Abstract
The heat shock protein 90 (HSP90) and cell division cycle 37 (CDC37) chaperones are key regulators of protein kinase folding and maturation. Recent evidence suggests that thermodynamic properties of kinases, rather than primary sequences, are recognized by the chaperones. In concordance, we observed a striking difference in HSP90 binding between wild-type (WT) and kinase-dead (KD) glycogen synthase kinase 3β (GSK3β) forms. Using model cell lines stably expressing these two GSK3β forms, we observed no interaction between WT GSK3β and HSP90, in stark contrast to KD GSK3β forming a stable complex with HSP90 at a 1:1 ratio. In a survey of 91 ectopically expressed kinases in DLD-1 cells, we compared two parameters to measure HSP90 dependency: static binding and kinase stability following HSP90 inhibition. We observed no correlation between HSP90 binding and reduced stability of a kinase after pharmacological inhibition of HSP90. We expanded our stability study to >50 endogenous kinases across four cell lines and demonstrated that HSP90 dependency is context dependent. These observations suggest that HSP90 binds to its kinase client in a particular conformation that we hypothesize to be associated with the nucleotide-processing cycle. Lastly, we performed proteomics profiling of kinases and phosphopeptides in DLD-1 cells to globally define the impact of HSP90 inhibition on the kinome.
INTRODUCTION
As protein kinases regulate a multitude of cellular functions, it is imperative that their activity be highly regulated. This meticulous control is achieved through a variety of mechanisms, including phosphorylation, proteolytic processing, and direct engagement with other molecules, including chaperones. The heat shock protein 90 (HSP90) chaperone, in particular, is recruited to kinase clients to assist in their nascent folding and, depending on the kinase, to further contribute to its stability and function (1). A recent kinome-wide study of pairwise HSP90-kinase interactions observed various strengths of binding affinities between protein kinases and HSP90 (2), consistent with HSP90 assuming different roles depending on the client.
HSP90 typically requires its cochaperone cell division cycle 37 (CDC37) to engage a kinase client, but the mechanism for kinase selection is not fully understood. There is no particular kinase class that forms stronger interactions with HSP90 (2), and there are even distinct binding specificities for closely related kinases. For example, epidermal growth factor receptor (EGFR) does not associate with HSP90, but close family member Erb-b2 receptor tyrosine kinase 2 (ERBB2) forms strong interactions with HSP90, and inhibition of HSP90 by geldanamycin induces rapid proteasome-mediated degradation of ERBB2 (3–5). Thus, amino acid sequences or structural features conserved in close family members are unlikely to be primary determinants for chaperone recognition, which is not surprising given that protein kinases represent a fraction of HSP90 clients (6). Instead, it appears that HSP90 recognizes kinases in a particular conformation. Reduced binding of HSP90 to the tyrosine kinase BCR-ABL was observed not only in the presence of imatinib, a nonhydrolyzable ATP analog inhibitor of ABL, but also with compounds that allosterically locked kinase domain conformations into either active or inhibited states, suggesting that HSP90 does not bind to a particular site but rather recognizes kinases as they become thermally and conformationally unstable (2). Structural evidence suggests that cochaperone CDC37 directly competes with nucleotide or analog inhibitors for binding to BRAF, ERBB2, and EGFRG179S, highlighting the importance of the ATP cleft for chaperone recognition (7).
Kinase catalysis is a dynamic process regulated by both intrinsic and extrinsic signals. In the oncogenic tyrosine kinase fusion nucleophosmin-anaplastic lymphoma receptor tyrosine kinase (NPM-ALK) model, downstream kinase AKT displayed increased activity as well as an increased dependency on HSP90 for its stability (8). This model suggests that attributes of AKT associated with its activity are recognized by the chaperone complex and is consistent with previous and striking observations that numerous cancer-promoting kinases, whether activated through mutation, aberrant upstream activation, or gene upregulation, are more dependent on HSP90 activities than their counterparts in normal cells (9). As a result, pharmacological inhibition of HSP90 has been exploited as a potential cancer treatment strategy (10–12) to selectively inactivate oncogenic kinases that are HSP90 clients (13, 14). In contrast to the use of specific kinase inhibitors to treat a specific type of tumor, the indirect approach of targeting oncogenic kinases via HSP90 inhibition does not require the identification of key tumor-promoting kinases for the intended cancer types. This is because cancer-promoting kinases rely on catalytic efficiency and therefore support from chaperones (14).
Unfortunately, the transient nature of the interaction between kinases and HSP90 often hinders the mechanistic study of their dynamic interplay in normal physiology and in malignancy. Coimmunoprecipitation does not capture transient interactions, and for stable complexes, it can only measure the average binding strength of a population of HSP90 and kinase molecules at particular yet unspecified states of activity. Therefore, a different parameter to assess HSP90 dependency is often used, which is the rate of kinase degradation through the ubiquitin proteasome pathway following HSP90 inactivation (2, 3, 15, 16). The key determinant in the action of chaperone binding for kinase degradation remains unclear.
This study started from a striking and unexpected observation that wild-type glycogen synthase kinase 3β (GSK3βWT) did not bind HSP90 but a kinase-dead (KD) mutant (GSK3βK85A) with an impairment to an ATP-anchoring nucleotide formed a stoichiometric interaction with HSP90, suggesting that HSP90 recognized attributes in GSK3β that were associated with distinct nucleotide states that may even appear transiently during its natural catalytic cycle. We then performed an extended study to compare HSP90 binding and kinase stability for >130 stably transfected and/or endogenous kinases. From this, we confirmed that there is no clear link between binding and stability. Importantly, we showed that kinase stability is cell context dependent, suggesting that the effect of HSP90 inhibition varies between cell types and conditions. We performed a proof-of-principle proteomic screen to monitor kinase levels after HSP90 inhibition by measuring differences in kinase capture by ATP analogs (Kinobeads) and changes in the phosphoproteome. This screen, in DLD-1 cells, was intended to profile kinase dependency on HSP90 in an unbiased manner and can subsequently be extended to characterize individual tumor types, with the anticipation that such an index may ultimately provide useful guidance for finding candidate tumor-promoting kinases.
MATERIALS AND METHODS
Cell lines and cDNAs.
Human DLD-1, HEK293, Jurkat, and mouse NIH 3T3 cells were obtained from the ATCC. 17-AAG (17-N-allylamino-17-demethoxygeldanamycin, a specific HSP90 inhibitor) was used at a 1 μM concentration in cell media for 3 h unless otherwise indicated. GSK3 inhibitor CHIR99021 (Chiron, Emeryville, CA) was used at 2.5 μM in cell culture media for 1 h. mTOR inhibitor rapamycin (from Sigma-Aldrich) was used at 1 μM for 1 h. Wild-type and mutant (K85A and R96A) forms of GSK3β were expressed from a pcDNA3 vector with an in-frame fusion of the FLAG tag sequence at their carboxy termini. Site-directed mutagenesis was performed using the standard DpnI method, and results were verified by sequencing of the region. These GSK3β constructs were used to generate stable expression clones in DLD-1 and PC12 cells. GSK3β and indicated point mutants were cloned using the Gateway system by following the manufacturer's standard protocol (Life Technologies Inc.) and were transfected into the Flp-In T-Rex cell line to create stable cell lines according to the manufacturer's protocol. Kinases were cloned into pMX pie pDEST 3X Flag and stably expressed in DLD-1 cells as described in reference 17.
Antibodies.
