ABSTRACT
Mycobacterium tuberculosis is a leading cause of death worldwide. The M. tuberculosis TAT (twin-arginine translocation) protein secretion system is present at the cytoplasmic membrane of mycobacteria and is known to transport folded proteins. The TAT secretion system is reported to be essential for many important bacterial processes that include cell wall biosynthesis. The M. tuberculosis secretion and invasion protein RipA has endopeptidase activity and interacts with one of the resuscitation antigens (RpfB) that are expressed during pathogen reactivation. MoxR1, a member of the ATPase family that is associated with various cellular activities, was predicted to interact with RipA based on in silico analyses. A bimolecular fluorescence complementation (BiFC) assay confirmed the interaction of these two proteins in HEK293T cells. The overexpression of RipA in Mycobacterium smegmatis and copurification with MoxR1 further validated their interaction in vivo. Recombinant MoxR1 protein, expressed in Escherichia coli, displays ATP-enhanced chaperone activity. Secretion of recombinant RipA (rRipA) protein into the E. coli culture filtrate was not observed in the absence of RipA-MoxR interaction. Inhibition of this export system in M. tuberculosis, including the key players, will prevent localization of peptidoglycan hydrolase and result in sensitivity to existing β-lactam antibiotics, opening up new candidates for drug repurposing.
IMPORTANCE
The virulence mechanism of mycobacteria is very complex. Broadly, the virulence factors can be classified as secretion factors, cell surface components, enzymes involved in cellular metabolism, and transcriptional regulators. The mycobacteria have evolved several mechanisms to secrete its proteins. Here, we have identified one of the virulence proteins of Mycobacterium tuberculosis, RipA, possessing peptidoglycan hydrolase activities secreted by the TAT secretion pathway. We also identified MoxR1 as a protein-protein interaction partner of RipA and demonstrated chaperone activity of this protein. We show that MoxR1-mediated folding is critical for the secretion of RipA within the TAT system. Inhibition of this export system in M. tuberculosis will prevent localization of peptidoglycan hydrolase and result in sensitivity to existing β-lactam antibiotics, opening up new candidates for drug repurposing.
INTRODUCTION
The TAT (twin-arginine translocation) protein secretion system is present at the cytoplasmic membrane of mycobacterium and is known to transport folded proteins. The TAT secretion system consists of three integral membrane proteins, TATA, TATB, and TATC. The target proteins secreted by the TAT secretion system bind to the TATBC complex in an energy-dependent manner, which then associates with TATA in a ΔpH-dependent manner and enables the substrate molecule to be transported across the membrane (1, 2). A twin-arginine signal peptide and the transmembrane pH gradient have been shown to activate the reversible assembly of the thylakoid ΔpH/TAT system, which can form a translocase to transport folded proteins (2). The signal peptide of the TAT substrate protein is cleaved by the signal peptidase LepB (3). The TAT signal peptides are characterized by the presence of the typical twin-arginine motif [S/T] RRxFLK or more general Z-R-R-x-Φ-Φ, where the arginine residues are almost conserved, whereas the frequency of the other amino acids is more than 50% (4). The length of TAT signal peptides is approximately 37 amino acid residues, whereas the Sec signal peptides have an average length of 23 residues. The TAT secretion system had been reported to be essential for many of the important bacterial processes that include cell wall biosynthesis (5) and virulence (6). In mycobacteria, four phospholipase C enzymes are known to be secreted by the TAT secretion system and have been shown to have a role in virulence (7). Another protein, Rv2525c, a homolog of transglycosidase enzyme, is involved in peptidoglycan metabolism and virulence and is secreted by the TAT secretion system (8).
Previous studies on resuscitation-promoting factors (Rpf), expressed during the reactivation stage of Mycobacterium tuberculosis infection, suggested that they play a role in growth or reactivation from a dormant stage. Rpf proteins function as peptidoglycan glycosidases. Among five of the resuscitation-promoting factors that are present in M. tuberculosis, RpfB has an N-terminal membrane lipoprotein lipid anchor and an Mce-like domain preceding the Rpf domain (9). RpfB was found to interact with Rpf-interacting protein A (RipA) of M. tuberculosis H37Rv. Furthermore, RipA together with RpfB showed hydrolytic activity targeting the M. tuberculosis cell wall peptidoglycan (10). Functionally, RipA is a secretory protein that also has endopeptidase activity (11, 12).
The MoxR1 protein belongs to ATPase family and is associated with various cellular activities. AAA+ proteins have been reported as novel molecular chaperone in bacteria and are involved in maturation or refolding of specific protein complexes (13). The MoxR protein homolog p618 has been found to play important roles in extracellular viral tail formation in the Acidianus two-tailed virus (14). Another MoxR protein homolog, CoxD, from Oligotropha carboxidovorans OM5, is involved in carbon monoxide dehydrogenase maturation, which is required by the bacteria to utilize CO as the sole source of carbon (15). The mechanism of secretion of RipA and the role of its interaction partner(s), however, remain elusive.
In the present study, we show that the N-terminal region of RipA consists of a signal peptide sequence with a conserved twin-arginine residue. We further show that RipA needs to be properly folded before secretion into the culture filtrate. The mycobacterial MoxR protein displays ATP-enhanced chaperone activity, and RipA-MoxR interaction leads to proper folding of RipA, resulting in its secretion.
RESULTS
In silico analysis of RipA identified MoxR as one of the possible interaction partners of RipA.
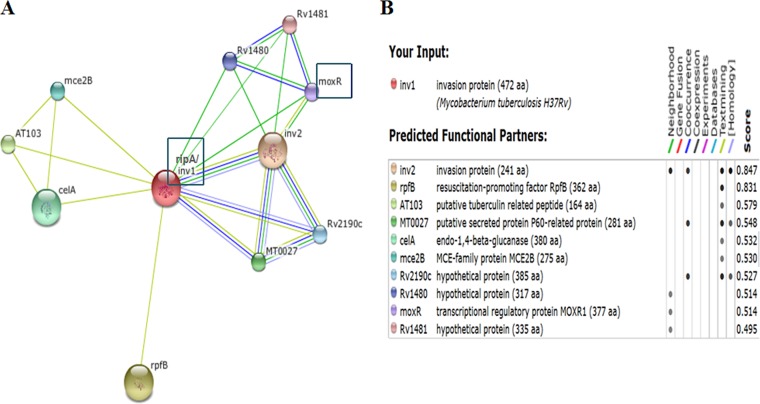
To identify proteins that could interact with RipA, STRING (Search Tool for the Retrieval of Interacting Genes/Proteins) analysis was performed using M. tuberculosis H37Rv database. inv1 (i.e., invasion1) is the other name for RipA, as suggested in earlier studies (16), and also RipA is present as inv1 in the STRING database. Some of the known interaction partners for RipA such as RpfB or RipB (inv2) were also observed (10), along with other proteins, such as CelA, which is predicted to possess cellulase activity cleaving β-1,4-glucosidic bond, Mce2B, which belongs to a mammalian cell entry protein with diverse role, including invasion property (17, 18), and proteins such as Rv1481,which contain the VWA motif. The VWA motif has been reported to act as a helper for the MoxR AAA+ homolog protein to function as a chaperone (14) and Rv2190c, a hypothetical protein predicted to possess a p60 hydrolase domain similar to RipA, also displayed a probable interaction with RipA after STRING analysis (Fig. 1). In silico analysis predicted MoxR1 protein as a novel target for interaction with RipA protein. MoxR1 was predicted to have a transcriptional regulatory domain and also a chaperone activity, the latter becoming evident after InterProScan analysis. It was therefore considered interesting to characterize such a multifunctional protein and its association with RipA, which is one of the important virulence proteins in M. tuberculosis H37Rv. Experiments were designed to gain further insights into the likely functional significance of the in silico predictions of MoxR1-RipA protein-protein interaction.
FIG 1 .
(A) STRING analysis reveals the top 10 interaction partners, both known and putative, of Mycobacterium tuberculosis H37Rv RipA (inv1) protein. (B) The score for each interaction partner is assigned and given in the table. The highest score for RipA association was found to be 0.847 for inv2/RipB. The confidence score for association of RipA and MoxR1 corresponds to 0.514, and that for Mce2B was found to be 0.547.
Bimolecular fluorescence complementation showed that RipA interacts with MoxR1.
