B220−/lo CD19+ ckit+). (E) B220/CD19-labeled cells in newborn and mice at day 3. Bar graphs show means and SD of relative contents in Lin− CD19+ B220−/lo and Lin− CD19− B220med populations of six pups/experiment from two independent experiments. (F, left) Schematic representation of hematopoietic transplantation of 5 × 106 total bone marrow CD45.2+ Rybpf/f Cre− or Cre+ cells into lethally irradiated CD45.1+ mice; 4 weeks later, hosts in which ≥90% of circulating cells were CD45.2+ were treated with poly(I·C) and sacrificed for analysis 4 weeks later. (Right) Representative plot of B220/CD19-labeled cells. (G) Absolute number of donor B220−/lo CD19+ and B220+ CD19− compartments containing B1 progenitors and pre-proB cells, respectively, of the indicated genotypes. Data are from two independent experiments (n = 6 and n = 4 for each genotype in transplants 1 and 2, respectively). Bar graphs show mean values and SD. In panels B and C, ten mice were analyzed of each genotype. **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; n.s., not statistically significant (P ≥ 0.05).
For the quantification of B cells and immature cells, bone marrow cells were immunomagnetically depleted of cells expressing lineage-specific markers except B220. The unlabeled population (Lin−/B cells) was stained with anti-B220 and CD19 antibodies (Fig. 2A, right). The ratio of B-1P- and B-2P-containing pools was inverted within mutant Lin−/B-cell populations (Fig. 2B), and CD19− B220+ and CD19+ B220−/lo cell numbers were equally shifted (Fig. 2C, left and middle bar graphs). In fact, only a small proportion of wild-type CD19+ cells expressed little or no B220 (less than 10%), and nearly 50% of mutant CD19+ cells were B220−/lo (Fig. 2C, right bar graphs). Thus, RYBP deficiency profoundly modified two B-cell compartments, decreasing by 80% the size of the pre-proB-containing population and enlarging by at least 5-fold the physiologically scarce B-1P-containing B220−/lo CD19+ population. In fact, using a more stringent definition of B-1P as Lin− AA4.1+ B220−/lo CD19+ ckit+ cells (38–40), we confirmed that Rybp mutant bone marrow was enriched in these early progenitors, as shown by the 10-fold increase of B-1P compared to the level in control bone marrow (Fig. 2D). Interestingly, the inverted B-2P-to-B-1P ratio in Rybp mutant mice seemed reminiscent of that of perinatal B-cell progenitors before the postnatal shift toward adult B-2 dominance (Fig. 2E).
We also asked whether the effects of Rybp inactivation were cell autonomous. Rybpf/f MxCre+ or MxCre− CD45.2+ bone marrow cells were transplanted into CD45.1 lethally irradiated mice. Rybp inactivation then was induced by poly(I·C) injections in recipient mice with >90% reconstitution (Fig. 2F, diagram). Rybp-deficient B cells (derived from recombined donor CD45.2+ stem cells) showed an identical altered distribution of CD19− and B220-expressing B-cell populations (Fig. 2F, right), with B-1P increase and concomitant B-2P decrease similar to those described above (Fig. 2G). We conclude that RYBP contributes, in a cell-autonomous manner, to the development of the adult B-cell compartment.
Absence of genetic interactions between Rybp, Ring1A, Ring1B, and Ink4a.
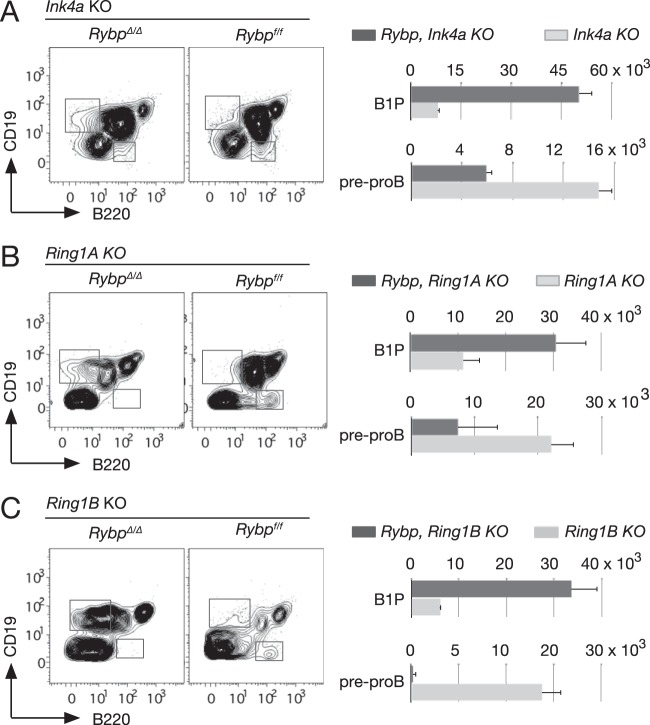
One of most paradigmatic Polycomb targets, the Ink4 locus, well known for its role in products of aging and cancer development, negatively affects B-2 progenitor development (41). Thus, we asked whether the observed phenotype in Rybp-deficient cells was related to a hypothetical upregulation of this inhibitor. The shifted B-1P-to-B-2P ratio in Rybp mutant mice also was seen in double RybpΔ/Δ Ink4a−/− mice but not in single Ink4a knockout (KO) mice (Fig. 3A). On the other hand, since RYBP associates directly with RING1A and RING1B in noncanonical PRC1 complexes (6), we analyzed single-mutant mice for Ring1A or Ring1B and double mutants also lacking RYBP. The results (Fig. 3B and C) showed that the enlargement of the B-1P-containing B-cell subpopulation is an Rybp-specific alteration not found in Ring1A or Ring1B single-mutant mice. Pre-proB populations in both double-mutant mice also were reduced compared to those of single mutants. In addition, the expressivity of the Rybp mutation was not affected in the compound mutants, suggesting the lack of genetic interaction. Thus, the genetic analysis indicated that RYBP activity regulating B-cell differentiation was mostly independent of Polycomb Ring1 products and of their target, Ink4.
FIG 3.
B-cell progenitor alterations associated with Rybp inactivation are independent of Ring1A, Ring1B, and Ink4a loci. (Left) Representative flow cytometry plots of CD19- and B220-labeled lineage-depleted (except for B220) bone marrow cells from mice of the indicated genotypes. (Right) Absolute number of populations containing B-1P and pre-proB progenitors. Values are means and SD from three mice per genotype.
Rybp deletion leads to enhanced expansion of B-1 progenitors and B-1-biased differentiation of most immature progenitors.