The anti-FLAG antibody was purchased from Sigma-Aldrich, and anti-enhanced green fluorescent protein (anti-EGFP) antibody (ab290) was from Abcam. Other antibodies were from a variety of sources and included antibodies against the following: AKT (Cell Signaling Technology; catalog number 9272), ASK1 (Santa Cruz; sc7931), BTK (Santa Cruz; sc1108), CAK/DDR1 (Santa Cruz; sc532, C20), cyclin-dependent kinase 2 (CDK2) (Santa Cruz; sc163, M2), CDK5 (Santa Cruz; sc750, H291), CDK7 (Millipore; catalog number 06-377), CHEK1/CHK1 (Millipore; catalog number 04-207), EPHA2 (Santa Cruz; sc924, C20), ERBB2 (Santa Cruz; sc1763, C20), FLT1 (Santa Cruz; sc9029, H225), FLT4 (Santa Cruz; sc637, M20), FYN (Santa Cruz; sc434), GSK3 (Santa Cruz; sc7291, 0011A), HCK (Santa Cruz; sc1428), insulin-like growth factor receptor 1 (IGFR1) (Cell Signaling Technology; catalog number 3027), INSR (Santa Cruz; sc711, C19), c-KIT (Santa Cruz; sc168), protein kinase CI (PKCI) (Santa Cruz; sc727, N20), PYK2/FAK2/PTK2B (BD Biosciences; catalog number 610548), RAF1 (Santa Cruz; sc133, C12), RET (Santa Cruz; sc13104), ROCK2/ρ kinase β (BD BioSciences; catalog number 610623), ROCK1 (Santa Cruz; sc5560, H85), SYK (Santa Cruz; sc1077, N19), WEE1 (Santa Cruz; sc5285, B11), phospho-Tyr100 (Cell Signaling Technology; catalog number 9411), HSP70 (Santa Cruz; sc24, W27), CAMKII (Santa Cruz; sc13082, H300), CHK2 (Santa Cruz; sc9064, H300), CSK (Santa Cruz; sc286, C20), EPHA3 (Santa Cruz; sc919, C19), EPHA4 (BD Biosciences; catalog number 610471), EPHA5 (Santa Cruz; sc927, C16), ERBB3 (Santa Cruz; sc285, C17), ERBB4 (Abcam; ab32375), extracellular signal-regulated kinase 1 (ERK1)/mitogen-activated protein kinase (MAPK) and ERK2/MAPK (Santa Cruz; sc93, C16), FAK/PTK2 (BD Biosciences, catalog number 610088), fibroblast growth factor receptor 1 (FGFR1) (Cell Signaling Technology; catalog number D38B1), FLK1 (Santa Cruz; sc6251, A3), c-FMS (Millipore; catalog number 06-457), LCK (Millipore; catalog number 05-435), LYN (BD BioSciences; catalog number 610003), Jun N-terminal protein kinase (JNK; MAPK8, MAPK9, and MAPK10) (Santa Cruz; sc571), MEK1 (Santa Cruz; sc219, C18), h-MET (Santa Cruz; sc10, C12), p38 (Cell Signaling Technology; catalog number 9212), p70S6K (Cell Signaling Technology; catalog number 9202), p110-PI3K (Santa Cruz; sc8010, D4), PAK1 (Cell Signaling Technology; catalog number 2602), PDGFβ (Santa Cruz; sc432), PKCzeta (Santa Cruz; sc216, C20), B-RAF (Santa Cruz; sc166, C19), SRC (Cell Signaling Technology; catalog number 2108), c-YES (Santa Cruz; sc14), ZAP70 (Santa Cruz; sc574), pTyr/4G10 (Millipore; catalog number 05-321), HSP90 (Santa Cruz; sc13119), CDC37 (Santa Cruz; sc17758, C11), tubulin/DM1A (Abcam; ab7291), and actin (Abcam; ab3280, C4).
SDS-PAGE and Western blotting procedures.
Immunoprecipitation (IP) samples were harvested from cultured cells lysed in NP-40 lysis buffer containing 1% NP-40, 150 mM NaCl, 50 mM Tris (pH 7.5), 1 mM dithiothreitol (DTT), and protease and phosphatase inhibitor cocktails from Sigma-Aldrich. Total cell lysates were harvested by direct lysis of the cells with SDS sample buffer. Colloidal Coomassie (trade name, GelCode Blue; Thermo Scientific) was used to visualize total protein bands by SDS-PAGE. Western blotting was conducted by following the standard protocol, and commercial primary antibodies were used according to the manufacturers' recommendations unless indicated otherwise.
MS.
A gel-based mass spectrometry (MS) method for protein identification was performed as previously reported (18). For stable isotope labeling by amino acids in cell culture (SILAC), isotopic versions of arginine and lysine (“heavy” [H], l-[13C6, 15N4]arginine and l-[13C6 and 15N2]lysine; “medium” [M], l-[15N4]arginine and l-[2H4]lysine; and “light” [L], [12C]arginine and [14N]lysine) were purchased from Sigma Isotech. Cells were cultivated in RPMI media deficient in arginine and lysine (Caisson Lab), 10% dialyzed fetal bovine serum (FBS; Gibco), penicillin-streptomycin (Gibco), and SILAC arginine and lysine (50 mg/liter). After 7 passages, ∼99.5% cellular proteins were labeled with the desired Lys and Arg combinations. At this point, cells were either treated with HSP90 inhibitor 17-AAG for 3 h or left untreated. For kinase enrichment, cells were lysed in NP-40 lysis buffer, the lysates from the three SILAC conditions were combined, and kinases were purified using Kinobeads and subjected to trypsin digestion (19). The phosphoproteomics profiling was performed according to our previous report (20). Briefly, cells from the 3 SILAC conditions were lysed in lysis buffer (50 mM Tris-HCl [pH 7.4], 150 mM NaCl, 1% NP-40, protease inhibitor cocktail [complete mini; Roche], and phosphatase inhibitor cocktail [PhosSTOP; Roche]), and the lysates were combined. The lysate was then precipitated in −20°C acetone and resolubilized in 8 M urea sample buffer (8 M urea, 20 mM HEPES [pH 8], 1 mM sodium orthovanadate, 2.5 mM sodium pyrophosphate, 1 mM β-glycerophosphate). Tryptic digestion was conducted in 2 M urea, and the peptides obtained were fractionated by strong-cation exchange chromatography following by titanium dioxide enrichment (GL Sciences). The enriched peptides were analyzed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) on a high-resolution mass spectrometer (Orbitrap Velos by Thermo Scientific). For both analyses, raw spectra were processed using the open-source software MaxQuant, with a false-discovery rate set at 1% (http://www.maxquant.org; parameters are listed in Tables S2 and S3 in the supplemental material) (21). Only peptides with a MaxQuant posterior error probability [PEP] of ≤0.01 were retained for further analysis. Corrections for labeling inefficiencies and proline-to-arginine conversions were performed as previously described (29). For each peptide, three pairwise intensity ratios from the L, M, and H channels were obtained, reflecting the relative ratio in the amount of the peptide from the three treatment conditions. This three-channel design to measure the two states of the cells—treated versus not treated with 17-AAG—was intended to serve as internal controls for deriving statistical significance to distinguish technical variation. Only reproducible peptides whose deviations between the log2 H intensities and log2 L intensities were below 1 (which is equivalent to removing data outside ±1.2 standard deviations of the log2 ratios) were retained. For the enrichment with Kinobeads, proteins with at least 2 distinct peptides were retained and any protein whose median ratio of 17-AAG-treated to untreated cells was outside the 5th or 95th percentile was considered significant. Reproducibility was assessed by quantile-to-quantile comparison of log10 intensities (M versus L and H versus M) and calculating the correlation coefficient between the median H/M and L/M log2 ratios on the common proteins identified in two separate experiments with reversed labeling (17-AAG treatment in the M channel or control in the M channel). For the phosphoproteome analysis, we used, as estimation of the noise, the 5% to 95th percentiles of the distribution of the log2 ratios of 17-AAG-treated to untreated cells for unphosphorylated peptides lacking phosphorylatable residues (no S, T, or Y amino acids). The median ratio of any phosphopeptide that was outside this range was considered significant.
MS data.
Raw MS data are available through the MassIVE Web portal (http://massive.ucsd.edu; MassIVE code MSV000078989 for FLAG-tagged kinases in Fig. 2 and code MSV000079445 for global analysis in Fig. 5).