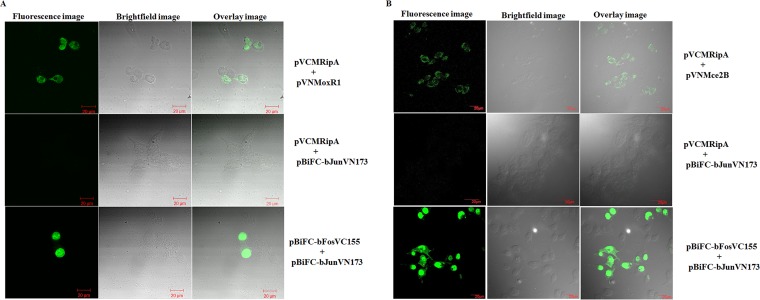
To experimentally validate in silico results pointing to possible interaction of RipA with MoxR, we performed a bimolecular fluorescence complementation (BiFC) assay. Genes encoding RipA and MoxR1 were cloned in pBiFC plasmid vectors harboring two nonfluorescent fragments of a fluorescent protein to construct recombinant plasmids pVCMRipA and pVNMoxR1, respectively. HEK293T cells were transfected with these recombinant plasmids expressing the predicted interaction partners (RipA and MoxR), and the protein-protein interaction was observed by fluorescence assay. As can be seen (upper left panel, Fig. 2A), HEK293T cells harboring pVCMRipA and pVNMoxR1 showed the presence of fluorescence, which is a consequence of the ability of two nonfluorescent yellow fluorescent protein (YFP) fragments to reassemble by complementation when they are in close proximity due to interaction between proteins fused to each fragment. Cells harboring pVCMRipA and pBiFC-bJunVN173 showed no fluorescence (middle left panel, Fig. 2A) and served as the negative control. HEK293T cells transfected with pBiFC-bFosVC155 and pBiFC-bJunVN173 expressing the anti-parallel leucine zipper, used as a positive control, gave a strong fluorescence due to YFP reassembly (lower left panel, Fig. 2A).
FIG 2 .
RipA interacts with MoxR1 and Mce2B, as is evident from the bimolecular florescence complementation assay. (A) Plasmids expressing the RipA and MoxR1 proteins in transfected HEK293T cells are shown. Fluorescence image for BiFC interaction was observed by imaging fixed cells (upper left panel) using confocal microscopy. The bright-field image for the same view is shown in the upper second panel. The overlay of the fluorescence and bright-field images is shown in the upper right panel. The plasmid expressing the negative control resulted in no fluorescence (middle left panel). Strong fluorescence intensity was observed for the positive control (lower left panel), and a merged image for fluorescence and the bright-field positive control is shown in the lower right panel (60× magnifications). (B) Fluorescence was also detected for interaction of RipA with Mce2B. The HEK293T cells transfected with plasmids that express both RipA and Mce2B exhibit fluorescence (upper left panel). The bright-field image of same view is shown in the upper second panel. The overlay of the fluorescence and bright-field images is shown in the upper right panel. In the negative control, no detectable fluorescence could be seen (middle left panel). The bright-field and merged images for the same view are also shown (middle second and right panels, respectively). The highest fluorescence intensity was detected for the positive control (lower left panel).
In silico analyses also predicted interaction of RipA with mammalian cell entry protein, Mce2B. To experimentally validate the interaction of RipA with Mce2B, a protein involved in virulence, a biomolecular fluorescence assay was carried out. The fluorescence image (upper left panel, Fig. 2B) for HEK293T cells harboring pVCMRipA and pVNMce2b showed the presence of an interaction as evident by fluorescence. The bright-field image (upper second panel, Fig. 2B) for the same view is also shown. The overlay of both fluorescence and the bright-field image (upper right panel, Fig. 2B) showed the localization of fluorescence within HEK293T cells, confirming that RipA indeed interacts with Mce2B protein. The fluorescence image of the negative control (middle left panel, Fig. 2B) showed an absence of interaction. These bimolecular fluorescence assay results clearly provide evidence of protein-protein interaction between RipA and MoxR and RipA and Mce2B.
Ni-NTA pulldown assay confirms RipA-MoxR1 interaction.
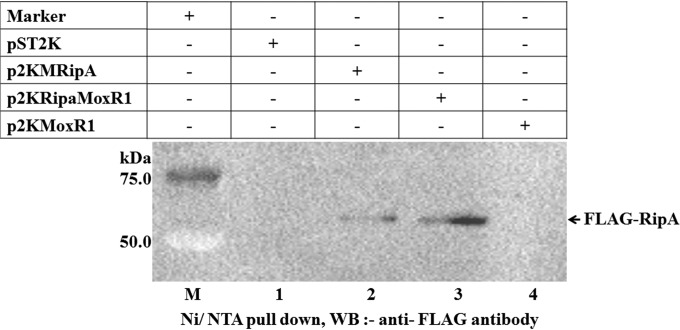
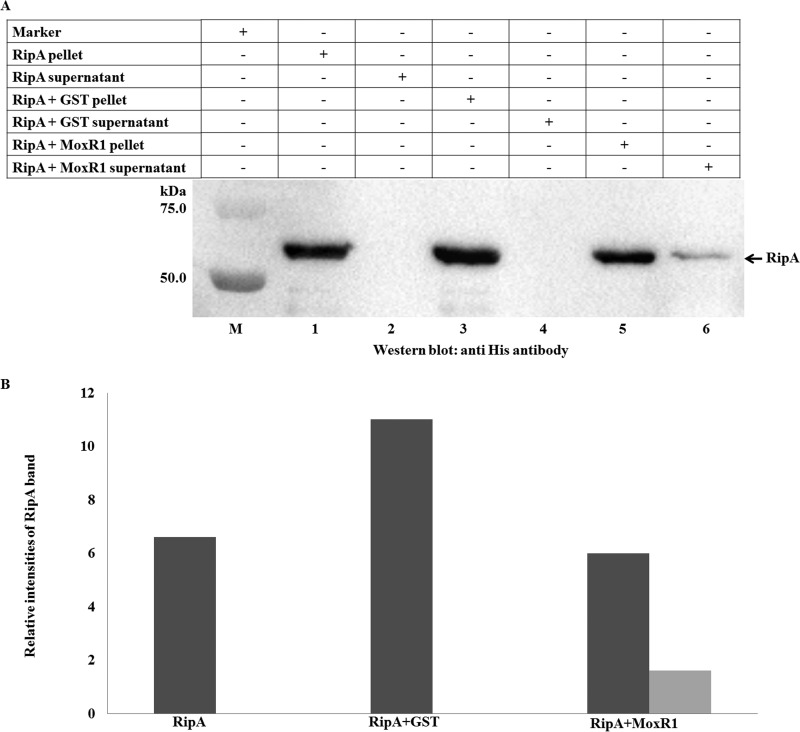
To further demonstrate that RipA and MoxR1 physically interact in vivo, we performed an Ni-nitrilotriacetic acid (NTA) pulldown assay. For this, we expressed recombinant proteins fused with tags for detection in Mycobacterium smegmatis. RipA was fused to a C-terminal FLAG tag, while MoxR1 was fused to the His tag at the N-terminal end. M. smegmatis cells were transformed with these recombinant constructs, and interactions between different recombinant proteins were assayed and compared with those of the controls. Soluble proteins from the lysates expressing (i) RipA, (ii) RipA and MoxR1, or (iii) MoxR1 alone were incubated with His6-tagged MoxR1 purified from Escherichia coli BL21(DE3) and bound to Ni-NTA beads. RipA protein was detected using anti-FLAG antibody. As can be seen, FLAG-RipA specifically copurified with His6-MoxR1; however, there was no detectable interaction of MoxR1 with control lysates (Fig. 3, lanes 1 and 4), thus confirming the RipA-MoxR interaction in vivo. These results further document the protein-protein interaction between the recombinant RipA and MoxR proteins expressed in M. smegmatis.
FIG 3 .
RipA protein interacts with MoxR1 in M. smegmatis. The vector alone was used as a negative control (lane 1). FLAG-tagged RipA binds in the presence of either MoxR1 protein purified from E. coli (lane 2) or MoxR1 expressed in both E. coli as well as M. smegmatis (lane 3). Note the absence of signal for MoxR1 alone (lane 4). Lane M, molecular mass marker lane.
MoxR1 possesses chaperone activity.