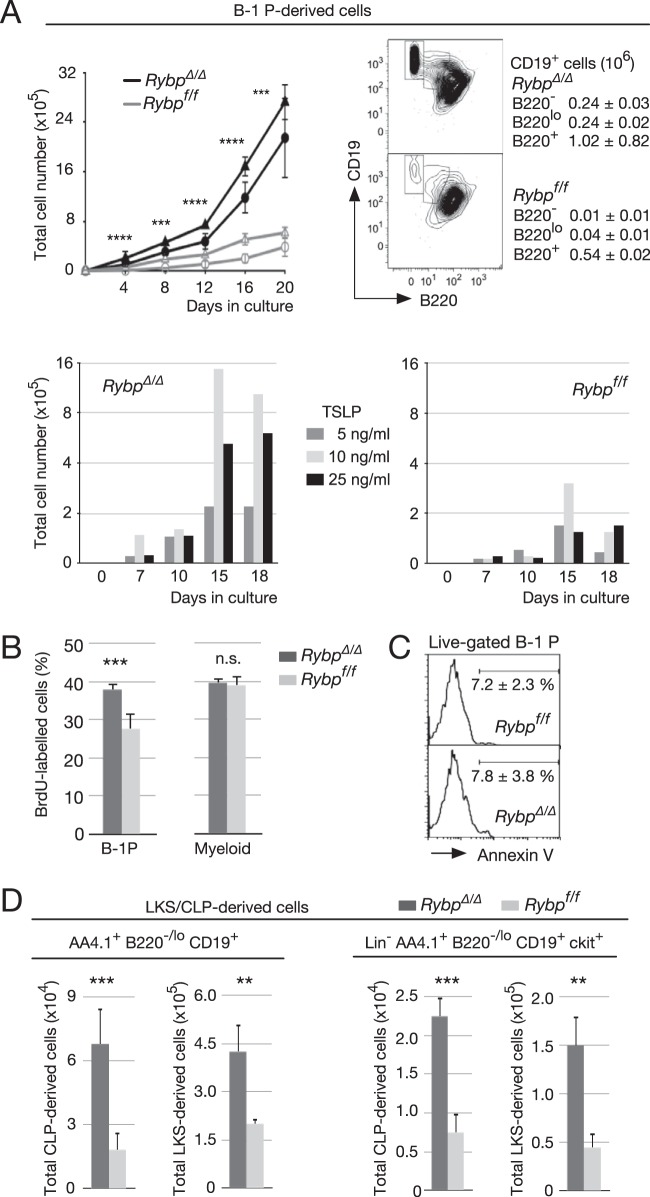
A characteristic feature of B-1P is the proliferative response to thymic stromal lymphopoietin (TSLP) when cocultured with stromal cells (38). We asked whether Rybp-deficient B-1P would proliferate under these conditions. A Transwell assay then was set up with RYBPΔ/Δ- or RYBPf/f-sorted B-1P (Lin− ckit+ AA4.1+ CD19+ B220−/lo). Indeed, both control and Rybp-deficient B-1P proliferated (Fig. 4A, top left, circles). However, Rybp ablation resulted in enhanced cell expansion. We also cultured B1-P isolated from RYBPf/f Cre-ERT2 mice, divided them into two identical aliquots, and cocultured them with S17 Transwell-seeded cells 1 day after treatment with 4′-OHT or the equivalent amount of vehicle (ethanol). As for in vivo recombined progenitors, tamoxifen-treated cells expanded to a larger extent than the nonrecombined cells (Fig. 4A, top left, triangles). After 20 days in culture, most control B-1P had differentiated into a homogenous population of B220int/+ cells (91% ± 3%) (Fig. 4A, top right). Mutant B-1P cultures also contained differentiated B220int/+ cells, in fact, twice as many as the control cultures. However, they still contained a considerable proportion of B220−/lo cells (16% each CD19+ B220− and CD19+ B220lo compared to 1.5 and 7%, respectively, in control cultures). In particular, the level of most immature CD19+ B220− cell accumulation was 24-fold higher in mutant than in control cultures (2.4 × 105 versus 0.1 × 105, respectively, recovered from 2 × 103 initially seeded CD19+ B220− cells). In order to explore whether the different expansion rates were due to an enhanced sensitivity to the lymphopoietin, we performed a similar assay with 5, 10, or 25 ng/ml of TSLP and assessed the number of generated cells over time in culture. As shown in Fig. 4A (bottom bar graphs), Rybp-deficient cells had an enhanced response to TSLP compared to that of wild-type cells at all concentrations and at any of the analyzed time points.
FIG 4.
Increased TSLP-activated self-renewal and expansion of isolated, HSC- or CLP-derived Rybp mutant B-1 progenitors. (A) Expansion and phenotype of cultured B-1 progenitors (B-1P; Lin− B220−/lo CD19+ ckit+). (Top left) Proliferation kinetics of 2 × 103 B-1P sorted from bone marrow of Rybpf/f MxCre+ or Rybpf/f MxCre− mice previously injected with poly(I·C) (circles) or of Rybpf/f; Cre-ERT2 mice after treatment with 4′-OHT (RybpΔ/Δ) or vehicle (ethanol; Rybpf/f) during the first 24 h in culture (triangles). Cells were cocultured with S17 stromal cells seeded in Transwells, with no cell contact, and medium supplemented with 10 ng/ml TSLP. (Top right) Representative flow cytometry plots of CD19- and B220-labeled cells after 20 days in culture; data correspond to mean absolute numbers and SD of CD19+ cells with different B220 labeling (boxes depicted in plots; triplicate cultures of three independent experiments). (Bottom) A total of 2 × 103 B-1P sorted from pooled RybpΔ/Δ (left) and Rybpf/f (right) bone marrow (n = 3 mice/genotype per experiment) were cultured in the presence of the indicated amounts of TSLP. Cells were collected and counted at the indicated time points (means of duplicate cultures from two independent experiments). (B) BrdU-labeled B-1P and myeloid progenitors (MyP) isolated from mice of the indicated genotypes 12 h after intraperitoneal injection of 1 mg BrdU. (C) Apoptosis within the B-1P population of the indicated genotypes, assessed by FITC-conjugated annexin V labeling of propidium iodide-impermeable cells. Horizontal bars denote apoptotic cells showing mean and SD values from triplicates of two independent experiments. (D) Enhanced production of B-1P from LKS and CLP pools in mice of the indicated genotypes. (Left) Total B220−/lo cells; (right) B-1P. Bar graphs show mean values and SD from triplicates of two independent experiments. **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001); n.s., not statistically significant (P ≥ 0.05).