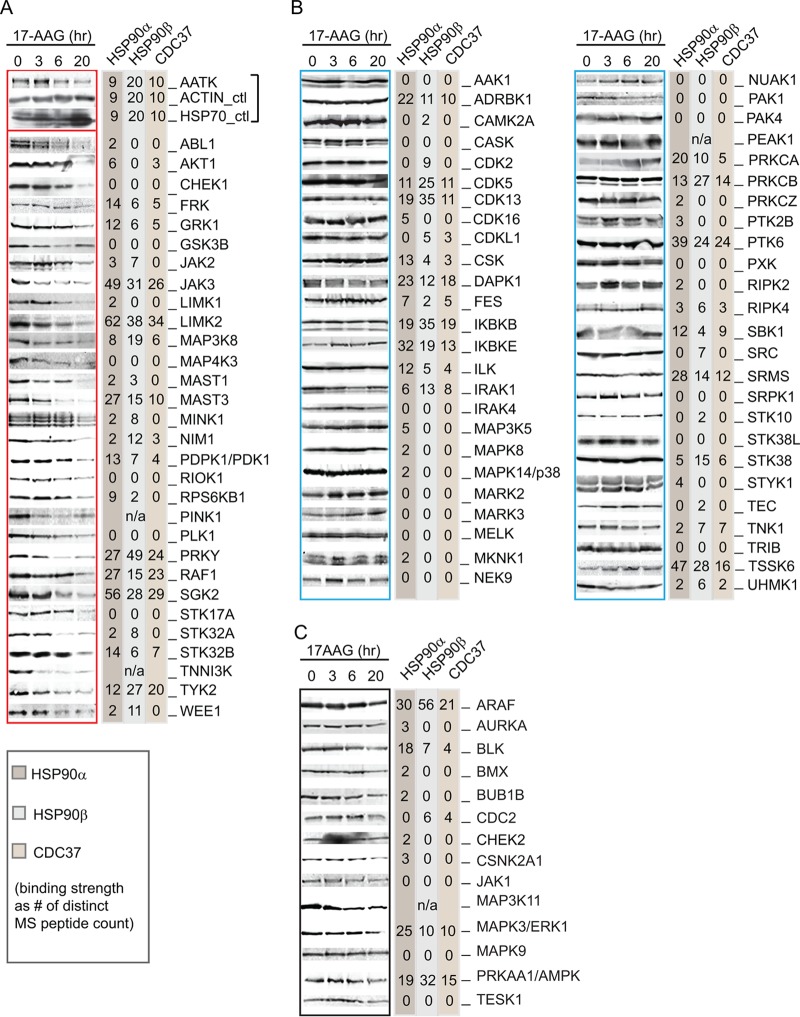
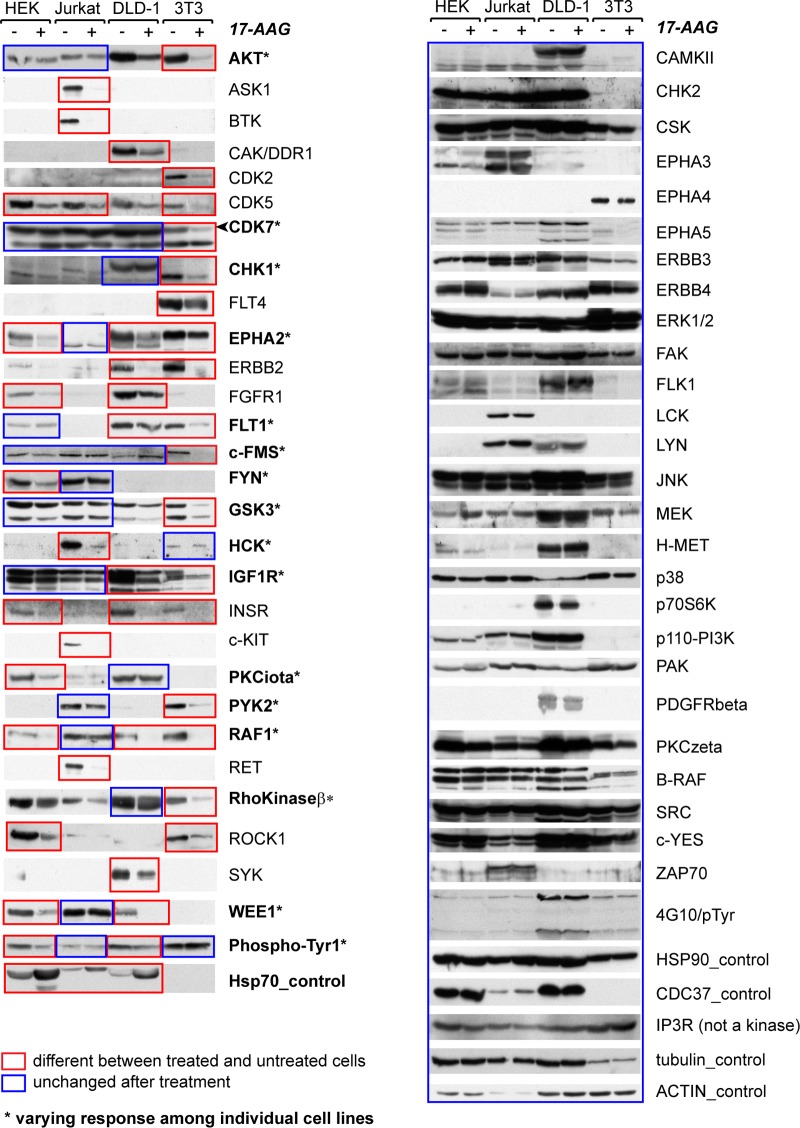
FIG 2.
HSP90 binding capacity and stability of protein kinases following 17-AAG treatment. Selected cytoplasmic protein kinases were individually tagged with an N-terminal FLAG and stably expressed in DLD-1 cells. Results from anti-FLAG Western blotting of cell lysate following a time course of 17-AAG inhibition are shown at the left of each panel (gel images were from a single experiment of cell treatment). Following anti-FLAG IP of the kinases from cell lysate, interacting proteins were identified by LC-MS/MS. The unique peptides identified by mass spectrometry for HSP90α, HSP90β, and CDC37 are shown at the right of each panel. For each membrane, anti-β-actin and anti-HSP70 blots were included as controls for sample loading and cellular response to inhibitor (representative control blots for AATK are shown). Kinases presented in panel A showed decreased expression upon 17-AAG treatment, while those listed in panel B remained at a constant expression level. (C) FLAG-tagged kinases whose stability after 17AAG treatment was inconclusive. These kinases are indicated as unclassified in Table S1 in the supplemental material.
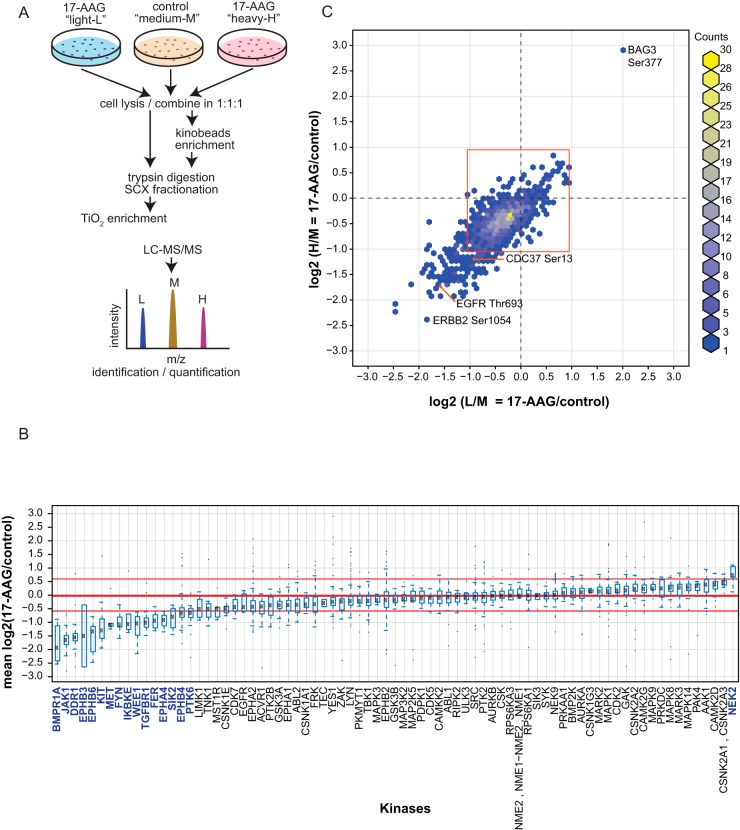
FIG 5.