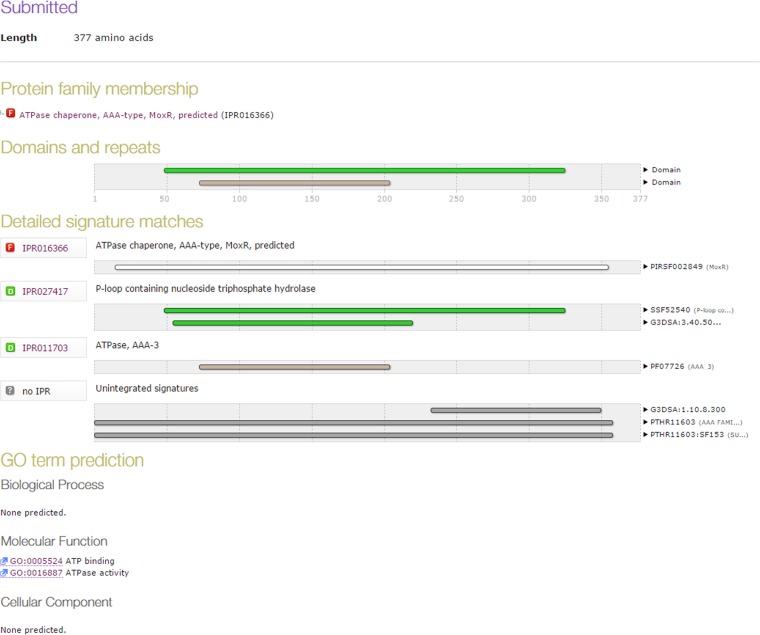
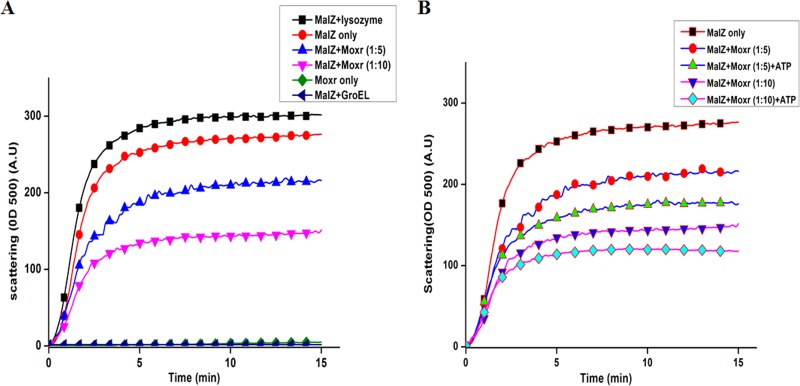
From the results presented so far, while it is clear that RipA and MoxR are involved in the protein-protein interaction the mechanisms, consequences, and functional implications of such cross talks are not clear. The InterProScan analysis of MoxR1 predicted that it contains an ATPase chaperone domain along with a nucleoside triphosphate hydrolase domain (Fig. 4), pointing to the possibility of MoxR being a chaperone. To experimentally demonstrate chaperone-like activity of MoxR1 in terms of its ability to prevent MalZ protein from thermal denaturation, a light-scattering assay to monitor aggregation was carried out as described previously (19) by measuring the absorbance of thermally denatured MalZ in the absence or presence of MoxR1 alone or MoxR1 plus ATP (Fig. 5A and B). The thermal stabilities of the MoxR1 and MalZ proteins were also checked individually. The MoxR1 protein was highly stable at 47°C for more than 15 min, showing negligible aggregation suggestive of a molecular chaperone (green diamonds, Fig. 5A), whereas MalZ protein showed aggregation that increased considerably with time (red circles, Fig. 5A). Chaperone activity of MoxR1 is evident from its ability to prevent the MalZ protein from thermal aggregation. MalZ in the presence of 5 µM MoxR1 (blue triangles, Fig. 5A) apparently resisted aggregation, and incubation with a higher concentration of MoxR1 (10 µM) resulted in increased stability of MalZ (pink triangles, Fig. 5A). To assess the role of the ATPase domain of MoxR1 in chaperone activity, 5 µM MoxR1 was incubated in the presence/absence of 1 mM ATP and magnesium. The aggregation prevention of MalZ was evident at a concentration of 5 µM MoxR1 with ATP (green triangles, Fig. 5B), and this was more than without ATP (red circles, Fig. 5B). The ability to protect MalZ from aggregation was higher at a concentration of 10 µM MoxR1 (blue triangles, Fig. 5B), and the ability to protect Malz from thermal denaturation/aggregation at a 10 µM concentration of MoxR1 increased in the presence of ATP (light blue diamond curve, Fig. 5B). These results experimentally confirm the InterProScan predictions about MoxR1 containing an ATPase chaperone domain along with a nucleoside triphosphate hydrolase domain.
FIG 4 .
Functional domain analysis of M. tuberculosis MoxR1 protein. The InterProScan analysis result shows the presence of the nucleoside triphosphate hydrolase, AAA type, and ATPase chaperone domain in MoxR1 protein.
FIG 5 .
(A) MoxR1 prevents thermal aggregation of MalZ protein. Shown are results from the light-scattering assay of thermal aggregation. Absorbance at 500 nm (OD500) was measured for MoxR1 protein at 47°C for 15 min. (B) MoxR1 prevents thermal aggregation of MalZ protein in the presence of 1 mM ATP and Mg2+, as a cofactor. The kinetics of prevention of thermal aggregation of MalZ by MoxR1 at a similar concentration was enhanced in the presence of ATP. GroEL was used as a control.
MoxR1 protein protects enzyme activity of NdeI from thermal denaturation.
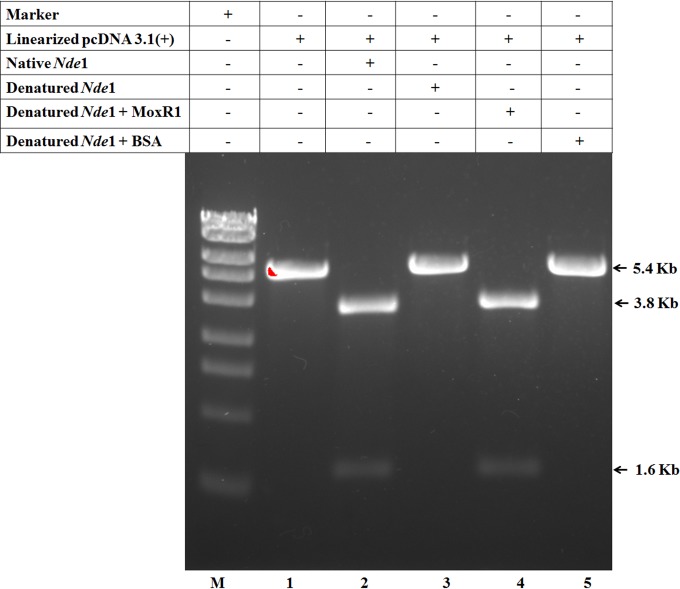
Experiments were designed to further prove that MoxR1 has the ability to act as a chaperone and can protect the functional activity of a protein from thermal denaturation. The enzymatic activity of restriction enzyme NdeI was assessed after heat denaturation at 60°C for 20 min in the presence or absence of MoxR1. Initially, pcDNA 3.1(+) vector, which has a single site for NdeI, was linearized using SmaI and was then used to investigate the enzyme activity of NdeI. Heat-denatured NdeI could not further cleave pcDNA 3.1(+); however, when NdeI was heat denatured in the presence of MoxR1, the enzyme activity was protected, as seen from the appearance of a cleavage product of 1.6 kb. In the presence of control protein bovine serum albumin (BSA), heat denaturation impaired the NdeI activity (Fig. 6). This ability of MoxR1 to protect the functional activity of restriction enzyme NdeI from thermal inactivation clearly demonstrates that it can act as a chaperone.
FIG 6 .
Restriction enzyme activity of NdeI enzyme is protected from thermal denaturation in the presence of MoxR1 protein. SmaI-linearized pcDNA 3.1(+) plasmid is present at 5.4 kb (lane 1). Digestion with NdeI restriction enzyme cuts pcDNA 3.1(+) to generate two bands of 3.8 kb and 1.6 kb, respectively (lane 2). These bands are seen in the case of native NdeI (lane 2) and denatured NdeI plus MoxR1 proteins (lane 4).
MoxR1 possesses a surface hydrophobic residue typical for chaperones.