The facilitated expansion of mutant cells could be due to the larger number of responsive B-1Ps or to a faster proliferative rate, as the response of ex vivo recombined progenitors suggested. Thus, we assessed in vivo BrdU incorporation into DNA. Mutant bone marrow cells contained a larger proportion of BrdU-labeled B-1P than controls (Fig. 4B), whereas similar numbers of mutant and control myeloid progenitors incorporated BrdU in similar proportions, indicating a cell stage-specific effect. Since Rybp-deficient B-1P cells and controls showed similar cell survival rates (as determined by annexin V labeling) (Fig. 4C), we conclude that the increased expansion of adult B-1Ps in vivo and in vitro is due, at least in part, to the higher proliferation rate in the absence of RYBP.
To assess whether Rybp deletion affected B-1P expansion only, we investigated the generation of B-1P from earlier progenitors. Noncommitted (LKS cells, containing HSCs and multipotent progenitors) and lymphoid-determined progenitors (CLP) were sorted from RybpΔ/Δ and Rybpf//f mice and cultured under the conditions described for B-1P with TSLP (for CLPs) or TSLP and IL-7 (required for lymphoid differentiation of the LKS pool) (42). In the absence of RYBP, in all cases a larger number of B-1Ps was generated, suggesting a biased differentiation of most immature cells toward the B-1 cell lineage. RYBP-deficient LKS or CLP yielded almost three times more B-1Ps than control progenitors after 7 days in culture (Fig. 4D), supporting an inhibitory role for RYBP early in the B-1P differentiation pathway. Together, the data suggest that the enlarged subset of B cells containing B-1P in mutant adult mice originates in shifted differentiation, skewed at very primitive stages.
Rybp depletion leads to decreased B-2 B-cell populations.
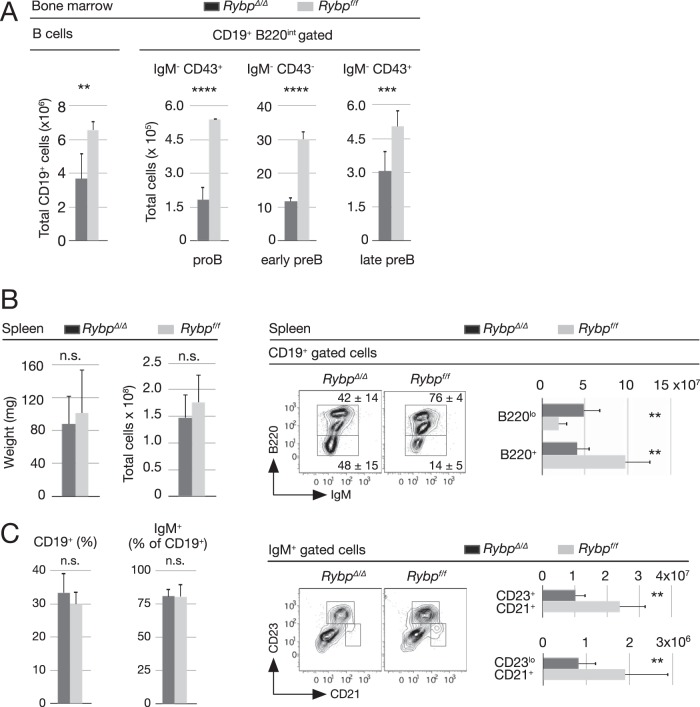
All bone marrow maturing B cells, i.e., total CD19+, proB (B220+ IgM− CD43+), early preB (B220+ IgM− CD43−), and late preB (B220+ IgM− CD43+) cells, were reduced in mutant mice (Fig. 5A). We then asked if these low levels of immature B cells, due to altered proportions of B-2P and B-1P in Rybp-deficient bone marrow, were reflected in spleen B cells.
FIG 5.
Decreased numbers of bone marrow and spleen Rybp-deficient B-2 B-cell precursors. (A) Absolute number of total bone marrow B cells (CD19+) and of subpopulations among early precursor CD19+ B220med populations, i.e., proB (IgM− CD43+), early preB (IgM− CD43−), and late preB (IgM+ CD43−) subsets from mice of the indicated genotypes. (B) Size of spleens (weight, cellularity) and content of CD19+ and IgM+ cells in control and mutant mice. (C) Splenic B-cell populations. Representative flow cytometry plots of control and mutant CD19+ cells (top) show mean percentages and SD of populations within drawn boxes and their absolute numbers (bar graph). Also shown are representative flow cytometry plots of control and mutant IgM+ cells and absolute numbers (bar graphs) of follicular (CD21lo CD23hi) and marginal-zone (CD21hi CD23lo) populations. Bar graphs show means and SD from analyzed mice (n = 8 [A and B] and n = 10 [C and D]). **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001; n.s., not statistically significant (P ≥ 0.05).
No major differences were found in spleen weight, cellularity, or frequencies of CD19+ or IgM+ cells (Fig. 5B). However, the ratio of B220+ to B220lo cells within CD19+ and IgM+ populations was inverted (5.2 in control cells versus 0.9 in mutant cells) (Fig. 5C, top left). In addition, the total number of B220+ mutant cells was less than half that of the control (Fig. 5C, top right), and both follicular (B220+ IgM+/lo CD23hi CD21lo) and marginal-zone (B220+ IgM+/lo CD23lo CD21hi) B-2 populations were decreased in RYBP-deficient mice (Fig. 5C, bottom). Thus, the data showed a correlation between decreased pre-proB progenitors and maturing and differentiated adult B-2 B-cell subpopulations.
Increased peritoneal B-1 B lymphocytes and kidney alterations in RybpΔ/Δ mice.