Mass spectrometry analysis of phosphoproteome and kinome changes following HSP90 inhibition. (A) General outline of the workflow for quantitative mass spectrometry analysis using SILAC. Cultured DLD-1 cells were differentially labeled using media containing either normal [12C]arginine and [14N]lysine (light), l-[15N4]arginine and l-[2H4]lysine (medium), or l-[13C6, 15N4]arginine and l-[13C6, 15N2]lysine (heavy). The light and heavy channels were treated with 17-AAG for 3 h, while the medium channel was left untreated (note that for sample A6, the medium channel was treated with 17-AAG, while the light and heavy channels were left untreated). (B and C) After lysis, the samples were enriched either for protein kinases using the Kinobead protocol (B) or for phosphopeptides following an SCX-TiO2 enrichment protocol (C). Kinase and phosphoproteome peptides were identified and quantified by mass spectrometry. This three-channel comparison scheme allowed for 2 biological replicates (light and heavy) to be directly compared in the same experiment. (B) After Kinobead enrichment in 2 independent experiments (A6 and E6 in Table S2 in the supplemental material), 77 protein kinases out of 232 proteins were identified. For each kinase, the ratio (mean and standard deviation) of 17-AAG treatment over no treatment was calculated using all peptides with a PEP score less than or equal to 0.01. Kinases were ranked based on their log2 mean ratios from low to high. The red lines represent the 5th (−0.58), 50th (−0.02), and 95th (0.60) percentiles, respectively. Significantly modulated kinases, outside the 5th and 95th percentiles, are colored blue. (C) In the phosphoproteomic analysis, the log2 ratios for the levels of phosphopeptides in 17-AAG-treated cells (both light and heavy) over that in the control (medium) are plotted in a two-dimensional graph. The red box approximates the 95th percentile for unphosphorylated peptides lacking S, T, and Y residues. Proteins discussed in the text are labeled.
RESULTS
A kinase-dead mutant of GSK3β is constitutively associated with HSP90.
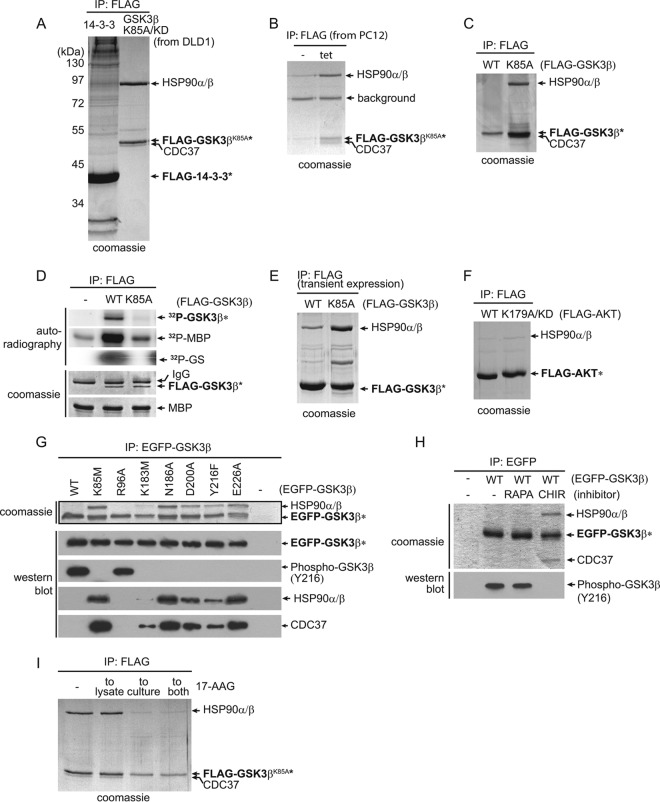
GSK3 family kinases mediate key signaling pathways that are implicated in a broad range of human diseases, such as cancer and neurodegenerative and metabolic disorders (23). To identify novel interactors of GSK3β, we performed immunoprecipitation (IP) of GSK3β, followed by mass spectrometry to identify bound proteins. We stably expressed a FLAG-tagged GSK3βKD, carrying a lysine 85-to-alanine (K85A) substitution of an ATP-anchoring residue, in the colorectal cancer cell line DLD-1 in the hope that a kinase-dead mutant would act as a substrate trap. The eluate of the GSK3βK85A complexes was resolved and then visualized by SDS-PAGE (Fig. 1A). Compared to a similar assay performed for 14-3-3ε, GSK3βK85A had a minimal number of binding partners: a dominant ∼90-kDa protein band confirmed by mass spectrometry as HSP90α and HSP90β chaperone proteins and an additional band that was identified as HSP90 cochaperone protein CDC37. To exclude the possibility that the interaction is DLD-1 cell specific, we stably expressed a tetracycline (Tet)-inducible FLAG-GSK3βK85A in rat pheochromacytoma-derived PC12 cells. As the expression of GSK3βK85A was induced by Tet, we saw a corresponding increase in binding partners HSP90α and HSP90β and CDC37 (Fig. 1B). Similar to the case with DLD-1 cells, GSK3βK85A bound HSP90 at an approximate 1:1 ratio. In a striking contrast, the wild-type (WT) GSK3β bait stably expressed in DLD-1 cells did not bind the chaperones (Fig. 1C). As a control, we confirmed through a series of in vitro kinase assays using glycogen synthase peptide (GS) or myelin basic protein (MBP) as a substrate that GSK3βK85A, but not the WT, lacked catalytic activity (Fig. 1D). Interestingly, when overexpressed in a transgene transfection system, GSK3βWT was able to bind HSP90, albeit at a much reduced level compared to that of the kinase-dead mutant, which remained tightly associated with HSP90 at an ∼1:1 ratio (Fig. 1E). Out of a concern that this may have been a result of inadvertent protein misfolding due to overexpression, in subsequent experiments, all kinases were expressed either endogenously or from stable transfections.
FIG 1.
GSK3β kinase-dead mutants, but not wild-type kinase, form stable complexes with HSP90. (A) Stably expressed FLAG-tagged GSK3βK85A/KD and FLAG-tagged 14-3-3ε were immunoprecipitated from DLD-1 cells, and the immunoprecipitates were resolved by SDS-PAGE. Bait (asterisks) and coprecipitating proteins were visualized by Coomassie staining. Proteins identified by mass spectrometry in the FLAG-tagged GSK3βK85A/KD immunoprecipitate are labeled. (B) GSK3βK85A, stably expressed under a tetracycline (tet)-inducible promoter, in PC12 cells also formed a stoichiometric complex with HSP90 and CDC37. Bands labeled as HSP90 and CDC37 were confirmed by mass spectrometry. (C) Stably expressed wild-type GSK3β (WT) did not bind HSP90 in DLD-1 cells. (D) Kinase assays confirmed the absence of GSK3βK85A activity toward in vitro substrates as measured by 32P-labeled MBP and GS. (E) Under transient-overexpression conditions, GSK3βK85A interacted with HSP90 in DLD-1 cells in an approximately 1:1 ratio, while wild-type GSK3b also bound HSP90 at a reduced but detectable level. (F) FLAG-tagged AKT1WT or KD AKTR179A was stably expressed in DLD-1 cells. Visualization of coprecipitating proteins by Coomassie staining showed no difference between WT and KD mutant AKT1 in binding HSP90. (G) EGFP-tagged GSK3β WT and a series of mutants were created and stably expressed in DLD-1 cells. Following anti-EGFP precipitation, their activities were measured by in vivo autophosphorylation of GSK3β on Y216. Binding of HSP90 and CDC37 was determined by comparing band intensities of Coomassie and Western blot protein bands. Wild-type and GSK3β R96A have similar activities, as measured by Y216 phosphorylation, but do not bind HSP90 or CDC37. (H) DLD-1 cells expressing EGFP-GSK3βWT were either left untreated (−) or treated with mTOR inhibitor rapamycin (RAPA) or GSK3-specific inhibitor CHIR99021 (CHIR). Following anti-EGFP IP of GSK3β and SDS-PAGE, major interacting proteins HSP90 and CDC37 in the CHIR99021-treated cell samples were visualized by Coomassie staining. (I) Treatment of the GSK3bK85A-expressing cells by adding 17-AAG to the culture medium drastically reduced the levels of HSP90 in the GSK3bK85A IP.