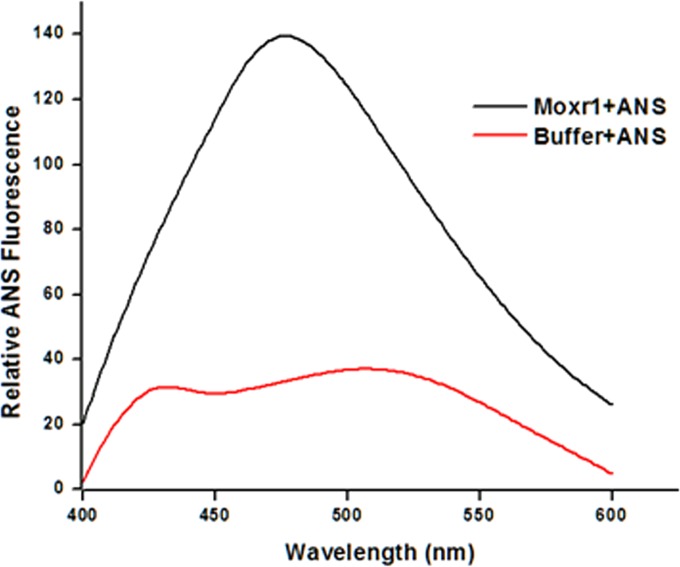
The presence of surface hydrophobicity patches is typical for chaperones and is considered a signature for such proteins. While the results presented so far clearly pointed to a chaperone-like activity in MoxR1, we carried out a simple experiment to investigate if it also carries such surface hydrophobic residues. 8-Anilino-1-naphthalenesulfonic acid (ANS) is a fluorophore that strongly binds to the cationic groups or the hydrophobic residue of proteins. Surface hydrophobicity of MoxR1 protein was determined by measuring emission fluorescence of ANS after binding to MoxR1. A very significant ANS binding by MoxR1, evident from the increase in fluorescence intensity and blue shift of fluorescence maxima, could be seen (Fig. 7). The presence of significant surface hydrophobicity, one of the striking features of a chaperone protein, in MoxR1 clearly explains the results obtained so far pointing to a chaperone-like function of MoxR1.
FIG 7 .
ANS fluorescence spectrum for MoxR1 protein shows the presence of hydrophobic residue at the protein surface. Shown are a comparison of the fluorescence spectra of MoxR1 and buffer at 40 µM ANS. The concentration of the MoxR1 protein was 5 µM. Each experiment was carried out in triplicate.
Coexpression of MoxR1 with RipA enhances the proper folding of recombinant RipA in E. coli.
Our in silico observations pointing to a protein-protein interaction between M. tuberculosis RipA and MoxR1 and the experimentally demonstrated chaperone-like activity of MoxR1, prompted us to investigate the functional consequence of coexpressing these two proteins in a heterologous system of E. coli. At 37°C, overexpression of RipA protein alone or with glutathione S-transferase (GST) showed the absence of RipA in the supernatant as soluble protein, and 100% of the protein was present in the pellet as inclusion bodies (Fig. 8A, lanes 1 to 4). However, when MoxR1 was overexpressed with RipA, solubility of RipA was enhanced (Fig. 8A, lane 6). SDS-PAGE analysis to ensure equal loading of GST plus RipA and MoxR1 plus RipA supernatant fractions is shown in Fig. S2 in the supplemental material. Densitometric analysis clearly showed that in the presence of MoxR1 protein, about 20% of the RipA protein was properly folded at 37°C and present in the soluble fraction (Fig. 8B). These results indicate that the predicted interaction between RipA and MoxR1 may have a functional significance in terms of proper folding and secretion of the former with the help of the chaperone function of the latter.
FIG 8 .
(A) In vivo folding of RipA protein in the presence of MoxR1 chaperone activity. The different lanes are as follows: Lane M, marker; lanes 1 and 2, RipA pellet (lane 1) and folded RipA (supernatant) (lane 2) in the absence of any chaperone. RipA proteins in the pellet (lane 3) and supernatant fraction (lane 4) in the presence of GST are also shown. Insoluble RipA and folded RipA (supernatant) in the presence of MoxR1 can be seen in lanes 5 and 6, respectively. (B) Change in the levels of folded RipA in the presence of GST and MoxR1 at 37°C in vivo. The dark gray bars represent the RipA concentration in the pellet, and light gray represents the RipA concentration in the supernatant fraction.
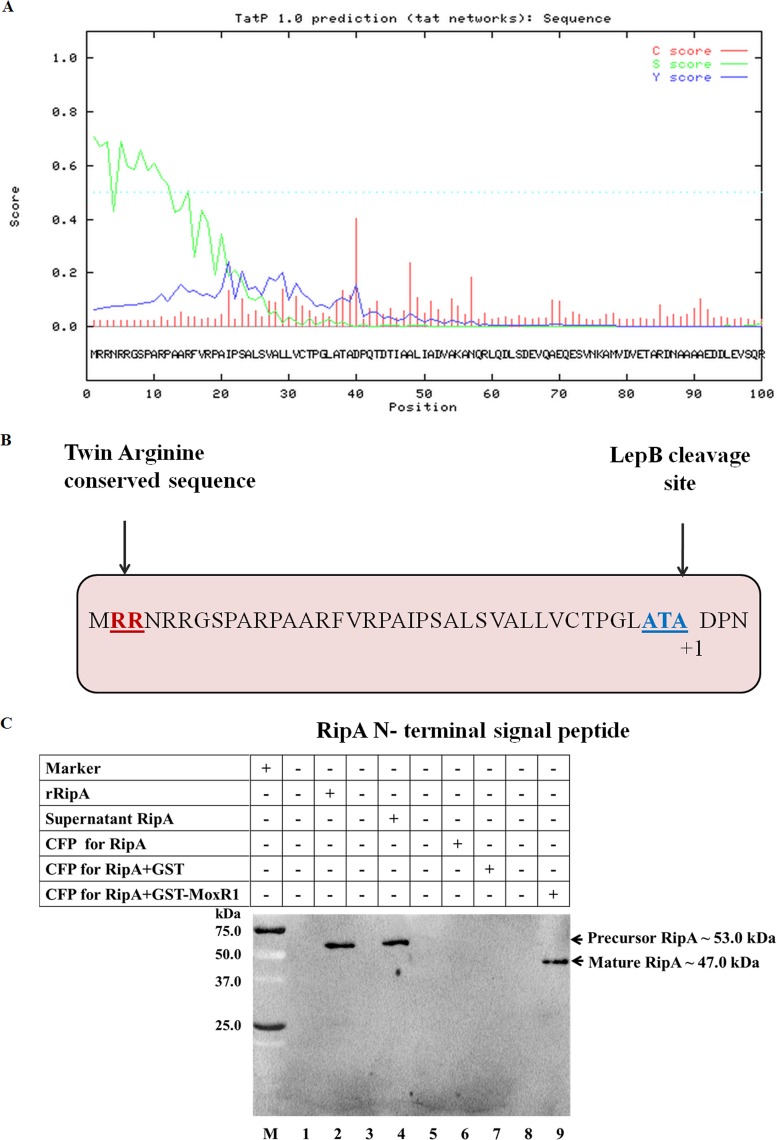
Importance of MoxR1 in the RipA secretion system.
The results presented so far have pointed to the ability of MoxR1protein to act as a chaperone and interact with the M. tuberculosis RipA protein. It is therefore arguable that this interaction enables correct folding of the RipA protein before its secretion through the TAT secretion system. That RipA protein contains a potential TAT signal peptide was clearly evident from the S-score value. C- and Y-scores indicate the positioning of a potential cleavage site (Fig. 9A). Moreover, the N-terminal region of RipA protein consists of signal peptide sequences, which were experimentally identified from the N-terminal mature protein. The N-terminal signal sequence of RipA consists of conserved twin-arginine sequence of Tat signal peptide and the signal cleavage site as depicted in Fig. 9B. Experiments were designed to identify the RipA secretion system and evaluate the role of MoxR1. Recombinant constructs encoding the His6-RipA and GST-MoxR1 proteins were coexpressed in E. coli BL21(DE3). These cells possess the TAT secretion system and are also deficient in proteases that protect cleavage of the recombinant proteins. E. coli BL21(DE3) cells expressing C-terminal His6-RipA and GST alone were used for expression of unfolded RipA. The presence of precursor His6-RipA expressed intracellularly in the E. coli BL21(DE3) cells at a predicted size of ~53 kDa (along with the His6 tag) was detected using anti-His-monoclonal antibody (1:2,500), and mature His6-RipA at the predicted size of ~47 kDa (Fig. 9C, lane 9) was found only in the culture filtrate when expressed with GST-MoxR1. These results point to the role of MoxR1 as a chaperone in folding RipA in the cytoplasm before its secretion.