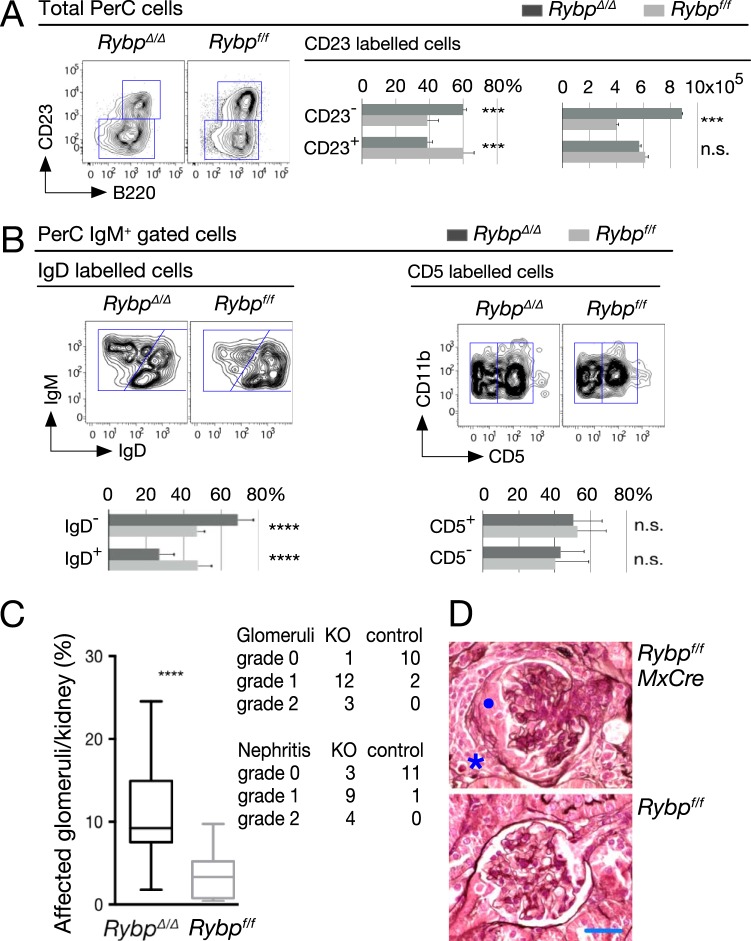
To assess if B-1P increase resulted in expanded numbers of B-1 B lymphocytes, we analyzed the peritoneal cavity (PerC), their most abundant reservoir. Rybp-deficient mice showed an increased ratio of B-1 (B220lo/+ CD23−) to B-2 (B220+/CD23+) cells, and the B-1 pool was nearly 2-fold larger in mutant than in wild-type PerC (Fig. 6A). Similarly, an increased B-1 to B-2 proportion was observed in IgD-labeled populations within IgM+ cells (Fig. 6B, left). In contrast, B-1a and B-1b subtype distribution (CD11b+ CD5+ and CD11b+ CD5− cells, respectively) was similar in both mutant and wild-type PerC compartments (Fig. 6B, right), indicating that both subpopulations were equally enlarged in the absence of RYBP.
FIG 6.
Peritoneal B-1 B cells and kidney disease in Rybp-mutant mice. (A and B) Representative flow cytometry plots and quantitation of the indicated subpopulations of the peritoneal cell cavity (PerC) in mutant and control mice showing increased absolute numbers of B-1 cells (A, bar graph on the right), increased B-1 to B-2 pool ratios (A, bar graph on the left, and B, left), and unaltered distribution of B-1a (CD5+) and B-1b (CD5−) subsets of CD11b+ cells (B, right). B-1 cells, B220lo/+ CD23− IgM+ IgDlo; B-2 cells, B220+ CD23+ IgM+ IgD+. Bar graphs show means and SD (n = 4 [A] and n = 10 [B]). *, P ≤ 0.05; ***, P ≤ 0.001; ****, P ≤ 0.0001; n.s., not statistically significant (P ≥ 0.05). (C) Glomerular lesions, box plot, and severity of interstitial nephritis foci in mice of the indicated genotypes. (D) Representative hematoxylin- and eosin-stained sections showing control (Rybpf/f) and unaffected glomerulus (RybpΔ/Δ). Note the tuft in the center of the mutant renal corpuscle compressed by a fibrocellular crescent (•), accompanied by a loss of capillary lumen. In addition, a mononuclear cell infiltrate (*) is found in the medullary interstitium. Scale bar, 25 μm. Bars in the box plot are median values (n = 16 [mutant]; n = 13 [control]); ****, P = 0.0003.
Mutant mice with increased numbers of PerC B-1 lymphocytes develop kidney disease related to immunoglobulin deposits (43, 44). Histological analysis of kidneys from poly(I·C)-treated mice showed glomerular damage in almost every Rybp-deficient mouse, varying both the number of glomeruli affected and the extent of alteration. Kidney abnormalities were seen in a small subset of control mice, affecting very few glomeruli and to a lower extent than that in mutant mice (Fig. 6C, left, and Table 1). Crescent formations were identified in almost 10% of glomeruli per mutant mice (a representative section of one moderately damaged is shown in Fig. 6D). Signs of nephritis, both periglomerular and interstitial, also were evident in a large subset of mutant kidneys. Nephritis degree, given by number and size of foci involved/section, also was more severe in mutant than in control mice (Fig. 6C, left, and Table 2).
TABLE 1.
Glomerular lesions
| Genotype and mouse | No. of glomerulia |
No. of altered glomeruli with a lesion extentb of: |
|||
|---|---|---|---|---|---|
| Total | Altered | Mild | Moderate | Severe | |
| RYBPf/f | |||||
| L870 | 62 | 2 | 2 | 0 | 0 |
| M183 | 68 | 1 | 1 | 0 | 0 |
| M476 | 62 | 3 | 3 | 0 | 0 |
| M488 | 82 | 0 | 0 | 0 | 0 |
| M543 | 106 | 0 | 0 | 0 | 0 |
| M546 | 75 | 0 | 0 | 0 | 0 |
| R968 | 75 | 7 | 5 | 1 | 1 |
| S254 | 65 | 4 | 2 | 1 | 1 |
| T266 | 65 | 2 | 1 | 0 | 1 |
| T268 | 63 | 3 | 3 | 0 | 0 |
| T272 | 72 | 2 | 2 | 0 | 0 |
| T273 | 67 | 1 | 1 | 0 | 0 |
| RYBPf/f MxCre | |||||
| L865 | 75 | 6 | 4 | 2 | 0 |
| L868 | 56 | 4 | 3 | 1 | 0 |
| M189 | 65 | 6 | 3 | 0 | 3 |
| M190 | 72 | 1 | 0 | 0 | 1 |
| M478 | 67 | 9 | 4 | 2 | 3 |
| M486 | 80 | 3 | 0 | 0 | 3 |
| M581 | 58 | 14 | 2 | 7 | 5 |
| M582 | 88 | 13 | 2 | 9 | 2 |
| M584 | 70 | 5 | 0 | 5 | 0 |
| M585 | 73 | 15 | 2 | 7 | 6 |
| S253 | 58 | 8 | 4 | 3 | 1 |
| S259 | 55 | 5 | 2 | 3 | 0 |
| T262 | 79 | 4 | 3 | 0 | 1 |
| T263 | 70 | 6 | 0 | 0 | 6 |
| T269 | 69 | 8 | 4 | 0 | 4 |
| T270 | 66 | 5 | 4 | 0 | 1 |
A minimum of 60 glomeruli were scored among 10 fields in cortical sections at ×20 magnification.
Mild, periglomerular fibrosis and reactive parietal epithelial cells lining Bowman's capsule; moderate, proliferation of the parietal epithelium belonging to Bowman's capsule mixed with scarce inflammatory cells; severe, half-moon-shaped fibrocellular mass formation with occasional glomerular tuft adhesion and sclerosis.
TABLE 2.