Given the contrast between GSK3βWT and its kinase-dead counterpart, we next tested if this was a more general phenomenon. We repeated the experiment on an unrelated serine/threonine kinase, AKT1 (also termed PKBα), that is known to bind HSP90 (24). AKT1WT and the catalytically inactive mutant AKT1K179A were observed to bind HSP90 at similar levels, albeit reduced compared to those of GSK3βK85A (Fig. 1F). Recognizing that GSK3β is distinct in its differential binding between wild-type and kinase-dead forms from at least one other kinase, we wanted to further characterize the binding of GSK3β to HSP90. We stably expressed a panel of EGFP-tagged GSK3β mutants in DLD-1 cells: the K85M (a variant of K85A), K183M, N186A, D200A, and E226A mutants, which are known based on structural evidence to affect nucleotide binding or transfer (25). A substitution of the activation loop autophosphorylation residue (Y216F) that renders GSK3β inactive (26) was also included. All these mutants bound HSP90 and CDC37 to various degrees, with strong binding by the K85M, N186A, E226A, and D200A mutants and weaker binding of the K183M and Y216F mutants (Fig. 1G). As before, there was no binding observed for the WT or for an R96A mutant that has normal activity as indicated by Y216 phosphorylation but has an altered substrate specificity (27). All HSP90-binding GSK3β mutants tested were unable to autophosphorylate Y216, consistent with them being kinase dead (Y216F was previously shown to have no activity [26]). To see if the binding to HSP90 was due to a potential steric effect in nucleotide binding that forced the kinase into an altered state, we inhibited EGFP-GSK3βWT with a nucleotide analog, GSK3-specific inhibitor CHIR99021. Treating the cells with CHIR99021 promoted HSP90 and CDC37 binding of GSK3βWT (Fig. 1H). No binding was observed from buffer or rapamycin treatment where GSK3βWT was active as measured by Y216 phosphorylation. Thus, HSP90-CDC37 formed a stable association with GSK3βWT when the nucleotide analog permanently bound the active site, thus locking GSK3β in a conformation that prevented the release of HSP90. Although we did not directly measure the nucleotide binding status of these mutants, the differences in HSP90 binding capacity among the kinase-dead mutants suggests that steric attributes associated with activity influence chaperone binding or release. HSP90 inhibition by 17-AAG (17-N-allylamino-17-demethoxygeldanamycin, a specific HSP90 inhibitor) treatment of the cells resulted in reduced levels of stable HSP90-GSK3βK85A interaction and, to a lesser extent, reduced expression of kinase-dead GSK3βK85A likely due to protein degradation (Fig. 1I), a well-known phenomenon for kinase clientele requiring HSP90 activities in cells.
Sensitivities of protein kinases to HSP90 inhibition.
Given the differences seen between GSK3β and AKT1, we wanted to more globally investigate the extent of kinase-HSP90 interactions in the human kinome. We had previously investigated kinase interaction networks in the human DLD-1 colorectal adenocarcinoma cell line by stably expressing a subset of human protein kinases as N-terminal triple FLAG tag fusions and performing immunoprecipitation followed by mass spectrometry to identify interacting proteins (17). This subset of kinases represented all major branches of the human kinome tree, except the receptor tyrosine kinase family. Endogenous HSP90α and -β, as well as CDC37, were present in 68 out of the 91 kinase precipitants (Fig. 2 and 3; see also Table S1 in the supplemental material). The unique peptide count, from a combined search of two replicates, is an approximate measure of HSP90 and CDC37 binding. Lim domain kinase 2 (LIMK2) had the highest associated counts for HSP90α and -β and CDC37, at 62, 38, and 34, respectively. Twenty-three kinases, including GSK3β (expressed in this study in its wild-type form), did not bind any of the three chaperones. The number of unique peptides for CDC37 tended to increase with the number of peptides identified for the HSP90s, consistent with CDC37 specifically mediating HSP90 interactions with kinase clientele.
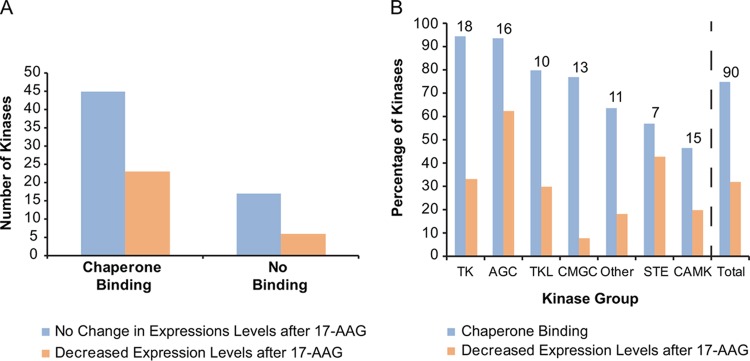
FIG 3.
Distribution of kinases in the HSP90-binding versus nonbinding and stable versus unstable categories. (A) The numbers of kinases are plotted according to their expression levels after 17-AAG treatment, separated by whether they bound chaperones (see Table S1 in the supplemental material). (B) The percentages of kinases within each kinase group that bound to chaperones or showed decreased expression levels after 17-AAG treatment are graphed. The number above each kinase group represents the number of kinases in each category. The atypical kinase group was not included, as there was only one member.
To further test the HSP90 dependency of this subset of protein kinases using the kinase degradation parameter, we performed a time course analysis on the stably transfected kinase cell lines following treatment with 17-AAG. Kinases that depend on HSP90 for stability generally undergo degradation in the presence of the inhibitor. For each cell line, lysates from 0, 3, 6, and 20 h of 17-AAG treatment were collected and subsequently subjected to anti-FLAG Western blotting to detect kinase levels (Fig. 2). Antiactin blots served as loading controls, and anti-HSP70 blots confirmed the cellular response to 17-AAG, as HSP90 inhibition increases HSP70 transcription through heat shock transcription factor 1 (HSF1) transactivation (28) (representative control blots for apoptosis-associated tyrosine kinase [AATK]-expressing cells are shown in Fig. 2A). Thirty-one out of 95 kinases (33%) exhibited decreased expression following treatment (Fig. 2A). The remaining ∼67% appeared stable in abundance (Fig. 2B) or could not be distinctively classified (Fig. 2C). It is important to note that this analysis provides only an approximate measurement of stability because the level of kinase expression is the net result from degradation and the rate of protein synthesis during the course of observation. Although the latter is less problematic in a stable transfection system, its effect cannot be discounted. Overall, there was no significant correlation between kinases that bind HSP90 and their stability in its absence (Fig. 3A) (Fisher exact test; P = 0.608). Some kinases (e.g., beta adrenergic receptor kinase 1 [ADRBK1], inhibitor of κB kinase beta [IKBKB], inhibitor of κB kinase epsilon [IKBKE], and protein tyrosine kinase 6 [PTK6]) displayed strong binding to the chaperones as measured by mass spectrometry but were not susceptible to HSP90 inhibition in terms of their protein stability (Fig. 2). Conversely, there was a subset of kinases whose expression was sensitive to 17-AAG treatment (e.g., ABL1, LIMK1, and checkpoint kinase 1 [CHEK1]) that displayed poor or no HSP90 binding. Thus, in this set of 91 representative kinases, we did not observe an apparent link between a kinase's strength of binding to HSP90 and its susceptibility to 17-AAG-induced destabilization.
Kinases whose expression decreased upon HSP90 inhibition were spread across families, with the CDK, MAPK, GSK3 and CLK (CMGC) kinases showing the least change in expression (1 out of 13 tested) and the AGC kinases showing the most (6 out of 16 tested) (Fig. 3B). Fifteen out of 16 members of the PKA, PKG, and PKC (AGC) family bound to HSP90, whereas only 3 out of 7 of the STE family bound, suggesting possible differences between kinase families (Fig. 3B). Within kinase families, we observed that some closely related kinases had distinct patterns in their affinities for binding to HSP90, such as LIMK1 and LIMK2 (2 and 62 peptides, respectively, for HSP90α) as well as Janus kinase 2 (JAK2) and JAK3 (3 and 49 peptides, respectively, for HSP90α), suggesting that any distinction between families is not at the primary amino acid level. These observations were consistent with the findings of other kinome-wide studies of kinase-HSP90 interactions (2, 3), suggesting that unspecified intrinsic properties other than the primary amino acid sequence of the kinases are the true determinants for HSP90 chaperoning.
Cell-specific responses of kinases to HSP90 inhibition.
To further pursue the relationship between kinases and HSP90, we investigated kinase responses to 17-AAG across different cell types and when exposed to different cell conditions. We studied endogenous kinases by immunoblotting in four cell lines: HEK293 from human embryonic kidney, Jurkat T, and DLD-1 and mouse NIH 3T3 fibroblasts. In total, about 150 antibodies against ∼100 endogenous kinases were tested on the same panel of cell lysates, of which 54 recognized bands of the expected molecular weights for the intended kinases (Fig. 4). Kinase expression levels after 20 h of 17-AAG treatment were compared to those with no treatment across all four cell lines. As expected for effective 17-AAG treatment, HSP70 levels markedly increased following inhibitor treatment (there was no signal for mouse NIH 3T3 cells, as the monoclonal HSP70 antibody does not recognize mouse HSP70). The amounts of HSP90 and CDC37 as well as control proteins inositol 1,4,5-trisphosphate receptor type 3 (IP3R), tubulin, and actin did not change upon treatment, suggesting that protein degradation following 17-AAG treatment is not a general phenomenon.