FIG 9 .
(A) The N-terminal region of RipA protein consists of TAT signal peptide. (B) Schematic of RipA N-terminal signal peptide consensus amino acid. The conserved twin arginine is underlined, and the conserved 3-amino-acid LepB cleavage site starts at the 37th amino acid. The mature protein starting at the amino acid aspartate (D) is shown as +1. (C) MoxR1 protein aids in secretion of mature RipA through the TAT secretion pathway. The different lanes show the protein marker (lane 1), recombinant C-terminal His6-RipA used as a positive control (lane 2), E. coli BL21 (DE3) cells expressing RipA protein in the soluble fraction used as an intracellular control (lane 5), and culture filtrate protein from transformed E. coli cells expressing RipA alone (lane 6), as well as both RipA and GST alone (lane 7), probed for mature RipA protein. Similarly, the culture filtrate proteins from cotransformed E. coli cells expressing both His6-RipA and GST-MoxR1 were probed for mature RipA peptide and can be seen as an ~47.0-kDa band (lane 9).
DISCUSSION
Comparative genomics and proteomics of M. tuberculosis identified several strategies, including gene cooption, adopted by this pathogen for virulence, pathogenesis, and survival (18, 20, 21). The virulence mechanism of mycobacteria is very complex. Broadly, the virulence factors can be classified as secretion factors, cell surface components, enzymes involved in cellular metabolism, and transcriptional regulators. The mycobacterium has evolved several mechanisms to secrete its protein. TAT-dependent transport and secretion derives its name from the presence of a conserved twin-arginine motif in the N-terminal signal peptide. The striking feature of the TAT secretion system is that it can secrete only those proteins that had been folded properly in the cytoplasm before being secreted. In most of the other secretion systems, such as the Sec-dependent or Sec-independent secretion system, the folding of the secretion substrate takes place in either the periplasmic space or the outer membrane of the mycobacterium. The core complex of TAT secretion consists of TATA, TATB, and TATC proteins that are present in almost all of the bacterial pathogens (22). In addition, other TAT proteins also have been reported: e.g., in Rhodobacter species, TATF, an AAA+ family protein, is found in association with TATC. TATF is not required for secretion but is believed to function as a chaperone (23).
RipA is a secretory protein and has been shown to possess the p60 domain, which exhibits peptidoglycan hydrolase activity and is capable of hydrolyzing the dipeptide d-glutamyl-meso-diaminopimelic acid (10). The p60 domains have been shown to have a role in formation of the cording morphology of a virulent strain of mycobacterium (24). The ability to gain entry and resist the antimicrobial intracellular environment of mammalian cells is an essential virulence property of M. tuberculosis. The properties of RipA protein such as virulence, invasion, secretion, and cell wall association make RipA an ideal candidate to evaluate its potential efficacy as a possible vaccine candidate (12, 25). In silico analysis showed MoxR1 to be a new candidate for RipA association (Fig. 1), and actual physical interaction of RipA with MoxR1 protein was demonstrated using bimolecular fluorescence complementation and the pulldown assay (Fig. 2 and 3).
The functional domain of MoxR1 belongs to the AAA+ protein family (ATPases associated with various cellular activities). AAA+ family proteins such as RavA from E. coli have been demonstrated to be assembled into hexameric rings (26) and have a role in maturation of NADH:ubiquinone oxidoreductase I (27). Prevention of aggregation of MalZ protein by MoxR1 is enhanced in the presence of ATP and magnesium, suggesting that MoxR1 protein possess a chaperonic activity (Fig. 5A and B) similar to what is shown by the metal chelatase family, which utilizes the hydrolysis of ATP to insert a transition cofactor such as Mg2+ and also remodel the substrate molecule (28). Moreover, MoxR1 protein was shown to have surface hydrophobic residues and was able to protect denaturation of NdeI enzyme from heat inactivation (Fig. 6 and 7).
RipA-MoxR1 interaction is a prelude to the role of MoxR1 in folding of RipA. Overexpression of RipA at 37°C resulted in the formation of insoluble aggregates in E. coli BL21(DE3) cells. Enhanced folding of RipA was observed primarily due to coexpression of MoxR1 at higher temperature, as significant amounts of RipA protein were found in the soluble fraction (Fig. 8). Also, proper folding by MoxR1 as a chaperone requires ATP-driven energy, which in turn reduces the overall rate of protein synthesis.
Our observation that RipA consists of conserved twin-arginine peptide at its N-terminal end and the direct observation of culture filtrate protein of E. coli BL21(DE3) cells expressing RipA along with MoxR1 indicate that MoxR1 is essential for secretion of mature RipA protein (Fig. 9C). E. coli BL21(DE3) cells harbor the TAT secretion system and also possess several chaperones. However, culture filtrate protein of E. coli BL21(DE3) cells expressing RipA alone did not show any mature protein, suggesting the specificity of MoxR1 in refolding and secretion of mycobacterial proteins such as RipA. This is consistent with the finding that when RipA protein from M. tuberculosis was overexpressed in M. smegmatis, only a single band corresponding to full-length precursor protein was detected, suggesting that RipA protein does not undergo efficient processing in M. smegmatis (29). AmiA and AmiC are peptidoglycan amidases present in E. coli and are functional homologs of mycobacterium RipA protein. They are required during cell division and have been shown to degrade the septal petidoglycan connecting the two daughter cells (5). Interestingly, both AmiA and AmiC were shown to be exported by the TAT system (5). The depletion of RipA results in branching or chaining of M. smegmatis (10). Similarly, deletion of both AmiA and TAT proteins leads to formation of the cell chaining phenotype (5, 30, 31). These striking similarities between the role and phenotype of homolog further favor the possibility of RipA secretion through TAT transport mechanism.
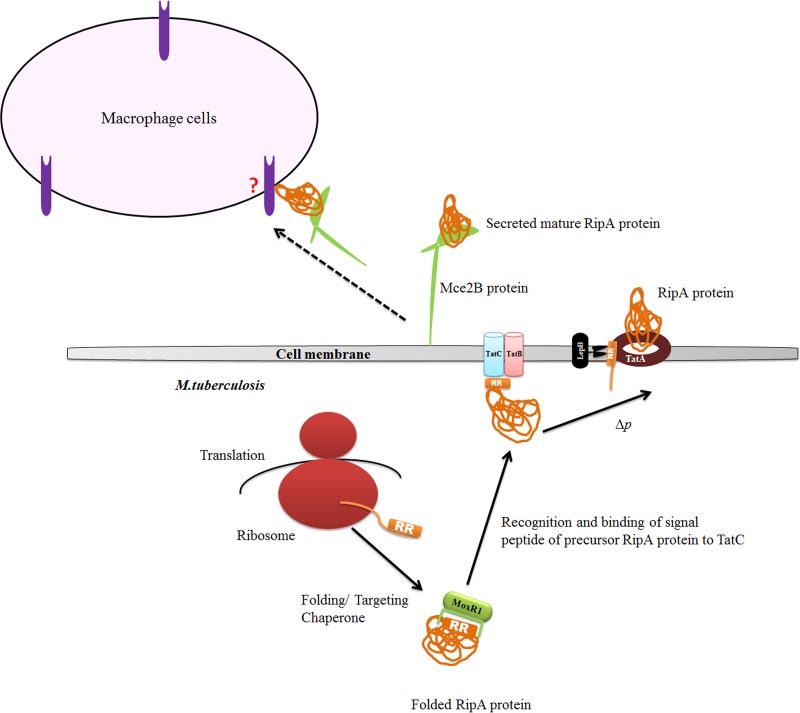
According to our proposed model (Fig. 10), MoxR1 protein folds RipA in the cytoplasm, making it incompatible to be secreted by the general Sec-dependent pathway. The RipA protein because of the chaperonic activity by MoxR1 is also protected from proteolysis to form an active truncated protein that is toxic to cells. Once RipA protein is secreted through TAT secretion, it interacts with other proteins and gets cleaved to start its peptidoglycan hydrolase activity. The RipA protein is known to consist of an RGD motif, and most likely the concerted effort along with Mce protein will facilitate the entry of RipA into host cells. This may then modulate the host immune response. That RipA also interacts (Fig. 2B) with mammalian cell entry protein Mce2B, another secretion protein known to be associated with virulence, reinforces the significance of MoxR1-mediated folding of RipA.