Nephritis data
| Genotype and mouse | No. of foci |
Lesion degreeb | ||||
|---|---|---|---|---|---|---|
| Total | With indicated focus sizea |
With indicated lesion location |
||||
| I | II | Periglomerular | Perivascular, interstitial | |||
| RYBPf/f | ||||||
| L870 | 3 | 3 | 0 | 1 | 2 | 0 |
| M183 | 3 | 3 | 0 | 1 | 2 | 0 |
| M476 | 4 | 3 | 1 | 3 | 1 | 0 |
| M488 | 4 | 2 | 2 | 0 | 4 | 0 |
| M543 | 2 | 0 | 2 | 0 | 2 | 0 |
| M546 | 2 | 1 | 1 | 1 | 1 | 0 |
| RYBPf/f MxCre | ||||||
| L865 | 6 | 4 | 2 | 4 | 2 | 1 |
| L868 | 9 | 8 | 1 | 8 | 1 | 1 |
| M189 | 5 | 4 | 1 | 4 | 1 | 0 |
| M190 | 5 | 4 | 1 | 2 | 3 | 0 |
| M478 | 11 | 3 | 8 | 6 | 5 | 2 |
| M486 | 8 | 5 | 3 | 2 | 6 | 1 |
| M581 | 11 | 3 | 8 | 6 | 5 | 2 |
| M582 | 14 | 6 | 8 | 9 | 6 | 2 |
| M584 | 10 | 9 | 1 | 4 | 6 | 1 |
| M585 | 9 | 7 | 2 | 6 | 3 | 1 |
Altogether, the data show that RYBP specifically controls adult B-cell development by inhibiting B-1P proliferation and promoting B-2 B subpopulation predominance.
DISCUSSION
Here, we show that the hematopoietic depletion of Rybp, a component of noncanonical PRC1, causes specific alterations in B-cell lineage by altering the adult bone marrow ratio of B-2 to B-1 B-cell progenitors. Rybp inactivation also reveals that adult B-cell lymphopoiesis is continuously regulated in adult life, as the postnatal B-1-to-B-2 shift must be actively perpetuated. RYBP appears to be a key actor maintaining B-2 progenitors as the dominant adult B lymphocyte precursors, and it also inhibits B-1P expansion. A number of genes in mouse mutant or transgenic lines have been associated with increased numbers of mature B-1 cells (39, 45–48), but to our knowledge, only Rybp has been involved in targeting adult B-1P expansion.
RYBP regulation of B-cell homeostasis and conventional Polycomb function.
Early B-cell lymphopoiesis implies lineage specification through the timely expression of instructive transcription factors (49–51). Their silent chromatin locus configuration often is enriched in histone modifications elicited by PRC1 and PRC2 enzymatic activity. Such an epigenetic state becomes modified when genes are activated. For example, repressed Ebf1, whose product is a key B-cell lineage specification instructor, is in an H2AUb state that is dependent on PRC1 H2A ubiquitin ligase-positive cofactor BMI1 (23, 52). Its activation requires the H2A deubiquitylase MYSM1, as indicated by the lack of B-2P (pre-proB cells) in Mysm1 mutants (53). In contrast with Rybp deficiency, where B-2P also are diminished, no B1-P increases were documented in Mysm1 mutants (53). Thus, RYBP appears to target the postnatal B-1-to-B-2 switch rather than the global B-cell transcriptional program. Moreover, although CLPs increased after Rybp inactivation, as in Bmi1 mutant mice (23), the overall phenotype is rather distinct, since Rybp mutant bone marrow contains fewer B cells and global H2AUb levels are hardly altered. The discrepancies suggest that mechanisms distinct from those involving BMI1 are involved and that RYBP acts independently of known PRC1 complexes, as we have seen in ES cells (54). Indeed, single Ring1A or Ring1B mutant mice do not show the described two side effects of Rybp-deficient B cell lymphopoiesis, and in fact, no genetic interactions are observed with the PRC1 Ub ligases or with their paradigmatic target, Ink4a. All of this points to a PRC1-independent function for RYBP in B-cell development.
B-1 biased differentiation of Rybp-deficient lymphoid progenitors at the expense of B-2 lineage.
Two models have been proposed to explain adult peritoneal B-1 cell origin: (i) a B-cell receptor-mediated positive selection model (B-1 or B-2 fate depending on the antigen exposure) and (ii) a layered model (B-1 or B-2 coming from distinct progenitors). The idea that a B-2 cell (or a B-2P) acquires B-1 characteristics under certain circumstances in a particular microenvironment (55, 56) or by genetic manipulation (43, 57, 58) has not been discarded. However, it is now evident that B-1P are generated in adult bone marrow (38, 39, 42). Both B-1 and B-2 B cell progenitors, as in fetal liver, arise from HSC-derived, more primitive progenitors (42). B-1 and B-2 cells generated from then on change during ontogeny so that a high B-1-to-B-2 ratio in fetal hematopoiesis turns into a low B-1-to-B-2 ratio in the adult. The postnatal lineage switch results in fewer progenitors with B-1 potential relative to those endowed with B-2 potential (42). Along this line of reasoning, the Rybp-deficient B-1P-to-B-2P inverted ratio, together with the low numbers of mutant B-2 cells, can be interpreted as an inverted flip toward a more juvenile developmental stage. Thus, RYBP appears to be a key positive regulator of definitive B-2 dominance throughout adult life.
B-1P also are produced more efficiently from isolated Rybp-deficient CLP and LKS. The existence of CLPs distinctly specified toward B-1 or B-2 has been proposed, as has the idea that B-1 CLP numbers and proliferative potential decline during postnatal development (42). In this context, we interpret that RYBP depletion provokes B-1 CLP expansion not only in vivo but also in vitro. This would explain the increased number of B-1P (and most probably B-1 CLPs) derived from mutant LKS. Interestingly, neonatal B-1 CLPs have a much higher proliferation rate than adult progenitors (42). RYBP then would be a positive regulator of B-1-to-B-2 switch by inhibiting B-1 CLP proliferation. Other important regulators, such as Lin28b, have been implicated in such a fetus-to-adult flip, since it is postnatally downregulated (59). In contrast, ectopic expression in adult stem and progenitor (59) or even in proB (58) cells results in increased peritoneal B-1a cells in transplanted mice. We tested the hypothesis that RYBP represses Lin28b, but the mRNA was not upregulated in Rybp-deficient proB cells (data not shown). Again, our data better support a role for RYBP in B-1 and B-2 CLP homeostasis, as well as in B-1P intrinsic proliferation rate (or their response to proliferative cues), than a role in B-cell specification. It is worth noting that Rybp-deficient B-1 progenitors have increased in vivo and in vitro proliferation. The higher proliferative rate of Rybp-deficient B-1 progenitors was observed regardless of whether cells were isolated from RybpΔ/Δ mice or whether they originated in vitro after tamoxifen-induced deletion. This strongly suggests an intrinsic role for RYBP within the progenitor response to the mitogen. Interestingly, a recently annotated map for TSLP signaling included three major proliferation-associated pathways, SRC/YES and STAT5 in normal cells and the AKT/mTOR axis in pre-ALL cells (60). Such a correlation merits subsequent work on the possible impact of RYBP on these signal transduction pathways.