FIG 4.
Individual kinases' differential responses to 17-AAG are cell type dependent. Endogenous kinases plus the indicated control proteins in four cell lines (HEK293, Jurkat T, DLD-1, and NIH 3T3) were detected by Western blotting after the cell lines were treated or not with 17-AAG for 20 h. Kinases showing either increased or decreased abundance in a cell line are highlighted in red boxes (and presented in the left panel). Kinases with unchanged expression upon 17-AAG treatment are marked with blue boxes. In cases in which no sensitivity was observed across all four cell lines, the proteins are shown on the right panel. In cases in which an individual kinase responded to 17-AAG in some, but not all, of the four cell types, the kinase is marked with an asterisk and the lanes are marked with mixed boxes of blue and red colors. The cell lysates used for each antibody blotting were aliquoted from the same master set harvested from a single experiment of inhibitor treatment. Some antibodies cross-reacted with multiple kinase targets, often closely related homologs, such as AKT (for at least AKT1 and -2), and GSK3 (both α and β isoforms [lower and upper bands, respectively, on the blot]) (see Table S1 in the supplemental material for details).
The survey revealed that the stability of 26 of the 54 kinases was insensitive to treatment across all four cell types (Fig. 4). Of the other 28 kinases whose protein levels were decreased, 15 exhibited cell-type-specific differential response (destabilized kinases are marked in red boxes), and this number would likely be higher if more cell lines had been tested, as several kinases (e.g., Bruton agammaglobulinemia tyrosine kinase [BTK] and spleen tyrosine kinase [SYK]) were expressed in only 1 cell line. This observation was particularly interesting because it again suggested that the determinants for HSP90 dependency are beyond the primary sequence of a kinase.
Proteomic profiling of kinase response to HSP90 inhibition.
As the dependency of individual kinases on HSP90 can vary between cells, it follows that an individual cell's kinome can be differentially affected by HSP90 function. Thus, it is possible that a collective kinome stability profile could be used to index a cell's state with respect to HSP90 function. As tumor cells subvert regulatory pathways to confer proliferation and survival advantages, their state of malignancy could be distinctively achieved, or at least supported, through changes in activation status of particular kinases (22). This proposed index of HSP90 dependency may provide guidance on tumor type-specific pathways mediated by kinases with high catalytic activities. We employed mass spectrometry to measure changes in the abundance of endogenous kinases and phosphopeptide substrates after HSP90 inhibition (Fig. 5A).
DLD-1 cells were grown in SILAC (stable isotope labeling by amino acids in cell culture) medium. Before lysis, cells from three labeling channels (light, medium, or heavy, depending on incorporated stable isotopes) were treated with 17-AAG for 3 h or left untreated (Fig. 5A). Endogenous kinases were enriched by binding to ATP analogs linked to Sepharose beads (Kinobeads) (19). After protein digestion, the samples were analyzed by mass spectrometry and spectral peak intensities were quantified using Maxquant (see Table S2 in the supplemental material). Only proteins with at least 2 distinct peptides with posterior error probabilities of ≤0.01 were retained. Corrections were made for nonincorporated label or arginine-to-proline conversions (29), nonreproducible outliers were removed, and the values from two independent experiments were combined (Pearson correlation coefficient [R2] = 0.54). We quantified 232 proteins, of which 77 were protein kinases. Mean ratios and standard deviations were then determined for 17-AAG treatment versus nontreatment conditions. Any protein outside the 5th or 95th percentile was considered to be significantly changed in its abundance. Differences in amounts of kinase binding to the Kinobeads reflect a change in either their expression or their activity (30). Sixteen showed significant decreases (Fig. 5B), with bone morphogenetic protein receptor type 1A (BMPR1A), JAK1, discoidin domain receptor tyrosine kinase 1 (DDR1), Eph receptor B3 (EPHB3), and EPHB6 showing the most reduced abundance. Only one kinase, NIMA-related kinase 2 (NEK2), showed a significant increase in Kinobead binding after 17-AAG treatment. This increase has been previously reported and is thought to be due to the arrest of these cells at the G2/M boundary (31). In contrast, the relative protein abundance of non-protein kinases from treated and untreated cells was more balanced, with 2 proteins showing decreased abundance and 5 showing increased abundance, including HSP70 family members HSPA8 and HSPH1 (see Fig. S1A in the supplemental material). Expanding this proteomics workflow to other cell lines would start to build cell-specific indices of kinase stability in response to HSP90 inhibition.
In a similar manner, we conducted a global phosphoproteomic analysis to determine if the impact on kinases was reflected in the levels of protein phosphorylation. We employed strong cation-exchange (SCX) fractionation and TiO2 to enrich phosphopeptides that were identified and quantified by mass spectrometry (Fig. 5C; see also Table S3 in the supplemental material). After data correction and outlier removal as described above for the Kinobeads, the ratios of treated to untreated phosphopeptides for the replicates were plotted (Fig. 5C) (R2 of 0.63). The data points were shifted from the center at 0/0 toward the double-negative direction, indicating a general reduction of the cellular phosphoproteome. The red box in Fig. S1B approximates the 95th percentile of copurifying unphosphorylated peptides that lack phosphorylatable residues. Phosphopeptides outside this box were considered significant. Of the 1,150 quantified phosphorylation sites, only 1 (Ser377 on BCL2-associated athanogene 3 [BAG3]) was significantly increased (see Table S3). BAG3 is a cochaperone of the stress-induced HSP70 protein (32) whose expression was increased after 17-AAG treatment (Fig. 4; see also Fig. S1A). In contrast, HSP90 cochaperone CDC37 showed decreased phosphorylation on Ser13. Ser13 is required for the binding of CDC37 to kinase clients and for HSP90 to form high-affinity stable complexes with its kinase clients (33). An additional 115 phosphopeptides also decreased upon 17-AAG treatment. This overall trend of decreased phosphorylation was likely due to the expression levels and perhaps the activities of protein kinases being negatively impacted by HSP90 inhibition. Ten of the phosphopeptides with attenuated phosphorylation were from protein kinases (Fig. 5C; see also Table S1). Ser1054 on tyrosine kinase ERBB2 had a 4-fold decrease. Activity of ERBB2 is associated with tumorigenesis (34); therefore, it is interesting to observe this marked change in ERBB2 phosphorylation upon HSP90 inhibition, although we cannot exclude the possibility that this was due to the reduction of ERBB2 protein levels. Thr693 on EGFR was also significantly decreased. Phosphorylation at this site is required for internalization of EGFR after EGF stimulation (35). Thus, in additional to defining global trends in phosphoproteome changes, this analysis can start to uncover functional relevancies of individual phosphosites.
DISCUSSION
Despite intensive research using various model systems, the exact mechanism for how the HSP90 chaperone dynamically engages its kinase clients remains unclear (36, 37). Serendipitously, we observed a striking all-or-nothing switch between GSK3βK85A, a mutant with impaired ATP binding, and the active wild-type kinase in their ability to stably bind HSP90 and the CDC37 cochaperone. GSK3bK85A and five other KD mutants (in contrast to the WT and the catalytically active R96A mutant) formed stable associations with HSP90. As HSP90 has previously been shown to be required transiently for GSK3 activation (38), we reasoned that since these KD mutants were unable to progress through their catalytic cycles, they were permanently locked in a thermodynamically unstable conformation that prevented the release of HSP90. Extending this further, it is possible that in general, HSP90-CDC37 is directly involved in facilitating nucleotide loading or exchange, or in correcting steric imperfections, as a kinase actively goes through ATP hydrolysis and phosphoric acid transfer cycles. Unlike most other kinases that exhibit heterogeneous levels of activation within individual cells, GSK3β becomes fully activated immediately following protein synthesis (38). Thus, GSK3β is expected to be a highly productive kinase, constantly going through its catalytic cycle of ATP uploading, hydrolysis, and phosphoric acid transfer, substrate release, and ADP unloading, where the need for HSP90 would be high but its association temporary. Perhaps this distinguishes it from AKT, as wild-type AKT is often held inactive in the cell and thus would not be such a striking contrast to its KD counterpart.