FIG 10 .
Proposed model of MoxR action. The nascent polypeptide of RipA protein after release from the ribosome interacts with the MoxR1 protein. MoxR1 protein acts as a chaperone that helps RipA fold properly to protect the signal peptide containing RR sequence from proteasomal degradation and target the protein-chaperone complex to the TAT secretion system. The binding of ATP to the AAA+ ATPase domain and divalent Mg cations as a cofactor to the metal ion-dependent adhesion site of MoxR1 enhances its chaperone activity. The processed and secreted RipA might then associate with the mammalian cell entry protein and gain entry into host cells to exhibit its virulence.
Recently, M. tuberculosis Rv1988 was identified as a secretory protein having histone methyltransferase activity and secreted by the TAT secretion system (32). The TAT secretion system is present in many bacteria and is involved in several aspects of bacterial regulation, including fine-tuning the export of virulence factors and immunogenic antigens of pathogenic mycobacteria. For example, β-lactamase is a known TAT substrate of M. tuberculosis that confers resistance to β-lactam-containing antibiotics. The consequence of deletion of the TAT system is that it will not allow the secretion of β-lactamase and thereby increase sensitivity to cell wall-targeting antibiotics. That RipA deletion leads to sensitivity to β-lactams such as carbenicillin (10) has a bearing on the development of new strategy for intervention against M. tuberculosis. Our findings that MoxR1 is involved in secretion of the virulent RipA protein through the TAT pathway, absent in mammalian systems, suggests that inhibition of this export system in M. tuberculosis will prevent localization of peptidoglycan hydrolase and result in sensitivity to existing β-lactam antibiotics, opening new candidates for drug repurposing. Increased understanding of the regulation of protein secretion through the TAT system and targeting it or its substrate will greatly aid in efforts to control tuberculosis.
MATERIALS AND METHODS
Bioinformatics tools.
The interaction partners for RipA protein from the M. tuberculosis H37Rv genome database were predicted using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (version 9.1) (http://string.embl.de). For STRING interaction analyses, the default parameters were used. All of the active prediction methods were selected: i.e., neighborhood, gene fusion, cooccurrence, coexpression, experiments, database, and text mining. Only the first 10 interacting proteins were selected to be displayed. The probability of interaction between two proteins is predicted in terms of the confidence score of the STRING database. The confidence score was set to a medium confidence level (S-score = 0.4), which corresponds to >50% possibility of association. In the network display, each node represents a protein, and each edge represents an interaction. TAT signal motif prediction for RipA protein was performed using TATP1.0 Server software. Protein domain analysis of MoxR1 was performed using InterProScan analysis (http://www.ebi.ac.uk/interpro/).
Expression of recombinant proteins.
The different constructs used in this study are described in Table S1 in the supplemental material, and the primers used are described in Table S2 in the supplemental material. All of the recombinant plasmid constructs were confirmed by restriction mapping and DNA sequencing. Recombinant proteins were expressed in E. coli BL21(DE3) cells after induction with 0.1 mM IPTG (isopropyl-β-d-thiogalactopyranoside), as described earlier (33). The cell pellet was collected by low-speed centrifugation and resuspended in lysis buffer (50 mM NaH2PO4 [pH 8.0], 1 mM phenylmethylsulfonyl fluoride, 600 mM NaCl) and disrupted on ice by ultrasonication. The cell debris was removed by centrifugation at 14,000 rpm at 4°C for 20 min, and supernatant was loaded onto a nickel-charged resin (Ni-Sepharose high-performance resin; Amersham Biosciences), and the bound protein was eluted using the elution buffer (NaH2PO4 [pH 8.0], 1 mM phenylmethylsulfonyl fluoride, 600 mM NaCl, 250 mM imidazole [pH 8.0]). The purified protein was dialyzed against sodium phosphate buffer (20 mM NaH2PO4, 20 mM NaCl [pH 7.4]). The protein concentration was determined by the Bradford method using a Bio-Rad kit (Bio-Rad Laboratories, Inc., Hercules, CA). Endotoxin-free status of the purified protein was ensured by treating the purified protein with polymyxin B as described earlier (34). Circular dichroism (CD) spectroscopy was performed for native MoxR1 protein to assess its secondary structure (see Fig. S1 in the supplemental material).
BiFC.
HEK293T cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) (Gibco, Carlsbad, CA), supplemented with 10% heat-inactivated fetal bovine serum (FBS) (Gibco, Carlsbad, CA) and 1% antibiotic-antimycotic (Gibco, Carlsbad, CA) at 37°C under humidified air containing 5% CO2 in a T75 flat-bottom flask. About 4 × 105 HEK293T cells were seeded in 2 ml of DMEM present in 6-well plates containing a poly-l-lysine (Sigma, Saint Louis, MO)-treated coverslip. After 60 to 70% confluence was achieved, the medium was removed, and 1 ml of incomplete medium (DMEM without FBS and antibiotic) was added to each well. About 1.5 µg of plasmid DNA for interaction partners was mixed and transfected using polyethyleneimine (PEI) reagent in 500 µl of incomplete DMEM in a separate tube. The 6-well plates were incubated at 37°C under humidified air containing 5% CO2 for 6 h. The incomplete medium was removed, complete DMEM was added, and the mixture was incubated for 16 h. Transfected HEK293T cells were fixed on the coverslip by treatment with 3.7% formaldehyde in phosphate-buffered saline (PBS) for 15 min at room temperature and mounted on glass slide. The slides were then observed with a Nikon A1R+ Eclipse Ti confocal microscope (Nikon Corp., Tokyo, Japan) with a setting of 60× (NA 1.4), and images were captured using photomultiplier tube (PMT) camera (Hamamatsu Photonics, Hamamatsu, Japan). Shutter and image acquisition control were performed using NIS-Element AR software (Nikon Corp.).
Pulldown assay.
M. smegmatis mc2155 was transformed with either pST2K vector or RipA alone or RipA along with MoxR1 or MoxR1 alone. Equal concentrations of whole-cell lysate (about 3 mg of total protein) of M. smegmatis mc2155 harboring either the empty pST2K vector (negative control), p2KMRipA, p2KRipaMoxR1, or p2KMoxR1 were incubated separately with 250 µg of His6-MoxR1 protein purified from E. coli BL21(DE3) in 50 µl of Ni-NTA beads for 4 h at room temperature. The resins were washed with 1× PBS to remove nonspecific proteins. The bound protein was electrophoresed by SDS-PAGE and transferred onto polyvinylidene difluoride (PVDF) membrane, and Western blotting was performed using anti-FLAG antibody (Sigma, Saint Louis, MO) as per the manufacturer’s instructions.
Aggregation prevention assay.
To determine the chaperone activity of MoxR1 protein, the aggregation prevention assay for maltodextrin glucosidase (MalZ) was performed. The MalZ protein is known to lose its native conformation and undergo thermal aggregation at 42°C. MalZ (0.4 µM), in both the absence and presence of 5 and 10 µM MoxR1, was mixed at room temperature, and light scattering was measured at 500 absorbance units (AU). GroEL protein at a 2 µM concentration was used as a chaperone control (35). The experiment was carried out at 47°C. A similar experiment was also performed with MalZ (0.4 µM) in the presence of 5 and 10 µM MoxR1 and 1 mM ATP. Each experiment was carried out in triplicate.
MoxR1 protein chaperone activity assay on heat-denatured NdeI restriction enzyme.
NdeI restriction enzyme was incubated with or without 30 µg of purified recombinant MoxR1 protein. BSA was used as a control protein. Heat denaturation of NdeI was performed at 65°C for 20 min as described earlier (19). The SmaI-digested and linearized pcDNA 3.1(+) plasmid was used for restriction digestion with denatured NdeI in the presence or absence of MoxR1 protein. Digestion with nondenatured NdeI was used as a positive control.
ANS fluorescence of recombinant MoxR1.