An additional difference between fetal and adult B-1 precursors is their potential to generate different B-1 B-cell types. B-1 cells comprise B-1a (CD5+) and B-1b cells (CD5−), present in both fetal and adult B-1 populations. B-1a cells are more efficiently produced by fetal (and neonatal) than by adult progenitors (42, 55, 56, 61). B-1a is also the main B-1 subtype generated when adult cells are reprogrammed by Lin28b transduction (58, 59). In contrast, adult B-1P give rise preferentially, but not exclusively, to B-1b cells (38–40, 55). That both PerC B1 populations are enlarged in RYBP mutant mice reinforces the notion that RYBP acts by inhibiting the expansion of adult B1 progenitors as a whole.
Altogether, our results further support the immune layered model by which B-1 and B-2 lineages are produced in waves of B lymphopoiesis arising from precursors changing during development (56, 62). We propose that RYBP, likely independently of other PRC1 subunits, is a key actor playing a lead part in adult B-cell development from multipotent progenitors.
ACKNOWLEDGMENTS
We are grateful to K. Dorshkind (UCLA) for S17 stromal cells, to M. Barbacid and M. Serrano (CNIO) for Polr2a::Cre-ERT2 and Ink4a mutant mice, respectively, and to Laura Molero, head of the UAM SIdI Flow Cytometry facility, for help with data analysis and cell sorting.
K.S. and M.B. are supported by FP7-PEOPLE-2011-ITN, grant 289611, and a MINECO fellowship. This work was supported by grants BFU2008-03386/BMC (MINECO) to C.C., BFU2010-18146, SAF2013-47997-P (MINECO), and the CAM OncoCycle Program (S2010/BMD-2470) to M.V.
REFERENCES
- 1.Schwartz YB, Pirrotta V. 2013. A new world of Polycombs: unexpected partnerships and emerging functions. Nat Rev Genet 14:853–864. doi: 10.1038/nrg3603. [DOI] [PubMed] [Google Scholar]
- 2.Laugesen A, Helin K. 2014. Chromatin repressive complexes in stem cells, development, and cancer. Cell Stem Cell 14:735–751. doi: 10.1016/j.stem.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 3.García E, Marcos-Gutiérrez C, del Mar Lorente M, Moreno JC, Vidal M. 1999. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J 18:3404–3418. doi: 10.1093/emboj/18.12.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang H, Wang L, Erdjument-Bromage H, Vidal M, Tempst P, Jones RS, Zhang Y. 2004. Role of histone H2A ubiquitination in Polycomb silencing. Nature 431:873–878. doi: 10.1038/nature02985. [DOI] [PubMed] [Google Scholar]
- 5.de Napoles M, Mermoud JE, Wakao R, Tang YA, Endoh M, Appanah R, Nesterova TB, Silva J, Otte AP, Vidal M, Koseki H, Brockdorff N. 2004. Polycomb group proteins Ring1A/B link ubiquitylation of histone H2A to heritable gene silencing and X inactivation. Dev Cell 7:663–676. doi: 10.1016/j.devcel.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 6.Gao Z, Zhang J, Bonasio R, Strino F, Sawai A, Parisi F, Kluger Y, Reinberg D. 2012. PCGF homologs, CBX proteins, and RYBP define functionally distinct PRC1 family complexes. Mol Cell 45:344–356. doi: 10.1016/j.molcel.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tavares L, Dimitrova E, Oxley D, Webster J, Poot R, Demmers J, Bezstarosti K, Taylor S, Ura H, Koide H, Wutz A, Vidal M, Elderkin S, Brockdorff N. 2012. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 148:664–678. doi: 10.1016/j.cell.2011.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang R, Taylor AB, Leal BZ, Chadwell LV, Ilangovan U, Robinson AK, Schirf V, Hart PJ, Lafer EM, Demeler B, Hinck AP, McEwen DG, Kim CA. 2010. Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure 18:966–975. doi: 10.1016/j.str.2010.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Endoh M, Endo TA, Endoh T, Isono K-I, Sharif J, Ohara O, Toyoda T, Ito T, Eskeland R, Bickmore WA, Vidal M, Bernstein BE, Koseki H. 2012. Histone H2A mono-ubiquitination is a crucial step to mediate PRC1-dependent repression of developmental genes to maintain ES cell identity. PLoS Genet 8:e1002774. doi: 10.1371/journal.pgen.1002774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eskeland R, Leeb M, Grimes GR, Kress C, Boyle S, Sproul D, Gilbert N, Fan Y, Skoultchi AI, Wutz A, Bickmore WA. 2010. Ring1B compacts chromatin structure and represses gene expression independent of histone ubiquitination. Mol Cell 38:452–464. doi: 10.1016/j.molcel.2010.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Isono K, Endo TA, Ku M, Yamada D, Suzuki R, Sharif J, Ishikura T, Toyoda T, Bernstein BE, Koseki H. 2013. SAM domain polymerization links subnuclear clustering of PRC1 to gene silencing. Dev Cell 26:565–577. doi: 10.1016/j.devcel.2013.08.016. [DOI] [PubMed] [Google Scholar]
- 12.Schoeftner S, Sengupta AK, Kubicek S, Mechtler K, Spahn L, Koseki H, Jenuwein T, Wutz A. 2006. Recruitment of PRC1 function at the initiation of X inactivation independent of PRC2 and silencing. EMBO J 25:3110–3122. doi: 10.1038/sj.emboj.7601187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Morey L, Aloia L, Cozzuto L, Benitah SA, Di Croce L. 2013. RYBP and Cbx7 define specific biological functions of polycomb complexes in mouse embryonic stem cells. Cell Rep 3:60–69. doi: 10.1016/j.celrep.2012.11.026. [DOI] [PubMed] [Google Scholar]
- 14.Blackledge NP, Farcas AM, Kondo T, King HW, McGouran JF, Hanssen LLP, Ito S, Cooper S, Kondo K, Koseki Y, Ishikura T, Long HK, Sheahan TW, Brockdorff N, Kessler BM, Koseki H, Klose RJ. 2014. Variant PRC1 complex-dependent H2A ubiquitylation drives PRC2 recruitment and polycomb domain formation. Cell 157:1445–1459. doi: 10.1016/j.cell.2014.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Konuma T, Oguro H, Iwama A. 2010. Role of the polycomb group proteins in hematopoietic stem cells. Dev Growth Differ 52:505–516. doi: 10.1111/j.1440-169X.2010.01191.x. [DOI] [PubMed] [Google Scholar]