An important experimental distinction in this study was that all our observations were from either endogenous or stably expressed kinases, as opposed to transient expressions that were used in other kinome-wide interaction studies of HSP90 (2, 3, 39). We made this choice after we compared HSP90 binding results for GSK3β from stably expressed versus transient overexpressed models. Because HSP90 is directed toward misfolded proteins, possibly through HSP70 as a cochaperone (40), it is important to separate the general folding functions of HSP90 from its specific regulatory functions on kinases. Transient expressions due to the level of overexpression tend to produce proteins that are poorly folded. We showed stably expressed GSK3β forms giving a clear-cut all-or-nothing binding of HSP90, while there was a substantial background level binding with transient transfected GSK3βWT (Fig. 1D).
For the FLAG-tagged kinases, we compared the two parameters for measuring HSP90 dependency: static binding strength in coimmunoprecipitation and time-dependent degradation of the kinases following HSP90 inhibition (Fig. 2). We did not observe a correlation between these two parameters, consistent with a previous study (2), confirming that these two methods captured distinct properties of HSP90 function. In our survey of 167 kinases (see Table S1 in the supplemental material), either in stably expressed forms (95 in Fig. 2) or in their endogenous forms (54 in Fig. 4 and 77 in Fig. 5), a total of 66 kinases displayed decreased stability in response to HSP90 inhibition. We also observed that for an individual kinase, degradation following HSP90 inhibition can be cell type dependent, suggesting that the contextual state of the kinase, instead of its intrinsic sequence properties, is targeted by HSP90. It was to our surprise that direct measure of HSP90 kinase-stable interactions by coimmunoprecipitation correlated poorly with 17-AAG-induced degradation responses (Fig. 2). With the caveat that protein synthesis over the time course of HSP90 inactivation may influence kinase expression levels or that the kinase may form aggregates (2), this suggests a more complex regulation than simply increased kinase stability reflecting a concomitant increase in HSP90 binding.
It has been proposed that HSP90-CDC37 associates with intrinsically unstable kinases, and by stabilizing the kinases with small molecular inhibitors, HSP90 association is often decreased (2). Interestingly, this decrease in association can lead to kinase degradation, which may be one route by which kinase inhibitors are effective as therapeutics (14). GSK3β seems to be an exception to this model, as its specific inhibitor CHIR99021 stabilized the association with both HSP90 and CDC37 (Fig. 1H). Whereas kinase inhibitors tend to be directed toward a particular tumor type, HSP90 inhibitors can target overproductive kinases in a wide variety of tumors. Since individual kinases can exhibit cell context-dependent responses to 17-AAG, as shown here, then it follows that each tumor type, or even individual tumors, would have a distinct kinome profile of HSP90 dependencies. Therefore, creating an index of these profiles might prove relevant to understanding response to therapies involving HSP90 inhibition. The advancements of proteomic tools have certainly made it possible to create these profiles, as shown in our experimental example (Fig. 5). In addition, since HSP90 inhibitors simultaneously inhibit multiple signaling pathways on which cancer cells depend for growth and survival, it has been speculated that HSP90 inhibitor response of kinases can validate the proteins as suitable molecular targets for anticancer drug development (14). Although we only performed the kinome analysis of one cell line to show technical feasibility of the proteomic workflow, we anticipate that there would be considerable value in extending this analysis further as a new metric for predicting cancer response to kinase inhibitor drugs.
Supplementary Material
ACKNOWLEDGMENTS
We commemorate the many contributions of our colleague, mentor, and friend Tony Pawson to the advancement of health science. During the course of this study, Pawson passed away.
This study was supported by Genome Canada through the Ontario Genomics Institute, by Ontario Research Fund GL2 to Tony Pawson, and by Cellzome (GSK) for Kinobead purification. Jing Jin and Ruijin Tian were recipients of Canadian Institutes of Health Research Fellowships. Cellzome was involved in data collection for Kinobead analysis.
The other funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication. Marcus Bantscheff and Mikhail M. Savitski are employees of GlaxoSmithKline.
J.J. conceived and coordinated the study, performed the Western blot analysis for Fig. 1 to 4, oversaw data analysis, contributed to the preparation of all figures, and wrote the paper. R.T. conceived the global proteome analysis and performed the phosphoproteomics purification and mass spectrometry analysis. A.Y.D. and K.W. cloned the FLAG-tagged constructs, created stable cell lines, and performed IP-MS for Fig. 2. A.P. performed the statistical analysis for the global proteome analysis in Fig. 5. L.T. helped design the global proteome analysis and assisted with data analysis. M.M.S. and M.B. contributed Kinobead samples and MS analyses. J.R.W. provided GSK3β constructs and guidance on data analysis. T.P. helped conceive and coordinate the study. K.C. helped coordinate the study, contributed to the preparation of all figures, performed data analysis for Fig. 5, and wrote the paper. All authors approved the final version of the manuscript.
Funding Statement
Ontario Government Genomic and Life Sciences Funding provided funding.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01045-15.
REFERENCES
- 1.Caplan AJ, Mandal AK, Theodoraki MA. 2007. Molecular chaperones and protein kinase quality control. Trends Cell Biol 17:87–92. doi: 10.1016/j.tcb.2006.12.002. [DOI] [PubMed] [Google Scholar]
- 2.Taipale M, Krykbaeva I, Koeva M, Kayatekin C, Westover KD, Karras GI, Lindquist S. 2012. Quantitative analysis of HSP90-client interactions reveals principles of substrate recognition. Cell 150:987–1001. doi: 10.1016/j.cell.2012.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Citri A, Harari D, Shohat G, Ramakrishnan P, Gan J, Lavi S, Eisenstein M, Kimchi A, Wallach D, Pietrokovski S, Yarden Y. 2006. Hsp90 recognizes a common surface on client kinases. J Biol Chem 281:14361–14369. doi: 10.1074/jbc.M512613200. [DOI] [PubMed] [Google Scholar]
- 4.Xu W, Mimnaugh E, Rosser MF, Nicchitta C, Marcu M, Yarden Y, Neckers L. 2001. Sensitivity of mature Erbb2 to geldanamycin is conferred by its kinase domain and is mediated by the chaperone protein Hsp90. J Biol Chem 276:3702–3708. doi: 10.1074/jbc.M006864200. [DOI] [PubMed] [Google Scholar]
- 5.Xu W, Yuan X, Xiang Z, Mimnaugh E, Marcu M, Neckers L. 2005. Surface charge and hydrophobicity determine ErbB2 binding to the Hsp90 chaperone complex. Nat Struct Mol Biol 12:120–126. doi: 10.1038/nsmb885. [DOI] [PubMed] [Google Scholar]
- 6.Taipale M, Tucker G, Peng J, Krykbaeva I, Lin ZY, Larsen B, Choi H, Berger B, Gingras AC, Lindquist S. 2014. A quantitative chaperone interaction network reveals the architecture of cellular protein homeostasis pathways. Cell 158:434–448. doi: 10.1016/j.cell.2014.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Polier S, Samant RS, Clarke PA, Workman P, Prodromou C, Pearl LH. 2013. ATP-competitive inhibitors block protein kinase recruitment to the Hsp90-Cdc37 system. Nat Chem Biol 9:307–312. doi: 10.1038/nchembio.1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Theodoraki MA, Kunjappu M, Sternberg DW, Caplan AJ. 2007. Akt shows variable sensitivity to an Hsp90 inhibitor depending on cell context. Exp Cell Res 313:3851–3858. doi: 10.1016/j.yexcr.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pearl LH. 2005. Hsp90 and Cdc37—a chaperone cancer conspiracy. Curr Opin Genet Dev 15:55–61. doi: 10.1016/j.gde.2004.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Carbonero R, Carnero A, Paz-Ares L. 2013. Inhibition of HSP90 molecular chaperones: moving into the clinic. Lancet Oncol 14:e358–e369. doi: 10.1016/S1470-2045(13)70169-4. [DOI] [PubMed] [Google Scholar]