To measure ANS fluorescence, 5 µM recombinant MoxR1protein was mixed with 40 µM ANS in buffer (20 mM sodium phosphate buffer, 20 mM NaCl [pH 7.4]), and the mixture was incubated for 2 h at 25°C. Spectra were obtained from 400 to 600 nm at an excitation wavelength of 370 nm using Cary eclipse fluorescence spectrophotometer (Agilent Technologies). The excitation and emission slit width of 10 nm each and data pitch of 0.5 nm were used. Each spectrum is the average of three measurements and was plotted after subtraction of ANS buffer (40 µM) for baseline correction.
Coexpression of chaperone MoxR1 with RipA.
A recombinant plasmid carrying a kanamycin resistance gene and the ripA gene cloned in the pET28b vector for expression as C-terminal His6-RipA and another recombinant pGEX6P1-based construct containing an ampicillin resistance gene and the moxR1 gene for expression as N-terminal GST-MoxR1 protein were cotransformed in E. coli BL21(DE3) cells. Cotransformants containing both recombinant plasmids were selected on an LB agar plate containing both kanamycin and ampicillin. Similarly, cotransformants for RipA cloned in pET28b expression vector and pGEX6P1 vector expressing glutathione S-transferase protein were obtained. These strains were cultured at 37°C in 10 ml LB medium supplemented with kanamycin and ampicillin until the optical density at 650 nm (OD650) reached 0.8. IPTG at a 100 µM concentration was added, and the mixture was incubated at 37°C for 5 h at 150 rpm. The cell pellet was harvested by centrifugation at 5,000 rpm for 10 min, resuspended in 2 ml of PBS, and sonicated. The supernatant and pellet were separated by centrifugation at 14,000 rpm for 20 min. The pellet was again resuspended in 2 ml PBS, and equal amounts from both the supernatant and pellet were resolved by 12% SDS-PAGE and transferred to PVDF membrane (Bio-Rad Laboratories, Inc.). Western blotting was performed using mouse monoclonal anti-His antibody (Sigma) as the primary antibody at a 1:3,000 dilution and goat anti-mouse IgG antibody conjugated to horseradish peroxidase (HRP) (Sigma) as the secondary antibody at a 1:10,000 dilution. The signal was developed using Clarity Western ECL enhanced chemiluminescence substrate (Bio-Rad Laboratories, Inc.).
The Chemidoc MP imaging system (Bio-Rad) was used for quantification of the relative quantities of RipA protein bands observed on the PVDF membrane. The Western blot image was first cropped to fit the size. Six lanes were selected manually using the lanes’ toolbar menu. Each lane was adjusted to fit the size of the lanes in the PVDF membrane. Lane background subtraction was enabled to remove the background intensity of the PVDF membrane. Bands were detected using the “detect band” option with selection of “low” as the detection setting. The areas of the band to be estimated were adjusted using the “adjust band” option and were similar for all six lanes. For quantification, the relative quantity parameter was selected, and lane 1 was selected as the reference band. Later, “analysis table” was selected, and the report of the relative quantification of bands in terms of percentage was generated as a PDF.
Secretion of rRipA protein from E. coli into the LB medium.
Different E. coli BL21(DE3) strains cotransformed with different plasmids as mentioned above were grown in 100 ml of LB broth separately. Upon induction with 10 µM IPTG and growth at 20°C for 14 h at 90 rpm, His6-RipA and GST-MoxR1 proteins and His6-RipA and GST were coexpressed optimally in the soluble fraction. RipA was also expressed alone in E. coli BL21(DE3) cells to check for intracellular expression. Cells were centrifuged at 3,000 rpm for 10 min at 4°C. Supernatant was collected in a fresh tube, again centrifuged at 14,000 rpm for 30 min at 4°C, and passed through Ni-NTA beads to enrich the His6-RipA secreted in the culture filtrate. The pellet was resuspended in lysis buffer (20 mM Tris-HCl [pH 8.0], 1 mM phenylmethylsulfonyl fluoride, 500 mM NaCl) and disrupted on ice by ultrasonication. The soluble protein was obtained by centrifugation at 14,000 rpm for 30 min at 4°C, and the remaining cell debris was collected as the pellet fraction. The culture filtrate protein of the cotransformants was resolved by 12% SDS-PAGE, transferred to PVDF membrane (Bio-Rad Laboratories, Inc.), and Western blotted using mouse monoclonal anti-His antibody (1:3,000) as the primary antibody and goat anti-mouse IgG antibody conjugated to HRP (1:10,000) as the secondary antibody. The signal was developed using the Clarity Western ECL substrate (Bio-Rad Laboratories, Inc.).
SUPPLEMENTAL MATERIAL
CD analysis of MoxR1 protein. Download
SDS-PAGE for total protein loading of RipA and GST or RipA and MoxR1. Download
Bacterial strains and plasmids used in this study.
Primers used in this study.
ACKNOWLEDGMENTS
M.B. thanks the Indian Council of Medical Research for a postdoctoral fellowship. M.K., A.S., and S.P. thank the Council for Scientific and Industrial Research for fellowships. This work was funded by a Centre of Excellence grant to S.E.H. and N.Z.E. from the Department of Biotechnology, Government of India.
N.Z.E. and S.E.H. conceptualized and designed the research, M.B. and N.A. performed the research, A.S., M.K., S.P., and T.K.C. carried out data analysis, and M.B., S.E.H., and N.Z.E. wrote the manuscript.
Funding Statement
This work was supported by DBT COE Phase 2 grant number BT/PR12817/COE/34/23/2015 to S.E.H. and N.Z.E.
Footnotes
Citation Bhuwan M, Arora N, Sharma A, Khubaib M, Pandey S, Chaudhuri TK, Ehtesham Hasnain S, Zafar Ehtesham N. 2016. Interaction of Mycobacterium tuberculosis virulence factor RipA with chaperone MoxR1 is required for transport through the TAT secretion system. mBio 7(2):e02259-15. doi:10.1128/mBio.02259-15.
REFERENCES
- 1.Alami M, Lüke I, Deitermann S, Eisner G, Koch HG, Brunner J, Müller M. 2003. Differential interactions between a twin-arginine signal peptide and its translocase in Escherichia coli. Mol Cell 12:937–946. doi: 10.1016/S1097-2765(03)00398-8. [DOI] [PubMed] [Google Scholar]
- 2.Mori H, Cline K. 2002. A twin arginine signal peptide and the pH gradient trigger reversible assembly of the thylakoid ΔpH/Tat translocase. J Cell Biol 157:205–210. doi: 10.1083/jcb.200202048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yahr TL, Wickner WT. 2001. Functional reconstitution of bacterial Tat translocation in vitro. EMBO J 20:2472–2479. doi: 10.1093/emboj/20.10.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berks BC. 1996. A common export pathway for proteins binding complex redox cofactors? Mol Microbiol 22:393–404. doi: 10.1046/j.1365-2958.1996.00114.x. [DOI] [PubMed] [Google Scholar]
- 5.Bernhardt TG, de Boer PA. 2003. The Escherichia coli amidase AmiC is a periplasmic septal ring component exported via the twin-arginine transport pathway. Mol Microbiol 48:1171–1182. doi: 10.1046/j.1365-2958.2003.03511.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voulhoux R, Ball G, Ize B, Vasil ML, Lazdunski A, Wu LF, Filloux A. 2001. Involvement of the twin-arginine translocation system in protein secretion via the type II pathway. EMBO J 20:6735–6741. doi: 10.1093/emboj/20.23.6735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Raynaud C, Guilhot C, Rauzier J, Bordat Y, Pelicic V, Manganelli R, Smith I, Gicquel B, Jackson M. 2002. Phospholipases C are involved in the virulence of Mycobacterium tuberculosis. Mol Microbiol 45:203–217. doi: 10.1046/j.1365-2958.2002.03009.x. [DOI] [PubMed] [Google Scholar]