- 16.Radulović V, de Haan G, Klauke K. 2013. Polycomb-group proteins in hematopoietic stem cell regulation and hematopoietic neoplasms. Leukemia 27:523–533. doi: 10.1038/leu.2012.368. [DOI] [PubMed] [Google Scholar]
- 17.Rossi L, Lin KK, Boles NC, Yang L, King KY, Jeong M, Mayle A, Goodell MA. 2012. Less is more: unveiling the functional core of hematopoietic stem cells through knockout mice. Cell Stem Cell 11:302–317. doi: 10.1016/j.stem.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Mönch K, Minucci S, Porse BT, Marine J-C, Hansen KH, Helin K. 2007. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev 21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jacobs JJ, Kieboom K, Marino S, DePinho RA, van Lohuizen M. 1999. The oncogene and Polycomb-group gene bmi-1 regulates cell proliferation and senescence through the ink4a locus. Nature 397:164–168. doi: 10.1038/16476. [DOI] [PubMed] [Google Scholar]
- 20.Voncken JW, Roelen BAJ, Roefs M, de Vries S, Verhoeven E, Marino S, Deschamps J, van Lohuizen M. 2003. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc Natl Acad Sci U S A 100:2468–2473. doi: 10.1073/pnas.0434312100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Agherbi H, Gaussmann-Wenger A, Verthuy C, Chasson L, Serrano M, Djabali M. 2009. Polycomb mediated epigenetic silencing and replication timing at the INK4a/ARF locus during senescence. PLoS One 4:e5622. doi: 10.1371/journal.pone.0005622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park I-K, Qian D, Kiel M, Becker MW, Pihalja M, Weissman IL, Morrison SJ, Clarke MF. 2003. Bmi-1 is required for maintenance of adult self-renewing haematopoietic stem cells. Nature 423:302–305. doi: 10.1038/nature01587. [DOI] [PubMed] [Google Scholar]
- 23.Oguro H, Yuan J, Ichikawa H, Ikawa T, Yamazaki S, Kawamoto H, Nakauchi H, Iwama A. 2010. Poised lineage specification in multipotential hematopoietic stem and progenitor cells by the polycomb protein Bmi1. Cell Stem Cell 6:279–286. doi: 10.1016/j.stem.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 24.Arranz L, Herrera-Merchan A, Ligos JM, de Molina A, Dominguez O, Gonzalez S. 2012. Bmi1 is critical to prevent Ikaros-mediated lymphoid priming in hematopoietic stem cells. Cell Cycle 11:65–78. doi: 10.4161/cc.11.1.18097. [DOI] [PubMed] [Google Scholar]
- 25.Calés C, Román-Trufero M, Pavón L, Serrano I, Melgar T, Endoh M, Pérez C, Vidal M. 2008. Inactivation of the polycomb group protein Ring1B unveils an antiproliferative role in hematopoietic cell expansion and cooperation with tumorigenesis associated with Ink4a deletion. Mol Cell Biol 28:1018–1028. doi: 10.1128/MCB.01136-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.van den Boom V, Rozenveld-Geugien M, Bonardi F, Malanga D, van Gosliga D, Heijink AM, Viglietto G, Morrone G, Fusetti F, Vellenga E, Schuringa JJ. 2013. Nonredundant and locus-specific gene repression functions of PRC1 paralog family members in human hematopoietic stem/progenitor cells. Blood 121:2452–2461. doi: 10.1182/blood-2012-08-451666. [DOI] [PubMed] [Google Scholar]
- 27.Pirity MK, Locker J, Schreiber-Agus N. 2005. Rybp/DEDAF is required for early postimplantation and for central nervous system development. Mol Cell Biol 25:7193–7202. doi: 10.1128/MCB.25.16.7193-7202.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trimarchi JM, Fairchild B, Wen J, Lees JA. 2001. The E2F6 transcription factor is a component of the mammalian Bmi1-containing polycomb complex. Proc Natl Acad Sci U S A 98:1519–1524. doi: 10.1073/pnas.98.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kuhn R, Schwenk F, Aguet M, Rajewsky K. 1995. Inducible gene targeting in mice. Science 269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 30.Mijimolle N, Velasco J, Dubus P, Guerra C, Weinbaum CA, Casey PJ, Campuzano V, Barbacid M. 2005. Protein farnesyltransferase in embryogenesis, adult homeostasis, and tumor development. Cancer Cell 7:313–324. doi: 10.1016/j.ccr.2005.03.004. [DOI] [PubMed] [Google Scholar]
- 31.Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. 1996. Role of the INK4a locus in tumor suppression and cell mortality. Cell 85:27–37. doi: 10.1016/S0092-8674(00)81079-X. [DOI] [PubMed] [Google Scholar]
- 32.del Mar Lorente M, Marcos-Gutiérrez C, Pérez C, Schoorlemmer J, Ramírez A, Magin T, Vidal M. 2000. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development 127:5093–5100. [DOI] [PubMed] [Google Scholar]
- 33.Herzenberg LA, Tung J, Moore WA, Herzenberg LA, Parks DR. 2006. Interpreting flow cytometry data: a guide for the perplexed. Nat Immunol 7:681–685. doi: 10.1038/ni0706-681. [DOI] [PubMed] [Google Scholar]
- 34.Montecino-Rodriguez E, Dorshkind K. 2006. Stromal cell-dependent growth of B-1 B cell progenitors in the absence of direct contact. Nat Protoc 1:1140–1144. doi: 10.1038/nprot.2006.163. [DOI] [PubMed] [Google Scholar]
- 35.Smeets B, Uhlig S, Fuss A, Mooren F, Wetzels JFM, Floege J, Moeller MJ. 2009. Tracing the origin of glomerular extracapillary lesions from parietal epithelial cells. J Am Soc Nephrol 20:2604–2615. doi: 10.1681/ASN.2009010122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shlomchik MJ, Madaio MP, Ni D, Trounstein M, Huszar D. 1994. The role of B cells in lpr/lpr-induced autoimmunity. J Exp Med 180:1295–1306. doi: 10.1084/jem.180.4.1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stokes MB, Markowitz GS, Lin J, Valeri AM, D'Agati VD. 2004. Glomerular tip lesion: a distinct entity within the minimal change disease/focal segmental glomerulosclerosis spectrum. Kidney Int 65:1690–1702. doi: 10.1111/j.1523-1755.2004.00563.x. [DOI] [PubMed] [Google Scholar]
- 38.Montecino-Rodriguez E, Leathers H, Dorshkind K. 2006. Identification of a B-1 B cell-specified progenitor. Nat Immunol 7:293–301. doi: 10.1038/ni1301. [DOI] [PubMed] [Google Scholar]