- 11.Neckers L, Trepel JB. 2014. Stressing the development of small molecules targeting HSP90. Clin Cancer Res 20:275–277. doi: 10.1158/1078-0432.CCR-13-2571. [DOI] [PubMed] [Google Scholar]
- 12.Kim YS, Alarcon SV, Lee S, Lee MJ, Giaccone G, Neckers L, Trepel JB. 2009. Update on Hsp90 inhibitors in clinical trial. Curr Top Med Chem 9:1479–1492. doi: 10.2174/156802609789895728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moulick K, Ahn JH, Zong H, Rodina A, Cerchietti L, Gomes DaGama EM, Caldas-Lopes E, Beebe K, Perna F, Hatzi K, Vu LP, Zhao X, Zatorska D, Taldone T, Smith-Jones P, Alpaugh M, Gross SS, Pillarsetty N, Ku T, Lewis JS, Larson SM, Levine R, Erdjument-Bromage H, Guzman ML, Nimer SD, Melnick A, Neckers L, Chiosis G. 2011. Affinity-based proteomics reveal cancer-specific networks coordinated by Hsp90. Nat Chem Biol 7:818–826. doi: 10.1038/nchembio.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neckers L. 2006. Using natural product inhibitors to validate Hsp90 as a molecular target in cancer. Curr Top Med Chem 6:1163–1171. doi: 10.2174/156802606777811979. [DOI] [PubMed] [Google Scholar]
- 15.Haupt A, Joberty G, Bantscheff M, Frohlich H, Stehr H, Schweiger MR, Fischer A, Kerick M, Boerno ST, Dahl A, Lappe M, Lehrach H, Gonzalez C, Drewes G, Lange BM. 2012. Hsp90 inhibition differentially destabilises MAP kinase and TGF-beta signalling components in cancer cells revealed by kinase-targeted chemoproteomics. BMC Cancer 12:38. doi: 10.1186/1471-2407-12-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kundrat L, Regan L. 2010. Balance between folding and degradation for Hsp90-dependent client proteins: a key role for CHIP. Biochemistry 49:7428–7438. doi: 10.1021/bi100386w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.So J, Pasculescu A, Dai AY, Williton K, James A, Nguyen V, Creixell P, Schoof EM, Sinclair J, Barrios-Rodiles M, Gu J, Krizus A, Williams R, Olhovsky M, Dennis JW, Wrana JL, Linding R, Jorgensen C, Pawson T, Colwill K. 2015. Integrative analysis of kinase networks in TRAIL-induced apoptosis provides a source of potential targets for combination therapy. Sci Signal 8:rs3. doi: 10.1126/scisignal.2005700. [DOI] [PubMed] [Google Scholar]
- 18.Jin J, Smith FD, Stark C, Wells CD, Fawcett JP, Kulkarni S, Metalnikov P, O'Donnell P, Taylor P, Taylor L, Zougman A, Woodgett JR, Langeberg LK, Scott JD, Pawson T. 2004. Proteomic, functional, and domain-based analysis of in vivo 14-3-3 binding proteins involved in cytoskeletal regulation and cellular organization. Curr Biol 14:1436–1450. doi: 10.1016/j.cub.2004.07.051. [DOI] [PubMed] [Google Scholar]
- 19.Bantscheff M, Eberhard D, Abraham Y, Bastuck S, Boesche M, Hobson S, Mathieson T, Perrin J, Raida M, Rau C, Reader V, Sweetman G, Bauer A, Bouwmeester T, Hopf C, Kruse U, Neubauer G, Ramsden N, Rick J, Kuster B, Drewes G. 2007. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat Biotechnol 25:1035–1044. doi: 10.1038/nbt1328. [DOI] [PubMed] [Google Scholar]
- 20.Tian R, Wang H, Gish GD, Petsalaki E, Pasculescu A, Shi Y, Mollenauer M, Bagshaw RD, Yosef N, Hunter T, Gingras AC, Weiss A, Pawson T. 2015. Combinatorial proteomic analysis of intercellular signaling applied to the CD28 T-cell costimulatory receptor. Proc Natl Acad Sci U S A 112:E1594–E1603. doi: 10.1073/pnas.1503286112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cox J, Mann M. 2008. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- 22.Proia DA, Bates RC. 2014. Ganetespib and HSP90: translating preclinical hypotheses into clinical promise. Cancer Res 74:1294–1300. doi: 10.1158/0008-5472.CAN-13-3263. [DOI] [PubMed] [Google Scholar]
- 23.Doble BW, Woodgett JR. 2003. GSK-3: tricks of the trade for a multi-tasking kinase. J Cell Sci 116:1175–1186. doi: 10.1242/jcs.00384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sato S, Fujita N, Tsuruo T. 2000. Modulation of Akt kinase activity by binding to Hsp90. Proc Natl Acad Sci U S A 97:10832–10837. doi: 10.1073/pnas.170276797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dajani R, Fraser E, Roe SM, Young N, Good V, Dale TC, Pearl LH. 2001. Crystal structure of glycogen synthase kinase 3 beta: structural basis for phosphate-primed substrate specificity and autoinhibition. Cell 105:721–732. doi: 10.1016/S0092-8674(01)00374-9. [DOI] [PubMed] [Google Scholar]
- 26.Hughes K, Nikolakaki E, Plyte SE, Totty NF, Woodgett JR. 1993. Modulation of the glycogen synthase kinase-3 family by tyrosine phosphorylation. EMBO J 12:803–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frame S, Cohen P, Biondi RM. 2001. A common phosphate binding site explains the unique substrate specificity of GSK3 and its inactivation by phosphorylation. Mol Cell 7:1321–1327. doi: 10.1016/S1097-2765(01)00253-2. [DOI] [PubMed] [Google Scholar]
- 28.Bagatell R, Paine-Murrieta GD, Taylor CW, Pulcini EJ, Akinaga S, Benjamin IJ, Whitesell L. 2000. Induction of a heat shock factor 1-dependent stress response alters the cytotoxic activity of hsp90-binding agents. Clin Cancer Res 6:3312–3318. [PubMed] [Google Scholar]
- 29.Pasculescu A, Schoof EM, Creixell P, Zheng Y, Olhovsky M, Tian R, So J, Vanderlaan RD, Pawson T, Linding R, Colwill K. 2014. CoreFlow: a computational platform for integration, analysis and modeling of complex biological data. J Proteomics 100:167–173. doi: 10.1016/j.jprot.2014.01.023. [DOI] [PubMed] [Google Scholar]
- 30.Duncan JS, Whittle MC, Nakamura K, Abell AN, Midland AA, Zawistowski JS, Johnson NL, Granger DA, Jordan NV, Darr DB, Usary J, Kuan PF, Smalley DM, Major B, He X, Hoadley KA, Zhou B, Sharpless NE, Perou CM, Kim WY, Gomez SM, Chen X, Jin J, Frye SV, Earp HS, Graves LM, Johnson GL. 2012. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell 149:307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu Z, Moghaddas Gholami A, Kuster B. 2012. Systematic identification of the HSP90 candidate regulated proteome. Mol Cell Proteomics 11:M111.016675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sherman MY, Gabai VL. 2015. Hsp70 in cancer: back to the future. Oncogene 34:4153–4161. doi: 10.1038/onc.2014.349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shao J, Prince T, Hartson SD, Matts RL. 2003. Phosphorylation of serine 13 is required for the proper function of the Hsp90 co-chaperone, Cdc37. J Biol Chem 278:38117–38120. doi: 10.1074/jbc.C300330200. [DOI] [PubMed] [Google Scholar]
- 34.Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, Fleming T, Eiermann W, Wolter J, Pegram M, Baselga J, Norton L. 2001. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med 344:783–792. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 35.Heisermann GJ, Wiley HS, Walsh BJ, Ingraham HA, Fiol CJ, Gill GN. 1990. Mutational removal of the Thr669 and Ser671 phosphorylation sites alters substrate specificity and ligand-induced internalization of the epidermal growth factor receptor. J Biol Chem 265:12820–12827. [PubMed] [Google Scholar]
- 36.Wayne N, Mishra P, Bolon DN. 2011. Hsp90 and client protein maturation. Methods Mol Biol 787:33–44. doi: 10.1007/978-1-61779-295-3_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Genest O, Reidy M, Street TO, Hoskins JR, Camberg JL, Agard DA, Masison DC, Wickner S. 2013. Uncovering a region of heat shock protein 90 important for client binding in E. coli and chaperone function in yeast. Mol Cell 49:464–473. doi: 10.1016/j.molcel.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lochhead PA, Kinstrie R, Sibbet G, Rawjee T, Morrice N, Cleghon V. 2006. A chaperone-dependent GSK3beta transitional intermediate mediates activation-loop autophosphorylation. Mol Cell 24:627–633. doi: 10.1016/j.molcel.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 39.Echeverría PC, Bernthaler A, Dupuis P, Mayer B, Picard D. 2011. An interaction network predicted from public data as a discovery tool: application to the Hsp90 molecular chaperone machine. PLoS One 6:e26044. doi: 10.1371/journal.pone.0026044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karagöz GE, Rudiger SG. 2015. Hsp90 interaction with clients. Trends Biochem Sci 40:117–125. doi: 10.1016/j.tibs.2014.12.002. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.