- 8.Saint-Joanis B, Demangel C, Jackson M, Brodin P, Marsollier L, Boshoff H, Cole ST. 2006. Inactivation of Rv2525c, a substrate of the twin arginine translocation (Tat) system of Mycobacterium tuberculosis, increases beta-lactam susceptibility and virulence. J Bacteriol 188:6669–6679. doi: 10.1128/JB.00631-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Downing KJ, Betts JC, Young DI, McAdam RA, Kelly F, Young M, Mizrahi V. 2004. Global expression profiling of strains harbouring null mutations reveals that the five rpf-like genes of Mycobacterium tuberculosis show functional redundancy. Tuberculosis (Edinb) 84:167–179. [DOI] [PubMed] [Google Scholar]
- 10.Hett EC, Chao MC, Deng LL, Rubin EJ. 2008. A mycobacterial enzyme essential for cell division synergizes with resuscitation-promoting factor. PLoS Pathog 4:e1000001. doi: 10.1371/journal.ppat.1000001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hett EC, Chao MC, Steyn AJ, Fortune SM, Deng LL, Rubin EJ. 2007. A partner for the resuscitation-promoting factors of Mycobacterium tuberculosis. Mol Microbiol 66:658–668. doi: 10.1111/j.1365-2958.2007.05945.x. [DOI] [PubMed] [Google Scholar]
- 12.Målen H, Berven FS, Fladmark KE, Wiker HG. 2007. Comprehensive analysis of exported proteins from Mycobacterium tuberculosis H37Rv. Proteomics 7:1702–1718. doi: 10.1002/pmic.200600853. [DOI] [PubMed] [Google Scholar]
- 13.Iyer LM, Leipe DD, Koonin EV, Aravind L. 2004. Evolutionary history and higher order classification of AAA+ ATPases. J Struct Biol 146:11–31. doi: 10.1016/j.jsb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 14.Scheele U, Erdmann S, Ungewickell EJ, Felisberto-Rodrigues C, Ortiz-Lombardía M, Garrett RA. 2011. Chaperone role for proteins p618 and p892 in the extracellular tail development of Acidianus two-tailed virus. J Virol 85:4812–4821. doi: 10.1128/JVI.00072-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pelzmann A, Ferner M, Gnida M, Meyer-Klaucke W, Maisel T, Meyer O. 2009. The CoxD protein of Oligotropha carboxidovorans is a predicted AAA+ ATPase chaperone involved in the biogenesis of the CO dehydrogenase [CuSMoO2] cluster. J Biol Chem 284:9578–9586. doi: 10.1074/jbc.M805354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Labò M, Gusberti L, Rossi ED, Speziale P, Riccardi G. 1998. Determination of a 15437-bp nucleotide sequence around the Inha gene of Mycobacterium avium and similarity analysis of the products of putative ORFs. Microbiology 144:807–814. doi: 10.1099/00221287-144-3-807. [DOI] [PubMed] [Google Scholar]
- 17.Chitale S, Ehrt S, Kawamura I, Fujimura T, Shimono N, Anand N, Lu S, Cohen-Gould L, Riley LW. 2001. Recombinant Mycobacterium tuberculosis protein associated with mammalian cell entry. Cell Microbiol 3:247–254. doi: 10.1046/j.1462-5822.2001.00110.x. [DOI] [PubMed] [Google Scholar]
- 18.Singh Y, Kohli S, Sowpati DT, Rahman SA, Tyagi AK, Hasnain SE. 2014. Gene cooption in mycobacteria and search for virulence attributes: comparative proteomic analyses of Mycobacterium tuberculosis, Mycobacterium indicus pranii and other mycobacteria. Int J Med Microbiol 304:742–748. doi: 10.1016/j.ijmm.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Suragani M, Aadinarayana VD, Pinjari AB, Tanneeru K, Guruprasad L, Banerjee S, Pandey S, Chaudhuri TK, Ehtesham NZ. 2013. Human resistin, a proinflammatory cytokine, shows chaperone-like activity. Proc Natl Acad Sci U S A 110:20467–20472. doi: 10.1073/pnas.1306145110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kohli S, Singh Y, Sharma K, Mittal A, Ehtesham NZ, Hasnain SE. 2012. Comparative genomic and proteomic analyses of PE/PPE multigene family of Mycobacterium tuberculosis H37Rv and H37Ra reveal novel and interesting differences with implications in virulence. Nucleic Acids Res 40:7113–7122. doi: 10.1093/nar/gks465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahman SA, Singh Y, Kohli S, Ahmad J, Ehtesham NZ, Tyagi AK, Hasnain SE. 2014. Comparative analyses of nonpathogenic, opportunistic, and totally pathogenic mycobacteria reveal genomic and biochemical variabilities and highlight the survival attributes of Mycobacterium tuberculosis. mBio 5:e02020. doi: 10.1128/mBio.02020-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ligon LS, Hayden JD, Braunstein M. 2012. The ins and outs of Mycobacterium tuberculosis protein export. Tuberculosis (Edinb) 92:121–132. doi: 10.1016/j.tube.2011.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindenstrauss U, Brüser T. 2006. Conservation and variation between Rhodobacter capsulatus and Escherichia coli Tat systems. J Bacteriol 188:7807–7814. doi: 10.1128/JB.01139-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gao LY, Pak M, Kish R, Kajihara K, Brown EJ. 2006. A mycobacterial operon essential for virulence in vivo and invasion and intracellular persistence in macrophages. Infect Immun 74:1757–1767. doi: 10.1128/IAI.74.3.1757-1767.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brennan MJ, Stone MR, Evans T. 2012. A rational vaccine pipeline for tuberculosis. Int J Tuberc Lung Dis 16:1566–1573. doi: 10.5588/ijtld.12.0569. [DOI] [PubMed] [Google Scholar]
- 26.El Bakkouri M, Gutsche I, Kanjee U, Zhao B, Yu M, Goret G, Schoehn G, Burmeister WP, Houry WA. 2010. Structure of RavA MoxR AAA+ protein reveals the design principles of a molecular cage modulating the inducible lysine decarboxylase activity. Proc Natl Acad Sci U S A 107:22499–22504. doi: 10.1073/pnas.1009092107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wong KS, Snider JD, Graham C, Greenblatt JF, Emili A, Babu M, Houry WA. 2014. The MoxR ATPase RavA and its cofactor ViaA interact with the NADH:ubiquinone oxidoreductase I in Escherichia coli. PLoS One 9:e85529. doi: 10.1371/journal.pone.0085529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reid JD, Hunter CN. 2002. Current understanding of the function of magnesium chelatase. Biochem Soc Trans 30:643–645 doi: 10.1042/BST0300643. [DOI] [PubMed] [Google Scholar]
- 29.Chao MC, Kieser KJ, Minami S, Mavrici D, Aldridge BB, Fortune SM, Alber T, Rubin EJ. 2013. Protein complexes and proteolytic activation of the cell wall hydrolase RipA regulate septal resolution in mycobacteria. PLoS Pathog 9:e1003197. doi: 10.1371/journal.ppat.1003197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Santini CL, Bernadac A, Zhang M, Chanal A, Ize B, Blanco C, Wu LF. 2001. Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J Biol Chem 276:8159–8164. doi: 10.1074/jbc.C000833200. [DOI] [PubMed] [Google Scholar]
- 31.Thomas JD, Daniel RA, Errington J, Robinson C. 2001. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol Microbiol 39:47–53. doi: 10.1046/j.1365-2958.2001.02253.x. [DOI] [PubMed] [Google Scholar]
- 32.Yaseen I, Kaur P, Nandicoori VK, Khosla S. 2015. Mycobacteria modulate host epigenetic machinery by Rv1988 methylation of a non-tail arginine of histone H3. Nat Commun 6:8922. doi: 10.1038/ncomms9922. [DOI] [PubMed] [Google Scholar]
- 33.Farhana A, Kumar S, Rathore SS, Ghosh PC, Ehtesham NZ, Tyagi AK, Hasnain SE. 2008. Mechanistic insights into a novel exporter-importer system of Mycobacterium tuberculosis unravel its role in trafficking of iron. PLoS One 3:e2087. doi: 10.1371/journal.pone.0002087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Banerjee S, Nandyala A, Podili R, Katoch VM, Murthy KJ, Hasnain SE. 2004. Mycobacterium tuberculosis (Mtb) isocitrate dehydrogenases show strong B cell response and distinguish vaccinated controls from TB patients. Proc Natl Acad Sci U S A 101:12652–12657. doi: 10.1073/pnas.0404347101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dahiya V, Chaudhuri TK. 2014. Chaperones GroEL/GroES accelerate the refolding of a multidomain protein through modulating on-pathway intermediates. J Biol Chem 289:286–298. doi: 10.1074/jbc.M113.518373. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
CD analysis of MoxR1 protein. Download
SDS-PAGE for total protein loading of RipA and GST or RipA and MoxR1. Download
Bacterial strains and plasmids used in this study.
Primers used in this study.