- 39.Esplin BL, Welner RS, Zhang Q, Borghesi LA, Kincade PW. 2009. A differentiation pathway for B1 cells in adult bone marrow. Proc Natl Acad Sci U S A 106:5773–5778. doi: 10.1073/pnas.0811632106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ghosn EEB, Sadate-Ngatchou P, Yang Y, Herzenberg LA, Herzenberg LA. 2011. Distinct progenitors for B-1 and B-2 cells are present in adult mouse spleen. Proc Natl Acad Sci U S A 108:2879–2884. doi: 10.1073/pnas.1019764108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Signer RAJ, Montecino-Rodriguez E, Witte ON, Dorshkind K. 2008. Aging and cancer resistance in lymphoid progenitors are linked processes conferred by p16Ink4a and Arf. Genes Dev 22:3115–3120. doi: 10.1101/gad.1715808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barber CL, Montecino-Rodriguez E, Dorshkind K. 2011. Reduced production of B-1-specified common lymphoid progenitors results in diminished potential of adult marrow to generate B-1 cells. Proc Natl Acad Sci U S A 108:13700–13704. doi: 10.1073/pnas.1107172108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kojima H, Kobata T. 2002. Abnormal B lymphocyte development and autoimmunity in hypoxia-inducible factor 1alpha-deficient chimeric mice. Proc Natl Acad Sci U S A 99:2170–2174. doi: 10.1073/pnas.052706699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jellusova J, Wellmann U, Amann K, Winkler TH, Nitschke L. 2010. CD22 x Siglec-G double-deficient mice have massively increased B1 cell numbers and develop systemic autoimmunity. J Immunol 184:3618–3627. doi: 10.4049/jimmunol.0902711. [DOI] [PubMed] [Google Scholar]
- 45.Hoffmann A, Kerr S, Jellusova J, Zhang J, Weisel F, Wellmann U, Winkler TH, Kneitz B, Crocker PR, Nitschke L. 2007. Siglec-G is a B1 cell-inhibitory receptor that controls expansion and calcium signaling of the B1 cell population. Nat Immunol 8:695–704. doi: 10.1038/ni1480. [DOI] [PubMed] [Google Scholar]
- 46.Györy I, Boller S, Nechanitzky R, Mandel E, Pott S, Liu E, Grosschedl R. 2012. Transcription factor Ebf1 regulates differentiation stage-specific signaling, proliferation, and survival of B cells. Genes Dev 26:668–682. doi: 10.1101/gad.187328.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Xiao C, Calado DP, Galler G, Thai T-H, Patterson HC, Wang J, Rajewsky N, Bender TP, Rajewsky K. 2007. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell 131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 48.Ludwig J, Federico G, Prokosch S, Küblbeck G, Schmitt S, Klevenz A, Gröne H-J, Nitschke L, Arnold B. 2015. Dickkopf-3 acts as a modulator of B cell fate and function. J Immunol 194:2624–2634. doi: 10.4049/jimmunol.1402160. [DOI] [PubMed] [Google Scholar]
- 49.Murre C. 2009. Developmental trajectories in early hematopoiesis. Genes Dev 23:2366–2370. doi: 10.1101/gad.1861709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yoshida T, Georgopoulos K. 2014. Ikaros fingers on lymphocyte differentiation. Int J Hematol 100:220–229. doi: 10.1007/s12185-014-1644-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Grosschedl R. 2013. Establishment and maintenance of B cell identity. Cold Spring Harbor Symp Quant Biol 78:23–30. doi: 10.1101/sqb.2013.78.020057. [DOI] [PubMed] [Google Scholar]
- 52.Seo W, Ikawa T, Kawamoto H, Taniuchi I. 2012. Runx1-Cbfβ facilitates early B lymphocyte development by regulating expression of Ebf1. J Exp Med 209:1255–1262. doi: 10.1084/jem.20112745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jiang X-X, Nguyen Q, Chou Y, Wang T, Nandakumar V, Yates P, Jones L, Wang L, Won H, Lee H-R, Jung JU, Müschen M, Huang XF, Chen S-Y. 2011. Control of B cell development by the histone H2A deubiquitinase MYSM1. Immunity 35:883–896. doi: 10.1016/j.immuni.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hisada K, Sánchez C, Endo TA, Endoh M, Román-Trufero M, Sharif J, Vidal M. 2012. RYBP represses endogenous retroviruses and preimplantation- and germ line-specific genes in mouse embryonic stem cells. Mol Cell Biol 32:1139–1149. doi: 10.1128/MCB.06441-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Baumgarth N. 2011. The double life of a B-1 cell: self-reactivity selects for protective effector functions. Nat Rev Immunol 11:34–46. [DOI] [PubMed] [Google Scholar]
- 56.Montecino-Rodriguez E, Dorshkind K. 2012. B-1 B cell development in the fetus and adult. Immunity 36:13–21. doi: 10.1016/j.immuni.2011.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ye M, Ermakova O, Graf T. 2005. PU.1 is not strictly required for B cell development and its absence induces a B-2 to B-1 cell switch. J Exp Med 202:1411–1422. doi: 10.1084/jem.20051089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhou Y, Li Y-S, Rao Bandi S, Tang L, Shinton SA, Hayakawa K, Hardy RR. 2015. Lin28b promotes fetal B lymphopoiesis through the transcription factor Arid3a. J Exp Med 212:569–580. doi: 10.1084/jem.20141510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yuan J, Nguyen CK, Liu X, Kanellopoulou C, Muljo SA. 2012. Lin28b reprograms adult bone marrow hematopoietic progenitors to mediate fetal-like lymphopoiesis. Science 335:1195–1200. doi: 10.1126/science.1216557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhong J, Sharma J, Raju R, Palapetta SM, Prasad TSK, Huang T-C, Yoda A, Tyner JW, van Bodegom D, Weinstock DM, Ziegler SF, Pandey A. 2014. TSLP signaling pathway map: a platform for analysis of TSLP-mediated signaling. Database (Oxford) 2014:bau007. doi: 10.1093/database/bau007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hardy RR, Kincade PW, Dorshkind K. 2007. The protean nature of cells in the B lymphocyte lineage. Immunity 26:703–714. doi: 10.1016/j.immuni.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 62.Herzenberg LA. 1989. Toward a layered immune system. Cell 59:953–954. doi: 10.1016/0092-8674(89)90748-4. [DOI] [PubMed] [Google Scholar]